Abstract
This paper presents a label-free affinity-based capacitive biosensor using interdigitated electrodes. Using an optimized process of DNA probe preparation to minimize the effect of contaminants in commercial thiolated DNA probe, the electrode surface was functionalized with the 24-nucleotide DNA probes based on the West Nile virus sequence (Kunjin strain). The biosensor has the ability to detect complementary DNA fragments with a detection limit down to 20 DNA target molecules (1.5 aM range), making it suitable for a practical point-of-care (POC) platform for low target count clinical applications without the need for amplification. The reproducibility of the biosensor detection was improved with efficient covalent immobilization of purified single-stranded DNA probe oligomers on cleaned gold microelectrodes. In addition to the low detection limit, the biosensor showed a dynamic range of detection from 1 µL−1 to 105 µL−1 target molecules (20 to 2 million targets), making it suitable for sample analysis in a typical clinical application environment. The binding results presented in this paper were validated using fluorescent oligomers.
Keywords: Affinity-based capacitive biosensor, Interdigitated electrodes, Nucleic acid-based biosensor, Viral diagnostics, Pathogen detection, Point-of-care diagnostics
Highlights
-
•
A capacitive DNA biosensor using interdigitated electrodes is presented.
-
•
It produced more than 70 nF in capacitance change in response to as few as 20 DNA targets (~1.5 aM).
-
•
It achieved a dynamic range between 1 μL−1 and 106 μL−1 molecules (20 to 2 million DNA molecules).
-
•
It achieved an excellent linearity within the dynamic range.
-
•
It is a good candidate for low-cost and rapid detection technology for POC pathogen detection.
1. Introduction
As universal biological information storage entities, nucleic acids (DNA and RNA) are unique biorecognition molecules, and the detection of pathogen genomic DNA or RNA provides one of the most reliable methods for viral infectious disease diagnostics. Emerging and remerging infectious pathogens, such as Ebola, Middle East Respiratory Syndrome (MERS), West Nile, dengue and Zika viruses create a strong need for a low-cost, point-of-care (POC) diagnostic platform that would enable rapid and sensitive pathogen detection (Woolhouse et al., 2015, Zumla et al., 2015, Daep et al., 2014, Petersen et al., 2016). Early and accurate detection of viral infectious diseases is of crucial importance in preventing epidemic disease outbreaks as well as in improving the efficacy of POC diagnostic technologies (Peeling and McNerney, 2014, Sin et al., 2014). In use since the early 1990s, quantitative real-time polymerase chain reaction (qPCR) amplification has been a gold standard in viral diagnostics (Yang and Rothman, 2004, Espy et al., 2006). While qPCR offers high specificity with low limits of detection, it and other similar existing diagnostic methods have their drawbacks, such as the use of expensive devices for repeated thermal cycling, specialized non-reusable reagents, the need for sensitive fluorescence detection optics, and laborious assay preparation steps requiring trained personnel (Timmer and Villalobos, 1993). For example, a typical qPCR will require highly trained technicians 4–8 h from sample preparation to completion using reagents such as specific TaqMan probes and matching master mixes with reverse transcriptase enzymes and DNA polymerase, costing up to hundreds of dollars per assay depending on sample number and origin. Consequently, such analytical methods do not align with the need for a rapid, inexpensive, highly specific and sensitive point-of-care platform (Craw and Balachandran, 2012, Peeling and McNerney, 2014, Sin et al., 2014).
One of the cost-effective alternatives to PCR-based detection of pathogen genomic DNA relies on measurement of electrical property changes (resistance, capacitance, and complex impedance) due to DNA-DNA hybridization at probe-target binding sites without labeling. Such a sensing modality has been used successfully to detect specific DNA molecules in complex mixtures in a number of different assays, making it attractive for reliable classification of target DNA (Boyd, 2013, Ventimiglia and Petralia, 2013; Drummond et al., 2003; Byron et al., 2016, Marzancola et al., 2016). Electrode configuration and geometry can have a significant effect on sensor performance (Pettine et al., 2012). One of the widely used electrode configurations, especially for sensors measuring capacitance changes due to DNA-DNA hybridization (capacitive biosensors), is the interdigitated microelectrode. Microelectrodes are often made using modern photolithographic and deposition techniques on glass, silicon, or other solid substrates (Blanda, 1991, Gawad et al., 2009, Pettine et al., 2012). When they are tightly integrated with the back-end measurement circuits, they have significant advantages over conventional carbon-based electrodes (Li and Miao, 2012) for analytical measurements, such as low resistance, high signal-to-noise ratio, rapid attainment of steady state, and the use of small solution volumes (Dandy et al., 2007, Yang and Bashir, 2008, Wydallis et al., 2015).
Operating modalities of biosensors using interdigitated electrodes (IDEs) can be non-faradaic (Stagni et al., 2006) or faradaic (Chornokur et al., 2011). The biosensor presented in this paper operates in the non-faradaic mode based on changes in capacitance between interdigitated electrodes to indicate molecular binding events at the electrode surface. Biosensors operating in the faradaic mode are often based on electrochemical impedance spectroscopy (EIS) (Daniels and Pourmand, 2007; Lisdat and Schäfer, 2008) by measuring electron transfer resistance and double layer capacitance within a frequency range. Comparing to capacitive biosensors, biosensors based on EIS have been widely explored for their ability to capture complex resistance changes due to binding events at biosensor's electrode sites. However, they are more complex from an electronics and experimental protocol perspective, requiring a wide-range frequency sweep and the use of a potentially hazardous redox couple (e.g. Fe(CN)6 3−/4−) for measuring faradaic current. With the simplicity of measuring capacitance change between electrodes due to DNA-DNA hybridization between targets and probes, combined with low limit of detection (LOD) and high specificity, the results from this paper show that label-free capacitive biosensors have the potential as a baseline technology for low-cost, low-power, easy to use rapid detection POC platform (Bracke et al., 2007).
