Abstract
Large multiscale neuronal network simulations are of increasing value as more big data is gathered about brain wiring and organization under the auspices of current major research initiative, such as Brain research through advancing innovative neurotechnologies. The development of these models requires development of new simulation technologies. We describe here the current use of the NEURON simulator with MPI (message passing interface) for simulation in the domain of moderately large networks on commonly available High Performance Computers (HPCs). We discuss the basic layout of such simulations, including the methods of simulation setup, the run-time spike passing paradigm and post-simulation data storage and data management approaches. Using the Neuroscience Gateway, a portal for computational neuroscience that provides access to large HPCs, we benchmark simulations of neuronal networks of different sizes (from 500 to 100,000 cells), and using different numbers of nodes (1 to 256). We compare three types of networks, composed either of Izhikevich integrate-and-fire neurons, single-compartment Hodgkin-Huxley cells, or a hybrid network with half of each. Results show simulation run time increased approximately linearly with network size and decreased almost linearly with the number of nodes. Networks with I&F neurons were faster than Hodgkin-Huxley networks, although differences were small since all tested cells were point neurons with a single compartment.
Keywords: Software, High Performance Computing, Neuronal networks, Simulation neurotechnology
1 Introduction
A number of recent national and international research initiatives have emphasized the importance of large neuronal network simulations for advancing our understanding of the brain. The U.S. initiative, Brain Research through Advancing Innovative Neurotechnologies (BRAIN), while emphasizing the physical neurotechnologies required to record from many neurons or many synapses, also encompasses the neurotechnologies required for simulation and data-mining. While some of these overlap with the comparable tools in other domains, the particular requirements of neural data interpretation and simulation necessitate specialized tools. Initiatives have also been developed by the E.U. – Human Brain Project based in Geneva – and privately – Allen Brain Institute in Seattle; additional projects are being developed in other countries.
In this paper we present development of large network simulations using parallel computing in the NEURON simulation environment for simulations of moderate size (order ~1 ·104 – 1 ·106 cells) neuronal networks.(Carnevale and Hines, 2006; Hines and Carnevale, 2001, 2008; Migliore et al., 2006). Some of the techniques presented are relevant at any scale – e.g., load balancing, data saving, independent random number streams, recomputation of partial simulation – while others do not scale past ~1000 nodes – e.g., use of a single Master node. The primary advance here involves the design of networks using coding in Python rather than in NEURON’s original hoc language. The Python interface involves several new NEURON calls but its primary advantage is for compatibility and portability with other simulators and analysis packages. The move to using Python as a lingua franca interpreter in neural simulation means that many of the methods presented will also be useful in other simulators (Brette et al., 2007; Davison et al., 2009; Hines et al., 2009a, 2007). Among other advantages, Python offers a large variety of data structures – lists, dictionaries, sets, etc – that can be used for data organization and data communication for organizing, saving and reloading simulations.
In this paper, we present the details of a NEURON implementation of three different network models featuring: a single compartment Hodgkin-Huxley model connected by excitatory synapses, simple integrate and fire cells (I&F cells cells based on the Izhikevich parameterization (Izhikevich, 2007)), and a third hybrid model composed of equal number I&F cells and Hodgkin-Huxley cells. We look at timing for the various stages of simulations, efficiency in parallelization with increasing number of nodes, and ease of use at different scales. In the interest of modularity, network design should be simulator independent and use data structures that can be loaded for different packages and for a variety of uses even outside of simulation (Djurfeldt et al., 2014). In a subsequent publication, we will explore further issues of network simulation design, setup and verification in NEURON.
Methods
NEURON is a hybrid system for running simulations across scales and thus must blend different simulation techniques. NEURON supports molecular dynamics McDougal et al. (2013) and large network simulations, giving it the flexibility to explore mathematical hypotheses for diverse experimental data. Instead of requiring the entire dynamical system to be expressed explicitly in terms of kinetic schemes or differential equations, NEURON provides domain-specific abstractions such as the notion of a Section, a continuous unbranched cable that may contain other mechanisms. These abstractions lend themselves to manipulation by graphical tools, such as the CellBuilder, for building and managing models.
In this paper we focus on NEURON’s ParallelContext which is instantiated in Python as pc = h.ParallelContext(), or alternatively, in the hoc language as pc = new ParallelContext(). All the code in this paper will be given in Python, which is the preferred language for use of the NEURON simulator Hines et al. (2009b). Note that most of the calls belonging to the pc object will be identical regardless of whether called from hoc or Python. (The exception to this involves the syntax of pointers in recording state trajectories.)
Parallel simulation in NEURON is carried out by using a separate Message Passing Interface(MPI) context, which sets up a language-independent communication structure for computing in parallel. This is generally provided on an HPC by an open source package such as MPICH or Open MPI. Launching NEURON on a specified number of processes is system dependent. In general, NEURON is asked to initialize the MPI libraries by adding the -mpi argument to nrniv’s argument list. An example of a typical call for these simulations would be :
mpiexec –n 4 nrniv –mpi –python init.py
Simulations can also be launched by a direct call to Python without changing the codebase. Basic usage then requires making all NEURON’s functionality available by importing the neuron module, typically with from neuron import h, and then using it to create the parallel context via pc = h.ParallelContext().
There is a wide variety of nomenclatures used for describing parallel computing. The CPUs, cores, or processes that are running in parallel are referred to as ranks (nodes or hosts are alternative names). The number of hosts is given in an mpiexec command using the –n (or –np) flag, and can be accessed in NEURON using e.g., nhost = int(pc.nhost()) (NEURON generally returns a float, but nhost is naturally an integer value). The individual ranks are numbered from 0, with this number referred to as the rank or host id: e.g., rank = int(pc.id()). The zero rank node is by convention considered the Master node. Although this node will not differ in any way from other processing nodes in many cases, the Master will generally be used for centralized, serialized operations such as progress and timing notices or for moderate nhost, saving data to disk. On a very large HPC, a single node cannot handle all of the traffic and the nodes may be broken up into subsets with a separate local Master for each.
A key concept for network parallelization is to associate a unique integer global identifier (gid) with each individual neuron (Carnevale and Hines, 2006). This gid will remain the same regardless of how many hosts are configured and which rank houses which cell. Providing this gid to functions that create cells, stimulators, and network wiring ensures that simulations will run the same despite different distributions of gids across different numbers of nodes. Importantly, the use of a gid to specify particular random streams ensures that randomized wiring, inputs, parameters, etc will always be based on the same pseudo-random values regardless of number of nodes.
Several major NEURON commands and structures are listed in Table 1. Each of these commands and structures are accessed from Python as a method of ‘h.’. Note that these structures, accessed from Python, are parts of the NEURON simulator written in C++. The ParallelContext commands listed represent only the subset of commands that are used in this paper.
1.
