Abstract
In the context of large-scale human system immunology studies, controlling for technical and biological variability is crucial to ensure that experimental data support research conclusions. Here, we report on a universal workflow to evaluate both technical and biological variation in multiparameter flow cytometry, applied to the development of a 10-color panel to identify all major cell populations and T cell subsets in cryopreserved PBMC. Replicate runs from a control donation and comparison of different gating strategies assessed technical variability associated with each cell population and permitted the calculation of a quality control score. Applying our panel to a large collection of PBMC samples, we found that most cell populations showed low intra-individual variability over time. In contrast, certain subpopulations such as CD56 T cells and Temra CD4 T cells were associated with high inter-individual variability. Age but not gender had a significant effect on the frequency of several populations, with a drastic decrease in naïve T cells observed in older donors. Ethnicity also influenced a significant proportion of immune cell population frequencies, emphasizing the need to account for these co-variates in immune profiling studies. Finally, we exemplify the usefulness of our workflow by identifying a novel cell-subset signature of latent tuberculosis infection. Thus, our study provides a universal workflow to establish and evaluate any flow cytometry panel in systems immunology studies.
Introduction
Flow cytometry allows rapid and simultaneous qualitative and quantitative analysis of multiple cell populations within a biological sample at the single-cell level (1). With recent technological advances, it is now routinely possible to perform flow cytometric experiments with 10 or more parameters (i.e. multiparameter flow cytometry) in most research infrastructures (2). Multiparameter flow cytometry has proven successful to identify disease signatures and prognostic markers in response to infection, immunization or treatment (3, 4), has led to the discovery of new cell types that contribute to protective immunity, such as polyfunctional T cells (5). Thus, flow cytometry is a key technique for human cellular immunophenotyping studies.
The systems biology approach is increasingly used in human immunology and often involves the analysis of samples from various human cohorts acquired by different researchers and research centers (6). In this context, experimental data must be quality controlled to ensure that differences reflect the biological variables of interest rather than being due to technical variation or biological covariates. Multiparameter flow cytometry results in particular are known to have low reproducibility when not adequately controlled (7, 8). Possible sources of technical variability in flow cytometry are diverse, ranging from differences in sample handling and staining procedures, to differences in assay reagents, data acquisition settings, cell analyzer performances and data analysis methods (9). The resolution of each cell population, defined by the expression profile of the markers used for phenotyping and their relative abundance within the sample of interest, can also affect technical variability (7, 10). Approaches aimed at evaluating and reducing technical variability in flow cytometry are therefore crucial to ensure that biological differences can be detected in the systems immunology settings.
Several approaches to assess and reduce technical variability in flow cytometry studies have been developed. Strategies to minimize variation in sample handling and staining procedures include the development of standardized protocols, reagents and flow cytometry panels (7, 9, 11, 12). The implementation of automated instrument set up templates from data acquisition softwares (i.e. BD Application Settings on BD Diva software), or the use of calibration beads (13) are helpful to reduce technical variation in data acquisition. Another major source of technical variation in flow cytometry is based on the manual gating of cell populations. This can be improved by performing manual gating following a defined standard operating procedure (SOP) by a centralized invariant operator, or based on automated gating pipelines. Such computational methods are currently being actively developed and benchmarked by the ‘flow informatics’ community, for instance through the FlowCAP (Flow Cytometry: Critical Assessment of Population Identification Methods) project (14).
While these standardizing efforts have tremendous value for studies that can directly re-use the standardized protocols and reagents that have been developed, they cannot be all encompassing as specific research questions often necessitate the use of custom staining panels for which specific standardization efforts will again be necessary. Furthermore, while all the previous standardizing efforts have been interested in the measurement of technical variation, none have developed specific metrics to control for it. Thus, the development of a universal workflow that control for technical variation and that could be easily applied to any flow cytometry panel would be of great interest for the clinical human immunology community.
Controlling and correcting for technical variability allows teasing out the true biological variation between samples. This in turn enables examining another important source of variability when assessing cell populations that is introduced by biological variations in the donor population introduced by co-variates such as different geographical locations, gender and age of the donors. These factors can contribute to large differences between samples and might mask immunological differences that were targeted by the study, or even lead to false positive results. Recent studies have found significant association between non-genetic factors such as geographic location with the cellular immune composition of human peripheral blood (15). This makes it important to establish procedures to assess the impact of co-variates in the population studied on the cell population under examination in order to separate the effect of these co-variates from the biological factor of interest.
For our studies of immune signatures associated with Dengue Virus and Mycobacterium tuberculosis infection and vaccination as part of the second round of the Human Immunology Project Consortium (HIPC) program (16), we wanted to design a single flow cytometry panel that could quantify naïve and memory T cell subsets, as well as delineate most of the major cell types circulating in the peripheral blood. We thus developed a 10-color flow cytometry panel for the quantification of 17 cell populations from cryopreserved peripheral blood mononuclear cells (PBMC). Based on the considerations above, we aimed to establish a re-usable approach to quantify technical variability and identify biological covariates associated with cell population frequencies. We devised quality control approaches that flag faulty batches of samples before they are used in resource intensive ‘omics assays. We also show that after correction for technical variability, it is possible to determine if and which cell population frequencies vary substantially as a function of such demographic variables, and that it is possible to either control for them during donor recruitment or correct for their presence in the final data analysis. Our experience has resulted in detailed specific recommendations for researchers interested in studying cell populations covered in our panel. More generally, our study provides a universal workflow that can be applied to establish and evaluate similar flow cytometry panels for systems immunology studies.
Material and Methods
Human samples
Blood samples were obtained from the University of California, San Diego Anti-viral Research Center clinic (UCSD, San Diego), Universidad Peruana Cayetano Heredia (UPCH, Peru), National Blood Center, Ministry of Health, Colombo (Sri Lanka) under approval from the respective Institutional Review Boards. All participants, except anonymously recruited blood bank donors in Sri Lanka, provided written informed consent prior to participation in the study. Latent tuberculosis (TB) infection status was confirmed by a positive IFN-γ release assay (IGRA) (QuantiFERON-TB Gold In-Tube, Cellestis or T-SPOT.TB, Oxford Immunotec) but no evidence of clinical symptoms of active TB. TB negative donors were defined as IGRA negative. Subjects born in areas where Bacillus Calmette–Guérin vaccination is prevalent were assumed to have been vaccinated. PBMC were obtained by density gradient centrifugation (Ficoll-Hypaque, Amersham Biosciences) from 100 ml leukapheresis or whole blood samples, according to the manufacturer’s instructions. Cells were resuspended at 50 to 100 million cells per mL in FBS (Gemini Bio-Products) containing 10% dimethyl sulfoxide (Sigma) or in Synth-a-Freeze cryopreservation medium (Life Technologies) and cryopreserved in liquid nitrogen.
