SUMMARY
Centrosome amplification is a common feature of human tumors, but whether this is a cause or a consequence of cancer remains unclear. Here, we test the consequence of centrosome amplification by creating mice in which centrosome number can be chronically increased in the absence of additional genetic defects. We show that increasing centrosome number elevated tumor initiation in a mouse model of intestinal neoplasia. Most importantly, we demonstrate that supernumerary centrosomes are sufficient to drive aneuploidy and the development of spontaneous tumors in multiple tissues. Tumors arising from centrosome amplification exhibit frequent mitotic errors and possess complex karyotypes, recapitulating a common feature of human cancer. Together, our data support a direct causal relationship between centrosome amplification, genomic instability and tumor development.
Keywords: Centrosome amplification, Plk4, genomic instability, aneuploidy, tumorigenesis
eTOC/In-Brief BLURB
Extra centrosomes are common in human cancers and are correlated with aneuploidy and poor patient prognosis. However, whether supernumerary centrosomes are a cause or consequence of tumorigenesis is still unclear. Levine et al. now demonstrate that centrosome amplification is sufficient to drive tumorigenesis in multiple tissues of mice.
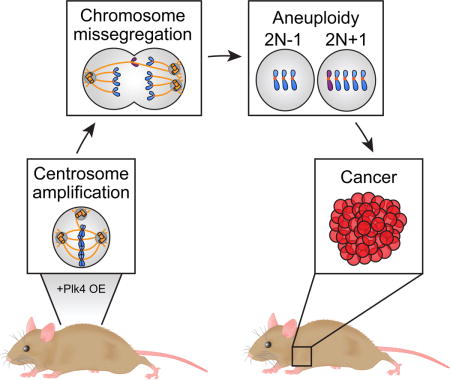
INTRODUCTION
The centrosome is a cellular organelle that plays a central role in coordinating most microtubule-related processes, including organizing the bipolar spindle that partitions the chromosomes during cell division. Faithful control of centrosome number is deregulated in a wide range of solid and blood-borne cancers, leading to the acquisition of extra copies of centrosomes, a feature known as centrosome amplification (Chan, 2011). Supernumerary centrosomes are observed early in the development of many tumors and often correlate with advanced tumor grade and poor clinical outcome (Godinho and Pellman, 2014; Nigg and Raff, 2009; Nigg, 2006). In cultured cells, centrosome amplification causes mitotic errors that can lead to chromosome missegregation (Ganem, et al., 2009; Silkworth, et al., 2009) and chromosomal rearrangements (Crasta, et al., 2012; Ganem and Pellman, 2012; Janssen, et al., 2011). Moreover, extra centrosomes can promote invasive phenotypes in a 3D culture model (Godinho, et al., 2014). These observations suggest that centrosome amplification could promote the initial stages of tumor development, but definitive evidence for this proposal is still lacking.
To examine the consequences of centrosome amplification in vivo, considerable attention has been focused on Plk4, a key regulator of centrosome duplication (Bettencourt-Dias, et al., 2005; Habedanck, et al., 2005). Overexpression of this kinase increases centrosome number in the absence of direct effects on cellular ploidy or oncogenes and tumor suppressor genes and provides an excellent experimental tool to study the long-term consequence of having cells with excess centrosomes. However, studies in animal models have so far provided contradictory views on the specific contribution of centrosome amplification to tumor development. Experiments in flies have shown that larval brain and wing disk tissues with supernumerary centrosomes are able to initiate tumors in transplantation assays (Sabino, et al., 2015; Basto, et al., 2008; Castellanos, et al., 2008). In mammals, however, centrosome amplification in embryonic neural progenitors results in aneuploidy, cell death and microcephaly, but does not promote tumorigenesis (Marthiens, et al., 2013). In addition, increasing centrosome number in the skin of mice failed to promote formation of spontaneous, or carcinogen-induced, skin tumors (Kulukian, et al., 2015; Vitre, et al., 2015). By contrast, centrosome amplification—either globally or in the skin—accelerates the onset of tumors caused by loss of p53 (Sercin, et al., 2016; Coelho, et al., 2015). Thus, while centrosome amplification can modify tumor outcome in a p53-null background, the interpretation is complicated by the fact that loss of p53 is associated with increased numbers of centrosomes in some contexts (Fukasawa, et al., 1996). Furthermore, it remains unclear if centrosome amplification can trigger tumor formation in the absence of direct effects on the p53 tumor suppressor pathway.
In this report, we describe the development of a doxycycline-inducible mouse model in which the levels of Plk4 can be increased to promote widespread and chronic centrosome amplification in vivo. This model allowed us to rigorously assess the long-term consequences of having cells with too many centrosomes and their contribution to tumor initiation. We show that despite being tolerated in many tissues, extra centrosomes increase tumor initiation in an intestinal cancer mouse model. Most importantly, we demonstrate that a chronic or transient increase in Plk4 promotes aneuploidy and centrosome amplification that drives the development of spontaneous tumors in multiple tissues. Tumors that form in the presence of extra centrosomes exhibit complex karyotypes similar to what is observed in the majority of human cancers. Together, these findings support the conclusion that centrosome amplification can promote genome instability and tumorigenesis.
RESULTS
Plk4 overexpression drives centrosome amplification in vitro
To drive centrosome amplification in a temporally-controlled manner in vivo, we developed a mouse model in which increased synthesis of Plk4 can be induced by addition of doxycycline. We integrated a single-copy Plk4-EYFP transgene, driven by a doxycycline-regulatable promoter, downstream of the Col1a1 locus in ES cells. Targeted ES cells were then used to produce the Plk4-EYFP transgenic mice, which were crossed with mice expressing the reverse tetracycline transactivator (rtTA) to allow doxycycline-inducible expression of Plk4-EYFP (Figure 1A). Mice and cells that harbor homozygous copies of the Plk4-EYFP and rtTA transgenes (Plk4-EYFPhom; rtTAhom) are referred to hereafter as Plk4Dox.
Figure 1. related to Figure S1. A modest increase in Plk4 promotes centrosome amplification and aneuploidy in vitro.
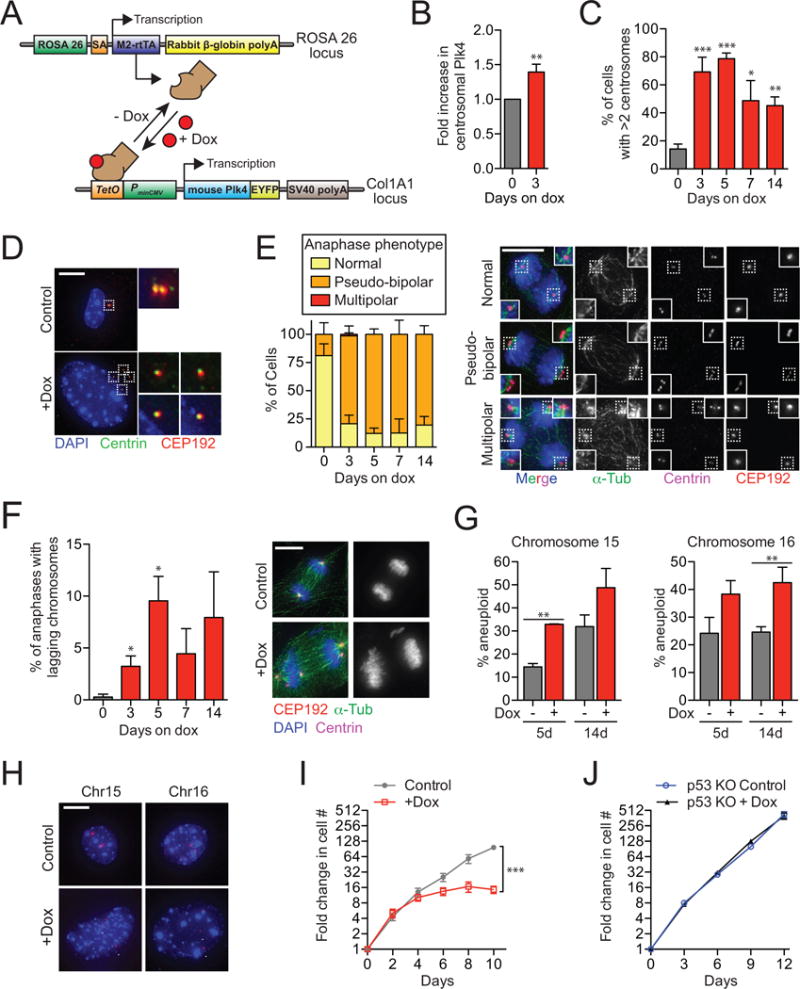
(A) System used for doxycycline-inducible expression of Plk4.
(B) Quantification of the level of centrosomal Plk4 in Plk4Dox MEFs. N = 3, >150 centrosomes per experiment.
