Abstract
Programmed Death-1 (PD-1) is a co-inhibitory receptor that down-regulates the activity of tumor-infiltrating lymphocytes (TIL) in cancer and of virus-specific T cells in chronic infection. The molecular mechanisms driving high PD-1 expression on TIL have not been fully investigated. We demonstrate that transforming growth factor-β1 (TGF-β1) directly enhances antigen-induced PD-1 expression through Smad3-dependent, Smad2-independent transcriptional activation in T cells in vitro and in TIL in vivo. The PD-1hi subset seen in CD8+ TIL is absent in Smad3-deficient tumor-specific CD8+ TIL, resulting in enhanced cytokine production by TIL and in draining lymph nodes and of anti-tumor activity. In addition to TGF-β1’s previously known effects on T cell function, our findings suggest that TGF-β1 mediates T cell suppression via PD-1 upregulation in the TME. They highlight bidirectional crosstalk between effector TIL and TGF-β-producing cells that upregulates multiple components of the PD-1 signaling pathway to inhibit anti-tumor immunity.
Keywords: TGF-β1, Smad3, programmed death-1 (PD-1), tumor infiltrating lymphocytes (TIL), tumor microenvironment (TME)
Introduction
Programmed death-1 (PD-1; known as Pdcd-1 by gene name) is a co-inhibitory receptor induced on T cells by antigenic stimulation (1). PD-1 expression on functional memory CD8+ T cells declines upon the resolution of inflammation and the clearance of antigen during acute infections (2). Conversely, PD-1 expression is maintained on exhausted T cells in chronic infections. In cancer, the PD-1 pathway is highly engaged within the TME, with tumor and immune system cells expressing high levels of the PD-1 ligands, PD-L1 (also known as B7-H1) and PD-L2 (also known as B7-DC), and tumor-infiltrating CD4+ and CD8+ T cells expressing high levels of PD-1 (3,4). Blockade of PD-1 has been effective in prolonging patient survival in melanoma, renal-cell carcinoma (RCC), non-small-cell lung cancers (NSCLC), Hodgkin’s Lymphoma and many other cancer types (5-8). Similarly, chronic infection with hepatitis C virus (HCV), hepatitis B virus (HBV) or human immunodeficiency virus (HIV) sustains high levels of PD-1 on viral-specific CD8+ T cells (9-11).
Binding of PD-1 on T cells to its ligands, PD-L1 and PD-L2, can inhibit T cell effector function (12). Pathogen- or tumor-driven inflammation can induce PD-L1 and –L2 expression. For example, PD-L1 is highly expressed on many human tumors (4,13) and its expression is highly co-localized with infiltrating CD8+ T cells in human melanoma patients (14). Similarly, patients with chronic liver disease from HCV and HBV infection also show increased levels of PD-L1 on hepatocytes and Kupffer cells in the liver (15). Elevated PD-L1 and –L2 expression may enhance engagement of PD-1 on T cells and pathogen evasion of host immune responses (4,16-19). The levels of PD-1 on TIL subsets in many cancers are much higher than those seen on normally activated or memory T cells in peripheral blood or in corresponding normal tissue (20). This induction of receptor, together with ligand up-regulation, is likely responsible for the profound inhibition of effector anti-tumor T cell activity in the TME. While interferon-γ (IFN-γ), a T cell effector cytokine, is known to enhance PD-L1 expression on tumor cells (13) and some cytokines have been shown previously to affect T cell expression of PD-1 (21,22), the molecular mechanisms that permit expression of PD-1 on human T cells at very high levels have not been fully elucidated. This is critical to our understanding of PD-1 inhibition of T cell control of tumors or chronic viral infections and modulation of that pathway through immunotherapy.
As part of a cytokine screen to identify those that regulate PD-1 induction on T cells, we found that transforming growth factor-β1 (TGF-β1) modified antigen-driven PD-1 induction to the greatest extent. TGF-β1 is a regulatory cytokine that suppresses immune function in cancers and in chronic viral infections (23-26). The Smad transcription factors transduce signals from TGF-β superfamily ligands that regulate cell proliferation, differentiation, and death through activation of receptor serine/threonine kinases. High serum levels of TGF-β are associated with poor prognosis in cancer (27,28) and TME-derived TGF-β can suppress anti-tumor T cell responses (29,30). Accordingly, the blockade of TGF-β1 signaling on T cells has been effective in restoring T cell effector functions (31,32). The known suppressive mechanisms of TGF-β1 include Smad2/3 dependent inhibition of effector cytokine production by CD8+ T cells in cancer (33) and development of CD4+ regulatory T cells (Tregs) that suppress neighboring effector cells through both contact-independent and - dependent mechanisms (34,35).
Here, we report a novel molecular mechanism of immunosuppression in which TGF-β directly enhances antigen-driven PD-1 gene transcription selectively through Smad3, resulting in enhanced surface expression of PD-1 protein. Utilizing mice with T cells conditionally deleted of Smad2 or Smad3, we found that TGF-β1-enhanced PD-1 expression is abrogated in Smad3-deficent T cells. In contrast, Smad2-deficient T cells expressed PD-1 at levels comparable to wild-type (WT) mice. This suggests that enhanced PD-1 expression on T cells is predominantly regulated by Smad3. The effect of Smad3 was specific to PD-1 since expression of other inhibitory receptors was not decreased by Smad3 deficiency. Mice with Smad3 deficient T cells more effectively controlled tumors in association with loss of the subset of antigen-specific TIL displaying the highest levels of PD-1 and increased TIL and draining lymph node (DLN) cytokine production. PD-1 blockade did not provide further anti-tumor activity beyond that produced by T cell-specific Smad3 knockout, demonstrating that PD-1 induction by the TGF-β1/Smad3 axis is critical in suppressing anti-tumor T cell function. Thus, our findings suggest that TME-derived TGF-β1 directly augments PD-1 expression on TIL, suppressing CD8+ T cells that engage tumor antigens and enhancing tumor immune resistance.
Results
TGF-β1 enhances PD-1 expression on activated human T cells
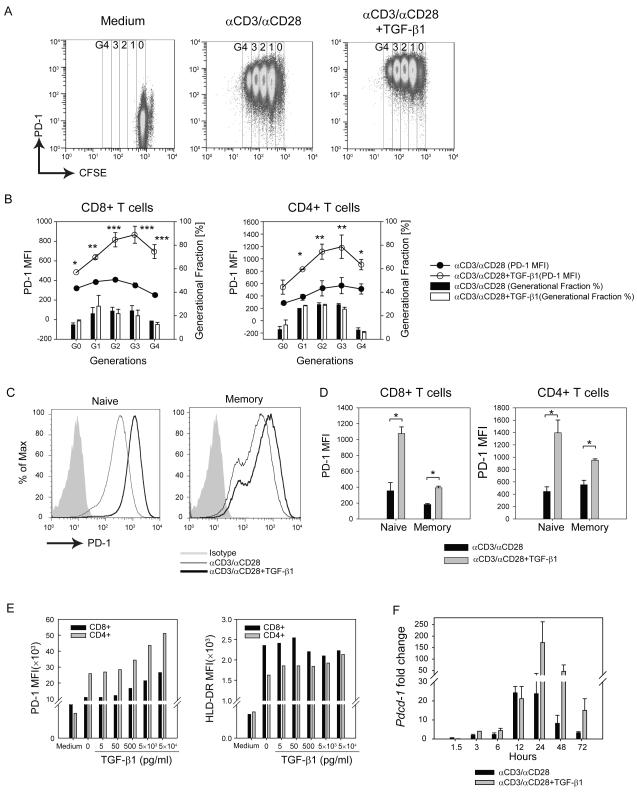
To assess the effects of cytokines known to alter T cell development, function, and/or proliferation on PD-1 expression, we isolated CD3+ T cells from healthy donor peripheral blood mononuclear cells (PBMC) and activated them with αCD3/αCD28-conjugated beads in the presence of one of 16 cytokines across a range of concentrations (Supplementary Figure S1a, data shown at 500 ng/mL). The cells were labeled with CFSE to monitor cellular proliferation and PD-1 expression was measured (Figure 1a, representative plots). αCD3/αCD28 induces higher PD-1 expression compared to resting CD8+ and CD4+ T cells, confirming TCR and co-stimulation dependent PD-1 expression (Figure 1a, left and middle graphs). While most of the cytokines tested had no effect or only a modest effect on PD-1 expression upon T cell activation, major enhancement of PD-1 expression was observed with TGF-β1 (Figure 1a middle and right graphs and Supplementary Figure S1a). In conjunction with T cell stimulation, interleukin (IL)-2, IL-6, IL-12, and TNFα induced only modest enhancement of αCD3/αCD28-induced PD-1 expression (Supplementary Figure S1a). Because increased TGF-β 1production is a hallmark of most TME, we chose to further explore its role in PD-1 expression. The co-culture of T cells with TGF-β1 further enhanced PD-1 expression on both CD8+ (Figure 1b, left panel; open symbols) and CD4+ (Figure 1b, right panel; open symbols) T cells versus αCD3/αCD28 alone (Figure 1b, closed symbols) on all generations (Figure 1b). TGF-β1 did not have any effects on cellular proliferation as measured by CFSE dilution (Figure 1b, black versus open bar graph), suggesting that enhanced PD-1 expression is not simply due to altered cellular proliferation.
Figure 1.
TGF-β1 enhances PD-1 expression on human T cells in a dose-dependent manner.
