Abstract
BACKGROUND & AIMS
The development of vaccines and other strategies to prevent hepatitis C virus (HCV) infection is limited by rapid viral evasion. HCV entry is the first step of infection; this process involves several viral and host factors and is targeted by host-neutralizing responses. Although the roles of host factors in HCV entry have been well characterized, their involvement in evasion of immune responses is poorly understood. We used acute infection of liver graft as a model to investigate the molecular mechanisms of viral evasion.
METHODS
We studied factors that contribute to evasion of host immune responses using patient-derived antibodies, HCV pseudoparticles, and cell culture–derived HCV that express viral envelopes from patients who have undergone liver transplantation. These viruses were used to infect hepatoma cell lines that express different levels of HCV entry factors.
RESULTS
By using reverse genetic analyses, we identified altered use of host-cell entry factors as a mechanism by which HCV evades host immune responses. Mutations that alter use of the CD81 receptor also allowed the virus to escape neutralizing antibodies. Kinetic studies showed that these mutations affect virus–antibody interactions during postbinding steps of the HCV entry process. Functional studies with a large panel of patient-derived antibodies showed that this mechanism mediates viral escape, leading to persistent infection in general.
CONCLUSIONS
We identified a mechanism by which HCV evades host immune responses, in which use of cell entry factors evolves with escape from neutralizing antibodies. These findings advance our understanding of the pathogenesis of HCV infection and might be used to develop antiviral strategies and vaccines.
Keywords: Virology, Liver Disease, Tissue Culture Model, Immunity
Hepatitis C virus (HCV) infection is a major cause of liver disease.1 A vaccine is not available and antiviral treatment is limited by resistance and adverse effects.2 HCV-induced liver disease is a leading indication for liver transplantation (LT).3 A major limitation of LT is the universal reinfection of the liver graft with accelerated recurrence of liver disease. A strategy to prevent reinfection is lacking.3 Thus, there is an urgent unmet medical need for the development of efficient and safe antivirals and vaccines.
HCV entry is required for initiation, maintenance, and dissemination of infection. Viral entry is a key target for adaptive host responses and antiviral strategies.4,5 Functional studies in clinical cohorts highlight that viral entry and escape from antibody-mediated neutralization play an important role in viral persistence and liver disease.6–12 HCV entry is a highly orchestrated process mediated by viral envelope glycoproteins E1 and E2 and several host factors including heparan sulfate, CD81, scavenger receptor BI (SR-BI), claudin-1 (CLDN1), occludin (OCLN) (reviewed by Zeisel et al5), and kinases.13 Although the role of E1E2 in antibody-mediated neutralization has been studied intensively,4,5,14 the role of host factors for viral evasion in vivo is only poorly understood.
Acute graft infection is an established in vivo model to study viral evasion because viral infection and host-neutralizing responses can be monitored precisely.8 Viral entry and escape from host-neutralizing responses are important determinants allowing the virus to rapidly infect the liver during transplantation.8 However, the molecular mechanisms by which the virus evades host immunity to persistently reinfect the liver graft are unknown.
To uncover viral and host factors mediating enhanced viral entry and escape, we functionally analyzed genetically closely related prototype variants derived from a well-characterized patient undergoing LT.8 In one variant, P01VL, reinfecting the liver graft was characterized by high infectivity and escape from neutralizing antibodies present in autologous pretransplant serum.8 The other closely related variants, P01VA and P01VC, were not selected during LT and were characterized by lower infectivity and high sensitivity to neutralization by autologous pretransplant serum.8 Previous studies had indicated that an E2 region comprising amino acids 425–483 most likely contained mutations responsible for the phenotype of enhanced entry and viral evasion of variants reinfecting the liver graft.8
Materials and Methods
Patients
Evolution and functional analysis of viral variants of patient P01 have been described.8 Anti-HCV–positive serum samples from patients undergoing transplantation and chronic HCV infection were obtained with approval from the Strasbourg University Hospital Institutional Review Board (ClinicalTrial.gov Identifiers NCT00638144 and NCT00213707).
Plasmids
Plasmids for HCV pseudoparticle (HCVpp) production of variants VL, VA, and VC have been described.8 E1E2-encoding sequences were used as templates for individual and combinations of mutations using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies, Massy, France). Mutations were confirmed by DNA sequence analysis (GATC Biotech, Mulhouse, France) for the desired mutation and for exclusion of unexpected residue changes in the full-length E1E2 encoding sequences. Mutated constructs were designated X#Y, where # is the residue location in H77c,15 X is the mutated amino acid, and Y is the original amino acid.
Antibodies
Monoclonal anti-E1 (11B7) and anti-E2 (AP33, IGH461, 16A6); human anti-HCV IgG10,16; human monoclonal antibodies (HMAbs) CBH-2, CBH-5, CBH-23, and HC-1 have been described.9,17 Anti-CD81 (JS-81) was from BD Biosciences (Heidelberg, Germany), AP33 was from Genentech (San Francisco, CA), and 11B7, IGH461, and 16A6 were from Innogenetics (Ghent, Belgium).
Cell Lines
HEK 293T and Huh7.5.1 cells were cultured as described.10,13,16 Huh7.5.1 cells overexpressing HCV entry factors were created by stable lentiviral gene transfer of CLDN1, OCLN, SR-BI, or CD81.18 Huh7.5 stably transduced with retroviral vectors encoding for CD81- and CD13-specific short hairpin (sh) RNAs have been described.19 Receptor expression was assessed by flow cytometry.13
HCV Pseudoparticle and Cell Culture-Derived HCV Production, Infection, and Neutralization
Lentiviral HCVpp bearing patient-derived envelope glycoproteins were produced as described.8,10,20 The amount of HCVpp was normalized after quantification of human immunodeficiency virus p24 antigen expression (Innotest Human Immunodeficiency Virus Antigen mAb Kit; Innogenetics) and HCVpp entry was performed as described.8,10,11,16 Chimeric HCVcc expressing patient-derived structural proteins were constructed and produced as described in the Supplementary Materials and Methods section. HCVcc infectivity was measured by determining the tissue culture infectious dose 50% (TCID50)21 or intracellular HCV-RNA levels as described.13,21,22 HCVpp and HCVcc neutralization were performed as described.8,10,11,16
Kinetic Assays
HCVpp kinetic assays were performed in Huh7.5.1 cells using anti-CD81 (JS-81) and anti-E2 (CBH-23) monoclonal antibodies (mAbs) as described.16,23
Statistical Analysis
Statistical analysis (repeated-measures analysis of variance) was performed using SPSS 16.0 software for Windows (SPSS, Inc, Chicago, IL).