Commands commonly used in this paper
| FUNCTION | USAGE | DESCRIPTION |
|---|---|---|
| Housekeeping functions | ||
| nhost | pc.nhost() | return the number of ranks; generally taken from mpiexec -n # |
| id | pc.id() | return identity (rank) of this process as number from 0 (the Master) to nhost-1 |
| barrier | pc.barrier() | hold execution till all ranks reach this point in program |
| Internode communication | ||
| py_alltoall | pc.py_alltoall(LIST) | send the ith object in the LIST (length nhost) to rank i (often only LIST[0] populated in order to send just to Master) |
| Simulation functions | ||
| set_maxstep | pc.set_maxstep(MAX) | interval between spike exchange computed as the smaller of MAX and the global minimum NetCon delay |
| psolve | pc.psolve(tstop) | integrate simulation up to tstop |
| Cell and synapse function | ||
| set_gid2node | pc.set_gid2node(GID, RANK) | global identifier (gid) exists on this rank (only if this rank == RANK) |
| gid_exists | val = pc.gid_exists(GID) | returns 0 if the gid does not exist on this rank |
| cell | pc.cell(GID, NETCON) | associate GID with a specific source of spikes defined by the source argument of h.NetCon() (can only be executed on the rank where both the GID and spike source exist) |
| gid_connect | nc=pc.gid_connect(PREGID,SYN) | connects the spike source PREGID with synapse SYN (this retained NetCon establishes the weight vector and delay and must exist on the rank where SYN exists) |
| spike_record | pc.spike_record(−1, VEC1, VEC2) | times go in VEC1, gids in VEC2, −1 means to record from all cells (on per node basis) |
| gid2cell | cell = pc.gid2cell(GID) | return cell associated with GID |
| Review of major variables, objects, functions, etc used in NEURON | ||
| timestep | h.dt | timestep for for numerical integration (preset for fixed timestep method) |
| time | h.t | current simulation time |
| Section | sec = h.Section() | an unbranched cylinder (cable) in NEURON |
| NetCon | nc = h.NetCon(SRC,TARG,sec=SEC) | network connection object |
| Random | r = h.Random() | r.Random123(gid,…) then provides independent per gid streams |
| Vector | vec = h.Vector(SIZE).resize(0) | Create (allocate) a vector and start with it empty for recording |
| event | h.cvode.event(t,Python_def_Name) | Python routine will be called at time t |
| state variable pointer |
soma(0.5)._ref_v | example of pointer to v at location 0.5 in section soma; in hoc: &soma.v(0.5) |
The main steps involved in the implementation and parallel simulation of networks in NEURON, together with the relevant ParallelContext functions are shown in Fig. 1. In the subsequent sections we discuss and provide example code for several of these stages, including network creation using a modular approach, load balancing, or data gathering and saving methods.
Figure 1.
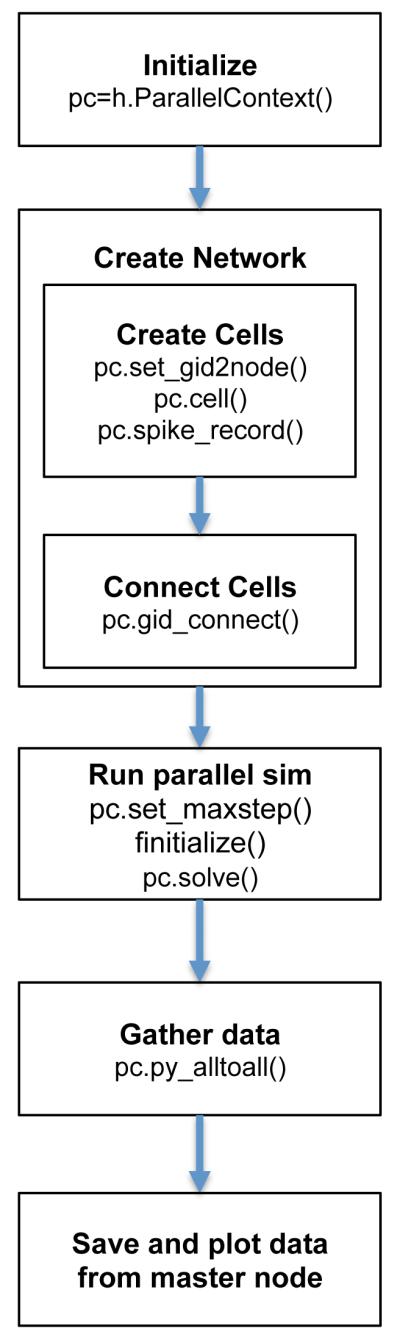
Main steps required to build and simulate a parallel network model, and NEURON ParallelContext functions employed at each stage.
Only fixed time-step numerical integration will be discussed in this paper. Reference is made to the CVODE object – the Livermore variable time-step integration solver for stiff and nonstiff ODE systems – because this object allows us to call a python function at a specific time (h.cvode.event(deliver, pycallable)).
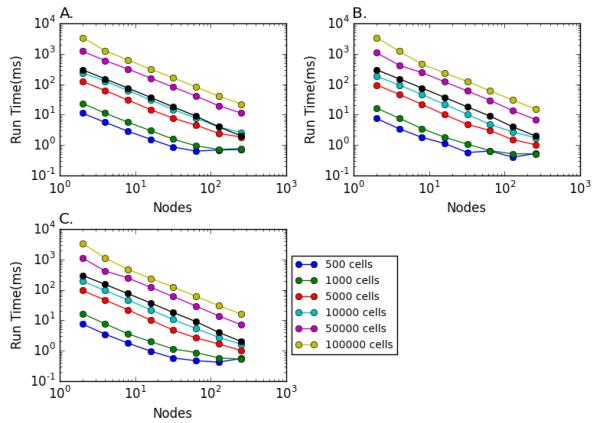
Growth of computational neuroscience and network computation will increasingly depend on the ease and accessibility of parallel computing. Simulations whose runtime performance is summarized Fig. 3 were carried out using the HPCs available under the Neuroscience Gateway Portal (NSGPortal; nsgportal.org), made freely available to investigators via a National Science Foundation grant (Sivagnanam et al., 2015). Using the NSGPortal, simulations can be run by: 1. uploading the model as a zip file, 2. choosing parameters (e.g. number of nodes, number of cores, language used), 3. running the simulation, and 4. downloading the output.
Figure 3.
Simulation run time as a function of number of cells and number of nodes in log-log plots. A. HH network, B. I&F network, C. Hybid network. (black: linear reference)
NetPyNE (Network development Python package for NEURON; www.neurosimlab.org/netpyne) was used here for benchmarking. NetPyNE hides the complexities described herein in order to give the user the ability to simply provide a high-level network specification using a Python-based declarative format. Code snippets shown below are taken from the package, simplified so as to be readily understandable independent of the full package. For reading convenience, the direct code model is provided along with the NetPyNE version on ModelDB (#188544).
1.1 Python modules
Several Python modules are mentioned in this paper, some of which are effectively obligatory. The pickle module is implicitly used to pass information between ranks using pc.py_alltoall(). It can also be explicitly included via import pickle, and then be used to serialize data for storage on disk. The gzip module can be used to compress the serialized data to reduce file size. The numpy package is a natural partner for NEURON’s Vector class, and numpy arrays can share memory with Vector objects avoiding any need to copy recorded values. Matplotlib can be used as an alternative or complement to the legacy NEURON graphics based on the Stanford Interviews package Linton et al. (1989). datetime or time or timeit can be helpful for benchmarking, along with the native h.startsw(). Such benchmarking techniques are thoroughly investigated in our simulations, and we will discuss in great detail exploration of the models as well as the results.
1.2 Modularity and subsidiarization
Here we describe a simplified version of the code required to implement the network using a Python/NEURON modular structure. A class ‘Cell’ that sets up basic functions for all cell models was created. Each cell model could then inherit from this class and adapt the methods based on its specific implementation requirements. The cell sections, topology, geometry, biophysics and synapses are defined within the methods of this class. The class constructor will then call these methods when the Cell object is created:
class Cell (object): def __init__(self): self.synlist = [] self.createSections() self.buildTopology() self.defineGeometry() self.defineBiophysics() |
Below are example methods to define the cell biophysics and geometry:
def defineBiophysics(self): """Assign the membrane properties across the cell.""" # Insert active Hodgkin–Huxley current in the soma self.soma.insert(’hh’) self.soma.gnabar_hh = 0.12 # Sodium conductance in S/cm2 self.soma.gkbar_hh = 0.036 # Potassium conductance in S/cm2 self.soma.gl_hh = 0.003 # Leak conductance in S/cm2 self.soma.el_hh = −70 # Reversal potential in mV |
def defineGeometry(self): """Set the 3D geometry of the cell.""" self.soma.L = 18.8 self.soma.diam = 18.8 self.soma.Ra = 123.0 |
Recording of cell-associated state variables can be done using the h.Vector() class to store the data. The Vector class is similar to Python’s Numpy Array. However, it offers the potential for use with direct pointers to internal state variables. Due to its use of “names” (untyped references with dynamic binding), Python lacks a C-style direct-address-access syntax – i.e., pointer = &value; value = *pointer. Therefore a special syntax was developed in NEURON to give a reference to a location: hocob j._ref_VARNAME. This syntax provides direct access for rapid recording to the Vector class. Vector also provides routines to fill with pseudo-random values from the NEURON Random class, which offers several special-purpose randomizers that provide improved randomization and independent streams, features not available from Python’s standard randomizers. (Python uses Mersenne-Twister by default and also supports an older pseudo-random number generator with a shorter period. random.SystemRandom uses environmental entropy to generate its bit stream which essentially makes the random numbers unpredictable, but is not useful for these application since it is seed-less – sequences cannot be reproduced.)
