Abstract
Homologous recombination (HR) is a central process to ensure genomic stability in somatic cells and during meiosis. HR-associated DNA synthesis determines in large part the fidelity of the process. A number of recent studies have demonstrated that DNA synthesis during HR is conservative, less processive, and more mutagenic than replicative DNA synthesis. In this review, we describe mechanistic features of DNA synthesis during different types of HR-mediated DNA repair, including synthesis-dependent strand annealing, break-induced replication, and meiotic recombination. We highlight recent findings from diverse eukaryotic organisms, including humans, that suggest both replicative and translesion DNA polymerases are involved in HR-associated DNA synthesis. Our focus is to integrate the emerging literature about DNA polymerase involvement during HR with the unique aspects of these repair mechanisms, including mutagenesis and template switching.
Keywords: DNA synthesis, genome stability, mutagenesis, template switching
Introduction
Homologous recombination (HR) is a critical pathway for maintaining genome stability, owing to its ability to repair complex DNA damage such as DNA double-strand breaks (DSBs) and interstrand cross-links (ICLs). Moreover, HR provides a mechanism for tolerating lesions that block the progress of replication forks. During meiosis, HR is critical in establishing the physical linkage between homologs by crossover formation that facilitates faithful chromosome segregation. Progress has been made in understanding the mechanisms of HR processes such as the initial processing of damage (e.g., DSB resection); assembly and function of the pivotal RAD51 nucleo-protein filament, which conducts homology search and DNA strand invasion; and enzymes that generate and process recombination junctions, including the hallmark intermediate for crossover formation, the double Holliday junction (dHJ) (Figure 1). The RAD51 filament also stabilizes stalled replication forks against nucleolytic degradation (128). This process does not appear to involve DNA strand invasion because RAD54 is not required (128); it is also unlikely to involve DNA synthesis. A collection of informative reviews documents these impressive advances (63).
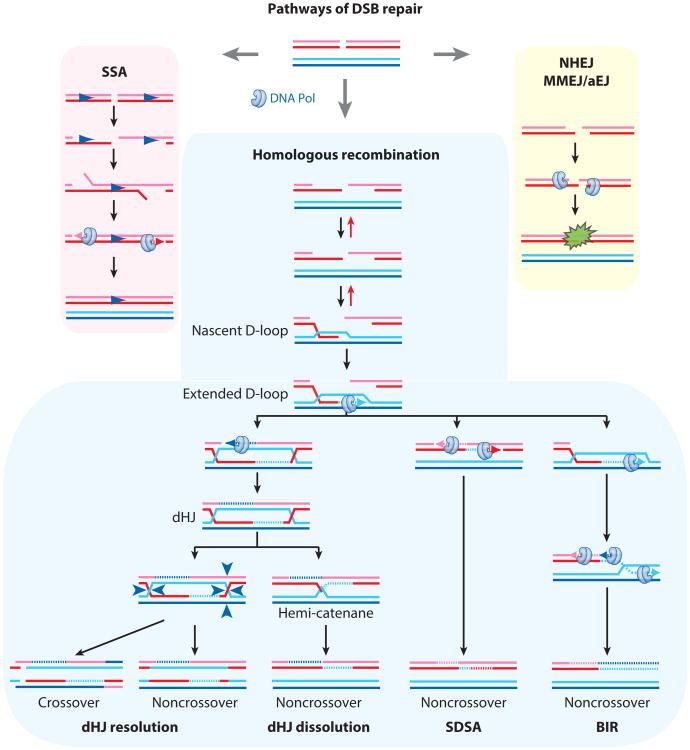
Figure 1.
Double-strand break (DSB) repair pathways. Different DNA synthesis steps are indicated by the DNA polymerase symbol. New DNA synthesis (broken lines) plays a role in all DSB repair pathways, including single-strand annealing (SSA), nonhomologous end joining (NHEJ), and its variants, microhomology-mediated end joining/alternative end-joining (MMEJ/alt-EJ) and homologous recombination (HR). Although HR is the only pathway that produces a double Holliday Junction (dHJ), it can also proceed by pathways that do not involve dHJ formation, such as synthesis-dependent strand annealing (SDSA) and break-induced replication (BIR). Abbreviation: D-loop, displacement loop.
Although DNA synthesis during HR is essential to restore the integrity of the chromosomes, this process had long been considered to be a rather mundane subject. Three distinct discoveries changed this perception and put HR-associated DNA synthesis, its mechanisms, and the proteins involved in the spotlight. First, Strathern and colleagues (142) discovered that HR-mediated DSB repair is associated with an ∼1,000-fold increase in mutation frequency, showing that HR is not error-free, as often posited. Second, the expanding complement of DNA polymerases in eukaryotes provides a rich stable of potential enzymes involved in DNA synthesis during HR, including translesion DNA polymerases. A list of all the eukaryotic DNA polymerases (reviewed in 151), their basic properties and subunit composition, and comments on their function in DSB repair by HR are provided in Table 1. Finally, recent genetic and molecular studies in Saccharomyces cerevisiae demonstrated that break-induced replication (BIR), a pathway for HR (Figure 1), involves long-range DNA synthesis with a mechanism that differs from canonical DNA replication (3, 123, 155).
Table 1. Eukaryotic DNA polymerases and their involvement in DSB repair and HRa.
| DNA polymerase | Human gene | Saccharomyces cerevisiae gene | Basic properties | DSB repair/HR function | Reference(s) |
|---|---|---|---|---|---|
| A family | |||||
| Pol γ |
POLG P0LG2 |
MIP1 | High fidelity, 3′–5′ exonuclease, processive (with POLG2 in humans), dRP lyase activity (humans) | Mitochondrial HR | 18, 58 |
| Pol θ | POLQ | NA | Low fidelity, bypass of abasic sites and thymine glycol, DNA-dependent ATPase activity | MMEJ/alt-EJ, regulation of HR efficiency in mammals | 16, 61, 84, 131 |
| Pol ν | POLN | NA | Low fidelity, similar to polymerase domain of Pol θ | Knockdown mildly decreases HR, interacts with HR factors, high testis expression | 92, 146 |
| B family | |||||
| Pol α |
POLA P0LA2 PRIM1 PRIM2A |
P0L1 P0L12 PRI1 PRI2 |
Relatively high fidelity, no proofreading, nonprocessive, no PCNA interaction | Unsettled role in DSB-induced gene conversion, BIR | 47, 153 |
| Pol δ |
P0LD1 P0LD2 P0LD3 P0LD4 |
P0L3 P0L31 POL32 |
High fidelity; 3′–5′ exonuclease, low DNA affinity, processivity increased by PCNA interaction, high PCNA affinity | Primary role in HR first-end synthesis, possible role in second-end synthesis, BIR | 26, 47, 71, 81 |
| Pol ε |
POLE P0LE2 P0LE3 P0LE4 |
P0L2 DPB2 DPB3 DPB4 |
High fidelity, 3′–5′ exonuclease, high DNA affinity, highly processive, PCNA dependent at high salt, low PCNA affinity, PCNA stimulates processivity | Role in HR, second-end synthesis?, BIR? | 153 |
| Pol ζ |
POLZ bREV7 P0LD3 P0LD4 |
REV3 REV7 P0L31 POL32 |
Lower fidelity than other family members, lacks a 3′–5′ exonuclease activity, powerful mismatch extender, interacts with REV1, functionally interacts with PCNA | Facilitate HR repair, ssDNA mutagenesis during HR repair, meiotic mutagenesis, ICL repair | 27, 69, 116, 121, 133 |
| X family | |||||
| Pol λ | POLL | P0L4 | Low fidelity for single base deletions, moderate fidelity for base substitutions, high affinity for dNTPs, Pol β-like domain dRP lyase activity | Increased expression during meiosis, NHEJ, MMEJ/alt-EJ | 7, 67, 108, 110 |
| Pol β | POLB | NA | Moderate fidelity, prefers substrates with short gaps, dRP lyase activity | Suggested role in meiosis | 15, 111 |
| Pol μ | POLM | NA | Low frameshift fidelity, can use primer and template that are on separate DNA molecules, sequence-independent nucleotidyl transferase, Pol β–like domain and dRP lyase activity | Immunoglobulin gene rearrangement, NHEJ | 21, 72, 125 |
| Terminal deoxynucleotidyl transferase | DNTT | NA | Template-independent, possesses Pol β–like domain dRP lyase activity | Immunoglobulin gene rearrangement, NHEJ | 33, 72 |
| Y family | |||||
| Pol η | POLH | RAD30 | Low fidelity, bypass of cis-syn thymine-thymine dimers, extends D-loop | Knockdown decreases HR efficiency in DT40 cells | 60 |
| Pol κ | POLK | NA | Low fidelity, profound mismatch-extending capacity, predominant T to G transversion | Possible role in HR | 130 |
| Pol ι | POLI | NA | Fidelity varying from extremely low to high depending on the type of error, weak dRP lyase activity, preferentially incorporates a G opposite a template T | No evidence for role in HR | 130, 148 |
| Revl | REV1L | REV1 | Deoxycytidyl transferase/G-template-specific polymerase, interacts with other specialized polymerases | Possible role in HR | 59, 91, 133 |
| PrimPol | PRIMPOL | NA | De novo synthesis of DNA, bypasses the most common oxidative lesions in DNA, both nuclear and mitochondrial | Lesion skipping, downstream priming after fork stalling | 93, 152 |
For a comprehensive review, see Reference 109.
Abbreviations: BIR, break-induced replication; D-loop, displacement loop; dNTP, deoxynucleoside triphosphate; dRP, deoxyribose phosphate; DSB, double-strand break; HR, homologous recombination; NA, not applicable; NHEJ, nonhomologous end joining; PCNA, proliferating cell nuclear antigen; ssDNA, single-stranded DNA.
In this review, we discuss the roles of DNA polymerases in HR, focusing on eukaryotes (S. cerevisiae, Drosophila melanogaster, Gallus gallus DT40 cells, Mus musculus, and Homo sapiens) with only brief mention of other DSB repair processes such as nonhomologous end joining (NHEJ), microhomology-mediated end joining [MMEJ; also called alternative end joining (alt-EJ)], and single-strand annealing (SSA) (see Figure 1). We highlight the different HR-associated DNA synthesis steps postulated by current models and attempt to elaborate common principles. The complement of available DNA polymerases varies from organism to organism (Table 2). Therefore, we resist projecting the data into a single model, but instead point out how the increasingly complex complement of DNA polymerases may lead to functional specialization. The reader is referred to other excellent reviews related to this topic and on DNA polymerases in bacteria and archaea (80, 104, 109).
Table 2. DNA polymerase complements in eukaryotes.
| DNA polymerase |

|

|

|

|
|
|---|---|---|---|---|---|
| α (alpha) | ✓ | ✓ | ✓ | ✓ | ✓ |
| δ (delta) | ✓ | ✓ | ✓ | ✓ | ✓ |
| ε (epsilon) | ✓ | ✓ | ✓ | ✓ | ✓ |
| γ (gamma) | ✓ | ✓ | ✓ | ✓ | ✓ |
| ζ (zeta) | ✓ | ✓ | ✓ | ✓ | ✓ |
| Rev1 | ✓ | ✓ | ✓ | ✓ | ✓ |
| η (eta) | ✓ | ✓ | ✓ | ✓ | ✓ |
| λ (lambda) | ✓ | ✓ | ✓ | ✓ | |
| θ (theta) | ✓ | ✓ | ✓ | ✓ | |
| ι (iota) | ✓ | ✓ | ✓ | ||
| β (beta) | ✓ | ✓ | ✓ | ||
| Terminal deoxynucleotidyl transferase | ✓ | ✓ | ✓ | ||
| κ (kappa) | ✓ | ✓ | ✓ | ||
| ν (nu) | ✓ | ✓ | ✓ | ||
| μ.(mu) | ✓ | ✓ | |||
| PrimPol | ✓ | ✓ | ✓ |
DNA polymerase data based on Genome Databases from Saccharomyces cerevisiae, Drosophila melanogaster, Gallus gallus, Mus musculus, and Homo sapiens. Pol σ (sigma) is a cytoplasmic poly-riboA terminal transferase and is not listed here (124).
