Abstract
α-1 Antitrypsin deficiency (A1ATD) can progress to cirrhosis and hepatocellular carcinoma; however, not all patients are susceptible to severe liver disease. In A1ATD, a toxic gain-of-function mutation generates insoluble ATZ “globules” in hepatocytes, overwhelming protein clearance mechanisms. The relationship between bile acids and hepatocytic autophagy is less clear but may involve altered gene expression pathways. Based on previous findings that bile duct ligation (BDL) induces autophagy, we hypothesized that retained bile acids may have hepatoprotective effects in PiZZ transgenic mice, which model A1ATD. We performed BDL and partial BDL (pBDL) in PiZZ mice, followed by analysis of liver tissues. PiZZ liver subjected to BDL showed up to 50% clearance of ATZ globules, with increased expression of autophagy proteins. Analysis of transcription factors revealed significant changes. Surprisingly nuclear TFEB, a master regulator of autophagy, remained unchanged. pBDL confirmed that ATZ globule clearance was induced by localized stimuli rather than diet or systemic effects. Several genes involved in bile metabolism were overexpressed in globule-devoid hepatocytes, compared to globule-containing cells. Retained bile acids led to a dramatic reduction of ATZ globules, with enhanced hepatocyte regeneration and autophagy. These findings support investigation of synthetic bile acids as potential autophagy-enhancing agents.
Key words: Hepatocyte, Autophagy, PiZZ, Cholestasis, Liver regeneration
INTRODUCTION
α-1 Antitrypsin deficiency (A1ATD) is the most common inherited pediatric liver disorder and the most frequent genetic cause of liver transplantation (reviewed in Ghouse et al.1). The classical form of A1ATD involves homozygosity for the Z allele (PiZZ) in the SERPINA1 gene, causing accumulation of misfolded ATZ protein monomers in the endoplasmic reticulum (ER) of hepatocytes. These toxic insoluble globules are periodic acid–Schiff (PAS)+/diastase resistant. Chronic hepatocyte injury from ER stress can progress to cirrhosis and hepatocellular carcinoma2. Although liver transplantation remains the only cure for patients with severe liver disease, prospective studies in PiZZ newborns found that only ∼8% of homozygotes develop clinically significant liver disease by adulthood, suggesting the role of other genetic or environmental modifiers of susceptibility3.
The PiZZ transgenic mouse overexpresses human ATZ protein, providing a useful model to study A1ATD-related liver disease4. Histologically, two main hepatocyte populations exist, described as “globule containing” (GC) or “globule devoid” (GD)5. In vivo studies have shown that stressed GC hepatocytes demonstrate decreased proliferation and increased apoptosis compared to healthier GD hepatocytes2,6–9. These differential capabilities can be exploited with autophagy-enhancing agents to promote ATZ globule clearance7,10.
At the transcriptional level, regulation of autophagy is tightly coordinated with induction of nuclear transcription factors11–13. Nuclear receptor pathways offer promising targets for pharmacologic agonists, such as synthetic bile acids, in treating chronic liver diseases14. The relationship of bile acids and hepatocytic autophagy is less clear15,16. A recent study found that although bile duct ligation (BDL) led to cholestatic liver injury in mice, activation of autophagy in hepatocytes was protective15. In human liver, biliary obstruction induces transcriptional reprogramming in hepatocytes17. Thus, altered gene expression may be a common survival mechanism following BDL. We therefore hypothesized that BDL may also lead to hepatoprotective effects in PiZZ mice. In this study, we show that retained bile acids trigger alternate transcriptional pathways and enhance hepatocyte regeneration and autophagy, leading to ATZ globule clearance.
MATERIALS AND METHODS
Animal Surgeries
We used 3-month-old mice, since ATZ globules in PiZZ mouse liver peak at ages 2–3 months. We used male PiZZ (c57/b6) mice, given sex-dependent phenotypic variations. Female PiZZ mice have significantly less ATZ globules compared to male PiZZ mice, making them less suitable for our studies18. Mice were maintained on 12-h light/dark cycles, with ad libitum mouse chow and water in an approved barrier facility. Surgery was performed under isoflurane anesthesia. The mouse was positioned on the surgical table under a surgical microscopy apparatus, and a midline abdominal incision exposed the hepatic hilum and hepatoduodenal ligament. For total BDL, the common bile duct was dissected from the hepatoduodenal ligament and ligated with 7-0 silk. A double ligature was made. Sham operations for BDL were performed in the same way without bile duct dissection and ligation. For partial BDL (pBDL), the unligated right lobe served as an internal control for the ligated left lobe, subjected to the same systemic effects, such as diet or circulating factors activated by BDL. Previous studies have observed less fibrosis and necrosis in the internal control lobe in pBDL19. For pBDL, the main biliary confluence of the right and left bile duct was carefully visualized at the hepatic hilum, and only the left bile duct was ligated with 10-0 nylon. The abdominal incision was closed in two layers using a continuous running suture. Liver tissues were harvested at days 3, 7, and 14 after surgery (n = 3–5 mice per time point). All procedures were in accordance with ethical guidelines of the Institutional Animal Care and Use Committee of the University of Pittsburgh (#15035202).
Histology and Globule Morphometry
Immunohistochemistry and confocal immunofluorescence microscopy were performed as previously described20. See Table 1 for the list of antibodies. PASD staining was performed according to the manufacturer’s protocol (Sigma-Aldrich). For quantitative ATZ globule morphometry, formalin-fixed paraffin sections were costained with PASD and DAPI nuclear stain, and whole slides of tissue sections from each liver lobe were imaged and reconstructed on a Nikon A1 tiling fluorescence microscope. The percent area of ATZ globules per cell nucleus was quantified blindly using an automated algorithm designed with the Nikon Elements software.
Table 1.
List of Antibodies Used in This Study
| Antibody | Clone (Source) | Dilution | Method |
|---|---|---|---|
| A1AT | Goat pAb #A80-122A (Bethyl) | 1:1,000 | IF |
| ATG5 | Rabbit mAb D5F5U (Cell Signaling) | 1:1,000 | WB |
| ATG12 | Rabbit mAb D888H11 (Cell Signaling) | 1:1,000 | WB |
| Activated caspase 3 | Rabbit pAb #9661 (Cell Signaling) | 1:200 | IHC |
| C/EBP-α | Rabbit pAb 14AA (Santa Cruz) | 1:1,000 | WB |
| C/EBP-β | Rabbit pAb C-19 (Santa Cruz) | 1:1,000 | WB |
| LC3A/BLC3B | Rabbit mAb D3U4C (Cell Signaling)Rabbit pAB #NB100-2220 (Novus Biologicals) | 1:1,0001:200 | WBIF |
| Ki-67 | Rabbit mAb SP6 (ThermoFisher) | 1:50 | IHC |
| TBP | Mouse mAb 1TBP18 (Abcam) | 1:200 | WB |
| TFEB | Goat pAb V-17 (Santa Cruz) | 1:1,000 | WB |
IF, immunofluorescence; IHC, immunohistochemistry; mAb, monoclonal; pAb, polyclonal; WB, Western blot.