def setRecording(self): self.soma_v_vec = h.Vector() # Membrane potential vector at soma self.tVec = h.Vector() # Time stamp vector self.soma_v_vec.record(self.soma(0.5)._ref_v) self.tVec.record(h._ref_t) |
We have presented here a basic model in a modular fashion. In this example, there would a Python class for each cell type, which inherits from a common Cell superclass; and there would be a class for the network. These classes will include attributes and methods which are common to all cells or networks, respectively.
The layout for the cell model requires a few additions from the usual serial implementation. Primarily, each cell must have a global identifier gid that identifies the particular cell across all ranks. Generally, a NetCon has a source and a target, but now the source and target may end up on different ranks. Therefore, the postsynaptic NetCon is created with the postsynaptic mechanism and subsequently associated with a presynaptic location. The presynaptic NetCon connection provides threshold detection at a particular location. Events will be delivered at a synapse on the postsynaptic side via weights and delays that have been defined via the NetCon created postsynaptically. Within the context of its class, a cell is registered to a node using the following method defined within the cell class:
def associateGid (self):
pc.set_gid2node(self.gid, idhost)
nc = h.NetCon(self.soma(0.5)._ref_v, None,
sec=self.soma)
nc.threshold = 10
pc.cell(self.gid, nc)
|
pc.set_gid2node is the key call that associates the unique gid as belonging to a specific rank. Then, the creation of the temporary NetCon has the side effect of making sure a unique and persistent internal threshold detection object exists at a specific cell location for which one can set the spike trigger threshold (here 10 mV). The target of this temporary NetCon is None since the only purpose of this NetCon is its side effect of having the internal spike detector exist at the desired location on the cell. Hence, weight and delay of this temporary NetCon are not relevant and are not set – they will be set for each synaptic target on the postsynaptic side. The seeming redundancy of sec=self.soma follows from the fact that self.soma(0.5)._ref_v returns a memory pointer and therefore does not itself contain information about which section is being referenced. Finally, the pc.cell() associates the gid with the spike detector. After the pc.cell() call, the NetCon is no longer needed and can be garbage collected. The spikes generated by a cell will be sent by default to every other node. However the option is available to make a particular cell project only within a node: it does not add unused spikes to the internode communication streams.
The object-oriented policy of subsidiarity (pushing all processing down to the lowest possible subroutine level) suggests making calls at as low a level of organization as possible. In the case of wiring, however, an argument can be made either to do the wiring at the subcircuit level (e.g., a nucleus or a neocortical column), or to push the wiring all the way down to the cell or even synapse level. (In the brain, neurites grow out as filopodia which determine a cell’s wiring using only local guidance from radial glia and local chemical gradients.) However, a peculiarity arises from the parallel computing context. Instead of a source connecting to a target (cf. axonal growth cone or the connect2target() routine in NEURON’s CellBuilder), a target (a synapse) must connect to a source by identifying the gid from which it will receive spike events – the pregid. This call also creates the postsynaptic NetCon:
nc = pc.gid_connect(pregid, syn)
which will also then be given the weight and delay: nc.weight[0]=weight; nc.delay=delay. Unlike the presynaptic half NetCon, the postsynaptic half is not discardable, and must be saved — otherwise the connection will disappear. We choose here to save these as Python tuples together with the presynaptic gid in a list belonging to the postsynaptic cell: postCell.nclist.append((preGid, nc)).
1.3 Independent pseudo-random streams
In order that simulations be identical regardless of number of ranks used, pseudo-random streams that are used to determine connectivity, delays, weights or other parameters must be the same regardless of number of ranks chosen. This is achieved by associating a stream with a particular function and with a particular gid, using the Random123 generators (Salmon et al., 2011). Specific randomizers are created for particular purposes:
randomizers=[randomdel,randomcon]=[h.Random(),h.Random()].
The identity of a particular simulation run can be established at the outset via a run identifier:
randomizers[0].Random123_globalindex(runid)
Using any instance to set the globalindex sets all the Random123 instances, establishing an identity for the simulation based on runid. The identity is used for subsequent replication, and also serves to distinguish this simulation from others done with the same parameters but different randomization for robustness testing (different random connectivities, delays, weights, etc). In the context of the individual cells, the randomizer stream is made distinct and reproducible by using the gid in the call e.g., randomdel.Random123(id32(’randomdel’), gid, 0). The id32(’randomdel’) provides a unique 32-bit numeric id by hashing the name, keeping this randomizer distinct from others: : def id32(obj):return hash(obj)&0xffffffff (bitwise AND to retain only the lower 32 bits for consistency with 32-bit processors).
In general, the type of the specific distribution will not need to be reset despite resetting the stream to a different gid. Thus, for example, randomdel.uniform(2,4) can be set once to provide a uniform distribution of synaptic delays. However, randomdel.normal(mean, var), providing a normal distribution, must be re-called each time the stream is reinitialized, because it uses a pre-picked value as its initial state. The required random values can either be picked one-by-one using e.g., randomdel.repick() or can be assigned to fill a vector en masse with vec.setrand(randomdel).
1.4 Spike exchange
In simulation of spiking neuronal networks, physically continuous state variables are simulated using ODEs while synaptic activations are simulated as Events (Hines and Carnevale, 2004). Contrasted with the typically single-type simulation problems in other domains, this mixed continuous and event-driven problem provides various advantages and disadvantages.
With regard to parallelization, this mixed nature offers an advantage over purely continuous simulations (Hines and Carnevale, 2008). One can determine a maxstep for internode communication that is considerably longer than the timestep being used for integration of the continuous ODEs. This maximum synchronization interval is only determined by the minimum interprocessor synaptic (NetCon) delay. In NEURON’s ParallelContext, this maximum time step that a single node can proceed forward without communicating with other nodes is set using
pc.set_maxstep(max).
This call established the minimum delay between event generation and delivery (i.e., axonal plus synaptic delay), based on the shortest delay present in the network (the max argument is only used if smaller than the shortest delay). Message exchange between nodes is only necessary after the delay since at that time events can be exchanged across ranks without risk of late events; an event generated immediately after the prior exchange will need to be delivered immediately after this exchange.
In the current implementation, the maxstep returned is a global value across all nodes. This global delay can be returned using
interval = pc.allreduce(pc.set_maxstep(minmax), 3)
In the future, we expect that cases will arise which would benefit from allowing different maxsteps. For example, if one node is housing a spinal cord simulation which communicates at relatively long delays with the brain, communication with that node could be done less frequently than needed for exchange within the nodes representing encephalic structures.
At the end of a maxstep, spike exchange takes place. The standard options used for spike-exchange is an allgather method based directly on MPI Allgather. This all-to-all exchange is generally superior to MPI point-to-point methods unless the number of ranks is significantly greater than the average number of connections per cell — often on the order of 10000 — since almost every spike needs to be delivered to almost every rank. Furthermore, large HPC machines generally have hardware optimized MPI collectives which are difficult to beat with MPI point-to-point methods even when the number of ranks is an order of magnitude larger than the number of connections per cell (see Figure 4 of (Hines et al., 2011)) multisend paper.