Double-Strand Break Repair Overview
HR is most commonly studied in the context of DSB repair (Figure 1). This is because of the importance of this pathway for the repair of this complex lesion and also because nucleases can easily induce site-specific DSBs, allowing a number of experimental approaches that are less readily employed in the study of other lesions that can also be repaired by HR, such as replication-associated gaps. Thus, the present discussion focuses on HR and insights learned from DSB repair. It is unclear whether gap repair by HR differs in its selection of DNA polymerases. As described below, three different pathways compete for DSB repair (Figure 1).
End-joining mechanisms (NHEJ and MMEJ/alt-EJ) require little to no resection and lead to direct ligation of the two ends of the DSB. The product is a contiguous chromosome and is often accompanied by deletions and insertions at the DSB site, depending on the initial chemical structure of the DSB. NHEJ and MMEJ/alt-EJ appear to require specialized DNA polymerases (Table 1), which may be distinguished by their ability to synthesize across noncontiguous template strands.
SSA requires extensive resection and the presence of tandem-repeated DNA sequences flanking the break, to allow annealing of complementary DNA sequences. In S. cerevisiae, Rad52 is the only activity shown to anneal single-stranded DNA (ssDNA) coated with replication protein A (RPA) acting inconcert with Rad59 (144). After the structure-selective endonuclease XPF-ERCC1 (Rad1-Rad10 in S. cerevisiae) trims the flaps, DNA synthesis fills in the remaining gaps (Figure 2). The product is a contiguous chromosome that lacks one repeat and the intervening sequence. It is currently not known which DNA polymerases are involved in SSA, but SSA may serve as a proxy for the second DNA synthesis step in synthesis-dependent strand annealing (SDSA) (see below) (also see Figure 2). We refer the reader to authoritative reviews discussing these processes and the associated enzymes (63, 72).
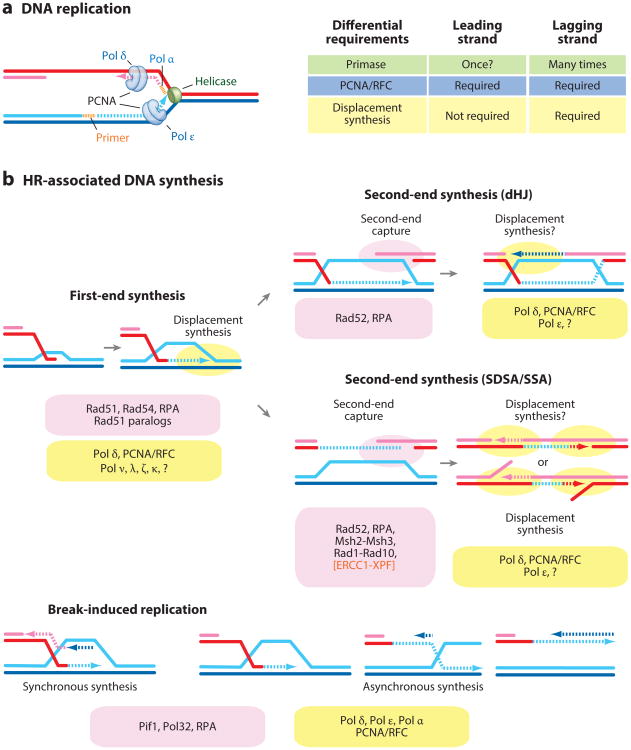
Figure 2.
Modes of DNA synthesis. (a) Replication fork. (b) Homologous recombination (HR)-associated DNA synthesis: first-end synthesis of invading strand at the displacement loop (D-loop), second-end synthesis in the double Holliday junction (dHJ) pathway, second-end synthesis during synthesis-dependent strand annealing (SDSA) and single-strand annealing (SSA), and break-induced replication. In Saccharomyces cerevisiae, SSA and, possibly, second-end capture require Rad59. Saw1 is a cofactor for the Rad1-Rad10 endonuclease, and S. cerevisiae Rad1-Rad10 corresponds to Homo sapiens XPF-ERCC1. Also shown are the proteins present at the HR intermediates (pink bubbles) as well as the DNA polymerases implicated in the steps of DNA synthesis (yellow bubbles). Abbreviations: PCNA, proliferating cell nuclear antigen; RFC, replication factor C; RPA, replication protein A.
HR involves the resection of DSBs to expose ssDNA. ssDNA is less chemically stable than double-stranded DNA (74), which has clear consequences for HR-associated mutagenesis (discussed below). Unlike NHEJ or MMEJ/alt-EJ, HR is a template-dependent process (Figure 1). During somatic DNA repair, the natural template is the sister chromatid (53, 57). Thus, HR is constrained to the S/G2 phase by several mechanisms, although HR in the G1 phase can be detected experimentally (25). The consequence of using the sister as a template for DSB repair by HR is that, in somatic cells, no mismatches are expected in DNA strand invasion intermediates (Figure 1), unless DNA strand invasion occurs between repeats with diverged sequences (also called homeologous DNA) at nonallelic sequences in a process referred to as homeologous recombination.
In contrast, during meiotic HR, the preferred template is the homolog, which can contain natural DNA sequence variations. This can potentially create mismatches at or close to the 3′ end of the invading strand, and these mismatches may affect extension by DNA polymerases. This fundamental difference between somatic recombinational DNA repair and meiotic recombination can be explored in experimentally tractable organisms such as yeast, flies, and mice, but it is not readily examined in other systems such as chicken DT40 cells and humans. Studies in S. cerevisiae are largely conducted in isogenic strains, where the homologs are identical except for the few genetic markers used, thus masking this potentially important biological feature of meiotic HR.
DNA Polymerases: General Mechanisms and Different Modes of DNA Synthesis During Homologous Recombination
All cells must accurately replicate their genomes during each cell division. DNA polymerases are at the core of DNA synthesis, a task that is constantly challenged by a variety of endogenous and exogenous DNA-damaging agents as well as unusual DNA structures. To do this, cells rely on a range of specialized DNA polymerases to properly replicate nuclear and organelle genomes (Table 1).
All DNA polymerases fold into a conformation composed of three distinct domains known as palm, thumb, and fingers. However, they differ in their polymerization properties, such as catalytic efficiency, processivity, fidelity, and preferred DNA substrate. Processivity defines the ability of DNA polymerases to incorporate more than one nucleotide at a primer template per binding event. Processivity is an intrinsic property of a polymerase that is highly influenced by whether and with what affinity a polymerase interacts with the processivity clamp proliferating cell nuclear antigen (PCNA) (79) (Table 1). This interaction can be modulated by PCNA modifications such as ubiquitylation and sumoylation. Repair synthesis of as little as 10 nucleotides or less during base-excision repair requires a PCNA-dependent polymerase, suggesting that PCNA-independent DNA polymerases, such as Pol β, which acts during base-excision repair, are specialized to fill short gaps and may be restricted to single nucleotide insertions. PCNA interaction and high processivity are expected to minimize template switching of DNA polymerases.
DNA polymerase fidelity is enforced during two aspects of the synthesis reaction. The first involves the insertion of a nucleotide, using Watson-Crick base-pairing rules, on undamaged or damaged templates. The second involves extension of a nonstandard primer template that contains either mismatches or damaged bases. The behavior of many DNA polymerases during these two aspects of synthesis has been painstakingly determined, and mutational profiles have been established (140). Proofreading by an intrinsic 3′–5′ exonuclease activity is another distinguishing feature of DNA polymerases δ and ε (Table 1), which greatly enhances their fidelity and allows them to engage a primer template with mismatches at or near its 3′ end. Participation by low-fidelity DNA polymerases determines the mutagenicity of the ensuing DNA synthesis steps in HR on damaged or undamaged DNA templates as well as during the extension of mismatched primer templates.
DNA polymerases are classified into different families according to their evolutionary conservation, as listed in Table 1. The A family includes DNA polymerases γ, θ, and ν. DNA polymerase γ is a high-fidelity polymerase with proofreading activity responsible for replication and repair of the mitochondrial genome (58). DNA polymerases θ and ν are low-fidelity polymerases involved in MMEJ/alt-EJ repair of DSBs and ICLs, respectively (16, 84, 164). The B family formed by DNA polymerases α, δ, ε, and ζ is involved in a large variety of DNA transactions, including chromosomal DNA replication, repair, translesion synthesis (TLS), and HR. Thus, not surprisingly, B-family polymerases are the most widespread polymerase family found in eukaryotes (109). DNA polymerases α, δ, and ε cooperate with other proteins in the replication fork, and DNA polymerases δ and ε are responsible for the bulk of chromosome replication (109). DNA polymerases β, λ, μ, and terminal deoxynucleotidyl transferase (TdT) form the X family of DNA polymerases and are involved in DNA repair. X-family DNA polymerases are strictly distributive and overall involved in short DNA fragment synthesis (32). DNA polymerase β is mainly involved in base-excision repair (109). DNA polymerases λ, μ, and TdT have been implicated in NHEJ, MMEJ/alt-EJ, and variable-diversity-joining [V(D)J] recombination, in addition to processes responsible for generating diversity in the immunoglobulin and T cell receptors during the early stages of B and T cell maturation, respectively (90, 109). The Y family comprises DNA polymerases η, ι, κ, and Rev1, which act primarily in lesion bypass (32, 109). PrimPol, the most recent addition to the complement of eukaryotic DNA polymerases (30), is a second DNA primase found in some but not all eukaryotes (Table 2).
During DNA replication, the DNA polymerase α-primase complex lays down a hybrid RNA-DNA primer for Pol δ and Pol ε. Leading-strand synthesis is, in concept, continuous, although repriming likely occurs in vivo (Figure 2a). This process does not require strand displacement, as the replicative DNA helicase unwinds the duplex template. In fact, Pol ε is incapable of strand displacement synthesis (31). Owing to the polarity of DNA synthesis, it is obligatory for the lagging strand to be synthesized in a discontinuous fashion. Priming is required for each Okazaki fragment, and Pol δ completes the synthesis involving displacement of the previous primer for lagging-strand processing (31). Both replicative polymerases engage with PCNA, albeit with different affinities (Pol δ higher than Pol ε) to achieve similar processivity and curtail template switching (19).
During HR, several different modes of DNA synthesis can be differentiated, and the various HR intermediates that DNA polymerases encounter are bound by different sets of proteins (see Figure 2). These conceptual considerations suggest that different DNA polymerases are involved during HR-associated DNA synthesis. After DNA strand invasion by the Rad51 filament, Rad54 is required to provide DNA polymerases access to the invading 3′ end to initiate DNA synthesis (70). DNA synthesis in the displacement loop (D-loop) is continuous, similar to leading-strand synthesis during DNA replication, but requires displacement synthesis in the bubble of the D-loop. Displacement synthesis is a feature of lagging-strand synthesis during DNA replication, although the displaced strand at the lagging strand is a short flap and not a bubble. Hence, only DNA polymerases capable of displacement synthesis are candidates for extending the invading strand during HR, termed here first-end DNA synthesis (Figure 2b).
A second mode of HR-associated DNA synthesis during the dHJ pathway engages the second end on the displaced strand of the D-loop (Figure 2b). This gap-filling reaction does not require displacement synthesis per se but may involve displacement of the 5′ end flanking the gap. Hence, any DNA polymerase incapable or capable of displacement synthesis is a potential candidate for this mode of DNA synthesis.
A third mode of DNA synthesis during HR occurs from the second end during SDSA (and SSA) (Figure 2b). What distinguishes both modes of second-end DNA synthesis is that during dHJ the displaced strand of the D-loop is topologically constrained, whereas during SDSA (or SSA) there is no such constraint. In S. cerevisiae, the second end in the dHJ and SDSA pathways is likely captured by Rad52-mediated reannealing, similar to SSA (65, 134). It is unclear whether second-end capture by Rad52 and potentially other factors (Rad59, flap processing factors) imposes specific requirements on the DNA polymerase or if it affects PCNA loading by RFC.
Finally, DNA synthesis associated with BIR requires specific factors (e.g., Pol32, Pif1) to ensure long-range DNA synthesis and to physically separate leading- from lagging-strand DNA synthesis, as discussed more fully below. DNA synthesis of >45 nucleotides (shorter extension could not be measured) from the invading strand during BIR starts with a significant ∼6-h delay compared with what appears to be the same reaction during SDSA (48). The processes involved are unclear, but coupling DNA synthesis to the recognition of a second DSB end is a distinguishing characteristic of SDSA. In sum, conceptual considerations suggest that different DNA polymerases are engaged during the distinguishable DNA synthesis steps in HR, and the type of DNA polymerase defines important properties affecting HR outcome, including associated mutations and template switching.
