Abstract
When Toxoplasma gondii egresses from the host cell, glyceraldehyde-3-phosphate dehydrogenase 1 (GAPDH1), which is primary a glycolysis enzyme but actually a quintessential multifunctional protein, translocates to the unique cortical membrane skeleton. Here we report the 2.25Å resolution crystal structure of the GAPDH1 holoenzyme in a quaternary complex providing the basis for the molecular dissection of GAPDH1 structure-function relationships Knockdown of GAPDH1 expression and catalytic site disruption validate the essentiality of GAPDH1 in intracellular replication but we confirmed that glycolysis is not strictly essential. We identify, for the first time, S-loop phosphorylation as a novel, critical regulator of enzymatic activity that is consistent with the notion that the S-loop is critical for cofactor binding, allosteric activation and oligomerization. We show that neither enzymatic activity nor phosphorylation state correlate with the ability to translocate to the cortex. However, we demonstrate that association of GAPDH1 with the cortex is mediated by the N-terminus, likely palmitoylation. Overall, glycolysis and cortical translocation are functionally decoupled by post-translational modifications.
Graphical Abstract
Introduction
The coccidian parasite, Toxoplasma gondii, infects over thirty percent of the world’s human population and causes severe neurological disorders and death in immunocompromised individuals1,2 The devastating and life threatening effects are a result of the lytic cycle of the parasite, which includes attachment to a host cell, invasion, intracellular replication and egress. Upon egress from the host cells, glycolytic enzymes that are cytosolic in intracellular parasites quickly translocate to the unique cortical membrane skeleton of Toxoplasma3,4. This unusual mechanism is postulated to localize the energy source for parasite cortical mediated motility, invasion and extracellular survival4. This cortical association is reversible as upon host cell invasion the enzymes are released to the cytoplasm.
The glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is an archetypical multifunctional protein with diverse activities and distinct subcellular localization5 and yet is relatively unexamined in Toxoplasma. This key enzyme (EC 1.2.1.12) of glycolysis catalyzes the oxidative phosphorylation of glyceraldehyde 3-phosphate (G3P) to D-glycerate-1, 3-bisphosphate in presence of NAD+ and inorganic phosphate. Additional moonlighting functions of GAPDH include membrane fusion and transport, cytoskeletal dynamics, iron uptake and transport, heme metabolism, post-transcriptional gene regulation, tRNA export, chromatin structure and maintenance of DNA integrity. These diverse functions are regulated by a combination of post-translational modifications, subcellular localization, conformation dynamicity and protein complex assembly6.
To unravel the complex GAPDH functions we performed X-ray crystallographic analysis of GAPDH1 protein structure and used this as the precise, structural reflection board for a thorough molecular dissection. The crystallographic structure of GAPDH is well documented across several biological kingdoms and the enzymatic mechanism is well studied but the structural basis of its non-glycolytic functions has not been completely explored. GAPDH is a homologous tetramer (~150 kDa) of O-R subunits that exhibits three asymmetric P, Q and R dyads7. This bilobal structure is universally conserved with the folding of an N-terminal NAD+-binding domain and a C-terminal G3P catalytic domain8. The flexible S-loop is pivotal for cofactor binding, allosteric activation and quaternary structure assembly9 but the cellular regulation of the dynamic S-loop is not understood.
Our study of Toxoplasma GAPDH1 uncovers and decouples two new features that have not been previously described. Firstly, glycolysis in the parasite is modulated by phosphorylation of the regulatory S-loop, which is a novel feature not described before for GAPDH in any species. Secondly, the unique association of GAPDH with the cortical membrane skeleton is conveyed by Cys3 at the N-terminus, likely through palmitoylation. Besides the two known mechanisms of GAPDH association with membranous structures, i.e. through Rab2 association10 or through spectrin-actin association11, our discovery provides a third and novel mechanism. These unique regulations might provide new drug targets to treat toxoplasmosis.
Results
Crystal quaternary structure of GAPDH1
The Toxoplasma genome database (ToxoDB, http://toxodb.org)12 indicates that GAPDH1 (TGME49_289690) and GAPDH2 (TGME49_269190) are both constitutively expressed in tachyzoite and bradyzoite stages. While the GAPDH2 isoenzyme is targeted to the apicoplast organelle13, GAPDH1 is cytosolic in intracellular parasites. Here we have focused on GAPDH1, the crystal structure of which was determined to 2.25Å resolution in the holoenzyme NAD+-bound state. The GAPDH1 fold is validated by DALI analysis of the Toxoplasma GAPDH1 structure and reveals its close similarity to the GAPDH oxidoreductase folds with the top scoring hit of Cryptosporidium parvum GAPDH (Z-score 55.4, PDB ID 3CIF)14. The secondary, tertiary and quaternary structures of Toxoplasma GAPDH1 are very similar to GAPDH structures that have been previously reported (Fig. 1, 2A–C).
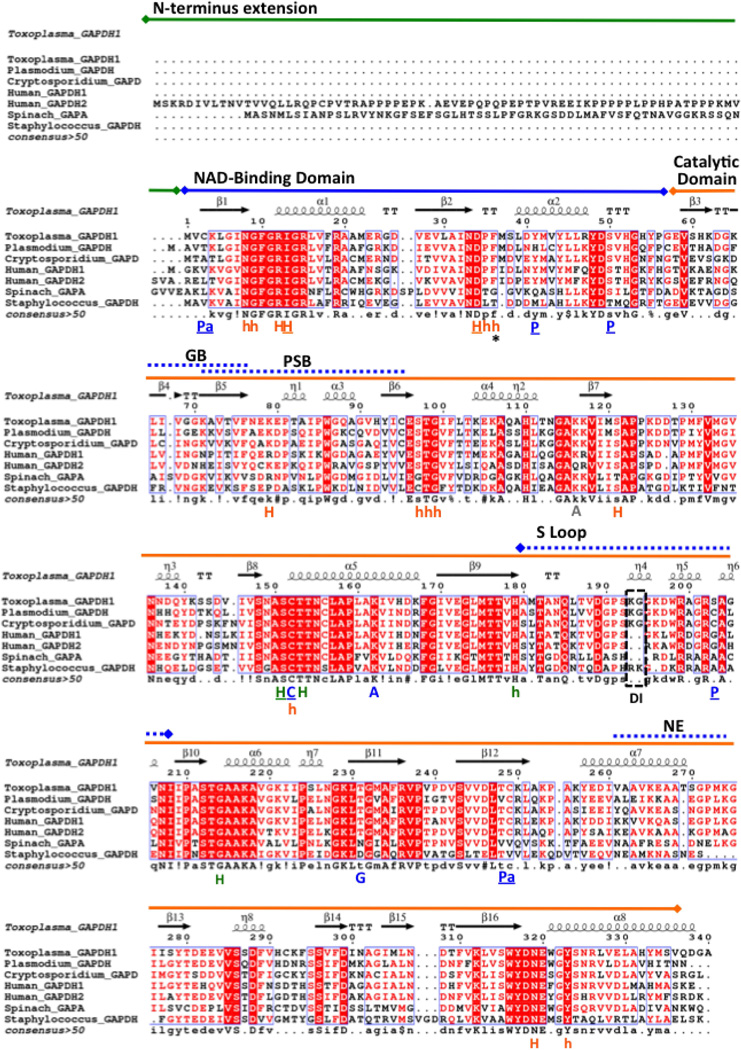
Figure 1.
Secondary structural alignment of GAPDH. Apicomplexan parasite (Toxoplasma gondii, Plasmodium falciparum, Cryptosporidium parvum), plant (spinach), bacterium (Staphylococcus areus) and human GAPDH are aligned to the Toxoplasma backbone using ESPript 3.0. The two domains as indicated function in NAD+ binding and catalysis. Plant and sperm GAPDH contain the N-terminal extension that is also observed in Toxoplasma GAPDH2 (not shown). Dashed box indicates the Lys-Gly dipeptide insertion (DI) in apicomplexans that is unique from eukaryotic GAPDH. Functional residues are identified as C (catalytic residues), P (Phosphorylation), Pa (Palmitoylation), A (Acetylation), G (O-linked N-acetyl Glucosamine), PSB (PhosphatidylSerine-rich Binding), NE (Nuclear Export), and GB (Glutathione Binding). P, Pa and C residues that have been mutagenized for functional assays in this study are underlined. Residues that form hydrogen, hydrophobic and both types of bonds with the cofactor NAD+ are labeled in orange, respectively as H, h and H. Similar labels in green are used for residues that bond with the substrate G3H. Sequences were extracted from UniprotKB database: T. gondii GAPDH1 (code Q9BKE2), C. parvum GAPDH (code Q7YYQ9), P. falciparum GAPHDH (code Q8T6B1), Homo sapiens (human) GAPDH1 (somatic, code P04406), H. sapiens (human) GAPDH2/GAPDHS (sperm, code O14556), Spinacia oleracea (spinach) chloroplast GAPDH (code P19866). Crystallographic structures of the seven selected GAPDH proteins are solved and deposited in PDB.
