Abstract
Mitochondria have specialized ribosomes (mitoribosomes) dedicated to the expression of the genetic information encoded by their genomes. Here, using electron cryomicroscopy, we have determined the structure of the 75-component yeast mitoribosome to an overall resolution of 3.3 Å. The mitoribosomal small subunit has been built de novo and includes 15S rRNA and 34 proteins, including 14 without homologs in the evolutionarily related bacterial ribosome. Yeast-specific rRNA and protein elements, including the acquisition of a putatively active enzyme, give the mitoribosome a distinct architecture compared to the mammalian mitoribosome. At an expanded mRNA channel exit, there is a binding platform for translational activators that regulate translation in yeast but not mammalian mitochondria. The structure provides insights into the evolution and species-specific specialization of mitochondrial translation.
Mitochondria are eukaryotic organelles that carry out aerobic respiration. Resulting from their likely ancestry as endosymbionts (1), mitochondria retain a vestigial genome of ~3–100 genes depending on the species. All mitochondrial DNA (mtDNA) encodes at least some of the essential transmembrane subunits of the oxidative phosphorylation complexes. To synthesize these proteins, mitochondria have dedicated ribosomes (mitoribosomes). Nearly all mitoribosomal proteins and all translational factors are encoded by nuclear DNA and imported from the cytoplasm, while mtDNA encodes the mitoribosomal RNA (rRNA) and mitochondria-specific transfer RNAs (mt-tRNAs).
Mitochondrial translation displays considerable species-specific specialization, largely dictated by the translational requirements of the mitochondrial genome (2). This specialization manifests in the diverse compositions and structures of mitoribosomes, which are distinct from one another and from all known ribosomes despite sharing an ancestor with modern bacterial ribosomes (2, 3).
Our current understanding of this diversity at the atomic level is limited to comparisons of the structure of the large subunit of the Saccharomyces cerevisiae mitoribosome (mt-LSU) (4) with the structures of the complete human (5) and porcine (6) mitoribosomes. These comparisons have revealed that the major evolutionary trajectory for mammalian mitoribosomes is the enlargement of the mitoribosomal proteome together with contraction of the mt-rRNA (2, 3, 7). In contrast, the yeast mitoribosome appears to be on an evolutionary path that has not experienced rRNA contraction (4). Furthermore, it differs functionally from the mammalian mitoribosome by synthesizing a soluble protein in addition to integral membrane proteins. The structure of the yeast mt-LSU showed a re-routed polypeptide exit channel that may be a response to this requirement (4).
