Abstract
The ribosome can change its reading frame during translation in a process known as programmed ribosomal frameshifting. These rare events are supported by complex mRNA signals. However, we found that the ciliates Euplotes crassus and Euplotes focardii exhibit widespread frameshifting at stop codons. 47 different codons preceding stop signals resulted in either +1 or +2 frameshifts, with the +1 frameshifting at AAA being the most frequent. The frameshifts show unusual plasticity and rapid evolution, and have little influence on translation rates. Proximity of a stop codon to the 3′-mRNA end rather than its occurrence or sequence context appeared to designate termination. Thus, a stop codon is not a sufficient signal for translation termination, and the default function of stop codons in Euplotes is frameshifting, whereas termination is specific to certain mRNA positions and likely requires additional factors.
There are several known mRNAs where translating ribosomes shift reading frame at specific locations with high efficiency that in very rare cases may even exceed the rate of concurrent standard translation. This phenomenon is known as programmed ribosomal frameshifting and is mostly observed in viruses1. While programmed ribosomal frameshifting is an omnipresent translation process, it is usually considered as a recoding mechanism. Recoding describes alterations in genetic decoding that take place at specific locations within particular mRNAs and is distinguished from codon reassignment 2. With an exception of 40% efficient programmed ribosomal frameshifting at a heptanucleotide site in Saccharomyces cerevisiae that is used during expression of the Ty1 transposon 3, complex stimulatory signals, such as RNA pseudoknots, are required for a high efficiency of programmed ribosomal frameshifting 4.
However, previous analyses of several sequenced genes of the ciliates Euplotes, suggested that +1 ribosomal frameshifting may be more common in these organisms (reviewed in 5). All frameshift motifs in Euplotes identified until recently consist of an AAA codon followed by a stop codon, either TAA or TAG. It has been hypothesized that frameshifting evolved as a consequence of TGA codon reassignment from stop to cysteine, which weakened release factor recognition of the remaining stop codons, TAA and TAG 5,6. Furthermore, it has been shown experimentally in a hybrid system that Euplotes release factors indeed recognize these stop codons inefficiently 6.
To understand this unusual case of frameshifting and the molecular mechanisms involved, we sequenced and analyzed the macronuclear genomes of two Euplotes species: E. crassus and E. focardii 7,8. We also sequenced the transcriptome of E. crassus and carried out ribosome profiling and proteomic analyses. The genomic and high-throughput biochemical analyses allowed us to identify and characterize over a thousand frameshift sites. This revealed that ribosomes of the Euplotes ciliates are characterized by inability to terminate at stop codons in internal positions of coding sequences and instead frameshift at these signals, whereas termination likely requires additional components in these organisms and occur only at specific mRNA positions.
Macronuclear genomes of E. crassus and E. focardii and their transcriptomes
Similar to other ciliates, Euplotes DNA is distributed among its two compartments: the macronucleus, which controls all cell functions during vegetative growth, and the micronucleus, which is needed for reproduction. The macronuclear genome consists of many small chromosomes. The copy number of individual chromosomes in ciliates may range from 100 to 10,000, with an average of 2,000 per macronucleus in Euplotes 9,10. These chromosomes are generated from the micronuclei DNA following sexual reproduction 11. It is the macronuclear DNA that is actively transcribed and is used as a template for mRNA synthesis, and therefore we were interested primarily in the macronuclear genomes.
To understand how Euplotes genes are translated, it was beneficial to examine at least two genomes, thereby allowing comparative sequence analysis. Thus, we sequenced macronuclear genomes of two related Euplotes. One is E. crassus, a sand-dwelling hypotrichous ciliate of the marine intertidal zone. The other is a recently isolated E. focardii, which is endemic to the Antarctic 7. The strain TN1 was obtained from the samples collected in Terra Nova Bay, and its psychrophilic phenotypes (optimal survival and multiplication rates at 4–5 °C) suggest adaptation to the stably cold Antarctic waters 7. The general properties of their genomes are described in Supplementary Figure 1-.
A large number of very short (20-30 nts) introns is a characteristic feature of macronuclear protein coding genes in some ciliates 12,13, but accurate prediction of introns is complicated by instances of alternative splicing and non-canonical splice junctions 14. Some short introns, if not detected by annotation pipelines, may result in ORF disruption and thus be misinterpreted as frameshift sites. To account for this possibility, we utilized experimentally confirmed rather than predicted mRNA transcripts (Supplementary Fig. 2).
Identification of ribosomal frameshifting using phylogenetics, ribosome profiling and proteomic analyses
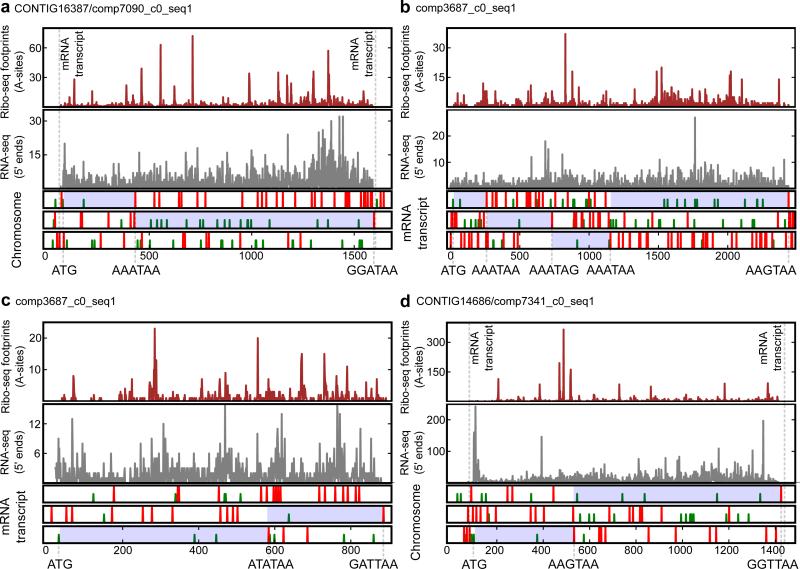
To identify sites of ribosomal frameshifting and estimate its efficiency, we first carried out ribosome profiling (Ribo-seq) in E. crassus. Ribosome profiling is based on sequencing of mRNA fragments protected by the translating ribosomes from nuclease digestion 15. It provides information on ribosome locations and their densities at the whole transcriptome level 16,17. Ribosome-protected fragments are expected to occur immediately downstream of stop codons only in cases of efficient stop codon readthrough or ribosomal frameshifting. To discriminate between readthrough and ribosomal frameshifting in −1 or +1 direction we compared the span of Ribo-seq coverage with ORF organization (Fig. 1). In certain cases, where unambiguous discrimination between potential events was difficult, we sought additional information. Using BLAST, we explored which of the potential products is more likely to have closely related homologs. Overall, we identified 1,765 putative frameshift sites spanning 1,326 transcripts from a total of 6,087, with at least 100 Ribo-seq reads per transcript. In a number of transcripts we found more than one site of ribosomal frameshifting (Fig. 1b). In addition to +1 frameshifting, we detected frameshifting into the −1/+2 frame (Fig. 1c). However, we did not find a single example of stop codon readthrough. The sequences of the transcripts were compared to the sequences of genomic contigs to exclude the possibility of identifying frameshifting as a result of misidentification of sequencing errors during RNA-seq analysis (Fig. 1a,d).