Western Blots
Total cell lysates or nuclear extracts were separated on SDS-PAGE, followed by Western blotting as previously described20 (antibodies listed in Table 1).
Laser Capture Analysis
Frozen unfixed liver tissue sections (n = 3) were stained with cresyl violet, and clusters of GC and GD cells were isolated via laser capture microdissection (LCM) for total RNA isolation, as previously described8. Gene array analysis of RNA expression was performed using Affymetrix mouse 2.0 ST arrays.
RESULTS
BDL Induces Clearance of ATZ Globules in PiZZ Mouse Liver
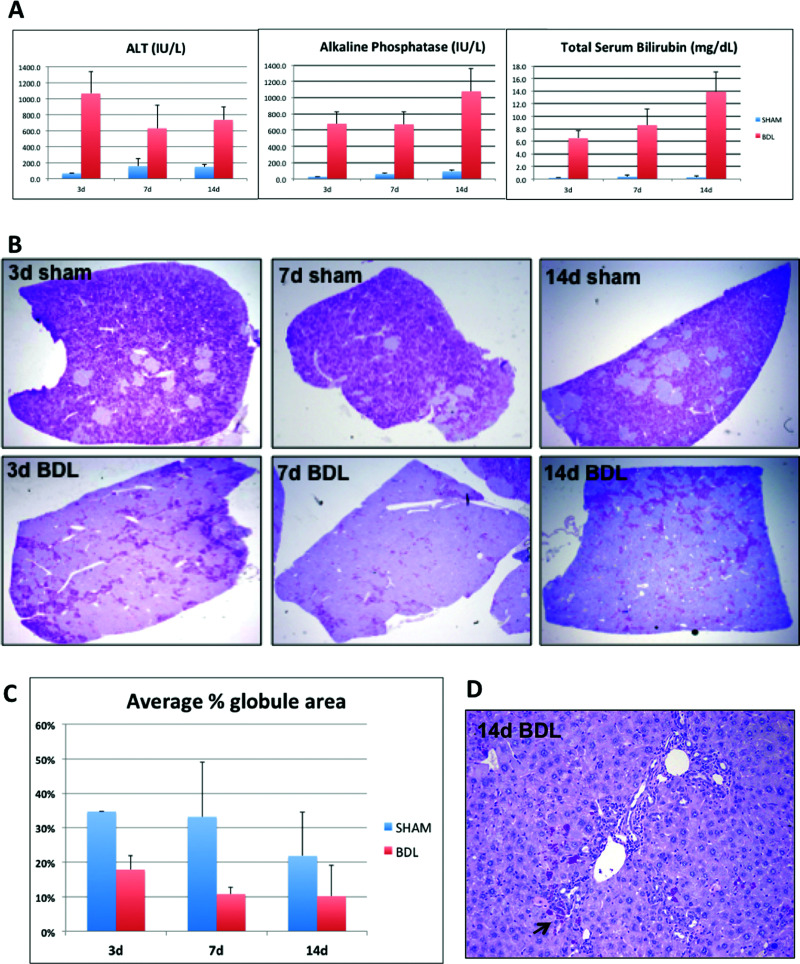
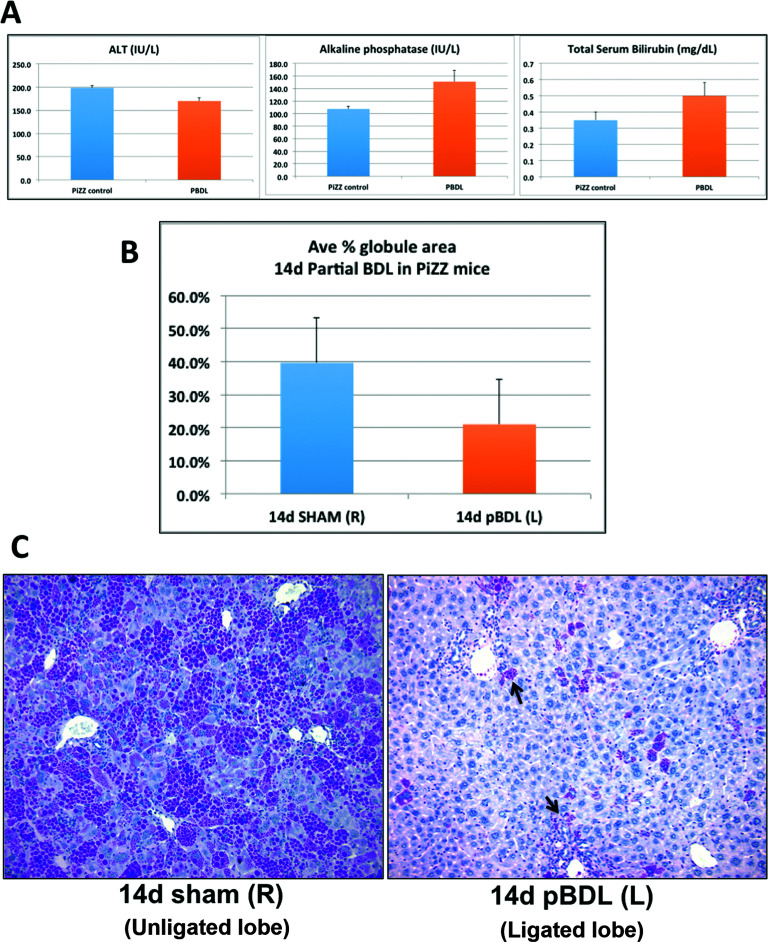
We performed BDL in PiZZ mice to provide a stimulus for biliary-driven regeneration. Serum biochemistries (Fig. 1A) confirmed acute liver injury at day 3, followed by chronic cholestasis at days 7 and 14 after BDL21. We observed a remarkable decrease in PASD staining in BDL livers compared to age- and sex-matched sham controls, as early as 3 days after BDL (Fig. 1B). We then performed blinded automated globule morphometry of whole-tissue sections from multiple liver lobes and found as high as 50% reduction in ATZ globules compared to sham controls (Fig. 1C). This indicates more accelerated globule clearance than expected with aging alone. Interestingly, residual GC cells localized to periportal regions between proliferating intralobular bile ducts (Fig. 1D).
Figure 1.
Bile duct ligation induces clearance of ATZ globules in PiZZ mouse liver. (A) Serum ALT, alkaline phosphatase, and total bilirubin levels at days 3, 7, and 14 after BDL or sham operations. (B) Low-power views of PASD stains show dramatic clearance of ATZ globules from the liver lobe (magnification: 40×). (C) Quantitative results of ATZ globule morphometry showing percent of PASD globules present in the tissue section. (D) PASD stain showing “cleared” region after 14d BDL (magnification: 100×). Arrow points to residual GC hepatocytes intermixed within areas of ductal proliferation.