Results
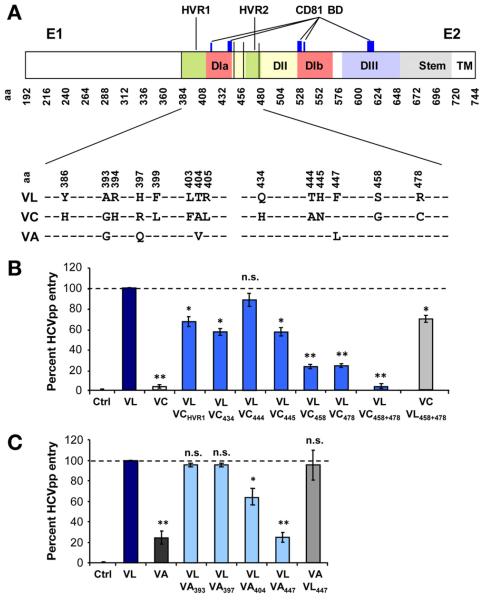
HCV E2 Residues at Positions 447, 458, and 478 Confer Enhanced Viral Entry of a High-Infectivity Variant Reinfecting the Liver Graft
To investigate the molecular mechanism of enhanced entry of the variant VL reinfecting the liver graft, we first introduced individual mutations of region E2425–4838 of the low-entry and neutralization-sensitive mutant VC into HCVpp expressing envelope glycoproteins of the highly infectious escape variant VL (Figure 1A). Previous studies indicated that this region most likely contains the mutations responsible for the high-infectivity phenotype of VL.8 After normalization of HCVpp levels by p24 antigen expression, viral entry was quantified relative to the escape variant VL. The entry level of the nonselected variant VC was 5% compared with the escape variant VL (Figure 1B). By introducing the mutations S458G and R478C into VC, chimeric HCVpp showed similar viral entry level as the paternal variant VL whereas introduction of individual or a combination of other mutations only had a partial effect (Figure 1B, Supplementary Figure 1). To explore the impact of other positions on viral entry we introduced mutations from another nonselected variant termed VA into VL (Figure 1A) and identified position F447 as an additional residue relevant for enhanced entry of the escape variant VL (Figure 1C). These results show that residues F447L, S458G, and R478C are largely responsible for the high infectivity of the escape variant VL.
Figure 1.
Positions 447, 458, and 478 confer enhanced viral entry of a high-infectivity variant reinfecting the liver graft. (A) Genomic organization and mutations of envelope glycoproteins of escape variant VL and nonselected variants VC and VA. HVR1 and HVR2 are depicted in green; E2 domains are depicted in red (DI), yellow (DII), and blue (DIII); and CD81 binding domains are depicted in dark blue.29,33,39 Positions 447, 458, and 478 are highlighted in black vertical lines. Differences between VL, VC, and VA in region E1E2384–483 are displayed. (B and C) Viral entry in Huh7.5.1 cells of the escape variant VL, the nonselected variants VC and VA, as well as chimeric variants containing defined mutations of VC and VA in VL or vice versa (Supplementary Figure 1). HCVpp infection was analyzed by luciferase reporter gene expression. Results are expressed as the percentage of viral entry compared with VL. Means ± standard deviation from at least 4 independent experiments performed in triplicate are shown. Significant differences in HCVpp entry between variants are indicated (*P ≤ .05; **P < .001). aa, amino acid; BD, binding domain.
Enhanced Viral Entry by Mutations F447L, S458G, and R478C of the Escape Variant Is the Result of Altered Use of CD81
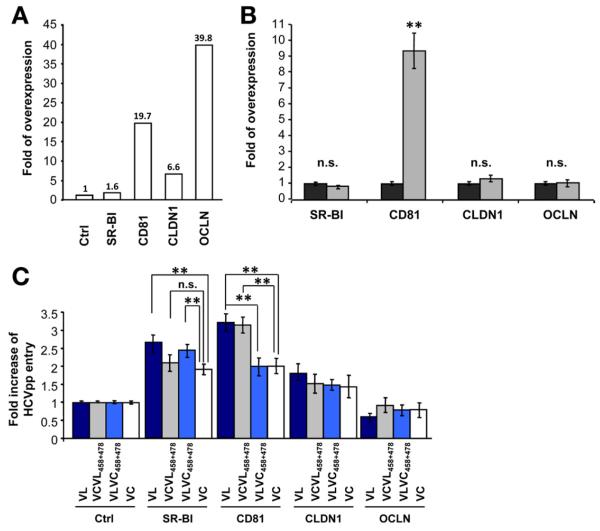
To address whether the mutations affect viral entry by different use of cell entry factors SR-BI, CD81, CLDN1, and OCLN, we studied viral entry of HCVpp derived from parental and chimeric variants in Huh7.5.1 cells stably overexpressing the 4 main entry factors individually (Figure 2A). Overexpression of either SR-BI, CD81, CLDN1, or OCLN did not affect the stability or proportion of other cell-surface HCV receptors (Figure 2B and data not shown).
Figure 2.
Altered use of CD81 is responsible for enhanced viral entry of the escape variant. (A) Entry factor expression in clones of SR-BI-, CD81-, CLDN1-, or OCLN-transduced Huh7.5.1 cells. The relative overexpression of each entry factor was determined by flow cytometry and is indicated as fold expression compared with parental Huh7.5.1 cells. (B) Entry factor expression in pools of CD81-overex-pressing Huh7.5.1 cells (grey bars). The relative entry factor expression was determined as described in panel A. (C) Receptor dependency of patient-derived HCVpp entry. Parental and transduced Huh7.5.1 cells were incubated with parental or chimeric HCVpp and viral entry was determined as described in Figure 1. Viral entry is expressed as the fold-change of viral entry compared with parental cells. Means ± standard deviation from 3 independent experiments performed in triplicate are shown. Significant differences in HCVpp entry between variants are indicated (**P < .001).
Overexpression of CD81 significantly enhanced viral entry of VL (3.2-fold) and VC (2-fold) compared with parental cells (P < .001) (Figure 2C). The fold-change in HCVpp entry was significantly higher for VL than for VC (P < .001). Exchanging the 2 residues at positions 458 and 478 similarly increased viral entry. This suggests that the combination of the 2 individual mutations modulates viral entry by altering CD81 dependency. Overexpression of SR-BI also increased viral entry of VL and VC, but no specific increase was observed for the chimeric strains containing substitutions at positions 458 and 478 (Figure 2C). These data confirm an important role for SR-BI as an entry factor for patient-derived variants, but also show that positions 458 and 478 do not significantly alter SR-BI dependency. Thus, increased entry efficiency of VL in SR-BI–overexpressing cells most likely is caused by other mutations (eg, in hypervariable region 1 [HVR1]). Viral entry enhancement was less pronounced in cells overexpressing CLDN1 or OCLN than CD81 and SR-BI (Figure 2C), and no specific modulation of viral entry was associated with the 2 variants or chimeric strains.