(C) Quantification of the level of centrosome amplification in Plk4Dox MEFs. N = 3, >150 cells per experiment.
(D) Immunofluorescent images of centrosomes in Plk4Dox MEFs.
(E) Quantification of anaphase phenotypes in Plk4Dox MEFs. N = 3, >150 cells per experiment.
(F) Quantification of anaphase lagging chromosomes in Plk4Dox MEFs. N = 3, >150 cells per experiment.
(G) Quantification of the fraction of cells having <2 or >2 copies of chromosome 15 or 16. N = 3, >150 cells per experiment.
(H) Immunofluorescent images of FISH performed on Plk4Dox MEFs using probes against chromosome 15 and 16. Arrowheads mark each copy of Chr15 or Chr16.
(I) Graph showing the fold increase in cell number for Plk4Dox MEFs. N = 3, performed in triplicate.
(J) Graph showing the fold increase in cell number for Plk4Dox MEFs expressing SpCas9 and an sgRNA against p53. N = 3, performed in triplicate.
All data represent the means ±SEM. *P < 0.05, **P < 0.01 and ***P < 0.001; two-tailed Student’s t-test. Scale bars represent 10 μm.
To characterize the effect of Plk4 overexpression in vitro, we derived primary mouse embryonic fibroblasts (MEFs) from control and Plk4Dox embryos. In doxycycline-treated Plk4Dox MEFs, Plk4 mRNA levels rose ~6-fold (Figure S1A) and the level of Plk4 protein at the centrosome increased ~2-fold (Figure 1B and Figure S1B and S1C). This modest elevation in the level of Plk4 induced substantial centrosome amplification; after 3 and 5 days the number of cells with increased centrosome amplification rose to 69% and 79%, respectively (Figure 1C and 1D and Figure S1D). As expected, centrosome amplification was not observed in doxycycline-treated MEFs that carried either the Plk4-EYFP or rtTA transgene alone (Figure S1E).
Cells that enter mitosis with centrosome amplification can either undergo multipolar divisions or cluster their centrosomes prior to division (Basto, et al., 2008; Quintyne, et al., 2005; Ring, et al., 1982). Examination of mitotic figures revealed that Plk4Dox MEFs avoided lethal multipolar divisions by clustering extra centrosomes into pseudo-bipolar spindles with high efficiency (Figure 1E). Consistent with previous reports (Ganem, et al., 2009; Silkworth, et al., 2009), centrosome clustering significantly increased the frequency of mitotic errors (Figure 1F). Although aneuploidy increased in primary MEFs with repeated passages in culture (Weaver, et al., 2007; Hao and Greider, 2004), cells with supernumerary centrosomes were more aneuploid than wildtype MEFs at both time points (as determined with fluorescence in situ hybridization for chromosome 15 or 16) (Figure 1G and 1H). Importantly, supernumerary centrosomes did not lead to an increase in DNA damage or tetraploidization (Figure S1F–I).
Previously, we showed that centrosome amplification elicits a durable p53-dependent proliferative arrest in non-transformed human cells (Holland, et al., 2012). Consistently, supernumerary centrosomes prevented the proliferation of primary MEFs (Figure 1I), and knocking out p53 alleviated this block (Figure 1J). The fraction of cells with 5 or more centrosomes declined in Plk4Dox MEFs after 5 days of doxycycline treatment, but continued to increase in cells lacking p53 (Figure S1D and Figure S1J–K). This suggests that cells with high levels of centrosome amplification are outcompeted in a p53-dependent manner in vitro. Together, our data demonstrate that modest overexpression of Plk4 in vitro drives centrosome amplification, mitotic errors and a p53-dependent cell cycle arrest.
Elevated Plk4 expression promotes formation of supernumerary centrosomes in tissues
To determine the effect of Plk4 overexpression on centrosome number in vivo, we treated Plk4Dox and control animals with doxycycline for 1 or 8 months and sacrificed animals to analyze centrosome number in tissues. With the exception of the brain (see below), there was an increase in Plk4 mRNA levels in all tissues analyzed in Plk4Dox mice (Figure 2A and Figure S2A). In line with the prior results in MEFs, we observed a modest (<2-fold) increase in Plk4 protein levels at the centrosome in the thymus of Plk4Dox mice (Figure S2B). Consistent with increased Plk4, we observed a chronic increase in centrosome number in the skin, spleen, intestine, thymus, liver, pancreas and stomach of Plk4-overexpressing mice (Figure 2B–C and Figure S2C). In almost every case where an increase in centrosome number was observed, cells contained at most three extra centrosomes (Figure 2D and 2E and Figure S2D). By contrast, there was no increase in centrosome amplification in the lung and kidney, despite the 11 and 338-fold increase in Plk4 mRNA levels in these tissues, respectively (Figure S2A and S2C).
Figure 2. related to Figure S2 and S3. Increased Plk4 levels promote chronic centrosome amplification in multiple tissues.
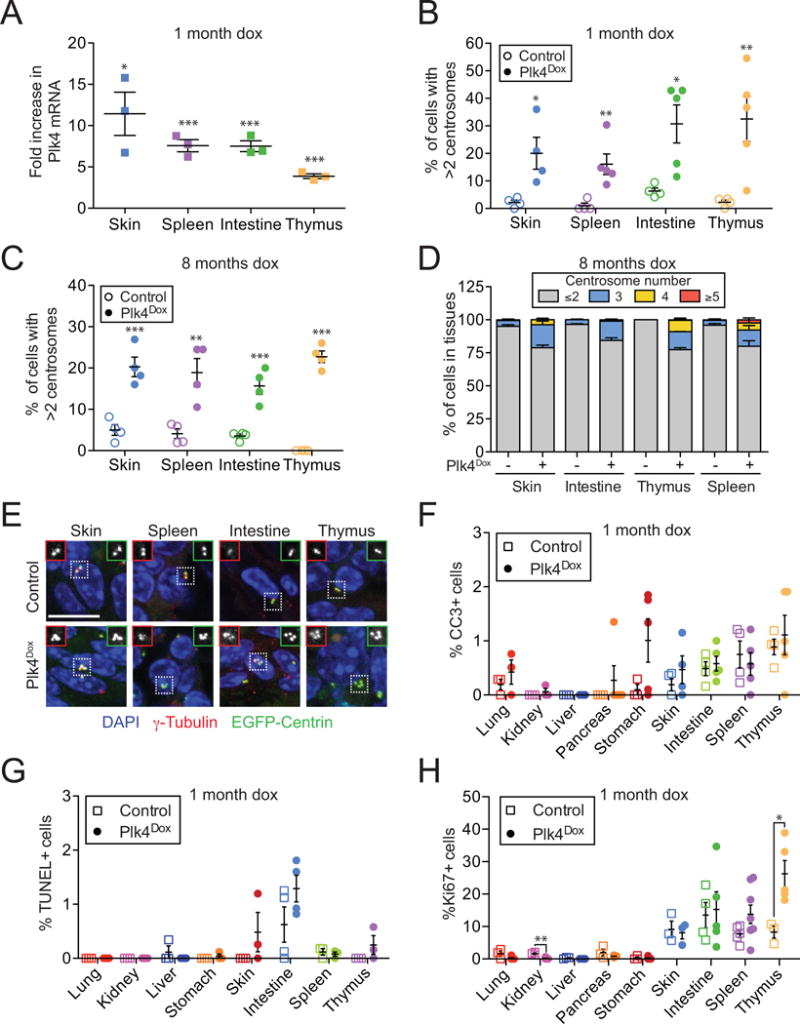
(A) Fold increase in Plk4 mRNA in tissues from Plk4Dox mice treated with doxycycline for 1 month. N = 3, performed in triplicate.
(B and C) Quantification of the level of centrosome amplification in tissues from Plk4Dox mice treated with doxycycline for 1 or 8 month. N ≥ 4.
(D) Quantification of centrosome number in tissues from Plk4Dox mice treated with doxycycline for 8 months. N = 4.
(E) Representative images of centrosomes in tissues from doxycycline treated Plk4Dox or control animals.
(F–H) Quantification of the fraction of cleaved caspase 3, TUNEL, or Ki67 positive cells in tissues from Plk4Dox mice treated with doxycycline for 1 month. Data are means ±SEM (N = ≥4).
All data represent the means ±SEM. *P < 0.05, **P < 0.01 and ***P < 0.001; two-tailed Student’s t-test. Scale bars represent 10 μm.
