Abstract
Myasthenia gravis (MG) is a prototypical B cell-mediated autoimmune disease affecting 20–50 per 100,000 people. The majority of patients fall into two clinically distinguishable types based on whether they produce autoantibodies targeting the acetylcholine receptor (AChR-MG) or muscle specific kinase (MuSK-MG). The autoantibodies are pathogenic, but whether their generation is associated with broader defects in the B cell repertoire is unknown. To address this question, we performed deep sequencing of the B cell receptor repertoire of AChR-MG, MuSK-MG and healthy subjects to generate approximately 518,000 unique VH and VL sequences from sorted naïve and memory B cell populations. AChR-MG and MuSK-MG subjects displayed distinct gene segment usage biases in both VH and VL sequences within the naïve and memory compartments. The memory compartment of AChR-MG was further characterized by reduced positive selection of somatic mutations in the H-CDR regions and altered H-CDR3 physicochemical properties. The VL repertoire of MuSK-MG was specifically characterized by reduced V/J segment distance in recombined sequences, suggesting diminished VL receptor editing during B cell development. Our results identify large-scale abnormalities in both the naïve and memory B cell repertoires. Particular abnormalities were unique to either AChR-MG or MuSK-MG indicating that the repertoires reflect the distinct properties of the subtypes. These repertoire abnormalities are consistent with previously observed defects in B cell tolerance checkpoints in MG, thereby offering additional insight regarding the impact of tolerance defects on peripheral autoimmune repertoires. These collective findings point toward a deformed B cell repertoire as a fundamental component of MG.
Introduction
Myasthenia gravis (MG) is a chronic autoimmune disorder of neuromuscular signal transmission characterized by muscle weakness and fatigability (1). The estimated annual incidence is 1–2 per 100,000 and the prevalence is as high as 20–50 per 100,000 (2, 3). Recent epidemiological studies indicate that, like other autoimmune diseases, its incidence is rising considerably (4). Approximately 80–85% of MG patients have circulating autoantibodies that target the acetylcholine receptor (AChR)(1) while up to 10% harbor autoantibodies that target muscle specific kinase (MuSK) (5). Autoantibodies to lipoprotein-related protein 4 (LRP4) are detected in a fraction (≈18%) of AChR and MuSK autoantibody double-negative MG patients, but also in a small fraction (<6%) that are AChR or MuSK autoantibody positive (6, 7). A minor fraction (<10%) of MG patients do not have detectable autoantibodies directed toward any of these recognized antigens. The majority of MG patients lacking thymomas have been commonly classified into two major distinct subtypes based on whether they test positive for anti-AChR (AChR-MG) or anti-MuSK (MuSK-MG) antibodies. While both AChR-MG and MuSK-MG demonstrate neuromuscular junction pathologies, these two subtypes differ in the degree of gender bias, clinical presentation, response to immunotherapies, and association with specific HLA alleles (1, 8–10). Thus, implying that non-thymoma AChR-MG and MuSK-MG may have different underlying disease mechanisms and genetics, despite broadly similar presentations.
AChR-MG and MuSK-MG are among the few autoimmune diseases for which the molecular immunopathology is well understood. The autoantibodies in AChR-MG, which are primarily IgG1, lead to the loss of AChR through internalization and localized complement-mediated postsynaptic tissue damage (11, 12). The majority of autoantibodies that recognize MuSK are from the IgG4 subclass. They inhibit the agrin-LRP4-MuSK pathway by preventing agrin-activated LRP4 binding to MuSK, leading to dispersal of the AChRs (13, 14). Transfer (both active and passive) of these autoantibodies from humans to animal models affect the disease, demonstrating the direct role these molecules play in its pathology (15–18). While the role of autoantibodies in both AChR-MG and MuSK-MG pathologies are clear, the immune dysfunction that precedes their production is less well understood.