However, without high-resolution structural information for complete mitoribosomes of non-mammalian species it has not been possible to observe the effects of distinct evolutionary pressures and species-specific mitochondrial translation on the mitoribosomal small subunit (mt-SSU). Recognized selective pressures on the mt-SSU include mRNA binding, the initiation of translation and decoding. Notably, translational initiation in yeast mitochondria involves long 5′ untranslated regions (5′ UTRs) of mitochondria-encoded mRNAs and transcript-specific translational activators, neither of which occur in mammalian mitochondria (8).
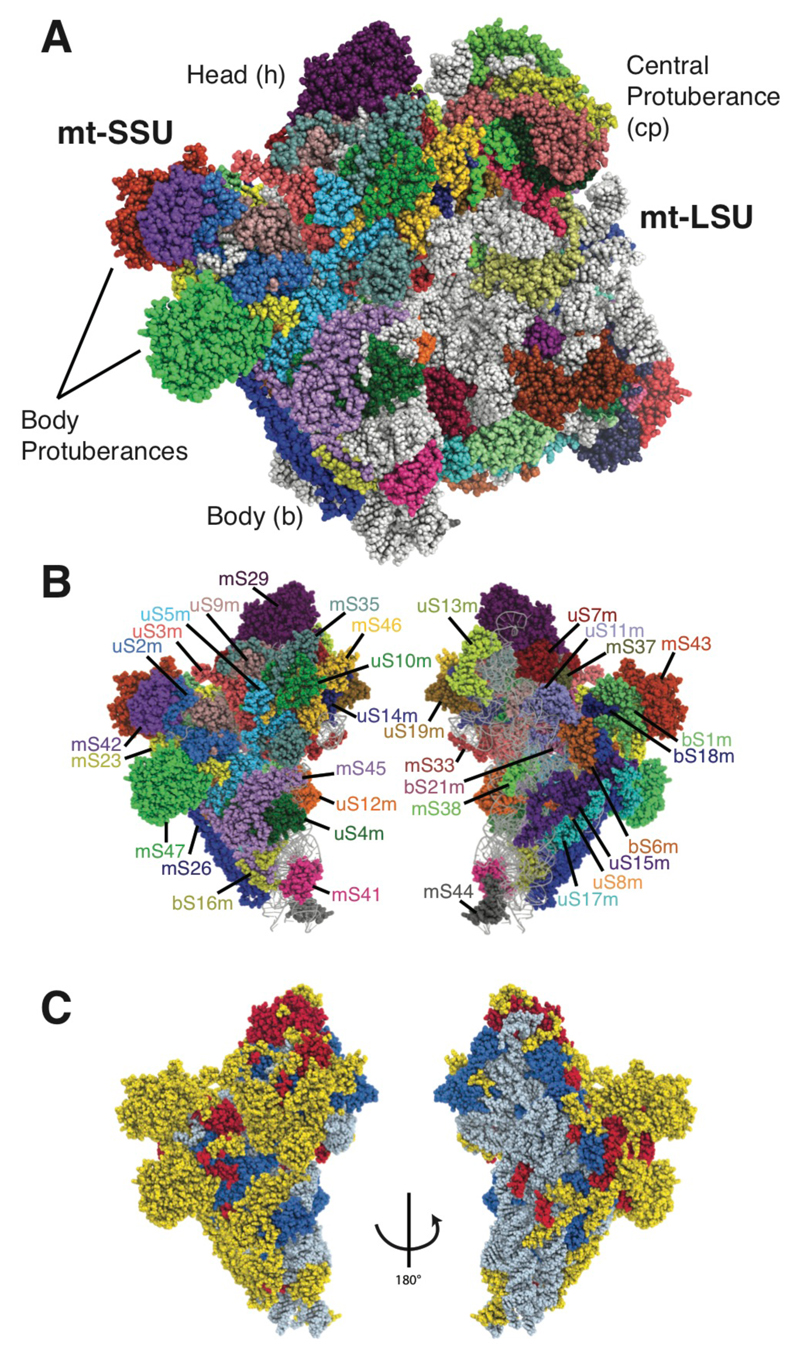
To improve our understanding of the effect of species-specific mitochondrial translation on mitoribosomal diversity, we have solved the structure of the complete yeast mitoribosome by electron cryomicroscopy (Fig. 1A and Fig. S1). The yeast mitoribosome adopts three well-populated conformations (classes A–C) that are resolved to between 3.3 and 5.0 Å resolution (Fig. S2A). To improve the quality of the maps further, masks were applied during processing which allowed the mt-LSU to be refined to 3.2 Å, the body of the mt-SSU to 3.3 Å and the head of the mt-SSU to 3.5 Å (Fig. S2B). These maps were used to improve the published model of the mt-LSU (4) (Fig. S3) and to build a de novo model of the mt-SSU.
Fig. 1. The structure of the yeast mitoribosome.
(A) The overall structure of the complete yeast mitoribosome. (B) The mitoribosomal proteins of the yeast mt-SSU. (C) Protein elements of the mt-SSU colored by conservation; elements conserved with the bacterial ribosome are blue, elements conserved with the human mitoribosome are red, and elements specific to the yeast mitoribosome are yellow. rRNA is colored grey.
The complete model of the yeast mitoribosome contains two rRNA molecules (21S rRNA in the mt-LSU and 15S rRNA in the mt-SSU), 73 proteins (table S2) and a single mt-tRNA bound at the E site (fig. S4) that is presumably a mixture of the 24 mt-tRNAs encoded by the yeast mitochondrial genome. The ribosomal proteins are named following the nomenclature adopted by the ribosomal community (9) with the standard yeast name according to the Saccharomyces Genome Database (10) in parentheses.
The 15S rRNA of the mt-SSU is longer than the equivalent 16S rRNA in E. coli (1,649 compared to 1,542 nucleotides). Of these, 1,501 nucleotides have been modeled revealing an rRNA core that adopts a similar domain structure to 16S rRNA (fig. S5). Discrepancies occur mostly at the periphery of the mitoribosome: helices h6, h8, h17, h21, h33 and h39 are shorter than their bacterial equivalents, but this is counteracted by three rRNA expansion segments (h16-ES, h17-ES and h41-ES) and extensions of h9, h44 and of the 5′ and 3′ tails. Many of the 148 nucleotides that could not be modeled occupy these peripheral expansion segments, which have poor density. Overall, there is less expansion of rRNA than in the yeast mt-LSU (4), but considerably more than in the 12S rRNA of human mitoribosomes which exhibits substantial rRNA reduction (5).