To determine whether the lack of centrosome amplification in the lung and kidney was caused by the death of cells with extra centrosomes, we assessed the expression of active caspase 3 and used TUNEL staining in tissues from Plk4Dox animals that were treated for 1 month with doxycycline. There was no significant increase in active caspase 3 or TUNEL staining in any of the tissues examined, suggesting that cells with extra centrosomes are not eliminated by cell death (Figure 2F and 2G). Plk4 overexpression does not promote centrosome amplification in quiescent cells (data not shown), suggesting that differences in proliferation rates could contribute to tissue specific differences in centrosome amplification in response to Plk4 overexpression. Concordantly, analysis of Ki67 staining in tissues revealed high rates of proliferation in the skin, intestine, spleen and thymus, where robust centrosome amplification was observed, and low turnover rates in the lung and kidney, where there was no increase in centrosome number (Figure 2B, 2H and S2C). Nevertheless, the liver, pancreas and stomach showed a significant increase in centrosome amplification despite a very small fraction of proliferating cells (Figure 2H and S2C). This suggests additional tissue-specific factors likely influence the relationship between Plk4 overexpression and centrosome amplification. Surprisingly, increased centrosome numbers correlated with hyperproliferation of cells in the thymus and decreased proliferation in the kidney (Figure 2H). These differences in cell proliferation highlight tissue specific differences in the response to centrosome amplification and may arise from alterations in growth signaling as a result of changes in centrosome or cilia number (Arquint, et al., 2014).
Overexpression of Plk4 in the brain of mice has been reported to cause microcephaly and behavioral defects (Coelho, et al., 2015; Marthiens, et al., 2013). However, transgenes integrated downstream of the Col1a1 locus are not expressed in major cells types of the brain (Hochedlinger, et al., 2005) (Figure S2E). Consistently, there was no increase in Plk4 RNA levels or centrosome amplification in the brain of doxycycline-treated Plk4Dox mice (Figure S2A and S2C). Moreover, Plk4-overexpressing animals did not show behavioral deficits or alterations in brain size (Data not shown and Figure S3A).
Centrosome amplification impairs epidermal architecture
A striking feature in mice overexpressing Plk4 was progressive hair loss that continued throughout the life of the animal and led to almost complete balding in one-year-old mice (Figure S3B). Consistent with previous reports in mice exhibiting centrosome amplification in the skin (Sercin, et al., 2016; Coelho, et al., 2015), mice overexpressing Plk4 exhibited a thickened epidermis and disrupted hair follicle morphology. Systematic histological examination of other tissues from Plk4-overexpressing mice revealed no major pathology (Figure S3C and S3D). We conclude that, with the notable exception of the skin, centrosome amplification is tolerated in many tissues in vivo.
Centrosome amplification causes aneuploidy in vivo
To evaluate whether centrosome amplification leads to aneuploidy in vivo, we assessed chromosome number in splenocytes from mice treated with doxycycline for 1 or 8 months. Centrosome amplification increased the fraction of aneuploid splenocytes at both time points (Figure 3A and Figure S4A and S4B), but did not promote cytokinesis failure or polyploidization (Figure S4C). To investigate whether extra centrosomes lead to the accumulation of aneuploid cells in aged mice, we isolated epidermal cells from 12–21 month old mice and determined their karyotype by low coverage genomic copy number analysis in single-cells. Analysis of 99 cells from 3 mice with centrosome amplification revealed 23 of the cells to be aneuploid (average of 23%), whereas 0 aneuploid cells were identified in the 78 single cells sequenced from two control animals (Figure 3B–C). In summary, supernumerary centrosomes promote chromosome segregation errors and aneuploidy, in the absence of polyploidization, in tissues.
Figure 3. related to Figure S4. Centrosome amplification drives aneuploidy in vivo.
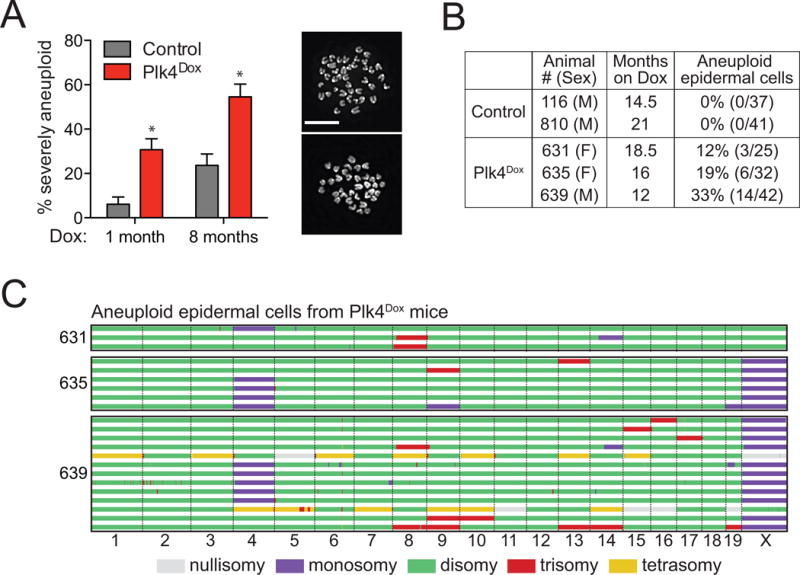
(A) Proportion of severely aneuploid (4N ± > 2 chromosomes) splenocytes from control and Plk4Dox mice treated with doxycycline for 1 or 8 months. N = 3, > 120 cells per experiment.
(B) Table shows the fraction of aneuploid cells determined by single cell sequencing of epidermal cells from doxycycline-treated control or Plk4Dox mice.
(C) Genome-wide copy number plots of aneuploid single cells sequenced from the epidermis of 3 Plk4Dox mice treated with doxycycline for 12–18.5 months. Individual cells are represented in rows with copy number states indicated in colors.
All data represent the means ±SEM. *P < 0.05; two-tailed Student’s t-test. Scale bars represent 10 μm.
Centrosome amplification increases the initiation of intestinal tumors
To test whether centrosome amplification is able to influence tumorigenesis, we first used a mouse model of intestinal neoplasia (Su, et al., 1992; Moser, et al., 1990). Mice that express a single truncated allele of the adenomatous polyposis coli (APC) tumor suppressor (APCMin) develop early onset adenomatous intestinal tumors with complete penetrance. To evaluate the effect of Plk4 overexpression on centrosome number in APCMin/+ cells, we derived MEFs from APCMin/+;Plk4Dox embryos. Doxycycline addition drove increased levels of Plk4 expression leading to sustained centrosome amplification, with 55% and 89% of APCMin/+;Plk4Dox cells containing extra centrosomes at day 3 and day 14 after doxycycline addition, respectively (Figure 4A and Figure S4D). As expected, extra centrosomes increased the frequency of chromosome segregation errors and micronuclei formation in APCMin/+;Plk4Dox MEFs and led to a cell cycle arrest in vitro (Figure 4B and 4C and Figure S4E).
Figure 4. related to Figure S4. Centrosome amplification promotes tumor initiation.
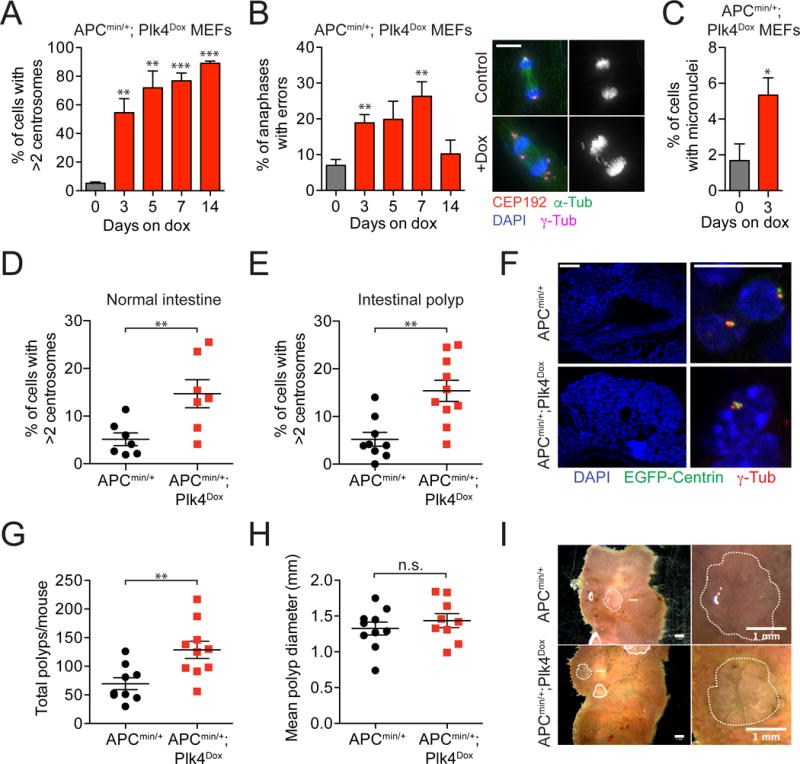
(A) Quantification of the level of centrosome amplification in APCMin/+; Plk4Dox MEFs. N = 3, >150 cells per experiment.