Human CD3+ T cells were isolated from healthy donor PBMCs and were activated with αCD3/αCD28-conjugated beads with or without TGF-β1 (50 ng/ml). (a) Representative plots PD-1 (Y-axis) vs. CFSE (X-axis) are shown for different conditions. (b) PD-1 MFI is shown as filled circle lines (αCD3/αCD28) and open-circle lines (αCD3/αCD28 + TGF-β1). The percentage of cells in each CFSE generation (G0, G1, G2, G3 and G4) is shown as black bar graphs (αCD3/αCD28) and white bar graphs (αCD3/αCD28 + TGF-β1). The data represent combined results of two independent trials. (c) Naïve and memory subset of T cells (CD4+ and CD8+ T cells) were isolated based on CCR7 and CD45RA expression and treated with αCD3/αCD28 activation in the presence or absence of TGF-β1. The representative histogram of PD-1 is shown: shaded histogram (Isotype); thin histogram (αCD3/αCD28); and bold histogram (αCD3/αCD28+TGF-β1). (d) PD-1 MFI was assessed on each subset (X-axis) of CD8+ (left) and CD4+ (right) T cells; αCD3/αCD28 alone (black bars); αCD3/αCD28 with TGF-β1 (grey bars). The data represent combined results of two independent trials. (e) Isolated human CD3+ T cells were activated with αCD3/αCD28-conjugated beads in the presence of varying concentrations of TGF-β1 (5 to 5 × 104 pg/ml). Mean fluorescent intensity (MFI) of PD-1 (left) and HLA-DR (right) expressions were assessed in CD4+ (grey bars) and CD8+ (black bars) T cells. The shown result is the representative of at least three independent trials. (f) Pdcd-1 transcript levels of human CD3+ T cells under different treatments were normalized to that of resting CD3+ T cells. The result is shown as mean +/− SEM of technical replicates and representative of at least three independent trials. The data were analyzed using Student’s t-test and considered significant if *P<0.05, **P<0.01, ***P<0.001.
While human memory T cell populations such as CMV and EBV-specific T cells express intermediate levels of PD-1, naïve T cells do not express PD-1 (36). To test whether TGF-β1-mediated enhancement of PD-1 expression depends on the basal level of PD-1 expression, we isolated naïve T cells (phenotype CCR7+ CD45RA+) and memory T cells (phenotype CCR7+ CD45RA− or CCR7− CD45RA+) from healthy donors. The cells were activated with αCD3/αCD28-conjugated beads with or without TGF-β1. Although TGF-β1 increased PD-1 expression on αCD3/αCD28 stimulated naive and memory CD4+ and CD8+ T cells (Figure 1c, representative plots), the effect was more pronounced on naïve T cells than on memory T cells for both CD4 and CD8 subsets (Figure 1d, dark and light grey bars). In the absence of αCD3/αCD28, TGF-β1 does not affect the basal levels of PD-1 expression on either naïve or memory T cell subsets (data not shown). This suggests that TGF-β1 enhancement of PD-1 expression is dependent on T cell activation. Furthermore, we found that TGF-β1 increased PD-1 surface expression in a concentration-dependent manner (Figure 1e, left). In contrast, TGF-β1 did not affect expression of the T cell activation marker HLA-DR on T cells (Figure 1e, right), demonstrating that the TGF-β1 effect on PD-1 expression does not simply reflect a general effect on T cell activation-induced antigens. Changes in intracellular and surface levels of PD-1 were positively and directly correlated (Supplemental Figure S1b). Finally, enhanced surface expression of PD-1 was preceded by increased transcription of PD-1, shown as kinetic changes of Pdcd-1 mRNA levels across different time points (Figure 1f).
TGF-β receptor I (TGF-βRI) kinase activity is critical for TGF-β-dependent enhancement of PD-1 expression
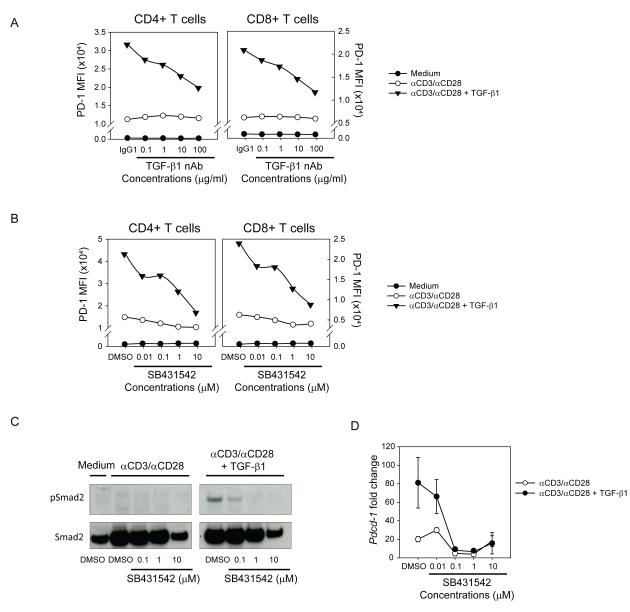
Next, we investigated whether blockade of TGF-β1 signaling can abrogate TGF-β-dependent PD-1 enhancement. TGF-β1 binds TGF-βRI and RII and acts through Smad-dependent and -independent mechanisms (37). Upon binding of the high affinity TGF-βRII by TGF-β1, TGF-βRI and RII heterodimerize and TGF-βRI, a serine-threonine kinase, phosphorylates Smad2/3. To address the role of TGF-β1 receptor signaling, the cells were activated in the presence of TGF-β1 with varying concentrations of either an antibody that blocks the activity of TGF-β1 but not TGF-β2 or TGF-β3 (neutralizing antibody, nAb) (Figure 2a) or a TGF-βRI kinase inhibitor (SB431542) (Figure 2b). Both TGF-β1 nAb and SB431542 decreased TGF-β1-dependent PD-1 expression in a dose-dependent manner, although SB431542 was more effective than TGF-β1 nAb. SB431542-mediated TGF-βR signaling inhibition was shown by the diminished phosphorylation levels of Smad2 (Figure 2c). Analogous to the effects on surface expression, SB431542 blocked the TGF-β1-dependent increase of Pdcd-1 mRNA levels (Figure 2d). Given the critical role of nuclear factor of activated T cell (NFATc1) during TCR dependent PD-1 induction (38), we tested whether TGF-β1-dependent PD-1 expression requires NFATc1 by treating cells with cyclosporine A (CsA), a calcineurin inhibitor that exerts its immunosuppressive effects by keeping the transcription factor NFATc1 inactive. We found that CsA completely abrogated not only TCR-dependent but also TGF-β1 enhanced PD-1 expression (Supplementary Figure S1c), suggesting that TGF-β1 requires TCR-induced NFATc1 activity to enhance the PD-1 expression. Based on the critical role of TGF-βR kinase activity, we next assessed downstream molecules in the TGF-βR signaling cascade.
Figure 2.
Anti-TGF-β1 neutralizing antibody and a TGF-βRI kinase inhibitor negate TGF-β1 mediated PD-1 enhancement
(a, b) Enriched human CD3+ T cells were activated with αCD3/αCD28-conjugated beads and TGF-β1 under varying concentrations of TGF-β1 nAb (a) or SB431542 (b) and PD-1 MFI was assessed: medium alone (closed circles) αCD3/αCD28 only (open circles); αCD3/αCD28 + TGF-β1 (closed triangles). The result shown is representative of at least three independent trials. (c) Western-blot analysis of phosphorylated-Smad2 (pSmad2) in human CD3+ T cells treated with varying concentrations of TGF-βRI kinase inhibitor (SB431542). (d) Enriched human CD3+ T cells were activated with αCD3/αCD28-conjugated beads and TGF-β1 under increasing concentrations SB431542. Pdcd-1 transcript levels in each condition were normalized to that of resting human CD3+ T cells. The result is shown as mean +/− SEM of technical replicates and representative of at least three independent trials.
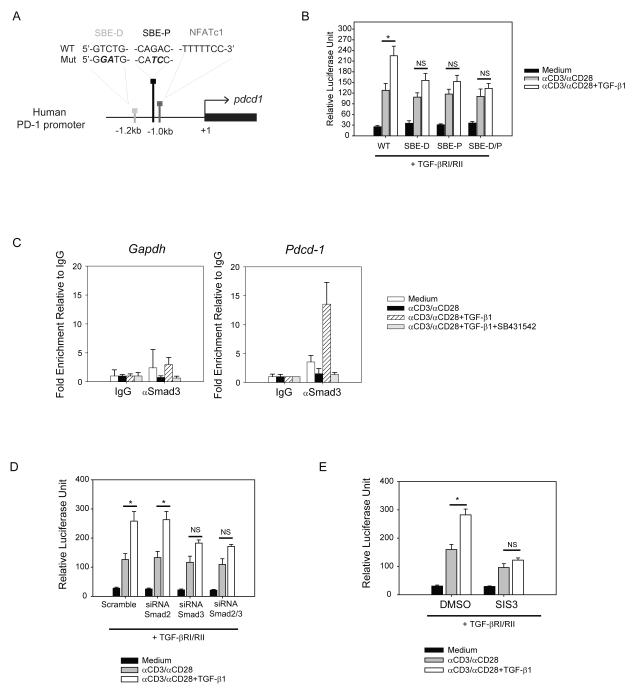
TGF-β1-dependent Smad3 regulates PD-1 promoter activity
Our data suggest that human PD-1 expression is under direct transcriptional control by TGF-β1 so we hypothesized that TGF-β1 directly modulates human PD-1 promoter activity. We identified putative Smad-binding elements (SBEs), one distal to (SBE-D) and the other proximal to (SBE-P) the Pdcd-1 transcription start site (Figure 3a). To test this hypothesis, Jurkat T cells were transfected with a luciferase vector containing the 1.9kb-long Pdcd-1 promoter region and luciferase activity was measured after different treatments. αCD3/αCD28 activation induced PD-1 promoter activity (Figure 3b, grey bars) and mutation in the NFATc1 binding site abrogated such induction (Supplementary Figure S1d, e). Because Jurkat T cells express minimal levels of the TGF-β receptors, the T cells were co-transfected with TGF-βRI and RII plasmids (Supplementary Figure S1f and S1g). The addition of TGF-β1 to αCD3/αCD28 enhanced NFATc1-dependent PD-1 promoter activity (Figure 3b WT and Supplementary Figure S1f). The introduction of site-directed mutations in SBEs (shown in bold letters in Figure 3a, named SBE-D and SBE-P) significantly diminished TGF-β1-driven PD-1 promoter activity and introduction of both mutations (SBE-D/P) further decreased the effect (Figure 3b). To further validate our luciferase-based reporter system, we utilized the chromatin immunoprecipitation (ChIP) assay to verify Smad3 binding to the human Pdcd-1 promoter. While αCD3/αCD28 did not induce Smad3 binding, addition of TGF-β1 significantly enhanced Smad3 binding to the human Pdcd-1 promoter (Figure 3c, right graph). This binding was specifically due to TGF-β1 receptor signaling as it was abrogated by treatment with the TGF-βRI kinase inhibitor, SB431542 (Figure 3c, right graph). There was no effect of TGF-β1 on binding of Smad3 to the Gapdh promoter (Figure 3c, left graph). We also confirmed that NFATc1 binds to the human Pdcd-1 promoter following αCD3/αCD28 stimulation and found that this binding was in fact enhanced by TGF-β1 (Supplementary Figure S1h).