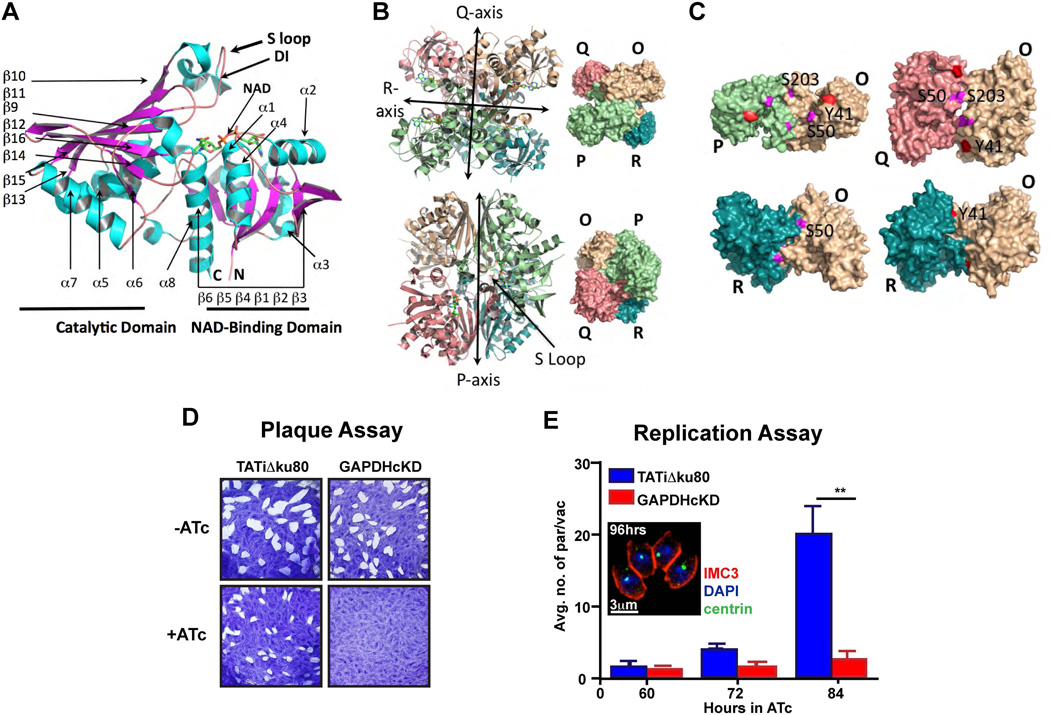
Figure 2.
Multifunctional protein GAPDH1 is essential for the lytic cycle. (A) Crystallographic structure of Toxoplasma GAPDH1 monomer showing the catalytic and NAD+-binding domain with a dipeptide insertion (DI) in the S-loop that is unique to apicomplexan parasites. (B) GAPDH1 tetramer (top two rows) exhibits a true axis of symmetry (P axis) and two pseudo symmetrical axes (Q and R axes). (C) Three configurations of dimer are shown (O-P, O-Q, O-R) in which the O-R dimer is optimally enzymatic active. Predicted tyrosine (Y41, in red) and serine (S50, S203, in magenta) phosphorylation sites are confirmed to be accessible to kinases on the surface of dimers (O-P, O-Q, O-R), with the only exception of S203 being buried in O-R dimer. (D) Plaque assay of parasites grown in the absence (−) and presence (+) of ligand anhydrotetracycline (ATc) for 10 days on HFF monolayer indicates that GAPDH1cKD parasites were not viable. (E) Replication assay by microscopic quantification of number of parasites per parasite vacuoles demonstrates that GAPDH1cKD parasites did not replicate. Parasites were first grown for 48 hrs in HFFs ±ATc, mechanically released and then allowed to invade new host cells on which counting was performed. Inset shows a vacuole containing four morphologically normal parasites at 96 hrs of ATc induction. N=3 ± s.e.m. statistical significance calculated by paired, two tailed t-test, **P < 0.02
The tetramer exhibits approximate 222 symmetry (Supplementary Table 1) with each bilobal subunit composed of a NAD+-binding domain (residue 4–152, 320–340, Pfam PF00044) and a catalytic domain (residue 153–319, Pfam PF02800). One NAD+ cofactor is bound per monomer and the average r.m.s.d. between monomers over all atoms is 0.02 Å as calculated by PISA. The GAPDH1 tetramer exhibits a dihedral symmetry with 3 axes of ‘symmetry’ (P, Q, R) in which only the P axis is a true symmetry (Fig. 2B). The O-R and P-Q dimers are however the most important for optimization of enzymatic activity, since the S-loop (residue 180–207) inserts into the NAD+-binding cavity of the adjacent subunit (Fig. 2A–C, 3A–C). The most distinct structural feature from human GAPDH is the Lys-Gly dipeptide insertion (Fig. 2A, 3B–C) in the S-loop that is also reported in other apicomplexan parasites (Fig. 1)14,15
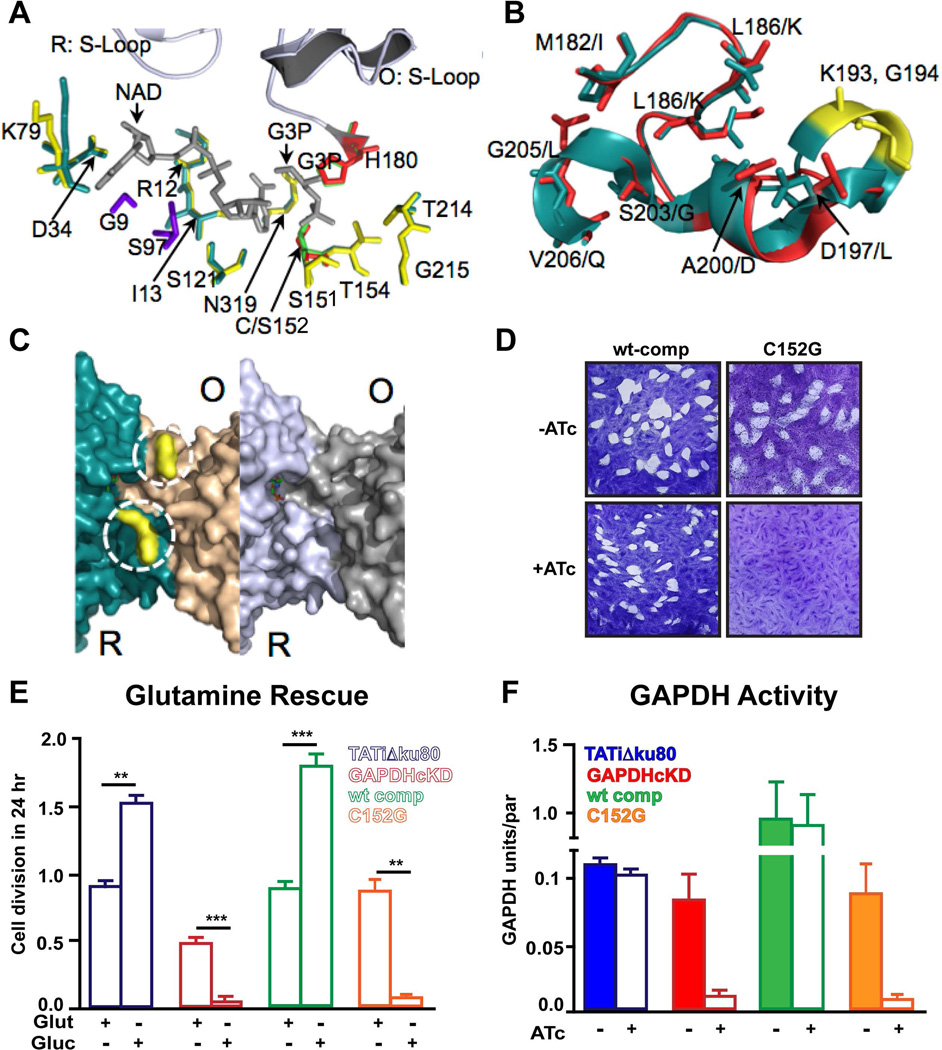
Figure 3.
Functionality of the Toxoplasma GAPDH1 catalytic site. (A) Superimposition of essential residues that interact with substrate glyceraldehyde-3 phosphate (G3P) and co-factor NAD+ in the active site of Toxoplasma and C. parvum (PDB code 3CIF) GAPDH. Residues that form hydrogen and hydrophobic bonds are colored yellow for C. parvum and deep teal for Toxoplasma. Catalytic residues of G3P are coded as red (C. parvum) and green (Toxoplasma). C. parvum structure has a C151S variant. Unique residues that can form hydrophobic interactions with NAD+ in Toxoplasma GAPDH are illustrated in purple. (B) Supposition of S-loops in Toxoplasma GAPDH shown in deep teal (R subunit) and human somatic GAPDH1 that is expressed in human brain cells are shown in red (PDB code 1ZNQ). (C) The prominent distinguishing feature between Toxoplasma parasite (left panel) and human brain (right panel) GAPDH is a dipeptide insertion of K193G194 (yellow, dashed circle); the insertion generates a positive surface charge as determined by surface electrostatic potentials (not shown). This dipeptide insertion is found uniquely in apicomplexan parasites (Toxoplasma, Cryptosporidium, Plasmodium) and not in human GAPDH. (D) Complementation of GAPDH1cKD was observed with wild type GAPDH but not with active site mutant C152G as shown by plaque assay for 10 days. (E) Parasite numbers per vacuole were counted and expressed as number of cell divisions post infection with ATc treatment in glutamine replete or glutamine-deplete media. N=3 ± s.e.m. t-test, **P < 0.02, ***P < 0.002 (F) GAPDH activity as calculated by measuring the reduction of NADH illustrates the conditional knockdown and catalytic residue mutation inhibit activity. To validate that the high level of activity compared to the parental line is not due to the presence of native GAPDH1 in the GAPDH1wt complemented line we ascertain that the native GAPDH1 is the down regulated in the complemented lines upon ATc treatment (Supplementary Fig. 1F).
GAPDH1 is essential for intracellular replication, but not for invasion and egress
To probe the essentiality of GAPDH1 for the Toxoplasma gondii lytic cycle, a GAPDH1 conditional knockdown line (GAPDH1cKD) was generated by replacing the endogenous promoter with anhydrous tetracycline (ATc) regulatable minimal TetO7Sag4 promoter (Supplementary Fig. 1A–B)16. Expression of GAPDH1 protein in the GAPDH1cKD parasite line decreases upon ATc addition and becomes undetectable within three days of ATc addition (Supplementary Fig. 1C). Plaque assays show that GAPDH1 is essential for completion of the lytic cycle (Fig. 2D). Invasion and egress of host cells are not significantly inhibited by absence of GAPDH1 (Supplementary Fig. 1D–E). Microscopic evaluation was used to pinpoint the exact point of arrest in parasite development. In the presence of ATc, GAPDH1cKD parasites replicate at a similar rate to the parent line control up to 60 hrs, at which point the mutant arrests and no further growth is observed (Fig. 2E). The parasites arrest while residing in the vacuole without any discernable morphological defects (Fig. 2E inset). These data demonstrate that GAPDH1 is essential for intracellular replication, but is not required to support invasion or egress.