DNA Polymerases and their Roles in Homologous Recombination
Below, we discuss the evidence for the involvement of each eukaryotic DNA polymerase in HR. Basic properties of these polymerases are presented in Table 1, and an overview of the DNA polymerases present in specific eukaryotic species is provided in Table 2; the discussion below follows the order in Table 2.
DNA Polymerase Alpha
DNA polymerase alpha (Pol α) is a two-subunit enzyme with a large, 167-kDa, catalytic subunit that forms a complex with the two-subunit primase complex to extend the RNA primer to allow de novo template-dependent DNA synthesis. Pol α is a relatively high-fidelity DNA polymerase but lacks proofreading activity. An initial genetic report suggested the involvement of Pol α in HR in the G1 phase of the cell cycle (37). Pol α primase is required in cycling cells for HO-induced mating-type switching, an SDSA-type HR event (47). Later analysis in G2-arrested cells concluded that neither Pol α nor primase is needed (153). It is unclear whether this discrepancy is due to cell cycle phase-specific effects on the temperature-sensitive mutant proteins used in these studies, as their protein levels were not monitored.
DNA Polymerase Delta
DNA polymerase delta (Pol δ) is a primary replicative polymerase that is believed to preferentially catalyze lagging-strand DNA synthesis, although this view has been recently challenged (54, 85). The human enzyme contains four subunits (POLD1–4); POLD1 is the largest (124 kDa), containing the catalytic polymerase activity and the proofreading 3′–5′ exonuclease function. S. cerevisiae Pol δ contains only three subunits, the catalytic subunit Pol3 as well as Pol31 and Pol32 (109). The noncatalytic subunits are required for processivity and optimal PCNA interaction. Interestingly, the Pol32 subunit in S. cerevisiae Pol δ is not essential for normal growth. Pol δ is capable of displacement synthesis and its processivity is strongly stimulated by PCNA, becoming essentially PCNA-dependent under conditions of physiological ionic strength (51). The interaction of S. cerevisiae Pol δ with PCNA is highly complex, involving the Fe-S cluster of the large subunit and PCNA-interaction sites, called PIP boxes, in all three subunits, such that Pol δ is likely occupying all three binding sites on the PCNA heterotrimer (2, 97).
Extensive genetic, molecular, and biochemical evidence shows that S. cerevisiae Pol δ is central to HR-associated DNA synthesis. Experiments using pol3 thermosensitive alleles showed that Pol δ is required for recombinational DNA repair (26). A complex screen for meiotic recombination mutants isolated pol3-ct, a short terminal truncation of the Pol δ catalytic subunit, which results in shortened conversion track lengths during meiotic and mitotic recombination as well as reduced meiotic crossovers and defective BIR (81, 82). The mutation likely affects processivity by impairing interactions with Pol31 and indirectly with PCNA (10). These results suggest that displacement synthesis associated with D-loop extension requires a high degree of processivity. A proofreading-deficient mutant of Pol δ, pol3-01, resulted in higher mutagenesis during gene conversion across the entire marker gene, suggesting that Pol δ synthesizes the bulk of the new DNA during this process (41). Remarkably, the pol3-01 mutation eliminated all –1 frameshift mutations and template switches (41). This may reflect the increased processivity and increased strand displacement of the proofreading-defective enzyme (31), consistent with a role of Pol δ in first-end synthesis. Pol δ appears to interact directly with the invading 3′ end, as its proofreading activity can trim short non-homologies prior to extension (106). Using molecular real-time assays to monitor DSB-induced gene conversion, Haber and colleagues showed a recombination defect in the thermosensitive pol3-14 strain, as the MAT switching product was reduced by approximately 30% at the restrictive temperature (47, 153). It is unclear whether the partial defects observed are due to functional overlap with additional DNA polymerases or if depletion of this essential protein was incomplete.
In vitro biochemical reconstitution experiments, using purified S. cerevisiae and human proteins, demonstrated that Pol δ can efficiently extend up to 80% of the invading strands in Rad51-mediated D-loops (69, 71, 129, 130, 139). In these studies, DNA synthesis was entirely dependent on PCNA, consistent with the need for displacement synthesis in the D-loop. The extent of synthesis was limited by the topology of the negatively supercoiled donor DNA because each 10 nucleotides synthesized introduce a superhelical turn. This topological constraint could be overcome by the addition of topoisomerase I, high levels of the ssDNA binding protein RPA, or the helicase Pif1 (71, 139, 155).
As noted in Figure 1, BIR requires long-range DNA synthesis that appears to be particularly sensitive to perturbations of Pol δ. In budding yeast, the pol3-ct mutation is entirely defective in BIR, as is the deletion of the nonessential subunit Pol32 (10, 75, 136). However, both mutants are proficient in gene conversion (44, 75, 81, 82). POLD3, the human POL32 ortholog, is also not essential, but it is required for BIR-like recombination events (20). It is unclear why the Pol3-ct and Pol32 defects are specific for BIR and do not show a significant defect in DNA replication. A possible explanation is that, compared with BIR-associated DNA synthesis, lagging-strand DNA synthesis is much shorter, estimated at 200 nucleotides (138). Also, gene conversion via SDSA involves significantly shorter DNA synthesis than does BIR. This suggests that Pol δ catalyzes processive DNA synthesis of longer than 200 nucleotides during BIR. Moreover, BIR requires a specific processivity factor, the Pif1 helicase, which migrates the extending D-loop to circumvent the topological constraint of D-loop extension discussed above (123, 155). The mechanism by which Pif1 contributes to BIR remains to be determined, in particular whether it directly interacts with Pol δ.
In sum, there is abundant evidence from genetic, molecular, and biochemical studies that Pol δ is a central player in HR-associated DNA synthesis. It is particularly adept at extending the 3′-OH end of the invading strand in the D-loop, performing first-end DNA synthesis in all HR pathways (Figure 1). Interestingly, Pol δ is also involved in MMEJ/alt-EJ-associated DNA synthesis in budding yeast (67, 90).
DNA Polymerase Epsilon
DNA polymerase epsilon (Pol ε) is a high-fidelity polymerase that may be primarily responsible for leading-strand replication, although this view has been challenged (54, 114). Human Pol ε is composed of four subunits: POLE (catalytic subunit A), POLE2 (subunit 2 or B), POLE3 (subunit 3 or C), and POLE4 (subunit 4 or D) and is best characterized in S. cerevisiae, in which the subunits are named Pol2, Dpb2, Dpb3, and Dpb4, respectively. The 3′–5′ proofreading exonuclease and DNA polymerase domains are located in the N-terminal domain of Pol2. However, the N-terminal domain is not essential in S. cerevisiae, and a mutant lacking these regions does not display hypersensitivity to DSBs (62). Interestingly, the C-terminal domain, responsible for protein-protein interactions, is essential for survival in budding yeast (23). Dpb3 and Dpb4 are nonessential subunits that affect Pol ε processivity and DNA binding (54, 150), whereas Dpb2 is also an essential protein in S. cerevisiae (24). Unlike Pol δ, Pol ε has high affinity for DNA, but low affinity for PCNA (19). However, Pol ε processivity can be stimulated by addition of PCNA (19). Mammalian and yeast Pol ε were reported to be incapable of displacement synthesis (28, 112), which is consistent with its role at the leading strand during replication (114) and with the alternate view that Pol ε acts in the repair of Pol δ–mediated errors (54). This property suggests that Pol ε is unlikely to be involved in first-end synthesis at the D-loop but leaves open the possibility that it acts during second-end synthesis.
Genetic studies in budding yeast have not provided a clear answer to the question of whether Pol ε is required for HR. Using the same approach to demonstrate the involvement of Pol δ in HR of monitoring recombination between noncomplementing thermosensitive alleles at the restrictive temperature (26), no evidence for a role of Pol ε was obtained (37). This is consistent with the observation that pol2-16 mutant cells, which lack the Pol ε catalytic polymerase and exonuclease domains, do not display repair defects (62). Even in a genetically sensitized background using the pol3-ct mutation, the pol2-16 Pol ε mutant did not enhance the phenotypes in UV-induced or IR-induced recombination of pol3-ct in hetero-allele recombination in diploid cells (82). Using a physical assay to monitor DSB-induced gene conversion at the MAT locus, the thermosensitive Pol ε allele pol2-18 showed a reduction in conversion products (∼30%) similar to that seen for the thermosensitive Pol δ allele pol3-14 (153). The relatively mild phenotype of both mutants could be due to functional overlap or incomplete inactivation of the affected enzyme. Independent evidence for the involvement of Pol ε in HR comes from the profile of HR-associated mutations in strains with the Pol ε proofreading-deficient allele pol2-01 (41). As with the analogous Pol δ allele discussed above, the mutations were spread over the entire marker gene. This finding might reflect a functional overlap between both polymerases, or it could be due to a possible division of labor between first-end synthesis (Pol δ) and second-end synthesis (Pol ε), consistent with the different biochemical properties of the two enzymes.
In sum, survival endpoint assays may lack the sensitivity to document a requirement for a specific enzyme in the context of partially overlapping function, and this could explain the failure of genetic studies to identify a role for Pol ε in HR. More sensitive endpoint assays, such as mutation profiles or molecular assays that also contain kinetic information, implicate Pol ε in HR. Biochemical studies with this enzyme are needed to address the hypothesis that Pol δ is specialized for D-loop extension (first-end synthesis) and Pol ε for second-end extension, as S. cerevisiae Pol ε is incapable of extending Rad51/Rad54-mdiated D-loops in vitro (28). These studies will address the likely functional overlap between these two and possibly additional DNA polymerases.
DNA Polymerase Gamma
DNA polymerase gamma (Pol γ) is a two-subunit enzyme in humans (POLG, POLG2) and a single-subunit enzyme in S. cerevisiae (Mip1). It functions in the replication and repair of mitochondrial DNA (18, 58). Recent evidence suggests that HR is utilized to repair DSBs in mitochondrial DNA in budding yeast and to a limited extent in mammals (18). Pol γ coimmunoprecipitates with the mitochondrial Rad52-type protein Mgm101 in S. cerevisiae, implying that Pol γ may function in DNA synthesis during HR-mediated DSB repair (89).
DNA Polymerase Zeta
DNA polymerase zeta (Pol ζ is a multisubunit, low-fidelity polymerase with a two-subunit core complex consisting of the catalytic subunits Rev3 and Rev7 (MAD2L2 or MAD2B in mice and humans) (96). S. cerevisiae Pol ζ forms a four-subunit complex with the addition of Pol31 and Pol32, shared with Pol δ, enhancing PCNA interaction and TLS activity (55). Pol ζ also works intimately with Y-family polymerase Rev1 to bypass DNA lesions and extend from terminally mismatched DNA primers and DNA secondary structures (39, 102). Pol ζ is responsible for the vast majority of UV- and methyl methanesulfonate (MMS)-induced mutagenesis observed in budding yeast, illustrating its role in TLS (32, 66).
Although Rev3 deletions in S. cerevisiae and Drosophila are viable (59), disruption of murine Rev3 polymerase activity results in embryonic lethality (64). Cells derived from Rev3 catalytically deficient mice spontaneously form DNA breaks and micronuclei; they also display chromosomal instability (64). Many of the chromosomal breaks and rearrangements observed are reminiscent of those seen in HR-deficient mouse embryonic fibroblasts, indicating that Rev3 may promote HR in mice (157).
The role of Pol ζ during HR is complex. Pol ζ confers γ-ray resistance in budding yeast, Drosophila, DT40 cells, mouse, and human cell lines, and it promotes DSB repair (133). The delayed DSB repair in Pol ζ-deficient cells is likely a consequence of defective HR, as rev3—/— DT40 cells show elevated sensitivity to irradiation in late S and G2, when HR is the predominant DSB repair pathway (105). S. cerevisiae Rev3 localizes to HO endonuclease-induced DSBs (42), and human Pol ζ promotes spontaneous and I-SceI –induced gene conversion (103, 133). In Drosophila and S. cerevisiae, the frequency of HR repair of DNA double-strand gaps is partially dependent on Pol ζ, but the amount of DNA synthesized during repair does not appear to be affected (36, 59). Biochemical data reveal that core S. cerevisiae Pol ζ catalyzes short extensions of undamaged oligonucleotide D-loop structures and linear templates more efficiently than does Pol δ in the absence of PCNA (69). Interestingly, PCNA mildly inhibits the ability of Pol ζ to extend D-loop structures (69). Together, these data suggest that Pol ζ may initiate first-end DNA synthesis at the D-loop prior to PCNA and Pol δ recruitment.