BDL Induces an Early Regenerative Response in GD Hepatocytes
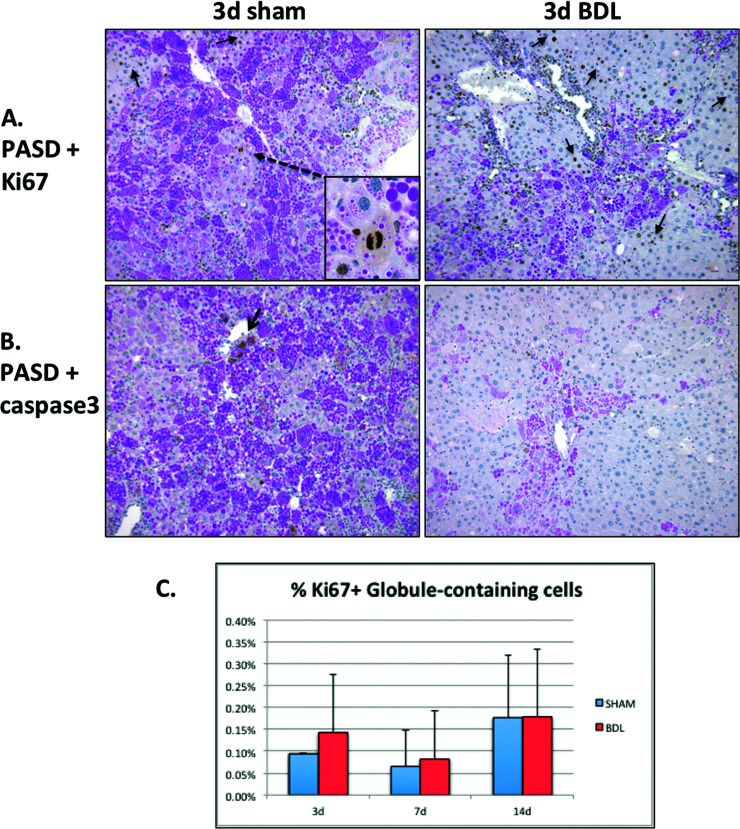
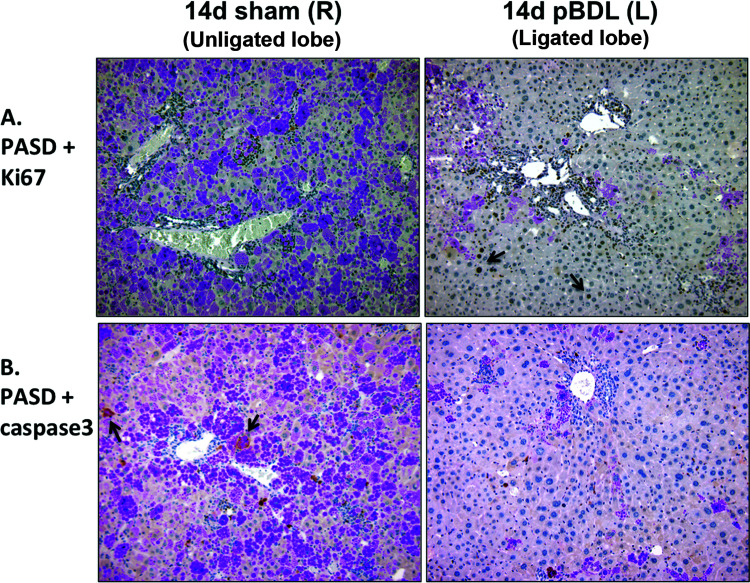
Given our findings of increased ATZ globule clearance as early as day 3 after BDL, corresponding to the period of acute liver injury, we investigated PASD costaining with markers of cellular proliferation and apoptosis (Fig. 2). In day 3 sham, Ki-67+ cells are an infrequent but distinguishing feature of small clusters of repopulating GD hepatocytes, which is a typical finding in PiZZ mice (Fig. 2A, left). In contrast, robust Ki-67+ proliferative activity was observed in GD hepatocytes 3 days after BDL in response to the acute liver injury (Fig. 2A, right), and a very small subset of GC hepatocytes were Ki-67+ (Fig. 2C). Similarly, activated caspase 3 colocalized with GC hepatocytes in day 3 sham but was surprisingly diminished 3 days after BDL (Fig. 2B). These results suggest that regeneration of new GD hepatocytes may contribute to the “cleared” regions of GD cells; however, this turnover is not completely balanced by increased apoptosis of GC hepatocytes.
Figure 2.
Bile duct ligation induces an early regenerative response in GD hepatocytes 3 days after surgery. (A) Ki-67 immunohistochemistry costained with PASD shows regenerating Ki-67+ GD hepatocytes (arrows). Inset shows a rare mitosis within a repopulating cluster of GD cells in sham control. (B) Activated caspase 3 immunohistochemistry costained with PASD does not show significant apoptosis in GC hepatocytes after BDL (magnification: 200×). (C) Quantitative immunofluorescence data for the cell proliferation marker Ki-67 and anti-human A1AT. Graph depicts a small subset of percentage of Ki-67+ cells colocalizing with ATZ GC cells (normalized to total number of cells).
BDL Induces Increased Expression of Autophagic Proteins in PiZZ Mouse Liver
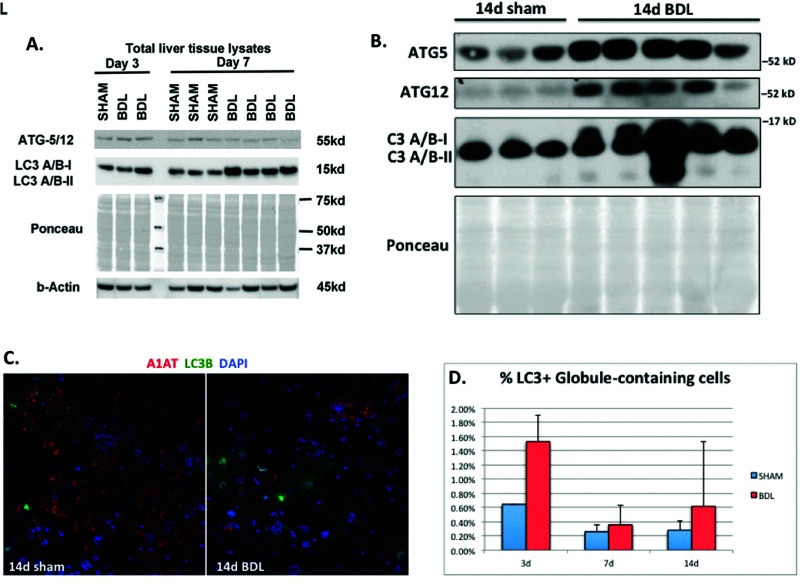
Given our findings of increased proliferation of GD cells without comparable increase in apoptosis of GC cells, we hypothesized that the reduction in ATZ globules may involve intracellular protein clearance pathways rather than exclusive proliferation of new GD hepatocytes. Autophagy involves conjugation of Atg proteins and conversion of LC3I to LC3II. To further investigate possible intracellular mechanisms for ATZ globule clearance, we performed Western blotting for members of the autophagy pathway, including beclin-1, Atg3, Atg7, Atg5, Atg12, Atg16L, and LC3A/B. BDL liver had increased expression of Atg5/12 (days 3 and 7) (Fig. 3A) and LC3A/B (days 3, 7, and 14) (Fig. 3A and B). These proteins signify later events in ubiquitin-like conjugation and the formation of autophagosomes. Specifically, Atg5 is an acceptor protein for the ubiquitin-like protein Atg1222. To further compare autophagic changes in GC and GD cells, we performed immunofluorescence microscopy for LC3 (green) and A1AT (red). As shown in Figure 3C and D, there was differential localization of LC3B in GD hepatocytes, compared to a very small subset of GC cells with A1AT globules. This suggests enhanced autophagy and protein clearance in GD hepatocytes after BDL.
