Abstract
Pancreatic ductal adenocarcinoma (PDAC) and other carcinomas are hierarchically organized, with cancer stem cells (CSC) residing at the top of the hierarchy, where they drive tumor progression, metastasis, and chemoresistance. As CSC and non-CSC share an identical genetic background, we hypothesize that differences in epigenetics account for the striking functional differences between these two cell populations. Epigenetic mechanisms, such as DNA methylation, play an important role in maintaining pluripotency and regulating the differentiation of stem cells, but the role of DNA methylation in pancreatic CSC is obscure. In this study, we investigated the genome-wide DNA methylation profile of PDAC CSC, and we determined the importance of DNA methyltransferases for CSC maintenance and tumorigenicity. Using high-throughput methylation analysis, we discovered that sorted CSCs have a higher level of DNA methylation, regardless of the heterogeneity or polyclonality of the CSC populations present in the tumors analyzed. Mechanistically, CSC expressed higher DNMT1 levels than non-CSC. Pharmacologic or genetic targeting of DNMT1 in CSCs reduced their self-renewal and in vivo tumorigenic potential, defining DNMT1 as a candidate CSC therapeutic target. The inhibitory effect we observed was mediated in part through epigenetic reactivation of previously silenced miRNAs, in particular the miR-17-92 cluster. Together, our findings indicate that DNA methylation plays an important role in CSC biology and also provide a rationale to develop epigenetic modulators to target CSC plasticity and improve the poor outcome of PDAC patients.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) represents the fourth most frequent cause of cancer-related death due to its extreme lethality and current lack of effective treatments (1). As incidence and death rates continue to increase, pancreatic cancer is predicted to become the second most frequent cause of cancer-related death by 2030 (2), making this disease a major unmet priority in public health care. Although multiple subclonal populations of cancer cells coexist within each tumor and are assumed to drive tumor adaptation and therapeutic failure through Darwinian selection (3), convincing evidence now demonstrates that cancer heterogeneity is also driven by phenotypic and functional heterogeneity within each of these subclones, resulting in a hierarchical tumor organization (4). At the apex of this hierarchy are populations of cancer stem cells (CSC) capable of self-renewal and long-term in vivo tumorigenicity. CSCs give rise to more differentiated progenies (non-CSCs), which, although sharing common mutation profiles, bear distinct and thus most likely epigenetically defined gene expression patterns (5, 6).
Identifying the epigenetic mechanisms that are responsible for the acquisition and preservation of these distinct CSC features could open up possibilities for the development of new and more effective therapeutic strategies for PDAC. Unlike genetic mutations, epigenetic changes are transient and reversible, and as such, therapies that convert the epigenetic balance of CSCs toward that of non-CSCs could provide the basis for developing more effective treatment strategies for cancer patients (7). Among the first epigenetic drugs proposed were inhibitors of DNA methylation, for example, 5-azacytidine (5-aza-CR, azacytidine) and 5-aza-2′-deoxycytidine (5-aza-CdR, decitabine), followed later by zebularine, which all incorporate into DNA and form covalent irreversible complexes with DNA methyltransferases (DNMT; ref. 8). These inhibitors have been shown to induce differentiation of cultured cancer cells (9), but our knowledge about their effects on CSCs is still sparse. Moreover, to date, only few studies have utilized the new DNA methylation inhibitor zebularine, which can be administered orally and is less toxic (10). Thus, we aimed to characterize the supposedly distinct methylation profile of primary pancreatic CSCs and subsequently studied the effects of genetic or pharmacologic targeting of DNMT1 on CSC phenotypes.
Materials and Methods
Primary human cancer cells
PDAC tumors were obtained with written consent from all pancreatic cancer patients, expanded in immunocompromised mice as patient-derived xenografts (PDX), and subsequently digested to establish low-passage primary cell cultures (11).
In vivo tumorigenicity
Serial dilutions of primary pancreatic cancer cells were resuspended in Matrigel (BD Biosciences), subcutaneously injected into the right and left flank of female NU-Foxn1nu nude mice (Harlan Laboratories), and tracked for up to 3 months. Experiments were approved by the Animal Experimental Ethics Committee of the Instituto de Salud Carlos III (Madrid, Spain; CBA 68_2013 & CBA 25_2009) and performed in accordance with the guidelines for Ethical Conduct in the Care and Use of Animals. CSC frequency was calculated using the extreme limiting dilution analysis (LDA) algorithm (http://bioinfo.wehi.edu.au/software/elda/index.html).
Sphere formation assay
Spheres were generated by culturing 2 × 103 PDAC cells/mL in ultra-low attachment plates (Corning) using serum-free DMEM/F12 supplemented with B27 (1:50, Invitrogen), 20 ng/mL bFGF, and 50 U/mL penicillin/streptomycin for 7 days. For serial passaging, sphere cultures were depleted for single cells and "spheres" measuring <40 μm using a 40-μm cell strainer. Retained spheres were dissociated into single cells, recultured for another 7 days. Spheres >40 μm were quantified with a CASY Cell Counter (Roche; ref. 11). Primary sphere-derived human PDAC cells were treated with zebularine (75 μmol/L) or decitabine (50 μmol/L) for 7days. The drugs were readministered every other day to the cell suspension.
Flow cytometry analysis and FACS
Primary pancreatic cells, dissociated spheres, or cells from tumor digestions were stained with anti-hCD133/1-APC or PE (Miltenyi Biotec), hEPCAM-APC (Miltenyi Biotec), hCD324-APC (BioLegend), hPan-Cytokeratin-FITC (Miltenyi Biotec), or appropriate control antibodies (all from BD Biosciences), counterstained with DAPI (2 μg/mL) for exclusion of dead cells, and analyzed using a FACSCanto II instrument (BD Biosciences). Data were analyzed with FlowJo 9.2 software (Tree Star). For FACS analysis, cells were adjusted to a concentration of 106 cells/mL in sorting buffer [1× PBS; 3% FBS (v/v); 3 mmol/L EDTA]. DAPI was added to exclude dead cells, and cells were sorted using a FACS Influx instrument (BD Biosciences).
DNA methylation analysis
Genomic DNA (1 μg) was treated by bisulfite conversion with the EZ DNA Methylation Kit (D5004, Zymo Research) according to the manufacturer's recommendations. The HumanMethylation450K BeadChip (Illumina, Inc.) was used for analysis of genome-wide DNA methylation according to the manufacturer's instructions. To identify differently methylated probes in paired autofluorescent-positive and -negative cells from all tumors, we used limma package (12). Probes were considered to be differentially methylated if the resulting adjusted P value was <0.05. The Benjamini–Hochberg method was used to adjust the P values and ensure that the FDR was <0.05. The genomic regions of the probes from the array were assigned according to their position relative to the transcript information obtained from the R/Bioconductor package FDb.InfiniumMethylation.hg19 (R package version 2.9.2). The CGI locations used in the analyses were obtained from the R/Bioconductor package FDb.InfiniumMethylation.hg19 (R package version 1.0.1). The definition of CGI was done as described previously (13).
Statistical analysis
Results for continuous variables are presented as means ± SEM unless stated otherwise, and significance was determined using the Mann–Whitney U test. All analyses were performed using SPSS 22.0 (SPSS). Significance was considered at P < 0.05.
Additional experimental details can be found in the Supplementary Materials and Methods.
Results
Pancreatic CSCs bear higher DNA methylation levels
We first performed genome-wide comprehensive methylation profiling using the 450K Illumina bead array (14) to gain insight into putative DNA methylation differences between CSCs and non-CSCs. CSCs can be separated from the tumor bulk population by several methods (11); however, we recently showed that PDAC CSCs can also be efficiently isolated using autofluorescence (an accumulation of riboflavin in ATP-dependent transporter ABCG2-coated vesicles exclusively found in CSCs; ref. 15). Using this CSC inherent marker, we separated CSCs from non-CSCs by FACS sorting (Fig. 1A; ref. 15), and the efficient enrichment for CSCs was validated by increased expression of pluripotency-associated genes in autofluorescent-positive (Fluo+) CSCs versus Fluo− cells (Supplementary Fig. S1A and S1B).