(B) Quantification of anaphase lagging chromosomes in APCMin/+; Plk4Dox MEFs. N = 3, >84 cells per experiment. Scale bar represents 10 μm.
(C) Frequency of micronuclei observed in APCMin/+; Plk4Dox MEFs. N = 3, >50 cells per experiment.
(D and E) APCMin and APCMin/+; Plk4Dox mice were treated with doxycycline from 10 days of age and sacrificed at 90 days old. Quantification shows the level of centrosome amplification in the intestines or intestinal polyps of APCMin and APCMin/+; Plk4Dox mice. N = 3, >150 cells per experiment.
(F) (Left) Immunofluorescence staining of an intestinal polyp and (Right) a magnified view of centrosomes in this tumor. Scale bars represent 200 μm (left) and 10μm (right).
(G and H) Quantification of tumor number (G) or size (H) in 90 day old APCMin and APCMin/+; Plk4Dox mice.
(I) Images show intestinal polyps in an APCMin and APCMin/+; Plk4Dox mouse.
All data represent the means ±SEM. *P < 0.05, **P < 0.01, ***P < 0.001 and NS (not significant) indicates P > 0.05; two-tailed Student’s t-test.
Next, we examined the size and number of tumors formed in the intestine of APCMin/+ and APCMin/+;Plk4Dox animals. Once again, centrosome number was significantly increased in both the normal intestine and in intestinal tumors from doxycycline-treated APCMin/+;Plk4Dox mice (Figure 4D–F). Importantly, tumor number was significantly increased in mice with centrosome amplification (average of 69 tumors in APCMin/+ animals compared to 129 tumors in APCMin/+;Plk4Dox mice; Figure 4G and 4I). However, tumor size remained unchanged (Figure 4H and 4I). Consistent with prior reports (Luongo, et al., 1994), we observed that intestinal APCMin/+ and APCMin/+;Plk4Dox tumors showed a reduced abundance of the wildtype allele of APC (Figure S4F). These data demonstrate that, in this context, centrosome amplification promotes the initiation, but not progression, of intestinal tumors.
Centrosome amplification drives spontaneous tumorigenesis
Despite the fact that centrosome amplification is a common feature of many cancer cells, it remains untested whether chronic centrosome amplification is sufficient to initiate tumorigenesis in mammals. To address this question, we aged cohorts of Plk4Dox and control mice that were fed doxycycline starting from 1–2 months of age. Strikingly, Plk4Dox mice succumbed to the development of spontaneous tumors starting at 36 weeks (median tumor-free survival of 55 weeks) (Figure 5A). Specifically, Plk4-overexpressing mice developed lymphomas, squamous cell carcinomas and sarcomas, whilst spontaneous tumors were not observed in Plk4-EYFP, rtTA or wild-type mice treated with doxycycline (Figure 5A and 5C). In contrast to lymphomas that developed in mice lacking p53, tumors from Plk4-overexpressing mice exhibited high levels of centrosome amplification (average of 44% amplification in lymphomas and squamous cell carcinomas in Plk4Dox mice) (Figure 5B). The vast majority of the tumor cells exhibiting centrosome amplification contained just one or two extra centrosomes (Figure S5A). Two of the lymphomas that developed in mice with centrosome amplification exhibited acute tumor lysis syndrome, a feature that was not observed in lymphomas that developed in p53-null animals (Figure S5B).
Figure 5. related to Figure S5. Centrosome amplification promotes spontaneous tumorigenesis.
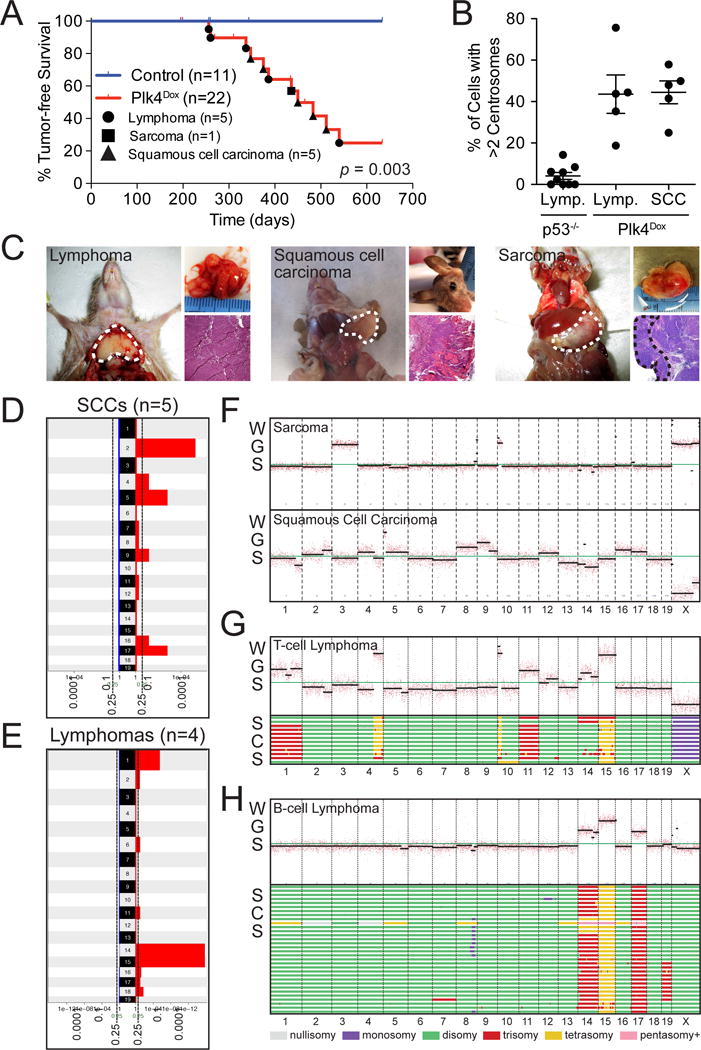
(A) Kaplan-Meier survival analysis of Plk4Dox and control (C57BL/6J) mice chronically fed doxycycline from 1–2 months of age. P value was calculated using the Log-rank test.
(B) Quantification of the level of centrosome amplification in tumors from Plk4Dox and p53−/− mice. Horizontal lines represent the mean and bars represent ±SEM.
(C) Representative examples of the different tumor types that develop in doxycycline-treated Plk4Dox mice.
(D and E) GISTIC analysis of low-coverage whole-genome sequencing of Squamous Cell Carcinomas (SCCs) and lymphomas from doxycycline-treated Plk4Dox mice shows gains of specific chromosomes. Scale represents Q values.
(F) Low-coverage whole-genome sequencing (WGS) plots for a Sarcoma and a Squamous Cell Carcinoma derived from Plk4Dox mice.
(G) (Top) WGS plots from a T-Cell Lymphoma derived from doxycycline-treated Plk4Dox mice. (Bottom) Genome-wide copy number plots of aneuploid single cells sequenced from the same T-cell lymphoma. 12/39 sequenced cells showed evidence of aneuploidy. Individual cells are represented in rows with copy number indicated in colors.
(H) (Top) WGS plots from a B-Cell Lymphoma derived from doxycycline-treated Plk4Dox mice. (Bottom) Genome-wide copy number plots of aneuploid single cells sequenced from the same B-cell lymphoma. 32/47 sequenced cells showed evidence of aneuploidy.
p53 has been shown to suppress the proliferation of cells with extra centrosomes in cell culture (Holland, et al., 2012). To examine whether spontaneous tumors that develop in mice with centrosome amplification exhibit inactivation of the p53 pathway, we analyzed the expression level of p53 target genes in thymic lymphomas that developed in p53−/− and Plk4Dox mice. As expected, p53−/− tumors had low expression of p53 and p53 transcriptional target genes (FAS, BCL2, BAX and PUMA) (Figure S5C). By contrast, thymic lymphomas that developed in Plk4Dox animals had a wide variation in the level of p53 expression. Despite the variation in p53 levels, the thymic tumors from Plk4Dox mice showed an overall reduction in the expression of p53 target genes, indicating the p53 pathway is at least partly comprised in spontaneous tumors that develop as a result of centrosome amplification (Figure S5C). Together, these data suggest that the p53 pathway acts as a barrier to the continued growth of cells with supernumerary centrosomes in vivo.
Since chronic increases in Plk4 could have consequences independent of centrosome amplification, we also tested whether a transient increase in Plk4 levels could trigger spontaneous tumor development. Remarkably, treatment with doxycycline for one month led to an increase in centrosome number in the spleen, intestine, liver and pancreas of 16–18 month old Plk4Dox mice (Figure S5D). Centrosome amplification can therefore persist in some tissues for long periods of time after transient Plk4 overexpression. Consistent with the observations in chronically-treated mice, Plk4Dox animals treated with doxycycline for one month also developed lymphomas, squamous cell carcinomas and sarcomas (Figure S5E). Moreover, tumors from these animals displayed high levels of centrosome amplification (Figure S5F). Together, these data establish a direct causal relationship between increased Plk4 levels, centrosome amplification and spontaneous tumor development.