Figure 3.
Bile duct ligation induces increased expression of autophagic proteins in PiZZ mouse liver. (A, B) Western blots of total liver tissue lysates showing increased Atg5, Atg12, and LC3A/B after BDL compared to sham livers. Ponceau is shown to confirm equal loading, since cytosketetal changes can affect both actin and tubulin in PiZZ mouse liver. (C) Confocal immunofluorescence microscopy showing the majority of LC3B (green) does not colocalize with GC cells labeled with anti-human A1AT (red) antibody (magnification: 100×). (D) Quantitative immunofluorescence data for LC3 and anti-human A1AT. Graph depicts a small subset of percentage of LC3+ cells colocalizing with ATZ GC cells (normalized to total number of cells).
BDL Induces Transcriptional Reprogramming in PiZZ Mouse Liver
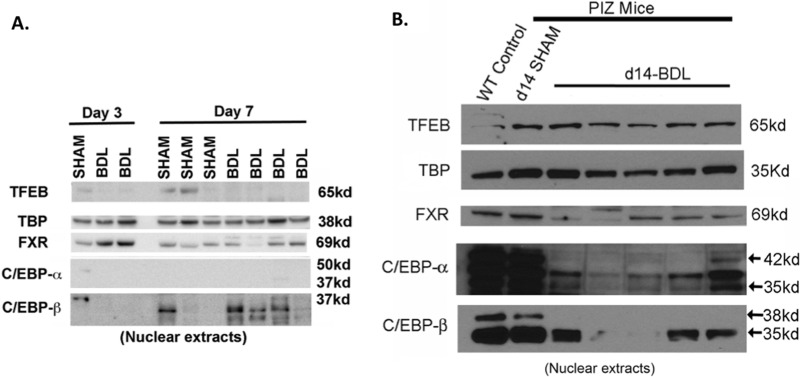
To further investigate altered gene expression pathways after BDL, we performed Western blots on nuclear extracts for selected transcription factors known to regulate autophagy (Fig. 4). As expected, increased nuclear FXR was observed early after BDL (Fig. 4A), which appeared to trend down at later time points (Fig. 4B)13. Surprisingly, nuclear TFEB, a master regulator of autophagy, was not significantly induced after BDL12. Compared to wild-type and sham PiZZ mice, we did observe a significant decrease in nuclear C/EBP-α after BDL, while the expression of C/EBP-β appears to fluctuate. Both C/EBP isoforms are well-described regulators of liver regeneration23.
Figure 4.
Bile duct ligation induces transcriptional reprogramming in PiZZ mouse liver. (A, B). Western blot of nuclear proteins showing decreased nuclear C/EBP-α and C/EBP-β isoforms in BDL livers compared to sham-operated and wild-type (WT) control liver. Nuclear FXR is decreased after BDL at later time points. Nuclear TFEB is not significantly changed in BDL and sham livers. TATA-binding protein (TBP) serves as the nuclear protein-loading control.
Partial BDL Induces Localized ATZ Globule Clearance in PiZZ Mouse Liver
To determine if ATZ globule clearance was induced by retained bile acids versus systemic effects, we performed pBDL on PiZZ mice, where only the left bile duct was ligated; thus, the unligated right lobe served as an internal control, subjected to the same dietary and systemic exposures as the left lobe. Although there was clear evidence of cholestasis in the ligated lobe after pBDL, serum biochemistries were less severely affected in pBDL mice (Fig. 5A), confirming compensatory hepatic function from the unligated lobes. We then analyzed both ligated and unligated (control) lobes by globule morphometry, which again demonstrated approximately 50% reduction in ATZ globules in the ligated lobe compared to the unligated lobe (Fig. 5B). Similar to total BDL, PASD stain (Fig. 5C) confirmed large areas of clearing and bile ductular proliferation in the ligated lobe. These results confirm that localized retained bile acids trigger a direct microenvironmental stimulus on signaling pathways leading to ATZ globule clearance. Given that pBDL mice were overall healthier appearing with less severe lab abnormalities than BDL mice, systemic effects such as diet/starvation-induced autophagy, gut–liver axis, or other circulating factors are less likely causes for the dramatic reduction in globules.
Figure 5.
Partial bile duct ligation induces localized ATZ globule clearance in PiZZ mouse liver. (A) Serum ALT, alkaline phosphatase, and total bilirubin levels in mice 14 days after pBDL are comparable to age- and sex-matched unoperated PiZZ (control) mice. As expected, biochemical parameters in BDL mice (shown in Fig. 1A) are significantly worse compared to pBDL values. (B) Quantitative results of ATZ globule morphometry showing decreased percentage of PASD globules present in the ligated left lobe (pBDL) compared to the unligated right control lobe. (C) PASD staining showing the cleared areas of ATZ globules in pBDL (ligated left lobe, right) compared to control (unligated right lobe, left). Arrows point to residual GC hepatocytes intermixed within areas of ductal proliferation, similar to total BDL (magnification: 200×).
