Abstract
Spongy degeneration with cerebellar ataxia (SDCA) is a severe neurodegenerative disease with monogenic autosomal recessive inheritance in Malinois dogs, one of the four varieties of the Belgian Shepherd breed. We performed a genetic investigation in six families and seven isolated cases of Malinois dogs with signs of cerebellar dysfunction. Linkage analysis revealed an unexpected genetic heterogeneity within the studied cases. The affected dogs from four families and one isolated case shared a ∼1.4 Mb common homozygous haplotype segment on chromosome 38. Whole genome sequence analysis of three affected and 140 control dogs revealed a missense variant in the KCNJ10 gene encoding a potassium channel (c.986T>C; p.Leu329Pro). Pathogenic variants in KCNJ10 were reported previously in humans, mice, and dogs with neurological phenotypes. Therefore, we consider KCNJ10:c.986T>C the most likely candidate causative variant for one subtype of SDCA in Malinois dogs, which we propose to term spongy degeneration with cerebellar ataxia 1 (SDCA1). However, our study also comprised samples from 12 Malinois dogs with cerebellar dysfunction which were not homozygous for this variant, suggesting a different genetic basis in these dogs. A retrospective detailed clinical and histopathological analysis revealed subtle neuropathological differences with respect to SDCA1-affected dogs. Thus, our study highlights the genetic and phenotypic complexity underlying cerebellar dysfunction in Malinois dogs and provides the basis for a genetic test to eradicate one specific neurodegenerative disease from the breeding population. These dogs represent an animal model for the human EAST syndrome.
Keywords: Canis familiaris, Kir4.1, potassium channel, EAST syndrome, SeSAME syndrome, Malinois, neurology, brain, central nervous system, animal model
EAST syndrome in humans is characterized by epilepsy, ataxia, sensorineural deafness, and renal salt wasting tubulopathy (Bockenhauer et al. 2009; OMIM#612780). It was also termed SeSAME syndrome for seizures, sensorineural deafness, ataxia, mental retardation, and electrolyte imbalance (Scholl et al. 2009). EAST syndrome is a rare disorder with an autosomal recessive mode of inheritance. Until now, it has been reported in 26 human patients with a total of 14 different pathogenic variants in the KCNJ10 gene (Sala-Rabanal et al. 2010; Williams et al. 2010; Abdelhadi et al. 2016).
KCNJ10 encodes an inward-rectifying potassium channel (Kir), also known as Kir4.1, which is expressed in the central nervous system (CNS), eye, inner ear, and kidney. In the kidneys, this channel is located in the distal nephron’s basolateral tubular epithelia. The KCNJ10 channels in the intermediate cells of the stria vascularis of the inner ear contribute to the endocochlear potential (Bockenhauer et al. 2009; Abdelhadi et al. 2016; Palygin et al. 2016). In the eye, KCNJ10 is expressed in Müller glia cells (Sala-Rabanal et al. 2010; Arai et al. 2015). Brain expression predominates in the cerebral and cerebellar cortex as well as in the caudate nucleus and putamen. KCNJ10 channels play a major role in modulating neuronal cells’ resting membrane potential through a process named potassium spatial buffering. Potassium spatial buffering, ensured by glial cells, mainly astrocytes, is a mechanism for the regulation of extracellular potassium (Djukic et al. 2007; Bockenhauer et al. 2009; Scholl et al. 2009; Sala-Rabanal et al. 2010).
The clinical hallmarks of EAST syndrome are nonprogressive neurological and renal symptoms. They include infantile-onset seizures, ataxia, tubulopathy, and sensorineural deafness (Bockenhauer et al. 2009; Cross et al. 2013; Abdelhadi et al. 2016). In the kidneys, KCNJ10 defects result in renal salt wasting and hypokalemic metabolic alkalosis (Bockenhauer et al. 2009; Abdelhadi et al. 2016). Additionally, the failure to generate a physiologic endocochlear potential can result in sensorineural deafness. Ocular abnormalities in EAST syndrome are only detectable with electroretinogram and are characterized by reduced amplitudes of the photopic negative response of light-adapted retinae (Thompson et al. 2011; Arai et al. 2015). Kcjn10-deficient knockout mice show defects in oligodendrocyte development and in vivo myelination (Neusch et al. 2001; Djukic et al. 2007).