Figure 1. Frequent frameshifting in Euplotes.
Ribo-seq profiles of individual mRNAs are shown in the upper panels, RNA-seq in the middle panels, and features of reading frames in the lower panels. Start (ATG, green vertical lines) and stop codons (TAA, TAG, red lines) are shown in each of the three reading frames for chromosomes (a, d) and transcripts (b, c). Inferred translated regions are highlighted in blue. ATG codons corresponding to translation initiation sites are indicated beneath each plot. Stop codons (and adjacent upstream codons) where termination or frameshifting occur are also indicated. (a) Example of +1 ribosomal frameshifting at AAA_TAA. (b) Example of mRNA with several ribosomal frameshifting sites. (c) Example of +2 frameshifting at the ATA_TAA. (d) Example of +1 frameshifting at AAC_TAA.
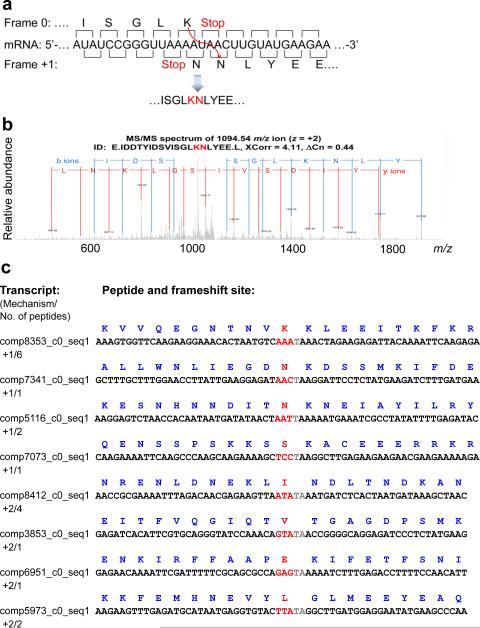
To verify putative sites of frameshifting and determine the associated mechanisms (i.e. direction and identity of amino acids incorporated at frameshift sites), we carried out LCMS/MS proteomics analyses of soluble E. crassus fractions, following trypsin and Glu-C digestions (the latter was used to preserve peptides with internal Lys). We examined if any of these peptides covered two different frames within the same gene and detected 13 such peptides with validated MS/MS spectra (Fig. 2, Supplementary Note 1, Supplementary Note 2). In addition to +1 frameshifting, some peptides were the products of +2 ribosomal frameshifting, consistent with our observation of ribosomal frameshifting into the −1/+2 frame based on Ribo-seq data.
Figure 2. Identification of amino acids inserted at frameshift sites.
(a) Lysine (K) and asparagine (N) are inserted at the AAA_TAA_C heptamer. Nucleotide sequence surrounding the AAA_TAA +1 frameshift site is shown in the middle. Amino acid sequence is shown above for the zero frame and below for the +1 frame. (b) Recorded MS/MS spectrum confirming the presence of a peptide derived from predicted frameshifting. (c) Peptides detected by MS/MS analysis that were derived from the translation of frameshift sites are shown along with the corresponding nucleotide templates. Nucleotides “skipped” as a result of frameshifting are highlighted in gray. Codons preceding stop codons are shown in red, and the amino acids inserted at frameshifting sites are indicated.
Sequence properties of +1 and +2 frameshifting sites
Among 1,765 putative frameshift sites detected with Ribo-seq, about three quarters (1,368) consisted of an AAA codon followed by a stop codon, and a quarter (397) contained other codons preceding stop. Altogether, we observed 47 out of 62 possible sense codons at the frameshift sites. The supporting information (ribosome footprint density and BLAST hit alignments) for various types of frameshifting sites is shown in Supplementary Note 3.