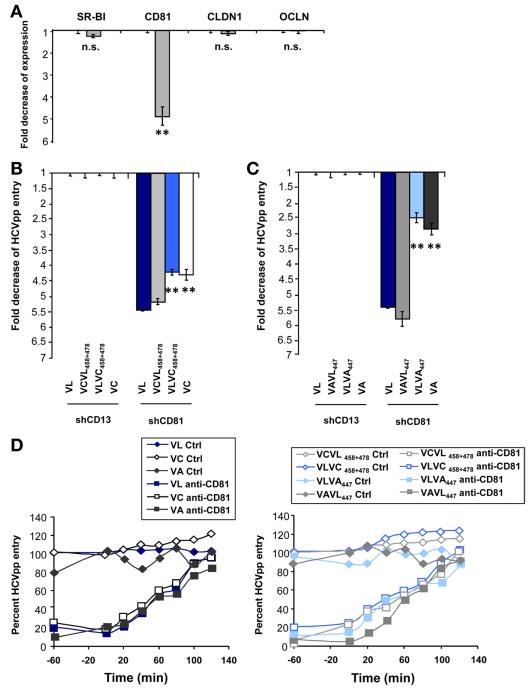
The CD81 use of viral variants VL, VC, and VA was investigated further using Huh7.5 cells with silenced CD81 expression (Figure 3A).19 The escape variant VL showed the highest decrease (5.4-fold) of viral entry in shCD81-Huh7.5 cells compared with the decrease of variants VC (4.3-fold; P < .001) and VA (2.9-fold; P < .001) (Figure 3B and C). Exchange of the mapped residues into chimeric expression plasmids conferred the phenotype of decreased entry of VL (Figure 3B and C), confirming that identified residues modulate viral entry by different CD81 use. Moreover, using a relevant model system for HCV-CD81 interactions occurring in vivo consisting of cell surface–expressed CD81, we show that E1E2 complexes of the escape variant VL bound less efficiently to shCD81-Huh7.5 cells than glycoproteins of variants VC and VA (Supplementary Figure 2A). Exchange of the mapped residues conferred similar phenotypes as the parental glycoproteins (Supplementary Figure 2B), suggesting that the residues at positions 447, 458, and 478 alter E1E2 interactions with cell surface CD81.
Figure 3.
Different CD81 use of viral variants in Huh7.5 cells with silenced CD81 expression. (A) Entry factor expression in Huh7.5 cells with silenced CD81 (grey bars) or CD13 (black bars) expression. CD81 expression was determined by flow cytometry and is indicated as fold expression compared with control shCD13-Huh7.5 cells. (B and C) Entry of patient-derived HCVpp VL, VC, and VA. Huh7.5 cells with silenced CD81 or CD13 expression were incubated with parental or chimeric HCVpp and viral entry was determined as described in Figure 1. Viral entry is expressed as the fold-change of viral entry compared with shCD13-Huh7.5 control cells. Means ± standard deviation from 3 independent experiments performed in triplicate are shown. Significant differences in HCVpp entry between wild-type and chimeric variants are indicated (**P < .001). (D) Entry kinetics of patient-derived variants. Kinetics of HCVpp entry was performed using anti-CD81 or isotype control antibody (5 μg/mL). HCV entry was determined as described in Figure 1. One representative experiment of 4 is shown.
Taken together, these data show the following: (1) the escape variant is characterized by markedly altered CD81 use, and (2) altered CD81 use of the variant is mediated by residues at positions 447, 458, and 478.
Because the levels of E1E2 incorporation into HCVpp and lentiviral p24 antigen expression were similar for all strains (Supplementary Figure 3A–D), it is unlikely that the differences in viral entry are the result of impaired HCVpp assembly or release.
Next, to assess whether enhanced entry is owing to more rapid internalization of viral particles, we investigated internalization kinetics of the parental and chimeric variants in the presence of anti-CD81 antibody.16,21,23,24 Because entry kinetics of parental and chimeric variants were similar (Figure 3D), it is unlikely that the mutant-induced modulation of CD81 dependency alters the velocity of viral entry.
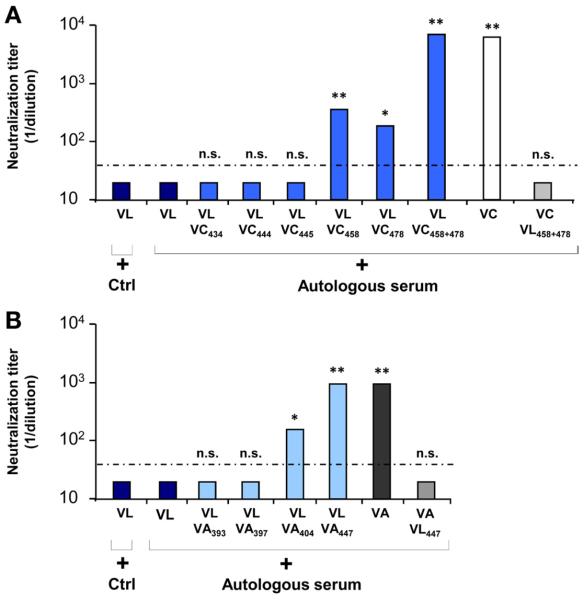
Positions 447, 458, and 478 Mediate Escape From Autologous Transplant Serum During Graft Reinfection
To assess whether the residues in region E2425–483 influencing viral entry (Figure 1) also were responsible for escape from antibody-mediated neutralization, we studied the impact of each single and combined substitution of the nonselected variant VC on neutralization by autologous pretransplant serum. Autologous pretransplant serum only poorly neutralized the selected variant VL as well as the variants substituted at positions 434, 444, and 445, whereas individual substitution at positions 458 and 478 significantly (P < .001 and P ≤ .05, respectively) increased the sensitivity of VLVC458 and VLVC478 to autologous neutralizing antibodies (1:400 and 1:200, respectively) (Figure 4A). It is noteworthy that only the variant VLVC458+478 showed a similar neutralization titer as the nonselected variant VC (1:6400; P < .001). To confirm that these mutations were indeed responsible for the phenotype of the parental variant VL, we investigated neutralization of VCVL458+478 by autologous serum. The variant VCVL458+478 escaped autologous neutralization similarly to the escape variant VL (Figure 4A). A similar phenotype was observed when mutation 447 of VA was introduced into the VL complementary DNA (Figure 4B). In contrast, the introduction of other residues into VL only had a minor effect on neutralization (Figure 4B). Taken together, these findings suggest that the residues at positions 447, 458, and 478 simultaneously are responsible for both enhanced viral entry and evasion from antibody-mediated neutralization.
Figure 4.
Positions 447, 458, and 478 mediate viral escape from neutralization by autologous transplant serum. Neutralization of the escape variant VL, variants VC and VA, and the chimeric strains. HCVpp were incubated with autologous anti-HCV–positive or control serum in serial dilutions for 1 hour at 37°C before incubation with Huh7.5.1 cells. Neutralization titers obtained by end point dilution are indicated. Dotted line indicates the threshold for a positive neutralization titer (1/40). Means ± standard deviation from at least 4 experiments performed in triplicate are shown. (A) Neutralization of variants VL, VL containing individual or combined mutations of VC, and VC with double substitutions of VL by autologous anti–HCV-positive pretransplant serum. (B) Neutralization of variants VL, VL containing individual mutations of VA, and VA with single substitution of VL by autologous anti-HCV–positive pretransplant serum. Significant differences in neutralization between variants are indicated (*P ≤ .05; **P < .001).