A KCNJ10 variant has also been reported in dogs. In several terrier breeds, the XM_545752.3:c.627C>G variant, predicted to result in p.Ile209Met, leads to hereditary ataxia, a disease characterized by lesions in the CNS, mainly in the spinal cord, but not in the cerebellum (Wessmann et al. 2004; Rohdin et al. 2010, 2015; Gilliam et al. 2014).
In the Belgian Shepherd breed, spongy degeneration with cerebellar ataxia (SDCA) was first described in 13 purebred Malinois puppies from five different litters. Clinical signs and histological findings were primarily localized to the cerebellum. The pedigree data suggested an autosomal recessive mode of inheritance (Kleiter et al. 2011). An earlier report of two cross-bred Malinois dogs may have described a similar disease (Cachin and Vandevelde 1991). However, to date, no causal variant for SDCA was described in Malinois dogs or any other of the varieties of the Belgian Shepherd breed.
The aim of this study was to identify the presumed causative genetic defect of SDCA in Malinois dogs using a positional cloning approach in combination with whole genome resequencing. The genetic analysis revealed an unexpected genetic heterogeneity and our findings strongly suggest that in the Belgian Shepherd breed more than one type of cerebellar dysfunction is present.
Materials and Methods
Ethics statement
All animal experiments were performed according to the local regulations. All dogs in this study were examined with the consent of their owners. The collection of blood samples was approved by the “Cantonal Committee for Animal Experiments” (Canton of Bern; permit 23/10).
Breed nomenclature
The Federation Cynologique Internationale (FCI) describes the Malinois, together with the Groenendael, the Laekenois, and the Tervueren, as a variety of the Belgian Shepherd dog breed. The American Kennel Club, however, officially recognizes the Belgian Malinois, the Belgian Sheepdog (FCI: Groenendael), the Belgian Laekenois, and the Belgian Tervuren (FCI: Tervueren) as four distinct breeds. In this paper, all references to the breed nomenclature correspond to the FCI standards.
Clinical examinations
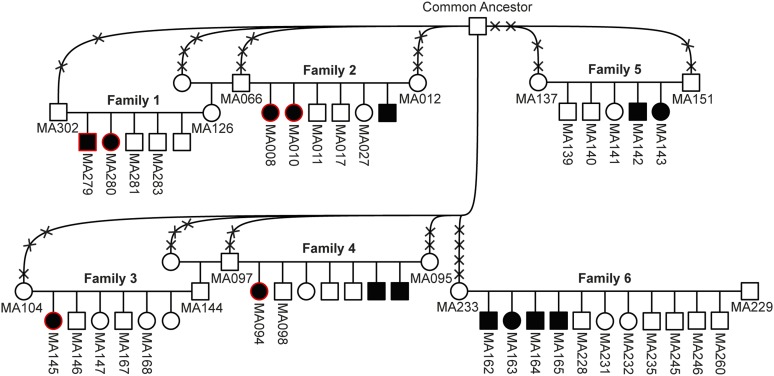
In this study, we performed neurological examination including video analysis of 12 Malinois puppies with clinical signs of cerebellar disease. These dogs belonged to six related families (Figure 1). Family 4 in the pedigree was one of the five families previously reported with SDCA (Kleiter et al. 2011).
Figure 1.
Pedigree of Malinois dogs used for genetic mapping of the disease locus. Filled symbols represent animals with cerebellar disorder. Numbers indicate dogs from which samples were available and which were genotyped on the SNP chip for linkage analysis. Six affected dogs indicated by red contours were selected for homozygosity mapping. A common ancestor in both maternal and paternal lineages could be identified in five of the six families. This dog had >1500 descendants in just three generations. Crosses intersecting the connection lines to the common ancestor represent the numbers of generations (e.g., MA302 is a great-grandson of the common ancestor). Family 4 was previously described (Kleiter et al. 2011).
Neurohistopathology examinations
Necropsy was performed in nine out of the 12 above described Malinois puppies with neurological symptoms (MA008, MA094, MA142, MA143, MA145, MA162, MA164, MA165, and MA280, Figure 1). We collected brain samples from all nine and spinal cord samples from seven puppies (MA142, MA143, MA145, MA162, MA164, MA165, and MA280). All tissues were fixed in 4% buffered formaldehyde solution, embedded in paraffin, and sectioned at 2–5 µm. Sections were stained with hematoxylin and eosin and examined by light microscopy.