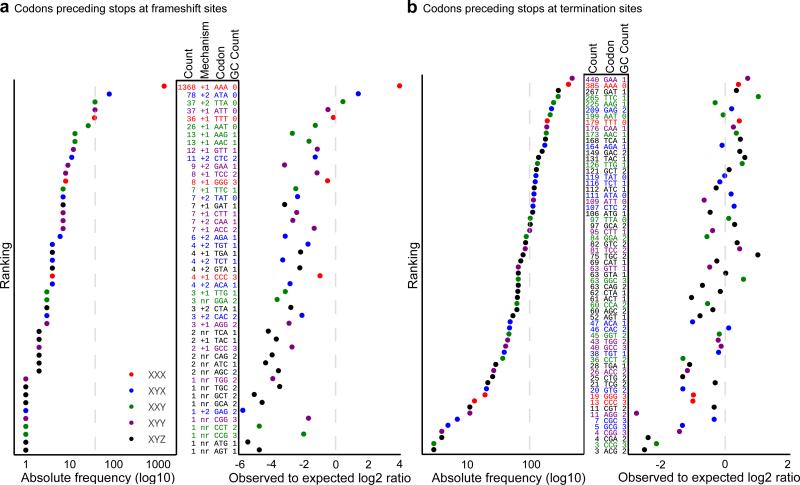
Earlier observations of frequent use of AAA_TAA and AAA_TAG as frameshifting sites in Euplotes prompted researchers to speculate that there is something special about AAA that allows frameshifting to take place at this codon 5. Our comparison of codon frequencies upstream of stop codons in the frameshift sites and in the sites of termination revealed that AAA was not only the most frequent codon at the frameshift sites (Fig. 3a), but also was the second most frequent codon at the termination sites (Fig. 3b). However, high frequency of AAA codons at frameshift sites cannot be explained simply by their high frequency upstream of stop codons. The AAA codon was overrepresented at the frameshift sites in comparison with its usage in internal positions of coding frames, occurring ~8 times more frequently than expected (Fig. 3a). Moreover, 6 out of 7 AT-only codons were the most frequent codons at the frameshift sites, and they were also overrepresented at the frameshift sites in comparison with internal positions (Fig. 3a). A higher frequency of AT-rich codons among frameshift sites suggests that weak interactions between P-site tRNA and its codon in the initial frame increases possibility of frameshifting. We also found that all XXX codons (i.e. codons with identical nucleotides) were also enriched (relative to most non-AAA codons) at the frameshift sites (Fig. 3a, right), even though CCC and GGG were not the most frequent ones, owing to a relatively low GC content of Euplotes genomes. This suggests that the ability of P-site tRNAs to form base pairing with a codon in +1 frameshifting also increases chances of frameshifting because XXX codons would re-pair with XXT forming perfect Watson-Crick interactions with the first two subcodon positions.
Figure 3. Distribution of codons upstream of stop codons at the frameshift sites and at the sites of translation termination.
(a) Frameshift sites. The plot on the left shows absolute frequency of each sense codon ranked based on its frequency. Identity of codons is given by Codon in the middle table. GC content and the inferred mechanism of frameshifting (+1 or +2) are also indicated (nr indicates that the mechanism was not resolved). The absolute number of frameshift sites is listed in Count. Plot on the right shows frequency of codons relative to their expected occurrence based on their usage in internal positions of coding regions. Rows are colored according to codon type. (b) Sites of translation termination. See panel (a) for details. Broken lines indicate average values for absolute frequencies and expected values for normalized frequencies.
Interestingly, XYX codons (same nucleotides at the 1st and 3rd subcodon position, but a different nucleotide in the 2nd subcodon position) supported +2 ribosome frameshifting. Figure 1c shows a ribosome density profile for an mRNA containing an ATA_TAA frameshift site. It appears that the ribosomes shifted into the −1 frame. However, the mechanism was found to be +2 frameshifting based on the MS/MS analysis (Supplementary Note 1). Also, +2 frameshifting seemed to be more likely because in this case the isoleucine tRNA decoding the ATA codon would re-pair with the same ATA codon. We found 9 XYX codons (out of 16 possible) in the +2 frameshift sites (Fig. 3a) with ATA being the most frequent. The other codons that seemed to support +2 frameshifting were XTA that have T and A in the +2 and +3 positions.
Surprisingly, we did not observe noticeable underrepresentation of “shifty” codons upstream of stop codons that are recognized as terminators. The AAA codon was the second most frequent codon preceding terminator stop codons (Fig. 3b). An example of termination at AAA_TAA is shown in Supplementary Fig. 3a. Therefore, it is clear that whether the ribosome terminates or not at a particular stop codon does not depend solely on the identity of a codon preceding it, and that additional signals should be in place. Examination of information content surrounding frameshift sites and termination sites did not reveal position-specific sequence signals (Fig. 4a). Instead, it appears that the translation machinery senses the end of the mRNA and terminates only at the stop codons close to polyA. This is consistent with Euplotes having very short 3’ UTRs. Some mRNAs require longer 3’UTRs, e.g. selenoprotein mRNAs need to accommodate SECIS elements (Supplementary Fig. 3b). However, the “distance” between the polyA tail and the genuine site of termination could be structural rather than sequence-based such that the SECIS structure could bring the polyA tail close to the position of the termination site. Indeed, we observed highly structured 3’UTRs in all selenoprotein genes and found only a single example of a long 3’UTR other than that coding for selenoproteins (Supplementary Fig. 3c), but even in this case there is a possibility of a functional RNA secondary structure in its 3’UTR.
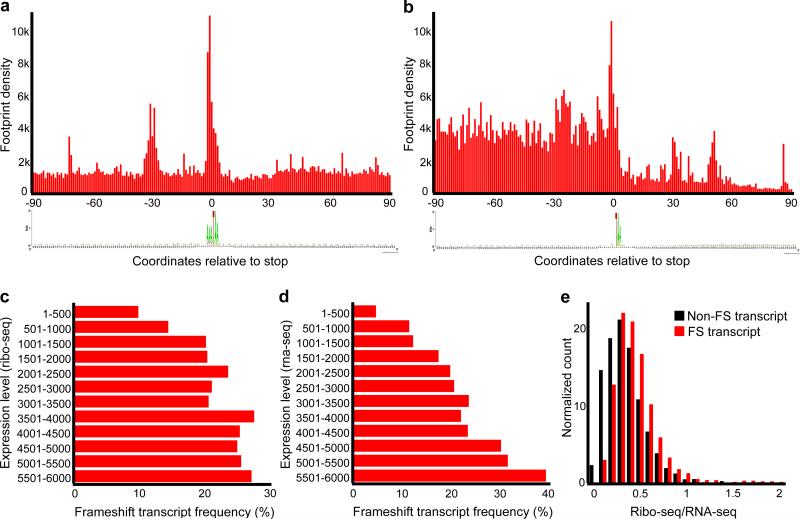
Figure 4. Metagene analysis of ribosome profiling and distribution of frameshifting according to transcript levels.