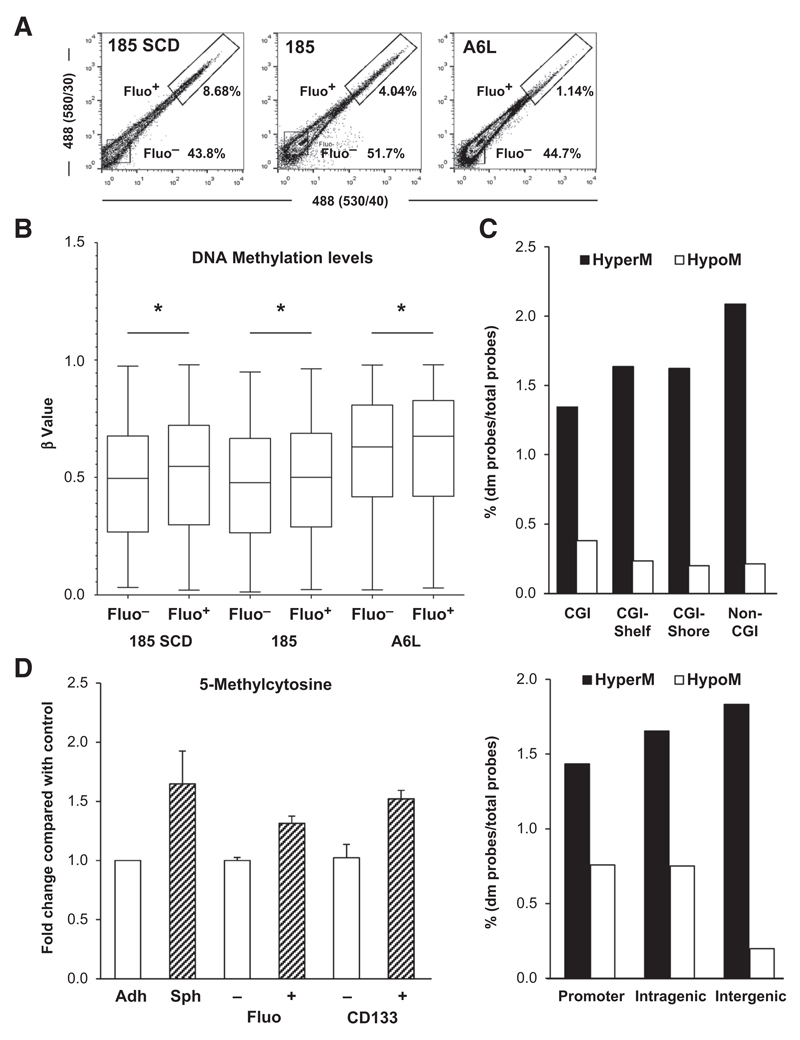
Figure 1. Pancreatic CSCs bear higher levels of DNA methylation.
A, flow cytometry analysis of autofluorescence in sphere-derived cells from PDAC-185 SCD (PDAC tumor derived from a single PDAC 185 autofluorescent cell), PDAC-185 (primary tumor), and PDAC-A6L (PDAC liver metastasis). B, box plots representing DNA methylation levels in representative pairs of autofluorescent-negative (Fluo–) and -positive (Fluo+) cells from the indicated primary PDAC sphere-derived cultures (*, P < 0.05). C, distribution of differently methylated (dm) probes based on their genomic location relative to CpG islands (CGI; top). M, methylated. CpG island shores represent regions 0 to 2 kb from CpG islands; shelves indicate regions 2 to 4 kb from CpG islands. Distribution relative to the promoter (upstream the transcription start site), and intragenic and intergenic nonpromoter regions (bottom). D, quantification of 5mC using the MethylFlash Quantification Kit in non-CSCs versus CSCs (adherent vs. sphere, Fluo– vs. Fluo+, and CD133– vs. CD133+). adh, adherent; sph, spheres. Data are shown as fold change compared with non-CSC (mean ± SD; n = 3).
As the vast intratumoral heterogeneity in PDAC might obscure distinct methylation profiles between CSCs and non-CSCs within each contained subclone, we did not only use cells derived from a heterogeneous primary pancreatic tumor (PDAC-185), but also analyzed a liver metastasis (PDAC-A6L) and a single cell–derived (SCD) tumor that was generated using a single CSC isolated from the primary tumor (PDAC-185 SCD; Supplementary Fig. S1C and S1D). We reasoned that the CSC heterogeneity should be highest in the primary tumor, less in the metastatic tumor, and homogenous in the CSCs isolated from the SCD tumor. DNA methylation levels were compared between CSCs and non-CSCs for each individual tumor, which revealed a slight, but significant increase in DNA methylation in the Fluo+ CSC compartment, regardless of the heterogeneity or polyclonality of the CSC populations present (Fig. 1B). These data suggest consistent differences in the methylation profile despite considerable intratumoral heterogeneity.
Furthermore, we looked at differentially methylated probes (hyper- or hypomethylated) in CSCs and found that their distribution was not universal. Hypermethylation was mostly located in non-CGI (OD = 1.43) and intergenic regions (OR = 1.17), and hypomethylation was mostly found in CGI (OR = 1.80) and promoter regions (OR = 1.43; Fig. 1C), indicating that the hypermethylation phenotype observed in the pancreatic CSC population is largely a result of methylation of regions that are outside of traditional CGI. To further confirm this observation, we measured the levels of the DNA methylation mark 5-methylcytosine (5 mC) using the MethylFlash Methylated DNA Kit or by manual dotblot analysis and show that regardless of the method used to isolate CSCs (sphere vs. adherent, Fluo+ vs. Fluo- or CD133+ vs. CD133-), the CSC population exhibited consistently higher levels of 5 mC, indicating that cytosine was methylated to 5-methylcytosine by DNMTs, which is in line with our Illumina Array data (Fig. 1D and Supplementary Fig. S2A).
Pancreatic CSCs overexpress DNMT1
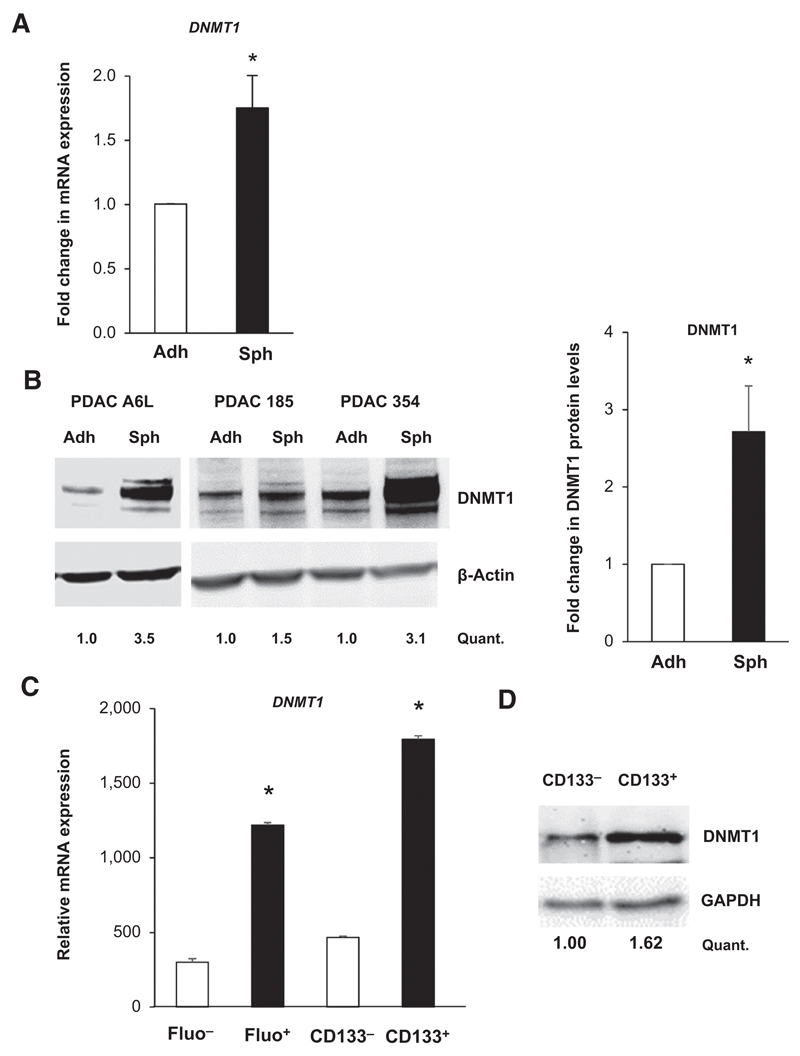
We next asked whether hypermethylation could be explained by differential expression of DNMTs. Most strikingly, we found higher mRNA expression levels of DNMT1 in the CSC population, regardless of the isolation method used (Fig. 2A and C and Supplementary Fig. S2B). Western blot analysis confirmed higher DNMT1 protein expression in CSCs versus non-CSCs (Fig. 2B–D). On the basis of the above data, we reasoned that DNMT1 overexpression plays a decisive role in preserving the stemness state of CSCs via maintaining their distinct methylation state (16).