Relationship of ATZ Globule Clearance to Regeneration and Autophagy in Partial BDL
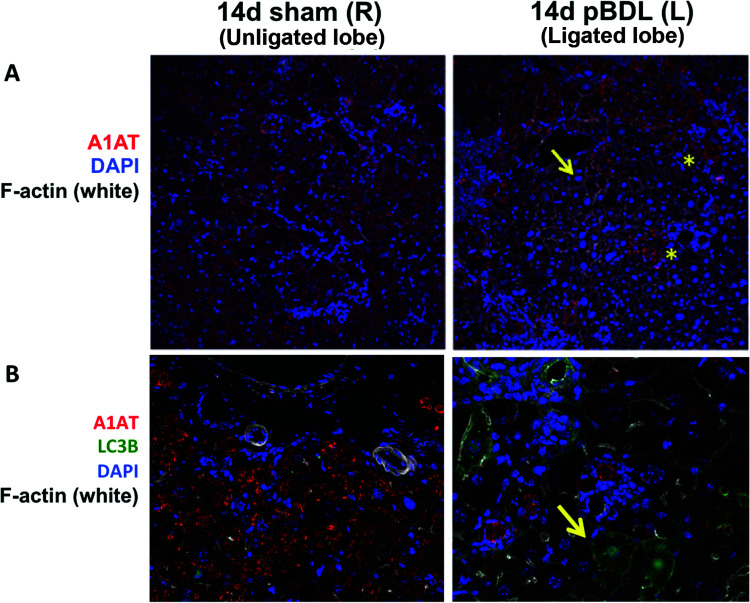
To identify mechanisms of ATZ globule clearance in the ligated lobe, we performed PASD costaining for Ki-67 and activated caspase 3 (Fig. 6), as well as immunofluorescence staining for LC3B (Fig. 7B). In pBDL, we observed Ki-67+ proliferating GD hepatocytes (Fig. 6A, right), as well as an absence of activated caspase 3 (Fig. 6B, right) in the ligated lobe. We observed increased LC3B staining in the ligated lobe (Fig. 7B, right) compared to the unligated lobe (Fig. 7B, left), confirming increased autophagosome activity in GD cells. Coupled to our findings in total BDL, hepatocyte regeneration of GD cells appears to be an early response driven by local stimuli.
Figure 6.
Relationship of ATZ globule clearance to regeneration in partial bile duct ligation. (A) Ki-67 immunohistochemistry costained with PASD shows persistence of regenerating Ki-67+ GD hepatocytes 14 days after pBDL (right, arrows). (B) Activated caspase 3 immunohistochemistry costained with PASD does not show significant apoptosis in GC hepatocytes 14 days after pBDL (right) compared to unligated right control lobe (left, arrows; magnification: 200×).
Figure 7.
Relationship of ATZ globule clearance to autophagy in partial bile duct ligation. (A) Confocal immunofluorescence microscopy showing the cleared areas of red ATZ globules in pBDL lobe (right, arrow) compared to unligated control lobe (left). *Residual GC hepatocytes intermixed within areas of ductal proliferation (magnification: 100×). (B) Increased LC3B staining (green, right, arrow) in GD hepatocytes in the ligated pBDL lobe (magnification: 200×).
Differential Expression of CYP-Related Genes in GC Versus GD Cells
To further investigate altered gene expression pathways in PiZZ mouse liver, we performed LCM on clusters of GC and GD hepatocytes, followed by gene array analysis. Interestingly, several genes involved in lipid metabolism, along with bile transport (Slc10a1), were upregulated in GD cells (Table 2). Surprisingly, CYP8b1, a key regulator of bile acid composition, was the second highest expressed gene in GD cells, with a 10.50-fold change over GC cells ( p = 0.000338). Increased expression was also noted for CYP2c54, CYP4a32, CYP2c50, and CYP2c44, all of which are involved in arachidonic acid pathways. The complete gene array data set was deposited in the NIH Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), accession ID# GSE84763.
Table 2.
Differential Expression of Bile-Related Genes in Microdissected PiZZ Mouse Liver
| Fold Change (GD/GC) | Gene ID | Gene Name |
|---|---|---|
| 10.50 | Cyp8b1 | Cytochrome P450, family 8, subfamily b, polypeptide 1 |
| 5.79 | Cyp2c54 | Cytochrome P450, family 2, subfamily c, polypeptide 54 |
| 4.66 | Cyp4a32 | Cytochrome P450, family 4, subfamily a, polypeptide 32 |
| 4.65 | Cyp2c50 | Cytochrome P450, family 2, subfamily c, polypeptide 50 |
| 4.32 | Cyp2c44 | Cytochrome P450, family 2, subfamily c, polypeptide 44 |
| 4.08 | Slc10a1 | Solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| 3.60 | Cyp2c37 | Cytochrome P450, family 2. subfamily c, polypeptide 37 |
| 3.23 | Cyp4a10 | Cytochrome P450, family 4, subfamily a, polypeptide 10 |
| 3.21 | Cyp2f2 | Cytochrome P450, family 2, subfamily f, polypeptide 2 |
| 3.03 | Cyp4a14 | Cytochrome P450, family 4, subfamily a, polypeptide 14 |
| 2.49 | Cyp4a31 | Cytochrome P450, family 4, subfamily a, polypeptide 31 |
| 2.23 | Cyp4f15 | Cytochrome P450, family 4, subfamily f, polypeptide 15 |
| 1.97 | Cyp2j9 | Cytochrome P450, family 2, subfamily j, polypeptide 9 |
| 1.94 | Cyp2d13 | Cytochrome P450, family 2, subfamily d, polypeptide 13; |
| 1.90 | Cyp2c68 | Cytochrome P450, family 2, subfamily c, polypeptide 68 |
| 1.89 | Cyp7b1 | Cytochrome P450, family 7, subfamily b, polypeptide 1 |
| 1.74 | Cyp2j5 | Cytochrome P450, family 2, subfamily j, polypeptide 5 |
| 1.70 | Cyp26a1 | Cytochrome P450, family 26, subfamily a, polypeptide 1 |
| 1.70 | Cyp2c67 | Cytochrome P450, family 2, subfamily c, polypeptide 67 |
| 1.68 | Klkb1; Cyp4v3 | Kallikrein B, plasma 1; cytochrome P450, family 4, subfamily v, polypeptide 3 |
| 1.67 | Cyp2u1 | Cytochrome P450, family 2, subfamily u, polypeptide 1 |
| 1.63 | Cyp2d40 | Cytochrome P450, family 2, subfamily d, polypeptide 40 |
| 1.61 | Cyp3a13 | Cytochrome P450, family 3, subfamily a, polypeptide 13 |
| 1.60 | Cyp2d26 | Cytochrome P450, family 2, subfamily d, polypeptide 26 |
| 1.59 | Cyp2d22 | Cytochrome P450, family 2, subfamily d, polypeptide 22 |
| 1.57 | Cyp4f14 | Cytochrome P450, family 4, subfamily f, polypeptide 14 |
| 1.57 | Cyp2c40 | Cytochrome P450, family 2, subfamily c, polypeptide 40 |
| 1.56 | Cyp39a1 | Cytochrome P450, family 39, subfamily a, polypeptide 1 |
| 1.54 | Cyp2a12 | Cytochrome P450, family 2, subfamily a, polypeptide 12 |
| 1.52 | Cyp2c69 | Cytochrome P450, family 2, subfamily c, polypeptide 69 |
| 1.41 | Cyp2g1 | Cytochrome P450, family 2, subfamily g, polypeptide 1 |
| 1.48 | Slc10a5 | Solute carrier family 10 (sodium/bile acid cotransporter family), member 5 |
DISCUSSION
In summary, we found that retained bile acids trigger alternate transcriptional pathways and enhanced autophagy, leading to ATZ globule clearance in PiZZ mouse liver. Mice subjected to pBDL showed lobe-specific reduction in ATZ globules independent of dietary or systemic exposures, suggesting a localized effect when bile flow is obstructed. The increase in GD hepatocytes after BDL is an early regenerative response, coinciding with stimulation of proliferation, but not with increased apoptosis of GC hepatocytes. Overall, our data point to a hepatoprotective role for bile acids in A1ATD.