The yeast mt-SSU has an almost complete complement of proteins with homologs in the bacterial SSU, with only bS20 absent (Fig. 1B). The mammalian mt-SSU lacks four additional homologs suggesting that this loss occurred after the mitoribosomes diverged from a common ancestor. The yeast mitochondrial homolog of uS3 (uS3m; Var1) is the only soluble protein encoded by mtDNA (11, 12). However, unlike nuclear-encoded uS3m in other species, yeast uS3m (Var1) has a ~30% asparagine content. These asparagine residues are distributed in solvent-exposed regions throughout the protein (Fig. 2B). The high asparagine content is likely a response to the propensity of mitochondrial genomes to be AT rich (yeast mtDNA has a 83% AT-content). Asparagine is the only hydrophilic residue specified by a codon formed exclusively of adenine and thymine nucleotides (AAT). Consistent with the AT-content determining the preferred amino acid, only 2 of the 127 asparagine residues in uS3m (Var1) are encoded by the alternative AAC codon.
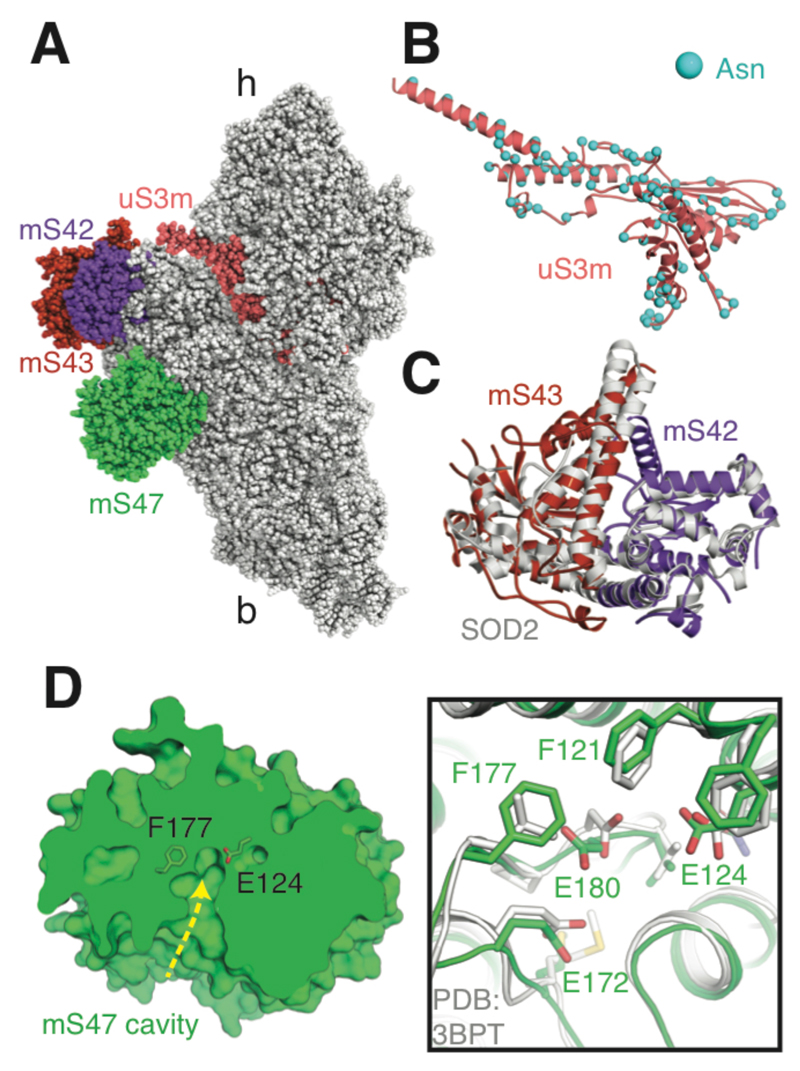
Fig. 2. Features of the yeast mt-SSU.
(A) The structure of the yeast mt-SSU with uS3m, mS42, mS43 and mS47 highlighted. (B) The asparagine residues of mitochondria-encoded uS3m are distributed on the protein surface. (C) mS42 and mS43 (related to panel A by a 90° rotation around the y axis) form a heterodimer that structurally resembles a yeast mitochondrial superoxide dismutase dimer (Protein Data Bank ID 3LSU). (D) mS47 (related to panel A by an approximate 180° rotation around the x axis) is a probable enzyme with a large cavity and catalytic residues conserved with human β-hydroxyisobutyryl-CoA hydrolase (Protein Data Bank ID 3BPT). Single-letter abbreviations for the amino acid residues are as follows: E, Glu; and F, Phe.