(a) Metagene analysis of ribosome density in the vicinity of frameshift sites. First nucleotide of a stop codon is shown as zero coordinate. Note that while ribosome density upstream and downstream of frameshift sites is similar, there is a peak of density at the frameshift sites and this is accompanied by another peak 30 nucleotides upstream. A sequence logo below represents the information content of sequences used for metagene alignment. The sequence AAA_TAA is predominant, and there are no other position-specific signals associated with frameshifting. (b) Metagene analysis of ribosome density in the vicinity of translation termination sites. A drop in ribosome density is evident downstream of stop codons. A sequence logo representing information content in the sequences used for metagene analysis is given below. Only mRNAs with 3’UTRs longer than 90 nts (polyA is not included) were used. (c) Frequency of transcripts with the sites of ribosomal frameshifting (axis X) versus the transcripts ranked based on the levels of protein synthesis (Ribo-seq density), axis Y. (d) Similar to (c), but ranking is based on RNA levels (RNA-seq density). (e) Distribution of transcripts with different Ribo-seq to RNA-seq ratios containing frameshift sites (red) and not containing frameshift sites (black).
The effect of frameshifting on gene expression
The high frequency of ribosomal frameshifting in Euplotes suggested that it was not as detrimental as in other organisms. Metagene analysis (Fig. 4a, see Supplementary Fig. 4 for corresponding RNA-seq density) revealed similar ribosome density upstream and downstream of frameshift sites. Therefore, the efficiency of frameshifting was comparable to that of standard decoding. On the other hand, there was a substantial drop of density relative to stop codons identified as termination sites (Fig. 4b). At the same time, a peak of ribosome density was also present about 30 nts upstream of frameshift sites (Fig. 4a), the distance roughly corresponding to the distance between A-sites of the two stacked ribosomes. Such stacking would be expected if ribosomal frameshifting is slower than standard decoding of sense codons. A slight depletion of ribosomes was also observed immediately downstream of the frameshift sites (Fig. 4a). Therefore, it is plausible that while ribosomal frameshifting does not impose considerable costs on the accuracy of synthesized proteins (e.g. AAA_TAA_A would be decoded in the same way as AAA_AAA), there is a cost to the speed of the ribosome and subsequently increased the number of ribosomes per mRNA. In this case frameshifting would be expected to be harmful in genes expressed at high levels.
To test this hypothesis, we explored how frameshifting relates to gene expression levels based on RNA-seq and Ribo-seq signals (Fig. 4c,d). Indeed, we found that frameshifting was less frequent in highly expressed genes, supporting the idea that frameshifting is somewhat harmful in highly expressed genes. However, when we measured frequency of frameshifting in genes with different translation efficiency (TE) measured as the ratio of Ribo-seq signal to RNA-seq signal, we found that frameshifting was more frequent in genes with high TE (Fig. 4e). The ribosome density at any given location is expected to positively correlate with translation initiation rates and anticorrelate with elongation rates at that location. Therefore, while we cannot exclude the possibility that frameshifting is more frequent in genes with high initiation rates, a much more likely explanation is that the high Ribo-seq to RNA-seq ratio in mRNAs expressed with ribosome frameshifting was due to increased ribosome density caused by ribosome pauses and queuing induced by ribosomal frameshifting.
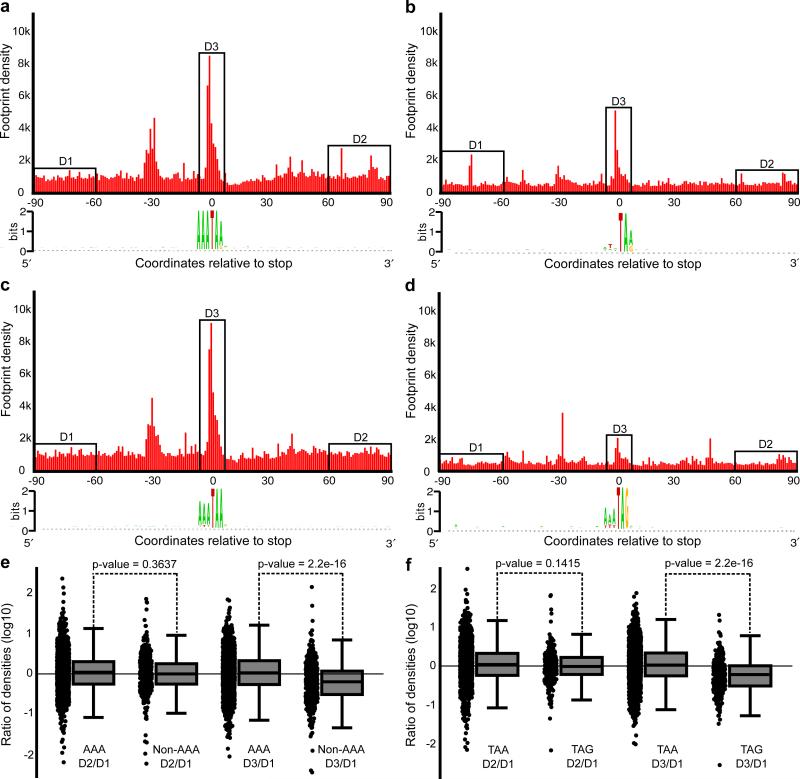
Since we found that particular codons are the most frequent at the frameshifting sites (mononucleotide and AT-rich with AAA being overrepresented the most), we hypothesized that frameshifting efficiency may vary depending on the identity of a codon upstream of a stop. To verify the hypothesis, we split frameshifting sites on AAA and non-AAA and analyzed the distribution of footprint densities (Fig. 5a,b). It appeared that the ribosome density does not change significantly downstream of frameshifting sites neither for AAA nor for non-AAA frameshifting sites (Fig. 5c), although the pause at non-AAA containing sites is less frequent (Fig. 5e). Why then are AAA codons preferred at frameshifting sites? A possible explanation is that the efficiency of frameshifting at non-AAA codons is context dependent and only efficient frameshifting sites are selected during evolution. While we have not observed a specific nucleotide context associated with non-AAA codons at the frameshifting sites, we noticed that TAG occurs almost three times more frequently (~29%) at non-AAA frameshifting sites than at AAA frameshifting sites (~12%) (Fig. 5a,b). To analyze how TAA and TAG stop codons affect frameshifting we compared footprint densities at the frameshifting sites depending on which stop codon is used (Fig. 5d,e). While we did not find significant difference in a change of density downstream of frameshifting sites, it appeared that the peak of density associated with presumed ribosome pausing at the frameshifting sites was significantly greater for TAA codons than for TAG codons (Fig. 5f).