Catalytic activity is essential for intracellular replication, but not for cortical translocation
The active site pocket of Toxoplasma and Cryptosporidium GAPDH is identical in the G3P residues that form hydrogen bonds with the substrate (S152, T154, T214, G215, N319 in Fig. 3A). The conserved NAD+-binding residues include I13, D34, R12, K79 and S121. The NAD+ binding mechanism in Toxoplasma GAPDH1 appears to be stronger than Cryptosporidium GAPDH in forming addition hydrogen bonds between residues K79 and N6A of NAD+ (3.1 Å) and hydrophobic interactions between N319 and N7N of NAD+ (2.8 Å). Highly conserved in Toxoplasma GAPDH1 are the catalytic C152 and H180, which mediate the nucleophilic attacks of carbonyl groups (Fig. 3A).
The essentiality of the Toxoplasma GAPDH1 enzymatic activity was evaluated by generating a point mutation of the catalytic cysteine (C152G)14. The GAPDH1cKD line was genetically complemented with tandem c-Myc (Myc2) epitope tagged alleles, encoding either wild-type (wt) or C152G mutant GAPDH1. Glycolysis is required for parasite viability since parasite growth is readily rescued by the wt construct but the C152G allele is unable to restore growth (Fig. 3D).
It has been reported that glycolysis can be bypassed by glutaminolysis as glycolytic intermediates are essential for lipid biosynthesis17. Indeed we show that glutamine can partly restore GAPDH1cKD parasite growth18 using two different assays (Fig. 3E and Supplementary Fig. 2). To further validate the loss of glycolytic activity we demonstrated dramatic drops in both ATP concentration19 and GAPDH activity upon loss of GAPDH1 altogether or complementation with the C152G allele (Supplementary Fig. 3 and Fig. 3F). Therefore, loss of GAPDH1 activity results in the loss of glycolysis, which is essential for parasite growth under normal conditions.
Serine phosphorylation of the S-loop blocks enzymatic activity and parasite growth
The GAPDH S-loops form a two-plier ridge that separates the NAD+-binding cavities of subunits across the R axis (Fig.2A and 3A–C). Binding of NAD+ increases the ordering of the S-loop and interactions between subunits that are positioned across each other in the tetramer, leading further to changes in the packing environment at the dyad interfaces (Fig. 2A). The opening to the NAD+ cavity is more constricted in Toxoplasma GAPDH1 in comparison to human GAPDH1 due to Lys196 and the bulge created by Lys193-Gly194 dipeptide insertion (Fig. 3B). Interestingly, the phosphoproteome of Toxoplasma provides evidence for the phosphorylation of two residues associated with the S-loop of GAPDH1, Ser50 and Ser20320. Ser50 is located on the surface loop that follows α-helix 2 (α2) and can form hydrogen bonds with S-loop of the neighboring subunits of the Q-R dyad (Fig. 4A). Ser50 in the O subunit can form contact with Ser287 in the Q subunit and Asp189 in the S-loop of R subunit. Serine 203 is located directly in the S-loop and can form hydrogen bonds with three S-loop residues (Thr187, Asp189, Asn207) to configure this flexible loop (Fig. 4B).
Figure 4.
Serine phosphorylation of regulatory S-loop is essential. (A) Carbon chain and surface rendering of dimer of the R and Q dyads (O-R, top panels, O-Q, bottom panels) reveals that phosphorylatable Ser50 (magenta) is located on a beta-turn that forms 3 hydrogen bonds with Asp189 residue in the S-loop of adjacent subunit in the O-R dimer. Ser50 of in the α 2 helix also forms a polar contact with Ser287 (cyan) at the interface of O-Q dimer. (B) Phosphorylatable Ser203 is positioned in the S-Loop and is critical to the loop stability by forming polar contacts with three residues (Thr187, Asp189, and Asn207). (C) Plaque assay shows the viability of the phospho-mimic and phospho-null mutants by genetic complementation. (D) GAPDH activity of the phospho-mutants as indicated measured after 60 hrs ± ATc. N=3 ± s.e.m. t-test, **P < 0.04. We verified that GAPDH activity was independent of GAPDH protein level in the experimental lines (Supplementary Fig. 4). Although expression levels for all mutant proteins were comparable, we detected a slightly shorter version of the S203A mutant as well as some smaller protein bands suggesting GAPDH1S203A might not be stable. (E) Representative example of cortical localization (green: α-Myc staining of tagged Myc2-GAPDH; red: α-IMC3 staining of cortical cytoskeleton). The yellow arrow marks the site of the intensity profile of both signals. (F) Representative image indicating the cytosolic localization of mutant GAPDH in extracellular parasites with its intensity profile across the yellow arrow. (G) Quantification of percentage of extracellular parasites displaying cortical GAPDH localization in the genetically complemented GAPDH1cKD line as indicated in three independent experiments for the wt and one for the mutant lines.
The novel observation of GAPDH S-loop phosphorylation was examined for potential regulatory effects. Phosphomimetic (Ser to Glu) and phosphonull (Ser to Ala) point mutations were evaluated for complementation capacity in the GAPDH1cKD line and also for cortical translocation. GAPDH1S50E and GAPDH1S203E did not support parasite growth (Fig. 4C). Parasites stably expressing the double phosphomimetic mutation (GAPDH1S50E, S203E) could not be established in the uninduced GAPDH1cKD line suggesting that this allele has a more severe phenotype and acts in a dominant negative fashion. GAPDH1S50A phosphonull mutation does not support viability but the S203A is completely viable (Fig. 4C). Interestingly, no decrease in activity for the S50E mutant compared to the S203E or S50A mutant was detected, which indicates that GAPDH activity is not disrupted due to constitutive phosphorylation of S50 (Fig. 4D). Since kinase activity is associated with egress21, we asked whether S-loop phosphorylation in involved in cortical translocation, but none of the mutants displayed a translocation defect (Fig. 4E–G). Overall, our observations show that serine phosphorylation of the S-loop in GAPDH1 regulates its enzymatic activity, probably by interference with oligomerization assembly and allosteric activation. However, this post-translational modification has no effect on cortical translocation of GAPDH1.
Rab2 binding mediated by S-loop tyrosine phosphorylation does not occur in Toxoplasma
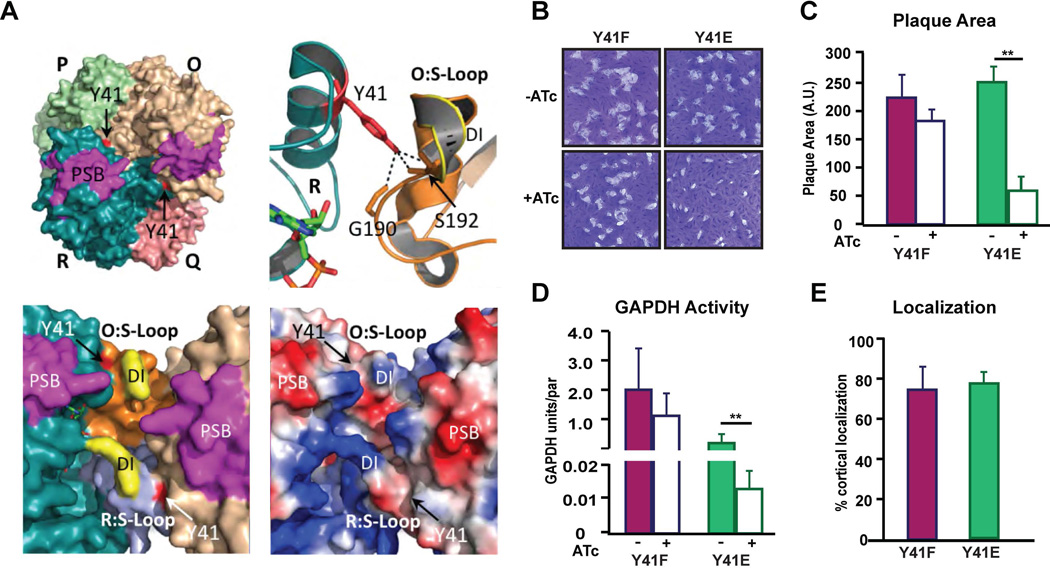
Phosphorylation of Tyr41 in human GAPDH1 by Src-dependent tyrosine phosphorylation contributes to the binding of Rab222,23 and there is evidence that this mechanism is also present in the malaria causing apicomplexan parasite Plasmodium24. Tyr41 is conserved in Toxoplasma GAPDH1 and is located in a surface helixα2 (Fig. 1). This residue can interact with the S-loop of the adjacent R dyad monomer; Tyr41 of the R subunit can form hydrogen bonds with Gly190 and Ser192 of S the loop in the O subunit (Fig. 5A). Phosphomimetic (Y41E) and phosphonull (Y41F) alleles were generated and in the GAPDH1cKD background to examine whether Tyr 41 phosphorylation is involved in GAPDH translocation to the cortical membrane skeleton. Both alleles support viability of the parasite as assessed by plaque assay (Fig. 5B), although we observe a nearly 4-fold reduction in plaque size for the Y41E allele (Fig. 5C). Reduction in plaque size is likely due to a dramatic reduction in GAPDH catalytic activity (Fig. 5D) by altering the dynamic S-loop. Importantly, neither allele alters cortical translocation (Fig. 5E) and thus negates the hypothesis that Y41 regulates GAPDH1 translocation to cortical membrane skeleton.
Figure 5.