Figure 3.
Smad3 directly binds to the Smad-binding elements (SBEs) and regulates PD-1 promoter activity
(a) Schematic illustration of the proximal region of human Pdcd-1 promoter. Two Smad-binding-elements (SBEs) are located at 1.2 kb (SBE-D) and 1.0 kb (SBE-P) upstream of the Pdcd-1 transcription start site. NFATc1 consensus sequence is located in immediate proximity to SBE-P. Wild-type (WT) and mutated (Mut) sequences of both SBE-D and SBE-P are shown. (b) Jurkat T cells were transfected with luciferase reporter vectors containing wild-type (WT), mutant SBE-D, mutant SBE-P, and mutant SBE-D/P sequences of PD-1 promoter (1.9 kb). After co-transfection with TGF-βRI and RII expression plasmids, the cells were activated with plate-bound αCD3 and soluble αCD28 in the absence (grey bars) or presence (white bars) of TGF-β1 (50 ng/ml). Luciferase activity was measured as described in the method. (c) Isolated human CD3+ T cells were activated under different conditions: medium alone (white bars); αCD3/αCD28 alone (black bars); αCD3/αCD28 with TGF-β1 (hatched bars); αCD3/αCD28 + TGF-β1 with SB431542 (grey bars). Immunoprecipitated DNA was subjected for qPCR and fold enrichment of binding relative to IgG is shown as mean +/− SEM of triplicate results. (d) Jurkat T cells were co-transfected with 1.5 μM of siRNA against Smad2 or Smad3 and PD-1 promoter-driven luciferase activity was measured in relative luciferase units (ratio of firefly to renilla luciferase activity). (e) Transfected Jurkat T cells were treated with 10μM of specific inhibitor of Smad3 (SIS3) and PD-1 promoter-driven luciferase activity was measured after activation. The results are shown as mean +/− SEM of technical replicates and is representative of at least three independent trials. The data were analyzed using two-way analysis of variance (ANOVA) and considered significant if *p<0.05.
TGF-βR1 has serine/threonine kinase activity that phosphorylates Smad2 and Smad3 (39). Smad2 and Smad3 bifurcate the signaling pathway by forming heterodimers with Smad4 (considered a co-Smad) (40,41). Thus, we further investigated whether Smad2 or Smad3 is the dominant regulator of PD-1 promoter activity by using siRNA (Supplementary Figure S2a, b). We found that knock-down of Smad3 expression (but not Smad2) abrogated TGF-β1 enhancement of Pdcd-1 promoter activity (Figure 3d). In addition, we tested whether specific inhibitor of Smad3 (SIS3), that inhibits phosphorylation of Smad3 but not Smad2, affects PD-1 promoter activity similarly to knock-down of Smad3 (42). SIS3 inhibited TGF-β1-enhancement of Pdcd-1 promoter activity (Figure 3e, white bars) without significantly altering NFATc1-dependent Pdcd-1 promoter activity (Figure 3e, grey bars). Thus, our data collectively showed that Smad3 is a key mediator of TGF-β1 enhanced Pdcd-1 promoter activity and increased Pdcd-1 transcription levels.
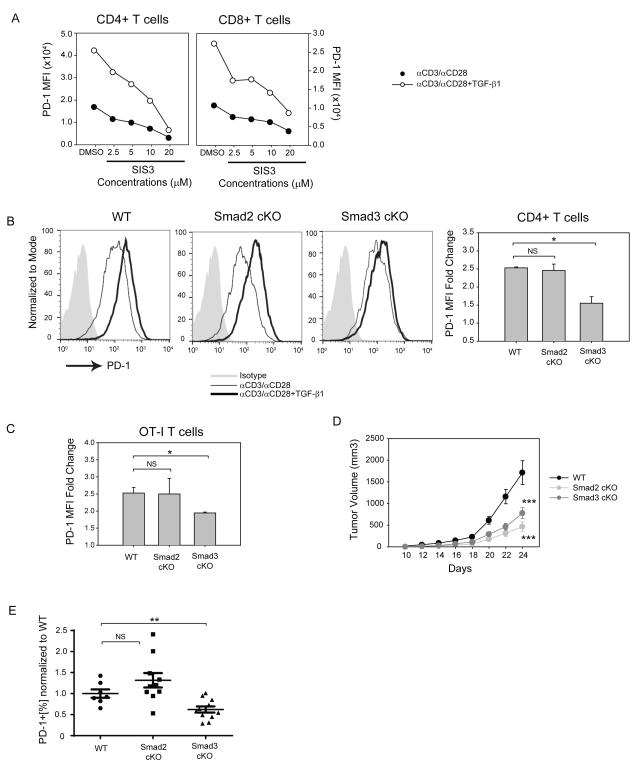
Smad3-dependent PD-1 enhancement is conserved in human and murine T cells
To investigate the role of Smad3 on PD-1 T cell surface expression, we treated human CD3+ cells with SIS3 and found that SIS3 treatment decreased PD-1 surface expression on both human CD4+ (Figure 4a, left panel) and CD8+ (Figure 4a, right panel) T cells in a dose-dependent manner. Next, we investigated whether Smad3 deficiency can abrogate TGF-β1-dependent PD-1 expression on murine T cells. CD4+ T cells were isolated from WT, Smad2 f/f; Cd4-cre (Smad2 cKO), and Smad3 f/f; Cd4-Cre (Smad3 cKO) mice and activated with αCD3/αCD28 with or without TGF-β1 (Figure 4b). Cre-mediated gene knock-out of Smad2 and Smad3 in CD4+ T cells was confirmed by western-blot analysis (Supplementary Figure S3a). Consistent with our human T cell findings, TGF-β1 minimally increased PD-1 expression on Smad3 cKO CD4+ T cells compared to WT CD4+ T cells as shown in both representative histogram (Figure 4b, left) and PD-1 MFI (Figure 4b, right). In contrast, Smad2 cKO CD4+ T cells maintained high PD-1 expression in response to TGF-β1 (Figure 4b). Similarly, when WT, Smad2 cKO, and Smad3 cKO OT-1 (ovalbumin-specific CD8+ T cells) cells were activated with type-I ovalbumin (Ova) in the presence of TGF-β1, Smad3 cKO OT-1 showed decreased PD-1 expression (Figure 4c). In contrast, the expression of lymphocyte-activation gene3 (LAG3), another inhibitory receptor, decreased on Smad3 cKO OT-I and OT-II (Ova-specific CD4+ T cells) cells activated in the presence of TGF-β1, suggesting that TGF-β1 has differential effects on inhibitory receptors (Supplementary Figure S3b). Taken together, the in vitro results in both human and murine T cells support the notion that TGF-β1 enhancement of PD-1 transcription is dependent selectively on Smad3.
Figure 4.
TGF-β1-dependent Smad3 enhances PD-1 expression on human and murine T cells
(a) Human CD3+ T cells from healthy donors were isolated and pretreated with SIS3 at varying concentrations. Subsequently, the cells were activated with αCD3/αCD28-conjugated beads with or without TGF-β1. Mean fluorescence intensity (MFI) of PD-1 expression in different conditions was assessed: αCD3/αCD28 (closed circles); αCD3/αCD28 + TGF-β1 (open circles). The result shown is representative of at least three independent trials. (b) CD4+ T cells were isolated from wild-type (WT), Smad2 f/f; Cd4-cre (Smad2 cKO), Smad3 f/f; Cd4-cre (Smad3 cKO) mice and activated with plated-coated αCD3 and soluble αCD28 with or without TGF-β1 (50 ng/ml). PD-1 expression is shown as overlaid histograms with shaded histogram (isotype control), thin histogram (αCD3/αCD28), bold histogram (αCD3/αCD28+TGF-β1). PD-1 MFI is also shown as mean +/− SEM and represents combined results of two independent trials (bar graphs). (c) Isolated WT, Smad2 cKO, and Smad3 cKO OT-1 cells were activated with type-1 ovalbumin in the presence of irradiated splenocytes. PD-1 MFI is shown as mean +/− SEM and represents combined results of two independent trials (bar graphs). (d) Growth kinetics of B16 melanoma in WT (n=7), Smad2 cKO (n=10), and Smad3 cKO (n=11) mice are shown as the mean volume +/− SEM on different days. The data represent the combined results of two independent experiments. (e) Average CD8+ PD-1+ percentages in Smad2 cKO and Smad3 cKO TIL are shown as normalized values to WT CD8+ PD-1+ percentages. The data were analyzed using Student’s t-test and considered significant if *p<0.05, **p<0.01, ***p<0.001.
Tumor-infiltrating Smad3 cKO CD8+ T cells have decreased PD-1 expression
PD-1 is highly expressed on TIL and its high expression is associated with decreased effector function in advanced stage human cancer (3,43-46). Given that tumor-microenvironment-derived TGF-β1 can suppress anti-tumor immunity (30,31), we hypothesized that Smad3 contributes to high TIL PD-1 expression. In order to investigate whether TGF-β1 regulates PD-1 expression through Smad3 in vivo, WT, Smad2 cKO, and Smad3 cKO mice were challenged with B16 melanoma and PD-1 expression was assessed on TIL. Smad2 and Smad3 are known suppressors of T cell function (33), and growth of B16 melanoma in Smad2 and Smad3 cKO mice was indeed significantly delayed (Figure 4d). Although both Smad2 cKO and Smad3 cKO mice had comparably decreased volumes of B16 melanoma versus WT, the PD-1hi subset population was significantly lower on Smad3 cKO CD8+ TIL, but not on Smad2 cKO CD8+ (Figure 4e). In contrast, LAG3 expression was significantly enhanced on Smad2 cKO CD8+ TIL but unaffected by Smad3 cKO (data not shown). PD-1 expression on CD4+ TIL was not significantly different among WT, Smad2 and Smad3 cKO groups although the level of PD-1 expression was much lower on CD4 T cells than on CD8 T cells in all three mouse groups (data not shown). The lack of a significant difference in PD-1 expression on CD4+ T cells in vivo could be due to the minimal fraction of antigen-specific CD4+ effector T cells in the tumor microenvironment. Alternatively, because PD-1 is expressed by Tregs (47,48) that constitute the majority of CD4+ TIL in B16 melanoma, most CD4+ TIL may not be specific for tumor antigens. Supporting this notion, we found that the majority of CD4+ PD-1+ TIL express Foxp3 (Supplementary Figure S3c). However, in the presence of TCR signaling in vitro, Foxp3+ and Foxp3− CD4+ T cells are capable of enhancing PD-1 expression to the same extent in response to TGF-β1 (Supplementary Figure S3d, ) and the FoxP3-CD4+ TIL did not express lower levels of PD-1 in the Smad3 cKO than in the WT mice (data not shown).