Centrosome amplification promotes the development of aneuploid tumors
In human tumors, centrosome amplification strongly correlates with genomic instability. To evaluate the degree of aneuploidy and genome instability in tumors caused by centrosome amplification, we performed whole genome sequencing of tumor DNA isolated from three spontaneous T-cell lymphomas, two B-cell lymphomas, five squamous cell carcinomas and one sarcoma from doxycycline-treated Plk4Dox mice. All tumors showed evidence of aneuploidy, and each tumor type showed evidence of clonal selection for recurring chromosomal abnormalities. In particular, gains of chromosome 2, 5 and 17 were observed in squamous cell carcinomas, while T and B cell lymphomas showed recurrent gains of chromosome 14 and 15 (Figure 5D–H and Figure S5G–I). Notably, chromosome 15 carries the Myc proto-oncogene and is frequently gained in murine blood cancers (Bakker, et al., 2016). To examine the extent of tumor heterogeneity, we performed whole genome sequencing of single cells isolated from a thymic and a splenic lymphoma that developed in two mice chronically overexpressing Plk4. In the T-cell lymphoma, 12 aneuploid cells were sequenced and many showed gains of chromosomes 1, 11, 15 as well as segments of chromosomes 4 and 10. 32 aneuploid cells were sequenced in the splenic lymphoma, with most cells having gains of chromosomes 14, 15 and 17 (Figure 5G and 5H). Importantly, while both of the tumor samples contained recurrent chromosomal alterations, these tumors also exhibited karyotypic diversity, with some cells in each tumor exhibiting different gains and losses of whole chromosomes. These data suggest ongoing chromosome segregation errors in tumors with extra centrosomes.
DISCUSSION
A causal association between centrosome amplification and tumorigenesis was originally proposed by Boveri over a century ago, but has yet to be firmly established (Boveri, 1914). Here, we have examined the long-term consequence of supernumerary centrosomes in mice. We demonstrate that centrosome amplification can increase tumor initiation events in a mouse model of intestinal cancer. Most importantly, we show that extra centrosomes cause aneuploidy and trigger spontaneous tumorigenesis in multiple tissues. We conclude that centrosome amplification is sufficient to promote tumorigenesis in mammals.
In our experiments, we used Plk4 overexpression as a tool to drive centrosome amplification in vivo. While roles of Plk4 outside of centrosome biogenesis have been proposed (Rosario, et al., 2015; Rosario, et al., 2010; Martindill, et al., 2007), multiple lines of evidence argue that centrosome amplification is responsible for triggering spontaneous tumorigenesis in mice that overexpress Plk4. First, modest increases in Plk4 protein are sufficient to promote persistent centrosome amplification and spontaneous tumor development. Second, centrosome number is elevated in all three tissues that exhibit a predisposition to tumor development; conversely, tissues with high levels of Plk4 expression, but no increase in centrosome amplification, do not show an increase in tumorigenesis. Third, tumors that develop in Plk4-overexpressing mice generally show higher levels of centrosome amplification than in the normal tissue from which they developed. Finally, even a transient increase in Plk4 promotes persistent centrosomes amplification and tumorigenesis. Therefore, although we cannot formally exclude that the effects we observe reflect roles of Plk4 outside of centrosome duplication, our evidence firmly argues that increases in centrosome number drive the effects we observe in vivo.
To our knowledge, our study provides the first demonstration that centrosome amplification is sufficient to drive aneuploidy in tissues with wildtype p53. However, the role of centrosomes are not restricted to mitosis and extra copies of centrosomes have been shown to disrupt cilia signaling (Mahjoub and Stearns, 2012) and promote alterations in the interphase cytoskeleton that could facilitate invasion (Godinho, et al., 2014). Since hematopoietic lineages lack primary cilia, alterations in ciliary signaling are unlikely to underlie lymphomagenesis in cells with supernumerary centrosomes (Finetti, et al., 2011). Instead, our study demonstrates that tumors with extra centrosomes exhibit recurrent aneuploidies. In addition, we show that centrosome amplification increases tumor initiation in the APCMin/+ mouse model. Since tumors in this model are proposed to be driven by loss of the wildtype allele of APC, we propose that centrosome amplification increases tumor initiation by facilitating the loss of the copy of chromosome 18 containing the wildtype APC allele (Luongo, et al., 1994). Therefore, while further studies will be required to determine the precise mechanism by which extra centrosomes promote tumorigenesis, our data is consistent with a model in which centrosome amplification drives aneuploidy that promotes tumor development.
A central question that arises is why have other studies that employed Plk4 overexpression not reported spontaneous tumorigenesis (Sercin, et al., 2016; Coelho, et al., 2015; Kulukian, et al., 2015; Vitre, et al., 2015; Marthiens, et al., 2013)? A key difference in the mouse model that we report here is that we use a single copy Plk4 transgene knocked into the Col1a1 locus to achieve a modest increase in Plk4 levels that typically leads to the creation of just one or two extra centrosomes per cell. This is similar to the extent of centrosome amplification observed in human tumors (Denu, et al., 2016; Kayser, et al., 2005). We propose that small increases in centrosome number are permissive for tumor development. By contrast, large numbers of extra centrosomes are likely to be detrimental to cell viability because they are clustered inefficiently prior to division and lead to an increase in the frequency of lethal multipolar divisions. Mouse models that are created by the random integration of Cre-inducible, Plk4 transgenes may express the kinase at higher levels than achieved in our animal model (Sercin, et al., 2016; Kulukian, et al., 2015; Vitre, et al., 2015). We predict that high levels of Plk4 overexpression, and thus larger increases in the number of centrosomes per cell, will be detrimental to long-term cell survival. This could explain silencing of Plk4 transgene expression that has been reported in the skin of one mouse model (Sercin, et al., 2016), and also why global overexpression of Plk4 in another mouse model did not achieve centrosome amplification in the majority of tissues without the removal of p53 (Vitre, et al., 2015). Finally, we note that a previous study that drove Plk4 overexpression using a single-copy transgene at the ROSA26 locus did not follow survival to the point at which we observe the development of spontaneous tumors in mice with centrosome amplification (Coelho, et al., 2015).
In summary, we demonstrate that mice with extra centrosomes develop spontaneous tumors with high levels of genomic instability. We conclude that extra centrosomes are not bystanders in tumor development, but actively promote tumorigenesis by provoking mitotic errors that facilitate the evolution of malignant karyotypes. These findings support the therapeutic targeting of cells with extra centrosomes in human tumors.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to the corresponding author, Dr. Andrew Holland (aholland@jhmi.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse lines
The Plk4Dox mouse line was created using previously described KH2 ES cells (Beard, et al., 2006). KH2 ES cells possess the M2-rtTA gene targeted to the ROSA26 locus under the control of the ROSA promoter. In addition, an FRT-flanked PGK-neomycin-resistance gene followed by a promoterless, ATG-less hygromycin-resistance is targeted downstream of the Col1a1 locus to allow site-specific integration of a single copy transgene. To FLP-IN the tetracycline responsive Plk4-EYFP construct into KH2 ES cells, the mouse Plk4 ORF C-terminally tagged with EYFP was cloned downstream of the tetracycline operator and CMV minimal promoter in the pBS31 FLP-IN vector. KH2 ES cells were electroporated with pBS31-Plk4-EYFP and the pCAGGS-FLPe-puro plasmid encoding the FLP recombinase. Cells were selected with Hygromycin B and clones were amplified and checked by PCR for correct targeting. Blastocysts were injected with the targeted KH2 ES cells and chimeric mice identified. Germline transmission was detected by polymerase chain reaction analysis of tail DNA obtained at weaning. Plk4-EYFP genotyping was performed with the following primers: ACT GTC GGG CGT ACA CAA AT, CAA CCT GGT CCT CCA TGT CT and TGC TCG CAC GTA CTT CAT TC. M2-rtTA genotyping was performed with the following primers: AAA GTC GCT CTG AGT TGT TAT, GCG AAG AGT TTG TCC TCA ACC and GGA GCG GGA GAA ATG GAT ATG. Plk4-EYFP; rtTA mice were maintained by mating with C57BL6/N mice. EGFP-Centrin mice were as previously described (Hirai, et al., 2016). APCMin/+ mice were purchased from the Jackson Laboratory (stock 002020) and genotyped using the following primers: GCC ATC CCT TCA CGT TAG, TTC CAC TTT GGC ATA AGG C and TTC TGA GAA AGA CAG AAG TTA. Embryos and adults from both genders were included in our analysis. Mice were housed and cared for in an AAALAC-accredited facility and all animal experiments were conducted in accordance with Institute Animal Care and Use Committee approved protocols.