Potential tyrosine phosphorylation of the S-loop modulates GAPDH activity. (A) Surface rendering of tetramer (left) illustrating the interaction of Tyr41 to S-Loop of neighboring subunit. Tyr41 (red) of R subunit forms 3 hydrogen bonds with Gly190 and Ser192 of the S-loop in the O subunit. Tyr41 is phosphorylated in human GAPDH23, but accessibility for kinase and Rab2 may be hindered sterically by the apicomplexan dipeptide insertion (DI) in the S-Loop. The apicomplexan dipeptide exhibits a positive surface charge as determined by surface electrostatic potential analysis (bottom, right panel), where positively charged residue is rendered as blue and negative charged is red. Attachment to membrane by the predicted PSB patch (magenta) may interfere with access to the S-loop and catalytic pocket. (B) Ten day plaque assay for genetically complemented Y41E and Y41F mutants of GAPDH1cKD. (C) Quantification of plaque size of the mutants as indicated after 10 days ± ATc. N=3 ± s.e.m. t-test **P < 0.02. (D) GAPDH activity of the phospho-mutants as indicated measured after 60 hrs ± ATc. N=3 ± s.e.m. t-test, **P < 0.02. (E) Percentage of extracellular parasites displaying cortical GAPDH localization of the genetically complemented GAPDH1cKD parasites as indicated. N=3 ± s.d.
Cortical translocation is inhibited by bulky polypeptides fused to GAPDH1
Targeted molecular disruption of the catalytic pocket and central core of the tetramer has not identified the translocation mechanism of GAPDH1 to the cortical membrane skeleton. In our optimization of protein reporter tags for GAPDH1, we confirmed the previous observation25 that fusion of bulky fluorophores (GFP: 238 amino acids, mCherryRFP: 226 amino acids) at the N-terminus block translocation of GAPDH1 to the cortex in extracellular parasites, whereas a smaller tandem Myc epitope tag (Myc2: 26 amino acid) does not (Fig. 6A). However, blocking translocation with bulky fusions impairs neither GAPDH activity of the parasites (Supplementary Fig. 6A), nor their viability (Fig. 6B). The N-terminus protrudes on the surface of GAPDH1 around a cleft that contains the N-terminal edge of the putative PSB domain composed of amino acids 70–94 (Fig. 6C)26,27 that could play a role in membrane association and might be blocked by steric hindrance due to a large tag.
Figure 6.
Targeting to cortical membrane skeleton is mediated by N-terminal palmitoylation.. (A) Cortical translocation of N-terminally tagged mCherry GAPDH1wt and Myc2 tagged GAPDH1wt and a GAPDH1C3S variant was quantified in the extracellular parasites. N=3 ± s.d. t-test, ****P < 0.0001, ***P < 0.0003. Inserts show most representative image corresponding to the N-terminally fused GAPDH1 with either mCherry or Myc2. The image of mCherry tagged GAPDH1 (red) is live in extracellular parasites and methanol fixed and probed using α-Myc (green) for GAPDH1wt and mutant GAPDH1C3S. (B) Plaque assay of parasites expressing N-terminally-tagged mCherryRFP Toxoplasma GAPDH1wt and Myc2 tagged GAPDH1C3S variants. (C) Crystallographic structure reveals that the N-terminus ends at surface cleft in which the predicted palmitoylation of C3 (red) is surface accessible. This residue is located near the N-terminus of a β-sheet (left). Surface electrostatic potential rendering (right) shows that the putatively palmitoylated residue lies in a positive surface patch that may facilitate protein-protein interaction. (D) Flowchart of fractionation and detergent extraction experiment. (E) Western blots of GAPDH1wt and GAPDH1C3S strains after the low speed and high speed spins. The membrane was probed with α-Myc. (F) Detergent extractions of GAPDH1wt and GAPDH1C3S strains using the indicated conditions followed by probing with α-Myc. Samples were probed with the following controls: α-IMC1 (cytoskeleton), α-CDPK1 (cytosolic), α-ACT1 (cytosolic), α-SAG1 (plasma membrane) and α-ISP1 (IMC membrane bound through myristoylation and palmitoylation). Protein sizes are as follows, GAPDH1 probed with α-Myc, 39 kDa; IMC1, 70 kDa; CDPK1, 65 kDa; ACT1, 42 kDa; SAG1, 35 kDa; ISP1, 18 kDa.
GAPDH1 cortical translocation is mediated by palmitoylation at the N-terminus
We considered other potential membrane skeleton translocation mechanisms. Membrane association by palmitoylation has been demonstrated for other proteins residing in the membrane skeleton21,28. Notably, S-palmitoylation is reversible and thus fits GAPDH1 release from the membrane skeleton upon invasion of a host cell25. Two putative palmitoylation sites are predicted for GAPDH1 by bioinformatics predictions using CSS Palm; Cys3, which is unique to Toxoplasma and other coccidians like Neospora, Eimeria etc., and Cys249, which is also found in human GAPDH129 (Fig. 1). The crystallographic structure of GAPDH1 indicates that Cys249 is not surface exposed in the GAPDH1 tetramer and its modification would block multimerization and thus enzymatic activity. However, Cys3 is surface exposed (Fig. 6C). We studied the role of Cys3 in cortical membrane skeleton by replacement it with Ser in a GAPDH1 allele fused at the N-terminus to a tandem Myc. To assess the role of GAPDH1C3S in the parasite, we tested the viability by plaque assay and GAPDH activity in the parasites. We observed that loss of Cys3 palmitoylation capacity has no effect on the lytic cycle (Fig. 6B) or GAPDH activity (Supplementary Fig. S6A). However, we observe that Myc2-GAPDH1wt is targeted to the cortical skeleton in extracellular parasites whereas this association is reduced by 80% in Myc2-GAPDH1C3S expressing parasites (Fig. 6A).
Using these mutants we attempted to obtain direct proof of GAPDH1 palmitoylation at C3S using a chemically manipulatable palmitate mimic, 17- octadecynoic acid (17-ODYA)30. In case C249 would also be palmitoylated, we also generated a C249S mutant parasite line so we can discern it from C3S palmitoylation. We observe strong signals for wild type, C3S and C249S mutants (Supplementary Fig. S6B) suggesting the presence of multiple sites of modification in GAPDH1. The observed signal was insensitive to hydroxylamine treatment suggesting that the modification does not occur on a cysteine residue. 17-ODYA can also detect O-palmitoylation31, N-palmitoylation32 as well as other post translational modifications such as GPI anchors33, which are insensitive to hydroxylamine. Bioinformatics predictions of other lipid modifications using online tools GPS lipid and NMT predictor (myristoylation, prenylation, farnesylation) were negative. Hence, these data reveal a complex modification picture preventing the direct assessment of whether C3S is palmitoylated.
To further test our hypothesis we determined whether GAPDH1 is membrane bound by differential centrifugation of wild type GAPDH1 and the predicted palmitoylation site mutant C3S according to the schematic in Fig. 6D. After osmotic and mechanical lysis of extracellular parasites in presence of minimal salt (5 mM) and low speed centrifugation, we observed slightly more material in the supernatant (S1) than in the pellet fractions (P1) for both wt and C3S parasites (Fig. 6E, F). This is somewhat unexpected since only non-lysed parasites and the cytoskeleton were expected to pellet. Probing for the cytoplasmic proteins actin (majority cytoplasmic) and CDPK1 (100% cytoplasmic) demonstrated that lysis was complete (Fig. 6F), thus suggesting that GAPDH1 associates with the cytoskeleton (IMC1 in Fig. 6F). Alternatively, we considered that the membrane skeleton and even plasma membrane could remain associated with the cytoskeleton. Therefore we assessed how plasma membrane (SAG1, GPI-anchored34) and IMC membrane (ISP2, palmitoylated35) fractionated. Like GAPDH1, roughly half SAG1 ends up in the pellet (P1) whereas 100% of ISP2 is in the pellet (P1) suggesting that half of the plasma membrane and all of the IMC membrane tightly associates with the cytoskeleton upon osmotic and mechanical parasite disruption.
Next we subjected the low-speed supernatant (S1) to high-speed centrifugation to pellet (P3) all membrane-associated proteins36. While GAPDH1wt resides in the supernatant, suggesting they are mostly cytoplasmic, the majority of GAPDH1C3S is in the supernatant, suggesting membrane association (Fig. 6E), Since, the majority of the membrane fraction of interest is present in the low-speed pellet, we further decipher the state of membrane associated proteins in the low speed pellet fraction. We resuspended the pellet in a physiological buffer (150 mM NaCl) and first performed a high-speed spin to release proteins that interact mostly through electrostatic interactions rather than through lipid membrane anchoring. GAPDH1wt remains in the pellet (P3) whereas nearly half of GAPDH1C3S becomes soluble (S3) (Fig. 6E). This suggests that the association of GAPDH1C3S with the membrane/cytoskeleton is based largely on electrostatic interactions.
In addition, we assessed whether we could remove GAPDH1 from the low speed pellet (P1) fraction using detergents. As expected, treatment with 1% SDS solubilizes all proteins probed (Fig. 6F S4 and P4). In the control where no detergent was added C3S again stood out by its significant release from the P1 fraction (Fig. 6F S7 and P7). We observed no major difference from the wild type construct using mild detergents (Fig. 6E, TX-100 and NP-40). As expected, both membrane-associated proteins SAG1 and ISP2 solubilized with mild detergent whereas the cytoskeletal protein IMC1 did not, which demonstrates that our assay works well. In conclusion, the interpretation of Toxoplasma membrane fractionation is complicated by strong association between the cytoskeleton and membrane structure of interest, but our results are consistent with a critical role for Cys3 in membrane association, potentially through palmitoylation.