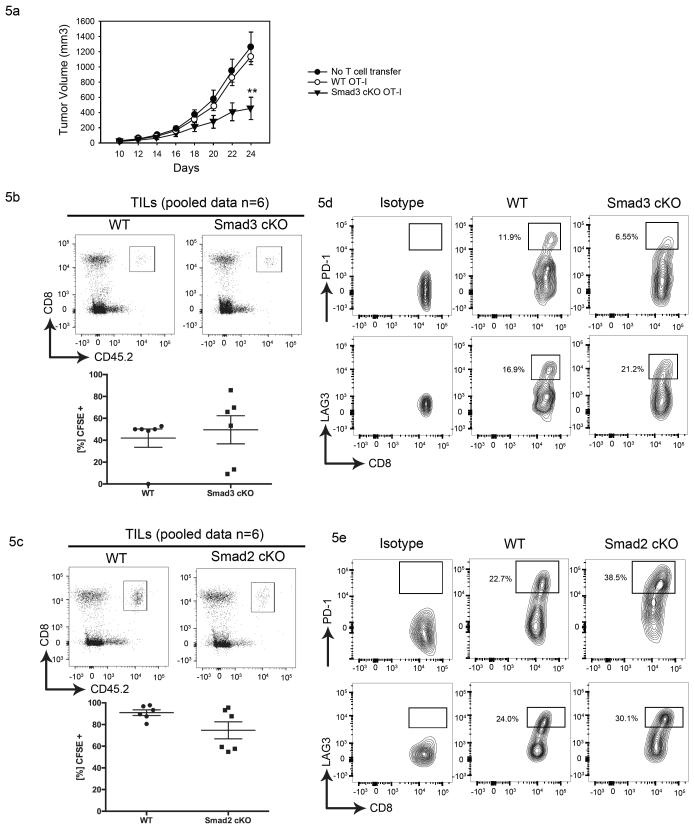
In order to test whether TGF-β1 induced Smad3 activation up-regulated PD-1 on tumor antigen-specific T cells, we utilized B16 melanoma cells stably expressing ovalbumin as a model tumor antigen (B16-Ova). CD45.1 WT mice were challenged with B16-Ova, and CD45.2 OT-1 cells from WT and Smad3 cKO OT-1 mice were adoptively transferred into the tumor-bearing mice after tumor cells became palpable. The tumor growth was monitored and lymphocytes infiltrating into tumors were harvested after 5 days. We found that transferred Smad3 cKO OT-1 cells limit tumor growth more effectively than WT OT-1 cells do (Figure 5a). Consistent with our in vitro data, neither Smad3 cKO TIL nor Smad2 cKO TIL showed significantly increased cellular proliferation by CFSE (Figure 5b, 5c) when gated on CD45.2+ (antigen experienced) donor cells. When a proliferated subset (i.e. CFSE negative subset) is further gated, Smad3 cKO TIL show significantly fewer of the PD-1hi expressing T cells highly characteristic of WT TIL (Figure 5d, top). This effect of Smad3 was specific to PD-1 since there was no reduction in LAG3hi subset in Smad3 cKO TIL (Figure 5d, bottom). PD-1hi expressing cells did not decrease in Smad2 cKO OT-1 (Figure 5e, top), further supporting that Smad3 is a critical mediator of PD-1 expression in the tumor microenvironment. In contrast, Smad2 cKO TIL maintained high levels of PD-1 (Figure 5e, top), which is consistent with our observation in polyclonal CD8+ T cells (Figure 4e). Similar to TIL, LAG-3 expression on OT-1 cells was not significantly affected in Smad3 cKO mice. Conversely, PD-1 and LAG3 expression on T cells in draining lymph nodes (DLN) and in non-draining lymph nodes (NDLN) was comparable between WT and Smad3 cKO OT-I (Supplementary Figure S4a,b) or Smad2 cKO OT-I (Supplementary Figure S4c,d), suggesting that the effect of TGF-β1 is specific to the tumor-microenvironment. To confirm the specificity to the tumor microenvironment, WT and DNTGFβRII Tg+ mice were challenged with B16 melanoma and tumor volume was measured. In association with enhanced anti-tumor immune responses in DNTGFβRII Tg+ mice (Supplementary Figure S4e), decreased PD-1 expression was also observed on CD8+ TIL, but not on T cells from DLN (Supplementary Figure S4f). As in Smad2 and Smad3 cKO mice, the DNTGFβRII Tg+ mice did not show significant decreases in LAG-3 expression on TIL or DLN T cells (Supplementary Figure S4g).
Figure 5.
Adoptive transfer of Smad3 cKO CD8+ T cells results in reduced tumor burden and PD1hi subset relative to transfer of WT CD8+ T cells
(a) Growth kinetics of B16-Ova in CD45.1 congenic mice that received no T cells (closed circles), WT OT-1 (open circles) or Smad3 cKO OT-1 (triangles). C57/BL6 expressing CD45.1 congenic markers were challenged with 1 × 105 B16-Ova melanoma cell line on Day 0. On Day 10, 107 CD45.2 CD8+ OT-1 T cells from WT (n = 7) or Smad3 cKO (n = 5) mice were adoptively transferred into the mice with comparable tumor sizes. Tumor volume (mm3) is shown as mean +/− SEM on different days, and the data represent combined results of two independent experiments. The data were analyzed using one-way ANOVA and considered significant if *p<0.05, **p<0.01. (b, c) CFSE-labelled tumor infiltrating WT OT-1 (top) or cKO OT-1 (bottom) were isolated from B16-Ova 5 days after adoptive transfer, and tumor infiltrating lymphocytes (TIL) proliferation was assessed: Smad3 cKO (b) and Smad2 cKO OT-1 (c). CD45.2+ donor population was gated from a plot of CD8 (Y-axis) and CD45.2 (X-axis) (left), and a representative histogram of CFSE (right) is shown from pooled TIL from n = 6 mice per group. (d, e) Contour plots of PD-1 (top) and LAG3 (bottom) among the proliferated cells (i.e CFSE negative populations) are shown as isotype (left), WT (middle) and cKO (right): Smad3 cKO (d) and Smad2 cKO (e). The data are representative of two independent experiments.
TGF-β1/Smad3-dependent enhanced PD-1 expression is associated with decreased T cell function
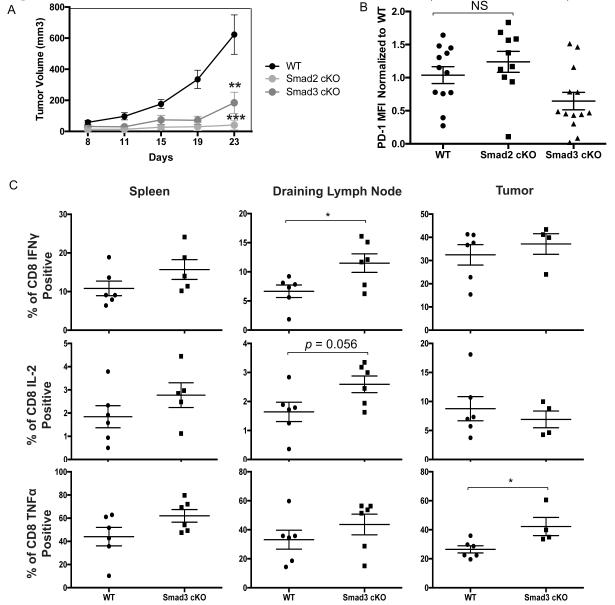
We next assessed the effect of Smad2 and Smad3 cKO on T cell function. The number of TIL obtained in the B16 model is smaller than in some other tumor models and isolation of TIL from the tumors harvested from the Smad2 and Smad3 cKO mice particularly challenging. To confirm our findings with B16 melanoma and to permit functional analysis of TIL and T cells from the DLN, we used a cancer model with more abundant TIL, the MC38 colon cancer model. WT, Smad2 cKO, and Smad3 cKO mice were challenged with MC38 colon cancer and PD-1 expression assessed. As with the B16 melanoma (Figure 4d), tumor growth in Smad2 and Smad3 cKO mice was significantly delayed (Figure 6a). In contrast to CD8+ TIL, the PDhi subset population was absent in CD4+ TIL in all groups and differences could not be assessed. As in B16 melanoma, the MFI of PD-1 on CD8 + TIL was significantly lower in the Smad3 cKO than in the WT or Smad2 cKO mice (Figure 6b). We performed intracellular cytokine staining (ICS) to examine CD8+ T cell production of IFN-γ, TNF-α, IL-2, Foxp3, and granzyme B in WT and Smad3 cKO mice, but had insufficient TIL to perform ICS analysis for the Smad2 cKO group. We saw no significant differences between the Smad3 group and the WT group in Foxp3 or granzyme B levels when examining TIL or DLN. However, in the Smad3 cKO group compared to the WT group, there was significantly higher production of (tumor necrosis factor-α (TNF-α) from CD8+ TIL and of IFN-γ and IL-2 in the DLN (Figure 6c). Thus, decreased PD-1 expression secondary to the loss of Smad3 signaling in TIL and T cells in the DLN is associated with increased production of multiple cytokines and functionality.
Figure 6.
Loss of Smad3 in CD8+ T cells leads to enhanced cytokine production.