Figure 2. Pancreatic CSCs overexpress DNMT1.
A, qRT-PCR analysis of DNMT1 mRNA in PDAC adherent (adh) and sphere (sph) cultures. Data are normalized to β-actin levels and represent pooled values from different primary PDAC cultures (A6L, 185, 354, and 215; *, P < 0.05; n = 5). B, representative Western blot images of DNMT1 protein expression in a panel of different primary adherent (adh) and sphere (sph)-derived cultures (A6L, 185, and 354) and densitometric quantification (Quant.; left). Changes in protein levels are depicted as fold change in pooled adherent cultures versus pooled sphere-derived cultures (* P < 0.05; n = 3). C, relative mRNA level of DNMT1 in CSCs (Fluo+ and CD133+) versus non-CSCs (Fluo– and CD133–; *, P < 0.05, n = 3; right). D, representative Western blot images of DNMT1 protein expression in CSCs (CD133+) versus non-CSCs (CD133–) and densitometric quantification.
DNMT1 inhibition decreases PDAC CSCs phenotypes
On the basis of the aforementioned results, we aimed to pharmacologically target DNMT1 using zebularine to reverse the distinct methylome signature of CSCs and assess whether DNMT1 inhibition could ablate PDAC CSC tumorigenicity. Three primary PDAC cell cultures were treated with zebularine over the course of 7 days in conditions that enrich for CSCs (Supplementary Fig. S3A and S3B). A concentration of 75 μmol/L was used in all of our subsequent experiments as cytotoxicity studies revealed that concentrations ≥100 μmol/L were potentially toxic (Supplementary Fig. S3C).
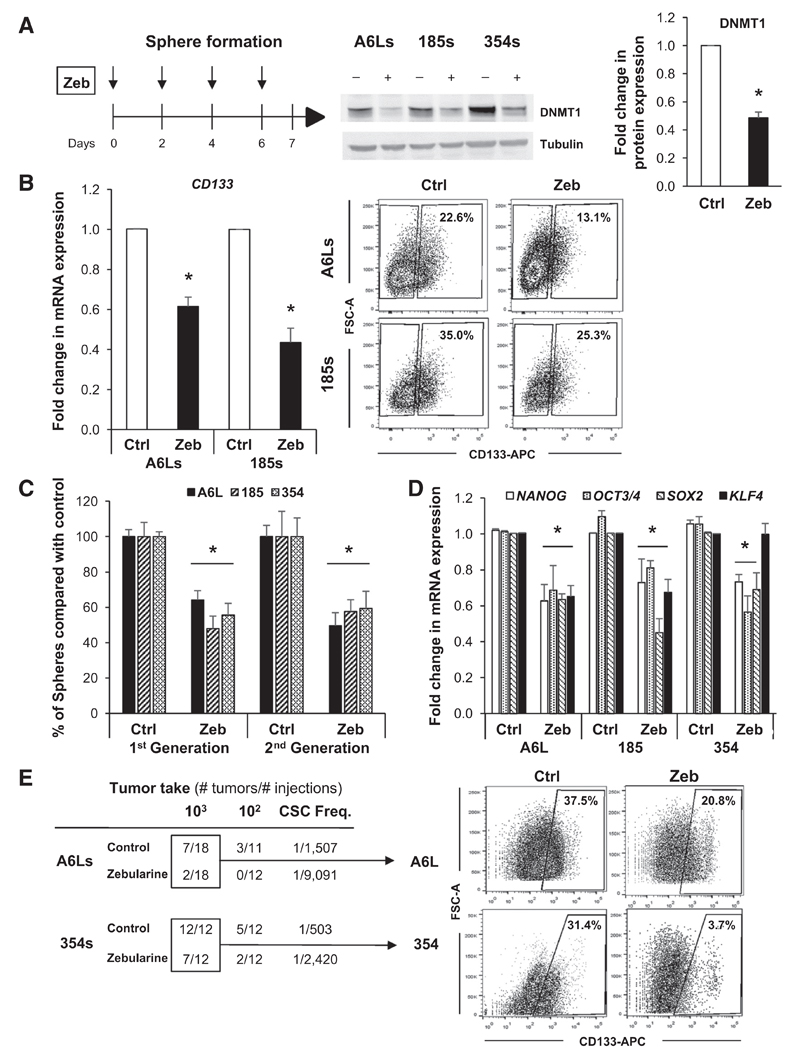
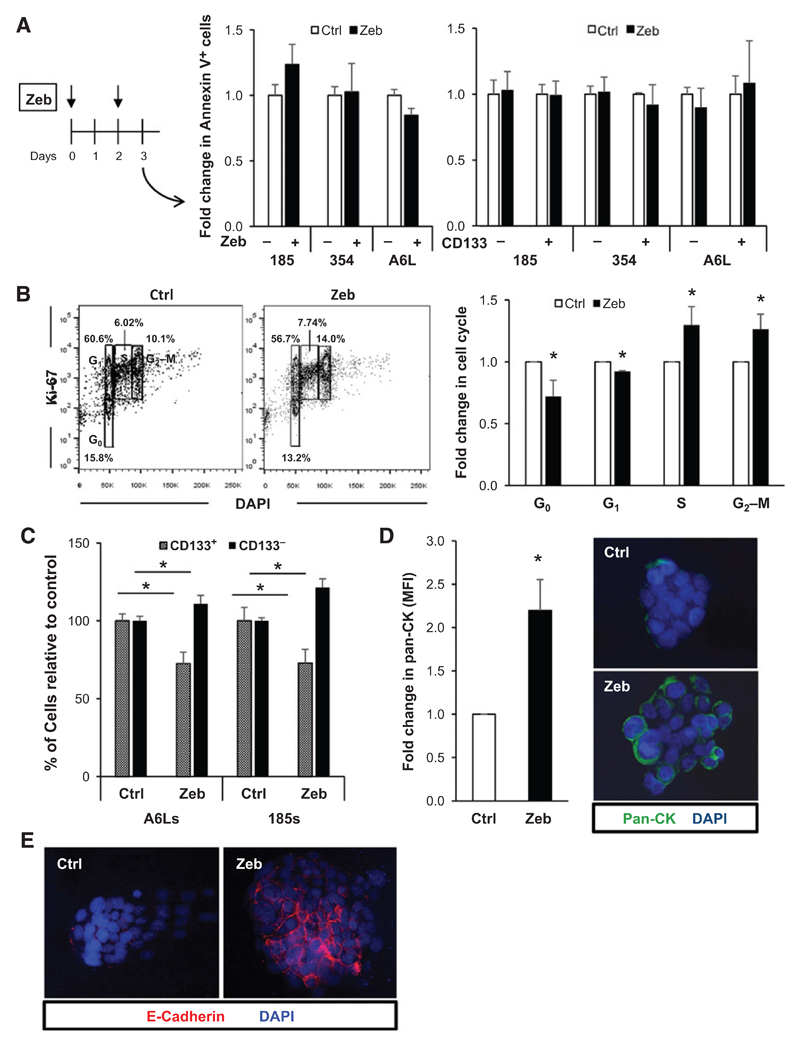
Although zebularine expectedly showed minor or no effect on DNMT1 mRNA levels (Supplementary Fig. S3D), we observed a consistent inhibition of DNMT1 at the protein level (Fig. 3A) and a marked reduction in CD133 mRNA and surface protein levels (Fig. 3B), suggesting preferential targeting of CSCs. A similar reduction was observed when we sorted cells for autofluorescence to identify CSCs (Supplementary Fig. S3E). At the functional level, zebularine reduced both CSC self-renewal in vitro (Fig. 3C) and the expression of pluripotency-associated genes (Fig. 3D). To further corroborate that pharmacologic inhibition of DNMT1 is suitable for targeting pancreatic CSCs, we used another DNMT inhibitor, decitabine. Indeed, nontoxic levels of DAC (Supplementary Fig. S4A) decreased DNMT1 protein levels (Supplementary Fig. S4B) and subsequently reduced the self-renewal capacity of CSCs in three different PDAC cultures (Supplementary Fig. S4C). Moreover, decitabine treatment significantly decreased the expression of the pluripotency-associated gene OCT3/4 (Supplementary Fig. S4D). The differential effects on the expression of pluripotency-associated genes between zebularine and decitabine may, at least in part, be related to differences in DNA-hypomethylating properties of the drugs as previously suggested (17). Nonetheless, at the functional level, both DNMT1 inhibitors significantly inhibited the CSC population. As the most defining feature of CSCs is their ability to form tumors in vivo, we also performed limiting dilution in vivo tumorigenicity assays with control or Zebularine-treated cells to assess their tumorigenic potential. Zebularine-treated cells produced significantly fewer (and smaller) tumors, resulting in a 5- to 6-fold lower calculated CSC frequency (Fig. 3E, left). Moreover, in tumors that did form, the percentage of CD133+ cells was markedly reduced (Fig. 3E, right), already suggesting enhanced epithelial differentiation as a possible mechanism of action.