Flow cytometry staining and acquisition
Cryopreserved PBMC were quickly thawed by incubating each cryovial at 37°C for 2 minutes, and cells transferred into 9 ml of cold medium (RPMI 1640 with L-Glutamin and 25 mM Hepes (Omega Scientific), supplemented with 5% human AB serum (GemCell), 1% Penicillin Streptomycin (Gibco) and 1% Glutamax (Gibco)) in a 15-ml conical tube. Cells were centrifuged at 1200rpm for 7 min, and resuspended in medium to determine cell concentration and viability using Trypan blue and a hematocytometer. 10 million cells were transferred into a 15-ml conical tube, centrifuged, resuspended in 200 μl of PBS containing 10% FBS and incubated for 10 minutes at 4°C. Cells were stained with 200 μl of PBS containing conjugated antibodies at previously determined optimum dilution (3 μl of anti-human CD3-AF700, anti-human CD4-APCeF780, anti-human CD8a-BV650, anti-human CD19-PE Cy7, anti-human CD14-APC, anti-human CD45RA eF450; 6 μl of anti-human CD56-PE and anti-human CD25-FITC; 12 μl of anti-human CCR7-PerCp Cy5.5; and 0.3 μl of live/dead eF506 stain; see Table SI for staining reagent sources) for 30 minutes at 4°C, protected from light. After 2 washes in PBS, cells were resuspended into 500 μl of PBS containing 0.5% FBS and 2mM EDTA (pH 8.0) (FACS Buffer), transferred into a 5-ml polypropylene FACS tube (BD Biosciences) and stored at 4°C protected from light for up to 4 hours until FACS acquisition. For samples requiring fixation, following staining and washes, cells were fixed in 200 μl of 0.5% or 4% paraformaldehyde (PFA) for 15 minutes at room temperature, washed twice in PBS, and resuspended into 500 μl of PBS containing 0.5% FBS and 2mM EDTA (pH 8.0) (FACS Buffer), transferred into a 5-ml polypropylene FACS tube (BD Biosciences) and stored at 4°C protected from light for up to 4 hours until FACS acquisition. FACS acquisition was performed on a BD LSR-I/II cell analyzer (BD Biosciences) or on a BD FACSAria III/Fusion cell sorter (BD Biosciences). Compensation was realized with single-stained beads (UltraComp eBeads, eBiosciences) in PBS using the same antibody dilution as for the cell staining. Fcs files for latent TB donors and healthy controls were deposited in the public database ImmPort (Study ID SYD820).
Gating
Manual gating was performed with FlowJo version 10.1. The automated gating method used in this manuscript is derived from the FLOCK (FLOw Clustering without K) density-based data clustering method for unsupervised identification of cell populations from polychromatic flow cytometry data (17). The FLOCK method is publicly available at ImmPort (Immunology Database and Analysis Portal, https://immport.niaid.nih.gov) and its source code is publicly accessible at sourceforge (https://sourceforge.net/projects/immportflock). Major extensions have been added to the FLOCK-based automated gating approach in this manuscript. User-provided constraints about the initial boundaries of the gates have been used to direct the unsupervised clustering of FLOCK as follows. When the centroid of a FLOCK-identified data cluster is outside the user-defined boundary, the whole data cluster will be excluded from the gate; when the centroids of multiple FLOCK-identified data clusters are within the user-defined boundary, the cell populations defined by these centroids will be merged into one a single cell population for comparison. Thus the user will not need to manually interpret the large number of FLOCK-identified cell populations, which is a common challenge for unsupervised clustering-based automated gating approaches. Further, the original FLOCK approach only outputs statistics for each individual data cluster. The automated gating approach used in this manuscript is able to output population statistics as in sequential manual gating steps by organizing the FLOCK-identified data clusters into the predefined cell type hierarchy. Therefore, the directed unsupervised clustering extension of FLOCK used here is specifically designed for scientific use cases when a sequential gating strategy (like Figure S1) is available.
Provoked fails
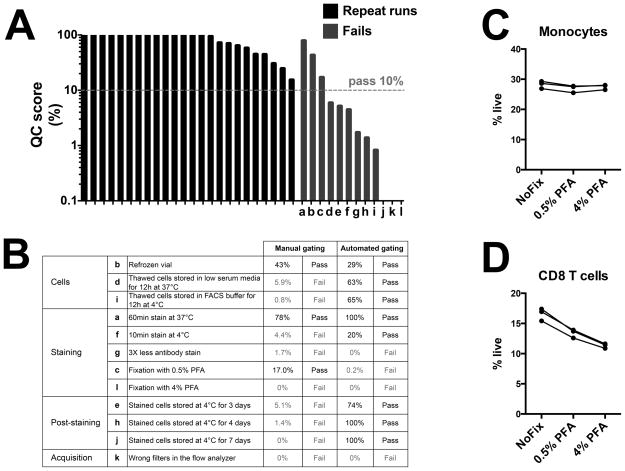
Each ‘provoked fail’ was performed as indicated in the flow cytometry acquisition section with the following specific changes: a) Staining for 60min at 37°C; b) Use of cryopreserved PBMC already thawed once and re-cryopreserved; c) Fixation of stained samples with 0.5% PFA; d) Thawed PBMC were stored in medium supplemented with 0.5% human AB serum for 12h at 37°C before staining; e) Stained cells were stored at 4°C for 3 days before acquisition; f) Staining for 10min at 4°C; g) Staining with 3-fold less antibody volumes; h) Stained cells were stored at 4°C for 4 days before acquisition i) Thawed PBMC were stored in FACS buffer for 12h at 4°C before staining; j) Stained cells were stored at 4°C for 7 days before acquisition; k) Acquisition on a BD LSR-II cell analyzer with 585/42 and 605/20 bandpasses on the violet laser instead of 525/50 and 660/20 bandpasses for the detection of eF506 and BV650 fluorescence emission, respectively; l) Fixation of stained samples with 4% PFA. Summary for each provoked fail is available on Figure 2B.
Figure 2. Development of a quality control score to evaluate and correct for technical variation between experiment runs.
Cell frequencies were determined with the 10-color flow cytometry panel and the centralized manual gating method on 24 repeat runs or 10 provoked fail runs of cryopreserved PBMC derived from a single blood donation of a control donor. Quality control (QC) score was calculated based on the deviation from average for each of the normally distributed lymphocyte populations and 10% defined as the pass/fail threshold. (A) Quality control score obtained with normal repeat runs (black) or provoked fail runs (blue). (B) Detailed protocol and associated QC score for each provoked fail run. Effect of fixation with paraformaldehyde (PFA) on the frequency of (C) monocytes and (D) CD8 T cells, from three independent repeat runs of the control donation.
Quality control score
Normality of cell populations was determined using the d’Agostino & Pearson omnibus normality test. The technical coefficient of variation associated with each cell population was calculated using the 24 repeat runs of the control donation. For each cell population that followed a normal distribution and a technical coefficient of variation lower than 35% (13 populations in total), a p value was calculated using a Z-score, corrected for multiple testing by the Bonferroni method and expressed as a percent. For each repeat run, the QC score was assigned to the lowest p-value from all 13 cell populations (thus reflecting the cell population that deviate the most from its expected coefficient of variation).
Statistics
The coefficient of variation (CV) of each cell population was defined as the ratio of its standard deviation and its arithmetic mean, expressed as a percent. The standard deviation of the CV was calculated by a bootstrapping procedure with 10,000 replications using the package boot for R, version 3.2.3. All statistical tests, correlations and linear regressions were performed using GraphPad Prism, version 6.0. The CV was compared between cell populations using an ordinary one-way ANOVA test. Inter-individual and technical variability for each cell population were assessed using multiple t-tests. The gender effect on cell population frequencies was determined using either a multiple non-parametric Kruskal-Wallis test or an equivalence test. The equivalence test was based on 90% confidence intervals calculated with a Mann-Whitney non parametric test between male and female and defining the zone of statistical indifference as 4-fold of the technical CV. The age effect on cell population frequencies was defined using a Pearson correlation. Multivariate linear regression analysis between age and latent tuberculosis signature was performed using the lm function in R. All p values were adjusted for multiple testing with the Bonferroni method.