(a) Growth kinetics of MC-38 in WT (n=7), Smad2 cKO (n=6), or Smad3 cKO cells (n=6) are shown as the mean volume +/− SEM on different days. Data is representative example from 2 independent experiments. (b) Average CD8+ CD44+ PD-1 MFI in Smad2 cKO and Smad3 cKO TIL are shown as normalized values to WT CD8+ PD-1+ percentages. The data were analyzed using Student’s t-test and considered significant if *p<0.05. (c) Percentage of CD8 T cells producing IFNγ, TNFα, or IL-2 in the spleen, draining lymph node, and tumor. The data were analyzed using Student’s t-test and considered significant if *p<0.05.
TGF-β1/Smad3-dependent enhanced anti-tumor effects involve PD-1 expression
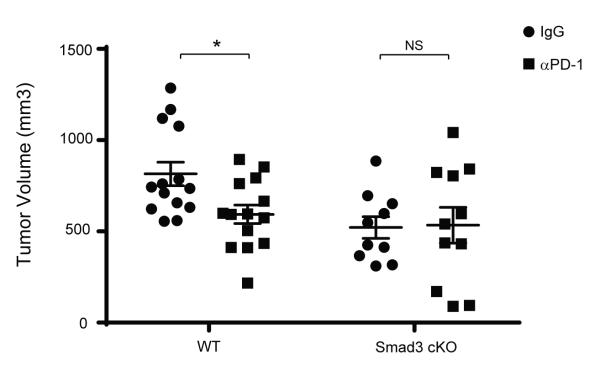
TGF-β1 is known to inhibit CD8+ T cell effector function through Smad2/3 (33) and many different mechanisms (37). Our data provide evidence that enhancement of PD-1 expression represents a newly defined mechanism through which TGF-β1/Smad3 suppresses T cell function. In order to address how significant the impact of Smad3-mediated PD-1 up-regulation is on tumor evasion of T cell responses, we treated Smad3 cKO mice bearing B16-melanoma with an anti-PD-1 blocking antibody (αPD-1) previously shown to have therapeutic efficacy in WT mice bearing B16-melanoma (49,50). If the effect of Smad3 cKO on tumor growth is mediated through a mechanism other than PD-1 or if the effects of Smad3 cKO on PD-1 expression are not sufficient to negate that mechanism of tumor evasion, treatment with αPD-1 would confer additional therapeutic benefits in Smad3 cKO mice. To assess this, WT and Smad3 cKO mice were challenged with B16 melanoma cells and were given either isotype-matched IgG or αPD-1. We found that the tumor volume was decreased with αPD-1 compared to IgG treated WT mice (Figure 7, WT) attesting to the general role of the PD-1 pathway in immune resistance in this tumor model. In contrast, αPD-1 had no effect on tumor growth in Smad3 cKO mice (Figure 7, Smad3).
Figure 7.
Anti-PD-1 blocking antibody enhances anti-tumor immune responses in WT mice, but has minimal effect in Smad3-deficient mice.
WT and Smad3 cKO were challenged with B16-melanoma cell line on Day 0. WT and Smad3 cKO mice were treated with either isotype-matched control IgG (circles) or anti-PD-1 antibody (squares) from the day of tumor implantation until Day 17. Tumor volume (mm3) on Day 17 is shown as mean +/− SEM on and the data represent combined results of two independent experiments. The data were analyzed using two-way analysis of variance (ANOVA) and considered significant if *p<0.05.
Discussion
We show here that TGF-β1, signaling selectively through Smad3 transcriptional regulation, significantly up-regulates PD-1 in the context of TCR engagement. We show that this mechanism is important in generating a PD-1hi population of T cells in the tumor microenvironment, where TGF-β1 expression is commonly very high. Thus, up-regulation of both PD-1 ligands and the PD-1 receptor itself contribute to PD-1 pathway mediated tumor immune resistance.
PD-1 expression can be differentially regulated by the environmental context in which a T cell encounters antigen. Upon activation, NFATc1 transiently induces PD-1 expression on T cells (38). Once PD-1 expression is induced, it is sustained in chronic infections or toleragenic environments (2), but high level PD-1 expression is not achieved when antigen is encountered in an inflammatory environment, such as in Listeria monocytogenes infection (51). Further supporting the notion that the level of PD-1 expression is context-dependent, there has been emerging evidence that cytokines can regulate NFATc1-induced PD-1 expression. Interferon-α (IFN-α) promotes PD-1 expression on murine T cells through signal transducer and activator of transcription-1 (STAT-1) mediated transcriptional regulation of PD-1 gene expression (21,22). IL-6 also increases PD-1 expression through a STAT3-dependent mechanism in murine CD8+ T cells in vitro (52), and we found the similar regulation in human CD4+ and CD8+ T cells (Supplementary Figure S1a). IL-12 has differential effects on PD-1 in vivo and in vitro. IL-12-conditioned tumor-specific memory CD8+ T cells have lower PD-1 expression in vivo with stronger anti-tumor immune responses (21). In contrast, we have found that IL-12 increases PD-1 expression on human CD4+ and CD8+ T cells in vitro, consistent with other’s findings on murine CD8+ T cells (52). Thus, while our data agree with the literature that IL-6 and IL-12 modulate PD-1 expression, TGF-β1 has the greatest effect on PD-1 expression, which has not been shown previously. The effects of some cytokines could be greater in vivo than we observed in vitro. However, our in vivo data demonstrating that loss of TGF-β1 signaling has a profound impact on high level PD-1 expression upon TCR engagement while signaling from other cytokines remains intact suggest that TGF-β1 signaling through Smad3 is the major regulator of T cell expression of PD-1 and function.
The data on the effect of other cytokines on PD-1 expression also collectively show that the regulatory mechanisms of PD-1 expression are highly conserved between human and mouse. This is further supported by high sequence homology between human and murine Pdcd-1 proximal promoter regions including the NFATc1 binding site (52). We demonstrate that Smad3-dependent PD-1 regulation is also conserved in showing that Smad3 has the greatest effects on PD-1 expression on both human and murine T cells. NFATc1 was previously shown to be critical for PD-1 induction in mice (38) and mutation of antigens such that the TCR is no longer engaged results in a decline in human PD-1 expression in chronic infection with HCV or HIV (10,53). Supporting previous findings that PD-1 expression depends on TCR engagement, we find that antigenic stimulation is required for TGF-β1 to enhance PD-1 expression and that NFATc1 binding to the human Pdcd-1 promoter following αCD3/αCD28 is enhanced by TGF-β1. Furthermore, TGF-β1 enhances PD-1 expression on both CD4+ and CD8+ T cells regardless of their naïve or memory status, although its effect was more pronounced on naïve T cells than on memory T cells.
Our proliferation assays showed that TGF-β1-mediated PD-1 enhancement is independent of cellular proliferation. In contrast to other’s findings for which Smads were proposed to play a role in TGF-β1 suppression of T cell proliferation (54), we found that TGF-βRI-dependent signaling, as demonstrated by phosphorylation of Smad2 (Figure 2c), did not result in suppression of T cell proliferation. TGF-β1-mediated suppression of proliferation can be overcome by CD28-mediated co-stimulation, and it is possible that αCD3/αCD28 used in our in vitro culture system masked inhibition (55). However, neither Smad2 cKO OT-1 nor Smad3 cKO OT-1 showed significantly altered proliferation in vivo (Figure 5bc). Nevertheless, we observed that isolated Smad3 cKO CD4+ T cells have increased IL-2 expression compared to WT litter mates when activated with αCD3/αCD28 (data not shown), consistent with previous reports (56).
Others have suggested a potential association between TGF-β1 signaling and high PD-1 expression, but the direct causal relationship, the molecular mechanism, and biological implications of this association have not ever been characterized (32,57,58). TGF-β1 signaling consists of Smad and non-Smad dependent pathways and Smad-dependent gene regulation (Smad2 and Smad3) has been well-characterized (59,60). Some genes are preferentially and exclusively regulated by Smad2 or Smad3 as with Id1 and Myc (61,62). On the other hand, Smad2 and Smad3 can redundantly regulate expression of many genes that are under control of TGF-β1 (63). Our luciferase assay and in vitro data suggest that PD-1 regulation is predominantly under the control of Smad3. Although our in vitro data support a minor role for Smad2 in TGF-β1-dependent PD-1 enhancement, our in vivo data clearly demonstrated no enhancement of PD-1 expression through Smad2 with Smad2 cKO mice showing a small increase rather than decrease in PD-1 expression. The in vivo data could reflect enhanced Smad3 activity in compensation for Smad2 deletion in T cells.
Our in vivo studies mainly focused on CD8+ T cells because the PD-1 expression difference was greater in CD8+ T cells than in CD4+ T cells in vivo and the levels of PD-1 on CD4+ T cells much lower than on CD8+ T cells. Supporting this notion, TGF-β1-suppression of anti-tumor immunity in vivo appears to be dependent on CD8+ T cells but not on CD4+ T cells in a murine mouse model (31). This discrepancy could be due to cellular intrinsic difference between CD4+ and CD8+ subset of T cells (64). Our observation that stimulated OT-I and OT-II T cells respond similarly to TGF-β1 in vitro (Supplementary Figure S3b) but CD4 and CD8 TIL in vivo do not may be explained by an absence of antigenic recognition by CD4+ TIL in vivo due to CD4+ T cells being primarily Tregs (Supplementary Figure S3c,d). Alternatively, mechanisms of PD-1 regulation unique to CD4+ T cells may exist in vivo given that the Foxp3 negative CD4+ T cells in the Smad3 cKO mice did not express lower levels of PD-1 than WT. (data not shown).
Interestingly, the effect of TGF-β1/Smad3 on PD-1 expression of CD8+ T cells was specifically on TIL, but not on those originating from tumor DLNs. We did not find the percentage of PD-1hi T cells isolated from the tumor-DLNs in WT mice to be significantly different from that of DNTGFβRII Tg+ and Smad3 cKO mice (Supplementary Figure S4a, b, f). This may be due to the fact that the PD-1hi CD8+ T cell population prominent in the tumor microenvironment was absent in DLNs in WT mice, and suggests that TGF-β1 levels could be much lower outside of the tumor microenvironment. Supporting this notion, others found that TGF-β1 expression is higher in human head and neck squamous cell carcinoma tissue than in adjacent mucosal tissue (65). Nevertheless, it is yet to be determined whether the dominant source of TGF-β1 in the tumor microenvironment is derived from tumor or T cells. In spite of extensive evidence that TGF-β1 suppresses anti-tumor immunity, tumor-specific deletion of TGF-β1 did not enhance anti-tumor immune responses (33). In contrast, others have reported that the deletion of T cell-derived TGF-β1 was sufficient to prevent tumor growth (32).