Discussion
The crystal structure of Toxoplasma GAPDH1 combined with post-translational modification data and allelic replacements permit the validation and uncoupling of GAPDH1’s role in glycolysis from its mechanism of cortical translocation. Uncoupling of functions is evidenced from the quaternary structure of GAPDH and our functional data. Glycolysis is mediated by the buried core center and is regulated by the S-loop phosphorylation whereas cortical membrane association is facilitated by palmitoylation at the surface exposed the N-terminus.
It is known that glycolysis is not strictly essential in Toxoplasma17,37, which is confirmed by our studies. Although our initial data indicated that GAPDH1 is essential, we show that a L-glutamine complementation rescues parasite viability to approximately 50%. This partial rescue is consistent with the contribution of glycolysis intermediates to lipid synthesis, which is growth limiting38,39. Since we observe a trend of increased rescue upon complementation of the complete knockdown with the C152G mutant (albeit not statistically significant), we conclude that there is a hint of critical moonlighting function(s) of GAPDH1 in the Toxoplasma lytic cycle.
In vertebrates, GAPDH has a function in control of apoptosis40. Acetylation of lysine 160 in helix α5 of nuclear GAPDH activates acetyltransferase P300/CBP-associated factor (PCAF), which blocks induction of apoptotic genes and decreases cell death41. This lysine is positionally conserved in Toxoplasma GAPDH1. We introduced the GAPDH1K162Q acetylation-mimic residue in our genetic complementation system, but this modification had no effect on parasite viability (Supplementary Fig. 5). Therefore, apoptosis in Toxoplasma is not mediated by GAPDH1 K162 acetylation.
The dynamic S-loop of Toxoplasma GAPDH1 is regulated by differential phosphorylation of Ser50, Ser203 and Tyr41 to trans-activate the catalytic pocket of other subunit in the R dyads. Phosphorylation of S50 and S203 are required for viability but only S203 phosphorylation is required for GAPDH activity. Our structure-function data show that dephosphorylated S203 is pivotal for reconfiguring the S-loop to fit into the neighboring NAD-binding pocket by forming atomic interactions with three other S-loop residues (Thr187, Asp189, Asn207). Dephosphorylated S50 contributes to homodimerization by hydrogen bonding across the dimer interface with S287 and stabilizes the neighboring S-loop by bonding with Asp189. Phosphorylation of Y41 is not required for viability but disrupts GAPDH enzymatic activity. Dephosphorylated Y41 also acts across the dimer interface by forming three hydrogen bonds with the neighboring S-loop. All three phosphorylation events modulate the dynamic motion of S-loop in regulating the NAD-binding pocket and oligomer assembly, but phosphorylation of S50 is essential for a non-glycolytic function of GAPDH1 that is yet to be described. GAPDH S-loop phosphorylation is possibly a universal feature since S50 is a widely conserved residue in the diversity of GAPDHs (Fig. 1).
The localized effect of catalytic loop phosphorylation on enzymatic activity is a widely conserved mechanism. Hallmark examples include the cis-phosphorylation in the activation loop of the Ephrin subfamily of tyrosine kinase receptors42, phosphorylation of the activation loop in the family of MAP kinases43, and the activation of Aurora A kinase by phosphorylation of its activation loop44. The allosteric regulation of the Toxoplasma GAPDH1 S-loop by multiple intra- and inter-phosphorylation events is a novel mechanism of catalytic loop activation in this class of enzymatic activity regulation.
Data from this study suggest that cortical translocation of GAPDH1 is not essential for parasite viability. It has been shown that after prolonged extracellular gliding stored ATP becomes depleted37 and it is therefore feasible that local ATP production at the cortex becomes much more critical in such situation. Alternatively, although our data demonstrate balanced expression of endogenous and complemented GAPDH alleles, it is possible that the spurious general cytoplasmic localization of GAPDH1 as a result of moderate overexpression is able to overcome specific membrane association, as has been shown for the CDPK3 kinase anchored in the IMC21. Overall, our data indicate that cortical translocation is decoupled from glycolytic essentiality.
Evidence for a novel cortical translocation mechanism of GAPDH is presented. IMC association is mediated by Cys3 residue of GAPDH1. Out attempt to validate the predicted palmitoylation of C3 residue by 17-ODYA labeling and click chemistry was not successful. Though we are able to show that 17-ODYA modifies GAPDH1, the modification is insensitive to hydroxylamine, suggesting there are residues other than the predicted C3 and C249 that may be palmitoylated and likely not mediated through thio-ester modification. As an alternative, we decided to pursue a role for C3S in membrane association through differential fractionation36 and saw a critical role of this residue in membrane association. Further evidence supporting the feasibility of our palmitoylation model is found in the recent identification of two different palmitoyl acyl transferases residing in the Toxoplasma IMC, TgDHHC2 and TgDHHC1445,46. In humans, it has been shown that BET3, the central unit of TRAPP complex, is palmitoylated at the Cys68 residue, which is involved in tethering of the transport vesicles to the cis-Golgi membrane47. This Cys residue is buried in a pocket that is similar to the palmitoylation crevice at Cys3 of Toxoplasma GAPDH1 (Fig. 6C). Overall, our structural, fractionation and extraction data together with the reversible cortical membrane association of GAPDH1 are consistent with palmitoylation at Cys3.
From the perspective of GAPDH1 as a therapeutic target we have generated some important insights in this study. As in other apicomplexan GAPDH1 enzymes, the Lys-Gly dipeptide insertion in the S-loop can potentially be exploited in specific drug development48. However, it is important to keep in mind that Toxoplasma and most other Apicomplexa contain GAPDH2, which is targeted to the apicoplast. How GAPDH2 functions in the parasite’s energy metabolism has not been studied, but given the sharp reduction in ATP levels we observe upon deletion of GAPDH1 it is unable to compensate for GAPDH1. Furthermore, our genetic complementation data with the catalytically dead C152G allele indicate that even if GAPDH1 can be pharmacologically inhibited, the parasite is still able to survive by switching to L-glutamine as an energy and carbon source. On the other hand, we highlight that the phosphorylation of S-loop residues S50 and S203 are critical to GAPDH1 and parasite survival. Thus, targeting the kinases mediating S-loop phosphorylation highlight a specific novel Toxoplasma drug target.
Methods
Parasites
RH strain parasites and transgenic derivatives were maintained in human foreskin fibroblasts (HFF) as previously described49. The RHΔHX and TATiΔku80 parasite strains (kindly provided by Vern Carruthers, University of Michigan) were used in this study. GAPDH1 conditional knockdown (cKD) was generated by double homologous promoter replacement under 1 µM pyrimethamine selection. Parasites stably expressing complementing transgenes or GAPDH mutations were selected under 20 µM chloramphenicol. All lines were cloned by limiting dilution. GAPDH mutations in parasites clones expressing mutant GAPDH1 alleles were validated by Sanger sequencing of their PCR amplified Myc tagged allele. The GAPDH1 conditional knockdown was induced at a concentration of 1.0 µg/ml anhydrous tetracycline (ATc).
Plasmids
All primer sequences can be accessed in the Supplementary Information Table S2. The plasmid ptub-Myc2-GAPDH1/sagCAT was generated by amplifying the GAPDH1 coding sequence using primers #1 and #2 The insert, digested with AvrII/EcoRV, was cloned into the ptub-YFP2(MCS)/sagCAT plasmid49.
Three-way Gateway cloning (Invitrogen) was employed to generate the GAPDH1 conditional knockdown vector; basically following published protocols50. In short, the entry clone plasmid pDONR221-5’GAPDH1R1/R4 was generated by amplifying approximately 1 kb upstream of the promoter region (1.5 kb upstream of start codon) with primers #5 and #6 and cloned into the pDONR221 P1/P4 plasmid. The entry clone plasmid pDONR221-GAPDH1 R3/R2 was generated by amplifying 0.6 kb beginning at the start codon using RH strain genomic DNA as a template with primers #7 and #8 and cloned into the pDONR221 P3/P2 plasmid. The conditional knockout vector pTgKO2-DHFR-T7S4GAPDH1 was generated by combining the pTgKO2, pDONR221-5’GAPDH1-R1/R4, pDONR221-DHFR-T7S4 R4/R3 and pDONR221-GAPDH1 R3/R2 plasmid in an LR reaction. The plasmid was linearized prior to transfection using restriction enzyme AflII. The stable lines were generated by pyrimethamine selection and screened for integration using #9– #10 and #11– #12 primer pairs. A single clone exhibiting the promoter replacement was selected by verifying the absence of native GAPDH1 promoter using primers #13- #12.
Megaprimer mutagenesis was used to generate point mutations as described51. PCR products were TOPO-cloned, sequence-validated by Sanger sequencing, and ligated to the ptub-Myc2-GOI/CAT backbone to generate the Myc2 tagged mutant GAPDH1 plasmid. 50 µg of plasmid was transfected in the GAPDH1cKD strain to generate the mutant lines under chloramphenicol selection.
Protein production and purification
Cloning, expression and purification were conducted as part of the Seattle Structural Genomics Center for Infectious Disease52,53 (SSGCID) following standard protocols described previously54–56. The full-length protein (Uniprot: Q9BKE2A) encoding amino acids 1–340 was PCR-amplified from Toxoplasma gondii ME49 genomic DNA that was kindly provided by Dr. David Roos. The gene was amplified using the following primer sequences #14 and # 15. The gene was cloned into the ligation independent cloning57 (LIC) expression vector pAVA042156 encoding a cleavable 6×His fusion tag followed by the human rhinovirus 3C protease-cleavage sequence (MAHHHHHHMGTLEAQTQGPGS-ORF). Plasmid DNA was transformed into chemically competent Escherichia coli BL21 (DE3) R3 Rosetta cells. The plasmid containing His-TgGAPDH1 was expression tested and two liters of culture were grown using auto-induction media58 in a LEX Bioreactor (Epiphyte Three Inc.) as previously described56.