The role of the thymus in the immunopathology of many AChR MG patients is conspicuous (19), this is further highlighted by a recent clinical trial demonstrating the clinical benefit following thymectomy (20). The thymus in a subset of MG patients includes (19) AChR expression by thymic epithelial cells and myoid cells, the presence of proinflammatory cytokines, and defective regulatory T cells. B cells often organize in the hyperplastic thymus within tertiary lymphoid organs, frequently exemplifying many characteristics of germinal centers. To better define the role of B cells in MG, a number of studies have focused on those which characteristically populate the thymus. B cells populating the hyperplastic thymus express markers of activation and display functional signs of activation (21). Limited-throughput Sanger-based sequencing and B cell immortalization studies have shown that the B cells resident in these MG thymi are broadly clonally heterogeneous; lacking a dominant clone(s) among the infiltrate (22, 23). They harbor the characteristics of antigen experience, including somatic hypermutation (24–26) and biased variable region gene segment usage, which include over-representation of the VH3 family at the expense of VH4 genes (22). Importantly, among these B cells are plasmablasts and plasma cells that produce autoantibodies directed toward AChR. However, they represent a minor fraction of the total B cell infiltrate, thus do not appear to be highly enriched (27, 28). While these studies focusing on the thymic tissue remain seminal, they are not applicable to the broader MG population given that the thymus of many AChR-MG patients is not hyperplastic and therefore contains few, if any, disease-associated B cells. Furthermore, the thymus tissue of MuSK-MG patients is consistently normal in morphology and lacks a B cell infiltrate (29). Therefore, to further profile and expand the evaluation of the MG B cell repertoire, sequencing studies of other compartments, namely the periphery, are needed.
Deep sequencing allows for the comprehensive evaluation of the B cell receptor (BCR) repertoire properties in health and disease and provides the depth necessary to adequately depict the circulating peripheral repertoire, which includes up to 1011 B cells in humans (30). We applied this approach to explore both the naïve and memory B cell compartments in AChR-MG and MuSK-MG patients and compared their features to healthy donors (HD). We focused our study on determining whether the AChR-MG or MuSK-MG repertoires included abnormal clonal expansions, skewed gene usage and distinctive antigen binding region properties.
Materials and Methods
Study subjects and specimens
The study was approved by the Human Research Protection Program at Yale School of Medicine. Peripheral blood was obtained from HD and MG subjects after acquiring informed consent. MG patients were recruited from the Yale Myasthenia Gravis Clinic, the University of Kansas, or the University of Rochester. Study subjects were selected based on their clear diagnosis of MG, conspicuous symptoms, the absence of immunotherapy or other treatment (AChR) and, in the case of MuSK-MG, the absence of aggressive immunotherapy. All of the MG patients harbored MG autoantibodies, and had clinical and electrodiagnostic features consistent with MG. Collected clinical data included demographics, duration of disease, immunosuppressive or other medications, AChR autoantibody titer, thymectomy status/thymus pathology, status of acute or chronic infections and Myasthenia Gravis Foundation of America (MGFA) Clinical Classification (Table 1). HDs were matched to MG patients as closely as possible with regard to age and gender. HDs reported no history of autoimmune disease or malignancies and no acute or chronic infections.
Table 1.
Study subject demographics and clinical status.
| Subject ID | Age (yrs) | Gender | Autoantibody status (titera) | Disease duration (yrs) | Diagnosis/Disease status (MGFA) | Thymus Status | Symptoms/signs of infectionc | Treatment |
|---|---|---|---|---|---|---|---|---|
| AR02 | 61 | M | anti-AChR+ (2.54) | <1 | IIIa | No thymoma, No thymectomy | none | None |
| AR03 | 18 | M | anti-AChR+ (0.09) | 2 | I | No thymoma, No thymectomy | none | None |
| AR04 | 13 | F | anti-AChR+ (NA) b | <1 | I | No thymoma, No thymectomy | none | None |
| AR05 | 67 | M | anti-AChR+ (35.2) | <1 | I | No thymoma, No thymectomy | none | None |
| none | ||||||||
| MK02 | 28 | F | anti-MuSK+ | 8 | IIIb | No thymoma, No thymectomy | none | Pred 15 |
| MK03 | 51 | M | anti-MuSK+ | 27 | IIIb | No thymoma, No thymectomy | none | Pyridostigmine, Pred 20 |
| MK04 | 54 | F | anti-MuSK+ | 19 | IIb | No thymoma, No thymectomy | none | None |
| MK05 | 38 | F | anti-MuSK+ | 8 | I | No thymoma, No thymectomy | none | None |
| MK08 | 33 | F | anti-MuSK+ | 8 | IIb | No thymoma, No thymectomy | none | Pyridostigmine, Pred 10, PEd |
| none | ||||||||
| HD07 | 33 | M | NA | NA | NA | NA | none | NA |
| HD09 | 31 | M | NA | NA | NA | NA | none | NA |
| HD10 | 31 | M | NA | NA | NA | NA | none | NA |
| HD13 | 51 | F | NA | NA | NA | NA | none | NA |
Pred, prednisolone (dosage in mg); PE, plasma exchange. All data presented in the table correspond with the time of sample acquisition, unless otherwise indicated.