Numerous studies have reported the role of bile acid signaling in liver injury and protection. Elevated bile acids have been shown to accelerate liver regeneration24. Bile acid composition has been used as a tool to assess graft function following liver transplantation25–30. Hydrophilic bile acids are cytoprotective. BDL in wild-type mice resulted in altered bile acid composition, with a significant increase in hydrophilic taurine-conjugated bile acids31. Similarly, the bile acid receptor TGR5 maintains hepatoprotective effects by limiting bile acid hydrophobicity32. In contrast, hydrophobic bile acids have been implicated in ER stress-induced apoptosis33. In relation to this, we found CYP8b1 to be highly expressed in GD cells compared to GC cells, unlike in normal liver, where it is diffusely expressed34. CYP8b1 catalyzes sterol hydroxylation, which in turn determines the ratio of primary bile acids and solubility35. Whether such changes would affect the solubility of polymerized ATZ protein is not yet known, and our attempts to compare bile acid composition in BDL versus sham-operated PiZZ mice were unsuccessful due to technical issues.
Regulation of bile acid signaling also involves interaction of hepatic nuclear transcription factors, growth factors (FGF15/19), and bile acid receptors (FXR and TGR5) with the gut–liver axis11,13,32,36. BDL also upregulates the nuclear transcription factor PPARα, although the latter is not directly affected by bile acids. PPARα is a master regulator of hepatic lipid metabolism37. Furthermore, the nuclear receptors FXR and PPARα both regulate autophagy, making these promising pharmacologic targets for enhancing ATZ clearance11,13. It was recently reported that FXR suppresses autophagy by repressing CREB in the “fed” state, while PPARα can reverse this suppression in the “fasted” state11,13. Our analysis of key transcription factors after BDL (Fig. 4) revealed several interesting findings. First, with increased exposure to retained bile acids after BDL, we observed downtrending of nuclear FXR, a transcription factor that acts as a bile acid receptor. Unfortunately, fasting in PiZZ mice can be lethal due to impaired glycogen stores38. Since our mice were not fasted prior to sacrifice, our results may support FXR’s role in suppressing autophagy13. Decreased FXR in the setting of BDL is intriguing, since that would, in theory, cause a decrease in SHP-1-mediated repression of CYP7a1 (cholesterol 7α-hydroxylase), which catalyzes the first step of bile acid synthesis39.
Given our findings of increased Atg5, Atg12, and LC3A/B after BDL (Fig. 3), we were also surprised to see no increase in nuclear translocation of TFEB after BDL, as this transcription factor is a master regulator of autophagy. Cytoplasmic TFEB is dephosphorylated in the fasted state and translocates to the nucleus to induce autophagy-related gene expression40. TFEB gene transfer was previously shown to correct the liver disease in PiZZ mice, leading to increased degradation of polymerized ATZ in autolysosomes and decreased expression of ATZ monomers12. Additional studies on PPARα and other transcriptional regulators (mTORC1 and NCoR1) may help to define the transcriptional program associated with autophagy in A1ATD.
Perhaps the most striking transcriptional change we observed was the fluctuation of both C/EBP-α and C/EBP-β isoforms after BDL (Fig. 4), which is intriguing given the increased proliferation of GD hepatocytes. Normal liver regeneration following partial hepatectomy is regulated by the ratio of these two isoforms. During liver regeneration, C/EBP-β increases and favors hepatocyte proliferation, while C/EBP-α decreases and has an opposing effect23,41. C/EBP-α was recently shown in liver to be suppressed in conditions favoring autophagy and fibrosis42. Similar to our findings, Tao et al. found a reduction in hepatocyte C/EBP-α and an increase in Atg5 with autophagy42. Taken together, the C/EBP isoforms may represent novel regulators of autophagy; therefore, further investigation of hepatocyte regeneration in this setting is warranted.
In human liver, biliary obstruction activates cholangiocyte-associated transcription factors, and hepatocytes undergo gene expression reprogramming17. Thus, altered gene expression may be a common survival mechanism following BDL-induced liver injury. BDL causes cholestatic injury, ductular proliferation, and activation of liver progenitor cells in rodents43. Our data support those of Gao et al., who recently found that in mice subjected to BDL, activation of autophagy in hepatocytes was protective against cholestatic liver injury15. It is interesting to note that our findings differ from a previous report of BDL in PIZZ mice by Mencin et al.44. They found increased fibrosis, apoptosis, and stellate cell activation in PiZZ mouse liver after BDL. However, since their study focused on fibrosis as an endpoint, the mice were sacrificed 21 days after BDL, when more chronic severe liver injury was apparent. In contrast, we used earlier endpoints in our studies (days 3, 7, and 14) to better characterize the dynamics of ATZ globule load and transcriptional changes. At these earlier time points, we observed increased ductular proliferation as expected, but less fibrosis.
In our experiments, the few remaining GC hepatocytes consistently resided adjacent to areas of proliferating bile ducts (Figs. 1D, 5C, and 7A). This is interesting, since developmentally ATZ globules first appear in periportal hepatocytes and then progress pericentrally, and the pattern of globule clearance appears to be a reversal of this. Although the significance of this varied localization is unknown, it may represent active remodeling of the liver lobule in response to intercellular crosstalk.
Since the hepatic manifestations and clinical prognosis of A1ATD are highly variable, understanding these early changes after BDL may identify which factors predispose some patients to develop severe liver disease while sparing others. Although BDL is obviously not a viable option in patients, it does provide mechanistic clues for future investigation. Most children diagnosed with A1ATD present with neonatal cholestasis that peaks, but over time it can spontaneously resolve without overt liver disease45. Bile acid derivatives and their receptor agonists/antagonists are already in clinical development and could potentially be repurposed for their autophagy-enhancing properties37,46. Similarly, as a cytoprotective bile acid, ursodeoxycholic acid has been beneficial in children with mild to moderate A1ATD liver disease47. Tauroursodeoxycholic acid also inhibited apoptosis induced by mutant ATZ protein48. While our manuscript was in preparation, Tang et al. published that PiZZ mice treated with the exogenous bile acid nor-ursodeoxycholic acid showed a 32% increase in hepatic autophagy and >70% reduction of ATZ protein, with reduced apoptosis and liver injury49. Our findings are consistent with those of Tang et al. and support comparable hepatoprotective effects. Clearly, a more focused approach to metabolomics and transcriptional regulation will offer a better understanding of A1ATD liver disease, so that novel therapeutic strategies can be tested.