Results
Development of a 10-colour flow cytometry panel to identify circulating lymphocyte populations in humans
Based on previously defined panels from the first round of the HIPC program (9) and our own research goals, we selected 6 lineage markers, 3 differentiation markers and 1 viability marker to combine into one 10-color flow cytometry panel (Table SI). Using fluorescence minus one controls, we ensured that the combination of antibodies and fluorochromes selected had no detrimental effect on the individual staining for each antigen (data not shown). Using the gating strategy illustrated in Figure S1, we used this panel to identify the 17 lymphocyte and monocyte populations listed in Table I comprising 6 major cell subsets, 5 CD4 T cell subsets, 4 CD8 T cell subsets, and 2 CD56 T cell subsets. Gates 1 to 4 were used as common baseline filters to identify all 17 populations (Figure S1). Throughout this manuscript, we are using the population names listed in Table I as shorthand rather than the more exact, but less readable, cell population definitions based on marker patterns. Based on the resolution sensitivity and the level of expression of each marker, cell populations were segregated into ‘clearly defined’ cell populations (clear distinction between marker pattern positive and negative populations) and ‘poorly resolved’ cell populations (dim expression and/or poor segregation between positive and negative populations) (Table I).
Table I.
Lymphocyte populations derived from the 10-color flow cytometry panel.
| Category | Gate ID | Population Name | Gating strategy | Resolution sensitivity |
|---|---|---|---|---|
| Major cell subsets | 5 | Monocytes | CD14+ | Clearly defined |
| 7 | B cells | CD14−/CD19+CD3− | Clearly defined | |
| 8 | NK cells | CD14−/CD56+CD3− | Clearly defined | |
| 11 | T cells | CD14−/CD56−CD3+ | Clearly defined | |
| 12 | CD4 T cells | CD14−/CD56−CD3+/CD4+CD8− | Clearly defined | |
| 13 | CD8 T cells | CD14−/CD56−CD3+/CD4−CD8+ | Clearly defined | |
| CD4 T cell subsets | 14 | Tregs | CD14−/CD56−CD3+/CD4+CD8−/CD25+ | Poorly resolved |
| 15 | Naïve CD4 T cells | CD14−/CD56−CD3+/CD4+CD8−/CD45RA+CCR7+ | Clearly defined | |
| 16 | Tcm CD4 T cells | CD14−/CD56−CD3+/CD4+CD8−/CD45RA−CCR7+ | Poorly resolved | |
| 17 | Tem CD4 T cells | CD14−/CD56−CD3+/CD4+CD8−/CD45RA−CCR7− | Poorly resolved | |
| 18 | Temra CD4 T cells | CD14−/CD56−CD3+/CD4+CD8−/CD45RA+CCR7− | Poorly resolved | |
| CD8 T cell subsets | 19 | Naïve CD8 T cells | CD14−/CD56−CD3+/CD4−CD8+/CD45RA+CCR7+ | Clearly defined |
| 20 | Tcm CD8 T cells | CD14−/CD56−CD3+/CD4−CD8+/CD45RA−CCR7+ | Poorly resolved | |
| 21 | Tem CD8 T cells | CD14−/CD56−CD3+/CD4−CD8+/CD45RA−CCR7− | Poorly resolved | |
| 22 | Temra CD8 T cells | CD14−/CD56−CD3+/CD4−CD8+/CD45RA+CCR7− | Poorly resolved | |
| CD56 T cell subsets | 9 | CD3+ CD56 T cells | CD14−/CD56+CD3+ | Poorly resolved |
| 10 | CD3hi CD56 T cells | CD14−/CD56+CD3hi | Poorly resolved |
Poorly resolved cell populations are associated with a higher technical variability compared to clearly defined cell populations
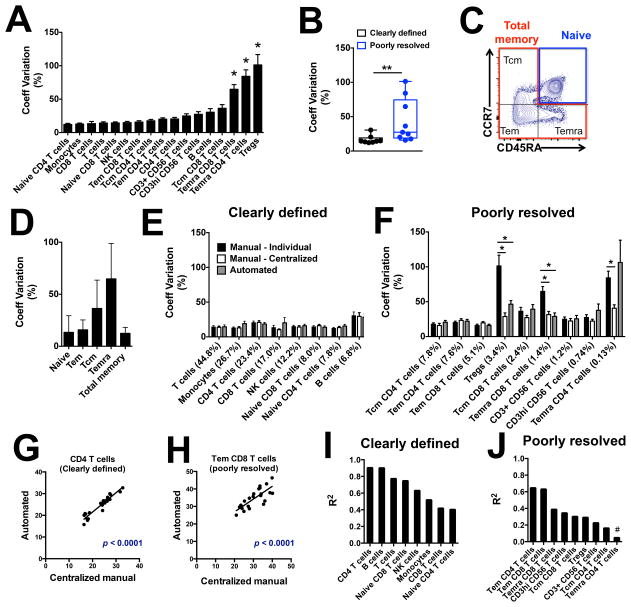
In order to estimate the technical variability associated with each cell population, we utilized multiple vials of cryopreserved PBMC obtained from a single blood donation and repeatedly analyzed them with the 10-color flow cytometry panel. A total of 24 repeat runs were performed on samples from this ‘control donation’ on various days throughout a 7-month period by 3 different operators, and acquired on 4 different cell analyzers. Technical variability differed greatly between cell populations, ranging from CV = 12 to 101% (Figure 1A). Three populations were identified as highly variable (significant higher coefficient of variation compared to the other cell populations, Figure 1A): regulatory T cells (Tregs), effector memory cells re-expressing CD45RA (Temra) CD4 T cells and Temra CD8 T cells. All three populations were previously defined as poorly resolved cell populations (Table I). When separating cell populations according to their resolution sensitivity, poorly resolved cell populations were associated with a significant higher technical variation compared to clearly defined cell populations (Figure 1B). To assess the extent to which the cell type resolution explained the increased technical variability, we examined gating of memory CD8 T cells, which can be considered either as three distinct poorly resolved cell populations (central memory (Tcm), effector memory (Tem) and Temra), or as one clearly defined cell population (Figure 1C). Technical variability in total memory CD8 T cells was lower compared to Tcm, Tem and Temra CD8 T cells, and of similar level to the clearly defined ‘naïve CD8 T cells’. (Figure 1D). Thus, the higher technical variability for memory CD8 T cell subpopulations is a direct consequence of them being poorly resolved.
Figure 1. Assessment of technical variability within lymphocyte populations using a control donation.