Figure 3. The DNMT1 inhibitor zebularine decreases CSC phenotypes.
A, scheme showing treatment strategy for control (Ctrl) versus zebularine (Zeb) in spheres (PDAC-A6L, -185, and -354; left) and Western blot analysis of DNMT1 protein levels following treatment (middle). Densitometric quantification analysis (right; *, P < 0.05; n = 3). B, qRT-PCR analysis of CD133 mRNA in PDAC sphere-derived cultures treated with zebularine for 7 days. Data are normalized to β-actin and represented as fold change compared with untreated cells (left; *, P < 0.05; n = 3). Representative flow cytometry showing the percentage of CD133-positive and -negative cells from PDAC spheres treated for 7 days with zebularine (right). C, number of spheres per mL in first- and second-generation cultures from primary PDAC tumors (A6L, 185, and 354; *, P < 0.05; n = 4). D, qRT-PCR analysis of pluripotency-associated genes in first-generation spheres. Data are normalized to β-actin and presented as fold change in comparison with untreated cells (*, P < 0.05; n = 4). E, summary of in vivo tumorigenicity of subcutaneously injected control and zebularine-treated sphere-derived cells 12 weeks postinjection (left). CSC frequencies (Freq.) were determined using the extreme LDA algorithm. Representative flow cytometry plots showing the percentage of CD133 expression in digested tumors derived from control and zebularine-treated cells (right).
Knockout of DNMT1 decreases CSCs phenotypes
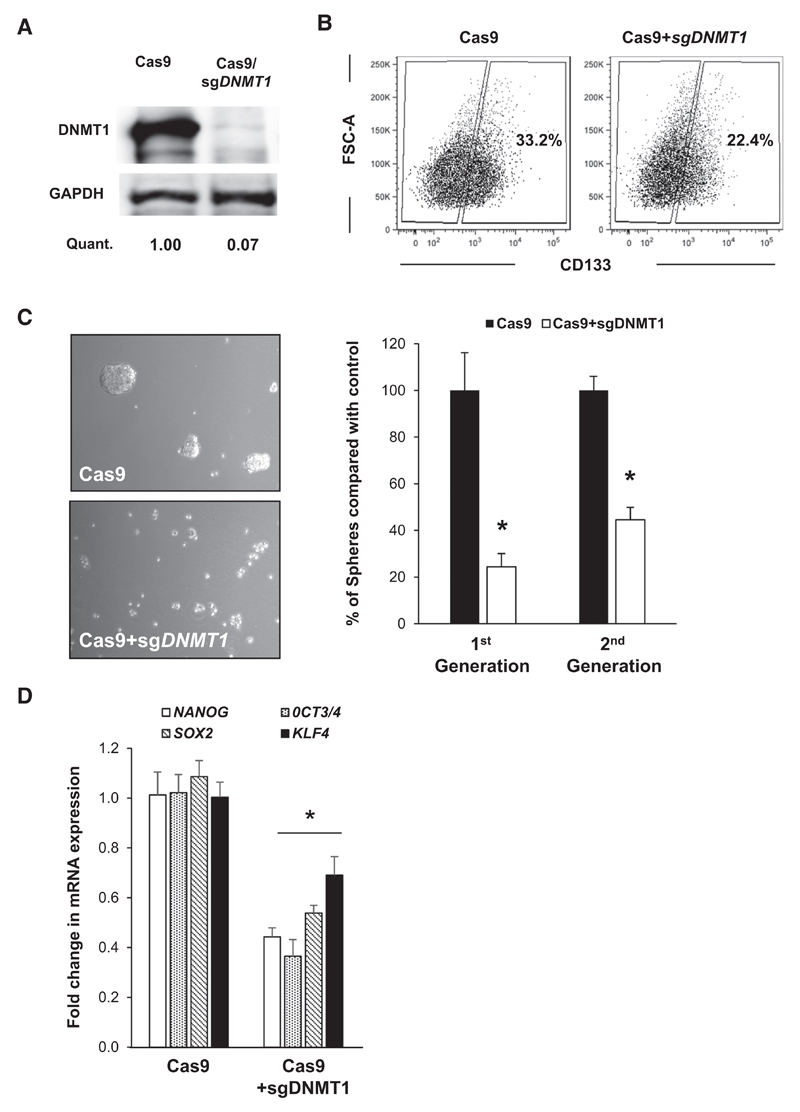
DNMT1 inhibitors, including zebularine and decitabine, have a similar mode of action (incorporation into DNA as cytosine analogues). The formation of covalent adducts between DNA and trapped DNA methyltransferase protein can induce toxic effects, making it difficult to separate demethylating activity from cytotoxicity (18). Therefore, to genetically verify our hypothesis that DNMT1 is indeed crucial for maintaining the CSC status, we generated PDAC cells lacking DNMT1 via CRISPR/Cas9 editing. After two weeks under selection, the 185 DNMT1-KO cells showed complete loss of DNMT1 expression at the protein level (Fig. 4A). The subsequent loss of DNMT1 (i) decreased the percentage of CD133+ CSCs (Fig. 4B), (ii) significantly abrogated the in vitro self-renewal capacity of CSCs as measured by multiple generation sphere formation potential (Fig. 4C), and (iii) decreased the expression of pluripotency-associated genes (Fig. 4D).
Figure 4. Knockout of DNMT1 decreases CSC phenotypes.
A, Western blot analysis of DNMT1 protein levels in control (Cas9) and DNMT1-KO cells and densitometric quantification (Quant.). B, representative flow cytometry plots of CD133 cell surface expression in control (Cas9) and DNMT1-KO cells. C, representative images of spheres (left) and sphere counts (right) in first- and second-generation in control (Cas9) and DNMT1-KO cells (*, P < 0.05; n = 3). D, qRT-PCR analysis of pluripotency-associated genes in DNMT1-KO cells. Data are normalized to β-actin and are presented as fold change in comparison with control (Cas9) cells (*, P < 0.05; n = 4).