Positions 447, 458, and 478 Define a Conformational Epitope Involved in Evasion From Host-Neutralizing Responses
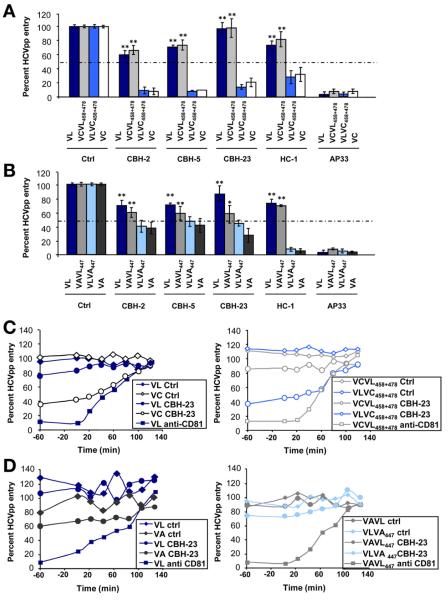
To further elucidate the mechanism of viral evasion of the escape variant VL from patient-derived neutralizing antibodies, we investigated whether the identified mutations F447L, S458G, and R478C confer resistance or sensitivity to a panel of mAbs directed against conformational9,17 and linear E2 epitopes.16 The conformational HMAbs (CBH-2, CBH-5, CBH-23, and HC-1) have shown a broad cross-neutralizing activity by interfering with E2-CD81 interaction9,17 and their epitopes only partially are defined (Supplementary Table 1). AP33 is directed against a conserved epitope comprising amino acids 412–423.25 Although the escape variant VL was neutralized poorly by several HMAbs directed against conformational epitopes, VC and VA were neutralized efficiently by all HMAbs (Figure 5A and B). Moreover, by substituting the residues at positions 458 and 478 or 447, the well-neutralized nonselected variants VC (VCVL458+478) and VA (VAVL447) became neutralization resistant as the escape variant VL. Introducing the residues of VC or VA into VL (VLVC458+478 and VLVA447) restored neutralization by HMAbs, suggesting that these residues are part of the HMAbs epitopes. In contrast, anti-E2 antibodies (AP33, 16A6, IGH461) targeting linear epitopes similarly neutralized parental and chimeric variants (Figure 5A and B and Supplementary Table 1).
Figure 5.
Mechanisms of viral evasion from neutralizing antibodies. (A and B) Escape from neutralization by HMAbs directed against conformational and linear epitopes. HCVpp produced from isolates shown in Figure 1 were incubated with HMAbs (Supplementary Table 1) or control Ab (10 μg/mL) for 1 hour at 37°C before incubation with Huh7.5.1 cells. Results are expressed as the percentage of viral entry relative to HCVpp incubated with control mAb. Means ± standard deviation from at least 4 experiments performed in triplicate are shown. Significant differences in HCVpp entry between variants are indicated (**P < .001). (C and D) Escape from neutralization of anti-E2 antibody CBH-23 in kinetic assays. Kinetics were performed as described in Figure 3 (HMAb, 10 μg/mL; JS-81, 5 μg/mL). One representative experiment of 4 is shown.
Antibody-mediated neutralization occurs at binding and postbinding steps during viral entry.16 To map the entry step involved in viral evasion from neutralizing antibodies by VL, we investigated the neutralization kinetics of parental and chimeric variants.16,21,23 The anti-E2 HMAb CBH-23 inhibited viral entry of VC and VLVC458+478 at postbinding steps during time points closely related to HCV-CD81 interaction (Figure 5C). Partial inhibition at postbinding steps by CBH-23 also was observed for VA and VLVA447 (Figure 5D). The VL variant escaped antibody-mediated neutralization at the same steps.
Interestingly, purified HCVpp expressing envelope glycoproteins of the escape variant bound similarly to neutralizing anti-E2 antibody CBH-23 as the envelope glycoproteins of nonselected variants or variants containing mutations of the identified escape residue (Supplementary Figure 4). Thus, it is likely that viral evasion is not caused by decreased antibody binding to circulating virions but rather occurs during postbinding steps of viral entry in which E2-host entry factor interactions result in conformational changes of the envelope and failure of antibodies to inhibit entry. Taken together, these data indicate that positions 447, 458, and 478 mediate viral evasion from neutralizing antibodies at postbinding steps and time points closely related to HCV-CD81 interaction.
Positions 447, 458, and 478 Mediate Escape From Antiviral Antibodies in Nonrelated Patients With Chronic HCV Infection
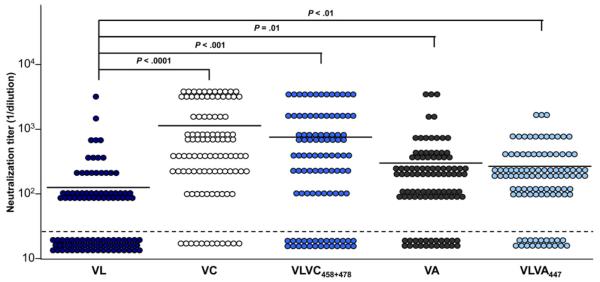
To investigate whether these mutations result not only in escape from antibodies from the same patient but also confer resistance to antiviral antibodies of nonrelated HCV-infected patients, we studied the neutralization of the parental variants by a large panel of sera randomly selected from chronically infected patients (n = 102). While VL was not neutralized by 53 of 102 patient sera (mean neutralizing titer, 1:144), VC was neutralized significantly by 90 of 102 patient sera (mean neutralizing titer, 1:1088; P < .001) (Figure 6 and Supplementary Tables 2 and 3). Similar results were obtained for VA (neutralization by 80 of 102 patient sera; mean neutralizing titer, 1:322; P = .01). Functional analysis of HCVpp expressing chimeric envelope glycoproteins showed that neutralization of VC and VA was mediated predominantly by the identified mutations in residues 447, 458, and 478 (Figure 6).
Figure 6.
The HCV VL strain is poorly neutralized by antibodies present in sera from a large panel of nonrelated patients with chronic HCV infection. Parental HCVpp (VL, VC, and VA) and chimeric HCVpp (VLVC458+478 and VLVA447) strains, adjusted for p24 antigen expression, were preincubated for 1 hour with serial dilutions of anti-HCV–positive sera from randomly selected patients with chronic hepatitis C before incubation with Huh7.5.1 target cells. Patient number, sex, HCV genotype, and viral load are indicated in Supplementary Tables 2 and 3. Neutralization was determined as in Figure 4. Mean neutralization titers are marked by lines. Means from at least 3 independent experiments performed in triplicate are shown. Significant differences in neutralization are indicated.