To date, capacitive biosensors developed for DNA/RNA classification and pathogen detection have focused on improving the electrode surface modification process, achieving better transducer sensitivity, and increasing detection circuit sensitivity and signal-to-noise ratio (SNR). Berggren et al. (1999) reported a label-free capacitive detection method for DNA detection, and pushed the limit of detection down to 25 complementary DNA targets per µL. Moreno-Hagelsieb et al. (2004) demonstrated the use of an inexpensive Al/Al2O3 hybrid electrode to achieve good sensitivity. Guiducci et al. (2006) elucidated a number of important details relevant to charge-based capacitance measurements, and provided insights into CMOS compatible implementation for integrated electronics. Stagni et al. (2006) demonstrated a capacitive biosensor design with a detection range of 330 pF to 10 µF with good linearity. Lee et al. (2010) developed CMOS circuitry based on charge/discharge theory for detecting capacitive signals to demonstrate its ability to detect 2 nM of target nucleic acid from H5N1 Influenza viruses. Qureshi et al. (2010) achieved multiplexed detection using an interdigitated electrode array, with 25 pg/mL sensitivity to the complementary target. Eberhardt et al. (2011) reported a bio-inspired artificial whisker to monitor fluid motion using capacitive sensing. Kallempudi and Gurbuz (2011) proposed a capacitance detection method using Fourier transform infrared spectroscopy (FT-IR) at high frequency, and achieved a 1 ng/mL detection limit. Wright and Chen (2015) demonstrated an ultra-sensitive read-out circuit for measuring capacitance changes capable of sub-fF detection limit.
However, even with more than a decade of progress in capacitive biosensor development, significant challenges remain. A number of studies (Berggren et al., 1999, Qureshi et al., 2010) reported poor sensor-to-sensor reproducibility. Non-uniformity of the self-assembled monolayer (SAM) and the covalently attached DNA probes on the sensor surface is a major contributing factor to poor sensor-to-sensor reproducibility. Sensor surface cleanliness greatly affects the uniformity of SAM layers (Love et al., 2005). Covalently attached ssDNA probes are largely affected by the presence of common contaminants in commercial thiolated DNA probes (Lee et al., 2010). The lack of emphasis on electrode cleaning prior to assay functionalization may have also contributed to significant variations of the reported results. Stagni et al. (2006) elucidated the importance of electrode surface cleaning in order to achieve uniform covalent binding of thiolated DNA probes. In addition, the total surface area of microelectrodes and the geometric relationship between two electrodes in a biosensor are other factors which directly affect sensor sensitivity and reproducibility, as measured capacitance output represents averaged values from all the fingers of the microelectrodes (Guiducci et al., 2004).
Here, a label-free, affinity-based capacitive IDE sensor is developed for unamplified nucleic acid detection, with high sensitivity and reproducibility. The proposed platform technology uses capacitance changes resulting from the solid-phase hybridization of nucleic acid targets with ssDNA probes immobilized on microelectrodes as the means of detection and identification. A 24-nucleotide DNA probe and target set was designed based on the West Nile virus sequence (Kunjin strain) and was applied as a model for nucleic acid based viral recognition and detection on the proposed capacitive biosensor (Steel et al., 2000, Oliveira et al., 2015). The process of DNA probe preparation for the biosensor has been optimized to include steps to minimize the effect of contaminants associated with commercial thiolated DNA probes. It is demonstrated here that this new biosensor produces an output of more than 70 nF in capacitance change in response to as few as 20 complementary DNA targets (0.25 attogram) at a concentration of ~1.5 aM. Due to the optimized cleaning process for the capacitive biosensor, including a pre-cleaning protocol, to improve covalent immobilization of purified single-stranded DNA probe oligomers onto the microelectrodes, the biosensor's reproducibility was improved compared to the reported results. In addition to the low detection limit, the biosensor showed a good dynamic range of detection from 1 µL−1 to 105 µL−1 target molecules (20 to 2 million total targets), making it suitable for sample analysis in a typical clinical application environment. The capacitance results obtained in this study were verified using fluorescently labeled oligonucleotides on a fluorescence scanner. Capacitive affinity-based assays have long been considered highly sensitive (Mattiasson and Hedstrom, 2016), but low specificity and reproducibility (Berggren et al., 1999) hinder further improvement in their sensitivity. The results presented in this paper provide a potential path for practical use of capacitive sensing technology for viral pathogen detection in clinical settings.
2. Materials and methods
2.1. DNA oligonucleotides and reagents
Validated HPLC purified DNA oligonucleotides were purchased from Integrated DNA Technologies (IDT, Inc., Coralville, IA). The sequences for single-strand DNA (ssDNA) oligonucleotides were based on the sequence of the West Nile Kunjin strain (Genbank Accession # AY274504) and were as follows:
DNA probe (Oligo 1):
5′-ThioMC6-D-TAGTATGCACTGGTGTCTATCCCT-3′
Complementary DNA target – 100% complementarity (Oligo 2):
5′-AGGGATAGACACCAGTGCATACTA-3′
Noncomplementary DNA target (Oligo 3):
5′-GCAATATAGATAACGCCAGATGGC-3′
Probe and target DNAs were resuspended in 1TE-MgSO4 buffer (TE stands for Tris–HCI and EDTA), which contained 100 µM magnesium sulfate (MgSO4), 1 µM Tris-Hydrochloride (Tris–HCl) and 0.1 µM Ethylenediaminetetraacetic acid (EDTA), and were filtered through a 0.45 µm syringe filter (Millipore) prior to use. The 24-mer oligonucleotide DNA probe (Oligo1) was selected to form a stable duplex with its complementary target (Oligo 2) at room temperature, with minimal interference due to self-complementarity or secondary structure. The non-complementary target (Oligo 3) has 1 out of 24 base pairs complementary with oligo1 (4.16% complementarity).
For fluorescence detection, a second probe was generated that was 5′-thiolated (C6 propyl spacer) with a 3′-fluorescent Alexa Fluor 488 label (5′-ThioMC6-D/TAGTATGCACTGGTGTCTATCCCT/AlexF488N/-3′), while complementary and non-complementary targets were 5′-labeled with fluorescent Alexa Fluor 594 dye (5′-Alex594N/AGGGATAGACACCAGTGCATACTA-3′ or 5′-Alex594N/GCAATATAGATAACGCCAGATGGC-3′). The 11-Mercapto-1-undecanol (MCU) (97%; Sigma-Aldrich, St. Louis, MO) was dissolved in water and filtered through a 0.45 µm syringe filter (Millipore) prior to use. Potassium hydroxide (KOH) and hydrogen peroxide (H2O2, 30% w/w in H2O) (Sigma-Aldrich, St. Louis, MO) were used as received. Milli-Q water from Millipore (Billerica, MA) (18.2 MΩ cm) was used in all experiments.