METHOD DETAILS
Doxycycline Induction
Mice were fed 1mg/mL doxycycline (RenYoung Pharma) in water supplemented with 25 mg/ml sucrose (Sigma). Water was changed twice per week for the duration of the treatment.
Spontaneous tumorigenesis studies
Plk4Dox and C57BL6/J animals were dosed chronically with doxycycline from 1 or 2 months of age. Mice were monitored daily during the course of the study. Mice were euthanized when signs of distress or when visible tumors grew to > 2cm in size as per the Johns Hopkins University ACUC guidelines.
Histological Analysis
A full necropsy was performed on every mouse sacrificed. Mouse tissues were harvested and fixed overnight in 4% paraformaldehyde at 4°C and then stored in 10% Neutral Buffered Formalin. The Johns Hopkins University, School of Medicine phenotyping core performed tissue processing, paraffin embedding, and Hematoxylin & Eosin staining. All pathology and tumors were analyzed by a certified veterinary pathologist.
Intestinal sample collection, tumor counts, and measurements
Mice were maintained in a C57BL6/J genetic background. Intestines from 90 day old mice were collected, opened lengthwise and laid flat on Whatman paper (GE Healthcare Life Sciences). Intestines were imaged on a Zeiss dissecting microscope with Zen imaging software. Polyp number and size was quantified using FIJI. Intestines were fixed on Whatman paper in 4% PFA overnight. After fixation, polyps were cut in half and processed for histology or immunofluorescence.
APC locus PCR-based assay
Analysis of the loss of the wildtype APC locus was performed as described using a quantitative APC locus PCR assay (Luongo, et al., 1994). Briefly, >15 intestinal polyps or areas of normal intestine from a single animal were pooled together and DNA extracted using the GenElute Mammalian Genomic DNA extraction kit (Sigma) following the manufacturer’s instructions. Each DNA sample was amplified in two separate PCR reactions using the following primers: For: TCT CGT TCT GAG AAA GAC AGA AGC T and Rev: TGA TAC TTC TTC CAA AGC TTT GGC TAT. The PCR product digested overnight with HindIII and then separated on a 3% Agarose gel. The integrated intensity of the APC+ and APCMin bands quantified using Fiji. Each band was background subtracted and the intensity of the APC+ bands multiplied by 1.17 (144 bp/123 bp) to correct for the smaller size, and proportionally reduced incorporation of ethidium bromide, in the digested APC+ allele. The mean ratio of the corrected APC+/APCMin band intensities was calculated for each sample.
Cell culture
Mouse embryonic fibroblasts (MEFs) were harvested as previously described (Xu, 2005). Briefly, embryos were harvested at E13.5 and incubated in trypsin overnight at 4°C. The following day, the embryos incubated at 37 °C for 30 minutes and cells dissociated by pipetting. Cells were plated in DMEM media (Corning Cellgro) supplemented with 10% fetal bovine serum (Sigma), 100 U/mL penicillin and 100 U/mL streptomycin. Cells were maintained at 37°C in an a tmosphere with 5% CO2 and 3% O2. For the growth assays, 2 × 105 cells/well were plated in 6 well dishes and cells counted every 3 days. Each condition was run in triplicate and each growth assay repeated at least 3 times. MEFs were passaged a maximum of 8 times before being discarded. Doxycycline (Sigma) was dissolved in H2O and used at a final concentration of 1 μg/ml and doxorubicin (Sigma) was dissolved in DMSO and used at 200 ng/ml unless otherwise stated.
Antibody production
Full-length γ-Tubulin or a fragment of CEP192 (amino acids 1-211) was cloned into a pET-23b bacterial expression vector (EMD Millipore) containing a C-term 6-His tag. Recombinant protein was purified from Escherichia coli using Ni-NTA beads (QIAGEN) and used for immunization (ProSci Incorporated). Goat immune sera were affinity-purified using standard procedures. A custom made Plk4 peptide (aa 564-580 was synthesized and conjugated to KLH for immunization (ProSci Incorporated). Rabbit immune sera were affinity-purified using standard procedures. Affinity-purified antibodies were directly conjugated to DyLight 550 and DyLight 650 fluorophores (Thermo Fisher Scientific) for use in immunofluorescence.
Immunofluorescence
For immunofluorescence in mouse tissues (with the exception of the brain sections for Figure S3A), samples were harvested and fixed overnight in 4% paraformaldehyde at 4°C. Tissues were washed 3 times for 30 minutes eac h with 1 × Phosphate Buffered Saline (PBS). Tissues were incubated in 30% sucrose overnight, embedded in OCT compound (Tissue-Tek) and frozen in a dry ice-ethanol bath cooled to −80°C. Tissues were cut in 12 μm sections using a Leica cryostat (Leica Biosystems, CM3050) and placed on Superfrost Plus treated microscope slides (Fisher Scientific). For staining, slides were rehydrated with PBS supplemented with 0.5% Triton X-10 (PBST), and incubated in primary antibody diluted in blocking solution (10% donkey serum in PBST) for 2 hours at room temperature or overnight at 4°C. Slides were washed 3 times with PBST and incubated for 1 hour at room temperature in secondary antibody with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) diluted in blocking solution. Slides were washed 3 more times with PBST and mounted in ProLong Gold Antifade (Invitrogen). For brain sections (for quantification of cortical thickness in Figure S3A), brains were harvested from 4 month old mice, fixed in 1% PFA overnight and 4°C, washed three times in PBS for 30 minutes each, then dehydrated in methanol overnight at −20°C. The next day, brains were rehydrated in PBS and embedded in 3% agarose. Once set, brains were cut in 120 μm sections using a Leica vibratome (Leica Biosystems) and kept in 1× PBS until staining. Sections were stained with 1 μg/ml DAPI diluted in PBS for 1 hour at room temperature and mounted in Flouromount-G (SouthernBiotech).
For immunofluorescence, primary MEFs were grown on 18-mm glass coverslips and fixed for 10 minutes in 100% ice cold methanol at −20°C for 10 minutes. Cells were blocked in 2.5% FBS, 200 mM glycine, and 0.1% Triton X-100 in PBS for 1 hour. Primary and secondary antibodies were incubated in the blocking solution for 1 hour at room temperature. DNA was stained with DAPI for 1 minute and cells were mounted in ProLong Gold Antifade (Invitrogen).
Staining was performed with the following primary antibodies: Pericentrin (rabbit, Abcam 1:1000), CEP192-647 (directly-labeled goat, raised against CEP192 a.a. 1-211, custom made by ProSci Incorporated, 1:100), γ-Tubulin-555 (directly-labeled goat, raised against full-length γ-Tubulin, custom made by ProSci Incorporated, 1:100), CEP192-650 (directly-labeled goat, raised against CEP192 a.a. 1-211, custom made by ProSci Incorporated, 1:100), Cleaved Caspase 3 (rabbit, Cell Signaling Technologies, 1:500), p-Histone H2A.X (Ser139) (rabbit, Cell Signaling Technologies, 1:1000), Centrin (mouse, Millipore, 1:1000), CEP192 (rabbit, raised against CEP192 a.a. 1-211, a kind gift from Karen Oegema, Ludwig Institute for Cancer Research, 1:1000), Plk4#1 (directly-labeled rabbit, raised against Plk4 a.a. 510-970, custom made by ProSci Incorporated, 1:1000 (Moyer, et al., 2015)) and Plk4#3 (directly-labeled rabbit, raised against Plk4 a.a. 564-580, custom made by ProSci Incorporated, 1:1000). TUNEL staining was performed using the in situ cell death detection kit (Sigma) following the manufacturer’s instructions. Secondary donkey antibodies were conjugated to Alexa Fluor® 488, 555 or 650 (Life Technologies).
Immunofluorescence images of MEFs were collected using a Deltavision Elite system (GE Healthcare) controlling a Scientific CMOS camera (pco.edge 5.5). Acquisition parameters were controlled by SoftWoRx suite (GE Healthcare). Images were collected at room temperature (25°C) using an Olympus 40× 1.35 NA, 60× 1.42 NA or Olympus 100× 1.4 NA oil objective at 0.2 μm z-sections. Images were acquired using Applied Precision immersion oil (N=1.516).
Immunofluorescence images of tissues were collected using a Zeiss LSM700 confocal microscope. Acquisition parameters were controlled by ZEN (Zeiss). Images were collected at room temperature (25°C) using a Zeiss 63× 1.4 NA oil objective at 0.3 μm z-sections. Images were acquired using Zeiss immersion oil (N=1.518).
Image Analysis
Quantification of Plk4 levels at the centrosome was performed as previously described (Lambrus, et al., 2015). Imaris software (Bitplane) was used to quantify of total number of nuclei per field of view in the tissues stained with CC3 or Ki67.