For very large numbers of ranks, however, MPI Allgather does not scale well either in terms of performance or in the size of the spike receive buffer which, on each rank, must have a size equal to the sum of the sizes of all the send buffers of every rank. In this case, at the memory cost of each cell having a list of spike target ranks, point to point communication becomes useful. Other benefits are that communication overlaps computation and that all spikes received are in fact needed by the target rank. NEURON implements this ”multisend” method using nonblocking MPI Isend on the source rank along with non-blocking MPI Iprobe polling on the target rank. However far higher performance is obtainable with HPC specific hardware aware implementations, e.g. (Hines et al., 2011).
1.5 Load balancing
The ordinary differential equations (ODEs) for single-cell simulations can in many cases be solved faster by moving from a fixed timestep methods to a variable timestep method. This can be particularly helpful in some single neuron simulations where long periods of quiescence are followed by a burst of spikes, e.g., as seen with thalamocortical cells (Lytton et al., 1997). In such cases, simulation during the slow ramp depolarization phase can be done using timesteps of many milliseconds, which then switches to the short ~25 microsecond timestep required to simulate the fast (1 ms duration) action potential.
Unfortunately, this variable timestep advantage is lost in most large network simulations because of the many spike events from other cells that will interrupt any attempt at a long timestep in a given cell (Lytton and Hines, 2005). In fact, each such event is very expensive in the variable timestep context: each input event triggers a re-initialization and restart of the integrator. Subsequently the integrator will fastidiously follow the consequences of this event discontinuity using a series of very small, though rapidly increasing, time steps. For this reason, network simulations are typically performed using a fixed timestep method. It remains plausible that a benefit to variable timestep could be demonstrated in some specialized simulations, e.g., a thalamic network showing coordinated burst-quiescence activity across the network, where a hybrid solver with periods of variable timestep alternating with periods of fixed timestep might be used. Note that the global variable time-step method is effective in gap junction coupled networks, which give continuous activations rather than event-based inputs.
This problem with variable, hence need for fixed, timestep has the fortuitous side-effect of simplifying the load balancing problem, because none of the neurons are going to race ahead. All cells will move forward at the same pace so that load balancing can be achieved by matching the number of neurons per node when cells are identical. In most simulations the neurons are all of similar design, all event-drive (no compartments), all one-compartment, or all multi-compartment with the same number of compartments per neuron. In these cases, the standard round-robin (card dealing) approach is used to assign neurons sequentially to each node. In practice, everything is done from the node perspective so that the node chooses cells.
In some cases, one will want to run hybrid networks of various types (Lytton and Hines, 2004). These may contain a mixture of event-driven cells (EDCs, represented as an ArtCell in NEURON’s NMODL language), compartmental cells, integrate-and-fire cells, and compartmental cells of varying numbers of compartments (termed segments in NEURON) or different densities of ion channels in particular dendritic compartments or in the soma. In order to demonstrate this, we ran a Hybrid model that included equal number of HH and integrate-and-fire cells that were randomly connected together. This hybrid model can be widely implemented as long as the varieties of neurons in the hybrid network are randomly assorted and no single cell type is extreme in size, round-robin should still be an effective means of deployment. However, one may be combining models taken from ModelDB or other source where one brain area is modeled with EDCs and another with compartmental cells. In such a case it would be advisable to split the round-robins and do each subset separately. Other situations will arise where a hybrid network involves use of a few cells with far greater complexity than the rest of the network, either by virtue of the large number of compartments or through inclusion of reaction-diffusion mechanisms in the cell. In these cases, load balancing might involve giving a complex cell its own node or in any case reducing the number of cells that share that node. In some cases, the single cell may be so large that a multisplit method should be considered, splitting the individual cell across nodes (Hines et al., 2008).
In the case of cells of varying size, a general estimate of load is obtained by simply noting the number of compartments and balancing these across ranks. A more precise estimate is available through use of the h.LoadBalance() object available from h.load_file(’loadbal.hoc’) which counts numbers of state variables. The most precise estimate available is to ask h.LoadBalance() to measure the complexity factor of each membrane mechanism by timing it in a test cable. These load balance calculations may be inaccurate in cases where there are significant dynamic spike handling imbalances or when the performance of the model is limited by memory bandwidth instead of cpu speed. In any case, the actual load balance should be subsequently measured at run time by evaluating on each rank:
computation_time = pc.step_time() max_comp_time = pc.step_time() + pc.step_wait() avg_comp_time = pc.allreduce(computation_time, 1)/nhost load_balance = avg_comp_time/max_comp_time |
If the load balance is less than 0.9 it may be worth revisiting and revising the gid distribution, always of course noting that one must successfully amortize time sunk into code rewrites with time then saved through faster runs. Note that one may want to consider both your time and the HPC time; a minute of walltime saved on a simulation running on 1000 processors saves over 16 hours of HPC time that is being dedicated to your job.
1.6 Timing
In order to analyze efficiency while running large network simulations in a parallel environment, the timing of numerous stages in the simulation was recorded. To do so we used a ‘stop watch’which was included at various locations throughout the code and collected in a dictionary, that was later evaluated (eg. (timing[’runTime’] = time() - startTime). The timing values were grouped organized for interpretation as: 1) pre-simulation timing, which included the amount of time to create the network, connect the network cells, set up the recordings, and initialize the simulation; 2) simulation time; and 3) the post-simulation time, composed of the time to gather all data from other nodes, and the time to save all of the ouput data. The timing values were returned as a component of the output file.
1.7 Gathering data in the master node
With hundreds or thousands of nodes, doing file writes, particularly intermediate writes, can produce a file management nightmare with the potential for many files per simulation clogging up the file system. Therefore, if rank 0 memory capacity is not a problem, it is straightforward to provide partial or full consolidation of file writing by having all ranks pass their data to rank 0 from where it will be saved to disk. Passing data between nodes is most easily handled in python using pc.py_alltoall() which uses pickle to allow any type of data to pass from a source to a chosen destination. In the present context, this call will be used for one-way many-to-one communication from a set of nodes to a master. NEURON provides an efficient version of alltoall by minimizing the amount of processing in cases where the target is filled with the Python None object. In the present case, saving is done by transfer to the Master alone so that all other targets are None.
pc.py_alltoall() uses a Python list of length nhost as the framework. This list is set up on each rank so that the ith object in the list is destined for rank i:, e.g., dataout = data = [None]*nhost. This produces a list of the form [None, None, None, None, None, …]. The nhost locations on the list each determines a destination for the corresponding node. It is important to initialize the list to be all None rather than filling with initializations of arrays, dictionaries or lists so as to avoid unnecessary sends of empty arrays, etc.
Once dataout is provisioned, any rank can send any desired data structure to a destination rank i by setting the corresponding destination location: dataout[i] = localdata. If using only a single master, that master will typically be rank 0 so that each node will set dataout[0]; e.g., dataout[0]=rank would be used to send the rank number from this node to the master. On the destination side, a list, also of length nhost, will be assembled out of the pieces gathered from nhost source nodes. The place locations in this new list now correspond to the rank of the source from which the data came. Putting this together results in the following routine to be run on every node including the master:
def put_data (): global gather dataout = [None]*nhost data[0] = localdata gather = pc.py_alltoall(dataout) pc.barrier() |
We place an explicit barrier after gathering the data in order to make sure that every node is caught up before we proceed. At this point the master will have access to the data from all the nodes, while all the non-master nodes will simply see a list of multiple Nones. In the example above, with dataout[0]=rank, the master will have a gather list of [0, 1, 2, …, nhost-1]. Immediately after the barrier, the gathered data can be arranged from the master:
if rank == 0: consolidate_data(gather)
Assuming that the master node is also used for simulation, this idiom explicitly creates data on the master and uses the pc.py_alltoall() to send this data to itself, However, there is minimal overhead associated with this self-to-self send.