Figure 5. Comparison of ribosomal frameshifting at AAA vs non-AAA frameshifting sites and TAA vs TAG frameshifting sites.
Aggregated densities of ribosome footprints around frameshift sites containing AAA codon preceding stop (a), non-AAA codons (b), TAA stop codons (d) and TAG stop codons (e). Comparison of footprint density changes observed at frameshift sites at each mRNA (D3 region) and downstream of frameshift sites (D2) relative to footprint density upstream of frameshift sites (D1). D1 and D3 regions were chosen 60 nts upstream and downstream of frameshift sites in order to avoid aberrant densities inflicted by ribosome pauses at frameshifting sites. Box plots represent ratio distributions with horizontal line corresponding to the median, box representing 25th and 75th percentiles and whiskers 5th and 95th percentiles. The comparison was carried out for AAA (n=1368) vs non-AAA (n=397) containing frameshift sites (e) and TAA (n=1488) vs TAG (n=277) containing frameshifting sites (f). P-values were calculated using unpaired Wicloxon rank-sum test on log ratios. The data suggest that the frameshifting efficiencies are similar at all frameshift sites, but strong pauses (D3/D1) are less frequent in non-AAA and TAG containing sites.
Frameshift patterns do not evolve under strong purifying selection
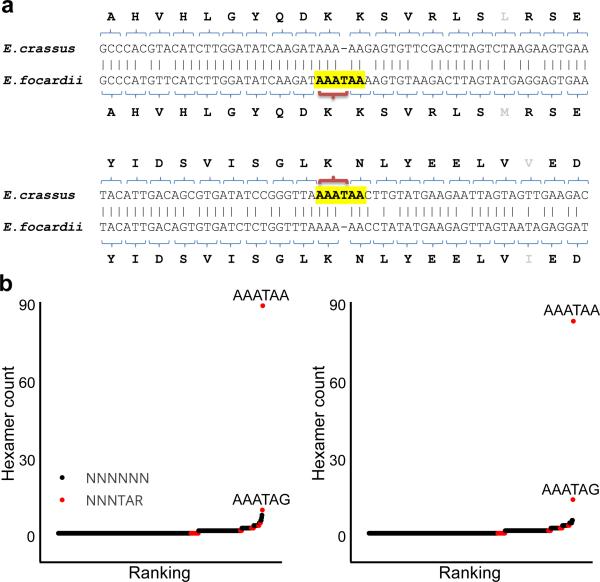
In most well-studied cases of programmed ribosomal frameshifting (e.g. eukaryotic antizymes and bacterial release factor 2), the frameshift sequence and its occurrence are remarkably conserved 18,19. In fact, evolutionary conservation of frameshift patterns is frequently used for the detection of recoded genes 20. In all these cases, the efficiency of frameshifting is below 100%, and two protein products are usually synthesized from the same mRNA, one being decoded according to the rules of standard genetic decoding and another being a product of frameshifting. The ratio between these two products is functionally important and is often tightly regulated1. Therefore, there is a strong evolutionary pressure to preserve the frameshift site and its regulatory capacity, leading to strong stabilizing selection acting on the sequences of frameshift sites and stimulatory signals. In contrast, frameshifting in two Euplotes species was often characterized by cases where only one of the two orthologous sites used frameshifting (a typical example is shown in Fig. 6a). While the amino acid sequences of two orthologous genes were conserved, the corresponding nucleotide sequences differed by a single indel. Thus, frameshifting in Euplotes is not regulatory and the phenotypic difference between gene variants with and without frameshift sites is unlikely to be high.
Figure 6. Cross-species comparison and frequency of nucleotide deletions in different hexamers.
(a) Two typical pairwise alignments containing single nucleotide gaps in one of two orthologous sequences in E. crassus and E. focardii. (b) Frequency analysis of all possible hexamer patterns corresponding to deletions (as highlighted in yellow in a) in pairwise alignments for E. crassus (left) and E. focardii (right). The Y axis shows the frequency of each hexamer found in the pairwise alignments with a gap corresponding to the fourth position of the hexamer. Hexamers that end with either TAA or TAG are shown in red. Two most frequent hexamers, AAATAA and AAATAG, are indicated.
Normally, there is a strong negative selection acting on single nucleotide indels inside protein coding regions due to their dramatic effects on the sequence of synthesized protein. In Euplotes, however, it could be expected that certain indels that likely create an efficient site of ribosomal frameshifting irrespective of nucleotide context (e.g. AAA_AAA to AAA_TAA_A mutation) would have no effect on the sequence of the synthesized protein. Therefore, indels would be expected to evolve under different evolutionary selection depending on where they occur. To explore evolution of indels, we analyzed the frequency of sequences surrounding single nucleotide indels. We generated pairwise alignments of orthologous sequences from the transcriptomes of both species using FASTA 21 and counted occurrences of each hexamer where a gap in the alignment corresponded to the fourth position (from the 5’ end) of the hexamer (highlighted sequence in Fig. 6a). Then, we normalized the frequency of such patterns in gapped alignments to the total number of their occurrence in the two transcriptomes (Fig. 6b,c). The abundance of patterns matching AAATAA was striking (Fig. 6b,c). Indels in the center of the AAATAA pattern were strongly overrepresented in comparison with other patterns in both species, suggesting that frameshifting in Euplotes evolves essentially neutrally to produce AAA-stop frameshifting sites, though this is unlikely to be the case for non-AAA frameshifting sites.
Conclusions
In this work, we provide manifold evidence for the frequent occurrence of ribosomal frameshifting during translation in Euplotes ciliates. Ribosomal frameshifting occurs at the stop codons where tRNAs in the P-site slip forward predominantly either by 1 or 2 nucleotides. The most frequent type of frameshifting is +1 at AAA codons preceding stop; however, frameshifting also occurs at many other sense codons. While this work was under review, a study of two other Euplotes was published where frameshifting sites were predicted based on genomic and transcriptomic sequences 22, supporting our findings. Our analyses further show that ribosomal frameshifting in Euplotes is plastic and rapidly evolves, that it is the predominant process at stop codons and that it has no or low impact on the accuracy of protein synthesis, though it likely affects ribosome speed. Interestingly, sequences that trigger ribosomal frameshifting are also found as genuine termination sites. The data suggest that the function of stop codons as frameshifters or terminators is determined by their proximity to polyA tails and that additional mechanisms are required for efficient termination. Thus, the presence of a stop codon is not a sufficient feature for translation termination in Euplotes. Instead, the default function of stop codons is ribosomal frameshifting. This is consistent with recent findings of reassignment of all stop codons in Condylostoma magnum where stop codons function as terminators only in close proximity to mRNA 3’ ends23,24. A significant evolutionary distance between Euplotes and Condylostoma suggests an intriguing possibility that it may be a general property of ciliate decoding. If so, it may explain high frequency of changes in the genetic code in these species. A degree of positional preference of translation termination in other eukaryotes requires further exploration.