The yeast mt-SSU also contains 14 mitochondria-specific proteins, of which 7 have homologs in the mammalian mitoribosome and 7 are specific to the yeast mitoribosome. mS38 (Cox24) had not previously been assigned as a yeast mitoribosomal protein and was identified using a sequence obtained directly by interpreting the density (fig. S6). A role for Cox24 in mitochondrial translation is consistent with a previous report that Cox24 null mutants have reduced levels of mitochondrially encoded proteins (13), although a second proposed role for Cox24 in mitochondrial RNA processing (13) cannot be explained from our structure alone. Two reported constituents of the yeast mitoribosome, Rsm22 (14) and Yms2 (15), were not located. This adds to evidence that Rsm22 is an RNA methyltransferase that only transiently interacts with the mitoribosome (16, 17).
The majority of yeast mitoribosomal proteins with homologs in other ribosomes have N- and C-terminal extensions. These extensions increase the interconnectivity of the mt-SSU (fig. S7), but rarely show conservation in length, sequence or structure with the extensions of the related proteins in the mammalian mitoribosome (fig. S8A). Therefore, most protein extensions appear to have occurred after the mitoribosomes diverged, but regions of conservation will help to reconstruct the evolution of the mitoribosome.
Overall, the preferential addition of mitochondria-specific proteins and protein extensions to the mt-SSU periphery creates a distinctive morphology compared with other ribosomes (Fig. 1C and fig. S8B). The solvent-exposed side of the mt-SSU body is dominated by two protein-rich protuberances. The largest protuberance adjacent to the mRNA channel exit is formed by a heterodimer of mS42 (Rsm26) and mS43 (Mrp1), which share structural homology with iron/manganese-binding superoxide dismutases (Fig. 2C). However, the metal-binding cores of each of these proteins have diverged in sequence rendering a likely inactive hydrophobic environment. This supports the idea that mitoribosomes have expanded their proteomes by acquiring proteins from the mitochondrial matrix with the original functions of these proteins then subsequently lost (2).
An exception to this is mS47 (Ehd3), which forms the other protuberance of the yeast mitoribosome and appears to be a catalytically active enzyme. mS47 is structurally similar to human 3-hydroxyisobutyryl-CoA hydrolase (HIBCH) (Fig. 2D), a mitochondrial protein with a putative role in valine catabolism and hereditary mitochondrial disease (18) but not a constituent component of the human mitoribosome (5). Consistent with mS47 being an active enzyme as well as an integral part of the yeast mitoribosome, mS47 retains a solvent-exposed cavity likely capable of accommodating a substrate and active site residues that are either preserved or show conservative mutations (Fig. 2D). In addition, recombinant yeast mS47 has been shown to be capable of hydrolyzing 3-hydroxyisobutyryl-CoA although the identity of its in vivo substrate is unknown (19). At its peripheral location, mS47 is unlikely to play an essential role in mitochondrial translation, consistent with viable null mutants, but may benefit from co-localization with the mitoribosome or act as a link between translation and the catabolic state of the mitochondrion.