Evidence also exists that Pol ζ plays only a minor role in HR and may primarily be used during second-end synthesis in a mutagenic capacity. DT40 cells lacking either Rev3 or Rev7 have normal rates of immunoglobulin gene conversion (105). Several studies in S. cerevisiae show that Pol ζ is dispensable for efficient mitotic gene conversion (41, 46, 117), BIR (22), and meiotic recombination (116), but the mutation frequency and signatures of the events in these studies indicate that Pol ζ can perform DNA synthesis during HR. Mutagenesis in S. cerevisiae rev3 mutants during meiotic recombination and mitotic gene conversion is decreased twofold and 1,000-fold, respectively, when the reversion reporter is adjacent to the DSB (116, 117, 142). In contrast, Pol ζ does not affect mutagenesis during budding yeast mating-type switching, a type of gene conversion assay wherein the repaired region consists of two newly synthesized strands of DNA (41). Together, these studies suggest a role for Pol ζ in ssDNA gap filling, but not in first-end synthesis. This model is supported by the Pol ζ–dependent mutagenesis detected during SSA, which requires only gap filling (162), and during BIR, which requires extensive amounts of second-end synthesis (22). Furthermore, in situations in which BIR is genetically impaired, Pol ζ facilitates microhomology-mediated BIR (MMBIR) by synthesizing microhomologies templated from the newly synthesized ssDNA behind the D-loop (126). The mutagenic consequences of Pol ζ synthesis during these various HR mechanisms are discussed in detail in the section Recombination-Associated Mutagenesis below.
Interestingly, Rev7 also antagonizes HR independently of Pol ζ. Human Rev7 localizes to DSBs, interacting with 53BP1 to prevent 5′-end resection and promote NHEJ (8, 159). Rev7 knockdown in human cells lacking BRCA1 and p53 restores HR function by allowing 5′-end resection and subsequent HR (159). Thus Rev7 apparently has two disparate roles: singly antagonizing HR by preventing resection at DSBs and facilitating DNA synthesis during HR as part of Pol ζ.
Rev1
Rev1 is a single-subunit Y-family polymerase that possesses dCMP transferase activity, a unique mode of DNA synthesis that allows it to insert a deoxycytosine across from abasic sites, adducts, uracil, and templated guanines (95). It also promotes spontaneous and DNA damage-induced mutagenesis by recruiting Pol ζ (66). Even though Rev1 contains a highly conserved catalytic domain, several studies have demonstrated that its activity is dispensable for most TLS (38, 156). Instead, Rev1 acts as a scaffolding protein, coordinating TLS by recruiting other polymerases through interactions with its noncatalytic domains. TLS polymerases η, ι, κ, and ζ bind the C-terminal domain of Rev1 (1, 34, 113, 158). The Rev1-PCNA interaction is enhanced by the Rev1 ubiquitin-binding motif, which interacts with monoubiquitylated PCNA (35).
Multiple studies indicate that Rev1 is needed for resistance to DSB-inducing agents and localizes to sites of DSBs. S. cerevisiae, Drosophila, DT40, and HeLa cells lacking Rev1 are sensitive to γ-rays (59, 88, 105, 133), and γ-ray-induced mutagenesis in S. cerevisiae depends on Rev1 (88). Additionally, REV1-knockout mouse embryonic fibroblasts are sensitive to camptothecin (161). In budding yeast, Rev1 localizes to HO-induced DSBs via its BRCT domain (42). In contrast to TLS, where Rev1 recruitment requires PCNA ubiquitylation (35), localization of budding yeast Rev1 to DSBs is independent of PCNA ubiquitylation (42), highlighting lesion-specific recruitment mechanisms. By contrast, microirradiation of human cell lines and murine mouse embryonic fibroblasts with UVA lasers also results in localization of Rev1, and this is partially dependent on the ubiquitin ligase Rad18 as well as on other factors associated with fork stabilization and HR (161). Although these results appear contradictory, the mechanistic differences of Rev1 DSB localization may be due to distinct modes of DSB induction and/or the absence, from budding yeast, of factors such as FANCD2, BRCA1, and BRCA2.
The data supporting a role for Rev1 in HR are somewhat inconsistent. Loss of Rev1 significantly reduces I-SceI –induced gene conversion in human cells (133, 161) as well as immunoglobulin gene conversion in DT40 cells (105). Rev1 also functions in an enigmatic way during HR in Drosophila, as loss of Rev1 leads to enhanced repair synthesis during double-strand gap repair (discussed in detail in the next section) (59). By contrast, in budding yeast, gene-conversion efficiency is unaffected by loss of Rev1 (41, 162), but Rev1 promotes mutagenesis during second-end synthesis of gene conversion, likely via Pol ζ recruitment (162). Similarly, budding yeast Rev1 promotes MMBIR in concert with Pol ζ, and Rev1 catalytic activity is partially required to synthesize the microho-mologies that are utilized during this process (126). In sum, on the basis of studies to date, Rev1 affects the efficiency and fidelity of HR by coordinating recruitment of other TLS polymerases during second-end synthesis, and it may also play a regulatory role in first-end synthesis.
DNA Polymerase Eta
DNA polymerase eta (Pol η) is a single-subunit enzyme that catalyzes the bypass of UV damage through the incorporation of two adenines opposite a pyrimidine dimer (56, 83). This property explains its involvement in xeroderma pigmentosum (XP), a human nucleotide excision repair deficiency and cancer predisposition syndrome that leads to skin cancer at sun-exposed parts of the body. The Pol η gene (human RADH, budding yeast POL30) is also known as XP variant and is the only XP gene not encoding a protein active in nucleotide excision repair, which targets UV-induced DNA damage. Pol η is also involved in the bypass of other lesions and associates preferentially with ubiquitylated PCNA (32, 109).
Pol η has been implicated in HR in chicken DT40 cells. DSB-induced gene conversion was reported to be reduced more than 15-fold in a Pol η mutant cell line, a defect that was fully complemented by the wild-type gene (60). Unexpectedly, Pol η–deficient DT40 cells do not display the DNA damage sensitivity profile typical for HR deficiency, as they are resistant to IR and cis-platin, an ICL-inducing agent (43). Also XP-variant patient cells that are deficient in Pol η and HeLa cells depleted for Pol η by small interfering RNA do not exhibit an HR defect (73, 99, 130). Likewise, Pol η–deficient Drosophila are IR resistant but do show an HR defect when the quality of the HR product is analyzed (59). In S. cerevisiae, mutations in Pol η do not cause an HR defect even in the sensitized pol3-ct background (82, 91). It is difficult to reconcile these data into a single interpretation. One difference is the comparison between endpoint data (survival, HR) with more sensitive assays, for example, examining the quality of the HR products. In addition, the diverse complement of DNA polymerases in different organisms (Table 2) may result in varying functional overlap between DNA polymerases in HR. Finally, chicken DT40 cells express activation-induced cytidine deaminase, which may result in additional requirements for TLS DNA polymerases; cytidines present in the ssDNA of the resected DSB may be targeted by this enzyme, resulting in base damage that requires TLS DNA synthesis.
In vitro biochemical studies using budding yeast or human proteins to reconstitute HR-associated DNA synthesis demonstrated the ability of Pol η to extend Rad51-mediated D-loops (11, 69, 71, 86, 129, 130, 139). Pol η was surprisingly efficient in utilizing 3′ ends in the D-loop and efficiently extended several hundreds of nucleotides. Unlike Pol δ, D-loop extension by Pol η did not depend on PCNA, but primer utilization was stimulated by PCNA. In sum, the in vitro studies suggest a potential for the involvement of Pol η in D-loop extension and open the possibility that Pol η could extend the invading strand prior to loading of PCNA. However, more in vivo and genetic evidence is needed to determine if this biochemical property of Pol η has biological significance.
DNA Polymerase Lambda
DNA polymerase lambda (λ) is a single-subunit enzyme lacking proofreading activity that has a major function in NHEJ and MMEJ/alt-EJ in budding yeast and mammals (7, 67, 90, 108, 110). Pol λ processivity and affinity for 3′-hydroxyl ends, but not its fidelity, are stimulated by RPA and PCNA (77). The human Pol λ allele, R438W, revealed a defect in sister chromatid exchange and sensitivity to the topoisomerase inhibitor camptothecin. This led authors to infer a function of Pol λ in HR (14), which had also been inferred from its induced expression during meiosis in fungi and mammals (29, 125, 135). The S. cerevisiae pol4 mutant reveals a meiotic hyper-recombination phenotype, which could be the result of an HR role of Pol λ, but the genetic analysis remains incomplete (68). There is no information on the function of Pol λ in reconstituted HR reactions. In sum, more studies are needed to evaluate the role of Pol λ in HR.
DNA Polymerase Theta
DNA polymerase theta (Pol θ) is a single-subunit, error-prone polymerase with a 10- to 100-fold-higher misincorporation rate relative to other A-family polymerases (5, 132). Pol θ can bypass and extend from AP sites (131) and extend from mismatches and bases inserted opposite 6-4 photoproducts (5). This flexibility is due to three unique sequence inserts found in the active site region of the polymerase (45). A notable feature of Pol θ is that it possesses both a translesion polymerase domain and a helicase-like domain, although the latter does not have demonstrable unwinding activity in vitro (131).
Pol θ was originally characterized in Drosophila as a protein important for resistance to ICL-inducing agents (9). However, there is no evidence that it participates in HR. Recently, it was shown to participate in an alternative form of NHEJ in flies, C. elegans, mice, and humans (6). The polymerase domain can bridge two broken DNA ends, promote annealing of microhomologous sequences, and extend from the 3′ ends (61). A recently published crystal structure of the helicase-like domain suggests that it may also assist in strand annealing during MMEJ/alt-EJ (98). Mammalian Pol θ possesses three RAD51 interacting domains and can antagonize HR by preventing RAD51 nucleofilament assembly (16). Loss of Pol θ in combination with mutation of BRCA1, BRCA2, or FANCD2 in mammalian cells results in extreme genomic instability and synthetic lethality (16, 84), suggesting that targeting Pol θ in HR-deficient cancer cells may be a viable chemotherapeutic option.
DNA Polymerase Iota
DNA polymerase iota (Pol ι) is a single-subunit, error-prone polymerase that can bypass a subset of UV photoproducts in Pol η mutants (49). Overexpression or knockdown of Pol ι in HeLa cells has no effect on HR frequency (130). In addition, purified Pol ι does not extend D-loops in vitro, although PCNA dependency has not been tested (130). Therefore, it seems unlikely that Pol ι plays a significant role in any aspect of HR synthesis.
DNA Polymerase Beta
DNA polymerase beta (β) is involved in vertebrate base-excision repair and is highly expressed in mouse testes, where it promotes the removal of the DSB-inducing endonuclease Spo11 during meiosis (15, 111). Although overexpression of Pol β stimulates HR in mammalian cells (13), there is currently no evidence that it is involved in DNA synthesis during HR.
Terminal Deoxynucleotidyl Transferase and DNA Polymerase Mu
There is currently no evidence for involvement of TdT and DNA polymerase mu in HR besides their established roles in immunoglobulin rearrangements and NHEJ (72).
DNA Polymerase Kappa
DNA polymerase kappa (κ) is an extremely error-prone TLS polymerase that can bypass a variety of lesions (32, 109). Depletion of Pol κ in HeLa cells by small interfering RNA has no effect on DSB-induced gene conversion. Codepletion of both Pol κ and Pol η results in a twofold reduction in gene conversion, in contrast to Pol κ overexpression, which stimulates HR (130). In vitro, Pol κ extends RAD51-mediated D-loops in a PCNA-stimulated manner with shorter synthesis tracts than those of Pol δ in the same system (130). More evidence is needed to ascertain the context in which Pol κ operates during HR.