Titers (nmol/L) were measured by RIA for AChR only, not for MuSK. Values above 0.02nmol/L are positive.
The specimen derived from subject AR04 at the time of the study tested positive for AChR autoantibodies in a clustered AChR cell-based assay.
Signs of infection were evaluated by physical exam/vital signs and through review of systems.
Subject MK08 had received plasma exchange treatment 2 weeks prior to the specimen collection.
Cell staining and sorting
B cells subsets were isolated using flow cytometry (Fig. S1A). Briefly, peripheral B cells from MG patients and healthy donors were first enriched to >90–95% by positive selection using CD20 magnetic beads (Milteny). The cells were then stained with, LIVE/DEAD™ Fixable Near-IR Dead Cell Stain (ThermoFisher) to evaluate viability; samples <85% viable were not included in the study. Enriched B cells were then stained with perCP-Cy5.5 anti–human CD27 (clone O323), Pacific Blue anti-human CD19 (clone HIB19) (both from Biolegend). Naïve (CD19+, CD27−) or memory B cells (CD19+, CD27+) were then sorted on a FACSAria flow cytometer (BD) into PCR tubes and immediately frozen on dry ice.
Library preparation and BCR sequencing
RNA (250 ng) was reverse-transcribed into cDNA using a biotinylated oligo dT primer. An adaptor sequence was added to the 3′ end of all cDNA, which contains a universal priming site and a 17-nucleotide unique molecular identifier (UMI). Products were purified using streptavidin-coated magnetic beads followed by a primary PCR reaction using a pool of primers targeting the IGHA, IGHD, IGHE, IGHG, IGHM, IGKC and IGLC regions, as well as a primer against the universal adapter sequence. The immunoglobulin-specific primers contained tail sequences for later Illumina adaptor addition. PCR products were then purified using AMPure XP beads. A secondary PCR was then performed to add the Illumina P5 adaptor to the constant region end and a sample-indexed P7 adaptor to the universal adaptor end. The number of secondary PCR cycles was tailored to each sample to avoid entering plateau phase, as judged by a prior quantitative PCR analysis. Final products were purified, quantified with a TapeStation (Agilent Genomics) and pooled in equimolar proportions, followed by high-throughput 2x300 base-pair paired-end sequencing with a 20% PhiX spike on the Illumina MiSeq platform according to manufacturer’s recommendations, except for performing 325 cycles for read 1 and 275 cycles for read 2. Sequencing data was deposited in the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under BioProject accession PRJNA338795; sequencing runs used are denoted A79HP, AAYHL and AB8KB.
Raw read quality control and assembly
Raw reads were filtered in several steps to identify and remove low quality sequences. Conservative thresholds were applied in all cases, to increase the reliability of mutation calls and clonal assignments, at the potential expense of excluding some genuine variants. Preprocessing was carried out using pRESTO v0.5.0 (http://presto.readthedocs.io) (31), as follows.
Reads with a mean Phred quality score below 20 were removed.
Reads without valid constant region primer or template switch sequences were removed, with a maximum primer match error rate of 0.2 and a maximum template switch error rate of 0.5. Both template switch additions and constant region primer sequences were deleted from the reads. A preliminary isotype assignment for each read was made according to its constant region primer match.
Reads with identical unique molecule identifiers (UMIs) were collapsed into a single consensus sequence for each UMI. UMI read groups with error scores exceeding 0.1 or majority isotype frequency under 0.6 were discarded. In cases where multiple isotypes were identified in a single UMI read group, the consensus sequence was based only upon the subset of reads in the UMI read group assigned to the majority isotype.
UMI consensus sequence mate-pairs were first assembled de novo into full length Ig sequences with a minimum allowed overlap length of 8 base pairs, a maximum error rate of 0.3, and p-value threshold of 1x10−5. UMI mate-pairs failing de novo assembly (eg, due to long CDR3 regions preventing sequence overlap) were assembled by alignment against the August 28th, 2014 IMGT V-segment germlines (32) with a minimum allowed identity of 0.5 and a E-value threshold of 1x10−5.