Although Smad3 cKO mice did not mount immune responses as potent as those of DNTGFβRII Tg+ mice, B16-melanoma growth in Smad3 cKO mice appeared comparable to that in Pdcd-1 KO mice or anti-PD-1 antibody treated WT mice (Figure 7). While adoptive transfer of naïve antigen-specific T cell is known to confer minimal anti-tumor effects, transferred Smad3 cKO OT1 effectively controlled tumor growth (Figure 5a). Collectively, these results provide direct evidence that PD-1-mediated anti-tumor immunity depends in part on Smad3 activation and that Smad3-driven PD-1 up-regulation is relevant to tumor immune evasion. Our data clearly show that αPD-1 treatment decreases B16 tumor growth in WT mice (49,50) but not in Smad3 cKO mice.
In sum, our data demonstrate a novel immunosuppressive function of TGF-β1 in regulating high-level PD-1 expression on T cells encountering cognate antigen. In addition to other suppressive roles for TGF-β1, TGF-β1 enriched in the tumor microenvironment may induce high levels of PD-1 on T cells as they encounter antigens on the tumor surface, reducing T cell effector function and limiting the anti-tumor T cell response. In addition, our data provide mechanistic understanding of the regulation of high-level PD-1 expression. While it is well known that T cells against intact antigen in the setting of chronic viral infections such as HCV and HIV or malignancy express very high levels of PD-1, it is not known how those high levels are induced. This study elucidates a mechanism through which the highest levels of PD-1 are induced. Indeed, high TGF-β1 serum levels are associated with worse disease outcome in HCV infection (66) and TGF-β1 expression is high in the tumor microenvironment of advanced stages of cancer, which may further limit the efficacy of T cells against disease in those settings (67-69). Given the potential for autoimmunity with PD-1 therapy, it is worth investigating whether inhibitors of Smad3 used in combination with other immunotherapeutic agents activate T cells expressing the highest levels of PD-1 rather than all T cells bearing PD-1. Since the PD-1hi subset of TIL may in fact contain the highest proportion of true tumor-specific cells, these may be the most important target population for Smad3 blockade.
Methods
Mice
All animals were housed and handled in compliance with Johns Hopkins Animal Care and Use policy. C57BL/6 DNTGFβRII Tg+ and C57BL/6 Cd4-Cre transgenic mice were purchased from the Jackson Laboratory. CD45.1 congenic mice were purchased from National Cancer Institute (NCI) at Frederick. Smad2 flox/flox (fl/fl) and Smad3 fl/fl mice were generated by Se-Jin Lee’s Laboratory at Johns Hopkins School of Medicine and backcrossed to C57BL/6 at least 6 generations. OT-I and OT-II mice were generous gifts from Drs. Charles Drake and Hyam Levitsky at Johns Hopkins School of Medicine. Age-matched female mice were utilized in all in vivo experiments.
Human and murine primary T cell Isolation and Culture
Human peripheral blood mononuclear Cells (PBMCs) were isolated from leukopheresis by Ficoll-Hypaque density gradient. Isolated human PBMCs were subjected for CD3+ T cell isolation by using the Pan T cell Isolation Kit (Miltenyi) as instructed in the manual. The isolated cells were activated for 72 hr with αCD3/αCD28 Dynabeads (Invitrogen) at a cell to bead ratio of 1:1 in RPMI+10% Fetal Bovine Serum (supplemented with HEPES buffer, Penicillin/Streptomycin, and L-glutamine). Murine CD4+ and CD8+ T cells were isolated from the spleen and lymph nodes using the Negative Selection Kit (Invitrogen), and were activated with plate-coated αCD3 (10μg/ml) and soluble αCD28 (2μg/ml) or with cognate Ova peptides in the presence irradiated splenocytes for 72 hr.
Transient Transfection and Luciferase Assay
Jurkat T cells (clone E6-1) were purchased from the ATCC and were kept as a frozen stock. 1.5×107 Jurkat T cells were transfected with 10 μg pGL-3 Firefly Luciferase Vector (Promega) and 1μg of pRL-TK Vector (Promega) by electroporation using Nucleofector II (Amaxa/Lonza). The cells were rested in a 6-well plate overnight and activated with plate-coated αCD3 (10 μg/ml) and soluble αCD28 (5 μg/ml) with or without rhTGF-β1 (50 ng/ml). After 24 hr, the cells were harvested and lysed followed by luminescence measurement using Dual-Luciferase Assay (Promega). Where indicated, the cells were co-transfected with empty vector (pSG-V5), TGF-βRI-His (Addgene plasmid #19161), TGF-βRII (Addgene plasmid #11766). For siRNA-mediated knock-down, the cells were co-transfected with 1.5 μM of siRNA for Smad2 (Santa Cruz Biotechnology, SC-38374) and Smad3 (Santa Cruz Biotechnology, SC-38376).
Cytokine and Drug Treatments
Human recombinant IL-1α, IL-2, IL-4, IL-6, IL-10, IL-12, IL-13, IL-15, IL-17, IL-18, IL-21, IL-23, INF-α, IFN-γ, TGF-β1, and TNF-α were purchased from Peprotech and used at 5, 50, and 500 ng/mL. The data shown in Supplemental Figure 1a are using 500 ng/mL only because the relative effects of cytokines were not different at other doses. Primary human T Cells were treated with neutralizing TGF-β1 antibody (Abcam, 2Ar2) and with small molecule inhibitors, SB431542, Cyclosporin A (Sigma-Aldrich), and SIS3 (Calbiochem) for 1 hr prior to activation at the indicated concentration range.
Flow Cytometry
After indicated time of culture, human T cells were harvested and centrifuged at 400g (or 1500 rpm) for 5 minutes. The cells were washed in FACS buffer (1× PBS + 2% Fetal Bovine Serum) and stained with Aqua Viability Dye (Invitrogen) as instructed in the manual. After wash, the cells were stained with PD-1 PE (Biolegend), CD8 PerCP, CD4 Pacific Blue, CD3 FITC (eBioscience), and HLA-DR APC (eBioscience) or qDot605 (Invitrogen). The similar protocol was used for murine T cells and PD-1 PE, CD4 or CD8 PerCP, FITC (eBioscience), LAG3 APC or PacBlue, CD4 BV605, CD8 BV570, CD3 AF700, PD-1 PE Cy7, CD44 AF700 (Biolegend) were used for flowcytometery. For intracellular staining, the cells were treated with Foxp3/transcription factor staining buffet set (eBioscience) and stained with Foxp3 FITC, TNFα PE (eBioscience), IL-2 PE-CF594, or IFNγ APC (BD Biosciences).
Real-time qPCR assay
Total RNA was extracted from primary T cells under the indicated conditions using RNEasy Plus Kit (Qiagen). 100 ng of extracted RNA was reverse-transcribed using SuperScriptIII First-Strand Synthesis System (Invitrogen). Generated cDNA was subjected for real-time PCR assay. Pdcd-1 primer sequences are Forward 5′- CACTGAGGCCTGAGGATGG-3′; Reverse 5′-AGGGTCTGCAGAACACTGGT-3′. All target genes were normalized to 18s rRNA or 28s rRNA as previously described.
Molecular Cloning and Site-directed Mutagenesis
Human PD-1promoter (1.9 kbases) was cloned from genomic DNA of isolated CD3+ T cells and the sequence was confirmed. The amplified clones were ligated to SacI/XhoI digested pGL3-Basic Vector (Promega) using In-Fusion Cloning Kit (Clonetech). Site-directed mutagenesis was carried out using following primers for NFAT, SBE-D, SBE-P sites using QuickChange Lightning Kit (Agilent Technologies). Forward: 5′-GATGCTCTTTTTGGACTGTTTCGG-3′ Reverse 5′-CCGAAACAGTCCAAAAAGAGCATC-3′ (NFAT); Forward: 5′-ACCTTAGCTGGATGGCAGCA-3′ Reverse 5′-TGCTGCCATCCAGCTAAGGT-3′ (SBE-D), Forward: 5′-CGCGCCTCGCATCCATCATCTT-3′ Reverse: 5′-AAGATGATGGATGCGAGGCGCG-3′ (SBE-P).
Chromatin Immunoprecipitation (ChIP) assay
ChIP assay was performed according to the manufacturer’s guidance (Invitrogen MAGnify ChIP system). Briefly, isolated CD3+ T cells were activated with αCD3/αCD28-conjugated beads for 24 hr and fixed with 2% formaldehyde. Sonicated DNA was immunoprecipiated with αSmad3 (Cell Signaling Technology), and αNFATc1 (Santa Cruz Biotechnology). The immunoprecipated chromatin was analyzed on Roche Light Cycler 480 by SYBR green using the following primers for PD-1 promoter. Pdcd-1: Forward 5′-CCTCACATCTCTGAGACCCG-3′; Reverse 5′-CCGAAGCGAGGCTAGAAACC-3′ Gapdh: 5′-TACTAGCGGTTTTACGGGCG-3′ 5′-TCGAACAGGAGGAGCAGAGAGCGA-3′
Western Immunoblotting
Human or murine T cells were activated as indicated and harvested and lysed in RIPA Buffer (Cell Signaling Technology). Protein-extract concentrations were measured using BCA protein assay kit (Thermo Scientific) and followed by heating under reducing conditions. The equal amounts of extracts were loaded/run on NuPAGE Precast gels (Invitrogen) and transferred membranes were blotted with following antibodies: p-Smad2, total Smad2, total Smad3 (Cell Signaling Technology), and β-actin (Sigma).