DNA Polymerase Nu
DNA polymerase nu (Pol ν) shares extensive similarity with the polymerase domain of Pol θ and is also error-prone, with a heightened tendency to insert thymine opposite a template guanine (145). Pol ν has been reported to promote both HR and ICL repair in HEK293T cells by interacting with the Fanconi core complex and HelQ helicase, which has high similarity to the helicase-like domain of Pol θ (92). According to another report, Pol ν interacts with FANCJ and BRCA1 in HEK293T cells, but its previously reported interactions were not detected (146). Different overexpression levels or tags may be responsible for this variance, and it remains unclear what the exact contribution of Pol ν is to HR.
PrimPol
PrimPol is the latest addition to the complement of DNA polymerases in eukaryotes (93, 152) (Table 2). The single-subunit enzyme is an archeo-eukaryotic primase that can synthesize DNA de novo in a template-dependent fashion without requirement for an RNA primer. PrimPol functions in the nucleus as well as in mitochondria and is involved in lesion-skipping at stalled replication forks. It will be of interest to test the involvement of PrimPol in BIR and potentially chromothripsis (see below) in mammalian cells.
Competition and Collaboration Between DNA Polymerases
It is well established that translesion DNA polymerases can collaborate during bypass of various DNA lesions (32). The specific nature of these collaborations varies, depending on the organism, cellular context, and type of lesion. For example, human Rev1 collaborates with polymerases η, ι, and κ, but not ζ, to replicate UV-damaged templates (163), whereas XP-variant cells lacking Pol η utilize pol ζ, ι, and κ in bypass of cyclobutane pyrimidine dimers (165). Recently, polymerases δ and λ were shown to work together to promote efficient MMEJ/alt-EJ repair of DSBs (90).
Emerging evidence supports the idea that multiple TLS polymerases may also be utilized for first-end synthesis during HR repair in certain contexts, potentially in competition with replicative polymerases. As mentioned above, the failure of many studies to find evidence for substantial TLS polymerase involvement in HR could be due to functional overlap and/or differences in model systems, cellular contexts, and the nature of the DSB. For example, whereas TLS polymerases do not play a substantial role in gene conversion in budding yeast (41), Pol η and Rev1/Pol ζ are independently required for repair of an 8-nucleotide gapped plasmid from a chromosomal template (36). S. cerevisiae pol3-01 (Pol δ) cells display increased rates of spontaneous recombination; this can be suppressed by mutation of REV1, suggesting that Rev1 may promote a recombination-based replication coping mechanism when a primary replicative polymerase is impaired (91).
In Drosophila, deletion of either POL η or REV3 has no effect on HR-mediated repair of a 14-kb gap, but the polη rev3 double mutant shows an increase in the extent of repair synthesis (59). This phenotype is also observed in rev1 mutants, suggesting that the C terminus of Rev1, which interacts with both Pol ζ and Pol η, may coordinate the recruitment of TLS polymerases. Based on these results, a model was proposed in which TLS polymerases may initiate first-end synthesis during gap repair. Because the initial synthesis is nonprocessive, repair proceeds through multiple cycles of strand invasion, synthesis, and D-loop dissociation. Eventually, a replicative polymerase (most likely Pol δ) binds to the D-loop and engages in extensive DNA synthesis (59). A similar phenomenon may occur during BIR, which at its onset is not processive and can involve many template switches (137). In cases where BIR is impaired, such as in pif1 mutants, TLS polymerases ζ and Rev1 promote repair by error-prone MMBIR (126).
Involvement of Proliferating Cell Nuclear Antigen in Homologous Recombination
Studies of S. cerevisiae mating-type switching using the cold-sensitive PCNA allele pol30-52 established an essential role for PCNA in HR (47, 153). An important question is whether the presence or absence of PCNA may regulate the access of various polymerases to the D-loop template. The presence of PCNA in vitro stimulates D-loop extension by Pol δ, Pol η, and Pol κ, but not Pol ζ, to various degrees, suggesting that the effect of PCNA is likely to be polymerase specific (69, 71, 130, 139). However, PCNA does not affect recruitment of Pol δ to D-loop intermediates, suggesting that its main role with Pol δ is to stimulate its strand displacement activity (69). Interestingly, in the absence of PCNA, D-loop extension by Pol η and Pol ζ is more efficient than extension by Pol δ. The presence of the Rad51 strand exchange protein inhibits PCNA loading and primer extension by Pol δ (69). These data are consistent with the replicative/TLS polymerase competition model described above.
PCNA has also been implicated in BIR. The PCNA FF248, 249AA mutation suppresses the cold-sensitive growth of a pol32 mutant but not its BIR defect (76). Indeed, this PCNA mutation has a dominant negative effect on BIR, although it has no significant effect on gene conversion or normal DNA replication. This suggests that BIR requires a specialized feature of PCNA not required for its other functions.
During TLS, monoubiquitylation of PCNA assists in the recruitment of S. cerevisiae Pol η and Rev1 to lesions (87). However, monoubiquitylation of PCNA is not required for the function of Rev1/Pol ζ in HR (133). Sumoylation of PCNA prevents unscheduled HR in S. cerevisiae (50), but whether and how sumoylated PCNA affects polymerase recruitment are presently unknown. Clearly, the relationship of PCNA modification to polymerase recruitment/retention in HR needs further exploration.
Recombination-Associated Mutagenesis
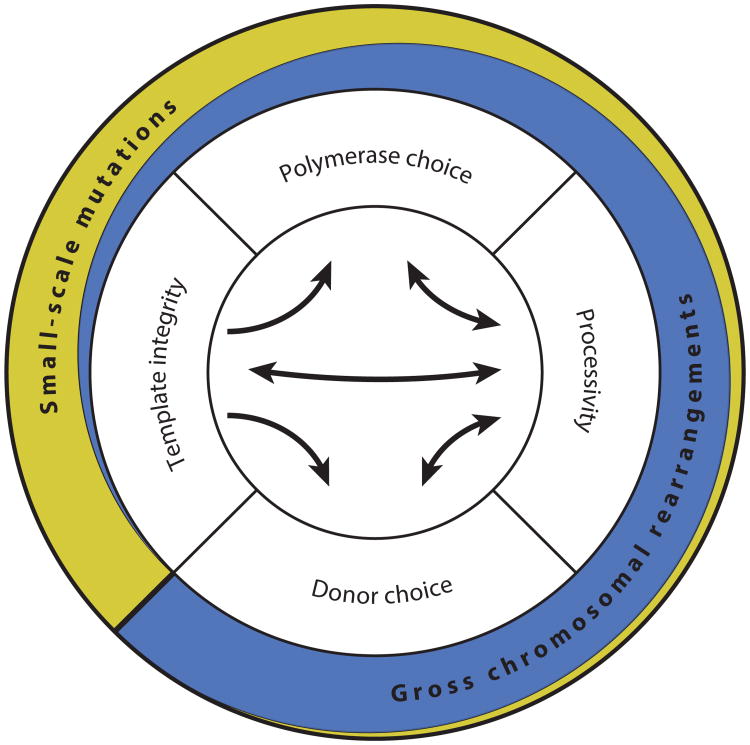
Although HR has historically been considered to be error-free, an expanding body of work disputes this notion, revealing the mutagenic nature of HR. Both small-scale mutations (such as point mutations, frameshifts, and small deletions and insertions) and gross chromosomal rearrangements have been linked to HR. Four broadly defined components underlie HR-associated mutagenesis: polymerase choice, processivity, donor choice, and template integrity (Figure 3). In addition, HR involving DNA sequences with the potential to form secondary structures may lead to genetic and epigenetic instability (154). Together, these components mediate mutagenesis and have their own respective propensities for generating small-scale mutations and/or gross chromosomal rearrangements.
Figure 3.
The four elements of mutagenic homologous recombination. Interactions of these elements are indicated by arrows. Double-headed arrows indicate that the elements influence each other. The wider the outer yellow and blue circles, the higher is the propensity of each element to contribute to each type of mutation.
Small-scale mutations are observed in all HR pathways and predominantly result from a combination of polymerase synthesis errors and copying of damaged templates. In budding yeast, meiotic recombination is associated with up to a 50-fold increase in mutagenesis, mostly point mutations (78, 101). The increased mutagenesis is dependent on Spo11, indicating that DSB formation and repair are responsible for the increased mutagenesis. Furthermore, Pol ζ is partially responsible (116). As with meiosis, mitotic DSB-induced gene conversion and SSA are also mutagenic. Gene conversion and SSA are 50 to > 1,400 times more mutagenic than replication, predominantly giving rise to single nucleotide mutations, but also to indels and more complex rearrangements (41, 46, 117, 143, 162). Studies using mutator alleles of S. cerevisiae Pol δ and ε suggest that they, but not Pol ζ, are responsible for the majority of frameshift mutations and template switches observed during gene conversion in the segment where both DNA strands are newly synthesized (41).
However, earlier studies concluded that Pol ζ is nearly wholly responsible for the observed nonsense mutations and partially responsible for the frameshift reversions near DSBs (46, 117). In addition, small-scale mutations generated during BIR are created by both Polζ and Pol δ, although Polζ does not contribute to mutagenesis at all distances from the break or to the same extent as Polδ (22). The observed difference in polymerase dependency on recombination-mediated mutagenesis may be related to reporter position and the extent of ssDNA surrounding the DSBs. Nonetheless, in general it appears that both replicative and TLS polymerases can be linked to spontaneous mutagenesis during HR synthesis, although their exact contributions are probably context dependent.
One prominent feature of HR-associated mutagenesis is likely the result of the greater chemical instability of ssDNA (74). Small-scale mutations are significantly more frequent in regions surrounding DSBs that require ssDNA fill-in synthesis during SSA (162). Exposure to UV-light, MMS, or expression of activation-induced cytidine deaminases after DSB induction further increases mutation frequencies and generates mutation clusters spanning up to 100 kb around DSBs. These clusters are reminiscent of kataegis observed in cancer genomes (100, 147) (see sidebar, Homologous Recombination, DNA Synthesis, Kataegis, and Chromothripsis). The mutation signatures in these clusters are characteristic of those that result from error-prone TLS past mutagen-specific ssDNA lesions (12, 121, 147, 160, 162). They are dependent on ubiquitylated PCNA recruitment of Pol ζ and Rev1 (162). Low levels of MMS exposure during BIR similarly result in mutation clusters up to 120 kb spanning the entire replicated chromosomal arm. However, the polymerases responsible for these clusters have not been determined (127). Taken together, these data support a model of small-scale mutation generation by both replicative and TLS polymerases, wherein Pol δ misincorporates nucleotides across from undamaged templates and TLS polymerases mediate error-prone TLS following recruitment to ssDNA lesions.
Genome rearrangements resulting from HR are largely dependent on polymerase processivity and donor choice. Although the sister chromatid is a preferred template for HR in somatic cells (53, 57), it is well documented that homologous and homeologous sequences present throughout the genome can be utilized (4, 149). Many assays designed to study DSB-induced HR take advantage of this promiscuity. The consequences of using homologous chromosomes and ectopic homology include localized loss of heterozygosity (LOH) and translocations (52, 94, 118).
A number of recent studies illustrate that ectopic donor sequences can be used even after strand invasion and initiation of first-end synthesis, resulting in complex mutagenesis events. Spontaneous DSBs in S. cerevisiae readily lead to HR-mediated template switching and rearrangements at repeated elements (17, 115), whereas induced BIR and gene conversions lead to template switches and rearrangements at ectopic homologous and homeologous sequences (4, 107, 122, 136, 137, 141, 149). Template switching during BIR is particularly mutagenic because of the sheer quantity of ssDNA that is generated. A single template switch during BIR can result in large regions of copy number variation, with a duplication of an entire chromosome arm and subsequent loss of another, as well as LOH and nonreciprocal translocations (107, 122, 136, 137). Furthermore, multiple template switches, involving a return to the original donor or to additional ectopic templates, lead to substantial genome rearrangements with complex copy number variations and aneuploidy (107, 136, 137, 141).