A secondary isotype assignment was made by local alignment of the tail (J segment end) of the UMI consensus sequences against a set of sequences expected to be upstream of the C region primer (sequences available on request) which were derived from the IMGT C region and J segment consensus reference sequences (32). Alignment was restricted to a 100 base pair window with a maximum allowed error rate of 0.4.
V(D)J gene annotation
Following preprocessing, V(D)J germline segments were assigned with IMGT/HighV-QUEST (http://imgt.org) (32) using the July 7, 2015 version of the IMGT gene database.
Additional quality control and clonal clustering
Post-processing of IMGT/HighV-QUEST output, additional quality control, and clonal clustering was performed using Change-O v0.3.0 (http://changeo.readthedocs.io) (33), Alakazam v0.2.3 (http://alakazam.readthedocs.io) (33), SHazaM v0.1.2 (http://shazam.readthedocs.io) (33), and custom scripts within the R statistical computing environment (34), as follows.
Non-functional sequences were removed from further analysis.
To remove sample contamination caused by multiplex indexing errors, sequences with the same UMI, V segment gene, J segment gene, and junction length were removed from all samples except one. Multi-sample sequences were assigned to the sample with the maximum number of reads for the given UMI read group.
Sequences having greater than 10% missing data (N characters) were discarded.
Duplicate V(D)J sequences were discarded, with the exception of retention of duplicate sequences derived from different biological samples, cell sorts, and/or which were assigned to different isotypes by internal C region alignment. Each sequence was assigned a total read count equal to the sum of member UMI counts contributing to the UMI consensus sequences and a preliminary mRNA copy number value based on the total number of UMIs having an identical sequence. The internal C region alignment was used for all further steps dependent upon isotype assignment.
Functional VH V(D)J sequences were assigned to clonal groups by first partitioning sequences based on common IGHV gene annotations, IGHJ gene annotations, and junction region lengths. IGHV and IGHJ gene annotations for each partition were determined by the union of ambiguous gene assignments within each junction length partition having at least one overlapping gene annotation amongst all gene assignments. Within these larger groups, sequences differing from one another by a distance of less than 0.25 within the junction region were defined as clones by single-linkage clustering. Distance was measured as the number of nucleotide differences weighted by a human single-nucleotide substitution model derived from the mutation model previously described (33, 35), and normalized by the length of the junction region. The clonal distance threshold was determined by manual inspection to identify the local minima between the two modes of the within-sample bimodal distance-to-nearest histogram (36).
Full length germline sequences were reconstructed for each clonal cluster with D segment and N/P regions masked (replaced with Ns), with any ambiguous gene assignments within clonal groups resolved by majority rule.
Sequences with inconsistent V segment, J segment and/or isotype assignments were removed from further analysis (e.g., IGLV paired with IGKJ).
To remove chimeric sequences, sequences having more than 5 mismatches from the germline reference in any 10 base pair window were discarded.
Artificial diversity introduced by errors in UMI sequences was corrected using two stages of single-linkage hierarchical clustering with a Hamming distance threshold of 4/17 base pairs. First, the UMI sequences within sets of identical V(D)J sequences, as determined during the duplicate removal process described above, were clustered to correct inflated mRNA copy numbers. A consensus UMI sequence was determined by majority rule for each UMI cluster, and the mRNA copy number was corrected to the number of UMI clusters for the given sequence. In the second stage of correction, unique variants were removed from clonal clusters which bore similar consensus UMI and V(D)J sequences. The consensus UMIs for sequences sets belonging to the same cell sort and VH clonal cluster, determined as described above, were clustered using the same approach as the first stage. For each consensus UMI cluster within a clonal group only the sequence variant with the maximum read count was retained. Second stage correction for the VL sequences was identical to VH sequences, except that correction was performed within groups having the same cell sort, V segment gene, J segment gene, and junction length, as clonal clustering was not performed for VL sequences.
All unique sequences that were not represented by at least two reads were removed from further analysis, where the two read threshold was either two reads within a single UMI read group or two distinct UMIs, using the UMI and read count corrections described above.