Confirmation of Differential Cell Entry Factor Use and Viral Evasion Using Chimeric HCVcc
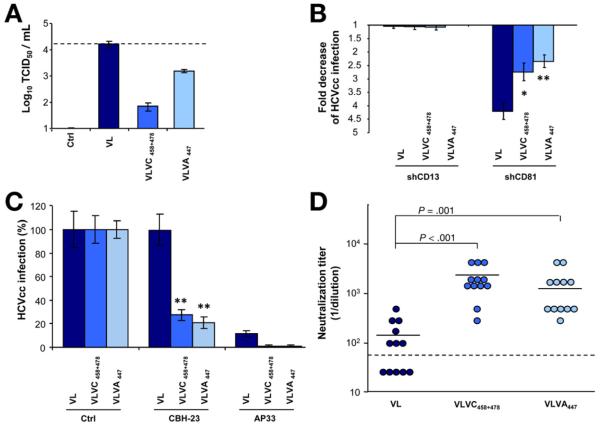
Finally, we confirmed the functional impact of the 3 residues on virus–host interactions using the HCVcc system. To address this issue we constructed chimeric JFH-1–based HCVcc expressing the VL wild-type envelope or VL-containing VC- and VA-specific functional residues. Viruses containing patient-derived envelopes showed similar levels of replication and envelope production (data not shown). Phenotypic analyses of infection and neutralization of chimeric HCVcc confirmed the relevance of the identified residues for enhanced entry, differential CD81 use, and viral evasion (Figure 7). While the escape variant VL was poorly neutralized, the identified mutations at positions 447, 458, and 478 restored its sensitivity to conformational HMAb CBH-23 (Figure 7C) as well as to heterologous sera from chronically infected patients (Figure 7D). These data confirm the functional relevance of the obtained results in the HCVcc system expressing authentic patient-derived envelopes.
Figure 7.
Viral entry and escape from neutralization of chimeric HCVcc expressing patient-derived viral envelopes. (A) Infectivity of HCVcc expressing envelopes of variant VL and functional residues of VA and VC is indicated by TCID50. Means ± standard deviation from 1 representative experiment are shown. (B) Relative infectivity of chimeric HCVcc expressing patient-derived viral envelopes in Huh7.5 cells with silenced CD81 or CD13 expression. Means ± standard deviation from 3 independent experiments performed in triplicate are shown. (C) Escape from neutralization by HMAb CBH-23. Neutralization was performed as described in Figure 5. Results are expressed as the percentage of viral infectivity relative to HCVcc incubated with control mAb. Means ± standard deviation (SD) from at least 3 experiments performed in triplicate are shown. (D) Inhibition of HCVcc infection by anti-HCV–positive sera described in Supplementary Table 3. Neutralization was performed as described in Figure 6. Means from 1 representative experiment performed in triplicate are shown. Significant differences in HCVcc infection between wild-type and chimeric variants are indicated (*P ≤ .05; **P < .001).
Discussion
By using acute infection of the liver graft as an in vivo model, we identified a novel clinically and therapeutically important mechanism of viral evasion, where co-evolution simultaneously occurs between cellular entry factor use and escape from neutralization.
Several host selection forces operate concomitantly during HCV infection. These include proviral host factors resulting in selection of most infectious viruses best adapted to host factors and antiviral host immune responses leading to escape from immune responses. Antibody-mediated selective pressure is thought to be an important driver of viral evolution.8,11 The immune response may fail to resolve HCV infection because neutralizing antibody-mediated response lags behind the rapidly and continuously evolving HCV glycoprotein sequences.11 However, continuous generation of escape mutations during chronic HCV infection also may compromise virus infectivity: indeed, it has been reported that structural changes in E2 leading to complete escape from neutralizing antibodies simultaneously compromised viral fitness by reducing CD81 binding.9 Moreover, escape from T-cell responses has been associated with impaired viral replication.26,27 We show that clinically occurring mutations simultaneously lead to enhanced viral infectivity by optimizing host factor use and escape from host immune responses. Because this mechanism was uncovered in patient strains isolated during acute liver graft infection it is likely that the novel and unique mechanism of co-evolution between host factor use and viral evasion ensures optimal initiation, dissemination, and maintenance of viral infection in the early phase of liver graft infection. In addition, because the VL strain escapes autologous antibodies from the transplant patient (Fig. 4) and resists monoclonal and polyclonal antibodies of heterologous patients (Figures 5, 6, and 7, and Supplementary Tables 1 and 2), and given the high prevalence of the identified mutations in a large genomic database of viral isolates (Supplementary Figure 5 and Supplementary Results section), the co-evolution of receptor use and escape from neutralizing antibodies also may play an important role for viral evasion in chronic HCV infection in general.
Our mechanistic studies show that the identified viral evasion factors are part of a conformational neutralizing epitope modulating E2-CD81 interactions at postbinding entry steps.28,29 It is noteworthy that the same mutations also were responsible for immune escape of VL. Neutralization studies using HMAbs directed against discontinuous envelope glycoprotein regions termed domain B and domain C30,31 show that the 3 positions are part of an epitope that plays a key role for neutralization and viral evasion. Because the mutations are outside the known contact residues within the epitopes of the HMAbs CBH-2, CBH-5, CBH-23, and HC-19,17 (Supplementary Table 1), and complementary to previously identified regions associated with escape from neutralizing monoclonal antibodies,25 positions 447, 458, and 478 either modulate the interaction of the majority of antibodies directed against domain B and C epitopes or are part of a novel E2 epitope mediating evasion from host neutralizing antibodies.
Based on previous functional observations and structural predictions, Krey et al29 proposed a model for a potential tertiary organization of E2. In this model, E2 comprises 3 subdomains with the CD81 binding regions located within domain I (W420, A440LFY, Y527, W529, G530, and D535) and potential CD81 binding sites over-lapping with domain III (Y613RLWHY).28,29,32,33 In this model, positions 447, 458, and 478 are located outside but in close proximity of the previously suggested CD81 binding domains. Moreover, position 447 is located immediately downstream of a conserved motif between HVR1 and HVR2, which has been shown to play an important role in CD81 recognition as well as pre- or post-CD81–dependent stages of viral entry.32 Position 478 is located within HVR2, which modulates, by a complex interplay with HVR1, binding of E2 glycoprotein to CD81.34
Because mutations F447L, S458G, and R478C (1) modulate CD81 dependency of HCV entry (Figures 2 and 3), (2) alter the interaction with cell surface CD81 (Supplementary 2), (3) mediate viral evasion from antibodies at postbinding steps closely related to HCV-CD81 interactions (Figure 5), and (4) are located within E2 loops of the predicted E2 secondary structure and tertiary organization,29 positions 447, 458, and 478 may be part of 2 loops belonging to a larger cluster of closely related surface-exposed E2 loops. These loops most likely are involved in E2-CD81 binding either directly or indirectly as a key point for structural rearrangement during viral entry.34,35
The polar S and R residues present in the escape variant can form nonbonded interactions with other residues by hydrogen bonds and salt bridge, respectively. These interactions could increase the stability of the interacting E2-CD81 interface, allowing efficient entry of the VL escape variant through E2-CD81-CLDN1 co-receptor complexes, which are key determinants for viral entry.13,23,36 Further-more, the E2 cluster of loops containing the mutations bears linear epitopes but also defines at least one conformational epitope that is a target of neutralizing antibodies. According to residue physical-chemical properties, the VL variant S458 and R478 residues enhance the hydro-philicity of the loops they belong to and may promote the surface exposure of the loops. This change could modulate E2-CD81 interactions further and impair the binding of neutralizing antibodies by blocking access to their target epitopes. The F to L substitution present in the VA strain most likely does not profoundly alter the tertiary or quaternary structure of E2. This is suggested by the fact that this position is located in a loop as predicted by the proposed E2 model.29 Thus, it is conceivable that this mutation, which increases E2 hydrophobicity, may reduce accessibility of the loop and its interactions with CD81 or CD81-CLDN1 co-receptor complexes. Alternatively, allosteric mechanisms may play a role in the observed virus-antibody-host interactions.