ACKNOWLEDGMENTS
The authors appreciate the excellent and generous technical assistance of Anne Orr, Meagan Haynes, John Stoops, Peng Zhang, Callen Wallace, Morgan Jessup, Mark Ross, Devin Boyles, and the Center for Biologic Imaging (Core A). The authors are grateful to Dr. David Geller for providing access to the transplant/microsurgery lab. Affymetrix analysis was performed by the Genomics Core, led by Dr. Jian-hua Luo. The authors also appreciate critical discussions with Dr. David Perlmutter and Dr. Sarangarajan Ranganathan. Financial support provided to Z.K. (K12HD052892; the Alpha-1 Foundation, the Hillman Foundation) and G.K.M., D.B.S., and A.W.B. (P01DK096990).
Footnotes
The authors declare no conflicts of interests.
REFERENCES
- 1. Ghouse R, Chu A, Wang Y, Perlmutter DH. Mysteries of alpha1-antitrypsin deficiency: Emerging therapeutic strategies for a challenging disease. Dis Model Mech. 2014;7(4):411–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Teckman JH, An JK, Blomenkamp K, Schmidt B, Perlmutter D. Mitochondrial autophagy and injury in the liver in alpha 1-antitrypsin deficiency. Am J Physiol Gastrointest Liver Physiol. 2004;286(5):G851–62. [DOI] [PubMed] [Google Scholar]
- 3. Piitulainen E, Carlson J, Ohlsson K, Sveger T. Alpha1-antitrypsin deficiency in 26-year-old subjects: Lung, liver, and protease/protease inhibitor studies. Chest 2005;128(4):2076–81. [DOI] [PubMed] [Google Scholar]
- 4. Sifers RN, Carlson JA, Clift SM, DeMayo FJ, Bullock DW, Woo SL. Tissue specific expression of the human alpha-1-antitrypsin gene in transgenic mice. Nucleic Acids Res. 1987;15(4):1459–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khan Z. Pathogenesis of alpha-1 antitrypsin deficiency in the liver: New approaches to old questions. J Liver Res Disord Ther. 2016;2(2):00023. [Google Scholar]
- 6. Lindblad D, Blomenkamp K, Teckman J. Alpha-1-antitrypsin mutant Z protein content in individual hepatocytes correlates with cell death in a mouse model. Hepatology 2007;46(4):1228–35. [DOI] [PubMed] [Google Scholar]
- 7. Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, Maurice N, Mukherjee A, Goldbach C, Watkins S, Michaelopoulos G, Perlmutter DH. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science 2010;329(5988):229–32. [DOI] [PubMed] [Google Scholar]
- 8. Ding J, Yannam GR, Roy-Chowdhury N, Hidvegi T, Basma H, Rennard SI, Wong RJ, Avsar Y, Guha C, Perlmutter DH, Fox IJ, Roy-Chowdhury J. Spontaneous hepatic repopulation in transgenic mice expressing mutant human alpha1-antitrypsin by wild-type donor hepatocytes. J Clin Invest. 2011;121(5):1930–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rudnick DA, Liao Y, An JK, Muglia LJ, Perlmutter DH, Teckman JH. Analyses of hepatocellular proliferation in a mouse model of alpha-1-antitrypsin deficiency. Hepatology 2004;39(4):1048–55. [DOI] [PubMed] [Google Scholar]
- 10. Pastore N, Blomenkamp K, Annunziata F, Piccolo P, Mithbaokar P, Maria Sepe R, Vetrini F, Palmer D, Ng P, Polishchuk E, Iacobacci S, Polishchuk R, Teckman J, Ballabio A, Brunetti-Pierri N. Gene transfer of master autophagy regulator TFEB results in clearance of toxic protein and correction of hepatic disease in alpha-1-anti-trypsin deficiency. EMBO Mol Med. 2013;5(3):397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, Moore DD. Nutrient-sensing nuclear receptors coordinate autophagy. Nature 2014;516(7529):112–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pastore N, Ballabio A, Brunetti-Pierri N. Autophagy master regulator TFEB induces clearance of toxic SERPINA1/alpha-1-antitrypsin polymers. Autophagy 2013;9(7):1094–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seok S, Fu T, Choi SE, Li Y, Zhu R, Kumar S, Sun X, Yoon G, Kang Y, Zhong W, Ma J, Kemper B, Kemper JK. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature 2014;516(7529):108–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Asgharpour A, Kumar D, Sanyal A. Bile acids: Emerging role in management of liver diseases. Hepatol Int. 2015;9(4):527–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao L, Lv G, Guo X, Jing Y, Han Z, Zhang S, Sun K, Li R, Yang Y, Wei L. Activation of autophagy protects against cholestasis-induced hepatic injury. Cell Biosci. 2014;4:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Manley S, Ni HM, Kong B, Apte U, Guo G, Ding WX. Suppression of autophagic flux by bile acids in hepatocytes. Toxicol Sci. 2014;137(2):478–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Limaye PB, Alarcon G, Walls AL, Nalesnik MA, Michalopoulos GK, Demetris AJ, Ochoa ER. Expression of specific hepatocyte and cholangiocyte transcription factors in human liver disease and embryonic development. Lab Invest. 2008;88(8):865–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rudnick DA, Shikapwashya O, Blomenkamp K, Teckman JH. Indomethacin increases liver damage in a murine model of liver injury from alpha-1-antitrypsin deficiency. Hepatology 2006;44(4):976–82. [DOI] [PubMed] [Google Scholar]
- 19. Heinrich S, Georgiev P, Weber A, Vergopoulos A, Graf R, Clavien PA. Partial bile duct ligation in mice: A novel model of acute cholestasis. Surgery 2011;149(3):445–51. [DOI] [PubMed] [Google Scholar]
- 20. Khan Z, Michalopoulos GK, Stolz DB. Peroxisomal localization of hypoxia-inducible factors and hypoxia-inducible factor regulatory hydroxylases in primary rat hepatocytes exposed to hypoxia-reoxygenation. Am J Pathol. 2006;169(4):1251–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vartak N, Damle-Vartak A, Richter B, Dirsch O, Dahmen U, Hammad S, Hengstler JG. Cholestasis-induced adaptive remodeling of interlobular bile ducts. Hepatology 2016;63(3):951–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Romanov J, Walczak M, Ibiricu I, Schuchner S, Ogris E, Kraft C, Martens S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31(22):4304–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Michalopoulos G. Terminating hepatocyte proliferation during liver regeneration: The roles of two members of the same family (CCAAT-enhancer-binding protein alpha and beta) with opposing actions. Hepatology 2015;61(1):32–4. [DOI] [PubMed] [Google Scholar]