Quantitative real time PCR
Total RNA was isolated from cells or homogenized tissue using Trizol Reagent (Thermo Fisher Scientific) and prepared for reverse transcription using SuperScript III/IV Reverse transcriptase (Thermo Fisher Scientific). Quantitative real time PCR was performed using SYBRGreen qPCR Master Mix (Thermo Fisher Scientific) on iQ5 multicolor real time PCR detection system (Bio-Rad). Analysis was performed using iQ5 optical system software (Bio-Rad). Reactions were carried out in triplicate using the following primers: Plk4 Fow: 5′-GAA ACA CCC CTC TGT CTT GG-3′ and Rev: 5′-GCA TGA AGT GCC TAG CTT CC-3′; p53 Fow: 5′-CCC GAG TAT CTG GAA GAC AG-3′ and Rev: 5′-ATA GGT CGG CGG TTC ATG CC-3′; FAS Fow: 5′-GGA AAA GGA GAC AGG ATG ACC-3′ and Rev: 5′-CTT CAG CAA TTC TCG GGA TG-3′; BCL2 Fow: 5′-TTC GCA GCG ATG TCC AGT CAG CT-3′ and Rev: 5′-TGA AGA GTT CTT CCA CCA CCG T-3′; BAX Fow: 5′-ATG CGT CCA CCA AGA AGC TGA-3′ and Rev: 5′-AGC AAT CAT CCT CTG CAG CTC C-3′; PUMA Fow: 5′-GCA GCA CTT AGA GTC GCC-3′ and Rev: 5′-GTC GAT GCT GCT CTT CTT GT-3′. Expression values for p53 target genes (Figure S5C) were normalized to GAPDH, amplified with GAPDH Fow: 5′-AAT GTG TCC GTC GTG GAT CTG A-3′ and Rev: 5′-GAT GCC TGC TTC ACC ACC TTC T-3′. Plk4 overexpression values in MEFs and tissues (Figure 2A and S1A and S2A) were normalized to β-actin, amplified with β-actin Fow: 5′-GGC TGT ATT CCC CTC CAT CG-3′ and β-actin Rev: 5′-CCA GTT GGT AAC AAT GCC ATG T-3′ primers, with the exception of the APCmin/+ MEF experiment in Figure S4D, which were normalized to HPRT, amplified with HPRT Fow: 5′-TGA TCA GTC AAC GGG GGA CA-3′ and HPRT Rev: 5′-TTC GAG AGG TCC TTT TCA CCA-3′. The fold changes in mRNA expression were calculated using the 2−ΔΔCt method, and expression values were expressed as fold increase in the average expression compared with non-transgenic tissues.
Metaphase spreads and FISH analysis
To harvest splenocytes, freshly harvested spleens were minced and filtered through a 40 μm cell strainer. Cells were resuspended in RPMI media (Corning Cellgro) supplemented with 10% fetal bovine serum (Sigma), 100 U/mL penicillin,100 U/mL streptomycin, 1% HEPES (Sigma), 1% Sodium Pyruvate (Corning Cellgro), 1% Nonessential amino acids (Sigma), 10 U/mL Interleukin-2 (Roche), 5 μg/mL Concanavalin A (Sigma), 10μg/mL Lipopolysaccharides (Sigma) and grown overnight at 37°C in an atmosphere of 5% CO2 and 3% O2. Cell were treated with 100 ng/ml Colcemid (Sigma) for 4 hours, trypsinized and resuspended in 75 mM KCl for 15 minutes at room temperature. Five drops of freshly prepared Carnoy’s fixative (75% Methanol: 25% Acetic Acid) was added, the cells pelleted and resuspended in fixative overnight at 4°C. Cells were dropped onto slides pr etreated with acetic acid. Dried slides were incubated with DAPI for 1 minute and imaged using a Deltavision Elite system.
Mouse FISH probes for 10 cM loci on chromosome 15 or 16 were purchased from Empire Genomics. Cells were fixed with Carnoy’s fixative (75% Methanol: 25% Acetic Acid) for 15 minutes at room temperature and stored at −20°C until needed. DNA and probes were denatured at 69°C for 2 minutes, and hy bridization was performed at 37°C overnight. The next day, cells were washed with 0.4× SSC buffer (Sigma) for 2 minutes at 72°C, then washed with 2× SSC (0.05% Tween-20) a t room temperature for 30 seconds. Cells were briefly washed with dH2O, air dried and mounted with VectaShield containing 150 ng/mL DAPI.
Flow cytometry
Cell pellets were fixed in cold 70% EtOH for 24 hours, washed once in PBS and resuspended in PBS supplemented with 10 μg/ml RNAse A and 50 μg/ml Propidium Iodide (PI). Samples were incubated at room temperature for 30 minutes and analyzed on a flow cytometer (FACSCalibur; Becton Dickinson).
Single cell sequencing
Single cells were isolated from thymic or B-cell lymphomas by dissecting the tumor and mincing the tissue through a 70 μm cell strainer. To isolate single epidermal cells, the skin was removed and floated on 0.25% trypsin with 1 mM EDTA in DMEM (Gibco) overnight at 4°C. The epidermis was scraped off using a scalpel and tissue was dissociated into single cells by pipetting. Trypsin was neutralized by addition of 7% FBS diluted in PBS. This suspension was then passed through a 70 μm (BD Biosciences) filter followed by a 40 μm (BD Biosciences) filter. Isolated single cells from the thymus, spleen and epidermis were washed twice in PBS and stored in FBS with 10% DMSO at −80 °C until sorted. Single cell karyotype analysis was performed and analyzed as previously described (Bakker, et al., 2016).
Whole genome sequencing
Genomic DNA was extracted from tissue samples using the GenElute Mammalian Genomic DNA extraction kit (Sigma) following the manufacturer’s instructions. Shallow Whole Genome Sequencing (WGS) was performed as previously described (Nassar, et al., 2015). Briefly, whole-genome DNA libraries were created using the Illumina TruSeq DNA sample preparation kit V2 according to the manufacturer’s instructions, and resulting whole-genome libraries were sequenced at low coverage on a HiSeq2500 (Illumina) using a V3 flow cell generating 50-bp reads. Raw sequencing reads were mapped to the mouse reference genome (GRCm38/mm10) using Burrows-Wheeler Aligner (Li and Durbin, 2009). We removed PCR duplicates with Picard (v1.32 and v1.43) and obtained an average of 7,788,246 unique mapped reads per sample. The number of reads was counted in windows of 50 Kb and corrected for the genomic wave. Segmentation was performed by the Ascat algorithm (Van Loo, et al., 2010). GISTIC 2.0 (Genomic Identification of Significant Targets in Cancer) (Beroukhim, et al., 2007) was used to identify recurrent Copy Number Alterations in Figure 4d,e.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed using GraphPad Prism software. Differences between samples were tested using a two-tailed Student’s t-test or a Log-rank test for survival analysis. Error bars represent SEM unless otherwise indicated. Please refer to figures and figure legends for number of cells or animals used per experiment.
DATA AND SOFTWARE AVAILABILITY
Raw sequencing reads for whole genome sequencing are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-5043. Raw sequencing reads for single-cell whole genome sequencing are available in the European Nucleotide Archive database (www.ebi.ac.uk/ena) under accession number PRJEB1854.
Supplementary Material
HIGHLIGHTS.
Plk4 overexpression promotes persistent centrosome amplification in vivo
Centrosome amplification promotes aneuploidy in vivo
Extra centrosomes promote tumor initiation in a model of intestinal neoplasia
Centrosome amplification drives spontaneous tumorigenesis
Acknowledgments
We thank Dr. Randall Reed for comments on this manuscript. We thank Dr. Cory Brayton, D.V.M, for pathological analysis of all mouse tissues in this manuscript. This work was supported by a Pew-Stewart Scholar Award (to A.J.H), a Kimmel Scholar Award (to A.J.H), a Johns Hopkins School of Medicine Innovation Award (to A.J.H), a P30 grant from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK, P30DK09086) (to A.J.H), a American Cancer Society Scholar Award (RSG-16-156-01-CCG) (to A.J.H), a Dutch Cancer Society grant (2012-RUG-5549) (to F.F) and research grants GM 114119 (to A.J.H) and GM 29513 (to D.W.C) from the National Institutes of Health. We thank Nancy Halsema, Inge Kazemier and Karina Hoekstra-Wakker for technical help with single cell sequencing. Financial support for single cell sequencing was provided in part by a European Research Council Advanced grant (ROOTS-Grant Agreement 294740) to P.M.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
A.J.H. and M.S.L. conceived the project. M.S.L. performed the experimental work. Bj.B. performed the single cell sequencing (SCS) and analysis. Br. B. performed the whole genome sequencing and analysis. J.M. and J.L. performed data quantification. D.C.S. and P.M.L provided technical assistance with the SCS. B.V. and D.W.C. assisted in the creation of the Plk4Dox mouse model. A.J.H. and M.S.L. wrote the manuscript and prepared the figures. All authors edited the manuscript. A.J.H. supervised all aspects of the project.