BCR repertoire analysis: exclusion criteria and sequence filtering
To reduce artifacts and noise due to sample quality and numerical discreteness, all samples having less than 1,900 unique VH sequences were excluded from the final analyses. The 1,900 sequence threshold was chosen on the basis of no observed correlation between sequence count and relative clonal abundance. All analyses were performed on the set of unique sequences (with duplicates removed) within each sample, allowing retention of duplicate sequences represented by more than one isotype. UMI copy numbers, estimating the number of mRNA templates, were not considered during abundance analyses, including clonal diversity and gene usage analyses.
BCR repertoire analysis: clonal abundance and diversity
Repertoire diversity analysis was performed using Alakazam (33). Initial relative clonal abundance, prior to inference of the complete clonal abundance distribution, was calculated as the number of unique clonal variants within a clonal cluster divided by the total number of unique variants for the given patient. A minimum bound on the estimate of unseen clones was determined using the Chao1 estimator (37). The complete clonal abundance distribution was inferred using the one-parameter variant of the Chao et al, 2015 method (38), with confidence intervals derived via bootstrapping (resampling with replacement) from the inferred complete distribution using 2,000 realizations. Repertoire diversity and evenness was characterized using the generalized diversity index proposed by Hill (39). To allow comparison of all subjects on the same scale regardless of sequencing yield, variability in sequencing depth was accounted for using rarefaction to a uniform sequence count of 1,959 for each sample, with 2,000 realizations of repeated random sampling with replacement from the inferred complete clonal abundance. The diversity index for each subject was determined from the mean value over all resampling realizations, with a 95% confidence interval about the diversity estimate derived via Z score using the bootstrap distribution variance. Evenness was calculated at diversity order q=4 as 4D/0D, which heavily up-weights abundant clones.
BCR repertoire analysis: V gene analysis
Individual genotypes were inferred using TIgGER v0.2.3 (http://tigger.readthedocs.io) (33, 40). V gene family usage was calculated as the percentage of unique sequences assigned to a given gene family within the total number of sequences for each isotype in each cellular compartment. V gene families IGHV6, IGHV7, IGKV5 through IGKV7, and IGLV4 through IGLV10 were excluded from gene usage analysis due to low total abundance. The Repertoire Dissimilarity Index (RDI) for each set of samples was calculated as previously described (41), with subsampling and 2,000 bootstrap realizations, except that analysis was performed at the level of V gene family and the arcsinh transformation on abundance was excluded. V-J gene proximity was determined by assigning the relative position of each V and J gene using the IGH, IGL and IGK loci annotations from IMGT (42) coupled with additional reported data (43, 44). Positions were fractionally ranked from most 5′ V gene (position 0.0) to the most 3′ J gene (position 1.0), excluding from consideration any non-V/ J genes and any V genes annotated as pseudogenes or ORFs.
BCR repertoire analysis: mutation and selection analysis
Mutational frequencies were calculated as the number of mismatches from the germline sequence, excluding the N/P and D regions, using SHazaM. Lineage trees were constructed for each clone using PHYLIP v3.69 (45) and Alakazam (33, 46). The selection strength of expanded memory compartment lineages was quantified with the BASELINe (47) implementation provided in SHazaM, using the focused test statistic. Each clonal lineage was represented by the most recent common ancestory (MRCA) of the lineage, and the probability density function (PDF) for each status (HD, AChR-MG, MuSK-MG) was determined by unweighted convolution of the PDFs for each subject in the status group. Selection analysis was restricted to lineages represented by at least 3 unique observed sequences. The MRCA of each lineage was defined as the member of the lineage having the minimum mutational distance from germline; inferred intermediates were allowed as MRCA sequences. For analysis of selection by physicochemical property, a replacement mutation was defined as a mutation that introduces a change in amino acid property class, according to the classes recommended by IMGT (http://www.imgt.org/IMGTeducation/Aide-memoire/_UK/aminoacids/IMGTclasses.html).