Online Methods
Genome sequencing and assembly
The nucleotide sequence of the E. crassus strain CT5 macronuclear genome was obtained by using a combination of Roche 454 (a total of 2,550,648 reads covering 577,513,019 bp, with an average read length of 236 bp) and Illumina (27,092,578 reads with an average read length of 77 bp, totaling 2,086,128,506 bp) sequencing. The macronuclear genome of E. focardii was generated through Illumina paired-end sequencing (a total of 43,588,788 reads covering 4,402,467,588 bp, with an average read length of 100 bp).
To identify sequences of other organisms within the dataset, we utilized DeconSeq 25. The following datasets were used: bacterial genomes (2,206 unique genomes, 02/12/11), archaeal genomes (155 unique genomes, 02/12/11), Salmonella enterica genomes (52 strains, 12/16/10), bacterial genomes HMP (76,337 WGS sequences, 02/12/11), and viral genomes in RefSeq 45 (3,761 unique sequences, 02/12/11). Whereas very little contamination was observed in E. crassus samples, bacterial sequences were found in E. focardii samples. To filter them out, we applied the following procedure: for E. crassus threshold values were left at default values (80% coverage and 95% identity), whereas for E. focardii they were changed to 50% coverage and 80% identity. Bacterial sequences in the genome data are not unexpected, considering that both ectosymbionts and endosymbionts have been reported in ciliates 26.
Several assembly programs were used to generate independent whole-genome assemblies, including ABYSS 27, SOAP 28, SSAKE 29, Velvet 30, Celera 31, 454 Newbler v.2.7, and PCAP 32,33. To perform the assembly, we followed the instruction manuals for Newbler and Celera and the published protocols for other programs. A hybrid assembly (short reads pre-assembled using Velvet, with the final assembly done using Newbler) was chosen for further analyses (designated as “Newbler” in Supporting data Table 1). The E. crassus genome assembly consisted of 56,588 contigs, with N50 of 1.6 kb. The E. focardii genome assembly consisted of 109,492 contigs, of which 36,663 contigs (59M) were larger than 500 bp with the N50 of 2.1 kb.
Separately, selenoprotein genes were analyzed as described 34. tRNA prediction was carried out using tRNAscan-SE 35 and ARAGORN 36.
Transcriptome analysis
Frozen E. crassus pellets were cryogenically ground in a Biospec bead homogenizer. Cell powder was lysed in 1 ml of lysis buffer (20 mM Tris-HCl, pH 7.5, 140 mM KCl, 10 mM MgCl2, 0.25% Triton, 100 mg/l cycloheximide, protease inhibitors from Roche). Lysate was loaded on a 2 ml cushion of 1 M sucrose in 20 mM Tris-HCl, pH 7.5, 140 mM KCl, 5 mM MgCl2, 100 mg/l cycloheximide). Samples were centrifuged for 2 h at 45,000 rpm in a SW55 rotor. Pellets were recovered and resuspended in lysis buffer, and then incubated for 1 h with 750 U of RNAse I (Ambion) per 30 U of lysate (measured at A260). Following RNA digestion, sequencing libraries were prepared as described 37, starting with gradient ultracentrifugation. There were several additional changes to the procedure. Instead of polyadenylation, we attached a 3′ adapter (IDT, miRNA linker #1) as a handle for subsequent reverse transcription step using T4 RNA ligase 2 (NEB). The reverse transcription primer was changed accordingly: (5’-GATCGTCGGACTGTAGAACTCTGAACCTGTCGGTGGTCGCCGTATCATT/iSp18/CAAGCAGAAGACGGCATACGAATTGATGGTGCCTACAG-3′), which allowed us to keep the 3′ ends of footprints unperturbed. The following are the sequences of forward and reverse primers for the final PCR: CAAGCAGAAGACGGCATACGA and AATGATACGGCGACCACCGA. Sequencing was performed on an Illumina HiSeq2000 platform. The transcriptome assembly was carried out using de novo assembler Trinity 38, producing 33,701 unique transcripts.
Identification of frameshift sites
Sequences of ribosome footprint cDNAs (Ribo-seq) from E. crassus obtained in three replicates were aggregated producing 9,620,943 reads. They were aligned to the transcriptome using Bowtie software v.0.12.839 allowing ambiguous mapping and up to 3 mismatches per read (-v 3). 8,353,221of reads (86.2%) were aligned to the transcriptome. The Integrative Genomics Viewer (IGV) 40 was used to visualize reads aligned to each transcript. Using IGV we visually analyzed all transcripts where the number of mapped footprints was ≥ 100 reads. Supplementary Note 4 shows examples of IGV screenshots in the vicinity of frameshifting sites whose productive translation was directly supported by peptides matching mass spectra (shown in Supplementary Note 1b). The obtained alignments were used to determine the boundaries of translated segment within a transcript. Frameshift sites were identified by analyzing ORF organization within the translated region at internal stop codons using maximum parsimony as a guiding principle in determining the direction of frameshifting to yield the minimal number of frameshift sites per transcript in most cases. Transcripts with frameshift sites were aligned to corresponding genomic contigs to verify sequence identity and avoid misinterpretation of indel sequencing errors as ribosomal frameshifting sites.