Animal selection for the genetic analysis
For the genetic analysis, we initially applied liberal phenotypic inclusion criteria and considered all available Malinois dogs with neurological abnormalities. Specifically, we used six Malinois families. Samples were available from the 12 Malinois puppies with cerebellar disorder, 20 nonaffected full siblings, and 12 nonaffected parents (Figure 1). We additionally investigated seven Malinois puppies with reported cerebellar signs, for which no relatives were available.
In addition to the individuals belonging to the six families and the seven isolated cases, we genotyped 187 other Malinois, 25 Groenendael, two Laekenois, and 34 Tervueren dogs whose blood samples were donated to the biobank of the Institute of Genetics at the University of Bern (Table 2). Furthermore, we analyzed 486 samples from 89 genetically diverse dog breeds (Supplemental Material, Table S1).
Table 2. Association of the KCNJ10:c.986T>C genotypes with cerebellar dysfunction.
| Genotype KCNJ10:c.986T>C | T/T | C/T | C/C |
|---|---|---|---|
| Malinois cases (families 1–4, MA152) | — | — | 7 |
| Malinois cases (families 5 and 6, six isolated dogs) | 9 | 3 | — |
| Malinois controls | 176 | 43 | — |
| Groenendael controls | 25 | — | — |
| Laekenois controls | 2 | — | — |
| Tervueren controls | 33 | 1a | — |
| Control dogs from other breedsb | 486 | — | — |
This Tervueren dog had Malinois parents.
These dogs were specifically genotyped for the KCNJ10:c.986T>C variant. The genome sequences of 140 independent control dogs were also homozygous T/T at this variant. Therefore, the number of control dogs totals 626.
DNA extraction and genotyping
Genomic DNA was isolated from EDTA blood samples with the Maxwell RSC Whole Blood DNA Kit, which were used with the Maxwell RSC Instrument (Promega). Genotyping was done on Illumina canine_HD chips containing 173,662 genome-wide single nucleotide polymorphisms (SNPs) by GeneSeek/Neogen. Genotypes were stored in a BC/Gene database version 3.5 (BC/Platforms).
Linkage and homozygosity mapping
For linkage analysis, Illumina canine_HD SNP chip genotypes from 44 dogs in six families were used (Figure 1). We analyzed the dataset for parametric linkage under a fully penetrant, recessive model of inheritance with the Merlin software (Abecasis et al. 2002).
PLINK v1.07 (Purcell et al. 2007) was used as described (Wiedmer et al. 2016) to search for extended intervals of homozygosity with shared alleles across the affected animals.
Reference sequences
The dog CanFam 3.1 genome assembly was used for all analyses. All references to the canine KCNJ10 gene correspond to the accessions XM_545752.5 (mRNA) and XP_545752.3 (protein). XP_545752.3 has the same length as the human protein (NP_002232.2; 379 amino acids) and 373 out of 379 amino acids (98%) are identical between dog and human.
Whole genome resequencing
PCR-free fragment libraries were prepared from three affected Malinois dogs (MA008, MA094, and MA152) with 300 bp (MA008, MA094) and 400 bp (MA152) insert sizes. The libraries were sequenced to roughly 16×–22× coverage on an Illumina HiSeq2000 (MA008, MA094) or on an Illumina HiSeq3000 (MA152) instrument using 2 × 100 bp paired-end reads (MA008, MA094) or 170 bp + 130 bp paired-end reads (MA152), respectively.
The reads were mapped to the dog reference genome assembly CanFam3.1 and aligned using Burrows-Wheeler Aligner version 0.7.5a with default settings (Li and Durbin 2009). The generated SAM file was converted to a BAM file and the reads were sorted using samtools (Li 2011). Picard tools (http://sourceforge.net/projects/picard/) was used to mark PCR duplicates. To perform local realignments and to produce a cleaned BAM file, we used the Genome Analysis Tool Kit (GATK version 2.4.9, 50; McKenna et al. 2010). GATK was also used for base quality recalibration with canine dbSNP data as training set.
Putative single nucleotide and small indel variants were identified in each sample individually using GATK HaplotypeCaller in gVCF mode, and subsequently genotyped per-chromosome and genotyped across all samples simultaneously (Van der Auwera et al. 2013). Filtering was performed using the variant filtration module of GATK. To predict the functional effects of the called variants, SnpEff software together with the ENSEMBL (version 72) annotation CanFam 3.1 was used (Cingolani et al. 2012). For variant filtering we used 140 control genomes, which were either publicly available (Bai et al. 2015) or produced during other projects of our group.