His-TgGAPDH1 was purified in a four-step protocol consisting of a Ni2+-affinity chromatography (IMAC) step, cleavage of the N-terminal His tag with 3C protease, removal of the cleaved His tag by passage over a second Ni2+-affinity chromatography column and size-exclusion chromatography9. All chromatography runs were performed on an ÄKTApurifier 10 (GE) using automated IMAC and SEC programs according to previously described procedures55. Thawed bacterial pellets were lysed by sonication in 200 ml buffer containing 25 mM HEPES pH 7.0, 500 mM NaCl, 5% Glycerol, 0.5% CHAPS, 30 mM Imidazole, 10 mM MgCl2, 1 mM TCEP, 250 µg/ml AEBSF, and 0.025% Na-Azide. After sonication, the crude lysate was clarified with 20 µl (25 U/µl) of benzonase and incubated while mixing at room temperature for 45 min. The lysate is then clarified by centrifugation at 10,000 rev/min for 1 hr using a Sorvall centrifuge (Thermo Scientific). The clarified supernatant was then passed over a Ni-NTA His-Trap FF 5 ml column (GE Healthcare) which was pre-equilibrated with loading buffer composed of 25 mM HEPES pH 7.0, 500 mM NaCl, 5% Glycerol, 30 mM Imidazole, 1 mM TCEP, and 0.025% Na-Azide. The column was washed with 20 column volumes (CV) of loading buffer and was eluted with loading buffer plus 250 mM imidazole in a linear gradient over 7 CV. Peak fractions, as determined by UV at 280 nm, were pooled and transferred into a 50 ml falcon tube. The eluted protein was transferred into a dialysis bag and mixed with His-MBP-3C protease at a ratio of 1:50 to cleave the His-tag. The sample was dialyzed overnight at 4°C in 1 l of 3C dialysis buffer containing 25 mM HEPES pH 7.5, 200 mM NaCl, 5% Glycerol, and 1 mM TCEP. After overnight dialysis, the concentration of imidazole and sodium chloride in the reaction mix was adjusted to 30 mM and 500 mM respectively and the dialysate was passed over a second Ni2+-affinity column to remove His-MBP-3C protease, uncleaved protein and cleaved His tag. The cleaved protein was recovered in the flow through and concentrated prior to loading onto the final column. A SEC column (Superdex 75, GE) was equilibrated with running buffer composed of 500 mM NaCl, 25 mM HEPES pH 7.0, 5% Glycerol, 0.025% Na-Azide, 2 mM DTT. The peak fractions were collected and analyzed for the presence of the protein of interest using SDS–PAGE. The SEC peak fractions eluted as a single large peak in the molecular-mass range 120–210 Da, suggesting multimeric complexes of tetramers (data not shown). The peak fractions were pooled and concentrated to 24 mg/ml using an Amicon purification system (Millipore). Aliquots of 200 µl were flash-frozen in liquid nitrogen and stored at −80°C until use for crystallization.
Crystallization and data collection
Purified TgGAPDH1 was screened for crystallization in 96-well sitting-drop plates against the JCSG-plus and PACT premier (Molecular Dimensions) crystal screens. Equal volumes of protein solution (0.4 µl) and precipitant solution were set up at 289.15 K against reservoir (80 µl) in sitting-drop vapor-diffusion format. The final crystallization precipitant was PACT condition F11 consisting of 200 mM Na-citrate tribasic, 100 mM Bis-tris propane, and pH 6.5 and 20% w/v PEG 3350. The crystals were cryoprotected in crystallant plus 25% ethylene glycol and flash-frozen by dipping into liquid nitrogen. Data were collected at 100°C on Advanced Light Source beamline 5.0.1 using an ADSC Quantum 315 CCD detector with 1° oscillations at a wavelength of 0.9774 Å. Data were reduced with HKL200059.
Structure solution and refinement
The structure was solved by molecular replacement with Phaser60 from the CCP4 suite of programs61 using PDB entry 1YWD48 as a search model. The structure was refined using iterative cycles of Refmac562 followed by manual rebuilding of the structure using Coot63. The quality of all of the structures was checked using MolProbity64. All data-reduction and refinement statistics are shown in Supplement Table 1. The structure was refined to a resolution of 2.25Å. Unambiguous electron density was visible for the placement of sodium ions, NAD, and ethylene glycol, which was used as a cryoprotectant during freezing of the crystals. Ambiguous electron density was also present which could not be definitively interpreted. One ambiguous piece of electron density was adjacent to Cys292 from molecule A and is an oxidized cysteine residue of either sulfenic or sulfinic acid. Another ambiguous piece of electron density sits adjacent to the ribose of NAD in molecules A and B of the asymmetric unit. A similar anomaly was identified in the structure of P. falciparum GAPDH115 (PDBID: 2B4R) also in only 2 of the 4 molecules of the asymmetric unit. This anomaly was identified in PfGAPDH1 as the protease inhibitor AEBSF. However, only the sulfonyl-fluoride moiety of AEBSF could be placed into the electron density of TgGAPDH1. HEPES also contains a similar sulfonyl moiety, which could be positioned similarly to that of AEBSF. Insufficient electron density prevented the discrimination between AEBSF and HEPES. Therefore, the ambiguous electron density near the ribose of NAD is either the sulfonyl moiety of partially disordered AEBSF or HEPES which come from either the lysis buffer or the SEC buffer respectively. Structure figures were analyzed and prepared using PyMOL (v.1.5; Schrodinger) and PISA65. Coordinates and structure factors have been deposited with the Protein Data Bank with accession number 3STH.
Immunofluorescence
Immunofluorescence assays were performed as previously described51. Parasite strains of choice were inoculated in six-well plates containing coverslips with HFF cells and fixed with 100% methanol and blocked in 1% BSA in PBS. Alternatively, extracellular parasites were adhered on coverslips coated with poly-Lysine for 1 hr at 37°C. The following primary antibodies were used: rat α-IMC3 1:200049, mouse MAb 45:36 α-IMC1 1:1000 (kindly provided by Gary Ward, University of Vermont), α-Myc conjugated with A-488 (Cell Signaling) and rabbit α-human centrin 1:1000 (kindly provided by Iain Cheeseman, Whitehead Institute). Alexa fluorophores A488 and A594 (Invitrogen) conjugated to α-rat, α-rabbit, and α-mouse secondary antibodies were used. Nuclear material was co-stained with 4′6-diamidino-2-phenylindole (DAPI). A Zeiss Axiovert 200 M wide-field fluorescence microscope equipped with a α-Plan-Fluar 100×/1.45 NA oil objective and a Hamamatsu C4742-95 CCD camera was used to collect images, which were deconvolved and adjusted for phase-contrast using Volocity software (Improvision/Perkin Elmer).
Replication assay
For intracellular replication, parasite division in HFF infected with 1.8×105 par/well was measured in regular medium ± ATc via IFA using α-IMC3 and α-human centrin as development markers66. More than 100 vacuoles per sample were counted in 3 independent experiments and expressed as average number of parasite/vacuole. Similarly, for assessing division in glutamine supplementation media, HFF monolayer was infected with parasites, after washing with DMEM without glucose or glutamine and incubating for 2 hrs in same media, to make them glucose starved. Also the parasites prior to infection were depleted of residual glucose. The ATc pretreated parasites were washed in DMEM without glucose containing 6 mM L-glutamine (or DMEM without glutamine containing 4.5 g/L glucose as control) and incubated at 37°C for 2 hrs. 1.8×105 parasites per well were inoculated in a 6 well plate with the corresponding media ± ATc. Following 24 hrs of incubation, monolayers were fixed with 100% methanol and subjected to IFA using rat α-IMC3 and rabbit α-human centrin antibodies. More than 100 vacuoles were counted in each experiment and values were expressed as cell division.
Cell monolayer assay
To monitor parasite growth in glutamine-supplemented media (see section above), monolayer lysis was done as described earlier18. Essentially, parasite and HFF monolayer were starved in absence of glucose (of L-glutamine as control) for 2 hrs before inoculation as described in the above section. Subsequently, confluent HFF cells in 96 well plates were inoculated with parasite dilutions in triplicate and allowed to grow for 48 hrs under the indicated conditions. Lysis was assessed by crystal violet staining and measuring absorbance at 570 nm using Molecular Dynamics M5 plate reader by well scanning.
Invasion assay
The ‘Red-Green-Assay’ was done as described previously21. Briefly, 2.5×105 parasites were allowed to invade in HH/FBS (Hank’s balanced salt solution, 1 mM Hepes pH 7.0, 1% FBS) medium ± ATc in black 96 well plate in triplicates at 37°C for 1 hr. A488 conjugated mouse α-SAG1/P30 T41E5 (kind gift from J.F. Dubremetz, University of Montpellier) was used to stain all the parasites and post 16% formaldehyde, 8% glutaraldehyde fixation and 0.25% Triton X-100 permeabilization, A594 conjugated mouse α-SAG1/P30 T41E5 was used to stain all extracellular parasites. Three images per well were recorded using EVOS FS Cell Imaging System (Life Technologies) and more than 100 parasites were counted to determine percentage of invaded parasites using Fiji software67. Parasites were treated with 1 µM Cytochalasin D for 15 min as positive control for the assay.
Egress assay
Parasite egress from host cells was assayed as previously described in21. Essentially, 6 well plate-containing coverslips confluent with HFF cells were infected with 6×106 parasites ± ATc. 30–34 hrs post-inoculation, 2 µM of A23187 or DMSO vehicle control was used to trigger egress for 5 min at 37°C. Methanol fixation followed by Hemacolor (EMD Chemicals) staining was done as per manufacturer’s instructions and over 15 fields were counted for intact vacuoles in each set.
Western blot
Western blot was performed as previously described51. Lysate from 5×106 parasites was loaded in 12% Bis-Tris gel (Invitrogen), transferred to PVDF membrane (Bio-rad), blocked in 5% milk and probed with rabbit α-human GAPDH antibody conjugated with horse radish peroxidase (HRP) (TGR BioSciences) at 1:500, or α-Myc (9E10) conjugated with HRP (Santa Cruz Biotechnologies) or α-IMC1. Following 1 hr incubation the membrane was washed 3 times with PBS containing 0.1% Tween40 for 10 min and once with PBS for 5 min. Signals were detected using chemiluminescent HRP substrate (Millipore) and exposure on X-ray film.