The manner in which get_data() gathers the gather will naturally depend on how the data has been stored. We have saved the data as a Python dictionary whose values are vectors. We will now create a consolidated gather dictionary gdict = {} out of the dictionaries in the gather list. Consolidating the state variable recordings is relatively easy since each of these recordings is already complete, having been confined to the node where the particular cell (a particular state variable) was calculated:
for d in gather: gdict.update(d)
By contrast, spikes from the various nodes must be collected and put together. This is done using numpy.concatenate() which can operate directly on hoc Vectors (here shown with import numpy as np):
gdict.update(
{’spkt’ : np.concatenate([d[’spkt’] for d in gather]),
’spkid’: np.concatenate([d[’spkid’] for d in gather])})
|
The final gdict dictionary should also save basic run information such as:
gdict.update({’walltime’:walltime, ’runid’:runid,
’tstop’:h.tstop, ’ncell’:ncell, …})
(walltime = datetime.datetime.now().ctime()).1.8 Data saving
The end product of simulation is data. Big simulations produce big data. Data management encompasses several stages. First, the data must be managed at its node of origin. Then, data from different nodes may be combined, something that can be done either during the simulation, or afterwards, or only virtually through accessing multiple files when later doing data analysis. Finally, data must be saved to disk in a fashion that makes the data subsequently accessible for data analysis and viewing. Choices at all of these stages lead to many variant strategies that will depend on exact circumstances. Additional complexity arises in NEURON due to the existence of preexisting storage strategies based on the h.Vector() whose functionality complements strategies based on Python data structures such as numpy arrays. There therefore exist multiple permutations for a complete data handling scheme. We will here provide some suggested idioms at the various stages out of which such a scheme can be built. We have generally built all of these formats atop Python’s dictionary dataformat so as to provide all data subsets with readily accessible and interpretable labels, such that identity and provenance can be readily tracked across the stages of data transfers, writes and reads.
If a simulation is small enough and short enough, then all the data saving can be put off until the end of an entire simulation. Recording in NEURON is done via the native Vector functionality, accessed as h.Vector(). The vector can be readily pre-sized and initialized using h.Vector(maxsize). Pre-sizing is not required but doing so avoids the necessity for expensive re-allocations of vector memory as data comes in and exceeds vector size. Two types of recording are supported in the ParallelContext: event-based recording of spike times and continuous recording of state variables. Events are recorded onto a pair of vectors using pc.spike_record(−1, spiketime_vector, gid_vector). Giving −1 as the first argument indicates that all cells are to be recorded. Alternatively one can record from individual neurons by giving a gid as the first argument, generally in a loop that would provide a list of neurons to record from. The spiketime and gid vectors will of course grow in parallel as each generated spike adds a spike time and cell identity (gid) to the corresponding vector. Since each node is simulating a different set of cells, a node will be recording spikes only from its own set of cells.
State variables are recorded “continuously,” that is to say at each time step or some multiple thereof. Values to be recorded typically include membrane voltage at particular locations in the cell, as well as ion concentrations, states of voltage-sensitive ion channels, synaptic conductances etc. The call is e.g., vec.record(cell.dend53(0.5)._ref_v, rec_step) which, in this example, provides saving at time steps rec_step (integer multiple of the simulation time step h.dt), of voltage v from the segment (individual compartment) at the center of the ‘dend53’ Section (section being an unbranched neurite made up of nseg segments). Note that the syntax _ref_StateVar provides the required pointer to the given StateVar in the simulator, necessary because variables in Python are not implemented using memory references. (In hoc, the equivalent pointer is &cell.dend53.v(0.5) – note the migration of the (0.5).)
We can use a Python dictionary to provide labels for the various recordings. For example, we set up the event recording in a dictionary acc to accumulate the data on each node:
acc = {} # create dictionary
for name in [’spkt’, ’spkid’]: acc[name] = h.Vector(1e4)
pc.spike_record(−1, acc[’spkt’], acc[’spkid’])
|
This creates two Vectors, each pre-allocated to store ten thousand values. Spike recording is done on a per-node basis. These vectors will later be consolidated after pulling together the output from different nodes and times.
State variable recordings can be setup in the context of the individual cell for modularity. A tuple may be used as the dictionary key in order to identify both the state variable being recorded and the gid of the cell being recorded from, e.g., acc.keys() might be
[(’ina’, 7), (’Vsoma’, 2), …
set up via:
acc[(key, self.gid)] = h.Vector(h.tstop/rec_step+1)
acc[(key, self.gid)].record(ptr, rec_step)
|
where the ptr pointers are themselves retrieved from a dictionary that relates labels, here ’ina’ and ’vsoma’ to corresponding pointers. The flexibility of dictionary keys in Python is used here to provide information about datasets without resorting to string manipulation or dictionaries of dictionaries.
So far we have described the data format being used for transfers from nodes to master. The next step is to save the data on disk for later access. There are several competing requirements that determine how these data might best be stored. 1. From a practical stand-point, it is desirable that storage protocols be readily accessed by a wide variety of programs that users may wish to use for further analysis. These would include numpy/scipy (Python packages), MATLAB, Mathematica, native NEURON (legacy hoc packages), IDL (Interactive Data Language) and others. The requirement of multi-tool access will generally be taken care of by Python, for which translation routines exist for several common formats. For example, MATLAB can import HDF5, which is a common data format also handled by Python. 2. Storage and retrieval should both be fast. This constraint tends to work against the prior constraint since the most portable formats, for example a comma-separated value (CSV) format, will tend to be the slowest. For this reason, we will give examples of the use of native Python formats (dictionary and pickle), which have been optimized in Python. However, we also note the value of providing random access to data, something that is not available with these formats but is available through direct numpy array or h.Vector binary saves. 3. Data from dynamic system simulations is typically indexed by time, providing parallel streams of state variable data which should be coordinated with each other (Lytton, 2006). 4. Size: Big data can often be made smaller with compression algorithms. In practice, voltage traces may be compressible since they typically involve minor alterations in voltage from one time step to the next. In general, data is compressible after pickling.
A standard way of saving data from Python is via the pickle routines. The idea of Python pickling is to allow most Python objects to be readily saved to a file in a single pickle.dump call as a pickled lump of data. Multiple disparate data types can be thrown together into a dictionary to make a single object before pickling. As noted above, pickling is also the way in which pc.py_alltoall() handles multiple data types for sending from 1 node to another. This is useful to know since errors in pc.py_alltoall() can be difficult to debug and it is worth checking that everything that is being sent can be readily pickled. An alternative to standard pickle is to use the more efficient cPickle module and compress the output data using gzip.
For large datasets there are advantages to avoiding pickle so as to provide access to voltage or other state variable from any location on any cell at any given time – random-access data. This functionality is provided by numpy.memmap() which provides read and write access to on-disk data using array indexing. For example, a NEURON Vector can be saved using vec.fwrite() which will write double precision values to a NEURON file handle. The equivalent save command for a numpy array is array.tofile(), which would also permit saving a higher dimensional array so as to save many state variables in one file. In either case, an index file or notebook entry should be used to identify the data set, data shape, precision, and perhaps the endian (the order of bytes for a machine). The data can then be accessed in-place using e.g., data = np.memmap(filename, dtype=’float64’, mode=’r’, shape=datashape) Here again, the use of fixed timestep integration and associated fixed timestep saving makes it easy to identify particular times in continuously recorded state variable streams. There is a further advantage to the use of index file when saving to multiple files since the index file will also indicate the various filenames which can then be made into a Python dictionary for seamless access.