Interestingly, template switching during BIR often takes place within 10 kb of the initial D-loop synthesis, which is within the region postulated to be synthesized by an inherently less-processive process (136, 137, 141). Some studies show a Pol32 requirement for the template switch events, whereas others only implicate a partial dependency on Pol32 (4, 107, 122, 136). The difference in Pol32 dependency is likely due to the varying amounts of synthesis necessary for either the first or second template switch to occur in each system. It remains to be determined what initiates spontaneous template switches or which polymerase is responsible. Rdh54 is required for template switching in BIR and gene conversion, but the involvement of Rad51, Rad54, and other factors required for the initial invasion cannot be tested, as absence of the initial invasion precludes template switching (4, 149).
Template switching in BIR can be increased by disrupting polymerase processivity. Treating budding yeast undergoing BIR with a high dose of MMS leads to half-crossover-initiation cascade rearrangements. These rearrangements result from polymerase stalling at MMS lesions formed in double-stranded DNA, leading to half-crossovers and a broken donor chromosome, which can then undergo a secondary BIR event (127). Half-crossover-initiation cascade events can also be initiated by decreasing processivity via genetic manipulation (10, 75, 136, 141). One of the most drastic consequences of nonprocessive BIR is MMBIR, which occurs when BIR stalls and a template switch occurs at microhomologous sequences, followed by limited synthesis by Pol ζ and Rev1 (126). These findings are particularly interesting because they suggest a mechanism by which TLS polymerase-mediated MMBIR could act as a potential driver of the gross chromosomal rearrangement that often occurs in cancer.
Homologous Recombination, DNA Synthesis, Kataegis, and Chromothripsis.
Kataegis and chromothripsis are novel forms of genome instability identified by next-generation sequencing of cancer genomes. Analysis of the mutation signatures and translocation junctions from these genomes suggests that HR could, in part, drive both kataegis (120) and chromothripsis (40). The observed mutation clusters display a mutation signature characteristic of APOBEC-mediated cytidine deamination (119). Studies in S. cerevisiae suggest that APOBECs likely act on ssDNA during HR and that the mutations are dependent on TLS polymerases. Chromothripsis, by contrast, has been linked in some instances to MMBIR. Nonprocessive BIR from either a DSB or collapsed replication fork may result in successive template switches, leading to extensive copy number variations with accompanying microhomology junctions. Although the proteins responsible for MMBIR observed in cancers remain unknown, a recent report suggests that Pol ζ and Rev1 can promote MMBIR in S. cerevisiae (126). Future studies are needed to delineate how various DNA polymerases are involved in error-prone HR repair that leads to the complex mutations observed in cancer genomes.
Summary Points.
HR involves several distinct modes of DNA synthesis that likely involve different, potentially overlapping, DNA polymerases.
The large and to some degree diverse complement of DNA polymerases in eukaryotes complicates comparisons between organisms, in particular for the involvement of non-replicative DNA polymerases.
The likely overlap between DNA polymerases necessitates more sophisticated, real-time, and qualitative product analysis in addition to survival and genetic HR endpoint assays.
DNA polymerase δ is the primary DNA polymerase to extend the invading strand in RAD51-mediated D-loops for first-end DNA synthesis.
Several DNA polymerases, including Pol ε, η, λ, ζ, and κ, have been implicated in HR, but more studies are needed to firmly establish their exact functions and potentially organism-specific roles.
Both small-scale mutations and genomic rearrangements can result from HR-mediated repair. These arise from a combination of template switching and generation of ssDNA that is prone to further mutational processes.
Acknowledgments
We thank Shunichi Takeda, Sergei Mirkin, Jim Haber, Auréle Piazza, and Sujeta Mukherjee for helpful discussions and critical comments. P.G.C. was partially supported by the Brazilian Science without Frontiers Postdoctoral Program. D.M. was partially supported by fellowship 15IB-0109 from the CBCRP and the Training Grant in Oncogenic Signals and Chromosome Biology CA108459. Work in M.M.'s laboratory is supported by National Institutes of Health (NIH) grants GM092866 and GM105473. Work in W.D.H.'s laboratory is supported by NIH grants GM58015, CA92276, and CA154920 as well as Department of Defense grant W81XWH-14-1-0435. We apologize that not all of the outstanding work in this area could be discussed or cited because of space constraints.
Glossary
- DSB
DNA double-strand break
- ICL
interstrand cross-link
- Crossover/noncrossover
Two outcomes of HR in which the flanking markers either remain in parental (noncrossover) or assume nonparental (crossover) configuration
- Double Holliday junction (dHJ)
HR intermediate whose resolution can lead to crossover; also used here to identify the HR pathway that involves this intermediate
- Break-induced replication (BIR)
a pathway of HR in which a single-ended DSB invades a duplex template to trigger long-range conservative DNA synthesis
- NHEJ
nonhomologous end joining
- MMEJ/alt-EJ
microhomology-mediated end joining/alternative end joining
- Single-strand annealing (SSA)
a mode of homology-directed DNA repair involving DNA reannealing but not Rad51-mediated DNA strand invasion
- RPA
replication protein A
- Synthesis-dependent strand annealing (SDSA)
a pathway of HR with dedicated noncrossover outcome
- Homeologous recombination
recombination between highly similar but not identical sequences, as found in DNA repeats
- PCNA
proliferating cell nuclear antigen
- Translesion synthesis (TLS)
DNA synthesis performed by specialized DNA polymerases, which tolerate template lesions and typically exhibit high error rates
- Displacement loop (D-loop)
primary DNA strand invasion product of the Rad51-ssDNA filament, from which different HR pathways diverge
- MMBIR
microhomology-mediated BIR
- Chromothripsis
complex chromosomal rearrangements and copy number variations that occur in a specific genomic region and temporal window
- Kataegis
regions of localized hypermutation often observed in cancer genomes and frequently associated with somatic genome rearrangements
Footnotes
Disclosure Statement: The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Related Resources: Haber JE. 2016. A life investigating pathways that repair broken chromosomes. Annu. Rev. Genet. 50:1–28
Gray S, Cohen PE. 2016. Control of meiotic crossovers: from double-strand break formation to designation. Annu. Rev. Genet. 50:175–210
Loidl J. 2016. Conversation and variability of meiosis across the eukaryotes. Annu. Rev. Genet. 50:293–316
Yazinski SA, Zou L. 2016. Functions, regulation, and therapeutic implications of the ATR checkpoint pathway. Annu. Rev. Genet. 50:155–73
Wu CI, Wang H-Y, Ling S. 2016. The ecology and evolution of cancer. Annu. Rev. Genet. 50:347–69
Literature Cited
- 1.Acharya N, Haracska L, Johnson RE, Unk I, Prakash S, Prakash L. Complex formation of yeast Rev1 and Rev7 proteins: a novel role for the polymerase-associated domain. Mol Cell Biol. 2005;25:9734–40. doi: 10.1128/MCB.25.21.9734-9740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acharya N, Klassen R, Johnson RE, Prakash L, Prakash S. PCNA binding domains in all three subunits of yeast DNA polymerase δ modulate its function in DNA replication. PNAS. 2011;108:17927–32. doi: 10.1073/pnas.1109981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anand RP, Lovett ST, Haber JE. Break-induced DNA replication. Cold Spring Harb Perspect Biol. 2013;5:a010397. doi: 10.1101/cshperspect.a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anand RP, Tsaponina O, Greenwell PW, Lee CS, Du W, et al. Chromosome rearrangements via template switching between diverged repeated sequences. Genes Dev. 2014;28:2394–406. doi: 10.1101/gad.250258.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36:3847–56. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beagan K, McVey M. Linking DNA polymerase theta structure and function in health and disease. Cell Mol Life Sci. 2016;73:603–15. doi: 10.1007/s00018-015-2078-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bebenek K, Pedersen LC, Kunkel TA. Structure-function studies of DNA polymerase λ. Biochemistry. 2014;53:2781–92. doi: 10.1021/bi4017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boersma V, Moatti N, Segura-Bayona S, Peuscher MH, van der Torre J, et al. MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5′ end resection. Nature. 2015;521:537–40. doi: 10.1038/nature14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyd JB, Sakaguchi K, Harris PV. mus308 mutants of Drosophila exhibit hypersensitivity to DNA cross-linking agents and are defective in a deoxyribonuclease. Genetics. 1990;125:813–19. doi: 10.1093/genetics/125.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brocas C, Charbonnier JB, Dherin C, Gangloff S, Maloisel L. Stable interactions between DNA polymerase δ catalytic and structural subunits are essential for efficient DNA repair. DNA Repair. 2010;9:1098–111. doi: 10.1016/j.dnarep.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Bugreev DV, Hanaoka F, Mazin AV. Rad54 dissociates homologous recombination intermediates by branch migration. Nat Struct Mol Biol. 2007;14:746–53. doi: 10.1038/nsmb1268. [DOI] [PubMed] [Google Scholar]
- 12.Burch LH, Yang Y, Sterling JF, Roberts SA, Chao FG, et al. Damage-induced localized hyper-mutability. Cell Cycle. 2011;10:1073–85. doi: 10.4161/cc.10.7.15319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canitrot Y, Capp JP, Puget N, Bieth A, Lopez B, et al. DNA polymerase β overexpression stimulates the Rad51-dependent homologous recombination in mammalian cells. Nucleic Acids Res. 2004;32:5104–12. doi: 10.1093/nar/gkh848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capp JP, Boudsocq F, Bergoglio V, Trouche D, Cazaux C, et al. The R438W polymorphism of human DNA polymerase lambda triggers cellular sensitivity to camptothecin by compromising the homologous recombination repair pathway. Carcinogenesis. 2010;31:1742–47. doi: 10.1093/carcin/bgq166. [DOI] [PubMed] [Google Scholar]
- 15.Carreras Puigvert J, Sanjiv K, Helleday T. Targeting DNA repair, DNA metabolism and replication stress as anti-cancer strategies. FEBS J. 2015;283:232–45. doi: 10.1111/febs.13574. [DOI] [PubMed] [Google Scholar]
- 16.Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, et al. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature. 2015;518:258–62. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan JE, Kolodner RD. A genetic and structural study of genome rearrangements mediated by high copy repeat Ty1 elements. PLOS Genet. 2011;7:e1002089. doi: 10.1371/journal.pgen.1002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen XJ. Mechanism of homologous recombination and implications for aging-related deletions in mitochondrial DNA. Microbiol Mol Biol Rev. 2013;77:476–96. doi: 10.1128/MMBR.00007-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chilkova O, Stenlund P, Isoz I, Stith CM, Grabowski P, et al. The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA. Nucleic Acids Res. 2007;35:6588–97. doi: 10.1093/nar/gkm741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, et al. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Covo S, Blanco L, Livneh Z. Lesion bypass by human DNA polymerase μ reveals a template-dependent, sequence-independent nucleotidyl transferase activity. J Biol Chem. 2004;279:859–65. doi: 10.1074/jbc.M310447200. [DOI] [PubMed] [Google Scholar]
- 22.Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, et al. Break-induced replication is highly inaccurate. PLOS Biol. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol ε and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 1999;274:22283–88. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- 24.Dubarry M, Lawless C, Banks AP, Cockell S, Lydall D. Genetic networks required to coordinate chromosome replication by DNA polymerases α, δ, and ε in Saccharomyces cerevisiae. G3. 2015;5:2187–97. doi: 10.1534/g3.115.021493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fabre F. Induced intragenic recombination in yeast can occur during the G1 mitotic phase. Nature. 1978;272:795–97. doi: 10.1038/272795a0. [DOI] [PubMed] [Google Scholar]
- 26.Fabre F, Boulet A, Faye G. Possible involvement of the yeast POLIII DNA polymerase in induced gene conversion. Mol Gen Genet. 1991;229:353–56. doi: 10.1007/BF00267455. [DOI] [PubMed] [Google Scholar]