Taken together, our data identified key determinants of immune evasion in vivo. Mutations conferring neutralization escape altered CD81 receptor use and enhanced cell entry. Moreover, our data suggest that mutations in HVR1, which may modulate entry and neutralization by altering SR-BI dependency (Figures 1, 2, and 4, and data not shown), may contribute to the high entry and escape phenotype of the escape variant. Furthermore, interfering non-neutralizing antibodies may constitute another mechanism of escape (data not shown).
Although proof-of-concept studies in animal models have shown a potential role for HMAbs in prevention of HCV infection,37,38 the partial or complete escape of the VL variant from autologous and heterologous serum-derived antibodies as well as many broadly cross-neutralizing HMAbs (Figure 5; Supplementary Table 1) shows the ability of the virus to evade cross-neutralizing anti-envelope mAbs. By identifying viral and host factors mediating immune evasion in the HCV-infected patient, our results may open new perspectives for the development of broadly cross-neutralizing anti-envelope or antibodies overcoming viral escape.
Supplementary Material
Supplementary Figure 1. Actual viral infectivity of HCVpp derived from variants VL, VC, and VA shown as relative light units (RLU) of luciferase reporter gene expression. (A and B) Comparative analysis of viral entry of HCVpp shown in Figure 1. Results are expressed in RLUs plotted in a logarithmic scale. The threshold for a detectable infection in this system is indicated by dashed lines. The detection limit for positive luciferase reporter protein expression was 3 × 103 RLU/assay, corresponding to the mean ± 3 standard deviations of background levels (ie, luciferase activity of naive noninfected cells or cells infected with pseudotypes without HCV envelopes).1,12,13 Background levels of the assay were determined in each experiment. Means ± standard deviation from at least 4 independent experiments performed in triplicate are shown. Significant differences in HCVpp entry VC, VA, and VL wild-type and mutant variants are indicated (*P ≤ .05; **P < .001). Ctrl, control; HVR, hypervariable region; V, viral variant.
Supplementary Figure 2. Positions 447, 458, and 478 modulate binding of envelope glycoproteins to CD81 expressed at the cell surface. Binding of native E1E2 complexes expressed from patient-derived complementary DNAs to Huh7.5 cells with silenced CD81 expression (described in Figure 3) was detected by flow cytometry. Results are expressed as the percentage of E1E2 binding compared with shCD13-Huh7.5 control cells. Means ± standard deviation from 3 independent experiments performed in triplicate are shown. Significant differences in binding between variants are indicated (**P < .001).
Supplementary Figure 3. Differences in viral entry are not caused by impaired HCVpp production. (A) Analysis of envelope glycoprotein expression. Protein expression was analyzed by immunoblotting as described in the Materials and Methods section. Molecular markers (in kilodaltons) are indicated on the right. (B) Transfection efficiency during HCVpp production. Transfection efficiency was analyzed for each variant and quantified by determining luciferase expression in HEK 293T producer cells expressed as a normalized percentage compared with control transfected cells. (C) Envelope glycoprotein expression in HCVpp. HCVpp were purified as described previously1,2 and subjected to immunoblot as described in panel A. (D) Lentiviral p24 antigen expression was analyzed by enzyme-linked immunosorbent assay (ELISA) and is indicated as optical density (OD) values at 450 nm. (E) Cellular binding of E2 derived from patient-derived or H77 and HCV-J strains. Binding of native E1E2 complexes to Huh7.5.1 cells was detected as described in Supplementary Materials and Methods. Results are expressed as delta mean fluorescence intensity (ΔMFI) ± standard deviation. One representative experiment of 3 is shown. Da, dalton; MW, molecular weight.
Supplementary Figure 4. Binding of neutralizing anti-E2 HMAb CBH-23 to patient-derived envelope glycoproteins expressed on HCVpp as capture antigens in an enzyme-linked immunosorbent assay (ELISA). HCVpp expressing envelope glycoproteins of variants VL, VA, VC, VLVA447, and VLVC458+478 were used as capture antigens on GNA-coated ELISA plates. Control (Ctrl) pseudoparticles with absent HCV envelope glycoprotein expression and recombinant soluble E2 (sE2 derived from strain H77)14 served as negative and positive controls, respectively. Anti-E2 CBH-23 reactivity was detected as described in the Supplementary Materials and Methods section and is indicated as optical density (OD) values at 450 nm. Means ± standard deviation from 1 representative experiment are shown.
Supplementary Figure 5. Distribution of residues at positions 447, 458, and 478 of HCV E2 sequences in the European HCV databases. Distribution of residues at positions 447, 458, and 478 for HCV complete E2 sequences from all subtypes (black) and from subtype 1b only (white) within the European Hepatitis C Virus databases7 (available: http://euhcvdb.ibcp.fr). F and S are the predominant residue at positions 447 and 458 (F447, 98.4%; 1b, 96.2%; S458 all, 94%; 1b, 90.3%). The position 478 is variable (it belongs to HVR2) but R (all, 2.4%; 1b, 10.8%) is more frequent than C (all, 0.2%; 1b, 0.9%).
Supplementary Table 1. Neutralization of Patient-Derived and Chimeric HCVpp by Monoclonal Anti-Envelope Antibodies
Supplementary Table 2. Characteristics of Patients and Viruses Used for Neutralization Studies
Supplementary Table 3. HCVcc Neutralization Titers
Acknowledgments
The authors thank F. Chisari (The Scripps Research Institute, La Jolla, CA) for the gift of Huh7.5.1 cells, J. A. McKeating (University of Birmingham, Birmingham, UK), C. Rice (Rockefeller University, New York, NY), C. Schuster (Inserm U748, Strasbourg, France), M. Heim (University of Basel, Basel, Switzerland), F. Wong-Staal (Itherx, San Diego, CA), J. Dubuisson (Inserm U1019, Lille, France), and F. Rey (Institut Pasteur, Paris, France) for helpful discussions. The authors acknowledge the excellent technical assistance of Michèle Bastien-Valle (Inserm U748, Strasbourg, France).
Funding
This work was supported by Inserm, the European Union (ERC-2008-AdG-233130-HEPCENT and Interreg IV FEDER-Hepato-Regio-Net 2009), the Agence Nationale de la Recherche chair of excellence program (ANR-05-CEXC-008), Agence Nationale de Recherches sur le Sida et les Hépatites Virales (2007/306, 2008/354, 2009/183, 2011/132), the Région d’Alsace (2007/09), the Else Kröner-Fresenius Stiftung (EKFS P17//07//A83/06), the Ligue Contre le Cancer (CA 06/12/08), INCA (2009-143), Canceropôle du Grand-Est (30/03/09), the Finovi Foundation, the Infrastrutures en Biologie Santé et Agronomie, the Société Française d’Exportation des Ressources Educatives program of Higher Education Commission of Pakistan, and Public Heatlh Service grants HL079381 and AI081903.