The DNMT1 inhibition promotes CSC proliferation and differentiation
The effects of DNMT1 inhibition on CSCs may be related to (i) apoptosis induction, (ii) cell-cycle arrest, or (iii) promotion of differentiation. Zebularine treatment during sphere formation did not significantly alter the percentage of early or late apoptotic cells in the entire tumor cell population (Fig. 5A, left) nor was there any evidence of apoptosis induction specifically in the CD133+ CSC population (Fig. 5A, right). Cell-cycle analysis, however, revealed reduced numbers of zebularine-treated cells residing in G0 and an increased number of actively cycling cells (Fig. 5B). Moreover, not only did we observe a decrease in the percentage of CD133+ CSCs upon treatment (Fig. 3B), but by comparing ratios between the CD133− and the CD133+ cell populations, we found that the decrease in CD133+ CSCs was followed by an increase in their CD133− non-CSC counterparts, suggesting that zebularine treatment potentially induced the "differentiation" of CSCs to non-CSC (Fig. 5C). This result was confirmed by assessing the cell surface levels of pan-cytokeratin and E-cadherin (Fig. 5D and E and Supplementary Fig. S5A and S5B) as markers of more differentiated PDAC cells (6). Higher expression of pan-cytokeratin and E-cadherin could also be observed following decitabine treatment (Supplementary Fig. S4E) and DNMT1 knockout (Supplementary Fig. S5C). Together, our data suggest a quiescence-inhibiting and differentiation-promoting effect of DNMT1 inhibition on pancreatic CSCs.
Figure 5. Zebularine promotes CSC proliferation and differentiation.
A, scheme of the treatment time course (left), Annexin V expression in control (Ctrl) and zebularine (Zeb)-treated PDAC-185, PDAC-354, and PDAC-A6L (n = 3; middle). Annexin V expression determined within the CD133+ and CD133− fractions, using CD133-PE for zebularine and control-treated PDAC-185, PDAC-354, and PDAC-A6L cultures (right). B, representative flow cytometry plots for Ki-67 staining in control versus zebularine-treated PDAC-354 cultures (left) and combined quantification (right; *, P < 0.05; n = 6). C, quantification of flow cytometry analysis of CD133 expression in control versus zebularine-treated spheres from PDAC-185 and PDAC-A6L (*, P < 0.05; n = 3). D, quantification of the mean fluorescence intensity (MFI) of pan-cytokeratin (pan-CK) staining in control versus zebularine-treated primary PDAC cultures (left; * P, < 0.05; n = 8). Representative confocal images are shown (right). E, representative confocal images of E-cadherin staining for control versus zebularine-treated cultures.
DNMT1 inhibition affects CSCs via hypomethylation of the miR-17-92 cluster
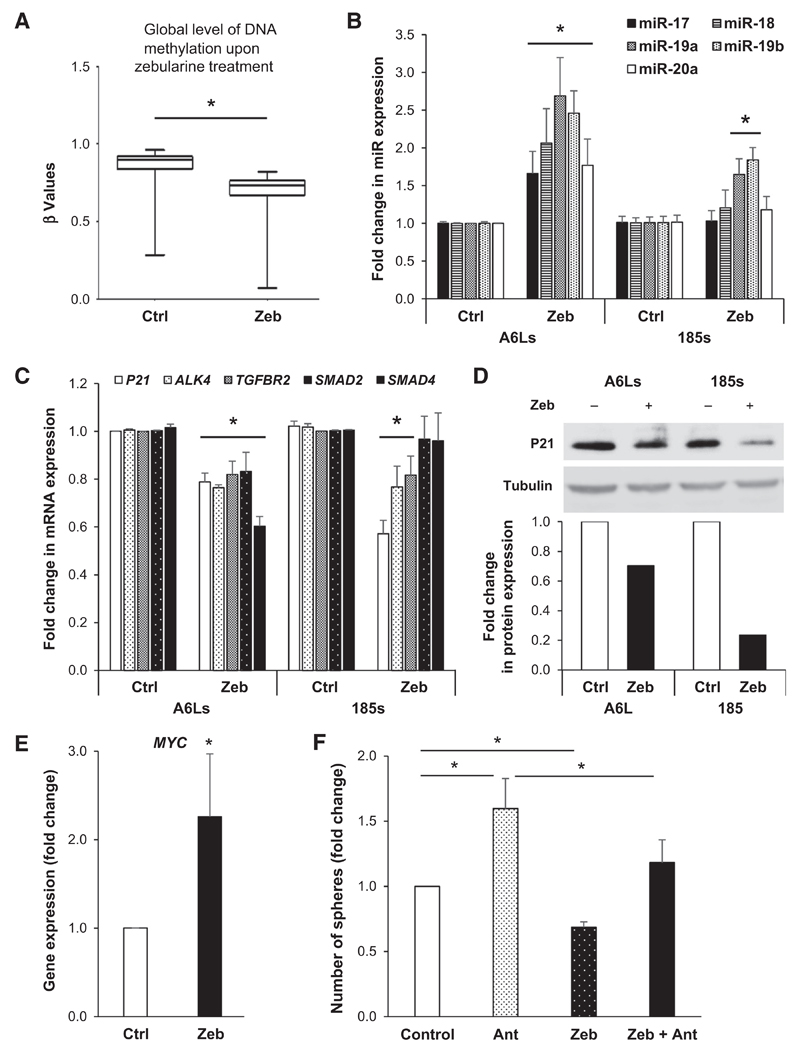
To determine the molecular mechanism(s) responsible for the effect of zebularine on CSCs, we analyzed putative changes in DNA methylation using the Illumina Infinium HumanMethylation450 Bead Chip. As expected, zebularine treatment decreased DNA methylation in PDAC CSCs (Fig. 6A). Interestingly, while methylation of many genes was changed upon treatment, others even gained methylation. Using a stringent analysis approach (fold change in β-methylation values less than 0.5 and fold change higher than 2), we found 97 and 548 genes demethylated and methylated, respectively (Supplementary Tables S1 and S2). While these genes are currently under investigation, previous work by our group indicated that miRNAs have an important role in PDAC CSCs, and thus, we initially focused our analysis on modulated miRNAs. Indeed, as shown in Table 1, we identified several miRNA more methylated in CSC-enriched spheres versus non-CSC adherent cultures, and using a less stringent analysis (fold change in β-methylation values 0.8), many were subsequently hypomethylated following zebularine treatment (Supplementary Table S3).
Figure 6. The effect of zebularine is mediated via hypomethylation of the miR-17-92 cluster.
A, box plots representing the decrease in DNA methylation levels in pooled control versus zebularine-treated PDAC cultures. *, P < 0.05. B, qRT-PCR analysis of members of the miR-17-92 cluster (miR17, 18a, 19a, 19b, 20a) in PDAC-A6L and PDAC-185 sphere-derived cultures (*, P < 0.05; n = 4). C, qRT-PCR analysis of miR-17-92 target genes in PDAC-A6L and PDAC-185 sphere-derived cultures. Data are normalized to β-actin levels and are represented as fold change compared with untreated cells (*, P < 0.05; n = 3). D, Western blot analysis of P21 protein levels in control versus zebularine-treated cultures (top) and subsequent densitometric quantification (bottom). Changes in protein levels are represented as fold change compared with untreated cells. E, qRT-PCR analysis for MYC expression in PDAC-185 cells. Data are normalized to β-actin and are represented as fold change in comparison with untreated cells (*, P < 0.05; n = 4). F, PDAC adherent cells were treated with antagomiR (Ant) for miR-17, 18a, 19a/b, and 20a for 24 hours. Cells were plated for sphere formation assay, treated with zebularine for 7 days, and number of spheres/mL were determined (*, P < 0.05; n = 6).
Table 1.
Hypermethylated miRs in CSC-enriched spheres
| has-miRNA | FC_sph vs. adh |
|---|---|
| hsa-miR548N | 1.45 |
| hsa-miR1281 | 1.42 |
| hsa-miR1259 | 1.41 |
| hsa-miR1225 | 1.40 |
| hsa-miR130B/hsa-miR301B | 1.40 |
| hsa-miR17HG | 1.34 |
| hsa-miR1224 | 1.31 |
| hsa-miR22 | 1.26 |
| hsa-miR1227 | 1.26 |
| hsa-miR1226 | 1.25 |
| hsa-miR330 | 1.24 |
| hsa-miR135B | 1.24 |
| hsa-miR585 | 1.21 |
| hsa-miR600 | 1.21 |
| hsa-miR375 | 1.20 |
NOTE: Given is the fold change (FC) of β-methylation values for spheres versus adherent. Only data for FC ≥ 1.20 are shown.
Abbreviations: adh, adherent; sph, spheres.