PCR and Sanger sequencing
Sanger sequencing was used to confirm the variant identified from whole genome sequencing. For these experiments we amplified PCR products from genomic DNA using AmpliTaqGold360Mastermix (Life Technologies). The PCR primers used for the genotyping of the KCNJ10:c.986T>C variant were AGCTGGTGCTGATCCTCAGT (forward primer) and TCCCTTAACGACTCCTCCAA (reverse primer). PCR products were directly sequenced on an ABI 3730 capillary sequencer (Life Technologies) after treatment with exonuclease I and shrimp alkaline phosphatase. Sanger sequence data were analyzed with Sequencher 5.1 (GeneCodes).
Data availability
File S1 is a video illustrating the clinical phenotype of an affected Malinois dog with the KCNJ10:c.986T>C variant at 5, 7, and 8 wk of age (MA008). Table S1 contains KCNJ10:c.986T>C genotypes of 486 control dogs from 89 diverse dog breeds. Table S2 lists genome regions ≥1 Mb that showed positive log of odds (LOD) scores in the linkage analysis. Table S3 illustrates the homozygous genome regions with shared alleles among the six analyzed affected Malinois puppies from families 1 to 4 that exhibited phenotypic homogeneity. Table S4 lists the accession numbers of all whole genome sequencing data, which were deposited in the European Nucleotide Archive. Table S5 shows the 23 variants in the critical interval on chromosome 38 that were absent from 140 other dog genomes.
Results
Clinical presentation
The 12 Malinois puppies with cerebellar dysfunction had early onset of clinical signs (4.5–8.5 wk of age). During presentation 10 puppies were bright, alert, and responsive. Two puppies were less alert but were responsive. All puppies showed a wide-based ataxic gait, which was more obvious in the hind limbs. Exaggerated gait movements were observed in 50% of the affected puppies. Less consistent clinical signs were stumbling, staggering, intention tremor, bunny hopping, as well as balance loss and falling. Decelerated eye ball coordination during fast head motion was noted in three puppies. Circling episodes or short episodes of muscle spasms together with aggravation of cerebellar symptoms were reported in two puppies after stress or exercise (File S1). The four affected Malinois puppies belonging to family 6 were reported to have seizures, to run into obstacles, and to show rapid progression of clinical signs. Due to the severity of the clinical signs all affected puppies were euthanized by the 17th wk of age.
Neuropathological findings
Lesions in the cerebellum and brain stem of four Malinois puppies belonging to families 1–4 were similar to those described in Kleiter et al. (2011). In these puppies, we observed mild to marked vacuolation of the cerebellar nuclei and granule cell layer, as well as vacuoles and spheroids in the reticular formation. In two of these four puppies, the spinal cord was examined and showed vacuoles and spheroids in the white matter as well.
The affected puppies belonging to family 6 had comparable vacuolation of the cerebellar nuclei and the reticular formation. However, no vacuoles were detected in the cerebellar granule cell layer and no spheroids were seen in the reticular formation. In contrast to the Malinois puppies belonging to families 1–4, all affected puppies of family 6 additionally showed necrotic neurons and marked gliosis in the spinal cord gray matter.
The neuropathological changes noted in family 5 differed from the other families. The two affected puppies in this family showed very few vacuoles in the CNS, but marked gliosis in the cerebellar nuclei, in selected medullary nuclei, and in the spinal cord gray matter.
Pedigree analysis
Pedigrees were available from six Malinois families and were consistent with a monogenic autosomal recessive mode of inheritance. A common ancestor in both maternal and paternal lineages was identified in five of the six studied families within a maximum of five generations. In the remaining family (family 6), this common ancestor was found on the maternal side, but not on the paternal side (Figure 1).
Genetic mapping of the causative variant
The mapping of the causative locus was performed by investigating six Malinois families with a total of 12 Malinois puppies with cerebellar dysfunction, 20 nonaffected offspring, and 12 nonaffected parents (Figure 1).
An initial linkage analysis including all animals did not reveal any linked segments. As the histopathological examinations had already suggested subtle phenotypic differences between some of the families, we then performed the linkage analysis separately for each of the six families (Table S2). This revealed that families 1–4 shared two tentatively linked intervals on chromosomes 8 and 38, both reaching a LOD score of 2.449. We assumed that the lack of linkage to chromosome 8 and/or 38 in families 5 and 6 was due to locus heterogeneity and that the affected puppies in these families had possibly genetically distinct diseases.