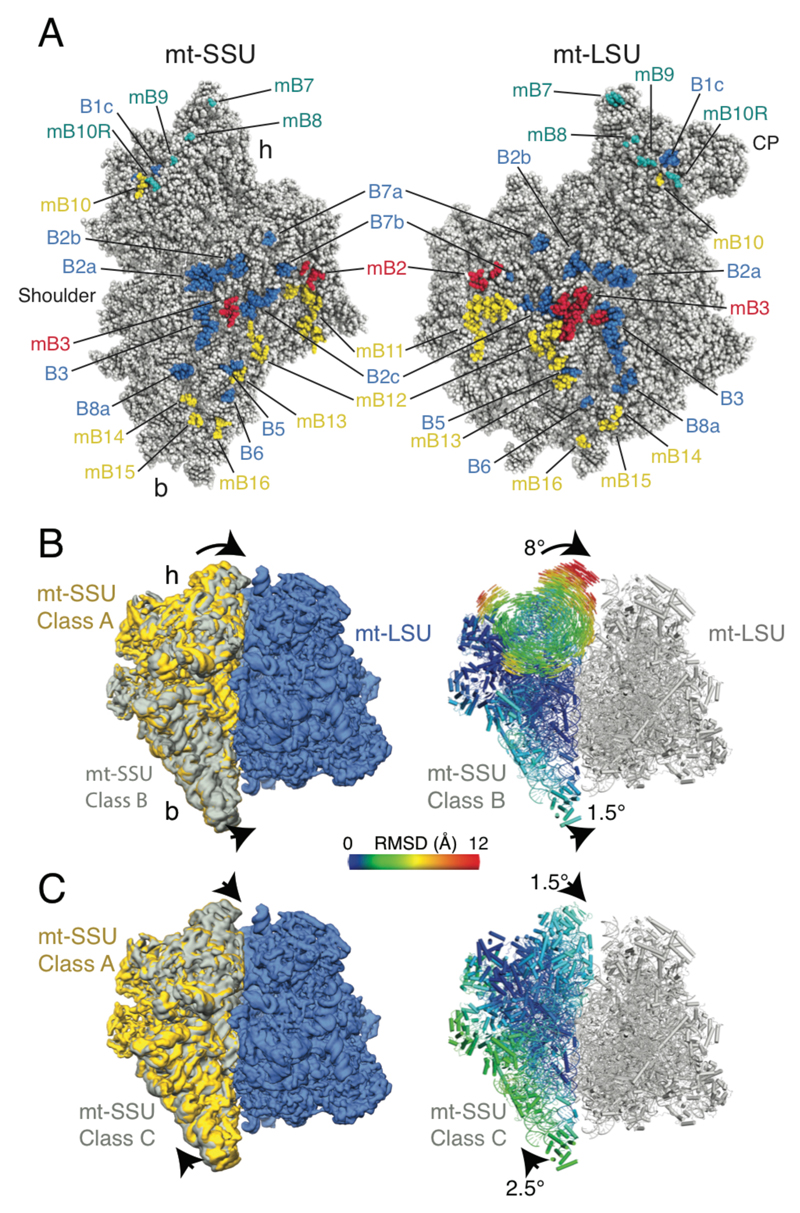
The mitoribosomal subunits are connected through a series of plastic intersubunit bridges (Fig. 3A, fig. S9-10 and table S3), which regulate the relative movement of the subunits. Most of the bridges present in the bacterial ribosome, with the exception of bridges B1a/b and B4, are also present in the yeast mitoribosome (fig. S9). In addition, there are 9 mitochondria-specific bridges in Class A, of which only two are also present in the mammalian mitoribosome (fig. S10). The extensive intersubunit bridges may restrain the movement of the yeast mitoribosomal subunits with the three observed conformations related by subtle movements only (Fig. 3B). This contrasts with the mammalian mitoribosome which, presumably as a result of fewer intersubunit contacts due to a remodeled helix 44 (5, 6), can sample a more extensive conformational space than bacterial ribosomes (3). The “ratcheted” and “rolled” conformations seen in the human mitoribosome (5) and the hyper-rotated state of the yeast mitoribosome seen by electron cryotomography (20) were not observed.
Fig. 3. Bridges and mitoribosomal dynamics.
(A) Intersubunit interfaces with residues that contribute to bridges highlighted. Bridges also present in the bacterial ribosome are in blue; mitoribosome-specific bridges conserved in the human mitoribosome are red and yeast-specific bridges are in yellow. Residues that form additional bridges in class B are shown in teal. (B) Class B is related to class A by a small rotation of the body and an 8° rotation of the head. The body atoms and head vectors (right) are colored by root-mean-square displacement from their positions in class A. (C) Class C is related to class A by small rotations of the body and head.
An 8° rotation of the head in class B results in the formation of additional bridges between the head of the mt-SSU and the central protuberance of the mt-LSU. In particular, it brings the guanosine triphosphatase, mS29 (Rsm23), into contact with an expansion segment (H82-ES4) of the 21S rRNA (fig. S11). The location of this protein at the intersubunit interface in the mammalian mitoribosome in a GDP-bound state (5, 6) together with assays showing a higher affinity for GTP by the mt-SSU over the intact monosome (21) had led to suggestions that nucleotide hydrolysis is linked to subunit association. However, the transitory location of mS29 at the interface of the yeast mitoribosome suggests that the role for mS29 in the mitoribosome may be more complex or species-dependent than previously thought. Furthermore, we cannot exclude the possibility that GTP is present given the ambiguous density for the nucleotide in the yeast mt-SSU.