Since there are so many choices, chosing a way to save data is often a point of contention. Pros and cons are weighed to determine how the data will be handled in any computational simulation. Some of the most popular output data formats are pickle, JSON, HDF5 and mat (Matlab format, which can also be saved from Python via a package). A comparison of saving time and disk space for different formats is included in the Results section. The function for saving thus includes the standard pickle calls necessary to pack the data into a dictionary and then store the data in an open file:
def saveData(self): print ’Saving data…’ dataSave = {’tVec’: self.tVecAll, ’idVec’: self.idVec} with open(’output.pkl’, ’wb’) as f: pickle.dump(dataSave, f) |
1.9 Multisave: saving repeatedly during a run
Running a large network, or even running a small networks with long simulation time, necessitates the use of intermediate saves (multisave method). This comes up with simulations of over one million cells running for long periods – e.g., 1 million cells for 1 hour would require an additonal 576 GB of internal memory if saved in RAM before dumping to disk. By saving every 100 ms, this requires only 16 MB RAM. During the simulation, writing to a file will be done at fixed intervals. Writing at intervals is both easier and faster than providing callbacks to be done when vectors fill up.
The pointer resets are done after the barrier at the end of the put_data() routine described above:
for v in acc.itervalues(): v.resize(0)
Writing to file is done as before except that the filename must be updated each time. A name of the following form suffices to provide a trackable, albeit long, filename with date, walltime, version, runnum, simtime (e.g.,
15Mar03_1800_4d45b8b_35_5000.dat
filename = ’{}_{:d}_{:s}_{:d}_{:d}.dat’.format( datetime.datetime.today().strftime(’%y%b%d_%H%m’), version, runid, int(round(h.t))) |
Where version is an identifier from a version control system such as mercurial or git (see getversion() routine in accompanying code).
Depending on the sizes and numbers of files full or partial consolidation may be desirable. Given a list of datafiles returned by glob.glob() or other Python routines, one can load datasets by reversing prior zipping and pickling:
datasets = [pk.loads(gzip.open(f,’rb’).read()) for f in datafiles]
This then should be sorted by accessing the simulation time available in each of the dictionaries:
datasets.sort(key = lambda d:d[’t’])
and concatenate the state variable vectors from statevars names into a new dictionary ddict using
ddict.update({k:np.concatenate([d[k] for d in
datasets]) for k in statevars})
similar to what was previously done to concatenate spike vectors from the different nodes. Spike time concatenations were handled in a similar manner.
At this point, we are looking at a third stage of data consolidation. First, the data recorded in a node was pulled together in a dictionary on that node for communication to the master. Second, the data was pulled together in the master so a common dictionary was prepared, with spikes from cells on all nodes represented together. Third, these data, saved to a file at different times during the simulation, were put together so that state variable and spike vectors were made available in single vectors of advancing time in a single dictionary. This dictionary might then be similarly saved to a file, with the intermediate files deleted.
Given the complexity of the multiple stitching processes and the potentially large sizes of the internal data structures required to do this stitching, it may be desirable to avoid this by leaving the data in multiple files and providing an index file that virtually stitches together a set of files by providing pointers to where vectors for a particular state variable or spike save begins in the individual files containing the simulation during one time interval. Multiple considerations may determine how much one wants to consolidate and how much index, including the availability of internal RAM memory for handling the data, the nature of disk access (e.g., NFS, NAS, local disk), and the desirability of later moving sections or all of the data over the internet to remote machines.
1.10 Cell re-simulation as an alternative to saving
An alternative approach for data management is to save the minimal information that is required for recreating all of the state variables – the spike times and cell gids. Because these network simulations are event-driven, event information can be used to re-drive one or more cells from a single node in order to recreate all of the activity on just those cells. Because full replication will require running a simulation from the beginning, this approach is most useful with a simulation that does not take long to run, or that utilizes many neurons on a single node. If each node is only 1 or 2 cells (or part of a cell using multisplit) and the simulation took 2 days to run, then the replication may also take 2 days to run, although it will be sped up somewhat if run on a computer that is faster than the node on which it originally ran. This strategy is particularly important when running large multicompartment cells with significant internal complexity (e.g., Ca2+ and second messenger cascades as well as voltages and ion channels state variables). In order to provide a full description of cell activity in the network, one might want to save multiple state-variables (10-20) from multiple compartments (100-1000) sampled at 0.1ms over seconds at double precision, bringing us up to 10-20 GBytes of storage.
Setting up the re-simulation utilizes PatternStim to feed the spikes back into the single cell. The setup code is largely unchanged from the full simulation except that we run only on cell instead of a range: e.g., chosen = [40] for cell #40. This “network” of one cell is now set up as usual for running on a single node. If running with more than 1 cell, set an unreachable firing threshold to prevent events from being generated within this stub network (nc.threshold = 1e9), since all events will now come from the prerecorded vectors. Having restored these vectors from disk into tvec for times, and idvec for cell gid’s, we can then setup playback using:
pattern = h.PatternStim() pattern.fake_output = 1 pattern.play(tvec, idvec) |
The simulation can be run as batch or interactively and does not need to be run under MPI, either h.run() or pc.psolve() can be used. It is valuable to record the spikes from the cell being resimulated so that the accuracy of the resimulation can be confirmed. In order to do this, these cells must have their threshold reset to the standard value.
2 Results
2.1 Simulation runtime
To evaluate the performance of network parallelization we benchmarked the simulation runtime under 3 different design parameters: 1. Types of cells in the network (HH, I&F, hybrid), 2. Number of nodes on HPC: powers of 2 from 2-256, 3. Number of cells in the network: 500, 1000, 5000, 1 ·104, 5 ·104, 10 ·104.
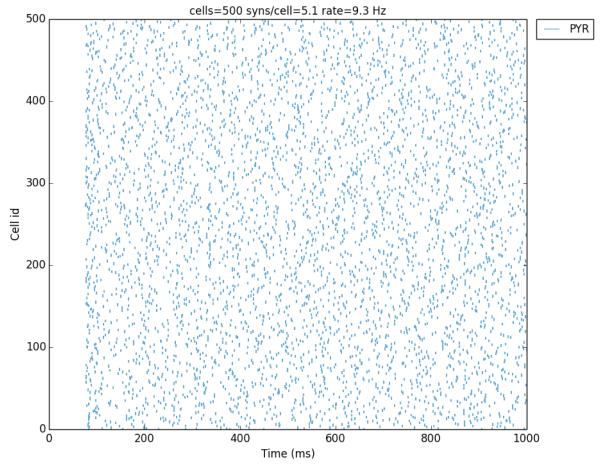
In order to focus on network design issues, we used two highly simplified cell models, the 4-dimensional Hodgkin-Huxley (HH) cell model Hodgkin (1952) and the 2-dimensional Izhikevich Integrate-and-fire (I&F) cell model Izhikevich (2007). In this way, most compute time was taken up by spike handling rather than being devoted to simulation of the individual neuron. Both cells responded with single spikes to synaptic inputs at the same delay, provided by spike generators (NetStims) with average firing rates of 10 Hz, associated with distinct statistically independent but reproducible random number generators. Cells were connected randomly with cell convergence obtained from a uniform distribution between 0 and 10. Connection weights were set to very low values to eliminate any connection dependent firing behavior that might arise due to the network or specific random connectivity. This resulted in identical firing patterns generated, regardless of the number of hosts, number of cells and cell types used (Fig. 2). Hybrid network models were run using half of each cell type, and also produced the same spike sequence. This demonstrates the flexibility of using NEURON to simulate very different types of networks simplified integrate-and-fire cells, biophysically realistic cells or hybrid simulations using some I&F cells for speed while setting up other cells as detailed simulations.
Figure 2.
Raster plot for a 500 cell network. The same sequence of spikes was produced with HH, I&F or hybrid networks.
Results were graphed and analyzed to display the relationship between cell type, network number and number of nodes (Fig. 3). Time decreased approximately linearly, with increased numbers of nodes for all simulation types. Flattening at high node-count was due to reduced improvement for the smaller networks at higher number of nodes, due to the increased dominance of internode communication with each node now only hosting 5-10 cells. Due to the smaller dimensionality of each cell the I&F network ran somewhat faster compared to the HH network. The hybrid network ran only slightly slower compared to the I&F network.