- 27.Gan GN, Wittschieben JP, Wittschieben BO, Wood RD. DNA polymerase zeta (pol ζ in higher eukaryotes. Cell Res. 2008;18:174–83. doi: 10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- 28.Ganai RA, Zhang XP, Heyer WD, Johansson E. Strand displacement synthesis by yeast DNA polymerase ε. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Diaz M, Dominguez O, Lopez-Fernandez LA, de Lera LT, Saniger ML, et al. DNA polymerase lambda (Pol λ), a novel eukaryotic DNA polymerase with a potential role in meiosis. J Mol Biol. 2000;301:851–67. doi: 10.1006/jmbi.2000.4005. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Gomez S, Reyes A, Martinez-Jimenez MI, Chocron ES, Mouron S, et al. PrimPol, an archaic primase/polymerase operating in human cells. Mol Cell. 2013;52:541–53. doi: 10.1016/j.molcel.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase δ maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–73. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodman MF, Woodgate R. Translesion DNA polymerases. Cold Spring Harb Perspect Biol. 2013;5:a010363. doi: 10.1101/cshperspect.a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gouge J, Rosario S, Romain F, Poitevin F, Beguin P, Delarue M. Structural basis for a novel mechanism of DNA bridging and alignment in eukaryotic DSB DNA repair. EMBOJ. 2015;34:1126–42. doi: 10.15252/embj.201489643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, et al. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBOJ. 2003;22:6621–30. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, et al. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol. 2006;26:8892–900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo X, Jinks-Robertson S. Roles of exonucleases and translesion synthesis DNA polymerases during mitotic gap repair in yeast. DNA Repair. 2013;12:1024–30. doi: 10.1016/j.dnarep.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halas A, Ciesielski A, Zuk J. Involvement of the essential yeast DNA polymerases in induced gene conversion. Acta Biochim Pol. 1999;46:862–72. [PubMed] [Google Scholar]
- 38.Haracska L, Unk I, Johnson RE, Johansson E, Burgers PM, et al. Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–54. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hashimoto K, Cho Y, Yang IY, Akagi J, Ohashi E, et al. The vital role of polymerase ζ, and REV1 in mutagenic, but not correct, DNA synthesis across benzo[α]pyrene-dG and recruitment of polymerase ζ by REV1 to replication-stalled site. J Biol Chem. 2012;287:9613–22. doi: 10.1074/jbc.M111.331728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLOS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirano Y, Sugimoto K. ATR homolog Mec1 controls association of DNA polymerase ζ-Rev1 complex with regions near a double-strand break. Curr Biol. 2006;16:586–90. doi: 10.1016/j.cub.2006.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirota K, Sonoda E, Kawamoto T, Motegi A, Masutani C, et al. Simultaneous disruption of two DNA polymerases, Polη and Pol ζ, in avian DT40 cells unmasks the role of Polη in cellular response to various DNA lesions. PLOS Genet. 2010;6:e1001151. doi: 10.1371/journal.pgen.1001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirota K, Yoshikiyo K, Guilbaud G, Tsurimoto T, Murai J, et al. The POLD3 subunit of DNA polymerase δ can promote translesion synthesis independently of DNA polymerase ζ. Nucleic Acids Res. 2015;43:1671–83. doi: 10.1093/nar/gkv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hogg M, Seki M, Wood RD, Doublie S, Wallace SS. Lesion bypass activity of DNA polymerase θ (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts. J Mol Biol. 2011;405:642–52. doi: 10.1016/j.jmb.2010.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holbeck SL, Strathern JN. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics. 1997;147:1017–24. doi: 10.1093/genetics/147.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holmes AM, Haber JE. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 1999;96:415–24. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- 48.Jain S, Sugawara N, Lydeard J, Vaze M, Tanguy Le Gac N, Haber JE. A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev. 2009;23:291–303. doi: 10.1101/gad.1751209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jansen JG, Temviriyanukul P, Wit N, Delbos F, Reynaud CA, et al. Redundancy of mammalian Y family DNA polymerases in cellular responses to genomic DNA lesions induced by ultraviolet light. Nucleic Acids Res. 2014;42:11071–82. doi: 10.1093/nar/gku779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jentsch S, Psakhye I. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu Rev Genetics. 2013;47:167–86. doi: 10.1146/annurev-genet-111212-133453. [DOI] [PubMed] [Google Scholar]
- 51.Jin YH, Ayyagari R, Resnick MA, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast. II Cooperation between the polymerase and 3′-5′-exonuclease activities of Pol δ in the creation of a ligatable nick. J Biol Chem. 2003;278:1626–33. doi: 10.1074/jbc.M209803200. [DOI] [PubMed] [Google Scholar]
- 52.Jinks-Robertson S, Petes TD. Chromosomal translocations generated by high-frequency meiotic recombination between repeated yeast genes. Genetics. 1986;114:731–52. doi: 10.1093/genetics/114.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson RE, Klassen R, Prakash L, Prakash S. A major role of DNA polymerase δ in replication of both the leading and lagging DNA strands. Mol Cell. 2015;59:163–75. doi: 10.1016/j.molcel.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson RE, Prakash L, Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. PNAS. 2012;109:12455–60. doi: 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Science. 1999;283:1001–4. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 57.Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaguni LS. DNA polymerase γ, the mitochondrial replicase. Annu Rev Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- 59.Kane DP, Shusterman M, Rong YK, McVey M. Competition between replicative and translesion polymerases during homologous recombination repair in Drosophila. PLOS Genet. 2012;8:e1002659. doi: 10.1371/journal.pgen.1002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kawamoto T, Araki K, Sonoda E, Yamashita YM, Harada K, et al. Dual roles for DNA polymerase η in homologous DNA recombination and translesion DNA synthesis. Mol Cell. 2005;20:793–99. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 61.Kent T, Chandramouly G, McDevitt SM, Ozdemir AY, Pomerantz RT. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase θ. Nat Struct Mol Biol. 2015;22:230–37. doi: 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kesti T, Flick K, Keranen S, Syvaoja JE, Wittenberg C. DNA polymerase ε catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 1999;3:679–85. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- 63.Kowalczykowski SC, Hunter N, Heyer WD, editors. DNA Recombination. Cold Spring Harbor, NY: Cold Spring Harb. Lab Press; 2016. [Google Scholar]
- 64.Lange SS, Tomida J, Boulware KS, Bhetawal S, Wood RD. The polymerase activity of mammalian DNA pol ζ is specifically required for cell and embryonic viability. PLOS Genet. 2016;12:e1005759. doi: 10.1371/journal.pgen.1005759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lao JP, Oh SD, Shinohara M, Shinohara A, Hunter N. Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol Cell. 2008;29:517–24. doi: 10.1016/j.molcel.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lawrence CW. Cellular roles of DNA polymerase ζ, and Rev1 protein. DNA Repair. 2002;1:425–35. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- 67.Lee K, Lee SE. Saccharomyces cerevisiae Sae2- and Tell-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007;176:2003–14. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leem SH, Ropp PA, Sugino A. The yeast Saccharomyces cerevisiae DNA polymerase IV: possible involvement in double-strand break DNA repair. Nucleic Acids Res. 1994;22:3011–17. doi: 10.1093/nar/22.15.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li J, Holzschu DL, Sugiyama T. PCNA is efficiently loaded on the DNA recombination intermediate to modulate polymerase δ, η, and ζ, activities. PNAS. 2013;110:7672–77. doi: 10.1073/pnas.1222241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li X, Heyer WD. RAD54 controls access to the invading 3′-OH end after RAD51-mediated DNA strand invasion in homologous recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:638–46. doi: 10.1093/nar/gkn980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li X, Stith CM, Burgers PM, Heyer WD. PCNA is required for initiating recombination-associated DNA synthesis by DNA polymerase δ. Mol Cell. 2009;36:704–13. doi: 10.1016/j.molcel.2009.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Limoli CL, Giedzinski E, Cleaver JE. Alternative recombination pathways in UV-irradiated XP variant cells. Oncogene. 2005;24:3708–14. doi: 10.1038/sj.onc.1208515. [DOI] [PubMed] [Google Scholar]
- 74.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–15. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 75.Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–23. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 76.Lydeard JR, Lipkin-Moore Z, Sheu YJ, Stillman B, Burgers PM, Haber JE. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev. 2010;24:1133–44. doi: 10.1101/gad.1922610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maga G, Villani G, Ramadan K, Shevelev I, Tanguy Le Gac N, et al. Human DNA polymerase λ functionally and physically interacts with proliferating cell nuclear antigen in normal and translesion DNA synthesis. J Biol Chem. 2002;277:48434–40. doi: 10.1074/jbc.M206889200. [DOI] [PubMed] [Google Scholar]
- 78.Magni GE. The origin of spontaneous mutations during meiosis. PNAS. 1963;50:975–80. doi: 10.1073/pnas.50.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Majka J, Burgers PM. The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol Biol. 2004;78:227–60. doi: 10.1016/S0079-6603(04)78006-X. [DOI] [PubMed] [Google Scholar]
- 80.Malkova A, Haber JE. Mutations arising during repair of chromosome breaks. Annu Rev Genet. 2012;46:455–73. doi: 10.1146/annurev-genet-110711-155547. [DOI] [PubMed] [Google Scholar]
- 81.Maloisel L, Bhargava J, Roeder GS. A role for DNA polymerase δ in gene conversion and crossing over during meiosis in Saccharomyces cerevisiae. Genetics. 2004;167:1133–42. doi: 10.1534/genetics.104.026260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maloisel L, Fabre F, Gangloff S. DNA polymerase δ is preferentially recruited during homologous recombination to promote heteroduplex DNA extension. Mol Cell Biol. 2008;28:1373–82. doi: 10.1128/MCB.01651-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, et al. The XPV(xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999;399:700–4. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 84.Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–57. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McElhinny SAN, Gordenin DA, Stith CM, Burgers PMJ, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–44. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC. Human DNA polymerase η promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol Cell. 2005;20:783–92. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 87.McIntyre J, Woodgate R. Regulation of translesion DNA synthesis: posttranslational modification of lysine residues in key proteins. DNA Repair. 2015;29:166–79. doi: 10.1016/j.dnarep.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McKee RH, Lawrence CW. Genetic analysis of gamma-ray mutagenesis in yeast. I. Reversion in radiation-sensitive strains. Genetics. 1979;93:361–73. doi: 10.1093/genetics/93.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meeusen S, Nunnari J. Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J Cell Biol. 2003;163:503–10. doi: 10.1083/jcb.200304040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meyer D, Fu BXH, Heyer WD. DNA polymerases δ and λ cooperate in repairing double-strand breaks by microhomology-mediated end-joining in Saccharomyces cerevisiae. PNAS. 2015;112:E6907–16. doi: 10.1073/pnas.1507833112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mito E, Mokhnatkin JV, Steele MC, Buettner VL, Sommer SS, et al. Mutagenic and recombinagenic responses to defective DNA polymerase δ are facilitated by the Rev1 protein in pol3-t mutants of Saccharomyces cerevisiae. Genetics. 2008;179:1795–806. doi: 10.1534/genetics.108.089821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moldovan GL, Madhavan MV, Mirchandani KD, McCaffrey RM, Vinciguerra P, D'Andrea AD. DNA polymerase POLN participates in cross-link repair and homologous recombination. Mol Cell Biol. 2010;30:1088–96. doi: 10.1128/MCB.01124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mouron S, Rodriguez-Acebes S, Martinez-Jimenez MI, Garcia-Gomez S, Chocron S, et al. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat Struct Mol Biol. 2013;20:1383–89. doi: 10.1038/nsmb.2719. [DOI] [PubMed] [Google Scholar]
- 94.Moynahan ME, Jasin M. Loss of heterozygosity induced by a chromosomal double-strand break. PNAS. 1997;94:8988–93. doi: 10.1073/pnas.94.17.8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–31. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 96.Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science. 1996;272:1646–49. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 97.Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, et al. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol. 2012;8:125–32. doi: 10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Newman JA, Cooper CD, Aitkenhead H, Gileadi O. Structure of the helicase domain of DNA polymerase theta reveals a possible role in the microhomology-mediated end-joining pathway. Structure. 2015;23:2319–30. doi: 10.1016/j.str.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nicolay NH, Carter R, Hatch SB, Schultz N, Prevo R, et al. Homologous recombination mediates S-phase-dependent radioresistance in cells deficient in DNA polymerase η. Carcinogenesis. 2012;33:2026–34. doi: 10.1093/carcin/bgs239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–93. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nishant KT, Wei W, Mancera E, Argueso JL, Schlattl A, et al. The baker's yeast diploid genome is remarkably stable in vegetative growth and meiosis. PLOS Genet. 2010;6:e1001109. doi: 10.1371/journal.pgen.1001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Northam MR, Moore EA, Mertz TM, Binz SK, Stith CM, et al. DNA polymerases ζ, and Rev1 mediate error-prone bypass of non-B DNA structures. Nucleic Acids Res. 2014;42:290–306. doi: 10.1093/nar/gkt830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.O'Connor MJ. Targeting the DNA damage response in cancer. Mol Cell. 2015;60:547–60. doi: 10.1016/j.molcel.2015.10.040. [DOI] [PubMed] [Google Scholar]