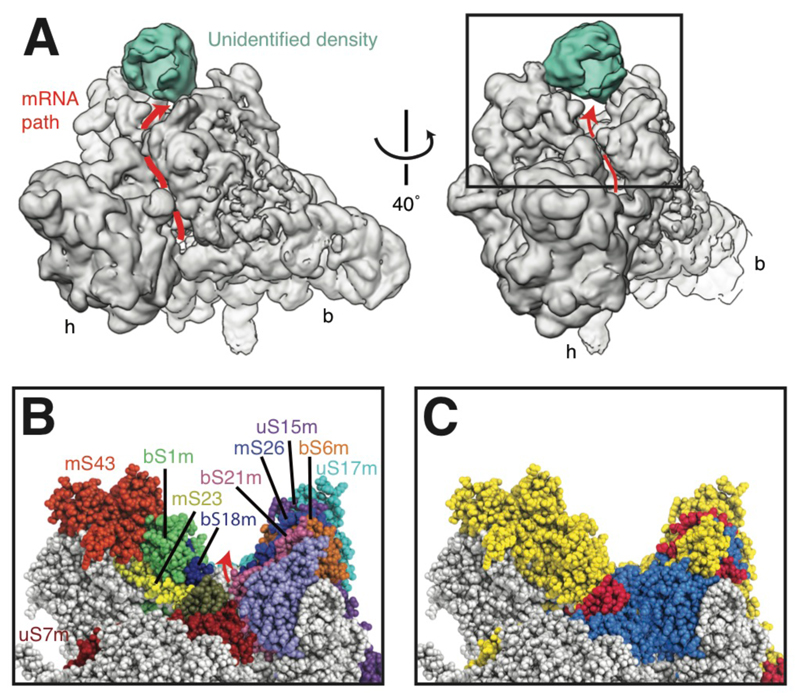
The path taken by mRNA through the yeast mitoribosome can be traced based on comparison with mRNA-bound structures of the bacterial ribosome (22, 23) (Fig. 4). The path curves around helix 28 of the 15S rRNA that forms the neck connecting the head and body of the mt-SSU in a non-covalently closed channel. As in bacteria, the mRNA enters the channel between the head and shoulder of the mt-SSU through a narrow entry site formed by uS3m (Var1), uS4m (Nam9) and uS5m (Mrps5). The constriction and the reported helicase activity of uS3 and uS4 in bacteria (24) are thought to ensure that only unpaired mRNA enters the channel. Although mitochondria-specific proteins mS35 (Rsm24) and mS45 (Mrps35) contribute to the architecture of the outer mRNA entrance, the large-scale remodeling that occurs in the mammalian mitoribosome has not occurred (5).
Fig. 4. The extended mRNA channel exit.
(A) View from the mt-LSU, showing the path of the mRNA channel (red line with arrow) around the neck of the yeast mt-SSU. Additional density is located above a canyon at the mRNA channel exit. The boxed section is shown in panels B-C. (B) The proteins that form the canyon. (C) The canyon walls are formed predominantly by mitoribosome-specific protein elements (colored red and yellow as in Fig. 1C).
The path exposes mRNA at the intersubunit interface for recognition by mt-tRNAs. At the A site the mRNA threads through the major groove of the upper part of helix 44. Here, the nucleotides G644, A1584 and A1585 (the equivalent of G530, A1492 and A1493 in bacteria) and a loop of uS12m (Mrps12) form the decoding center of the mt-SSU. The similarity of this region with the decoding centers of other ribosomes indicates a conserved mechanism of decoding. There appears to be no equivalent to the P-site finger that extends from the central protuberance to contact both and A- and P-site tRNAs in the mammalian mitoribosome (5, 6).
In contrast to the conserved early and intermediate parts of the mRNA channel, the channel exit shows considerable remodeling adjacent to the E site. In bacteria, this region includes the 3′ tail of the 16S rRNA that facilitates start-codon selection during translational initiation by base pairing with the Shine-Dalgarno sequence of the mRNA 5′ UTR. As yeast mitochondrial transcripts lack Shine-Dalgarno sequences, the 3′ end of the 15S rRNA is not constrained to a position at the mRNA channel exit and instead extends into the mt-SSU body where it is sequestered by mitoribosomal proteins. A second possible adaptation to the absence of Shine-Dalgarno sequences and the need for alternative mechanisms of initiating translation is a wide V-shaped canyon at the mRNA channel exit (Fig. 4). The canyon is flanked on one side by the mS42/mS43 (Rsm26/Mrp1) heterodimer protuberance and on the other by series of extensions to a number of ribosomal proteins including bS6m (Mrp17), uS15m (Mrps28), uS17m (Mrps17), bS18m (Rsm18) and bS21m (Mrp21). mRNA occupying this canyon would help to explain ribosome-profiling data (25) that have shown that the yeast mitoribosome protects longer mRNA stretches during active translation than cytosolic ribosomes (approximately 38 nucleotides compared to 28).