B16 melanoma and adoptive T cell transfer experiments
B16 melanoma cell lines were purchased from the ATCC and kept as a frozen stock. 1 × 105 B16 melanoma cell lines were injected on a flank in 100 μl volume. Tumor volumes were measured every other day using a caliper and assessed using the formula ½ (Length×Width2). CD45.1 host mice were injected with 1 × 105 B16-Ova melanoma cell lines on a flank. 8 × 106 WT OT-1 or cKO OT-1 were labeled with CellTrace CFSE Cell Proliferation Kit (Life Technologies) and were adoptively-transferred in to tumor-bearing mice by retroorbital injection on Day 12. The tumors were harvested on Day 5 after the adoptive transfer and lymphocytes were purified using Percoll (GE Healthcare) gradient. In a blocking experiment, 5 × 105 B16 melanoma cells were injected on a flank and 100 μg Armenian hamster IgG isotype control (Rockland) or anti-PD1 antibody (G4) were injected intraperitoneally two times a week from Day 0.
MC38 colon adenocarcinoma experiments
MC38 colon adenocarcinoma cell lines were purchased from the ATCC and kept as a frozen stock. 4 × 105 B16 melanoma cell lines were injected on a flank in 100 μl volume. Tumor volumes were measured using a caliper and assessed using the formula ½ (Length×Width2). The tumors were harvested on Day 23 and lymphocytes were purified using Percoll (GE Healthcare) gradient. For intracellular cytokine staining, lymphocytes were stimulated with phorbol 12- myristate 13- acetate (PMA, 50 ng/ml) and inomycin (500 ng/ml) in the presence of Brefeldin A and Monensin (eBiosciences) for 4 hours prior. After stimulation, cells permeablizied and stained for intracellular cytokines.
Supplementary Material
Significance.
Engagement of the co-inhibitory receptor PD-1 of its ligand, PD-L1, dramatically inhibits the anti-tumor function of TIL within the TME. Our findings represent a novel immunosuppressive function of TGF-β and demonstrate that TGF-β1 allows tumors to evade host immune responses in part through enhanced Smad3-mediated PD-1 expression on TIL.
Acknowledgements
We would like to acknowledge Ada Tam and Richard L. Blosser from the Cancer Research Flow-cytometry Core, and Tricia L. Nilles from School of Public Health Flow-cytometry Core at Johns Hopkins University for their technical help. We are grateful for Juan Fu and Young Kim for providing anti-PD-1 antibody reagents. Also, we want to thank the Jeff Wrana and Joan Massagué Laboratory for their generous donation of plasmids to Addgene. This work was supported by NIH grant R01AR060636, U10 AI088791 and P30CA006973.
Footnotes
Note: authors declare no potential conflict of interest.
Authors’ Contributions
Conception and design: D.M. Pardoll, A.L. Cox, and B.V. Park
Development of methodology: D.M. Pardoll, A.L. Cox, and B.V. Park
Acquisition of data: A. Rutebemberwa, J. Steigner, M.A. Chattergoon, A. Ghasemzadeh, Z.T. Freeman, and B.V. Park
Analysis and interpretation of data: M.A. Chattergoon, A. Ghasemzadeh, Z.T. Freeman, B.V. Park and F. Pan
Writing, review and/or revision of the manuscript: D.M. Pardoll, A.L. Cox, and B.V. Park
Administrative, technical or material support: M. Winter, T.V. Huynh, S.M. Sebald., F. Pan and S. Lee
Study supervision: D.M. Pardoll, and A.L. Cox
References
- 1.Agata Y, Kawasaki A, Nishimura H, Ishida Y, Tsubat T, Yagita H, et al. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. International Immunology. 1996;8(5):765–72. doi: 10.1093/intimm/8.5.765. [DOI] [PubMed] [Google Scholar]
- 2.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439(7077):682–87. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 3.Ahmadzadeh M, Johnson LA, Heemskerk B, Wunderlich JR, Dudley ME, White DE, et al. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood. 2009;114(8):1537–44. doi: 10.1182/blood-2008-12-195792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8(8):793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 5.Lipson EJ, Sharfman WH, Drake CG, Wollner I, Taube JM, Anders RA, et al. Durable cancer regression off-treatment and effective reinduction therapy with an anti-PD-1 antibody. Clin Cancer Res. 2013;19:462–8. doi: 10.1158/1078-0432.CCR-12-2625. United States: 2012 Aacr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–7. doi: 10.1038/nature14011. England. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N Engl J Med. 2015;372(4):311–9. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443(7109):350–54. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 10.Rutebemberwa A, Ray SC, Astemborski J, Levine J, Liu L, Dowd KA, et al. High-programmed death-1 levels on hepatitis C virus-specific T cells during acute infection are associated with viral persistence and require preservation of cognate antigen during chronic infection. J Immunol. 2008;181:8215–25. doi: 10.4049/jimmunol.181.12.8215. United States. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boni C, Fisicaro P, Valdatta C, Amadei B, Di Vincenzo P, Giuberti T, et al. Characterization of hepatitis B virus (HBV)-specific T-cell dysfunction in chronic HBV infection. J Virol. 2007;81(8):4215–25. doi: 10.1128/JVI.02844-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keir ME, Butte MJ, Freeman GJ, Sharpel AH. PD-1 and its ligands in tolerance and immunity. Annual Review of Immunology. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. Annual Review of Immunology. Palo Alto: Annual Reviews. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curiel TJ, Wei S, Dong H, Alvarez X, Cheng P, Mottram P, et al. Blockade of B7-H1 improves myeloid dendritic cell-mediated antitumor immunity. Nat Med. 2003;9(5):562–7. doi: 10.1038/nm863. [DOI] [PubMed] [Google Scholar]
- 14.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, et al. Colocalization of Inflammatory Response with B7-H1 Expression in Human Melanocytic Lesions Supports an Adaptive Resistance Mechanism of Immune Escape. Science Translational Medicine. 2012;4(127):127ra37–27ra37. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kassel R, Cruise MW, Iezzoni JC, Taylor NA, Pruett TL, Hahn YS. Chronically inflamed livers up-regulate expression of inhibitory B7 family members. Hepatology. 2009;50(5):1625–37. doi: 10.1002/hep.23173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohigashi Y, Sho M, Yamada Y, Tsurui Y, Hamada K, Ikeda N, et al. Clinical significance of programmed death-1 ligand-1 and programmed death-1 ligand-2 expression in human esophageal cancer. Clin Cancer Res. 2005;11(8):2947–53. doi: 10.1158/1078-0432.CCR-04-1469. [DOI] [PubMed] [Google Scholar]
- 17.Thompson RH, Kuntz SM, Leibovich BC, Dong H, Lohse CM, Webster WS, et al. Tumor B7-H1 is associated with poor prognosis in renal cell carcinoma patients with long-term follow-up. Cancer Res. 2006;66(7):3381–5. doi: 10.1158/0008-5472.CAN-05-4303. [DOI] [PubMed] [Google Scholar]
- 18.Wu C, Zhu Y, Jiang J, Zhao J, Zhang XG, Xu N. Immunohistochemical localization of programmed death-1 ligand-1 (PD-L1) in gastric carcinoma and its clinical significance. Acta Histochem. 2006;108(1):19–24. doi: 10.1016/j.acthis.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43–51. doi: 10.1158/2159-8290.CD-14-0863. United States: 2014 American Association for Cancer Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerner MY, Heltemes-Harris LM, Fife BT, Mescher MF. Cutting Edge: IL-12 and Type I IFN Differentially Program CD8 T Cells for Programmed Death 1 Re-expression Levels and Tumor Control. J Immunol. 2013 doi: 10.4049/jimmunol.1300652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terawaki S, Chikuma S, Shibayama S, Hayashi T, Yoshida T, Okazaki T, et al. IFN-alpha directly promotes programmed cell death-1 transcription and limits the duration of T cell-mediated immunity. J Immunol. 2011;186(5):2772–9. doi: 10.4049/jimmunol.1003208. [DOI] [PubMed] [Google Scholar]
- 23.Shi Y, Massagué J. Mechanisms of TGF-β Signaling from Cell Membrane to the Nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 24.Alatrakchi N, Graham CS, van der Vliet HJ, Sherman KE, Exley MA, Koziel MJ. Hepatitis C virus (HCV)-specific CD8+ cells produce transforming growth factor beta that can suppress HCV-specific T-cell responses. J Virol. 2007;81(11):5882–92. doi: 10.1128/JVI.02202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cumont MC, Monceaux V, Viollet L, Lay S, Parker R, Hurtrel B, et al. TGF-beta in intestinal lymphoid organs contributes to the death of armed effector CD8 T cells and is associated with the absence of virus containment in rhesus macaques infected with the simian immunodeficiency virus. Cell Death Differ. 2007;14(10):1747–58. doi: 10.1038/sj.cdd.4402192. [DOI] [PubMed] [Google Scholar]
- 26.Penaloza-MacMaster P, Kamphorst AO, Wieland A, Araki K, Iyer SS, West EE, et al. Interplay between regulatory T cells and PD-1 in modulating T cell exhaustion and viral control during chronic LCMV infection. J Exp Med. 2014;211(9):1905–18. doi: 10.1084/jem.20132577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Yue ZC, Zhang YY, Bai J, Meng XN, Geng JS, et al. Elevated serum level and gene polymorphisms of TGF-beta1 in gastric cancer. J Clin Lab Anal. 2008;22(3):164–71. doi: 10.1002/jcla.20236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito H, Tsujitani S, Oka S, Kondo A, Ikeguchi M, Maeta M, et al. An elevated serum level of transforming growth factor-beta 1 (TGF-beta 1) significantly correlated with lymph node metastasis and poor prognosis in patients with gastric carcinoma. Anticancer Res. 2000;20:4489–93. [PubMed] [Google Scholar]
- 29.Liu VC, Wong LY, Jang T, Shah AH, Park I, Yang X, et al. Tumor evasion of the immune system by converting CD4+CD25− T cells into CD4+CD25+ T regulatory cells: role of tumor-derived TGF-beta. J Immunol. 2007;178(5):2883–92. doi: 10.4049/jimmunol.178.5.2883. [DOI] [PubMed] [Google Scholar]