Cell frequencies were determined with the 10-color flow cytometry panel on 24 repeat runs of cryopreserved PBMC derived from a single blood donation of a control donor. (A) Technical variability of each lymphocyte population obtained with the individual manual gating method. (B) Technical variability of clearly defined (black) and poorly resolved (blue) cell populations obtained with the individual manual gating method. (C) Representative staining and (D) technical variability of CD8 T cell subset frequencies obtained with the individual manual gating method. Comparison between technical variability of (E) clearly defined or (F) poorly resolved cell populations frequency obtained with individual manual gating, centralized manual gating or automated gating. Linear regression of cell frequencies obtained with centralized manual gating or automated gating for (G) CD4 T cells, (H) Tem CD8 T cells, (I) clearly defined or (J) poorly resolved lymphocyte populations. Technical variability between individual cell populations was compared with ordinary one-way ANOVA test; technical variability between clearly resolved and poorly defined cell populations and between each gating method was compared with the non-parametric Mann-Whitney test. * p < 0.05, ** p < 0.01.
Gating approaches influence technical variability of poorly resolved cell populations, but other factors dominate beyond that
It is intuitive that poorly resolved populations may be associated with higher technical variability when gated by different operators as was done in Figure 1A due to the subjectiveness when visually setting a gate for such populations. We were interested in assessing the impact of the gating method on the technical variability associated with our flow cytometry panel. To do so, we compared the cell frequencies obtained from the same set of repeat runs of the control donation using three different gating methods, ranging from the most to the least subjective approach: i) individual manual gating (gating done by different operators that performed the flow cytometry acquisition as was done for Figure 1A), ii) centralized manual gating (gating done by one unique operator) and iii) automated gating using directed unsupervised clustering using FLOCK as described in Materials and Methods. Technical variability of clearly defined cell populations was similar between the different gating methods (Figure 1E). However, technical variability of poorly resolved cell populations was significantly reduced with centralized manual gating (Tregs, Temra CD4 T cells, and Temra CD8 T cells) or automated gating (Tregs and Temra CD8 T cells) compared to individual manual gating (Figure 1F). Thus, the gating approach significantly impacts technical variability associated with poorly resolved cell populations, and centralized manual or automated gating can significantly reduce this technical variability.
Next, we wanted to determine to which extent gating approaches explained the remaining variability of cell population frequencies derived from aliquots of the same biological sample, versus other technical factors, such as aliquot sample quality, procedures, reagents or acquisition parameters (9). We took advantage of having population frequencies generated by two independently performed gating approaches on these samples - centralized manual gating and automated gating. When correlating the frequencies derived by the two gating approaches from the same FCS files, we found high correlations for both clearly defined (Figure 1G) and poorly resolved (Figure 1H) populations. This was broadly true for all clearly defined cell populations (r2 = 0.4 – 0.9, Figure 1I) and most poorly resolved populations (r2 = 0.2 – 0.6, Figure 1J) except for Temra CD4 T cells. Taken together, these results suggest that a significant proportion of the technical variability in flow cytometry is independent of the gating method used – especially for clearly resolved populations. Rather, this variability might be due to differences in samples (i.e. batch to batch variation in cryovials of the control donation), in reagents (i.e. batch to batch variation in antibody lots), or in cell analyzer daily performances.
While it is impossible to account and correct for each factor driving technical variability, overall we found that less subjective centralized manual gating can significantly reduce technical variability. Thus, in the following sections, the technical coefficient of variation based on centralized manual gating was chosen as a technical baseline for each cell population against which the effect of biological variables can be assessed.
Development of a quality control (QC) score to control for technical variation beyond gating
In our HIPC studies, flow cytometry based cell sorting is typically the first step in resource intensive experiments such as RNA-Seq or ChIP-Seq. Cell sorts are performed in batches on multiple days to process sufficient numbers of donors. To ensure that flow cytometry was performed equivalently for different batches, we set out to establish a quality score that assesses technical variability between runs using the control donation samples, and provides a pass or fail output. We focused the QC score based on the identification of cell populations that a) show a normal distribution (p > 0.05 with the D’Agostino and Pearson omnibus normality test) and b) have low technical variability (coefficient of variation < 35%) between runs. Thirteen out of the 17 cell populations met these thresholds when using centralized manual gating (data not shown). Based on the cell frequencies of these 13 cell populations, we calculated a quality control score for each of the repeat runs of the control donation (Figure 2A). We then provoked fail runs, by modifying either the sample preparation or data acquisition parameters. Comparing the quality control scores between the regular repeat runs, and the provoked fails, we found that a threshold pass/fail at 10% identified 9 out of the 12 provoked fail runs as problematic, while all regular runs had scores above this threshold (Figure 2A, 2B). The three provoked fail runs that were associated with a ‘pass’ QC score were the refrozen vial, the prolonged staining period at 37°C, and the fixation with 0.5% PFA (Figure 2A, 2B). Fixation affected cell population frequencies in a PFA dose-dependent manner: the QC score of the control donation dropped to 17% and 0%, when samples were fixed with 0.5% or 4% PFA, respectively (Figure 2A, 2B). Indeed, whereas most cell population frequencies remained unchanged (e.g. Monocytes, Figure 2C), the staining of some cell populations was significantly affected by the fixation process (e.g. CD8 T cells, Figure 2D). However, repeat runs of fixed or unfixed samples of the control donation generated similar technical variation for each cell population (data not shown).
Interestingly, when using the automated gating method, the provoked fail runs obtained a higher quality control score, often greater than the threshold pass/fail (Figure 2B). It appears therefore that whereas a human eye can successfully detect and flag missing or fluorescence shifted cell populations, the DAFi algorithm for the automated gating which aims to be robust to data shifts across samples, is still able to rescue those populations.
In all experiments reported in the following sections, one sample from the control donation was included to ensure technical reproducibility between batches of samples. Runs in which the samples had a QC score higher than 10%, were considered acceptable and those with scores below were considered failing our quality control, and were thus excluded.
Biological inter-individual variability of cell populations
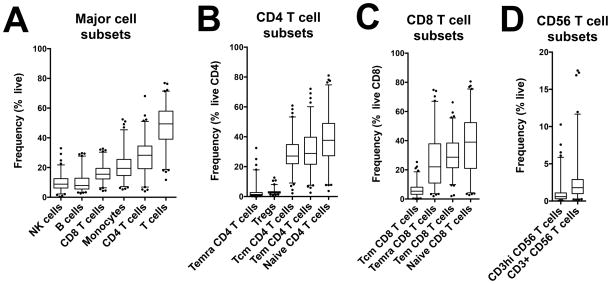
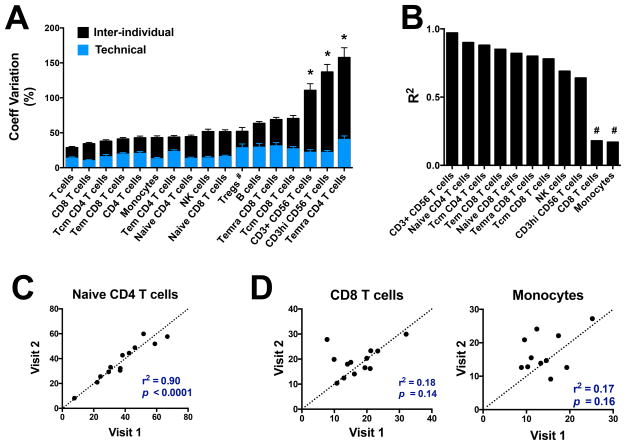
Following our investigation of technical variation, we assessed inter-individual variability of cell populations by analyzing cryopreserved PBMC samples collected from various human cohorts with our 10-color flow cytometry panel (total of 176 subjects from San Diego (124), Peru (8) and Sri Lanka (44)). For each lymphocyte population, we defined a normal range based on the average and 2.5th-97.5th percentiles of cell frequencies obtained with the manual centralized gating method (Figure 3 and Table SII). Average frequencies varied greatly amongst all major cell populations as well as within T cell subsets (Figure 3 and Table SII). The coefficient of variation associated with each cell population also varied greatly, ranging from 28.7% to 157% (inter-individual variability, Figure 4A). Temra CD4 T cells, CD3hi CD56 T cells and CD3+ CD56 T cells had the highest inter-individual variability, significantly higher compared to the other populations. Importantly, for all cell populations except Tregs, the biological inter-individual variability was significantly higher compared to the technical variability previously calculated (Figure 4A).