Based on the results from the parametric linkage analysis and the pedigree records we hypothesized that the six affected puppies from families 1 to 4 most likely were inbred to one single founder animal. Under this scenario the affected individuals were expected to be identical by descent for the causative genetic variant and flanking chromosomal segments. We used a homozygosity mapping approach to fine-map the region of interest and analyzed the six affected Malinois puppies for extended regions of homozygosity with simultaneous allele sharing. We identified three genome regions with a total of 2.74 Mb that fulfilled our search criteria (Table S3). By intersecting the linked segments from families 1 to 4 and the homozygous intervals from the six cases of these families, only one segment on chromosome 38 remained (Figure 2). Therefore, the combined linkage and homozygosity analysis defined an exact critical interval of 1,414,646 bp at Chr38:21,060,597–22,475,242. Upon inspection of the SNP chip genotypes of isolated Malinois cases with unknown relationships to our families, we identified one additional puppy, which also carried the disease-associated haplotype in homozygous state (MA152). We therefore assumed that this puppy was affected by the same genetic disease as the cases in family 1–4.
Figure 2.
Combined linkage and homozygosity mapping. We performed parametric linkage analysis for a recessive trait in families 1–4 and homozygosity analysis across six selected cases. Two linked genome segments are indicated in orange and three homozygous segments with shared alleles are indicated in red. Only one region on chromosomes 38 showed both linkage and homozygosity and was considered the critical interval (arrow). Specifically, this ∼1.4 Mb region corresponded to Chr38:21,060,597–22,475,242.
Identification of the causative variant
A total of 30 genes was annotated in the 1.4 Mb critical interval on chromosome 38. To obtain a comprehensive overview of all variants in this region we resequenced the whole genome of three affected Malinois puppies and called single nucleotide as well as indel variants with respect to the reference genome of a presumably nonaffected Boxer (CanFam 3.1). The genotypes of the affected Malinois puppies were further compared with 140 dog genomes from various breeds that had been sequenced during other studies (Table S4). We hypothesized that the causative variant should be completely absent from all dog breeds in the sample set except the Belgian Shepherd breed. After applying this filter, 23 disease-associated variants including two missense variants remained (Table 1 and Table S5).
Table 1. Variants detected by whole genome resequencing of three affected Malinois dogs.
| Filtering Step | Number of Variants |
|---|---|
| Variants in the whole genomea | 938,586 |
| Variants in the critical 1.4 Mb interval on chromosome 38 | 3558 |
| Variants in the critical interval that were absent from 140 other dog genomes | 23 |
| Nonsynonymous variants in the whole genomea | 6007 |
| Nonsynonymous variants in the 1.4 Mb critical interval on chromosome 38 | 48 |
| Nonsynonymous variants in the critical interval, absent from 140 other dog genomes | 2 |
The sequences were compared to the reference genome (CanFam 3.1) from a Boxer. Only variants that were homozygous in all three affected Malinois puppies (MA008, MA094, and MA152) are reported. Nonsynonymous variants were classified based on the ENSEMBL annotation (version 72).
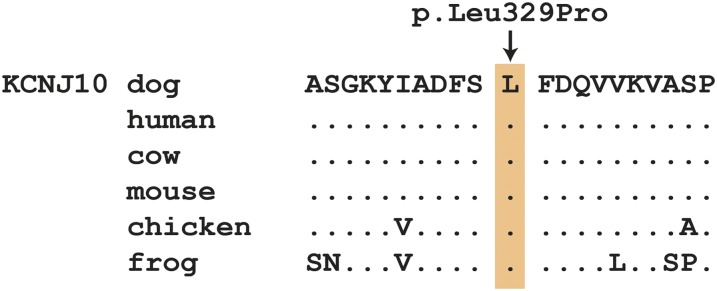
We genotyped the two missense variants in additional dogs and found the variant allele also in healthy control dogs from other breeds. Thus, we considered these variants as unlikely candidates and focused on the remaining 21 variants, which had been predicted to be noncoding by our automated pipeline. When we double-checked these variants with respect to the National Center for Biotechnology Information (NCBI) annotation, we recognized that one, Chr38:22,140,659T>C, is actually a missense variant located within exon 2 of the KCNJ10 gene, which is not correctly annotated in ENSEMBL. The variant can therefore also be described as KCNJ10:c.986T>C and is predicted to lead to a nonconservative amino acid exchange from leucine to proline at codon 329 (p.Leu329Pro). Residue 329 is located in the C-terminal cytoplasmic domain of KCNJ10. The region of the variant is highly conserved across vertebrates (Figure 3).