In a subset of our particles (~60%), additional density could be observed above the mRNA exit canyon (Fig. 4A). Similarly placed density was also observed by subtomogram averaging of yeast mitoribosomes in situ (20). Despite focused-classification approaches (fig. S12) this density could not be resolved further, likely due to compositional and conformational heterogeneity.
The density contacts both walls of the mRNA exit canyon: bS1m (Mrp51), bS6m (Mrp17) and mS43 (Mrp1) on one side, and uS15m (Mrps28), uS17m (Mrps17) and mS26 (Pet123) on the other. bS1m (Mrp51) has been shown to functionally interact with the 5′ UTRs of yeast mt-mRNAs (26), fulfilling a similar role to bS1 in bacteria (27), while bS6m (Mrp17) (28), mS26 (Pet123) (29) and mS43 (Mrp1) (30) have all been shown to functionally interact with Pet122, a translational activator for cytochrome c oxidase subunit III.
In yeast, translational activators provide a physical link between the 5′ UTRs of mitochondrial transcripts, the mitoribosome and the inner mitochondrial membrane and are necessary for the translation of most, if not all, yeast mitochondrial mRNAs (2, 8). Translational activators have been proposed to compensate for the absence of a Shine-Dalgarno sequence by aligning the mitoribosome on the mRNA and defining the start codon. Furthermore, translational activators are specific to individual transcripts and regulate translation beyond initiation by establishing tailored microenvironments (2).
Based on the functional interaction studies, we speculate that the mRNA exit canyon acts as a platform for translational activators and that the density above the channel represents a heterogeneous mixture of translational activators that co-purified with the yeast mitoribosome (table S4).
In conclusion, the complete structure of the yeast mitoribosome reveals new insights into the diversity and evolution of mitoribosomes. As S. cerevisiae is one of the few organisms whose mtDNA can be genetically manipulated, the structure provides a framework to help understand the species-specific mechanisms that regulate mitochondrial translation.
Supplementary Material
Acknowledgements
We thank C. Savva and J. Murray for assistance with the electron microscopes; J.L. Llácer for help with model building and analysis; S. Tan for help with figure presentation; J. Grimmett and T. Darling for computing support; and M. Skehel for mass spectrometry. This work was funded by grants to V.R. from the UK Medical Research Council (MC_U105184332.); a Wellcome Trust Senior Investigator award (WT096570), the Agouron Institute and the Louis-Jeantet Foundation. N.D. is funded by the Wellcome Trust Clinical PhD Fellowship (110301/Z/15/Z). The maps have been deposited with the Electron Microscopy Data Bank under the accession codes EMD-3551, EMD-3552, EMD-3553, EMD-3554, EMD-3555, and EMD-3556. Atomic coordinates have been deposited with the Protein Data Bank under IDs 5MRC, 5MRE, and 5MRF.
References
- 1.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 2.Ott M, Amunts A, Brown A. Organization and Regulation of Mitochondrial Protein Synthesis. Annu Rev Biochem. 2016;85:77–101. doi: 10.1146/annurev-biochem-060815-014334. [DOI] [PubMed] [Google Scholar]
- 3.Greber BJ, Ban N. Structure and Function of the Mitochondrial Ribosome. Annu Rev Biochem. 2016;85:103–132. doi: 10.1146/annurev-biochem-060815-014343. [DOI] [PubMed] [Google Scholar]
- 4.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amunts A, Brown A, Toots J, Scheres SHW, Ramakrishnan V. Ribosome. The structure of the human mitochondrial ribosome. Science. 2015;348:95–98. doi: 10.1126/science.aaa1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grebe BJ, et al. Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science. 2015;348:303–308. doi: 10.1126/science.aaa3872. [DOI] [PubMed] [Google Scholar]