2.2. Gold interdigitated microelectrodes sensor
Commercial gold coated IDE sensors were purchased from DropSens (Asturias, Spain). Each IDE has a finger width and spacing of 10 µm, with a total number of 125 fingers, a total electrode length of 6760 µm, and electrode surface area of 8.45 mm2. Titanium is present as an adhesive layer.
2.3. Capacitance measurement setup
Capacitance measurement data were collected using the Instek LCR-821 benchtop LCR meter (New Taipei City, Taiwan), which interfaces with a PC for data acquisition. A graphical user interface (GUI) on the PC was used for sending command signals to the LCR meter. Since the measurement is obtained from non-faradaic current, a 0 V DC bias voltage was applied across the IDE sensor. A 20 mV root mean square (RMS) AC voltage with 20 Hz frequency was applied to the IDE sensors. All capacitance readouts were recorded under 20 µL of 100 µM 1TE-MgSO4 buffer on the interdigitated electrodes and 50 data points were collected per reading. Capacitance data were analyzed using Matlab (Mathworks) and statistical tests were carried out with (www.r-project.org). Only <0.05 values were considered as statistically significant.
2.4. Melt curve generation for buffer optimization
A CFX 96 Real Time system (C1000 Thermal cycler, Bio-Rad) was used to generate melt curves in order to follow DNA probe interactions with complementary and non-complementary target oligos through double stranded DNA (dsDNA) formation under different buffer types and concentrations. For each case fluorescence emission was detected every 5 s from 4 °C to 95 °C at 0.5 °C degree increments. The duplex (dsDNA) formation reaction was carried out under different buffer conditions, specifically, in 1 M and 100 mM 1TE-NaCl, and 1 mM and 100 µM 1TE-MgSO4. The DNA probe to DNA target ratio was fixed at 1:1 for 1 µg/µL oligos for all reactions. SYBR Green nucleic acid gel stain (Life Technologies, Carlsbad, CA) was included in all reactions as a real time indicator of the presence of dsDNA.
2.5. Pretreatment of the gold electrodes surface
Upon exposure to ambient conditions the gold microelectrode surface is subject to a variety of uncontrolled conditions and contaminants, which can affect thiol reaction kinetics and therefore the probe attachment chemistry; because of this variability, a cleaning step is required immediately prior to functionalization. To prepare the surfaces, the chips with the gold microelectrodes were immersed in a solution of 50 mM KOH and 25% H2O2 for 10 min (Fischer et al., 2009), and thoroughly rinsed in Milli-Q water to remove the reagents, followed by oxygen plasma treatment. Baseline capacitance was recorded immediately following chemical cleaning and prior to plasma cleaning and probe immobilization.
2.6. Pretreatment of thiolated single-stranded DNA oligomers
In order for thiol bond formation between the oligo and the gold surface to proceed as expected, the 5′ thiol-modified oligomers require a reduction of the disulfide bonds prior to application. Immobilized TCEP Disulfide Reducing Gel (Thermo Fisher Scientific, Waltham, MA) was washed several times with 100 µM 1TE-MgSO4 buffer prior to oligo addition in a 2:1 TCEP gel:oligo ratio, and the mixture was incubated for 1 h on a rocker platform at 23 °C. After incubation and a 1500 rpm spin for 2 min, the supernatant with reduced thiol-modified oligomers was transferred to a clean tube.
2.7. SH-modified DNA probe oligonucleotide immobilization
Gold IDE sensors were plasma-cleaned for 5 min in an O2 Plasma Etch PE-25 (Plasma Etch, Carson City, NV, USA) at a pressure of 200 mTorr and 150 W applied to the RF coil immediately before DNA probe incubation. The 10 µM ssDNA probe solution was prepared in 100 µM 1TE-MgSO4 buffer, and the gold microelectrodes were immersed overnight (15–17 h) in 30 µL of the solution, then rinsed copiously with 100 µM 1TE-MgSO4 buffer and Milli-Q water, and dried with N2. The IDE sensors were then passivated with 20 µL MCU (5 µM in water) for 45 min to fill vacant gold sites and promote linear orientation of the ssDNA probe molecules. After MCU passivation, the electrodes were rinsed with water and dried with N2. All reactions were carried out at 23 °C.
2.8. DNA target hybridization
The IDA sensors were immersed in 20 µL target DNA solutions for 30 min at room temperature, at target concentrations ranging from 1 µL−1 to 105 µL−1 in 100 µM 1TE-MgSO4 buffer. Following incubation, electrodes were copiously rinsed with 100 µM 1TE-MgSO4 buffer and dried with N2.
2.9. Fluorescence scanning
Gold IDA sensors were scanned using a FLUOstar Omega fluorescence scanner (BMG LABTECH, Cary, NC). Fluorescent Alexa Fluor 488 and Alexa Fluor 594 signals from the sensors were quantified and analyzed using the FLUOstar Omega software against experimental controls. The paired t-test was used for statistical analysis and only <0.05 values were considered to be statistically significant.
3. Results and discussion
3.1. Measurement setup
The experiments carried out using the functionalized IDE sensors focus on quantifying the key sensor performance metrics of sensitivity and selectivity. To obtain these data, capacitance values of the IDE sensors were measured at four different stages of the sensing process:
-
1.
Cleaned bare gold electrode surfaces. The initial cleaning step was always applied to provide a baseline capacitance response.
-
2.
Electrodes after covalent attachment of thiolated ssDNA probes. The layer of immobilized DNA probes has the intrinsic ability to capture complementary single-stranded target oligonucleotides.
-
3.
Electrodes after incubation with MCU. The measured capacitance at this stage is with the presence of buffer without any target DNA present in the buffer. The MCU is used to block any remaining unoccupied sites on the sensor surface and to align the probes perpendicular to the sensor surface in order to improve hybridization efficiency (Herne and Tarlov, 1997).
-
4.
Electrodes after incubation and hybridization with ssDNA targets, both complementary and non-complementary.
Capacitance between IDE pairs was obtained by applying a 20 mV RMS AC voltage with a fixed frequency (stimulus) and by measuring the resulting current through the sensor IDE pair (response). The measurements of capacitance with the bare clean electrodes and with immobilized probes are mainly intended for verifying the state of probe immobilization. Unless otherwise stated, the change in capacitance after hybridization with either complementary or non-complementary targets is referred to as the difference between the capacitances before and after hybridization (i.e. capacitance difference between stage 3 and stage 4).