Proteomic and Ribo-Seq analyses
Proteomics analysis employed conventional shotgun bottom-up approach described elsewhere41-43. Briefly, cells were resuspended in the lysis buffer (50 mM Tris-HCl pH 8.0, 8 M urea, 10 mM DTT, 1 mM EDTA), pulverized in liquid nitrogen followed by melting and sonication in a water bath for 1 min. The proteins were then digested using trypsin (samples 1 and 2) and Glu-C (sample 3, pH 7.5), followed by fractionation by SCX (trypsin sample, 25 fractions collected) and High-pH RP (trypsin and Glu-C samples, 24 concatenated fractions collected 44). Analysis by liquid chromatography coupled with LTQ Orbitrap (Thermo Fisher, San Jose, CA) mass spectrometry (LC-MS/MS) was performed using a 100 min LC gradient. The details on the gradient and mass spectrometer settings can be found elsewhere41. The data were pre-processed with DeconMSn 45 and DtaRefinery 46 tools, and analyzed using MS-GF+47. The raw, peak lists and MS/MS identification files were deposited at PRIDE (dx.doi.org/10.6019/PXD004333). Amongst the all peptide identifications, we retained only those that uniquely matched protein sequences originating from the frameshift events. The tolerances on parent ion mass measurement and MS/MS spectrum matching scores were optimized to achieve maximum number of identifications while not exceeding false discovery rate of 5%. Spectra for peptides spanning the frameshift locations were manually verified. The details on MS/MS data analysis along with parameter files and executable document reproducing all the post-search analysis steps were deposited as an R package at GitHub https://github.com/vladpetyuk/EuplotesCrassus.proteome.
For Ribo-Seq analysis, frozen E. crassus pellets were cryogenically ground in a Biospec bead homogenizer. Pellets were recovered and resuspended in lysis buffer, and then incubated for 1 h with 750 U of RNAse I (Ambion) per 30 U of lysate (measured at A260). Following RNA digestion, sequencing libraries were prepared as described 37, starting with gradient ultracentrifugation. Sequencing was performed on an Illumina HiSeq2000 platform.
E. crassus genome and transcriptome sequences were used as references for read alignments. The alignments were generated using Bowtie software v.0.12.7 39; up to two mismatches per read were allowed. We estimated positions of the ribosome A-sites with an offset of 15 nucleotides downstream of 5’ ends of Ribo-seq data. Visualization and further manual analysis were conducted by using SAMtools package 48, custom scripts and IGV 40.
Sequence patterns analysis
To analyze for frequency of indels that occurred since E. crassus and E. focardii split from their common ancestor we generated a set of pairwise alignments using FASTA 21. The alignments were generated by searching E. crassus sequences as query against E. focardii and also in a reverse order. The sequence pairs with the best scores were considered as true orthologous sequences and were used in further analysis. To minimize the potential effect from misalignments, or highly diverged sequence pairs, only those indels were analyzed that occurred exactly in the center of a 41-nucleotide stretch of the alignment containing no other indels. For each gap a hexamer pattern was registered whose fourth position (counting from the 5’ end) corresponds to a gap in the alignment, e.g. PPPPPP pattern in the schematic alignment below
NNPPPPPPNN
NNNNN-NNNN
The observed-to-expected ratio of deletions in hexamers was calculated as the following
where gi is the number of gaps corresponding to pattern i and fi is the number of patterns i in the fraction of the genome predicted as coding.
Statistics
For the data shown in Figure 5 to estimate statistical significance between distributions of changes in footprint densities downstream of, upstream of and at the frameshifting sites. log(D2/D1) and log(D3/D1) we used Wilcoxon rank test. The exact p-values and degrees of freedom are provided in figure legend.
Supplementary Material
Acknowledgements
Supported by NIH GM061603 to V.N.G. S.M.H. and P.V.B. are supported by the grants from Wellcome Trust [094423] and Science Foundation Ireland [12/IA/1335]. CM acknowledges the Italian PNRA and the COST action BM1102 for supporting a part of this work.
Footnotes
Author Contributions. A.V.L., S.M.H., P.V.B. and V.N.G. analyzed the data and wrote the paper with advice of D.L.H. and J.F.A.; A.A.T. and M.V.G. prepared samples for sequencing; S.P., R.R.D., C.M. and L.A.K. performed cell culture maintenance and growth, F.X., V.A.P., and R.D.S. conducted MS analysis. All authors discussed the results and implications and commented on the manuscript at all stages.
Accession Codes. PRJNA329413; SAMN05412464; SRP078897; PRJNA329414; SAMN05412809; SRP078901; MJUV00000000; MECR00000000; PXD004333;.
Data availability. Sequence data that support the findings of this study have been deposited in the following repositories: for E. crassus (BioProject: PRJNA329413; BioSample: SAMN05412464; SRA: SRP078897) and for E. focardii (BioProject: PRJNA329414; BioSample: SAMN05412809; SRA: SRP078901). Proteomics data were deposited to PRIDE (PXD004333) The interpretations of sequence data, such as coordinates of frameshifting sites are available upon request.
Author Information. The authors declare no competing financial interests.