Plaque assay
Six-well plates confluent with HFF cells were inoculated with 100 freshly lysed lines and incubated for 8–10 days ± ATc. Monolayers were fixed with 100% ethanol for 10 min, washed with PBS and stained with crystal violet50. Plaques were counted and plaque area quantified using Fiji67.
ATP measurement
ATP concentration was determined using the CellTiter-Glo Luminescent Cell Viability Assay (Promega; Madison, WI) according to manufacturer’s protocol. In general, intracellular parasites grown 60 hrs ± ATc, were filtered, centrifuged and resuspended in dye free DMEM supplemented with 1% Fetal Bovine serum, 10 units/ml penicillin, 100 µg/ml streptomycin and 1mM glutamine medium to 107 parasites/ml. 100 µL parasites were added to a 96 well plate and equilibrated at room temperature for 30 min An equal volume of CellTiter-Glo reagent was added and the plate was shaken for 5 min to allow cell lysis. An M5 plate reader (Molecular Dynamics) was used to record luminescence at 500 ms integration speed after 10 min. ATP (disodium salt) was used to generate the standard curve.
GAPDH activity assay
GAPDH activity in parasite lines grown ± ATc (60 hrs) was colorimetric measured by the reduction of NADH as per instructions from the manufacturer (ScienceCell Research Laboratories). Briefly, syringe lysed parasites were filtered and centrifuged at 1500g for 15 min PBS wash was done by gently resuspending the parasite pellet. Number of parasites in each condition was normalized and assay was performed in duplicate as per instructions. In general, the parasite pellet was resuspended in ice-cold lysis buffer provided in the kit and gently agitated at 4°C for 20 min. The lysate was centrifuged at 14000 rpm for 5 min and supernatant was carefully collected and stored at −80°C for detection of activity. Cell assay buffer was prepared as per instructions and initial absorbance was measured at 340 nm using a Molecular Dynamics M5 plate reader. Final absorbance was measured after addition of cell lysate supernatant and rate of decrease in absorbance over time was calculated to determine GAPDH activity in the sample.
Fractionation Assay
To fractionate the membrane bound and cytosolic proteins, parasite pellet (1×108 extracellular parasites) was cooled in liquid nitrogen for 5 min and then thawed at 37°C before resuspending in hypotonic buffer (10 mM Tris pH 7.8 and 5 mM NaCl). The resuspended pellet was mechanically lysed using 40 douncer strokes and centrifuged at low speed (1000g) for 15 min. The supernatant was carefully separated (Lo Sp-S1) and the pellet was resuspended in an equal volume of resuspension buffer (100 mM Tris pH 7.8 and 150 mM NaCl) and loaded as Lo Sp-P1. The resuspended pellet was equally divided in 5 parts and either centrifuged at 100,000g for 60 min (Hi Sp-P) or solubilized with 1% SDS for 10 min at 98°C, or 45 min on ice with 1% TX-100, 0.5% NP-40 or no detergent as control. An equivalent volume of the low speed supernatant in resuspension buffer without detergent was also centrifuged at 100,000g for 60 min (Hi Sp-S). All pellets were resuspended in an equal volume of SDS-PAGE loading buffer and corresponding amounts analyzed by western blotting.
Direct assessment of palmitoylation
Method essentially follows previously described methods68. Parasites were grown in ATc (1 µg/µl) for 48 hrs, at which time 17-ODYA was added to culture media for a final concentration of 25 µM. Parasites were allowed to grow over for an additional 20 hrs in the presence of 17-ODYA and ATc. Extracellular parasites were collected by syringe lysis, after which they were spun 1200g and washed once with 1X PBS. Parasites were re-suspended and incubated for 4 hrs in 500 µl of warm media with ATc and 25 µM 17-ODYA. Parasites were spun again at 1200g and washed with 1X PBS. Parasites were lysed in 1X PBS with 0.1% SDS and 1% NP-40 for 1 hr on ice and subsequently spun at 15,000 RPM for 10 min at 4°C. Protein concentration determined by BCA. 200 µg of protein lysate was used for each immunoprecipitation with the exception of C249S where 400 µg was used. Lysates were incubated overnight with 60 µl of protein A/G plus agarose (Santa Cruz biotechnology; Sc-2003) and 5 µl of anti α–Myc antibody 9E10 (Thermo Fischer scientific; 13–2500). Immunoprecipitation, rhodamine click reaction, hydroxylamine treatment and scanning were done as previously described68. Western blot for α–Myc was done using the 71d10 antibody from Cell Signaling Technology #2278S at 1:2000 dil.
Supplementary Material
Acknowledgments
We thank Dr. Moritz Treeck (Francis Crick Institute) and Dr. Klemens Engelberg for useful discussions, Dave Faugno-Fusci and Angela Lin for technical support and Patrick Autissier for FACS. This project has been funded in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract Nos.: HHSN272201200025C and HHSN272200700057C and individual grants AI081924 (M.-J.G.), AI110690 (M.-J.G.) and GM111703 (M.B.) as well as American Cancer Society RSG-12-175-01-MPC (M.-J.G.). H.M.N. is grateful to grant support from the Brain and Behavior Research Foundation. I.T.F was supported by the American Heart Association grant 14POST20280004.
Footnotes
Author contributions
H.M.N. selected and submitted Toxoplasma protein targets to SSGCID via Protein Structure Initiative: Biology. P.J.M. directs the expression, purification and crystallization of protein. B.L.S. analyzed and deposited the crystallographic data. H.M.N analyzed the protein structure. R.D. and M.-J.G. designed the molecular and cell biology experiments in collaboration with H.M.N.; R.D. performed these experiments. I.T.F. and M.B. performed and analyzed palmitoylation experiments. R.D., H.M.N and M.-J.G. analyzed the molecular and cell biological experiments and composed the manuscript. I.T.F., B.L.S. and P.J.M. edited the manuscript.
TgGAPDH1 PDBID: 3STH. The authors state no competing financial interests.
References
- 1.Hill DE, Chirukandoth S, Dubey JP. Biology and epidemiology of Toxoplasma gondii in man and animals. Anim Health Res Rev. 2005;6:41–61. doi: 10.1079/ahr2005100. [DOI] [PubMed] [Google Scholar]
- 2.Fabiani S, Pinto B, Bonuccelli U, Bruschi F. Neurobiological studies on the relationship between toxoplasmosis and neuropsychiatric diseases. J Neurol Sci. 2015;351:3–8. doi: 10.1016/j.jns.2015.02.028. [DOI] [PubMed] [Google Scholar]
- 3.Blader IJ, Coleman BI, Chen CT, Gubbels MJ. Lytic Cycle of Toxoplasma gondii: 15 Years Later. Annu Rev Microbiol. 2015;69:463–485. doi: 10.1146/annurev-micro-091014-104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pomel S, Luk FC, Beckers CJ. Host cell egress and invasion induce marked relocations of glycolytic enzymes in Toxoplasma gondii tachyzoites. PLoS Pathog. 2008;4:e1000188. doi: 10.1371/journal.ppat.1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sirover MA. On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochimica et biophysica acta. 2011;1810:741–751. doi: 10.1016/j.bbagen.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 6.Tristan C, Shahani N, Sedlak TW, Sawa A. The diverse functions of GAPDH: views from different subcellular compartments. Cell Signal. 2011;23:317–323. doi: 10.1016/j.cellsig.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roitel O, Vachette P, Azza S, Branlant G. P but not R-axis interface is involved in cooperative binding of NAD on tetrameric phosphorylating glyceraldehyde-3-phosphate dehydrogenase from Bacillus stearothermophilus. Journal of Molecular Biology. 2003;326:1513–1522. doi: 10.1016/s0022-2836(03)00049-4. [DOI] [PubMed] [Google Scholar]
- 8.Jenkins JL, Tanner JJ. High-resolution structure of human D-glyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2006;62:290–301. doi: 10.1107/S0907444905042289. [DOI] [PubMed] [Google Scholar]
- 9.Biesecker G, Harris JI, Thierry JC, Walker JE, Wonacott AJ. Sequence and structure of D-glyceraldehyde 3-phosphate dehydrogenase from Bacillus stearothermophilus. Nature. 1977;266:328–333. doi: 10.1038/266328a0. [DOI] [PubMed] [Google Scholar]
- 10.Tisdale EJ, Kelly C, Artalejo CR. Glyceraldehyde-3-phosphate dehydrogenase interacts with Rab2 and plays an essential role in endoplasmic reticulum to Golgi transport exclusive of its glycolytic activity. The Journal of biological chemistry. 2004;279:54046–54052. doi: 10.1074/jbc.M409472200. [DOI] [PubMed] [Google Scholar]
- 11.Waingeh VF, et al. Glycolytic Enzyme Interactions with Yeast and Skeletal Muscle F-Actin. Biophysical Journal. 2006;90:1371–1384. doi: 10.1529/biophysj.105.070052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gajria B, et al. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 2008;36:D553–D556. doi: 10.1093/nar/gkm981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fast NM, Kissinger JC, Roos DS, Keeling PJ. Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Molecular biology and evolution. 2001;18:418–426. doi: 10.1093/oxfordjournals.molbev.a003818. [DOI] [PubMed] [Google Scholar]
- 14.Cook WJ, Senkovich O, Chattopadhyay D. An unexpected phosphate binding site in glyceraldehyde 3-phosphate dehydrogenase: crystal structures of apo, holo and ternary complex of Cryptosporidium parvum enzyme. BMC Struct Biol. 2009;9:9. doi: 10.1186/1472-6807-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robien MA, et al. Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum at 2.25 A resolution reveals intriguing extra electron density in the active site. Proteins. 2006;62:570–577. doi: 10.1002/prot.20801. [DOI] [PubMed] [Google Scholar]
- 16.Meissner M, Schluter D, Soldati D. Role of Toxoplasma gondii myosin A in powering parasite gliding and host cell invasion. Science. 2002;298:837–840. doi: 10.1126/science.1074553. [DOI] [PubMed] [Google Scholar]