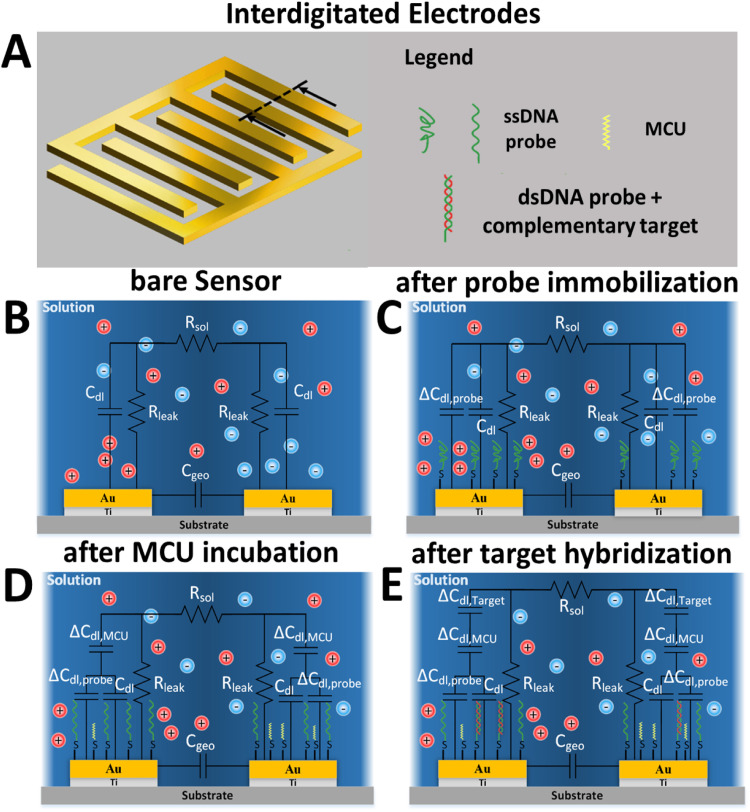
Fig. 1A shows a schematic of the IDE sensor and Fig. 1B–E show the equivalent circuits for stages 1, 2, 3, and 4, respectively. The quantity is the intrinsic buffer solution resistance; is an equivalent resistance representing a leakage current going through the electrode-electrolyte interface; is the double layer capacitance created between the IDEs and adjacent buffer (Fig. 1B) resulting from the applied voltage; and is the geometric capacitance which depends on the geometry of the electrodes and their configuration, which may be negligible relative to the double layer capacitance (Stagni et al., 2006). The quantity is the amount of change in double layer capacitance after probe immobilization, and is different from because the ssDNA attachment modifies the surface capacitive characteristics of the electrodes. The additional double layer capacitance results from incubation with MCU, and is the double layer capacitance change due to target hybridization.
Fig. 1.
Equivalent circuit models on interdigitated electrode (IDE) sensors. (A) Bare IDEs with multiple fingers. (B) Equivalent circuit model for bare IDE sensors. (C) Equivalent circuit model after probe immobilization. (D) Equivalent circuit model after MCU incubation and before target hybridization. (E) Equivalent circuit model after target hybridization for complementary targets.
Because the MCU blocking agent acts as an insulation layer that pushes mobile ions away from the electrode into the solution, the total double layer capacitance is expected to decrease after the MCU step in stage 3. Similarly, after incubation and hybridization with the complementary and non-complementary target DNA in stage 4, the resulting duplex structures should act to repel additional ions away from the electrode surface, resulting in further decreases in the double layer capacitance. These behaviors are illustrated in Fig. 1C and D, where the sequential decreases in total double layer capacitance after the MCU and target incubation are denoted by and in a series configuration, and connected to the baseline double layer capacitance, .
3.2. Effect of buffer composition and temperature on hybridization
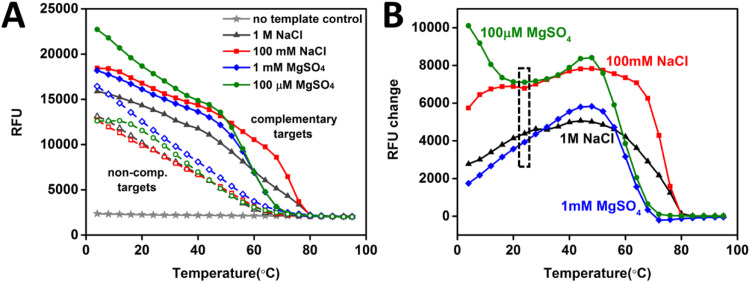
The specificity of an ssDNA probe is a critical factor that governs the performance characteristics of the biosensor, and which depends in part on electrolyte salt composition, concentration, and temperature. Two widely used salt buffers, each at two different ionic concentrations, 1 M and 100 mM 1TE-NaCl, and 1 mM and 100 µM 1TE-MgSO4, were tested to quantify their effect on the degree of hybridization between the probe and complementary and non-complementary targets. Intercalation of the planar SYBR Green fluorophore into dsDNA increases its fluorescence emission dramatically, allowing detection of dsDNA by monitoring fluorescence intensity. SYBR Green fluorescence intensity in RFU (Relative Fluorescence Unit) in the presence of 24-base synthetic probe and complementary/non-complementary targets with increasing temperature is shown in Fig. 2A, illustrating double stranded DNA (dsDNA) formation due to DNA probe-target interactions in different buffer conditions and within 4–95 °C temperature range. As temperature increases, dsDNA concentrations decrease resulting in decrease in SYBR green fluorescence intensity. To understand the effect of buffer composition on hybridization specificity, fluorescence intensity differences between complementary and non-complementary targets in each buffer type within the temperature range from 4 °C to 95 °C are shown in Fig. 2B. Non-complementary target RFU background seen in the melt curve assay, especially at the lower temperature range (Fig. 2A), can be attributed to two factors. First, lower temperatures favor non-complementary binding and specificity of duplex formation is increased with an increase in reaction temperature. Second, the conditions with very high target and probe oligo concentrations enhance the formation of primer dimers which contributed to higher SYBR Green signal at the beginning of the assay in the non-complementary target samples.