References
- 1.Atkins JF, Loughran G, Bhatt PR, Firth AE, Baranov PV. Ribosomal Frameshifting and Transcriptional slippage: from genetic steganography & cryptography to adventitious use. Nucl Acids Res. 2016 doi: 10.1093/nar/gkw530. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baranov PV, Atkins JF, Yordanova MM. Augmented genetic decoding: global, local and temporal alterations of decoding processes and codon meaning. Nature Rev Genetics. 2015;16:517–529. doi: 10.1038/nrg3963. [DOI] [PubMed] [Google Scholar]
- 3.Belcourt MF, Farabaugh PJ. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 2009;139:193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klobutcher LA, Farabaugh PJ. Shifty ciliates: frequent programmed translational frameshifting in euplotids. Cell. 2002;111:763–766. doi: 10.1016/s0092-8674(02)01138-8. [DOI] [PubMed] [Google Scholar]
- 6.Vallabhaneni H, Fan-Minogue H, Bedwell DM, Farabaugh PJ. Connection between stop codon reassignment and frequent use of shifty stop frameshifting. RNA. 2009;15:889–897. doi: 10.1261/rna.1508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valbonesi A, Luporini P. Biology of Euplotes focardii, an Antarctic ciliate. Polar Biology. 1993;13:489–493. [Google Scholar]
- 8.Pucciarelli S, et al. Molecular cold-adaptation of protein function and gene regulation: The case for comparative genomic analyses in marine ciliated protozoa. Marine Genomics. 2009;2:57–66. doi: 10.1016/j.margen.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Baird SE, Klobutcher LA. Differential DNA amplification and copy number control in the hypotrichous ciliate Euplotes crassus. J Protozool. 1991;38:136–140. doi: 10.1111/j.1550-7408.1991.tb06033.x. [DOI] [PubMed] [Google Scholar]
- 10.Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong LC, Landweber LF. Evolution of programmed DNA rearrangements in a scrambled gene. Mol Biol Evol. 2006;23:756–763. doi: 10.1093/molbev/msj089. [DOI] [PubMed] [Google Scholar]
- 12.Swart EC, et al. The Oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16,000 tiny chromosomes. PLoS Biol. 2013;11:e1001473. doi: 10.1371/journal.pbio.1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ricard G, et al. Macronuclear genome structure of the ciliate Nyctotherus ovalis: single-gene chromosomes and tiny introns. BMC Genomics. 2008;9:587. doi: 10.1186/1471-2164-9-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vinogradov DV, et al. [Draft macronuclear genome of a ciliate Euplotes crassus]. Molekuliarnaia biologiia. 2012;46:361–366. [PubMed] [Google Scholar]
- 15.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. doi:10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingolia NT. Ribosome profiling: new views of translation, from single codons to genome scale. Nature reviews. Genetics. 2014;15:205–213. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- 17.Michel AM, Baranov PV. Ribosome profiling: a Hi-Def monitor for protein synthesis at the genome-wide scale. RNA. 2013;4:473–490. doi: 10.1002/wrna.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baranov PV, Gesteland RF, Atkins JF. Release factor 2 frameshifting sites in different bacteria. EMBO Rep. 2002;3:373–377. doi: 10.1093/embo-reports/kvf065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanov IP, Atkins JF. Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: close to 300 cases reveal remarkable diversity despite underlying conservation. Nucl Acids Res. 2007;35:1842–1858. doi: 10.1093/nar/gkm035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma V, et al. A pilot study of bacterial genes with disrupted ORFs reveals a surprising profusion of protein sequence recoding mediated by ribosomal frameshifting and transcriptional realignment. Mol Biol Evol. 2011;28:3195–3211. doi: 10.1093/molbev/msr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson W. Finding protein and nucleotide similarities with FASTA. Curr Protol Bioinf. 2004 doi: 10.1002/0471250953.bi0309s04. Chapter 3, Unit3 9. [DOI] [PubMed] [Google Scholar]
- 22.Wang R, Xiong J, Wang W, Miao W, Liang A. High frequency of +1 programmed ribosomal frameshifting in Euplotes octocarinatus. Sci Rep. 2016;6:21139. doi: 10.1038/srep21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heaphy SM, Mariotti M, Gladyshev VN, Atkins JF, Baranov PV. Novel ciliate genetic code variants including the reassignment of all three stop codons to sense codons in C. magnum. 2016 doi: 10.1093/molbev/msw166. under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swart EC, Serra V, Petroni G, Nowacki M. Genetic codes with no dedicated stop codon: context-dependent translation termination. Cell. 2016;166:691–702. doi: 10.1016/j.cell.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 25.Schmieder R, Edwards R. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PloS One. 2011;6:e17288. doi: 10.1371/journal.pone.0017288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dziallas C, Allgaier M, Monaghan MT, Grossart HP. Act together-implications of symbioses in aquatic ciliates. Front Microbiol. 2012;3:288. doi: 10.3389/fmicb.2012.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simpson JT, et al. ABySS: a parallel assembler for short read sequence data. Genome research. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo R, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warren RL, Sutton GG, Jones SJ, Holt RA. Assembling millions of short DNA sequences using SSAKE. Bioinformatics. 2007;23:500–501. doi: 10.1093/bioinformatics/btl629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myers EW, et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 32.Huang X, Wang J, Aluru S, Yang SP, Hillier L. PCAP: a whole-genome assembly program. Genome research. 2003;13:2164–2170. doi: 10.1101/gr.1390403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang X, Yang SP. Generating a genome assembly with PCAP. Current protocols in bioinformatics / editoral board, Andreas D. Baxevanis ... [et al.] 2005 doi: 10.1002/0471250953.bi1103s11. Chapter 11, Unit11 13. [DOI] [PubMed] [Google Scholar]
- 34.Turanov AA, et al. Genetic code supports targeted insertion of two amino acids by one codon. Science. 2009;323:259–261. doi: 10.1126/science.1164748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucl Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerashchenko MV, Lobanov AV, Gladyshev VN. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc Natl Acad Sci USA. 2012;109:17394–17399. doi: 10.1073/pnas.1120799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature Protocols. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Depuydt G, et al. Reduced insulin/insulin-like growth factor-1 signaling and dietary restriction inhibit translation but preserve muscle mass in Caenorhabditis elegans. Mol Cell Prot. 2013;12:3624–3639. doi: 10.1074/mcp.M113.027383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Depuydt G, et al. LC-MS proteomics analysis of the insulin/IGF-1-deficient Caenorhabditis elegans daf-2(e1370) mutant reveals extensive restructuring of intermediary metabolism. J Proteome Res. 2014;13:1938–1956. doi: 10.1021/pr401081b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petyuk VA, et al. Characterization of the mouse pancreatic islet proteome and comparative analysis with other mouse tissues. J Proteome Res. 2008;7:3114–3126. doi: 10.1021/pr800205b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang F, Shen Y, Camp DG, 2nd, Smith RD. High-pH reversed-phase chromatography with fraction concatenation for 2D proteomic analysis. Exp Rev Prot. 2012;9:129–134. doi: 10.1586/epr.12.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayampurath AM, et al. DeconMSn: a software tool for accurate parent ion monoisotopic mass determination for tandem mass spectra. Bioinformatics. 2008;24:1021–1023. doi: 10.1093/bioinformatics/btn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petyuk VA, et al. DtaRefinery, a software tool for elimination of systematic errors from parent ion mass measurements in tandem mass spectra data sets. Mol Cell Prot. 2010;9:486–496. doi: 10.1074/mcp.M900217-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim S, Pevzner PA. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.