Further simulations were carried out to demonstrate the reproducibility of connected cell network activity in serial and parallel. A 5000 cell network was run on NSG using 1, 2, 4, 8, 16, 32, 64, 128 or 256 nodes. Increasing the network weights resulted in an average firing rate of 33.08 Hz, increased from 10 Hz due to spikes produced by network interaction. The output spike times of all cells in the connected network were identical in all simulations regardless of the number of nodes employed. As before, simulation time decreased linearly with the number of nodes.
Standard modern computers have multiple cores so that simulations can be greatly sped up by running under MPI, even on a laptop. We compared the 50,000 cell I&F cell network on the 4 cores of a MacBook Pro with a i7 2.6 GHz processor. Compared to the NSG simulations on the same number of nodes, the MacBook took 1.7 times longer to run, reflecting the better processors available on the NSG HPCs, as well as what is probably a slight slow-down from concurrent background processing on the MacBook.
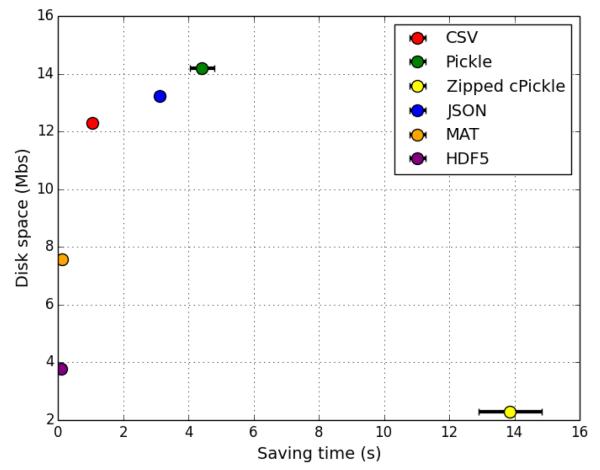
2.2 Saving time and disk space for different file formats
We benchmarked the saving time and disk space required to store the output of the simulation (spike times and cell gids) for 6 common file formats (Fig. 4). The fastest formats were HDF5 and MAT, both of which also required less disk space than CSV, Pickle or JSON. Although zipped cPickle saved the most disk space, the saving time required was over 100× than for HDF5. See the Methods section for a more in-depth discussion of the pros and cons of the different file formats. Saving time benchmarks were run on a MacBook Pro with 4 i7 2.6 GHz processors.
Figure 4.
Comparison of saving time and disk space to store the output of the 50,000 cell network simulation for 6 common file formats. 473,879 spikes were saved with saving of spike times and gids (cell global identifiers) for each.
3 Discussion
We have described NEURON techniques for building large network simulations, demonstrating some preferred idioms and approaches. Many of there discussed here are not specific to NEURON but would also be useful for other simulators that use Python as their interpreter language, such as NEST, MOOSE, Auryn, GENESIS, PyNN, BRAIN, and others (Cornelis et al., 2012; Davison et al., 2008; Eppler et al., 2009; Gewaltig and Diesmann, 2007; Goodman and Brette, 2009; Ray and Bhalla, 2008; Sneddon et al., 2011; Zenke and Gerstner, 2014). The use of Python as a common language is encouraging further development of common file formats not only for simulation output and subsequent data analysis, but also for network design (Djurfeldt et al., 2010; Gardner et al., 2003; Schmitt and Eipert, 2012).
We have primarily here focused on the small to midsize HPC clusters that are readily available to most researchers both through their university computing facilities and through the Neuroscience Gateway portal at San Diego Supercomputer Center (Sivagnanam et al., 2013). Extremely large simulations, of the type envisioned as part of the Human Brain Project, Allen Brain Project and U.S. BRAIN initiative, will need to be run on the very largest supercomputers and will require somewhat different techniques that are extensions of the techniques presented here. The very large datasets generated will likely never be moved off of their machine of origin due to size, so that analysis will need to be done locally on the same machine or a closely connected computer with access to the same tertiary data storage. Very large simulations require long setup times and may be run continuously for long periods of time with periodic data analysis and on-the-fly readjustment of parameters along with self-adjustment through plasticity rules (Helias et al., 2012; Lytton et al., 008b; Yamazaki et al., 2011; Zenke and Gerstner, 2014).
Our simulations consistently showed approximately linear speed-up with increase in number of nodes. This speed-up with node number was able to greatly offset the time increase due to larger number of cells in the network. In addition, we showed that type of cell model also correlated with simulation time. Overall, the results were able to demonstrate the ease at which HPC simulations can be carried out, timings for steps in the parallel model, and data storage. The models we used, as well as others are readily available on modeldb, and can be run on large HPCs such as provided on NSGportal in order to minimize time and simplify job submission.
Two related themes that bear repeated emphasis are the importance of modularity and the importance of provenance ((Bezaire and Soltesz, 2013; McDougal et al., 2016)). Simulations should be designed in a way that makes it easy to find particular parameters and to separate out structures at different scales. Our models were designed so that major parameters such as, number of cells, type of cell, and simulation time were easily able to be adjusted. At each scale, it is desirable to be able to track back a parameter or design decision to some experimental result which supports the design decision made. Modularity will assist in ensuring that provenance tracks along with parameters when they are taken to form parts of new simulations. (Unfortunately, not every parameter and design decision can be experimentally justified so there is a place in provenance for acknowledging that a ‘best guess’ has been used.) Having labels and meta-data, e.g., through the use of Python dictionaries with multiple fields, improves the chances of retaining information about data. These considerations hold both for input parameterization and design, and for output data storage and access. In both cases, one will be concerned with proper concordance with datasets obtained experimentally; experimental data-mining for network design is partnered with data-mining for simulation interpretation, and with experimental data-mining for exploring the validity of simulation predictions (Lytton and Stewart, 2007).
One of the overall projects of computational neuroscience is to use the computer not only as a tool for understanding the brain but also as a metaphor for developing concepts about the brain. Briefly, the problem of large network simulation on a computer cluster can be regarded as mirroring some of the problems that the brain must face in its organization. 1. saving/writing outputs to disk/spinal cord 2. use of randomization to produce flexibility in response, 3. balance between signals sent for organizing activity (barriers and clocking) and signals sent that have meaning, 4. provisioning of cells so that cells that must exchange messages frequently are close together and can signal rapidly. 5. Importance of hierarchical modularity in organizing activity and putting out “data” for downstream sites. 6. The problem of ranks (mirroring brain areas) that do not know which other places want their information, and do not know what places have information that they want.
The complexity of neuronal tissue yields problems that are multiscale in time and space as well as multi-conceptual, upwards to problems of information, cognition and behavior. The NEURON simulation environment has grown by incorporating new tools to manage these multiple scales and viewpoints. For example NEURON has added simulation of networks of integrate-and-fire or event-driven neurons with no cellular detail, and at the other end of the cell-complexity spectrum, now provides simulation of subcellular reaction-diffusion simulation in given parts of a cell or in the whole cell. This broad reach makes it a tool of many compromises that is thereby able to approach large problems across many scales.
Acknowledgments
Supported by National Insitutes of Health (R01MH086638, U01EB017695, T15LM007056, R01NS11613), ETH Domain for the Blue Brain Project (BBP), Human Brain Project (European Union Seventh Framework Program FP7/2007-2013 under grant agreement no. 604102, HBP). Thanks to the Whitman Center of the Marine Biological Laboratories, Woods Hole, MA where some of this work was performed.