Figure 3. Normal range of lymphocyte populations frequencies within humans.
Cell frequencies were determined with the 10-color flow cytometry panel and centralized manual gating on cryopreserved PBMC samples obtained from 176 human subjects. Box plots represent average and normal range (2.5th–97.5th percentiles) of (A) Major cell subsets, (B) CD4 T cell subsets, (C) CD8 T cell subsets, and (D) CD56 T cell subsets.
Figure 4. Inter-individual and intra-individual variability of lymphocyte populations frequencies within humans.
Cell frequencies were determined with the 10-color flow cytometry panel and centralized manual gating on cryopreserved PBMC samples obtained from 176 human subjects. (A) Comparison between technical variability calculated in Figure 2 (blue) and inter-individual variability (black). Linear regression of (B) lymphocyte subset frequencies, (C) naïve CD4 T cell frequencies, (D) CD8 T cell frequencies and monocyte frequencies obtained between two longitudinal samples for 13 healthy human subjects. Visit 2 ranged from 114 to 194 days following visit 1. Inter-individual variability between cell populations was compared with ordinary one-way ANOVA test. Inter-individual and technical variability for each cell population were compared with multiple comparison t test. * p < 0.05, # p >0.05.
Biological intra-individual variability of cell populations
In order to address whether cell population frequencies derived from our flow cytometry panel were stable over time in a given individual, we applied the 10-color flow cytometry panel on 13 subjects from San Diego for which a second blood sample was collected 3 to 7 months later (samples named visit 1 and visit 2, respectively). For 6 out of the 17 cell populations, the inter-individual variability within the longitudinal cohort at visit 1 was too low to be discriminated from the technical variability (fold change difference between inter-individual and technical variability < 2); these populations were therefore excluded from the subsequent analysis. For the remaining 11 populations, there was a very high correlation between the frequencies obtained at visit 1 and visit 2 for most cell populations, including both clearly defined and poorly resolved cell populations (Figure 4B and 4C). CD8 T cells and monocytes were the two populations for which no significant correlation was observed between the two visits (Figure 4B and 4D). Both CD8 T cells and monocytes are clearly defined cell populations by flow cytometry with two of the three lowest levels of technical variability in the control samples (Figure 1A). Thus, their lack of stability over time likely reflects immunological perturbations in some of the longitudinal samples between donors. In contrast, the other 9 of 11 cell populations that showed differential frequencies across donors largely maintained those differences, indicating that they are relatively stable markers of blood immune cell composition within each donor.
Age but not gender is an important correlate of inter-individual variability of cell populations in our panel
Having observed that for most cell types, inter-individual variability in their abundance in the peripheral blood was much greater than either technical variability or longitudinal variability across our human cohorts (Figure 4A), we were interested to see whether some of this biological variation could be accounted for by demographic variables such as gender and age. Amongst the 132 samples from which we had demographics data available (San Diego (124) and Peru (8)), there were no significant differences in cell population frequencies between male and female using the multiple comparison non-parametric Kruskal-Wallis test (data not shown). More stringently, the frequencies in male vs. females of all the assessed cell populations also passed a test for equivalence, meaning that there is statistical support to affirm that differences observed are contained within a range expected based on technical variability (p<0.05, see methods).
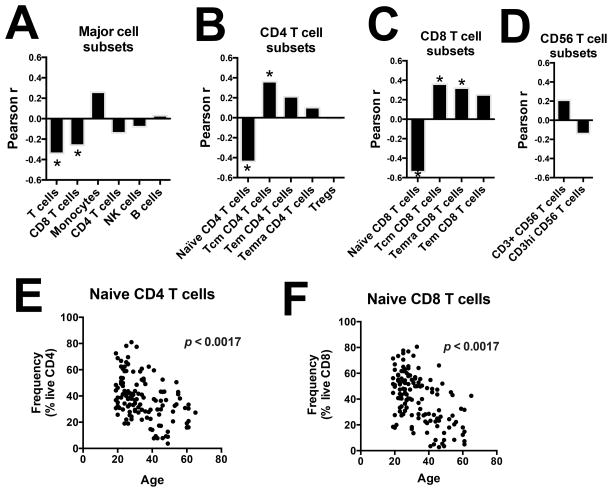
In contrast, there was a striking significant inverse correlation between age and the frequency of T cells and CD8 T cells (Figure 5A). Within T cell subsets, age was inversely correlated with the frequency of naïve CD4 T cells and naïve CD8 T cells, and positively correlated with the frequency of Tcm CD4 T cells, Tcm CD8 T cells and Temra CD8 T cells (Figure 5B and 5C). No association between age and CD56 T cell subsets frequencies was observed (Figure 5D). Scatter plots for the two strongest age-frequency correlations are shown in Figure 5E for naïve CD4 T cells and in Figure 5F for naïve CD8 T cells.
Figure 5. Demographic correlates of biological variability of lymphocyte populations frequencies.
Cell frequencies were determined with the 10-color flow cytometry panel and centralized manual gating on cryopreserved PBMC samples obtained from 132 human subjects. Pearson correlation coefficient between age and the frequency of (A) major cell subsets, (B) CD4 T cell subsets, (C) CD8 T cell subsets and (D) CD56 T cell subsets, (E) naïve CD4 T cells and (F) naïve CD8 T cells. P values were corrected for multiple testing. * p < 0.05.
Even after correcting for age and gender, a significant proportion of the inter-individual variability still remained above technical variability, indicating that other confounding factors are influencing immune cell population frequencies. Across our cohort of samples from San Diego, ethnicity was highly diverse (Figure S2A), thus we were interested to see whether this factor could explain a proportion of the inter-individual variability. While our study was not powered enough to fully dissect the effect of ethnic background on each cell population abundance, we found significant differences between ethnicity groups for the frequency of major cell types such as T cells and B cells (Figure S2B) as well as CD4 and CD8 T cell subsets (Figure S2C, S2D) but not CD56 T cell subsets (Figure S2E). Taken together, these results suggest that ethnicity is an important co-variate of blood immune cell composition and should therefore be taken into consideration whenever possible in human immunophenotyping studies, by ensuring that clinical groups being compared have a balanced ethnic composition.