Among these, we identified miR-203 and -205, which have previously been implicated in promoting cellular differentiation (19). Although the expression of these miRNAs could not be confirmed to be significantly altered by zebularine (Supplementary Fig. S6A), we did see a strong and significant increase in the expression of these two miRNAs in DNMT1-KO cells (Supplementary Fig. S6B). Next, we focused our attention on the miR-17HG (miR-17-92 cluster host gene) as a potentially methylation-regulated miRNA as zebularine treatment phenocopied previous data from our laboratory related to the miR-17-92 cluster in CSCs (20). We previously showed that suppression of this cluster was necessary for the maintenance of CSC phenotypes and artificial overexpression of miR-17-92 members forces quiescent CSCs into an active cell-cycle state (20). Likewise, previous reports have observed a CpG island in close proximity of the miR-17-92 promoter (21). Encouraged by these findings, we further investigate whether hypermethylation of this CpG island could be responsible for the apparent suppression of this important miRNA cluster in CSC-enriched spheres (20). Indeed, analysis of our methylation array data showed that CpG sites in close proximity of the miR-17-92 cluster were hypomethylated upon zebularine treatment (Supplementary Table S3). Using an independent set of CSC-enriched samples from various PDAC tumors, we observed a consistent and notable increase in the expression of several miR-17-92 members following zebularine treatment (Fig. 6B), with a particular increase in miR-19a and miR-19b. Consistently, the increased expression of miR-17-92 members, in particular miR-19b, was mimicked in cells lacking DNMT1 (Supplementary Fig. S6C). Of note, miR-92a, bearing a unique seed sequence distinguishing it from the other miR-17-92 family members, was not modulated by zebularine treatment (control vs. zebularine: 1.13 ± 0.02-fold change, n.s.). Known targets of the miR-17-92 cluster, such as P21 (CDKN1A), TGFBR2, ACVR1B (ALK4), SMAD2, and SMAD4, have also been implicated in pancreatic CSC phenotypes including self-renewal and chemoresistance (11, 20). We found that many of these genes were suppressed upon zebularine treatment at both the mRNA (Fig. 6C) and protein levels (e.g., P21; Fig. 6D), recapitulating our previous findings (20) and suggesting that DNMT1 inhibition is capable of unlocking the epigenetic repression of this cluster in CSCs to reactivate repressed "anti-CSC" miRs.
It has been shown that the MYC proto-oncogene family is also involved in the transcriptional regulation of miR-17-92 (22). MYC is generally overexpressed in PDAC, but we recently showed that MYC is actually suppressed in pancreatic CSCs (23), which may contribute to the specific reduced expression of the miR-17-92 cluster in these cells. Indeed, our methylation array data indicated that CpG sites annotated to MYC were more methylated in CSCs (fold change adherent vs. sphere: 1.57 ± 0.18), which could be reversed by zebularine treatment (fold change control vs. zebularine: 0.80 ± 0.02). We consistently found a significant increase in c-MYC mRNA levels in CSC-enriched spheres upon treatment with zebularine (Fig. 6E).
Finally, to provide more conclusive evidence that the effects of DNMT1 inhibition were primarily mediated via reactivation of the miR-17-92 cluster, we performed loss-of-function experiments in the presence or absence of zebularine. For this purpose, we plated primary PDAC cells in adherent conditions to obtain a predominantly differentiated PDAC culture and treated these cells for 24 hours with antagomiR targeting the various members of the miR-17-92 cluster to promote "stemness," as described previously (20). Following antagomiR treatment, we next plated the cells in ultra-low adhesion conditions (sphere culture) to foster the expansion of CSCs and then treated cultures with zebularine to competitively reverse the effects on CSC phenotypes mediated via miRNA downregulation. Zebularine reversed the antagomiR-mediated enhancement of sphere formation (Fig. 6F), supporting the conclusion that zebularine mediates its inhibitory effects on CSCs, at least in part, by inducing the expression of the members of the miR-17-92 cluster, which functions to repress the expression of CSC-promoting genes.
Discussion
Recent advances in our understanding of CSC epigenetics provide important insights into how these cells acquire their specific stem-like characteristics and, at the same time, shed light on how CSCs can be successfully targeted using epigenetic-modifying agents (24). Our data demonstrate that pancreatic CSCs (i) bear higher DNA methylation levels, (ii) express high levels of the DNMT methyltransferase protein DNMT1 and (iii) loose stemness upon pharmacologic or genetic inhibition of DNMT1. Although previous studies have already shown that DNMTs are overexpressed in other cancer tissues compared with normal tissue (25, 26), herein we report specific upregulation of DNMT1 in PDAC and provide data to support its role as an important epigenetic modifier in pancreatic CSCs. Intriguingly, demethylation of pancreatic CSCs using the DNMT1 inhibitors zebularine and decitabine markedly reduced their CSC functions and properties, an effect that was regulated via DNMT1-mediated demethylation of the miR-17-92 cluster promoter. These data are in line with reports for other cancers where DNMT1 was also found to be essential for the maintenance of stem cells (27, 28), but we now also provide a mechanistic link in the context of PDAC by demonstrating the modulation of a crucial miRNA cluster.
Epigenetic modifications are able to alter gene expression and have been shown to play a crucial role in stem cell function and maintenance (27, 29). As CSCs and their more differentiated progenies share the same genetic background, epigenetic changes should account for the striking functional differences between CSCs and their nontumorigenic progenies. Specifically, we recently demonstrated that even a single CSC could give rise to a tumor that recapitulated the functional heterogeneity of the original parental tumor at the pathologic, biologic, and genetic levels (15). Thus, epigenetics and not genetics must be the underlying drivers of this intraclonal heterogeneity, which is indeed supported by our present finding that the genome of CSCs is hypermethylated compared with non-CSCs. Strikingly, DNA hypermethylation could not only be found in single-CSC-derived tumors that lack intratumoral heterogeneity, but also in tumors derived from metastatic lesions and even highly heterogeneous primary tumors. This finding supports the notion that hypermethylation is a consistent and robust differential factor between pancreatic CSCs and non-CSCs. At the DNA level, this hypermethylation phenotype was restricted to regions outside traditional CpG islands, specifically non-CpG islands and intergenic regions. In general, it has been shown that cancer cells exhibit hypomethylation of intergenic regions (30), which consequently could contribute to activation of transposable elements and genome instability. On the other hand, promoter regions of many CpG islands of tumor suppressor genes become hypermethylated, resulting in their loss of function (31). Fortunately, using genome-scale methylation screening approaches, we have learned that a fraction of normally methylated CpG islands become hypomethylated and transcriptionally active in cancer cells (32). Moreover, some CpG islands located within the 3′ ends of genes (33) and in intergenic regions (34) exhibit hypermethylation in cancer cells. It is still unclear to what extent methylation of these nonpromoter CpG islands might affect gene expression and more importantly what is the subsequent result/phenotype in different populations of cancer cells. Although further studies are necessary to address these specific questions in the context of CSCs, our data allude to a possible mechanism by which PDAC CSCs may protect their genome from undesirable transcription or instability, achieving better fitness for survival and growth than their more differentiated non-CSC counterparts.
DNA methylation is evolutionarily ancient and associated with gene silencing in eukaryotes. It represents a key regulatory mechanism for the self-renewal and differentiation programs of embryonic stem cells and of adult stem cells (35). Maintenance of their "stemness" state is conferred to the set of developmental transcription factors (OCT4, NANOG, and SOX2), occupying promoters of genes associated with self-renewal. Expression of these transcriptional regulators is usually controlled by CpG promoter methylation, and differentiation is accomplished by partial or full methylation of pluripotency-associated genes, resulting in their downregulation (36). Thus, it is conceivable to predict that treatment of CSCs with demethylating agents would further upregulate pluripotency-associated gene expression; however, we observed a downregulation of pluripotency factors, indicating that the epigenetic wiring of CSCs is not only different than stem cells but that other epigenetic regulators are likely shaping the epigenetic landscape of CSCs.