Fig. 2.
Effect of different buffers on DNA hybridization. (A) SYBR Green fluorescence in the presence of complementary (solid line) and non-complementary (dashed line) targets incubated with 24-base ssDNA probe in 1 M 1TE-NaCl buffer (black), 100 mm 1TE-NaCl buffer (red), 1 mM 1TE-MgSO4 buffer (blue) and 100 µM 1TE-MgSO4 buffer (green) within temperature range from 4 °C to 95 °C. (B) Corresponding relative fluorescence change between the complementary and non-complementary targets in four different buffers. The black dashed box indicates the temperature used in all the experiments. RFU=Relative fluorescence units. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
For three of the four buffers tested, the specificity increased with temperature up to 45 °C with the trend reversing at temperatures above 45 °C, except for the 100 µM 1TE-MgSO4 buffer. The specificity from using the 100 µM 1TE-MgSO4 buffer decreased with temperature up to 23 °C, but it is still better than that for all other three salt buffers up to 45 °C. For each salt type the specificity increased with decreasing ionic concentration, and this behavior is consistent with other results reported in the literature (Petrovykh et al., 2003). At the temperature used for all subsequent hybridization studies (23 °C) the 100 µM 1TE-MgSO4 buffer demonstrates the best hybridization specificity. While low ionic concentrations are beneficial, in general, for achieving high specificity, a potential downside is that a low concentration of monovalent cations may hinder ssDNA probe immobilization on the IDE sensor surface (Petrovykh et al., 2003). The negatively charged DNA phosphate backbone, coupled with low cation concentration, may result in unacceptable electrostatic repulsion effects. However, the cation valence has a larger effect on probe immobilization than its concentration, so that bivalent cations, such as Mg2+, can mitigate the repulsion effect, and therefore result in a higher density of immobilized probes on the IDE sensor surface (Petrovykh et al., 2003). Furthermore, low ionic concentration serves to reduce measurement interference from the leakage current that often occur due to sodium deposits on the sensor surface. Overall, the 100 µM 1TE-MgSO4 buffer was found to be the best compromise of composition and concentration to achieve high hybridization specificity without compromising ssDNA probe immobilization, and it was used as the electrolyte buffer for all remaining experiments.
3.3. Optimization of stimulus signal frequency
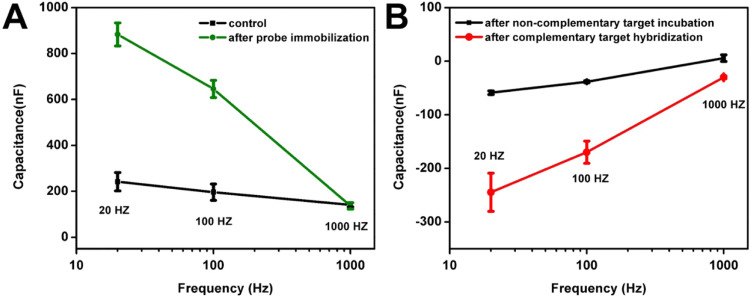
The amount of change measured in double layer capacitance due to target binding is strongly dependent on the frequency of the stimulus AC signal. Since the double layer capacitance at the electrode-electrolyte interface is established due to the displacement of mobile ions in the fluid, it is expected that increasing the stimulus signal frequency will result in gradual disappearance of the double layer capacitance (Guiducci et al., 2004). To gain a better understanding of the sensor's response characteristics as a function of the stimulus signal frequency, a 20 mV RMS AC voltage is applied at three separate frequencies (20 Hz, 100 Hz and 1 kHz). Capacitance values for all four stages were measured for all three frequencies. The results shown in Fig. 3A verify changes in capacitance due to probe immobilization compared to the bare clean electrodes (marked as “control” in Fig. 3A). The results in Fig. 3B illustrate that changes in capacitance before and after hybridization decrease with increase in operating frequency. To maximize sensor sensitivity, 20 Hz was used as the operating frequency for all remaining experiments.
Fig. 3.
Frequency effects on capacitance during each step of sensor preparation/usage. (A) Changes in capacitance after probe immobilization at 20 Hz, 100 Hz, and 1 kHz frequencies. (B) Changes in capacitance after complementary and non-complementary (control) target hybridization at 20 Hz, 100 Hz, and 1 kHz frequencies.
3.4. Immobilization of oligonucleotides probes on sensor surface
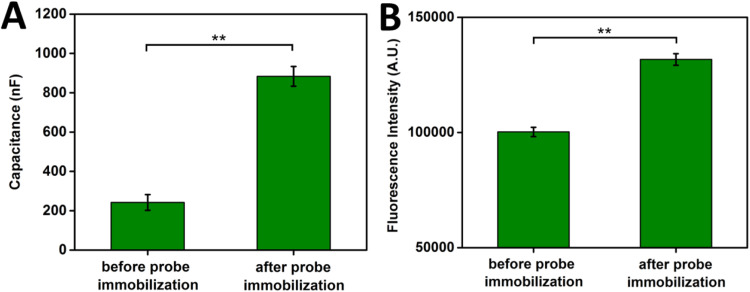
A protocol for deposition of 5′ thiol modified ssDNA probe molecules onto the gold IDE surface and passivation of the IDE surface was established. ssDNA immobilization can be significantly affected by the condition of the IDE surface, the incubation time of 5′ thiol-modified ssDNA with the gold surface, and the concentration of ssDNA. In general, the cleaner the IDE surface the better thiol-modified molecules will attach to the gold surface (Xue et al., 2014). The IDE sensor surface was cleaned with the protocol described in Section 2.5 and treated with oxygen plasma to promote ssDNA immobilization. In addition to surface cleanliness, ssDNA immobilization is also determined by time the thiol modified ssDNA is in contact with the gold surface. In general, twenty-four hour incubations are required to reach the saturation point for 1 µM ssDNA immobilization in 1 M salt conditions, and further exposure results in little additional adsorption (Herne and Tarlov, 1997). For ssDNA probe concentrations ranging from µM to mM, a surface density in the order of 103 cm−2 can be obtained after four hours of incubation (Guiducci et al., 2004). In order to maximize sensor functionalization and the number of probes present on the functionalized sensor based on our previous work (Dandy et al., 2007), overnight incubation was chosen for the 10 µM 5′ thiol-modified ssDNA probe in our experiments. Probe concentration was set in the µM range based on previous work on the gold electrode DNA biosensors (Dandy et al., 2007) and was further optimized with additional chemical and plasma oxygen cleaning steps to maximize sensor coverage and target capture in 100 µM 1TE-MgSO4 buffer. To validate 5′ thiol-modified ssDNA immobilization on the IDE sensor surface, capacitance measurements were obtained from 3 individual sensors after overnight ssDNA probe incubation, but before passivation with MCU. Fig. 4A demonstrates a statistically significant difference (p<0.01) between the average capacitance measurements before and after probe immobilization, indicating that ssDNA probe was immobilized on the IDE surface. To further validate the capacitance results, AlexF488N labeled ssDNA probes were immobilized on IDEs as a complementary assay to monitor probe deposition. Total IDE fluorescence intensities were obtained before and after probe immobilization (Fig. 4(B)), showing that incubation with fluorescent ssDNA probe resulted in a significant increase in fluorescence on the IDE (p<0.01) similar to what was observed by capacitance measurements.