Successful detection of a latent tuberculosis signature in PBMC
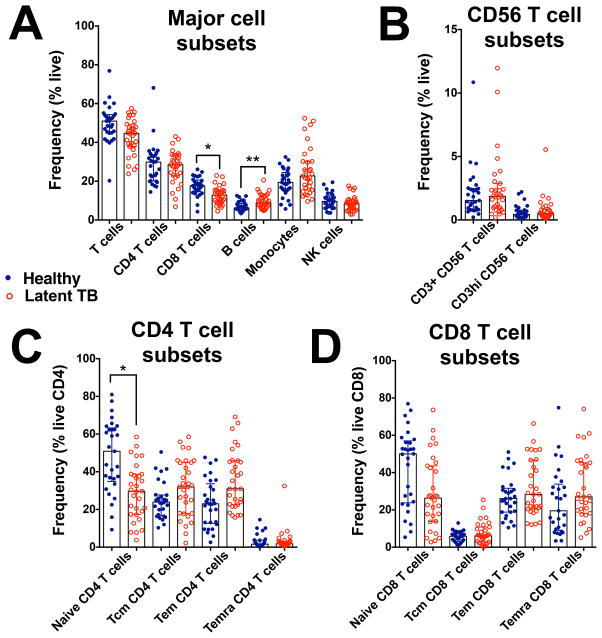
Finally, we applied our workflow to a cohort of donors with latent tuberculosis infection (LTBI) to check if we could detect a disease signature, even after correction for technical and biological variability. LTBI status is typically defined by positivity to IFN-γ release assays (IGRA) combined with absence of clinical symptoms of active TB (18). While IGRAs are associated with a relative good sensitivity (>75%), they require an in vitro stimulation step that increases the result variability and complicates logistics (19). An ex vivo immune signature specific of LTBI would facilitate diagnosis, but there is no evidence that LTBI and non-TB infected donors can be differentiated solely based on their ex vivo PBMC immune profile. We focused our analysis on the comparison of cell frequencies in PBMC obtained from 29 healthy controls (San Diego) and 31 individuals with LTBI (IGRA+) from two different geographic locations (San Diego (n=23) and Peru (n=8)). First, we confirmed that biological differences could be detected in our study by ensuring that the biological variability within our combined cohorts was above technical variability. Tregs was the only population for which inter-individual variability was not significantly higher than technical variability (p > 0.05 with a multiple comparison t-test), and was thus excluded from the subsequent analysis. Next, looking at demographics, we found that the LTBI cohort was significantly older than healthy controls (median age (range) of 31(19–60) and 43(20–65) for healthy controls and LTBI donors, respectively; p < 0.05 with the non-parametric Man-Whitney test), indicating the need to take in consideration this co-factor in our statistical analysis. After age correction, we found that individuals with latent tuberculosis infection were associated with a lower frequency of CD8 T cells, and a higher frequency of B cells compared to healthy controls (Figure 6A). No significant differences in CD56 T cell subsets and CD8 T cells frequencies were observed between the two cohorts (Figure 6B and 6D). However, within CD4 T cell subsets, individuals with latent tuberculosis infection had a lower frequency of naïve CD4 T cells compared to healthy controls (Figure 6C).
Figure 6. Identification of a latent tuberculosis signature within PBMCs.
Cell frequencies were determined with the 10-color flow cytometry panel and centralized manual gating on cryopreserved PBMC samples obtained from 31 subjects with latent tuberculosis and 29 tuberculosis negative subjects. Differences in the frequency of (A) major lymphocyte subsets, (B) CD56 T cell subsets, (C) CD4 T cell subsets and (D) CD8 T cell subsets between latent TB donors and healthy subjects, determined by multilinear regression analysis with age as a co-variate and multiple testing correction. * p<0.05, ** p < 0.01.
Discussion
Standardization of flow cytometry data acquisition and analysis is critical to ensure quality and reproducibility of large-scale human immunology studies. Previous studies have aimed at reducing technical variability by optimizing and standardizing reagents, protocols and data analysis for pre-designed panels (7, 9–13, 20) but none established a universal workflow that could be applied to any flow cytometry experiment. Furthermore, previous studies did not simultaneously assess the effect of technical variability on cell populations along with biological variability due to demographic co-factors. Here, we report on the development of a re-usable workflow to control for both technical and biological variation in flow cytometry. We applied this workflow to evaluate a customized panel measuring the frequency of 17 immune cell populations within cryopreserved human PBMC of major interest to our studies. We quantified the technical variation associated with each cell population by repeatedly applying this panel to PBMC samples obtained from the same control donation and comparing different analysis strategies and developing a quality control metric. In addition, we also studied the biological variability associated with our cell populations in different individuals and in a given individual over time, as well as the influence of demographics. Finally, we illustrated the usefulness of our approach by identifying a disease signature in volunteers infected with latent tuberculosis.
Technical variation differed greatly between cell populations identified in our panel. As expected, poorly resolved cell populations were associated with higher technical variability compared to clearly resolved cell populations. This is explained by the expression profile of our markers used for phenotyping that is either continuous (e.g. Tcm and Tem T cells) or dimly expressed (e.g. Tregs) in poorly resolved cell populations. Technical variability associated with those cell populations can be improved by using less subjective gating strategies (see paragraph below) or by choosing alternative markers to define the cell population (e.g. the inclusion of markers such as FoxP3 to more clearly separate the Treg population).
Our standard flow cytometry protocol for HIPC does not include cell fixation, as we are usually sorting cell populations for downstream transcriptomic analyses. However, some clinical immunophenotyping studies might require sample fixation, for instance if no acquisition is possible right away, or if the samples are considered infectious. Here we show that fixation significantly affect the staining of some cell populations and thus their frequencies, and that this effect is dependent on the concentration of PFA used. This observation reflects variable degree of sensitivity of fluorochromes and antibody clones to paraformaldehyde. Thus, the choice of antibodies and dilutions need to be carefully optimized for each given panel and staining protocol. For instance, the normal range reported here should only be used as a guideline when samples have been stained under similar conditions. Nevertheless, fixed repeat runs of the control donation generated similar technical variation compared to unfixed repeat runs, validating that our methodological framework can also be applied to flow cytometric experiments requiring sample fixation.
Our study compared three gating approaches: individual manual gating, centralized manual gating and automated gating. We found that for cell populations that are clearly resolved, all three approaches had a high degree of concordance and all can equally be applied. In contrast, individual manual gating proved to be problematic when applied to cell populations that are poorly resolved. Centralized manual gating and automated gating methods both significantly reduced technical variability of poorly resolved cell populations, which is in concordance with previous reports (7, 10). Comparing the technical variability achieved with centralized manual and automated gating, the two performed comparably except when applied to the poorly resolved and comparatively low frequency Temra CD4 T cell population. The automated gating pipeline, which requires the detected cell populations to either follow a predefined distribution model or form a data cluster with relatively abundant number of events, could not reliably detect this cell population. This highlights the need to verify robustness of automated gating pipelines to detect cell populations of interest before blindly accepting their output.
Relative abundance and inter-individual variability of each cell population differed widely between cell subsets. Our data allowed us to define an expected ‘normal range’ in human peripheral blood for the 17 immune cell populations derived from our panel. This normal range can be of use for any researcher interested in those same cell populations, in order to flag any technical issues associated with a given flow cytometry panel or acquisition workflow (e.g. use of wrong antibody clone or poor instrument laser efficiency) as well as to identify biological outliers within a large set of samples.
Remarkably, despite being associated with significant biological inter-individual variability, we found that within each individual, most lymphocyte populations have surprisingly stable frequencies over time. This confirms previous findings (4, 15), and suggests that the relative abundance of most lymphocyte subsets in an individual over a period of at least several months is an intrinsic feature of the individual, presumably regulated by invariant factors such as genetic background and infrequent immunological events with major impact that shape the individuals immune history. The observation that most lymphocyte populations display such low intra-individual variability, including poorly resolved and low frequency cell populations such as CD3+ CD56 T cells, reinforces the conclusion that important biological differences between individuals can reliably be detected using our flow cytometry panel.