- 30.Kao JY, Gong Y, Chen CM, Zheng QD, Chen JJ. Tumor-derived TGF-beta reduces the efficacy of dendritic cell/tumor fusion vaccine. J Immunol. 2003;170(7):3806–11. doi: 10.4049/jimmunol.170.7.3806. [DOI] [PubMed] [Google Scholar]
- 31.Gorelik L, Flavell RA. Immune-mediated eradication of tumors through the blockade of transforming growth factor-beta signaling in T cells. Nat Med. 2001;7:1118–22. doi: 10.1038/nm1001-1118. United States. [DOI] [PubMed] [Google Scholar]
- 32.Donkor MK, Sarkar A, Savage PA, Franklin RA, Johnson LK, Jungbluth AA, et al. T cell surveillance of oncogene-induced prostate cancer is impeded by T cell-derived TGF-beta1 cytokine. Immunity. 2011;35:123–34. doi: 10.1016/j.immuni.2011.04.019. United States: 2011 Elsevier Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas DA, Massague J. TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell. 2005;8(5):369–80. doi: 10.1016/j.ccr.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 34.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, et al. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–86. doi: 10.1084/jem.20030152. United States. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–8. doi: 10.1038/nature04753. England. [DOI] [PubMed] [Google Scholar]
- 36.Duraiswamy J, Ibegbu CC, Masopust D, Miller JD, Araki K, Doho GH, et al. Phenotype, function, and gene expression profiles of programmed death-1(hi) CD8 T cells in healthy human adults. J Immunol. 2011;186:4200–12. doi: 10.4049/jimmunol.1001783. United States. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 38.Oestreich KJ, Yoon H, Ahmed R, Boss JM. NFATc1 regulates PD-1 expression upon T cell activation. J Immunol. 2008;181:4832–9. doi: 10.4049/jimmunol.181.7.4832. United States. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wrana JL, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, et al. TGF beta signals through a heteromeric protein kinase receptor complex. Cell. 1992;71(6):1003–14. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 40.Nagaraj NS, Datta PK. Targeting the transforming growth factor-beta signaling pathway in human cancer. Expert Opin Investig Drugs. 2010;19(1):77–91. doi: 10.1517/13543780903382609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubtsov YP, Rudensky AY. TGFbeta signalling in control of T-cell-mediated self-reactivity. Nat Rev Immunol. 2007;7(6):443–53. doi: 10.1038/nri2095. [DOI] [PubMed] [Google Scholar]
- 42.Jinnin M, Ihn H, Tamaki K. Characterization of SIS3, a novel specific inhibitor of Smad3, and its effect on transforming growth factor-beta1-induced extracellular matrix expression. Mol Pharmacol. 2006;69(2):597–607. doi: 10.1124/mol.105.017483. [DOI] [PubMed] [Google Scholar]
- 43.Sfanos KS, Bruno TC, Meeker AK, De Marzo AM, Isaacs WB, Drake CG. Human prostate-infiltrating CD8+ T lymphocytes are oligoclonal and PD-1+ Prostate. 2009;69(15):1694–703. doi: 10.1002/pros.21020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson RH, Dong H, Lohse CM, Leibovich BC, Blute ML, Cheville JC, et al. PD-1 is expressed by tumor-infiltrating immune cells and is associated with poor outcome for patients with renal cell carcinoma. Clin Cancer Res. 2007;13:1757–61. doi: 10.1158/1078-0432.CCR-06-2599. United States. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Huang S, Gong D, Qin Y, Shen Q. Programmed death-1 upregulation is correlated with dysfunction of tumor-infiltrating CD8+ T lymphocytes in human non-small cell lung cancer. Cell Mol Immunol. 2010;7:389–95. doi: 10.1038/cmi.2010.28. China. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi F, Shi M, Zeng Z, Qi RZ, Liu ZW, Zhang JY, et al. PD-1 and PD-L1 upregulation promotes CD8(+) T-cell apoptosis and postoperative recurrence in hepatocellular carcinoma patients. Int J Cancer. 2011;128(4):887–96. doi: 10.1002/ijc.25397. [DOI] [PubMed] [Google Scholar]
- 47.Wang C, Li Y, Proctor TM, Vandenbark AA, Offner H. Down-modulation of programmed death 1 alters regulatory T cells and promotes experimental autoimmune encephalomyelitis. J Neurosci Res. 2010;88(1):7–15. doi: 10.1002/jnr.22181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, Kuchroo VK, et al. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med. 2009;206(13):3015–29. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng W, Liu C, Xu C, Lou Y, Chen J, Yang Y, et al. PD-1 blockade enhances T-cell migration to tumors by elevating IFN-gamma inducible chemokines. Cancer Res. 2012;72:5209–18. doi: 10.1158/0008-5472.CAN-12-1187. United States. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woo SR, Turnis ME, Goldberg MV, Bankoti J, Selby M, Nirschl CJ, et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res. 2012;72(4):917–27. doi: 10.1158/0008-5472.CAN-11-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldberg MV, Maris CH, Hipkiss EL, Flies AS, Zhen L, Tuder RM, et al. Role of PD-1 and its ligand, B7-H1, in early fate decisions of CD8 T cells. Blood. 2007;110:186–92. doi: 10.1182/blood-2006-12-062422. United States. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Austin JW, Lu P, Majumder P, Ahmed R, Boss JM. STAT3, STAT4, NFATc1, and CTCF regulate PD-1 through multiple novel regulatory regions in murine T cells. J Immunol. 2014;192(10):4876–86. doi: 10.4049/jimmunol.1302750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Streeck H, Brumme ZL, Anastario M, Cohen KW, Jolin JS, Meier A, et al. Antigen load and viral sequence diversification determine the functional profile of HIV-1-specific CD8+ T cells. PLoS Med. 2008;5:e100. doi: 10.1371/journal.pmed.0050100. United States. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McKarns SC, Schwartz RH. Distinct effects of TGF-beta 1 on CD4+ and CD8+ T cell survival, division, and IL-2 production: a role for T cell intrinsic Smad3. J Immunol. 2005;174(4):2071–83. doi: 10.4049/jimmunol.174.4.2071. [DOI] [PubMed] [Google Scholar]
- 55.Sung JL, Lin JT, Gorham JD. CD28 co-stimulation regulates the effect of transforming growth factor-beta1 on the proliferation of naive CD4+ T cells. Int Immunopharmacol. 2003;3(2):233–45. doi: 10.1016/S1567-5769(02)00276-X. [DOI] [PubMed] [Google Scholar]
- 56.McKarns SC, Schwartz RH, Kaminski NE. Smad3 is essential for TGF-beta 1 to suppress IL-2 production and TCR-induced proliferation, but not IL-2-induced proliferation. J Immunol. 2004;172(7):4275–84. doi: 10.4049/jimmunol.172.7.4275. [DOI] [PubMed] [Google Scholar]
- 57.Tinoco R, Alcalde V, Yang Y, Sauer K, Zuniga EI. Cell-Intrinsic Transforming Growth Factor-β Signaling Mediates Virus-Specific CD8+ T Cell Deletion and Viral Persistence In Vivo. Immunity. 2009;31(1):145–57. doi: 10.1016/j.immuni.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park HB, Paik DJ, Jang E, Hong S, Youn J. Acquisition of anergic and suppressive activities in transforming growth factor-beta-costimulated CD4+CD25− T cells. Int Immunol. 2004;16(8):1203–13. doi: 10.1093/intimm/dxh123. [DOI] [PubMed] [Google Scholar]
- 59.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-[beta] family signalling. Nature. 2003;425(6958):577–84. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 60.Gu A-D, Wang Y, Lin L, Zhang SS, Wan YY. Requirements of transcription factor Smad-dependent and -independent TGF-β signaling to control discrete T-cell functions. Proceedings of the National Academy of Sciences. 2012;109(3):905–10. doi: 10.1073/pnas.1108352109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang Y-Y, Brunicardi FC, Lin X. Smad3 mediates immediate early induction of Id1 by TGF-[beta] Cell Res. 2009;19(1):140–48. doi: 10.1038/cr.2008.321. [DOI] [PubMed] [Google Scholar]
- 62.Frederick JP, Liberati NT, Waddell DS, Shi Y, Wang XF. Transforming growth factor beta-mediated transcriptional repression of c-myc is dependent on direct binding of Smad3 to a novel repressive Smad binding element. Mol Cell Biol. 2004;24(6):2546–59. doi: 10.1128/MCB.24.6.2546-2559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takimoto T, Wakabayashi Y, Sekiya T, Inoue N, Morita R, Ichiyama K, et al. Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development. J Immunol. 2010;185(2):842–55. doi: 10.4049/jimmunol.0904100. [DOI] [PubMed] [Google Scholar]
- 64.Foulds KE, Zenewicz LA, Shedlock DJ, Jiang J, Troy AE, Shen H. Cutting edge: CD4 and CD8 T cells are intrinsically different in their proliferative responses. J Immunol. 2002;168(4):1528–32. doi: 10.4049/jimmunol.168.4.1528. [DOI] [PubMed] [Google Scholar]
- 65.Lu SL, Reh D, Li AG, Woods J, Corless CL, Kulesz-Martin M, et al. Overexpression of transforming growth factor beta1 in head and neck epithelia results in inflammation, angiogenesis, and epithelial hyperproliferation. Cancer Res. 2004;64:4405–10. doi: 10.1158/0008-5472.CAN-04-1032. United States. [DOI] [PubMed] [Google Scholar]
- 66.Guido M, De Franceschi L, Olivari N, Leandro G, Felder M, Corrocher R, et al. Effects of interferon plus ribavirin treatment on NF-kappaB, TGF-beta1, and metalloproteinase activity in chronic hepatitis C. Mod Pathol. 2006;19:1047–54. doi: 10.1038/modpathol.3800592. United States: 2006 S. Karger AG, Basel. [DOI] [PubMed] [Google Scholar]
- 67.Willimsky G, Czeh M, Loddenkemper C, Gellermann J, Schmidt K, Wust P, et al. Immunogenicity of premalignant lesions is the primary cause of general cytotoxic T lymphocyte unresponsiveness. J Exp Med. 2008;205:1687–700. doi: 10.1084/jem.20072016. United States. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Javle M, Li Y, Tan D, Dong X, Chang P, Kar S, et al. Biomarkers of TGF-beta signaling pathway and prognosis of pancreatic cancer. PLoS One. 2014;9:e85942. doi: 10.1371/journal.pone.0085942. United States. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Kruijf EM, Dekker TJ, Hawinkels LJ, Putter H, Smit VT, Kroep JR, et al. The prognostic role of TGF-beta signaling pathway in breast cancer patients. Ann Oncol. 2013;24:384–90. doi: 10.1093/annonc/mds333. England. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.