References
- Bezaire M, Soltesz I. Quantitative assessment of CA1 local circuits: Knowledge base for interneuron-pyramidal cell connectivity. Hippocampus. 2013;23:751–785. doi: 10.1002/hipo.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brette R, Rudolph M, Carnevale T, Hines M, Beeman D, Bower J, Diesmann M, Goodman P, Harris F, Zirpe M, Natschläger T, Pecevski D, Ermentrout B, Djurfeldt M, Lansner A, Rochel O, Vieville T, Muller E, Davison A, El Boustani S, Destexhe A. Simulation of networks of spiking neurons: a review of tools and strategies. J Comput Neurosci. 2007;23:349–398. doi: 10.1007/s10827-007-0038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevale N, Hines M. The NEURON Book. Cambridge University Press; New York: 2006. [Google Scholar]
- Cornelis H, Rodriguez A, Coop A, Bower J. Python as a federation tool for GENESIS 3.0. Plos One. 2012;7:e29018. doi: 10.1371/journal.pone.0029018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A, Brüderle D, Eppler J, Kremkow J, Muller E, Pecevski D, Perrinet L, Yger P. PyNN: a common interface for neuronal network simulators. Frontiers in neuroinformatics. 2008;2 doi: 10.3389/neuro.11.011.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A, Hines M, Muller E. Trends in programming languages for neuroscience simulations. Front Neurosci. 2009;3:374–380. doi: 10.3389/neuro.01.036.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djurfeldt M, Davison A, Eppler J. Efficient generation of connectivity in neuronal networks from simulator-independent descriptions. Front Neuroinform. 2014;8:43. doi: 10.3389/fninf.2014.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djurfeldt M, Hjorth J, Eppler J, Dudani N, Helias M, Potjans T, Bhalla U, Diesmann M, Kotaleski J, Ekeberg O. Run-time interoperability between neuronal network simulators based on the MUSIC framework. Neuroinformatics. 2010;8:43–60. doi: 10.1007/s12021-010-9064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppler J, Helias M, Muller E, Diesmann M, Gewaltig M. Pynest: a convenient interface to the nest simulator. Front Neuroinform. 2009;2:12. doi: 10.3389/neuro.11.012.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner D, Toga AW, Ascoli GA, Beatty JT, Brinkley JF, Dale AM, Fox PT, Gardner EP, George JS, Goddard N, et al. Towards effective and rewarding data sharing. Neuroinformatics. 2003;1(3):289–295. doi: 10.1385/NI:1:3:289. [DOI] [PubMed] [Google Scholar]
- Gewaltig M-O, Diesmann M. NEST (NEural Simulation Tool) Scholarpedia. 2007;2(4):1430. [Google Scholar]
- Goodman D, Brette R. The Brian simulator. Frontiers in neuroscience. 2009;3(2):192. doi: 10.3389/neuro.01.026.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helias M, Kunkel S, Masumoto G, Igarashi J, Eppler J, Ishii S, Fukai T, Morrison A, Diesmann M. Supercomputers ready for use as discovery machines for neuroscience. Front Neuroinform. 2012;6:26. doi: 10.3389/fninf.2012.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines M, Carnevale N. NEURON: a tool for neuroscientists. Neuroscientist. 2001;7:123–135. doi: 10.1177/107385840100700207. [DOI] [PubMed] [Google Scholar]
- Hines M, Carnevale N. Discrete Event Simulation in the NEURON Environment. Neurocomputing. 2004;58:1117–1122. [Google Scholar]
- Hines M, Carnevale N. Translating network models to parallel hardware in neuron. J Neurosci Methods. 2008;169:425–455. doi: 10.1016/j.jneumeth.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines M, Davison A, Muller E. NEURON and Python. Front Neuroinformatics. 2009a;3:1. doi: 10.3389/neuro.11.001.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines M, Eichner H, Schürmann F. Neuron splitting in compute-bound parallel network simulations enables runtime scaling with twice as many processors. J Comput Neurosci. 2008;25:203–210. doi: 10.1007/s10827-007-0073-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines M, Kumar S, Schürmann F. Comparison of neuronal spike exchange methods on a Blue Gene/P supercomputer. Front Comput Neurosci. 2011;5:49. doi: 10.3389/fncom.2011.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines M, Morse T, Carnevale N. Model structure analysis in neuron : toward interoperability among neural simulators. Methods Mol Biol. 2007;401:91–102. doi: 10.1007/978-1-59745-520-6_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines ML, Davison AP, Muller E. NEURON and Python. Frontiers in Neuroinformatics. 2009b;3:1. doi: 10.3389/neuro.11.001.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin AL, H. AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117(4):500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izhikevich EM. Dynamical systems in neuroscience. MIT press; 2007. [Google Scholar]
- Linton MA, Vissides JM, Calder PR. Composing User Interfaces with InterViews. Computer. 1989;22:8–22. [Google Scholar]
- Lytton W. Neural query system: data-mining from within the NEURON simulator. Neuroinformatics. 2006;4:163–176. doi: 10.1385/NI:4:2:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytton W, Contreras D, Destexhe A, Steriade M. Dynamic interactions determine partial thalamic quiescence in a computer network model of spike-and-wave seizures. J Neurophysiol. 1997;77:1679–1696. doi: 10.1152/jn.1997.77.4.1679. [DOI] [PubMed] [Google Scholar]
- Lytton W, Hines M. Hybrid neural networks - combining abstract and realistic neural units. IEEE Engineering in Medicine and Biology Society Proceedings. 2004;6:3996–3998. doi: 10.1109/IEMBS.2004.1404116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytton W, Hines M. Independent variable timestep integration of individual neurons for network simulations. Neural Comput. 2005;17:903–921. doi: 10.1162/0899766053429453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytton W, Neymotin S, Hines M. The virtual slice setup. J Neurosci Methods. 2008b;171:309–315. doi: 10.1016/j.jneumeth.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytton W, Stewart M. Data mining through simulation. Methods Mol Biol. 2007;401:155–166. doi: 10.1007/978-1-59745-520-6_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougal R, Bulanova A, WW L. Reproducibility in computational neuroscience models and simulations. IEEE Transactions on Biomedical Engineering. 2016 doi: 10.1109/TBME.2016.2539602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougal RA, Hines ML, Lytton WW. Reaction-diffusion in the NEURON simulator. Frontiers in neuroinformatics. 2013;7 doi: 10.3389/fninf.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliore M, Cannia C, Lytton W, Hines M. Parallel network simulations with NEURON. J. Computational Neuroscience. 2006;6:119–129. doi: 10.1007/s10827-006-7949-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Bhalla U. PyMOOSE: interoperable scripting in Python for MOOSE. Front Neuroinformatics. 2008;2:6. doi: 10.3389/neuro.11.006.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon J, Moraes M, Dror R, Shaw D. Parallel random numbers: as easy as 1,2,3. Proceedings of 2011 International Conference for High Performance Computing, Networking, Storage and Analysis; Seattle, Washington. November 12-18, 2011.2011. [Google Scholar]
- Schmitt O, Eipert P. Neuroviisas: approaching multiscale simulation of the rat connectome. Neuroinformatics. 2012;10:243–267. doi: 10.1007/s12021-012-9141-6. [DOI] [PubMed] [Google Scholar]
- Sivagnanam S, Majumdar A, Yoshimoto K, Astakhov V, Bandrowski A, Martone M, Carnevale N. Introducing the Neuroscience Gateway. CEUR Workshop Proceedings. 2013 doi: 10.1002/cpe.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivagnanam S, Majumdar A, Yoshimoto K, Astakhov V, Bandrowski A, Martone M, Carnevale NT. Early experiences in developing and managing the neuroscience gateway. Concurrency and Computation: Practice and Experience. 2015;27:473–488. doi: 10.1002/cpe.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneddon M, Faeder J, Emonet T. Efficient modeling, simulation and coarse-graining of biological complexity with NFsim. Nature Methods. 2011;8(2):177–183. doi: 10.1038/nmeth.1546. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Ikeno H, Okumura Y, Satoh S, Kamiyama Y, Hirata Y, Inagaki K, Ishihara A, Kannon T, Usui S. Simulation platform: a cloud-based online simulation environment. Neural Netw. 2011;24:693–698. doi: 10.1016/j.neunet.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Zenke F, Gerstner W. Limits to high-speed simulations of spiking neural networks using general-purpose computers. Front Neuroinform. 2014;8:76. doi: 10.3389/fninf.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]