The highest intra-individual variability of cell subsets over time was observed for monocytes and CD8 T cells. Monocytes might be more susceptible to minor immunological perturbations as they are known to be the first line of defense upon antigen exposure, and have a strong ability to migrate from the blood to a site of infection (21). Thus, it is likely that their relative abundance in the peripheral blood fluctuates over time depending on the recent antigenic load being experienced by the individual in a given point of time. For CD8 T cells, the low correlation observed between the two visits was driven by two subjects for which CD8 T cell frequencies dramatically increased at visit 2. It is possible that these two subjects contracted a minor infectious disease or inflammatory process between the two visits that perturbed the CD8 frequency in these individuals specifically. Larger panels of donors with longitudinal samples would be necessary to follow up these findings.
For each immune cell population identified with our panel, the comparison of the technical variability with its biological variability provides an estimate on how many samples will be required to detect significant differences between groups of samples with an effect size similar to the normal biological variation. We found marked differences amongst cell populations, with some having a high ratio of biological over technical variability (e.g. CD56 T cell subsets) vs. others for which a much greater number of samples will be necessary to be sufficiently powered to overcome technical variability (e.g. Tregs, for which our data would alternatively recommend utilizing a different staining panel). Additionally, depending on the study design, different sources of variability have to be taken into account. Because the intra-individual variability is relatively low for most cell populations, in a study comparing paired samples from the same donor cohort pre- and post perturbation, the technical variability of cell population detection becomes the dominant source of noise. In contrast, when samples from different groups of donors are compared, the much larger biological variability between donors has to be considered – which can easily lead to erroneous results if the donor cohort sizes are not sufficiently large.
Genetic heritability account for less than 50% of the immune system variation amongst humans, suggesting that other non-genetic factors (such as age and exposures) might be critical in explaining the remaining variability (22, 23). Interestingly, we found that while gender had no impact on our immune cell population frequencies, age was strongly associated with changes in the cellular immune composition of human peripheral blood. In particular, older individuals were characterized by a striking loss of naïve CD4 and CD8 T cells, which is in concordance with previous reports looking at circulating T cell subsets in young and elderly populations (24, 25). These results also corroborate a recent large-scale study that highlighted the importance of non-genetic factors such as age but not gender on the cellular composition of the circulating human immune system (15).
Additionally, we were able to examine the influence of ethnic background on lymphocyte subsets frequencies. Our results strongly suggest that ethnicity can be an important factor driving variation in the cellular immune composition of human peripheral blood. This proposal is also supported by previous studies reporting both qualitative and quantitative differences in the immune response to infection or vaccination in human cohorts of different ethnicities (26–28). Although the exact cause underlying those observations remain uncertain, the diversity in genetic background, microbiome communities, environmental exposures, and social differences are likely to be driving human immune variation between ethnicities. Another factor often related to ethnicity and important to take in consideration is the geographical location of the human subjects. Indeed, distinct geographic locations might indicate exposure to different microorganisms, resulting in different shaping of the immune system. Hence two individuals with the same ethnicity but living in different locations might have completely separate immune profiles.
Taken together, our results and those from previous reports highlight the critical importance in clinical immunophenotyping studies to take into consideration non-genetic features that are known or suspected to drive immune variability, by recruiting matching disease and healthy cohorts, or correcting for those differences between groups when performing data analysis.
Finally, our flow cytometry panel allowed us to identify a specific signature of latent tuberculosis infection. Previous T cell immunophenotyping studies have successfully discriminated active TB from latent TB or from non-TB infected individuals (29–31). However, the distinction between latent TB and non-TB infected subjects based solely on their PBMC profile has not been reported yet. Here we show that after controlling for technical variation and age, individuals with latent tuberculosis had a lower frequency of CD8 T cells, naïve CD4 T cells and a higher frequency of B cells, compared to TB negative subjects. We thus provide primary evidence that ex vivo immune signatures in PBMC can discriminate between LTBI and non-TB infected donors, which could potentially be applied as an additional tool to IGRAs for improving the specificity of LTBI diagnosis. Recent studies have also shown differences in whole blood cell counts of infants and adults with various TB status (32, 33). Specifically, a high monocyte/lymphocyte ratio was associated with an increased risk of developing active TB (32, 33). Thus, we predict that combining whole blood counts to our newly defined ex vivo PBMC signature of LTBI will improve its discriminatory power.
Overall, our study indicates that controlling for both technical and biological variability is crucial to appropriately design and power human immunology studies. Our findings can be directly applied to any flow cytometry study examining cell populations identified with our panel. In addition, we provide with a universal workflow for how to develop and evaluate pre-existing or new flow cytometry panels for large-scale immunology studies.
Supplementary Material
Acknowledgments
Research reported in this manuscript was supported by NIAID of the National Institutes of Health under award numbers U19AI118626 and R24AI108564. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank the National Blood Center, Ministry of Health, Colombo, Sri Lanka for providing buffy coat samples used in this study.
Footnotes
Author contributions
JGB, PV, RHS, AS and BP designed the experiments. JGB, YQ, CLA, DW and JZG conducted the experiments and/or analyzed the data. RT, RHG, MS, ADS provided samples. JGP and BP wrote the manuscript and all authors edited the manuscript.