- 24. Huang W, Ma K, Zhang J, Qatanani M, Cuvillier J, Liu J, Dong B, Huang X, Moore DD. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science 2006;312(5771):233–6. [DOI] [PubMed] [Google Scholar]
- 25. Vilca Melendez H, Rela M, Setchell KD, Murphy GM, Heaton ND. Bile acids analysis: A tool to assess graft function in human liver transplantation. Transpl Int. 2004;17(6):286–92. [DOI] [PubMed] [Google Scholar]
- 26. Ericzon BG, Eusufzai S, Kubota K, Einarsson K, Angelin B. Characteristics of biliary lipid metabolism after liver transplantation. Hepatology 1990;12(5):1222–8. [DOI] [PubMed] [Google Scholar]
- 27. Ericzon BG, Eusufzai S, Kubota K, Einarsson K, Angelin B. Biliary lipid secretion early after liver transplantation. Transplant Proc. 1990;22(4):1537–8. [PubMed] [Google Scholar]
- 28. Fujiwara S, Ku Y, Saitoh Y. Serum bile acid monitoring as an early indicator of allograft function in canine orthotopic liver transplantation. Kobe J Med Sci. 1992;38(4):217–31. [PubMed] [Google Scholar]
- 29. Hertl M, Harvey PR, Swanson PE, West DD, Howard TK, Shenoy S, Strasberg SM. Evidence of preservation injury to bile ducts by bile salts in the pig and its prevention by infusions of hydrophilic bile salts. Hepatology 1995;21(4):1130–7. [PubMed] [Google Scholar]
- 30. Xu HS, Pilcher JA, Jones RS. Physiologic study of bile salt and lipid secretion in rats after liver transplantation. Ann Surg. 1993;217(4):404–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y, Hong JY, Rockwell CE, Copple BL, Jaeschke H, Klaassen CD. Effect of bile duct ligation on bile acid composition in mouse serum and liver. Liver Int. 2012;32(1):58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pean N, Doignon I, Garcin I, Besnard A, Julien B, Liu B, Branchereau S, Spraul A, Guettier C, Humbert L, Schoonjans K, Rainteau D, Tordjmann T. The receptor TGR5 protects the liver from bile acid overload during liver regeneration in mice. Hepatology 2013;58(4):1451–60. [DOI] [PubMed] [Google Scholar]
- 33. Tamaki N, Hatano E, Taura K, Tada M, Kodama Y, Nitta T, Iwaisako K, Seo S, Nakajima A, Ikai I and others. CHOP deficiency attenuates cholestasis-induced liver fibrosis by reduction of hepatocyte injury. Am J Physiol Gastrointest Liver Physiol. 2008;294(2):G498–505. [DOI] [PubMed] [Google Scholar]
- 34. Wang J, Greene S, Eriksson LC, Rozell B, Reihner E, Einarsson C, Eggertsen G, Gafvels M. Human sterol 12a-hydroxylase (CYP8B1) is mainly expressed in hepatocytes in a homogenous pattern. Histochem Cell Biol. 2005;123(4–5):441–6. [DOI] [PubMed] [Google Scholar]
- 35. Gafvels M, Olin M, Chowdhary BP, Raudsepp T, Andersson U, Persson B, Jansson M, Bjorkhem I, Eggertsen G. Structure and chromosomal assignment of the sterol 12alpha-hydroxylase gene (CYP8B1) in human and mouse: Eukaryotic cytochrome P-450 gene devoid of introns. Genomics 1999;56(2):184–96. [DOI] [PubMed] [Google Scholar]
- 36. Naugler WE, Tarlow BD, Fedorov LM, Taylor M, Pelz C, Li B, Darnell J, Grompe M. Fibroblast growth factor signaling controls liver size in mice with humanized livers. Gastroenterology 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rakhshandehroo M, Knoch B, Muller M, Kersten S. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res. 2010;2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hubner RH, Leopold PL, Kiuru M, De BP, Krause A, Crystal RG. Dysfunctional glycogen storage in a mouse model of alpha1-antitrypsin deficiency. Am J Respir Cell Mol Biol. 2009;40(2):239–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Willson TM, Kliewer SA. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell 2000;6(3):517–26. [DOI] [PubMed] [Google Scholar]
- 40. Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31(5):1095–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jin J, Hong IH, Lewis K, Iakova P, Breaux M, Jiang Y, Sullivan E, Jawanmardi N, Timchenko L, Timchenko NA. Cooperation of C/EBP family proteins and chromatin remodeling proteins is essential for termination of liver regeneration. Hepatology 2015;61(1):315–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tao LL, Zhai YZ, Ding D, Yin WH, Liu XP, Yu GY. The role of C/EBP-alpha expression in human liver and liver fibrosis and its relationship with autophagy. Int J Clin Exp Pathol. 2015;8(10):13102–7. [PMC free article] [PubMed] [Google Scholar]
- 43. Omori M, Evarts RP, Omori N, Hu Z, Marsden ER, Thorgeirsson SS. Expression of alpha-fetoprotein and stem cell factor/c-kit system in bile duct ligated young rats. Hepatology 1997;25(5):1115–22. [DOI] [PubMed] [Google Scholar]
- 44. Mencin A, Seki E, Osawa Y, Kodama Y, De Minicis S, Knowles M, Brenner DA. Alpha-1 antitrypsin Z protein (PiZ) increases hepatic fibrosis in a murine model of cholestasis. Hepatology 2007;46(5):1443–52. [DOI] [PubMed] [Google Scholar]
- 45. Khan Z, Venkat VL, Soltys KA, Stolz DB, Ranganathan S. A challenging case of severe infantile cholestasis in alpha-1 antitrypsin deficiency. Pediatr Dev Pathol. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fiorucci S, Zampella A, Distrutti E. Development of FXR, PXR and CAR agonists and antagonists for treatment of liver disorders. Curr Top Med Chem. 2012;12(6):605–24. [DOI] [PubMed] [Google Scholar]
- 47. Lykavieris P, Ducot B, Lachaux A, Dabadie A, Broue P, Sarles J, Bernard O, Jacquemin E. Liver disease associated with ZZ alpha1-antitrypsin deficiency and ursodeoxycholic acid therapy in children. J Pediatr Gastroenterol Nutr. 2008;47(5):623–9. [DOI] [PubMed] [Google Scholar]
- 48. Miller SD, Greene CM, McLean C, Lawless MW, Taggart CC, O’Neill SJ, McElvaney NG. Tauroursodeoxycholic acid inhibits apoptosis induced by Z alpha-1 antitrypsin via inhibition of Bad. Hepatology 2007;46(2):496–503. [DOI] [PubMed] [Google Scholar]
- 49. Tang Y, Fickert P, Trauner M, Marcus N, Blomenkamp K, Teckman J. Autophagy induced by exogenous bile acids is therapeutic in a model of alpha-1-AT deficiency liver disease. Am J Physiol Gastrointest Liver Physiol. 2016;311(1):G156–65. [DOI] [PubMed] [Google Scholar]