Figure 3.
Evolutionary conservation of the leucine residue at position 329 in the KCNJ10 protein. Vertebrates share highly conserved acid sequences in the region of the variant. The sequences were derived from the following database accessions: C. lupus XP_545752.3, H. sapiens NP_002232.2, B. taurus NP_001075070.1, M. musculus NP_001034573.1, G. gallus XP_003643542.1, X. tropicalis NP_001072312.1.
The presence of this variant in homozygous state was confirmed by Sanger sequencing in the six affected Malinois puppies belonging to families 1–4 and in the isolated case MA152. We additionally genotyped 231 other Malinois, 25 Groenendael, two Laekenois, 34 Tervueren, and 486 dogs of genetically diverse other breeds for this variant. The variant was not found outside the Belgian Shepherd population. The variant was present in heterozygous state or absent in the remaining six cases from families 5 and 6 and the six remaining isolated cases (Table 2 and Table S1).
Discussion
In this study, we identified a missense variant of KCNJ10 as a candidate causative genetic defect for SDCA in the Belgian Shepherd breed, more specifically in Malinois dogs. We propose to call this particular phenotype spongy degeneration with cerebellar ataxia, subtype 1 (SDCA1).
We acknowledge the limitations of our linkage analysis, which did not reach a significant LOD score of three in the investigated families. The chosen short read resequencing methodology in combination with an incomplete reference genome was also not 100% sensitive. However, as we identified a nonsynonymous variant in a highly plausible functional candidate gene, we think that our mapping and genome resequencing data combined with the knowledge on KCNJ10 function in humans, mice, and dogs strongly support the causality of KCNJ10:c.986T>C in the Belgian Shepherd breed.
In dogs, a KCNJ10 variant (XM_545752.3:c.627C>G) has already been reported in several terrier breeds (Gilliam et al. 2014; Rohdin et al. 2015). This missense variant is predicted to result in p.Ile209Met and leads to hereditary ataxia, also described as spinocerebellar ataxia with myokymia, seizures, or both (SAMS, Gilliam et al. 2014). Although there are some similarities in the clinical findings of the terrier’s hereditary ataxia and the SDCA1 in Belgian Shepherd dogs, substantial neuropathological differences in these two distinct diseases are evident. In terriers, the neuropathological lesions are characterized by bilateral symmetrical axonopathy with secondary demyelination, most prominent in the spinal cord. Lesions in the peripheral nervous system and in the brain were also described. However, in contrast to Belgian Shepherd dogs with SDCA1, changes in the cerebellum and/or spongy degeneration in the CNS were not noted in the terrier breeds. Our findings highlight the phenotypic variability caused by different KCNJ10 variants in the dog, which has also been described in humans (Cross et al. 2013; Abdelhadi et al. 2016). The two canine variants (p.Ile209Met and p.Leu329Pro) are relatively far apart from each other and also relatively far away from all known human missense variants (Abdelhadi et al. 2016). Therefore, a detailed genotype–phenotype correlation with respect to specific amino acid exchanges cannot yet be established. Additional studies are necessary to better understand these rare and complex diseases in dogs and humans.
It remains unclear why seizures were not found in dogs with the KCNJ10:c.986T>C variant. However, it is possible that infrequent or subtle (focal) seizures were missed or misinterpreted by owners. Furthermore, future assessment of renal, auditory, and ocular functions in SDCA1-affected Belgian Shepherd dogs could reveal additional similarities to the human EAST syndrome.
Our findings strongly suggest that more than one form of cerebellar dysfunction occurs in the Belgian Shepherd breed. The KCNJ10:c.986T>C variant was not present in homozygous state in any of the dogs with cerebellar dysfunction in families 5 and 6, as well as in six isolated Malinois puppies with neurological abnormalities. Moreover, after retrospective revision of histopathology, we identified a subtle phenotype heterogeneity, which further supports our hypothesis of a different genetic basis in closely related litters. However, it also should be kept in mind that neuropathological differences between cases may have been caused by different intervals between onset of clinical signs and postmortem examination.
This study highlights the challenges that dog breeders are confronted with if they do not have access to genetic tests. Prior to our study, there was a strong suspicion that the common ancestor of our six families, a very popular sire, had been a carrier for one recessive neurological defect. Our study demonstrates that the common ancestry of this sire was most likely coincidental and and it remains unclear whether this dog was indeed a carrier for the KCNJ10 and/or the hypothetical defect(s) underlying the cases in families 5 and/or 6.