- 7.van der Sluis EO, et al. Parallel Structural Evolution of Mitochondrial Ribosomes and OXPHOS Complexes. Genome Biol Evol. 2015;7:1235–1251. doi: 10.1093/gbe/evv061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herrmann JM, Woellhaf MW, Bonnefoy N. Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochim Biophys Acta. 2013;1833:286–294. doi: 10.1016/j.bbamcr.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Ban N, et al. A new system for naming ribosomal proteins. Current Opinion in Structural Biology. 2014;24:165–169. doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherry JM, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Research. 2012;40:D700–5. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groot GS, Mason TL, Van Harten-Loosbroek N. Var1 is associated with the small ribosomal subunit of mitochondrial ribosomes in yeast. Mol Gen Genet. 1979;174:339–342. doi: 10.1007/BF00267808. [DOI] [PubMed] [Google Scholar]
- 12.Bullerwell CE, Burger G, Lang BF. A novel motif for identifying rps3 homologs in fungal mitochondrial genomes. Trends in Biochemical Sciences. 2000;25:363–365. doi: 10.1016/s0968-0004(00)01612-1. [DOI] [PubMed] [Google Scholar]
- 13.Barros MH, Myers AM, Van Driesche S, Tzagoloff A. COX24 codes for a mitochondrial protein required for processing of the COX1 transcript. J Biol Chem. 2006;281:3743–3751. doi: 10.1074/jbc.M510778200. [DOI] [PubMed] [Google Scholar]
- 14.Saveanu C, et al. Identification of 12 new yeast mitochondrial ribosomal proteins including 6 that have no prokaryotic homologues. J Biol Chem. 2001;276:15861–15867. doi: 10.1074/jbc.M010864200. [DOI] [PubMed] [Google Scholar]
- 15.Kitakawa M, et al. Identification and characterization of the genes for mitochondrial ribosomal proteins of Saccharomyces cerevisiae. Eur J Biochem. 1997;245:449–456. doi: 10.1111/j.1432-1033.1997.t01-2-00449.x. [DOI] [PubMed] [Google Scholar]
- 16.Petrossian TC, Clarke SG. Multiple Motif Scanning to identify methyltransferases from the yeast proteome. Mol Cell Proteomics. 2009;8:1516–1526. doi: 10.1074/mcp.M900025-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park Y, Bader JS. How networks change with time. Bioinformatics. 2012;28:i40–8. doi: 10.1093/bioinformatics/bts211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown GK, et al. beta-hydroxyisobutyryl coenzyme A deacylase deficiency: a defect in valine metabolism associated with physical malformations. Pediatrics. 1982;70:532–538. [PubMed] [Google Scholar]
- 19.Hiltunen JK, et al. The biochemistry of peroxisomal beta-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2003;27:35–64. doi: 10.1016/S0168-6445(03)00017-2. [DOI] [PubMed] [Google Scholar]
- 20.Pfeffer S, Woellhaf MW, Herrmann JM, Förster F. Organization of the mitochondrial translation machinery studied in situ by cryoelectron tomography. Nat Commun. 2015;6:6019. doi: 10.1038/ncomms7019. [DOI] [PubMed] [Google Scholar]
- 21.Denslow ND, Anders JC, O'Brien TW. Bovine mitochondrial ribosomes possess a high affinity binding site for guanine nucleotides. J Biol Chem. 1991;266:9586–9590. [PubMed] [Google Scholar]
- 22.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 23.Selmer M. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 24.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 25.Couvillion MT, Soto IC, Shipkovenska G, Churchman LS. Synchronized mitochondrial and cytosolic translation programs. Nature. 2016;533:499–503. doi: 10.1038/nature18015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green-Willms NS, Fox TD, Costanzo MC. Functional interactions between yeast mitochondrial ribosomes and mRNA 5' untranslated leaders. Molecular and Cellular Biology. 1998;18:1826–1834. doi: 10.1128/mcb.18.4.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hajnsdorf E, Boni IV. Multiple activities of RNA-binding proteins S1 and Hfq. Biochimie. 2012;94:1544–1553. doi: 10.1016/j.biochi.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Haffter P, Fox TD. Suppression of carboxy-terminal truncations of the yeast mitochondrial mRNA-specific translational activator PET122 by mutations in two new genes, MRP17 and PET127. Mol Gen Genet. 1992;235:64–73. doi: 10.1007/BF00286182. [DOI] [PubMed] [Google Scholar]
- 29.McMullin TW, Haffter P, Fox TD. A novel small-subunit ribosomal protein of yeast mitochondria that interacts functionally with an mRNA-specific translational activator. Molecular and Cellular Biology. 1990;10:4590–4595. doi: 10.1128/mcb.10.9.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haffter P, McMullin TW, Fox TD. Functional interactions among two yeast mitochondrial ribosomal proteins and an mRNA-specific translational activator. Genetics. 1991;127:319–326. doi: 10.1093/genetics/127.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.