Zebularine, a cytidine analogue, acts primarily as a trap for DNMT proteins by forming tight covalent complexes between DNMT proteins and DNA after zebularine incorporation (10). It is believed that the antitumor effects of DNMT inhibitors, such as zebularine or decitabine, are largely due to the reexpression of tumor suppressor genes (37), which are often silenced in cancer cells. Indeed, we observed that treatment with zebularine was able to alter methylation patterns of a number of genes in PDAC CSCs, including miRNA genes, which are regulated by methylation and are known to be involved in a wide range of biologic processes, including stem cell differentiation (29). In addition, several miRNAs have been shown to be involved in promoting/maintaining stemness in cancers. For example, miR-145 and miR-9, both well-known tumor suppressors, were found to be suppressed in several human cancers (38) due to aberrant DNA methylation of their promoters. Thus, we sought to focus our investigation on miRNA genes and to identify specific CpG sites located in the proximity of various miRNA promoter regions, in which zebularine-mediated hypomethylation reactivated genes that were originally silenced in CSCs. We observed that the miR-17-92 cluster (comprised of six members – miR17, 18a, 19a, 19b, 20a, and 92a) was hypermethylated in CSCs versus non-CSCs (Table 1). This finding is in line with recent studies from our laboratory showing that the miR-17-92 cluster is consistently downregulated in pancreatic CSCs (20). In this study, we showed that gain of function, using forced overexpression of miR-17-92, reduced CSC self-renewal capacity, in vivo tumorigenicity, and chemoresistance. On the other hand, downregulation of this cluster (i.e., inhibition of miR-17-92 using antagomiR) in more differentiated cells had the opposite effect, imparting non-CSCs with CSC-like phenotypes. This effect was mediated by suppressing multiple members of the NODAL/ACTIVIN/TGFβ1 signaling cascade as well as downstream targets, such as P21, P57, and TBX3, all of which have been shown to be crucial for maintaining the stem-like state of pancreatic CSCs (11, 20, 39). Our data now further validate and expand upon these previous findings and provide novel insights into the epigenetic mechanism(s) controlling the suppression of the miR-17-92 cluster in CSCs. We show, using different approaches, that reactivation of the miR-17-92 cluster following DNMT1 pharmacologic or genetic inhibition augments the phenotype of PDAC CSCs.
Unlike other DNMT inhibitors, zebularine is more stable in aqueous solution and is less toxic in vitro and in vivo (40). Continuous exposure of various cancer cell lines to zebularine has already been shown to selectively slow tumor cell growth, highlighting its potential value as a chemotherapeutic agent. It has previously been shown that zebularine has anticancer effects in established PDAC cell lines, supposedly via induction of apoptosis and subsequent suppression of tumor growth in vivo (41); however, using primary low-passage PDAC cultures derived from equally low-passage PDX tumors, we found no evidence for a direct proapoptotic effect. Instead, we show that zebularine treatment forces normally slow-cycling CSCs into a more proliferative fast-cycling state, which has been previously linked to chemosensitization via enhanced expression of miR-17-92 (42). Moreover, we observed a marked promotion of CSC differentiation as determined by the loss of "stemness" markers (e.g., CD133) and a gain in the expression of differentiation markers including cytokeratin and E-cadherin. More importantly, these phenotypes were recapitulated using not only a different DNMT inhibitor, decitabine, but also in DNMT1-KO cells, indicating that regardless of the approach used, the loss of DNMT1 results in the same phenotypic changes: loss of "stemness" and promotion of differentiation.
We would like to highlight that we cannot exclude the possibility that other CSC-inhibitory miRNAs could be reactivated by zebularine treatment (or DNMT1 inhibition). Indeed Supplementary Table S3 shows that upon treatment with zebularine, β methylation values of various miRNA genes were decreased. On the basis of this list and after careful review of the literature, we opted to focus on the two miRNAs, miR-203 and miR-205, as both miRNAs were previously described to play a role in the biology of pancreatic cancer (43, 44). miR-203 acts as a known suppressor of stem cell pluripotent factors; therefore, reactivation of miR-203 by hypomethylation might contribute to the reduced levels of pluripotency-associated genes observed following zebularine treatment in PDAC CSCs. Overexpression of miR-203 has also been demonstrated to induce expression of E-cadherin by inhibition of the E-cadherin repressors ZEB1/2, which correlates well with our E-cadherin immunofluorescence studies presented herein (45). Thus, the prodifferentiation effect of zebularine can also be potentially explained by zebularine-induced hypomethylation of miR-203, which has previously been described as an epithelial differentiation factor (19). Likewise, miR-205 is a well-known anticancer miRNA and is consistently downregulated in clinical pancreatic cancer samples including CSCs (44, 46). miR-205 replenishment reduces the expression of the pluripotency/stem cell marker OCT3/4, the CSC marker CD44, and resensitizes cells to chemotherapy. On the basis of these findings, we checked the expression of miR-203 and miR-205 in PDAC cells treated with zebularine and deficient in DNMT1 (i.e., DNMT1-KO cells). Although expression of these miRNAs was not significantly changed during short-term treatment with zebularine (Supplementary Fig. S6A), we noticed a strong and significant increase in DNMT1-KO cells (Supplementary Fig. S6B), suggesting that more potent and/or long-term inhibition of DNMT1 is necessary for miR-203 and miR-205 reactivation. Collectively, we cannot exclude the possibility that other anti-CSCs miR could be reactivated following DNMT1 inhibition; however, our data support the hypothesis that reactivation of the miR-17-92 cluster is a dominating driving factor responsible for the inhibitory effects observed in the PDAC CSC population.
We are still far from thoroughly understanding the role of DNMT1 in the context of CSC biology. Increasing evidence already suggests that DNMT1 protein expression promotes the development of PDAC, from normal tissue to precancerous lesions to PDAC (47). Moreover, DNMT1 has been demonstrated to be essential for the maintenance of hematopoietic stem cells (HSCs)/progenitor cells (48), epidermal progenitor cells and leukemia stem cells (49). More recently, DNMT1 was also shown to be indispensable for mammary stem/progenitor cells and CSC maintenance, and functional inactivation of this gene drastically reduces mammary tumor formation (50). The sum of prior evidence and new insights from our study certainly highlight the important role that DNMT1 plays in cancer biology and at the same time support the continued development of more effective methylation inhibitors as a means of improving the clinical outcome of PDAC patients.