Abbreviations used in this paper
- CLDN
claudin
- HCV
hepatitis C virus
- HCVcc
cell culture-derived HC
- HCVpp
hepatitis C virus pseudop-articles
- HMAb
human monoclonal antibody
- HVR
hypervariable region
- LT
liver transplantation
- mAb
monoclonal antibody
- OCLN
occludin SR-BI, scavenger receptor class B type I
- VA
variant A
- VC
variant C
- VL
variant L
Footnotes
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at doi: http://dx.doi.org/10.1053/j.gastro.
Conflicts of interest
This author discloses the following: Thomas Pietschmann is a member of the advisory board of Biotest AG and has received consulting fees. The remaining authors disclose no conflicts.
References
- 1.Alter H. Viral hepatitis. Hepatology. 2006;43:S230–S234. doi: 10.1002/hep.21030. [DOI] [PubMed] [Google Scholar]
- 2.Hofmann WP, Zeuzem S. A new standard of care for the treatment of chronic HCV infection. Nat Rev Gastroenterol Hepatol. 2011;8:257–264. doi: 10.1038/nrgastro.2011.49. [DOI] [PubMed] [Google Scholar]
- 3.Watt K, Veldt B, Charlton M. A practical guide to the management of HCV infection following liver transplantation. Am J Transplant. 2009;9:1707–1713. doi: 10.1111/j.1600-6143.2009.02702.x. [DOI] [PubMed] [Google Scholar]
- 4.Zeisel MB, Cosset FL, Baumert TF. Host neutralizing responses and pathogenesis of hepatitis C virus infection. Hepatology. 2008;48:299–307. doi: 10.1002/hep.22307. [DOI] [PubMed] [Google Scholar]
- 5.Zeisel MB, Fofana I, Fafi-Kremer S, et al. HCV entry into hepato-cytes: mechanism and potential therapeutic implications. J Hepatol. 2011;54:566–576. doi: 10.1016/j.jhep.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 6.Lavillette D, Morice Y, Germanidis G, et al. Human serum facilitates hepatitis C virus infection, and neutralizing responses inversely correlate with viral replication kinetics at the acute phase of hepatitis C virus infection. J Virol. 2005;79:6023–6034. doi: 10.1128/JVI.79.10.6023-6034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dowd KA, Netski DM, Wang XH, et al. Selection pressure from neutralizing antibodies drives sequence evolution during acute infection with hepatitis C virus. Gastroenterology. 2009;136:2377–2386. doi: 10.1053/j.gastro.2009.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fafi-Kremer S, Fofana I, Soulier E, et al. Enhanced viral entry and escape from antibody-mediated neutralization are key determinants for hepatitis C virus re-infection in liver transplantation. J Exp Med. 2010;207:2019–2031. doi: 10.1084/jem.20090766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keck ZY, Li SH, Xia J, et al. Mutations in hepatitis C virus E2 located outside the CD81 binding sites lead to escape from broadly neutralizing antibodies but compromise virus infectivity. J Virol. 2009;83:6149–6160. doi: 10.1128/JVI.00248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pestka JM, Zeisel MB, Blaser E, et al. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc Natl Acad Sci U S A. 2007;104:6025–6030. doi: 10.1073/pnas.0607026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Hahn T, Yoon JC, Alter H, et al. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology. 2007;132:667–678. doi: 10.1053/j.gastro.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Osburn WO, Fisher BE, Dowd KA, et al. Spontaneous control of primary hepatitis C virus infection and immunity against persistent reinfection. Gastroenterology. 2010;138:315–324. doi: 10.1053/j.gastro.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lupberger J, Zeisel MB, Xiao F, et al. EGFR and EphA2 are hepatitis C virus host entry factors and targets for antiviral therapy. Nat Med. 2011;17:589–595. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stamataki Z, Grove J, Balfe P, et al. Hepatitis C virus entry and neutralization. Clin Liver Dis. 2008;12:693–712. doi: 10.1016/j.cld.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 15.Kuiken C, Combet C, Bukh J, et al. A comprehensive system for consistent numbering of HCV sequences, proteins and epitopes. Hepatology. 2006;44:1355–1361. doi: 10.1002/hep.21377. [DOI] [PubMed] [Google Scholar]
- 16.Haberstroh A, Schnober EK, Zeisel MB, et al. Neutralizing host responses in hepatitis C virus infection target viral entry at post-binding steps and membrane fusion. Gastroenterology. 2008;135:1719–1728. doi: 10.1053/j.gastro.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 17.Hadlock KG, Lanford RE, Perkins S, et al. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J Virol. 2000;74:10407–10416. doi: 10.1128/jvi.74.22.10407-10416.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haid S, Windisch MP, Bartenschlager R, et al. Mouse-specific residues of claudin-1 limit hepatitis C virus genotype 2a infection in a human hepatocyte cell line. J Virol. 2009;84:964–975. doi: 10.1128/JVI.01504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koutsoudakis G, Herrmann E, Kallis S, et al. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J Virol. 2007;81:588–598. doi: 10.1128/JVI.01534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med. 2003;197:633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeisel MB, Koutsoudakis G, Schnober EK, et al. Scavenger receptor BI is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology. 2007;46:1722–1731. doi: 10.1002/hep.21994. [DOI] [PubMed] [Google Scholar]
- 22.Fofana I, Krieger SE, Grunert F, et al. Monoclonal anti-claudin 1 antibodies for prevention of hepatitis C virus infection. Gastroenterology. 2010;139:953–964. 964.e1–4. doi: 10.1053/j.gastro.2010.05.073. [DOI] [PubMed] [Google Scholar]
- 23.Krieger SE, Zeisel MB, Davis C, et al. Inhibition of hepatitis C virus infection by anti-claudin-1 antibodies is mediated by neutralization of E2-CD81-claudin-1 associations. Hepatology. 2010;51:1144–1157. doi: 10.1002/hep.23445. [DOI] [PubMed] [Google Scholar]
- 24.Koutsoudakis G, Kaul A, Steinmann E, et al. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J Virol. 2006;80:5308–5320. doi: 10.1128/JVI.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owsianka A, Tarr AW, Juttla VS, et al. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J Virol. 2005;79:11095–11104. doi: 10.1128/JVI.79.17.11095-11104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dazert E, Neumann-Haefelin C, Bressanelli S, et al. Loss of viral fitness and cross-recognition by CD8+ T cells limit HCV escape from a protective HLA-B27-restricted human immune response. J Clin Invest. 2009;119:376–386. doi: 10.1172/JCI36587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uebelhoer L, Han JH, Callendret B, et al. Stable cytotoxic T cell escape mutation in hepatitis C virus is linked to maintenance of viral fitness. PLoS Pathog. 2008;4:e1000143. doi: 10.1371/journal.ppat.1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Owsianka AM, Timms JM, Tarr AW, et al. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J Virol. 2006;80:8695–8704. doi: 10.1128/JVI.00271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krey T, d’Alayer J, Kikuti CM, et al. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 2010;6:e1000762. doi: 10.1371/journal.ppat.