Fig. 4.
Validation of probe immobilization on IDE sensors. (A) Capacitance measurements before and after probe immobilization. (B) The fluorescence intensity before and after probe immobilization. ** paired t-test: p<0.01.
3.5. Specificity
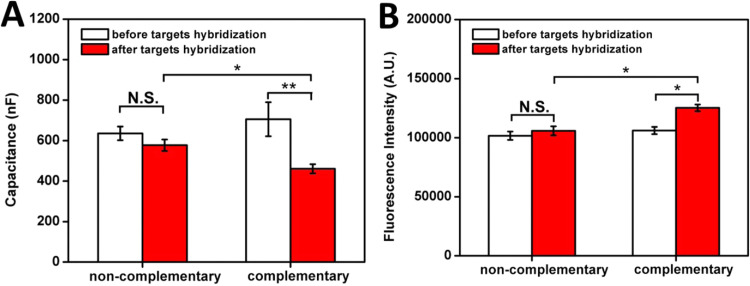
After probe immobilization and MCU incubation, specificity tests of complementary and non-complementary DNA targets were conducted. The rate of DNA hybridization is dependent on DNA length, with shorter duplex regions showing higher hybridization rates (Noble, 1995, Steel et al., 2000). Based on the 24-base DNA target used in the experiments, target incubation was set for 30 min at room temperature (23 °C). AlexF594N labeled complementary and non-complementary targets with the concentration of 105 µL−1 in 20 µL of 100 µM 1TE-MgSO4 buffer were incubated with the probe immobilized IDE surface, and excess target DNA was washed off. The average capacitance of sensors incubated with complementary and non-complementary target DNAs with the standard error of three individual sensors are shown in Fig. 5A. Non-complementary target DNAs did not induce a significant capacitance change, whereas complementary DNA induced a large capacitance change in the sensor. This is further validated using fluorescent target capture on the sensor surface (Fig. 5B). Decreased capacitance on IDE surfaces upon target binding has been previously reported in the literature. When ssDNA probes are immobilized on the IDE surfaces, the double layer of ions due to the polarized metal surface are displaced (Berggren et al., 1999). When complementary DNA strands bind with the probes, the distance between the charge on the surface of the electrode and the ions in the electrolyte increases, resulting in decreasing in the overall capacitance (Berggren et al., 1999, Guiducci et al., 2004).
Fig. 5.
Specificity of target hybridization. (A) Capacitance measurements before and after complementary and non-complementary target hybridization. (B) Fluorescence intensity before and after complementary and non-complementary target hybridization. ** paired t-test: p<0.01; * paired t-test: p<0.05.
3.6. Sensitivity
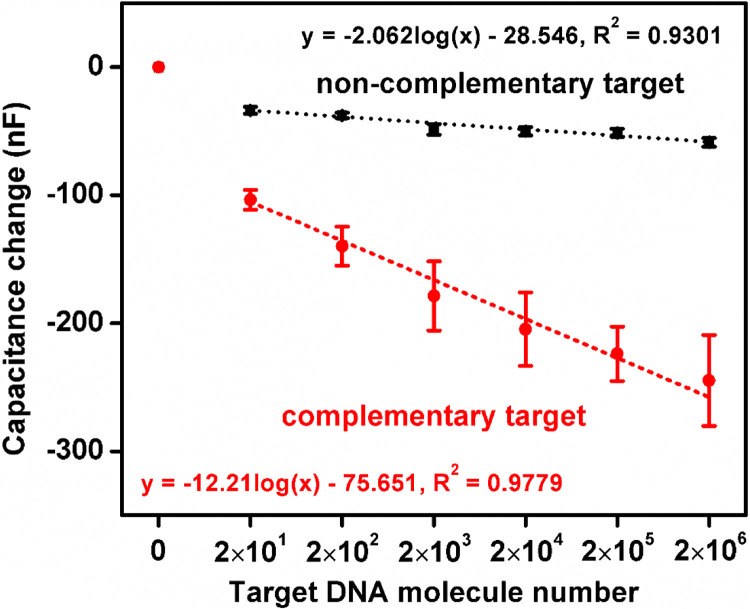
To determine the sensitivity of the sensor, DNA concentrations between 0 and 2 million molecules, consistent with the range seen in typical clinical settings, were incubated on the sensor and capacitance was determined ( Fig. 6). Complementary target DNAs displayed a significant capacitance change as few as 20 DNA molecules, whereas non-complementary DNAs did not significantly change capacitance even at 2106 molecules. For target concentrations between 20 and 2 million DNA molecules, complimentary targets showed a linear response in capacitance change with increasing target concentration, indicating an excellent correlation between low-range target concentrations and capacitance responses. These results demonstrate that the label-free capacitive detection limit for the complementary targets is at least as low as 20 DNA molecules (~1.5 aM). Increasing complementary target concentrations increased capacitance changes, whereas increasing non-complementary DNA concentrations did not significantly increase capacitance. There have been a number of ultra-high sensitive biosensors reported to date which achieved limits of detection similar to the results presented in this paper. Most of these ultra-high sensitive approaches relied on novel amplification methods using nano-materials, such as nano-particles (Li et al., 2010) and quantum dots (Shen and Gao, 2015). Other approaches using nano-structures include the use of a novel molecular gate structure with carbon-nanotubes to control sensor's conduction state (Nuzaihan et al., 2016), the use of carbon nanotubes (CNTs) as a highly conductive agent to enhance redox current sensitivity (Nie et al., 2012), and the use of CNTs combined with Au nano-particles to increase sensor surface area (Wang et al., 2013). However, these approaches incur additional costs related to additional sample preparation for nano-materials and amplification, and nano-scale device manufacturing. The biosensor presented in this paper achieved a high sensitivity comparable to those achieved using nano-materials for amplification and other enhancements, thus lowering the cost and simplifying sample processing and device manufacturing requirements.