References
- 1.Baumgarth N, Roederer M. A practical approach to multicolor flow cytometry for immunophenotyping. J Immunol Methods. 2000;243:77–97. doi: 10.1016/s0022-1759(00)00229-5. [DOI] [PubMed] [Google Scholar]
- 2.Chattopadhyay PK, Hogerkorp CM, Roederer M. A chromatic explosion: the development and future of multiparameter flow cytometry. Immunology. 2008;125:441–449. doi: 10.1111/j.1365-2567.2008.02989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, Germain RN, Wang E, Olnes MJ, Narayanan M, Golding H, et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157:499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darrah PA, Patel DT, De Luca PM, Lindsay RW, Davey DF, Flynn BJ, Hoff ST, Andersen P, Reed SG, Morris SL, et al. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med. 2007;13:843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- 6.Burel JG, Apte SH, Doolan DL. Systems Approaches towards Molecular Profiling of Human Immunity. Trends Immunol. 2016;37:53–67. doi: 10.1016/j.it.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Maecker HT, Rinfret A, D’Souza P, Darden J, Roig E, Landry C, Hayes P, Birungi J, Anzala O, Garcia M, et al. Standardization of cytokine flow cytometry assays. BMC Immunol. 2005;6:13. doi: 10.1186/1471-2172-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saeys Y, Gassen SV, Lambrecht BN. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat Rev Immunol. 2016;16:449–462. doi: 10.1038/nri.2016.56. [DOI] [PubMed] [Google Scholar]
- 9.Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol. 2012;12:191–200. doi: 10.1038/nri3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finak G, Langweiler M, Jaimes M, Malek M, Taghiyar J, Korin Y, Raddassi K, Devine L, Obermoser G, Pekalski ML, et al. Standardizing Flow Cytometry Immunophenotyping Analysis from the Human ImmunoPhenotyping Consortium. Sci Rep. 2016;6:20686. doi: 10.1038/srep20686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalina T, Flores-Montero J, van der Velden VH, Martin-Ayuso M, Bottcher S, Ritgen M, Almeida J, Lhermitte L, Asnafi V, Mendonca A, et al. EuroFlow standardization of flow cytometer instrument settings and immunophenotyping protocols. Leukemia. 2012;26:1986–2010. doi: 10.1038/leu.2012.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Streitz M, Miloud T, Kapinsky M, Reed MR, Magari R, Geissler EK, Hutchinson JA, Vogt K, Schlickeiser S, Kverneland AH, et al. Standardization of whole blood immune phenotype monitoring for clinical trials: panels and methods from the ONE study. Transplant Res. 2013;2:17. doi: 10.1186/2047-1440-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perfetto SP, Ambrozak D, Nguyen R, Chattopadhyay PK, Roederer M. Quality assurance for polychromatic flow cytometry using a suite of calibration beads. Nat Protoc. 2012;7:2067–2079. doi: 10.1038/nprot.2012.126. [DOI] [PubMed] [Google Scholar]
- 14.Aghaeepour N, Finak G, Flow CAPC, Hoos H, Mosmann TR, Brinkman R, Gottardo R, Scheuermann RH Consortium D. Critical assessment of automated flow cytometry data analysis techniques. Nat Methods. 2013;10:228–238. doi: 10.1038/nmeth.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carr EJ, Dooley J, Garcia-Perez JE, Lagou V, Lee JC, Wouters C, Meyts I, Goris A, Boeckxstaens G, Linterman MA, et al. The cellular composition of the human immune system is shaped by age and cohabitation. Nat Immunol. 2016;17:461–468. doi: 10.1038/ni.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poland GA, Quill H, Togias A. Understanding the human immune system in the 21st century: the Human Immunology Project Consortium. Vaccine. 2013;31:2911–2912. doi: 10.1016/j.vaccine.2013.04.043. [DOI] [PubMed] [Google Scholar]
- 17.Qian Y, Wei C, Eun-Hyung Lee F, Campbell J, Halliley J, Lee JA, Cai J, Kong YM, Sadat E, Thomson E, et al. Elucidation of seventeen human peripheral blood B-cell subsets and quantification of the tetanus response using a density-based method for the automated identification of cell populations in multidimensional flow cytometry data. Cytometry B Clin Cytom. 2010;78(Suppl 1):S69–82. doi: 10.1002/cyto.b.20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pai M, Behr MA, Dowdy D, Dheda K, Divangahi M, Boehme CC, Ginsberg A, Swaminathan S, Spigelman M, Getahun H, et al. Tuberculosis. Nat Rev Dis Primers. 2016;2:16076. doi: 10.1038/nrdp.2016.76. [DOI] [PubMed] [Google Scholar]
- 19.Petruccioli E, Scriba TJ, Petrone L, Hatherill M, Cirillo DM, Joosten SA, Ottenhoff TH, Denkinger CM, Goletti D. Correlates of tuberculosis risk: predictive biomarkers for progression to active tuberculosis. Eur Respir J. 2016 doi: 10.1183/13993003.01012-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalina T, Flores-Montero J, Lecrevisse Q, Pedreira CE, van der Velden VH, Novakova M, Mejstrikova E, Hrusak O, Bottcher S, Karsch D, et al. Quality assessment program for EuroFlow protocols: summary results of four-year (2010–2013) quality assurance rounds. Cytometry A. 2015;87:145–156. doi: 10.1002/cyto.a.22581. [DOI] [PubMed] [Google Scholar]
- 21.Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol. 2011;11:762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brodin P, Jojic V, Gao T, Bhattacharya S, Angel CJ, Furman D, Shen-Orr S, Dekker CL, Swan GE, Butte AJ, et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orru V, Steri M, Sole G, Sidore C, Virdis F, Dei M, Lai S, Zoledziewska M, Busonero F, Mulas A, et al. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155:242–256. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fagnoni FF, Vescovini R, Passeri G, Bologna G, Pedrazzoni M, Lavagetto G, Casti A, Franceschi C, Passeri M, Sansoni P. Shortage of circulating naive CD8(+) T cells provides new insights on immunodeficiency in aging. Blood. 2000;95:2860–2868. [PubMed] [Google Scholar]
- 25.Lazuardi L, Jenewein B, Wolf AM, Pfister G, Tzankov A, Grubeck-Loebenstein B. Age-related loss of naive T cells and dysregulation of T-cell/B-cell interactions in human lymph nodes. Immunology. 2005;114:37–43. doi: 10.1111/j.1365-2567.2004.02006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coussens AK, Wilkinson RJ, Nikolayevskyy V, Elkington PT, Hanifa Y, Islam K, Timms PM, Bothamley GH, Claxton AP, Packe GE, et al. Ethnic variation in inflammatory profile in tuberculosis. PLoS Pathog. 2013;9:e1003468. doi: 10.1371/journal.ppat.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haralambieva IH, Ovsyannikova IG, Kennedy RB, Larrabee BR, Shane Pankratz V, Poland GA. Race and sex-based differences in cytokine immune responses to smallpox vaccine in healthy individuals. Hum Immunol. 2013;74:1263–1266. doi: 10.1016/j.humimm.2013.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haralambieva IH, Salk HM, Lambert ND, Ovsyannikova IG, Kennedy RB, Warner ND, Pankratz VS, Poland GA. Associations between race, sex and immune response variations to rubella vaccination in two independent cohorts. Vaccine. 2014;32:1946–1953. doi: 10.1016/j.vaccine.2014.01.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harari A, Rozot V, Bellutti Enders F, Perreau M, Stalder JM, Nicod LP, Cavassini M, Calandra T, Blanchet CL, Jaton K, et al. Dominant TNF-alpha+ Mycobacterium tuberculosis-specific CD4+ T cell responses discriminate between latent infection and active disease. Nat Med. 2011;17:372–376. doi: 10.1038/nm.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arlehamn CL, Seumois G, Gerasimova A, Huang C, Fu Z, Yue X, Sette A, Vijayanand P, Peters B. Transcriptional profile of tuberculosis antigen-specific T cells reveals novel multifunctional features. J Immunol. 2014;193:2931–2940. doi: 10.4049/jimmunol.1401151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Q, Xu Q, Chen Q, Li J, Zhang M, Cai Y, Liu H, Zhou Y, Deng G, Deng Q, et al. Discriminating Active Tuberculosis from Latent Tuberculosis Infection by flow cytometric measurement of CD161-expressing T cells. Sci Rep. 2015;5:17918. doi: 10.1038/srep17918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naranbhai V, Kim S, Fletcher H, Cotton MF, Violari A, Mitchell C, Nachman S, McSherry G, McShane H, Hill AV, et al. The association between the ratio of monocytes:lymphocytes at age 3 months and risk of tuberculosis (TB) in the first two years of life. BMC Med. 2014;12:120. doi: 10.1186/s12916-014-0120-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rakotosamimanana N, Richard V, Raharimanga V, Gicquel B, Doherty TM, Zumla A, Rasolofo Razanamparany V. Biomarkers for risk of developing active tuberculosis in contacts of TB patients: a prospective cohort study. Eur Respir J. 2015;46:1095–1103. doi: 10.1183/13993003.00263-2015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.