- 17.Blume M, et al. Host-derived glucose and its transporter in the obligate intracellular pathogen Toxoplasma gondii are dispensable by glutaminolysis. Proc Natl Acad Sci U S A. 2009;106:12998–13003. doi: 10.1073/pnas.0903831106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen B, Sibley LD. Toxoplasma aldolase is required for metabolism but dispensable for host-cell invasion. Proc Natl Acad Sci U S A. 2014;111:3567–3572. doi: 10.1073/pnas.1315156111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Starnes GL, Coincon M, Sygusch J, Sibley LD. Aldolase is essential for energy production and bridging adhesin-actin cytoskeletal interactions during parasite invasion of host cells. Cell host & microbe. 2009;5:353–364. doi: 10.1016/j.chom.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Treeck M, Sanders JL, Elias JE, Boothroyd JC. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites' boundaries. Cell Host Microbe. 2011;10:410–419. doi: 10.1016/j.chom.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrison E, et al. A forward genetic screen reveals that calcium-dependent protein kinase 3 regulates egress in Toxoplasma. PLoS Pathog. 2012;8:e1003049. doi: 10.1371/journal.ppat.1003049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tisdale EJ, Azizi F, Artalejo CR. Rab2 utilizes glyceraldehyde-3-phosphate dehydrogenase and protein kinase C{iota} to associate with microtubules and to recruit dynein. Journal of Biological Chemistry. 2009;284:5876–5884. doi: 10.1074/jbc.M807756200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tisdale EJ, Artalejo CR. A GAPDH mutant defective in Src-dependent tyrosine phosphorylation impedes Rab2-mediated events. Traffic. 2007;8:733–741. doi: 10.1111/j.1600-0854.2007.00569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daubenberger CA, et al. The N'-terminal domain of glyceraldehyde-3-phosphate dehydrogenase of the apicomplexan Plasmodium falciparum mediates GTPase Rab2-dependent recruitment to membranes. Biol Chem. 2003;384:1227–1237. doi: 10.1515/BC.2003.135. [DOI] [PubMed] [Google Scholar]
- 25.Pomel S, Luk FC, Beckers CJ. Host cell egress and invasion induce marked relocations of glycolytic enzymes in Toxoplasma gondii tachyzoites. PLoS pathogens. 2008;4:e1000188. doi: 10.1371/journal.ppat.1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glaser PE, Gross RW. Rapid plasmenylethanolamine-selective fusion of membrane bilayers catalyzed by an isoform of glyceraldehyde-3-phosphate dehydrogenase: discrimination between glycolytic and fusogenic roles of individual isoforms. Biochemistry. 1995;34:12193–12203. doi: 10.1021/bi00038a013. [DOI] [PubMed] [Google Scholar]
- 27.Nakagawa T, et al. Participation of a fusogenic protein, glyceraldehyde-3-phosphate dehydrogenase, in nuclear membrane assembly. Journal of Biological Chemistry. 2003;278:20395–20404. doi: 10.1074/jbc.M210824200. [DOI] [PubMed] [Google Scholar]
- 28.Beck JR, et al. A novel family of Toxoplasma IMC proteins displays a hierarchical organization and functions in coordinating parasite division. PLoS Pathog. 2010;6:e1001094. doi: 10.1371/journal.ppat.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, et al. Submicromolar concentrations of palmitoyl-CoA specifically thioesterify cysteine 244 in glyceraldehyde-3-phosphate dehydrogenase inhibiting enzyme activity: a novel mechanism potentially underlying fatty acid induced insulin resistance. Biochemistry. 2005;44:11903–11912. doi: 10.1021/bi0508082. [DOI] [PubMed] [Google Scholar]
- 30.Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou C, et al. Acyl-CoA:lysophosphatidylcholine acyltransferase I (Lpcat1) catalyzes histone protein O-palmitoylation to regulate mRNA synthesis. J Biol Chem. 2011;286:28019–28025. doi: 10.1074/jbc.M111.253385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 33.Binder EM, Lagal V, Kim K. The prodomain of Toxoplasma gondii GPI-anchored subtilase TgSUB1 mediates its targeting to micronemes. Traffic. 2008;9:1485–1496. doi: 10.1111/j.1600-0854.2008.00774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burg JL, Perelman D, Kasper LH, Ware PL, Boothroyd JC. Molecular analysis of the gene encoding the major surface antigen of Toxoplasma gondii. J Immunol. 1988;141:3584–3591. [PubMed] [Google Scholar]
- 35.Beck JR, et al. A novel family of Toxoplasma IMC proteins displays a hierarchical organization and functions in coordinating parasite division. PLoS Pathog. 2010;6:e1001094. doi: 10.1371/journal.ppat.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taha MS, et al. Subcellular fractionation and localization studies reveal a direct interaction of the fragile X mental retardation protein (FMRP) with nucleolin. PLoS One. 2014;9:e91465. doi: 10.1371/journal.pone.0091465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacRae JI, et al. Mitochondrial metabolism of glucose and glutamine is required for intracellular growth of Toxoplasma gondii. Cell Host Microbe. 2012;12:682–692. doi: 10.1016/j.chom.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nitzsche R, Zagoriy V, Lucius R, Gupta N. Metabolic cooperation of glucose and glutamine is essential for the lytic cycle of obligate intracellular parasite Toxoplasma gondii. J Biol Chem. 2015 doi: 10.1074/jbc.M114.624619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramakrishnan S, et al. The intracellular parasite Toxoplasma gondii depends on the synthesis of long-chain and very long-chain unsaturated fatty acids not supplied by the host cell. Mol Microbiol. 2015;97:64–76. doi: 10.1111/mmi.13010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hara MR, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 41.Sen N, et al. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat Cell Biol. 2008;10:866–873. doi: 10.1038/ncb1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Overman RC, Debreczeni JE, Truman CM, McAlister MS, Attwood TK. Completing the structural family portrait of the human EphB tyrosine kinase domains. Protein Science. 2014;23:627–638. doi: 10.1002/pro.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akella R, Min X, Wu Q, Gardner KH, Goldsmith EJ. The third conformation of p38alpha MAP kinase observed in phosphorylated p38alpha and in solution. Structure. 2010;18:1571–1578. doi: 10.1016/j.str.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 44.Zorba A, et al. Molecular mechanism of Aurora A kinase autophosphorylation and its allosteric activation by TPX2. Elife. 2014;3:e02667. doi: 10.7554/eLife.02667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beck JR, et al. A Toxoplasma palmitoyl acyl transferase and the palmitoylated armadillo repeat protein TgARO govern apical rhoptry tethering and reveal a critical role for the rhoptries in host cell invasion but not egress. PLoS Pathog. 2013;9:e1003162. doi: 10.1371/journal.ppat.1003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frenal K, et al. Global analysis of apicomplexan protein S-acyl transferases reveals an enzyme essential for invasion. Traffic. 2013;14:895–911. doi: 10.1111/tra.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turnbull AP, et al. Structure of palmitoylated BET3: insights into TRAPP complex assembly and membrane localization. EMBO J. 2005;24:875–884. doi: 10.1038/sj.emboj.7600565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Satchell JF, et al. Structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum. Acta Crystallogr D Biol Crystallogr. 2005;61:1213–1221. doi: 10.1107/S0907444905018317. [DOI] [PubMed] [Google Scholar]
- 49.Anderson-White BR, et al. A family of intermediate filament-like proteins is sequentially assembled into the cytoskeleton of Toxoplasma gondii. Cellular microbiology. 2011;13:18–31. doi: 10.1111/j.1462-5822.2010.01514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farrell M, Gubbels MJ. The Toxoplasma gondii kinetochore is required for centrosome association with the centrocone (spindle pole) Cell Microbiol. 2014;16:78–94. doi: 10.1111/cmi.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gubbels MJ, Vaishnava S, Boot N, Dubremetz JF, Striepen B. A MORN-repeat protein is a dynamic component of the Toxoplasma gondii cell division apparatus. J Cell Sci. 2006;119:2236–2245. doi: 10.1242/jcs.02949. [DOI] [PubMed] [Google Scholar]
- 52.Myler PJ, et al. The Seattle Structural Genomics Center for Infectious Disease (SSGCID) Infect Disord Drug Targets. 2009;9:493–506. doi: 10.2174/187152609789105687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stacy R, et al. Structural genomics of infectious disease drug targets: the SSGCID. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:979–984. doi: 10.1107/S1744309111029204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Serbzhinskiy DA, et al. Structure of an ADP-ribosylation factor, ARF1, from Entamoeba histolytica bound to Mg(2+)-GDP. Acta Crystallogr F Struct Biol Commun. 2015;71:594–599. doi: 10.1107/S2053230X15004677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bryan CM, et al. High-throughput protein production and purification at the Seattle Structural Genomics Center for Infectious Disease. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:1010–1014. doi: 10.1107/S1744309111018367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi R, et al. Immobilized metal-affinity chromatography protein-recovery screening is predictive of crystallographic structure success. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:998–1005. doi: 10.1107/S1744309111017374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 59.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. in. In: Carter CW Jr, Sweet RM, editors. Methods in Enzymology. Vol. 276. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 60.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vagin AA, et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 63.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 66.Szatanek T, et al. Cactin is essential for G1 progression in Toxoplasma gondii. Molecular microbiology. 2012;84:566–577. doi: 10.1111/j.1365-2958.2012.08044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Foe IT, et al. Global Analysis of Palmitoylated Proteins in Toxoplasma gondii. Cell Host Microbe. 2015;18:501–511. doi: 10.1016/j.chom.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.