References
- Arquint C, Gabryjonczyk AM, Nigg EA. Centrosomes as signalling centres. Philos Trans R Soc Lond B Biol Sci. 2014;369 doi: 10.1098/rstb.2013.0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker B, Taudt A, Belderbos ME, Porubsky D, Spierings DC, de Jong TV, Halsema N, Kazemier HG, Hoekstra-Wakker K, Bradley A, et al. Single-cell sequencing reveals karyotype heterogeneity in murine and human malignancies. Genome Biol. 2016;17:115. doi: 10.1186/s13059-016-0971-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, Raff JW. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–42. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–8. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–12. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15:2199–207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Boveri T. Zur Frage der Entstehung maligner Tumoren. Fischer; Jena: 1914. [Google Scholar]
- Castellanos E, Dominguez P, Gonzalez C. Centrosome dysfunction in Drosophila neural stem cells causes tumors that are not due to genome instability. Curr Biol. 2008;18:1209–14. doi: 10.1016/j.cub.2008.07.029. [DOI] [PubMed] [Google Scholar]
- Chan JY. A clinical overview of centrosome amplification in human cancers. Int J Biol Sci. 2011;7:1122–44. doi: 10.7150/ijbs.7.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho PA, Bury L, Shahbazi MN, Liakath-Ali K, Tate PH, Wormald S, Hindley CJ, Huch M, Archer J, Skarnes WC, et al. Over-expression of Plk4 induces centrosome amplification, loss of primary cilia and associated tissue hyperplasia in the mouse. Open Biol. 2015;5:150209. doi: 10.1098/rsob.150209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–8. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denu RA, Zasadil LM, Kanugh C, Laffin J, Weaver BA, Burkard ME. Centrosome amplification induces high grade features and is prognostic of worse outcomes in breast cancer. BMC Cancer. 2016;16:47. doi: 10.1186/s12885-016-2083-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finetti F, Paccani SR, Rosenbaum J, Baldari CT. Intraflagellar transport: a new player at the immune synapse. Trends Immunol. 2011;32:139–45. doi: 10.1016/j.it.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude GF. Abnormal centrosome amplification in the absence of p53. Science. 1996;271:1744–7. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Pellman D. Linking abnormal mitosis to the acquisition of DNA damage. J Cell Biol. 2012;199:871–81. doi: 10.1083/jcb.201210040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho SA, Pellman D. Causes and consequences of centrosome abnormalities in cancer. Philos Trans R Soc Lond B Biol Sci. 2014;369 doi: 10.1098/rstb.2013.0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho SA, Picone R, Burute M, Dagher R, Su Y, Leung CT, Polyak K, Brugge JS, Thery M, Pellman D. Oncogene-like induction of cellular invasion from centrosome amplification. Nature. 2014;510:167–71. doi: 10.1038/nature13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–6. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Hao LY, Greider CW. Genomic instability in both wild-type and telomerase null MEFs. Chromosoma. 2004;113:62–8. doi: 10.1007/s00412-004-0291-7. [DOI] [PubMed] [Google Scholar]
- Hirai M, Chen J, Evans SM. Generation and Characterization of a Tissue-Specific Centrosome Indicator Mouse Line. Genesis. 2016;54:286–96. doi: 10.1002/dvg.22937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–77. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Fachinetti D, Zhu Q, Bauer M, Verma IM, Nigg EA, Cleveland DW. The autoregulated instability of Polo-like kinase 4 limits centrosome duplication to once per cell cycle. Genes Dev. 2012;26:2684–9. doi: 10.1101/gad.207027.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–8. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- Kayser G, Gerlach U, Walch A, Nitschke R, Haxelmans S, Kayser K, Hopt U, Werner M, Lassmann S. Numerical and structural centrosome aberrations are an early and stable event in the adenoma-carcinoma sequence of colorectal carcinomas. Virchows Arch. 2005;447:61–5. doi: 10.1007/s00428-004-1191-1. [DOI] [PubMed] [Google Scholar]
- Kulukian A, Holland AJ, Vitre B, Naik S, Cleveland DW, Fuchs E. Epidermal development, growth control, and homeostasis in the face of centrosome amplification. Proc Natl Acad Sci U S A. 2015;112:E6311–20. doi: 10.1073/pnas.1518376112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrus BG, Uetake Y, Clutario KM, Daggubati V, Snyder M, Sluder G, Holland AJ. p53 protects against genome instability following centriole duplication failure. J Cell Biol. 2015;210:63–77. doi: 10.1083/jcb.201502089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luongo C, Moser AR, Gledhill S, Dove WF. Loss of Apc+ in intestinal adenomas from Min mice. Cancer Res. 1994;54:5947–52. [PubMed] [Google Scholar]
- Mahjoub MR, Stearns T. Supernumerary centrosomes nucleate extra cilia and compromise primary cilium signaling. Curr Biol. 2012;22:1628–34. doi: 10.1016/j.cub.2012.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthiens V, Rujano MA, Pennetier C, Tessier S, Paul-Gilloteaux P, Basto R. Centrosome amplification causes microcephaly. Nat Cell Biol. 2013;15:731–40. doi: 10.1038/ncb2746. [DOI] [PubMed] [Google Scholar]
- Martindill DM, Risebro CA, Smart N, Franco-Viseras Mdel M, Rosario CO, Swallow CJ, Dennis JW, Riley PR. Nucleolar release of Hand1 acts as a molecular switch to determine cell fate. Nat Cell Biol. 2007;9:1131–41. doi: 10.1038/ncb1633. [DOI] [PubMed] [Google Scholar]
- Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–4. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- Moyer TC, Clutario KM, Lambrus BG, Daggubati V, Holland AJ. Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J Cell Biol. 2015;209:863–78. doi: 10.1083/jcb.201502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar D, Latil M, Boeckx B, Lambrechts D, Blanpain C. Genomic landscape of carcinogen-induced and genetically induced mouse skin squamous cell carcinoma. Nat Med. 2015;21:946–54. doi: 10.1038/nm.3878. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Origins and consequences of centrosome aberrations in human cancers. Int J Cancer. 2006;119:2717–23. doi: 10.1002/ijc.22245. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–78. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–9. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- Ring D, Hubble R, Kirschner M. Mitosis in a cell with multiple centrioles. J Cell Biol. 1982;94:549–56. doi: 10.1083/jcb.94.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario CO, Kazazian K, Zih FS, Brashavitskaya O, Haffani Y, Xu RS, George A, Dennis JW, Swallow CJ. A novel role for Plk4 in regulating cell spreading and motility. Oncogene. 2015;34:3441–51. doi: 10.1038/onc.2014.275. [DOI] [PubMed] [Google Scholar]
- Rosario CO, Ko MA, Haffani YZ, Gladdy RA, Paderova J, Pollett A, Squire JA, Dennis JW, Swallow CJ. Plk4 is required for cytokinesis and maintenance of chromosomal stability. Proc Natl Acad Sci U S A. 2010;107:6888–93. doi: 10.1073/pnas.0910941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino D, Gogendeau D, Gambarotto D, Nano M, Pennetier C, Dingli F, Arras G, Loew D, Basto R. Moesin is a major regulator of centrosome behavior in epithelial cells with extra centrosomes. Curr Biol. 2015;25:879–89. doi: 10.1016/j.cub.2015.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sercin O, Larsimont JC, Karambelas AE, Marthiens V, Moers V, Boeckx B, Le Mercier M, Lambrechts D, Basto R, Blanpain C. Transient PLK4 overexpression accelerates tumorigenesis in p53-deficient epidermis. Nat Cell Biol. 2016;18:100–10. doi: 10.1038/ncb3270. [DOI] [PubMed] [Google Scholar]
- Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, Gould KA, Dove WF. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–70. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- Van Loo P, Nordgard SH, Lingjaerde OC, Russnes HG, Rye IH, Sun W, Weigman VJ, Marynen P, Zetterberg A, Naume B, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;107:16910–5. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitre B, Holland AJ, Kulukian A, Shoshani O, Hirai M, Wang Y, Maldonado M, Cho T, Boubaker J, Swing DA, et al. Chronic centrosome amplification without tumorigenesis. Proc Natl Acad Sci U S A. 2015;112:E6321–30. doi: 10.1073/pnas.1519388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy Acts Both Oncogenically and as a Tumor Suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Xu J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol. 2005 doi: 10.1002/0471142727.mb2801s70. Chapter 28, Unit 28 1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