- 104.O'Donnell M, Langston L, Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol. 2013;5:a010108. doi: 10.1101/cshperspect.a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Okada T, Sonoda E, Yoshimura M, Kawano Y, Saya H, et al. Multiple roles of vertebrate REV genes in DNA repair and recombination. Mol Cell Biol. 2005;25:6103–11. doi: 10.1128/MCB.25.14.6103-6111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Paques F, Haber JE. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6765–71. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pardo B, Aguilera A. Complex chromosomal rearrangements mediated by break-induced replication involve structure-selective endonucleases. PLOS Genet. 2012;8:e1002979. doi: 10.1371/journal.pgen.1002979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pardo B, Ma E, Marcand S. Mismatch tolerance by DNA polymerase Pol4 in the course of nonhomologous end joining in Saccharomyces cerevisiae. Genetics. 2006;172:2689–94. doi: 10.1534/genetics.105.053512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pavlov YI, Shcherbakov VP, Rogozin IB. Rols of DNA polymerases in replication, repair, and recombination in eukaryotes. Int Rev Cytol. 2006;255:41–132. doi: 10.1016/S0074-7696(06)55002-8. [DOI] [PubMed] [Google Scholar]
- 110.Picher AJ, Garcia-Diaz M, Bebenek K, Pedersen LC, Kunkel TA, Blanco L. Promiscuous mismatch extension by human DNA polymerase λ. Nucleic Acids Res. 2006;34:3259–66. doi: 10.1093/nar/gkl377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Plug AW, Clairmont CA, Sapi E, Ashley T, Sweasy JB. Evidence for a role for DNA polymerase β in mammalian meiosis. PNAS. 1997;94:1327–31. doi: 10.1073/pnas.94.4.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Podust VN, Hubscher U. Lagging strand DNA synthesis by calf thymus DNA polymerases alpha, beta, delta and epsilon in the presence of auxiliary proteins. Nucleic Acids Res. 1993;21:841–46. doi: 10.1093/nar/21.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pozhidaeva A, Pustovalova Y, D'Souza S, Bezsonova I, Walker GC, Korzhnev DM. NMR structure and dynamics of the C-terminal domain from human Rev1 and its complex with Rev1 interacting region of DNA polymerase η. Biochemistry. 2012;51:5506–20. doi: 10.1021/bi300566z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase ε participates in leading-strand DNA replication. Science. 2007;317:127–30. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Putnam CD, Hayes TK, Kolodner RD. Specific pathways prevent duplication-mediated genome rearrangements. Nature. 2009;460:984–89. doi: 10.1038/nature08217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rattray A, Santoyo G, Shafer B, Strathern JN. Elevated mutation rate during meiosis in Saccharomyces cerevisiae. PLOS Genet. 2015;11:e1004910. doi: 10.1371/journal.pgen.1004910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rattray AJ, Shafer BK, McGill CB, Strathern JN. The roles of REV3 and RAD57 in double-strand-break-repair-induced mutagenesis of Saccharomyces cerevisiae. Genetics. 2002;162:1063–77. doi: 10.1093/genetics/162.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ray A, Machin N, Stahl FW. A DNA double-chain break stimulates triparental recombination in Saccharomyces cerevisiae. PNAS. 1989;86:6225–29. doi: 10.1073/pnas.86.16.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer. 2014;14:786–800. doi: 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–76. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roberts SA, Sterling J, Thompson C, Harris S, Mav D, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46:424–35. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ruiz JF, Gomez-Gonzalez B, Aguilera A. Chromosomal translocations caused by either pol32-dependent or pol32-independent triparental break-induced replication. Mol Cell Biol. 2009;29:5441–54. doi: 10.1128/MCB.00256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, et al. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502:389–92. doi: 10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Saitoh S, Chabes A, McDonald WH, Thelander L, Yates JR, Russell P. Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell. 2002;109:563–73. doi: 10.1016/s0092-8674(02)00753-5. [DOI] [PubMed] [Google Scholar]
- 125.Sakamoto A, Iwabata K, Koshiyama A, Sugawara H, Yanai T, et al. Two X family DNA polymerases, λ and μ, in meiotic tissues of the basidiomycete, Coprinus cinereus. Chromosoma. 2007;116:545–56. doi: 10.1007/s00412-007-0119-3. [DOI] [PubMed] [Google Scholar]
- 126.Sakofsky CJ, Ayyar S, Deem AK, Chung WH, Ira G, Malkova A. Translesion polymerases drive microhomology-mediated break-induced replication leading to complex chromosomal rearrangements. Mol Cell. 2015;60:860–72. doi: 10.1016/j.molcel.2015.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sakofsky CJ, Roberts SA, Malc E, Mieczkowski PA, Resnick MA, et al. Break-induced replication is a source of mutation clusters underlying kataegis. Cell Rep. 2014;7:1640–48. doi: 10.1016/j.celrep.2014.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–42. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sebesta M, Burkovics P, Haracska L, Krejci L. Reconstitution of DNA repair synthesis in vitro and the role of polymerase and helicase activities. DNA Repair. 2011;10:567–76. doi: 10.1016/j.dnarep.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sebesta M, Burkovics P, Juhasz S, Zhang S, Szabo JE, et al. Role of PCNA and TLS polymerases in D-loop extension during homologous recombination in humans. DNA Repair. 2013;12:691–98. doi: 10.1016/j.dnarep.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Seki M, Marini F, Wood RD. POLQ (Pol θ), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–26. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, et al. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBOJ. 2004;23:4484–94. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sharma S, Hicks JK, Chute CL, Brennan JR, Ahn JY, et al. REV1 and polymerase ζ, facilitate homologous recombination repair. Nucleic Acids Res. 2012;40:682–91. doi: 10.1093/nar/gkr769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shi I, Hallwyl SC, Seong C, Mortensen U, Rothstein R, Sung P. Role of the Rad52 amino-terminal DNA binding activity in DNA strand capture in homologous recombination. J Biol Chem. 2009;284:33275–84. doi: 10.1074/jbc.M109.057752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shimizu K, Santocanale C, Ropp PA, Longhese MP, Plevani P, et al. Purification and characterization of a new DNA polymerase from budding yeast Saccharomyces cerevisiae. A probable homolog of mammalian DNA polymerase β. J Biol Chem. 1993;268:27148–53. [PubMed] [Google Scholar]
- 136.Smith CE, Lam AF, Symington LS. Aberrant double-strand break repair resulting in half crossovers in mutants defective for Rad51 or the DNA polymerase δ complex. Mol Cell Biol. 2009;29:1432–41. doi: 10.1128/MCB.01469-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–5. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 138.Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–38. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sneeden JL, Grossi SM, Tappin I, Hurwitz J, Heyer WD. Reconstitution of recombination-associated DNA synthesis with human proteins. Nucleic Acids Res. 2013;41:4913–25. doi: 10.1093/nar/gkt192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.St Charles JA, Liberti SE, Williams JS, Lujan SA, Kunkel TA. Quantifying the contributions of base selectivity, proofreading and mismatch repair to nuclear DNA replication in Saccharomyces cerevisiae. DNA Repair. 2015;31:41–51. doi: 10.1016/j.dnarep.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Stafa A, Donnianni RA, Timashev LA, Lam AF, Symington LS. Template switching during break-induced replication is promoted by the Mph1 helicase in Saccharomyces cerevisiae. Genetics. 2014;196:1017–28. doi: 10.1534/genetics.114.162297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Strathern JN, Shafer BK, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–72. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Strathern JN, Weinstock KG, Higgins DR, McGill CB. A novel recombinator in yeast based on gene II protein from bacteriophage f1. Genetics. 1991;127:61–73. doi: 10.1093/genetics/127.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by Rad52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. PNAS. 1998;95:6049–54. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Takata K, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J Biol Chem. 2006;281:23445–55. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 146.Takata K, Tomida J, Reh S, Swanhart LM, Takata M, et al. Conserved overlapping gene arrangement, restricted expression, and biochemical activities of DNA polymerase ν (POLN) J Biol Chem. 2015;290:24278–93. doi: 10.1074/jbc.M115.677419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, et al. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tissier A, McDonald JP, Frank EG, Woodgate R. Polι, a remarkably error-prone human DNA polymerase. Genes Dev. 2000;14:1642–50. [PMC free article] [PubMed] [Google Scholar]
- 149.Tsaponina O, Haber JE. Frequent interchromosomal template switches during gene conversion in S. cerevisiae. Mol Cell. 2014;55:615–25. doi: 10.1016/j.molcel.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tsubota T, Maki S, Kubota H, Sugino A, Maki H. Double-stranded DNA binding properties of Saccharomyces cerevisiae DNA polymerase epsilon and of the Dpb3p-Dpb4p subassembly. Genes Cells. 2003;8:873–88. doi: 10.1046/j.1365-2443.2003.00683.x. [DOI] [PubMed] [Google Scholar]
- 151.van Loon B, Woodgate R, Hubscher U. DNA polymerases: biology, diseases and biomedical applications. DNA Repair. 2015;29:1–3. doi: 10.1016/j.dnarep.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 152.Wan L, Lou J, Xia Y, Su B, Liu T, et al. hPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity. EMBO Rep. 2013;14:1104–12. doi: 10.1038/embor.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang X, Ira G, Tercero JA, Holmes AM, Diffley JF, Haber JE. Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:6891–99. doi: 10.1128/MCB.24.16.6891-6899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wickramasinghe CM, Arzouk H, Frey A, Maiter A, Sale JE. Contributions of the specialised DNA polymerases to replication of structured DNA. DNA Repair. 2015;29:83–90. doi: 10.1016/j.dnarep.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 155.Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, et al. Pif1 helicase and Polδ promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013;502:393–96. doi: 10.1038/nature12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wiltrout ME, Walker GC. The DNA polymerase activity of Saccharomyces cerevisiae Rev1 is biologically significant. Genetics. 2011;187:21–35. doi: 10.1534/genetics.110.124172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wittschieben JP, Reshmi SC, Gollin SM, Wood RD. Loss of DNA polymerase ζ causes chromosomal instability in mammalian cells. Cancer Res. 2006;66:134–42. doi: 10.1158/0008-5472.CAN-05-2982. [DOI] [PubMed] [Google Scholar]
- 158.Wojtaszek J, Lee CJ, D'Souza S, Minesinger B, Kim H, et al. Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric polymerase (Pol) ζ, and Pol κ. J Biol Chem. 2012;287:33836–46. doi: 10.1074/jbc.M112.394841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xu G, Chapman JR, Brandsma I, Yuan J, Mistrik M, et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature. 2015;521:541–44. doi: 10.1038/nature14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yang Y, Gordenin DA, Resnick MA. A single-strand specific lesion drives MMS-induced hyper-mutability at a double-strand break in yeast. DNA Repair. 2010;9:914–21. doi: 10.1016/j.dnarep.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yang Y, Liu Z, Wang F, Temviriyanukul P, Ma X, et al. FANCD2 and REV1 cooperate in the protection of nascent DNA strands in response to replication stress. Nucleic Acids Res. 2015;43:8325–39. doi: 10.1093/nar/gkv737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLOS Genet. 2008;4:e1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yoon JH, Park J, Conde J, Wakamiya M, Prakash L, Prakash S. Rev1 promotes replication through UV lesions in conjunction with DNA polymerases η, ι, and κ but not DNA polymerase ζ. Genes Dev. 2015;29:2588–602. doi: 10.1101/gad.272229.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zietlow L, Smith LA, Bessho M, Bessho T. Evidence for the involvement of human DNA polymerase N in the repair of DNA interstrand cross-links. Biochemistry. 2009;48:11817–24. doi: 10.1021/bi9015346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ziv O, Geacintov N, Nakajima S, Yasui A, Livneh Z. DNA polymerase ζ cooperates with polymerases κ and ι in translesion DNA synthesis across pyrimidine photodimers in cells from XPV patients. PNAS. 2009;106:11552–57. doi: 10.1073/pnas.0812548106. [DOI] [PMC free article] [PubMed] [Google Scholar]