In conclusion, we identified the KCNJ10:c.986T>C missense variant as most likely causative for SDCA1 in the Belgian Shepherd breed. Cerebellar dysfunction in this breed is heterogeneous and the reported variant explains only a fraction of clinically comparable cases. Further investigations on the other forms of cerebellar dysfunction are needed to clarify their precise phenotype and the underlying genetic variants. The identified affected dogs may serve as a model for the human EAST syndrome. Our findings enable genetic testing in Belgian Shepherd dogs, so that the nonintentional breeding of affected puppies with this specific disease can be avoided in the future.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.038455/-/DC1.
Acknowledgments
We are grateful to all dog owners and breeders who donated samples and shared pedigree data and other valuable information about their dogs, especially R. Lutz and D. Huber of the veterinary service of the Swiss army and M. Wollschläger. We acknowledge collaborators of the Dog Biomedical Variant Database Consortium, Gus Aguirre, Catherine André, Danika Bannasch, Doreen Becker, Cord Drögemüller, Oliver Forman, Eva Furrow, Urs Giger, Christophe Hitte, Marjo Hytönen, Vidhya Jagannathan, Tosso Leeb, Hannes Lohi, Cathryn Mellersh, Jim Mickelson, Anita Oberbauer, Jeffrey Schoenebeck, Sheila Schmutz, and Claire Wade, for sharing dog genome sequence data from control dogs. We thank Nathalie Besuchet Schmutz and Muriel Fragnière for expert technical assistance, the Next Generation Sequencing Platform of the University of Bern for performing the whole genome sequencing experiments, and the Vital-IT high-performance computing center of the Swiss Institute of Bioinformatics for performing computationally intensive tasks (http://www.vital-it.ch/). This study was supported by a grant from the Albert-Heim Foundation. During the final production stages of this manuscript, another independent study also reported the KCNJ10:c.986T>C variant as being pathogenic for the SeSAME/EAST homolog in Malinois dogs (Van Poucke et al. 2016).
Footnotes
Communicating editor: D. L. Bannasch
Literature Cited
- Abdelhadi O., Iancu D., Stanescu H., Kleta R., Bockenhauer D., 2016. EAST syndrome: clinical, pathophysiological, and genetic aspects of mutations in KCNJ10. Rare Dis. 4: e1195043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis G. R., Cherny S. S., Cookson W. O., Cardon L. R., 2002. Merlin –rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30: 97–101. [DOI] [PubMed] [Google Scholar]
- Arai E., Baba Y., Iwagawa T., Kuribayashi H., Mochizuki Y., et al. , 2015. Ablation of Kcnj10 expression in retinal explants revealed pivotal roles for Kcnj10 in the proliferation and development of Müller glia. Mol. Vis. 21: 148–159. [PMC free article] [PubMed] [Google Scholar]
- Bai B., Zhao W. M., Tang B. X., Wang Y. Q., Wang L., et al. , 2015. DoGSD: the dog and wolf genome SNP database. Nucleic Acids Res. 43(Database issue): D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockenhauer D., Feather S., Stanescu H. C., Bandulik S., Zdebik A. A., et al. , 2009. Epilepsy, ataxia, sensorineural deafness, tubulopathy, and KCNJ10 mutations. N. Engl. J. Med. 360: 1960–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachin M., Vandevelde M., 1991. Congenital tremor with spongy degeneration of the central nervous system in two puppies. J. Vet. Intern. Med. 5: 87–90. [DOI] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross J. H., Arora R., Heckemann R. A., Gunny R., Chong K., et al. , 2013. Neurological features of epilepsy, ataxia, sensorineural deafness, tubulopathy syndrome. Dev. Med. Child Neurol. 55: 846–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djukic B., Casper K. B., Philpot B. D., Chin L.-S., McCarthy K. D., 2007. Conditional knock-out of Kir4.1 leads to glial membrane depolarization, inhibition of potassium and glutamate uptake, and enhanced short-term synaptic potentiation. J. Neurosci. 27: 11354–11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliam D., O’Brien D. P., Coates J. R., Johnson G. S., Johnson G. C., et al. , 2014. A homozygous KCNJ10 mutation in Jack Russell Terriers and related breeds with spinocerebellar ataxia with myokymia, seizures, or both. J. Vet. Intern. Med. 28: 871–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiter M., Högler S., Kneissl S., Url A., Leschnik M., 2011. Spongy degeneration with cerebellar ataxia in Malinois puppies: a hereditary autosomal recessive disorder? J. Vet. Intern. Med. 25: 490–496. [DOI] [PubMed] [Google Scholar]