Fig. 6.
Capacitance response from 0 to 2×106 complementary (red line) and non-complementary (black line) DNA targets. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Conclusion
In this paper, a capacitive biosensor with high sensitivity and a wide dynamic detection range is presented. The carefully designed protocol for sensor surface preparation, probe immobilization, and hybridization strikes a balance among the competing factors of ease of immobilization, specificity, and the overall capacitance measurement quality. The reported biosensor improves the state of the art on three fronts. First, the sensor was able to produce an output of more than 70 nF in capacitance change in response to as few as 20 complementary DNA targets (0.25 attogram) at a concentration of ~1.5 aM. The magnitude of capacitance change increases linearly with complementary target concentration in the range between 20 and 2 million target DNA molecules. The detection limit of this biosensor is among the lowest reported to date for a non-faradaic capacitive biosensor. Second, sensor specificity is clearly demonstrated by the capacitance change differential between complementary and non-complementary target binding. For non-complementary ssDNA target molecules, the measured change in capacitance is significantly less than that for the complementary ssDNA target and does not display an appreciable change in capacitance with non-complementary molecule concentration increases. Third, the sensor achieved a dynamic range for detection between 1 µL−1 and 106 µL−1 target molecules (20 to 2 million target DNA molecules) with excellent linearity in the measurement of capacitance changes. Combining its performance in sensitivity, specificity, and dynamic detection range, this capacitive biosensor demonstrates the potential to be a viable technology for low-cost, low-power, ease of use, and rapid detection technology that is a critical step toward POC pathogen detection. Areas where further investigations can provide more optimized conditions for the capacitive biosensor include optimized incubation time to decrease time for sample analysis. Furthermore, future development of this capacitive biosensor includes detection of viral nucleic acids (DNA and RNA) from complex biological samples, as well as platform multiplexing, with the ultimate goal of developing a POC device for rapid and multiplexed diagnosis of viral infections in clinical and field settings.
Acknowledgment
Part of this work was supported by National Institute of Health/National Institute of Allergy and Infectious Diseases [Grant 5 R01 AI114675]; National Science Foundation [GDE-0841259], and internal seed grant funding from the Colorado State University Coalition for Development and Implementation of Sensors and Sensing Systems (CDISS). The authors would like to thank Sam Wright, Yusra Obeidat, and Jessie Filer for their assistance in many experiments. The authors would also like to thank Dr. Brad Reisfeld for providing the use of a fluorescent scanner.
References
- Berggren C., Stålhandske P., Brundell J., Johansson G. Electroanalysis. 1999;11(3):156–160. [Google Scholar]
- Blanda, T. (Ed.), 1991. Microelectrodes: Theory and Applications. Nato Science Series E. (Inglés).
- Boyd S.D. Annu. Rev. Pathol. 2013;24(8):381–410. doi: 10.1146/annurev-pathol-020712-164026. [DOI] [PubMed] [Google Scholar]
- Bracke W., Merken P., Puers R., Van Hoof C. IEEE Trans. Circuits Syst. I Regul. Pap. 2007;54(1):130–140. [Google Scholar]
- Byron S.A., Van Keuren-Jensen K.R., Engelthaler D.M., Carpten J.D., Craig D.W. Nat. Rev. Genet. 2016;17(5):257–271. doi: 10.1038/nrg.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chornokur G., Arya S.K., Phelan C., Tanner R., Bhansali S. J. Sens. 2011;2011 [Google Scholar]
- Craw P., Balachandran W. Lab Chip. 2012;12(14):2469–2486. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- Daep C.A., Muñoz-Jordán J.L., Eugenin E.A. J. Neurovirol. 2014;20(6):539–560. doi: 10.1007/s13365-014-0285-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandy D.S., Wu P., Grainger D.W. Proc. Natl. Acad. Sci. USA. 2007;104(20):8223–8228. doi: 10.1073/pnas.0606054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels J.S., Pourmand N. Electroanalysis. 2007;19(12):1239–1257. doi: 10.1002/elan.200603855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond T.G., Hill M.G., Barton J.K. Nat. Biotechnol. 2003;21(10):1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- Eberhardt W.C., Shakhsheer Y.A., Calhoun B.H., Paulus J.R., Appleby M. IEEE Sens. 2011:982–985. [Google Scholar]
- Espy M.J., Uhl J.R., Sloan L.M., Buckwalter S.P., Jones M.F., Vetter E.A., Yao J.D., Wengenack N.L., Rosenblatt J.E., Cockerill F.R., 3rd, Smith T.F. Clin. Microbiol. Rev. 2006;19(1):165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer L.M., Tenjea M., Heiskanena A.R., Masudaa N., Castilloa J., Bentienb A., Émneusa J., Jakobsena M.H., Boisena A. Microelectron. Eng. 2009;86:1282–1285. [Google Scholar]