Supplementary Material
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Acknowledgments
Grant Support
This research was supported by the ERC Advanced Investigator Grant (Pa-CSC 233460 to C. Heeschen) and the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 256974 (EPC-TM-NET; to C. Heeschen) and no. 602783 (CAM-PaC; to Heeschen), the 2015 SU2C Lustgarten CRUK Pancreatic Cancer Dream Team Award (C. Heeschen), Pancreatic Cancer UK RIF2014_04 and RIF2015_03 (both to C. Heeschen), a R'amon y Cajal Merit Award from the Ministerio de Economía y Competitividad, Spain (B. Sainz Jr), a Clinic and Laboratory Integration Program (CLIP) grant from the Cancer Research Institute (B. Sainz Jr), and a Proyecto de Investigacio'n de Salud, ISCIII, Spain (no. PI15/01507 to B. Sainz Jr).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors' Contributions
Conception and design: S. Zagorac, T.B. Kheir, B. Sainz Jr, C. Heeschen
Development of methodology: S. Zagorac, T.B. Kheir
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): S. Zagorac, S. Alcala, T.B. Kheir, A. González-Neira, A. Aicher
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): S. Zagorac, S. Alcala, G.F. Bayon, T.B. Kheir, M. Schoenhals, M.F. Fraga, A. Aicher, C. Heeschen, B. Sainz Jr
Writing, review, and/or revision of the manuscript: S. Zagorac, M.F. Fraga, A. Aicher, C. Heeschen, B. Sainz Jr
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): C. Heeschen
Study supervision: T.B. Kheir, A. Aicher, C. Heeschen, B. Sainz Jr
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–21. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 3.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multi-region sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Silva S, Frias-Aldeguer J, Heeschen C. Stem cells & pancreatic cancer. Pancreatology. 2013;13:110–3. doi: 10.1016/j.pan.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717–28. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–23. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 8.Borodovsky A, Salmasi V, Turcan S, Fabius AW, Baia GS, Eberhart CG, et al. 5-azacytidine reduces methylation, promotes differentiation and induces tumor regression in a patient-derived IDH1 mutant glioma xenograft. Oncotarget. 2013;4:1737–47. doi: 10.18632/oncotarget.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Constantinides PG, Jones PA, Gevers W. Functional striated muscle cells from non-myoblast precursors following 5-azacytidine treatment. Nature. 1977;267:364–6. doi: 10.1038/267364a0. [DOI] [PubMed] [Google Scholar]
- 10.Cheng JC, Yoo CB, Weisenberger DJ, Chuang J, Wozniak C, Liang G, et al. Preferential response of cancer cells to zebularine. Cancer Cell. 2004;6:151–8. doi: 10.1016/j.ccr.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 11.Lonardo E, Hermann PC, Mueller MT, Huber S, Balic A, Miranda-Lorenzo I, et al. Nodal/Activin signaling drives self-renewal and tumorigenicity of pancreatic cancer stem cells and provides a target for combined drug therapy. Cell Stem Cell. 2011;9:433–46. doi: 10.1016/j.stem.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Smyth G. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Statistics for Biology and Health. 2005. limma: linear models for microarray data; pp. 397–420. Chapter. [Google Scholar]
- 13.Wu H, Caffo B, Jaffee HA, Irizarry RA, Feinberg AP. Redefining CpG islands using hidden Markov models. Biostatistics. 2010;11:499–514. doi: 10.1093/biostatistics/kxq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang T, Guan W, Lin J, Boutaoui N, Canino G, Luo J, et al. A systematic study of normalization methods for Infinium 450K methylation data using whole-genome bisulfite sequencing data. Epigenetics. 2015;10:662–9. doi: 10.1080/15592294.2015.1057384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miranda-Lorenzo I, Dorado J, Lonardo E, Alcala S, Serrano AG, Clausell-Tormos J, et al. Intracellular autofluorescence: a biomarker for epithelial cancer stem cells. Nat Methods. 2014;11:1161–9. doi: 10.1038/nmeth.3112. [DOI] [PubMed] [Google Scholar]
- 16.Jeltsch A, Jurkowska RZ. New concepts in DNA methylation. Trends Biochem Sci. 2014;39:310–8. doi: 10.1016/j.tibs.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Flotho C, Claus R, Batz C, Schneider M, Sandrock I, Ihde S, et al. The DNA methyltransferase inhibitors azacitidine, decitabine and zebularine exert differential effects on cancer gene expression in acute myeloid leukemia cells. Leukemia. 2009;23:1019–28. doi: 10.1038/leu.2008.397. [DOI] [PubMed] [Google Scholar]
- 18.Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci U S A. 1994;91:11797–801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taube JH, Malouf GG, Lu E, Sphyris N, Vijay V, Ramachandran PP, et al. Epigenetic silencing of microRNA-203 is required for EMT and cancer stem cell properties. Sci Rep. 2013;3:2687. doi: 10.1038/srep02687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cioffi M, Trabulo SM, Sanchez-Ripoll Y, Miranda-Lorenzo I, Lonardo E, Dorado J, et al. The miR-17–92 cluster counteracts quiescence and chemoresistance in a distinct subpopulation of pancreatic cancer stem cells. Gut. 2015;64:1936–48. doi: 10.1136/gutjnl-2014-308470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, et al. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–83. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 23.Sancho P, Burgos-Ramos E, Tavera A, Bou Kheir T, Jagust P, Schoenhals M, et al. MYC/PGC-1alpha balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab. 2015;22:590–605. doi: 10.1016/j.cmet.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 24.Wongtrakoongate P. Epigenetic therapy of cancer stem and progenitor cells by targeting DNA methylation machineries. World J Stem Cells. 2015;7:137–48. doi: 10.4252/wjsc.v7.i1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patra SK, Patra A, Zhao H, Dahiya R. DNA methyltransferase and demethylase in human prostate cancer. Mol Carcinog. 2002;33:163–71. doi: 10.1002/mc.10033. [DOI] [PubMed] [Google Scholar]
- 26.Oh BK, Kim H, Park HJ, Shim YH, Choi J, Park C, et al. DNA methyltransferase expression and DNA methylation in human hepatocellular carcinoma and their clinicopathological correlation. Int J Mol Med. 2007;20:65–73. [PubMed] [Google Scholar]
- 27.Broske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41:1207–15. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 28.Morita R, Hirohashi Y, Suzuki H, Takahashi A, Tamura Y, Kanaseki T, et al. DNA methyltransferase 1 is essential for initiation of the colon cancers. Exp Mol Pathol. 2013;94:322–9. doi: 10.1016/j.yexmp.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116–25. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988;48:1159–61. [PubMed] [Google Scholar]
- 31.Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–8. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 32.Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, et al. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023–36. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith JF, Mahmood S, Song F, Morrow A, Smiraglia D, Zhang X, et al. Identification of DNA methylation in 3′ genomic regions that are associated with upregulation of gene expression in colorectal cancer. Epigenetics. 2007;2:161–72. doi: 10.4161/epi.2.3.4805. [DOI] [PubMed] [Google Scholar]
- 34.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–62. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 35.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 36.Berdasco M, Esteller M. DNA methylation in stem cell renewal and multipotency. Stem Cell Res Ther. 2011;2:42. doi: 10.1186/scrt83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–95. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 38.Suh SO, Chen Y, Zaman MS, Hirata H, Yamamura S, Shahryari V, et al. MicroRNA-145 is regulated by DNA methylation and p53 gene mutation in prostate cancer. Carcinogenesis. 2011;32:772–8. doi: 10.1093/carcin/bgr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lonardo E, Frias-Aldeguer J, Hermann PC, Heeschen C. Pancreatic stellate cells form a niche for cancer stem cells and promote their self-renewal and invasiveness. Cell Cycle. 2012;11:1282–90. doi: 10.4161/cc.19679. [DOI] [PubMed] [Google Scholar]
- 40.Valenzuela MM, Neidigh JW, Wall NR. Antimetabolite treatment for pancreatic cancer. Chemotherapy. 2014;3 doi: 10.4172/2167-7700.1000137. pii:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neureiter D, Zopf S, Leu T, Dietze O, Hauser-Kronberger C, Hahn EG, et al. Apoptosis, proliferation and differentiation patterns are influenced by Zebularine and SAHA in pancreatic cancer models. Scand J Gastroenterol. 2007;42:103–16. doi: 10.1080/00365520600874198. [DOI] [PubMed] [Google Scholar]
- 42.Andrade AF, Borges KS, Castro-Gamero AM, Silveira VS, Suazo VK, Oliveira JC, et al. Zebularine induces chemosensitization to methotrexate and efficiently decreases AhR gene methylation in childhood acute lymphoblastic leukemia cells. Anticancer Drugs. 2014;25:72–81. doi: 10.1097/CAD.0000000000000028. [DOI] [PubMed] [Google Scholar]
- 43.Xu D, Wang Q, An Y, Xu L. MiR203 regulates the proliferation, apoptosis and cell cycle progression of pancreatic cancer cells by targeting Survivin. Mol Med Rep. 2013;8:379–84. doi: 10.3892/mmr.2013.1504. [DOI] [PubMed] [Google Scholar]
- 44.Singh S, Chitkara D, Kumar V, Behrman SW, Mahato RI. miRNA profiling in pancreatic cancer and restoration of chemosensitivity. Cancer Lett. 2013;334:211–20. doi: 10.1016/j.canlet.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 45.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Y, Brenn T, Brown ER, Doherty V, Melton DW. Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br J Cancer. 2012;106:553–61. doi: 10.1038/bjc.2011.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng DF, Kanai Y, Sawada M, Ushijima S, Hiraoka N, Kosuge T, et al. Increased DNA methyltransferase 1 (DNMT1) protein expression in precancerous conditions and ductal carcinomas of the pancreas. Cancer Sci. 2005;96:403–8. doi: 10.1111/j.1349-7006.2005.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5:442–9. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trowbridge JJ, Sinha AU, Zhu N, Li M, Armstrong SA, Orkin SH. Haploinsufficiency of Dnmt1 impairs leukemia stem cell function through derepression of bivalent chromatin domains. Genes Dev. 2012;26:344–9. doi: 10.1101/gad.184341.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pathania R, Ramachandran S, Elangovan S, Padia R, Yang P, Cinghu S, et al. DNMT1 is essential for mammary and cancer stem cell maintenance and tumorigenesis. Nat Commun. 2015;6:6910. doi: 10.1038/ncomms7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.