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keck ZY, Op De Beeck A, Hadlock KG, et al. Hepatitis C virus E2 has three immunogenic domains containing conformational epitopes with distinct properties and biological functions. J Virol. 2004;78:9224–9232. doi: 10.1128/JVI.78.17.9224-9232.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helle F, Goffard A, Morel V, et al. The neutralizing activity of anti-hepatitis C virus antibodies is modulated by specific glycans on the E2 envelope protein. J Virol. 2007;81:8101–8111. doi: 10.1128/JVI.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drummer HE, Boo I, Maerz AL, et al. A conserved gly436-trp-leuala-gly-leu-phe-tyr motif in hepatitis C virus glycoprotein E2 is a determinant of CD81 binding and viral entry. J Virol. 2006;80:7844–7853. doi: 10.1128/JVI.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boo I, Tewierek K, Douam F, et al. Distinct roles in folding, CD81 receptor binding and viral entry for conserved histidines of HCV glycoprotein E1 and E2. Biochem J. 2012;443:85–94. doi: 10.1042/BJ20110868. [DOI] [PubMed] [Google Scholar]
- 34.Roccasecca R, Ansuini H, Vitelli A, et al. Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain specific and is modulated by a complex interplay between hypervariable regions 1 and 2. J Virol. 2003;77:1856–1867. doi: 10.1128/JVI.77.3.1856-1867.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCaffrey K, Boo I, Poumbourios P, et al. Expression and characterization of a minimal hepatitis C virus glycoprotein E2 core domain that retains CD81 binding. J Virol. 2007;81:9584–9590. doi: 10.1128/JVI.02782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harris HJ, Davis C, Mullins JG, et al. Claudin association with CD81 defines hepatitis C virus entry. J Biol Chem. 2010;285:21092–21102. doi: 10.1074/jbc.M110.104836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Law M, Maruyama T, Lewis J, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med. 2008;14:25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 38.Vanwolleghem T, Bukh J, Meuleman P, et al. Polyclonal immuno-globulins from a chronic hepatitis C virus patient protect human liver-chimeric mice from infection with a homologous hepatitis C virus strain. Hepatology. 2008;47:1846–1855. doi: 10.1002/hep.22244. [DOI] [PubMed] [Google Scholar]
- 39.Rychlowska M, Owsianka AM, Foung SK, et al. Comprehensive linker-scanning mutagenesis of the hepatitis C virus E1 and E2 envelope glycoproteins reveals new structure-function relationships. J Gen Virol. 2011;92:2249–2261. doi: 10.1099/vir.0.034314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Actual viral infectivity of HCVpp derived from variants VL, VC, and VA shown as relative light units (RLU) of luciferase reporter gene expression. (A and B) Comparative analysis of viral entry of HCVpp shown in Figure 1. Results are expressed in RLUs plotted in a logarithmic scale. The threshold for a detectable infection in this system is indicated by dashed lines. The detection limit for positive luciferase reporter protein expression was 3 × 103 RLU/assay, corresponding to the mean ± 3 standard deviations of background levels (ie, luciferase activity of naive noninfected cells or cells infected with pseudotypes without HCV envelopes).1,12,13 Background levels of the assay were determined in each experiment. Means ± standard deviation from at least 4 independent experiments performed in triplicate are shown. Significant differences in HCVpp entry VC, VA, and VL wild-type and mutant variants are indicated (*P ≤ .05; **P < .001). Ctrl, control; HVR, hypervariable region; V, viral variant.
Supplementary Figure 2. Positions 447, 458, and 478 modulate binding of envelope glycoproteins to CD81 expressed at the cell surface. Binding of native E1E2 complexes expressed from patient-derived complementary DNAs to Huh7.5 cells with silenced CD81 expression (described in Figure 3) was detected by flow cytometry. Results are expressed as the percentage of E1E2 binding compared with shCD13-Huh7.5 control cells. Means ± standard deviation from 3 independent experiments performed in triplicate are shown. Significant differences in binding between variants are indicated (**P < .001).
Supplementary Figure 3. Differences in viral entry are not caused by impaired HCVpp production. (A) Analysis of envelope glycoprotein expression. Protein expression was analyzed by immunoblotting as described in the Materials and Methods section. Molecular markers (in kilodaltons) are indicated on the right. (B) Transfection efficiency during HCVpp production. Transfection efficiency was analyzed for each variant and quantified by determining luciferase expression in HEK 293T producer cells expressed as a normalized percentage compared with control transfected cells. (C) Envelope glycoprotein expression in HCVpp. HCVpp were purified as described previously1,2 and subjected to immunoblot as described in panel A. (D) Lentiviral p24 antigen expression was analyzed by enzyme-linked immunosorbent assay (ELISA) and is indicated as optical density (OD) values at 450 nm. (E) Cellular binding of E2 derived from patient-derived or H77 and HCV-J strains. Binding of native E1E2 complexes to Huh7.5.1 cells was detected as described in Supplementary Materials and Methods. Results are expressed as delta mean fluorescence intensity (ΔMFI) ± standard deviation. One representative experiment of 3 is shown. Da, dalton; MW, molecular weight.
Supplementary Figure 4. Binding of neutralizing anti-E2 HMAb CBH-23 to patient-derived envelope glycoproteins expressed on HCVpp as capture antigens in an enzyme-linked immunosorbent assay (ELISA). HCVpp expressing envelope glycoproteins of variants VL, VA, VC, VLVA447, and VLVC458+478 were used as capture antigens on GNA-coated ELISA plates. Control (Ctrl) pseudoparticles with absent HCV envelope glycoprotein expression and recombinant soluble E2 (sE2 derived from strain H77)14 served as negative and positive controls, respectively. Anti-E2 CBH-23 reactivity was detected as described in the Supplementary Materials and Methods section and is indicated as optical density (OD) values at 450 nm. Means ± standard deviation from 1 representative experiment are shown.
Supplementary Figure 5. Distribution of residues at positions 447, 458, and 478 of HCV E2 sequences in the European HCV databases. Distribution of residues at positions 447, 458, and 478 for HCV complete E2 sequences from all subtypes (black) and from subtype 1b only (white) within the European Hepatitis C Virus databases7 (available: http://euhcvdb.ibcp.fr). F and S are the predominant residue at positions 447 and 458 (F447, 98.4%; 1b, 96.2%; S458 all, 94%; 1b, 90.3%). The position 478 is variable (it belongs to HVR2) but R (all, 2.4%; 1b, 10.8%) is more frequent than C (all, 0.2%; 1b, 0.9%).
Supplementary Table 1. Neutralization of Patient-Derived and Chimeric HCVpp by Monoclonal Anti-Envelope Antibodies
Supplementary Table 2. Characteristics of Patients and Viruses Used for Neutralization Studies
Supplementary Table 3. HCVcc Neutralization Titers