- Gawad S., Guigliano M., Heuschkel M., Wessling B., Markram H., Schnakenberg U., Morgan H. Front. Neuroeng. 2009 doi: 10.3389/neuro.16.001.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiducci C., Stagni C., Fischetti A., Mastromatteo U., Benini L., Riccò B. IEEE Sens. J. 2006;6(5):1084–1093. [Google Scholar]
- Guiducci C., Stagni C., Zuccheri G., Bogliolo A., Benini L., Samorì B., Riccò B. Biosens. Bioelectron. 2004;19(8):781–787. doi: 10.1016/s0956-5663(03)00266-5. [DOI] [PubMed] [Google Scholar]
- Herne T.M., Tarlov M.J. J. Am. Chem. Soc. 1997;119(38):8916–8920. [Google Scholar]
- Kallempudi S.S., Gurbuz Y. Sens. Actuators B: Chem. 2011;15(160 (1)):891–898. [Google Scholar]
- Lee K.H., Lee J.O., Sohn M.J., Lee B., Choi S.H., Kim S.K., Yoon J.B., Cho G.H. Biosens. Bioelectron. 2010;26(4):1373–1379. doi: 10.1016/j.bios.2010.07.055. [DOI] [PubMed] [Google Scholar]
- Li, G., Miao, P., 2012. Theoretical Background of Electrochemical Analysis. In: Electrochemical Analysis of Proteins and Cells. Springer. doi: 10.1007/978-3-642-34252_2
- Li H., Sun Z.Y., Zhong W.Y., Hao N., Xu D.K., Chen H.Y. Anal. Chem. 2010;82:5477–5483. doi: 10.1021/ac101193e. [DOI] [PubMed] [Google Scholar]
- Lisdat F., Schäfer D. Anal. Bioanal. Chem. 2008;391:1555–1567. doi: 10.1007/s00216-008-1970-7. [DOI] [PubMed] [Google Scholar]
- Love J.C., Estroff L.A., Kriebel J.K., Nuzzo R.C., Whitesides G.M. Chem. Rev. 2005;1103–1169:105. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- Marzancola M.G., Sedighi A., Li P.C.H. Microarray Technology: Methods and Applications. 2016. pp. 161–178. [DOI] [PubMed] [Google Scholar]
- Mattiasson B., Hedstrom M. TrAC. 2016;79:233–238. [Google Scholar]
- Moreno-Hagelsieb L., Lobert P.E., Pampin R., Bourgeois D., Remacle J., Flandre D. Sens. Actuators B: Chem. 2004;15(98 (2)):269–274. [Google Scholar]
- Nie G.M., Bai Z.M., Chen J., Yu W.Y. ACS Macro Lett. 2012;1:1304–1307. doi: 10.1021/mz300341h. [DOI] [PubMed] [Google Scholar]
- Noble D. Anal. Chem. 1995;67:201A. doi: 10.1021/ac00101a728. [DOI] [PubMed] [Google Scholar]
- Nuzaihan M.N.M., Hashim U., Md Arshad M.K., Kasjoo S.R., Rahman S.F., Ruslinda A.R., Fathil M.F., Adzhri R., Shahimin M.M. Biosens. Bioelectron. 2016;15(83):106–114. doi: 10.1016/j.bios.2016.04.033. [DOI] [PubMed] [Google Scholar]
- Oliveira N., Souza E., Ferreira D., Zanforlin D., Bezerra W., Borba M.A., Arruda M., Lopes K., Nascimento G., Martins D., Cordeiro M., Lima-Filho J. Sensors. 2015;15(7):15562–15577. doi: 10.3390/s150715562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeling R.W., McNerney R. Expert Rev. Mol. Diagn. 2014;14(5):525–534. doi: 10.1586/14737159.2014.915748. [DOI] [PubMed] [Google Scholar]
- Petersen L.R., Jamieson D.J., Powers A.M., Honein M.A. N. Engl. J. Med. 2016;21(374 (16)):1552–1563. doi: 10.1056/NEJMra1602113. [DOI] [PubMed] [Google Scholar]
- Petrovykh D.Y., Kimura-Suda H., Whitman L.J., Tarlov M.J. J. Am. Chem. Soc. 2003;125:5219–5226. doi: 10.1021/ja029450c. [DOI] [PubMed] [Google Scholar]
- Pettine W., Jibson M., Chen T., Tobet S., Nikkel P., Henry C.S. IEEE Sens. J. 2012;5(12):1187–1192. [Google Scholar]
- Qureshi A., Niazi J.H., Kallempudi S., Gurbuz Y. Biosens. Bioelectron. 2010;25:2318–2323. doi: 10.1016/j.bios.2010.03.018. [DOI] [PubMed] [Google Scholar]
- Shen W., Gao Z. Biosens. Bioelectron. 2015;30(65C):327–332. doi: 10.1016/j.bios.2014.10.060. [DOI] [PubMed] [Google Scholar]
- Sin M.L., Mach K.E., Wong P.K., Liao J.C. Expert Rev. Mol. Diagn. 2014;14(2):225–244. doi: 10.1586/14737159.2014.888313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagni C., Guiducci C., Benini L., Riccò B., Carrara S., Samorí B., Paulus C., Schienle M., Augustyniak M., Thewes R. IEEE J. Solid-State Circuits. 2006;41(12):2956–2964. [Google Scholar]
- Steel A.B., Levicky R.L., Herne T.M., Tarlov M.J. Biophys. J. 2000;79(2):975–981. doi: 10.1016/S0006-3495(00)76351-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventimiglia G., Petralia S. BioNanoScience. 2013;3(4):428–450. [Google Scholar]
- Timmer W.C., Villalobos J.M. J. Chem. Educ. 1993;70:273. [Google Scholar]
- Wang S., Li L., Jin H.L., Yang T., Bao W.W., Huang S.M., Wang J.C. Biosens. Bioelectron. 2013;41:205–210. doi: 10.1016/j.bios.2012.08.021. [DOI] [PubMed] [Google Scholar]
- Woolhouse M.E., Rambaut A., Kellam P. Sci. Transl. Med. 2015;30(7 (307)) doi: 10.1126/scitranslmed.aab0191. (307rv5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S., Chen, T., 2015. In: Proceedings of the IEEE 58th International Midwest Symposium on Circuits and Systems (MWSCAS) 2015 Aug 2, pp. 1–4
- Wydallis J.B., Feeny R.M., Wilson W., Kern T., Chen T., Tobet S., Reynolds M.M., Henry C.S. Lab Chip. 2015;15(20):4075–4082. doi: 10.1039/c5lc00778j. [DOI] [PubMed] [Google Scholar]
- Xue Y., Li X., Li H., Zhang W. Nat. Commun. 2014;5:4843. doi: 10.1038/ncomms5348. [DOI] [PubMed] [Google Scholar]
- Yang S., Rothman R.E. Lancet Infect. Dis. 2004;4(6):337–348. doi: 10.1016/S1473-3099(04)01044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Bashir R. Biotechnol. Adv. 2008;26:135–150. doi: 10.1016/j.biotechadv.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Zumla A., Hui D.S., Perlman S. Lancet. 2015;5(386 (9997)):995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]