- Li H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The genome analysis toolkit: a mapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neusch C., Rozengurt N., Jacobs R. E., Lester H. A., Kofuji P., 2001. Kir4.1 potassium channel subunit is crucial for oligodendrocyte development and in vivo myelination. J. Neurosci. 21: 5429–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palygin O., Pochynyuk O., Staruschenko A., 2017. Role and mechanisms of regulation of the basolateral Kir4.1/Kir5.1 K+ channels in the distal tubules. Acta Physiol. (Oxf). 219: 260–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohdin C., Lüdtke L., Wohlsein P., Hultin Jäderlund K., 2010. New aspects of hereditary ataxia in smooth-haired fox terriers. Vet. Rec. 166: 557–560. [DOI] [PubMed] [Google Scholar]
- Rohdin C., Gilliam D., O’Leary C. A., O’Brien D. P., Coates J. R., et al. , 2015. A KCNJ10 mutation previously identified in the Russell group of terriers also occurs in Smooth-Haired Fox Terriers with hereditary ataxia and in related breeds. Acta Vet. Scand. 57: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Rabanal M., Kucheryavykh L. Y., Skatchkov S. N., Eaton M. J., Nichols C. G., 2010. Molecular mechanisms of EAST/SeSAME syndrome mutations in Kir4.1 (KCNJ10). J. Biol. Chem. 285: 36040–36048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl U. I., Choi M., Liu T., Ramaekers V. T., Häusler M. G., et al. , 2009. Seizures, sensorineural deafness, ataxia, mental retardation, and electrolyte imbalance (SeSAME syndrome) caused by mutations in KCNJ10. Proc. Natl. Acad. Sci. USA 106: 5842–5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D. A., Feather S., Stanescu H. C., Freudenthal B., Zdebik A. A., et al. , 2011. Altered electroretinograms in patients with KCNJ10 mutations and EAST syndrome. J. Physiol. 589: 1681–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera G. A., Carneiro M. O., Hartl C., Poplin R., Del Angel G., et al. , 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43: 11.0.1–11.0.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Poucke, M., K. Stee, S. F. M. Bhatti, A. Vanhaesebrouck, L. Bosseleret al. , 2016. The novel homozygous KCNJ10 c.986T>C (p.(Leu329Pro)) variant is pathogenic for the SeSAME/EAST homologue in Malinois dogs. Eur. J. Hum. Genet. DOI: 10.1038/ejhg.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessmann A., Goedde T., Fischer A., Wohlsein P., Hamann H., et al. , 2004. Hereditary ataxia in the Jack Russell Terrier—clinical and genetic investigations. J. Vet. Intern. Med. 18: 515–521. [DOI] [PubMed] [Google Scholar]
- Wiedmer M., Oevermann A., Borer-Germann S. E., Gorgas D., Shelton G. D., et al. , 2016. A RAB3GAP1 SINE insertion in Alaskan huskies with polyneuropathy, ocular abnormalities, and neuronal vacuolation (POANV) resembling human Warburg micro syndrome 1 (WARBM1). G3 6: 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. M., Lopes C. M., Rosenhouse-Dantsker A., Connelly H. L., Matavel A., et al. , 2010. Molecular basis of decreased Kir4.1 function in SeSAME/EAST syndrome. J. Am. Soc. Nephrol. 21: 2117–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
File S1 is a video illustrating the clinical phenotype of an affected Malinois dog with the KCNJ10:c.986T>C variant at 5, 7, and 8 wk of age (MA008). Table S1 contains KCNJ10:c.986T>C genotypes of 486 control dogs from 89 diverse dog breeds. Table S2 lists genome regions ≥1 Mb that showed positive log of odds (LOD) scores in the linkage analysis. Table S3 illustrates the homozygous genome regions with shared alleles among the six analyzed affected Malinois puppies from families 1 to 4 that exhibited phenotypic homogeneity. Table S4 lists the accession numbers of all whole genome sequencing data, which were deposited in the European Nucleotide Archive. Table S5 shows the 23 variants in the critical interval on chromosome 38 that were absent from 140 other dog genomes.