BCR repertoire analysis: somatic hypermutation profile analysis
Somatic hypermutation profiles in HD, AChR-MG and MuSK-MG subjects were analyzed using SHazaM using, separately, memory IGH sequences with IgM, IgG and IgA combined, memory IGK sequences, and memory IGL sequences. To reduce bias from clonal expansion, only one representative sequence was analyzed from each VH clonal group, with the representative sequence defined as the sequence with the highest mutational load within the clonal group. Using the synonymous, 5-mer, functional criteria (S5F) (35), mutability and substitution rates of 5-mer motifs were estimated for individual subjects. Mutability of each 5-mer was directly measured from the data when at least 200 mutations were observed for a given 5-mer. Comparisons between individual subjects were performed via pairwise Pearson correlation of 5-mer mutability scores between individual subjects. Similarly, comparisons between status groups were performed via pairwise Pearson correlation of the average 5-mer mutability scores for all subjects in a given status. For each pairwise comparison, the set of 5-mers used was restricted to those that were directly measured (non-inferred) in the subjects or statuses under comparison, with re-normalization of mutability scores following removal of non-intersecting 5-mers. To confirm comparison results between disease status groups, a background distribution of correlations between statuses was generated by performing all possible 35 permutations of the status labels of the subjects and recalculating the average mutability for each status using the same approach described above. The percentile method was then used to determine the statistical significance of the observed correlations, relative to the background distribution.
BCR repertoire analysis: H-CDR3 convergence and physicochemical property analysis
Public heavy chain complementarity determined region 3 (H-CDR3) sequences were identified by clustering identical length H-CDR3 amino acid sequences using the uclust algorithm (48) with a 90% identity threshold. Clustering was performed on the combined memory and naïve population, with public H-CDR3 sequences defined as sequences that clustered with at least one sequence observed in a different subject. Repertoire properties, including gene usage and H-CDR3 physicochemical properties, were calculated using Alakazam. The H-CDR3 grand average of hydrophobicity (GRAVY) was calculating using the Kyte & Doolittle scale (49), net charge was calculated using the EMBOSS pK values (http://emboss.sourceforge.net/apps/cvs/emboss/apps/iep.html) and Henderson-Hasselbalch equation at pH 7.4 (50), average bulkiness was calculated using the Zimmerman scale (51), average polarity was calculated using the Grantham scale (52), and the aliphatic index was calculated using the Ikai method (53) normalized by the number of informative (non-missing) positions. Acidic (Asp or Glu), basic (Arg, His or Lys) and aromatic content (His, Phe, Trp or Tyr) was calculated as the fraction of informative (non-missing) residues in the H-CDR3.
Statistical tests
Statistical tests were performed using ANOVA to determined significance, followed by one-tailed t-tests for determination of pairwise p-values. All confidence intervals are 95% confidence intervals.
Results
We performed deep sequencing of the B cell receptor repertoire in AChR-MG (n=4), MuSK-MG (n=5), and HD (n=4) subjects (Table 1). None of the MG subjects had undergone significant immunotherapy treatment, and three of the subjects (AR02, AR03 and MK02) were part of our earlier study (54), which identified B cell tolerance checkpoint defects. Sequencing of IGH, IGK and IGL chains from sorted naïve and memory B cells from each subject produced a total of 22 million raw reads. After quality control and processing, a high-fidelity data set was generated that consisted of 205,540 unique error-corrected IgM, IgG and IgA sequences and 311,979 unique error-corrected IgL and IgK sequences (Table 2). Within each subject, approximately half of the memory repertoire was IgM (49%–57%), with the remaining sequences composed of IgG (13–15%) and IgA (22–30%) (Fig. S1B). IGL made up 34–40% of the naïve compartment and 36–41% of the memory compartment (Fig. S1C), with no significant difference observed between MG and HD subjects.
Table 2.
Summary of sequencing and sequence processing results.
| Subject ID | Cells | Raw reads | Final analyzed naïve sequencesa | Final analyzed memory sequencesa | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Naïve | Memory | Naïve | Memory | IgM | IgK | IgL | IgM | IgG | IgA | IgK | IgL | |
| AR02 | 143K | 66K | 939,295 | 918,374 | 3,739 | 3,382 | 2,481 | 2,794 | 1,539 | 2,990 | 10,139 | 7,501 |
| AR03 | 154K | 12K | 921,768 | 770,573 | 3,194 | 1,713 | 1,099 | 368b | 253b | 366b | 1,032b | 1,124b |
| AR04 | 324K | 39K | 995,808 | 541,605 | 47,577 | 19,034 | 11,417 | 4,216 | 1,689 | 2,290 | 9,003 | 6,340 |
| AR05 | 140K | 77K | 741,561 | 743,956 | 30,648 | 16,579 | 11,008 | 13,502 | 1,485 | 4,234 | 19,022 | 11,681 |
| MK02 | 57K | 50K | 894,106 | 787,754 | 427b | 327b | 162b | 6,479 | 927 | 888 | 6,753 | 3,413 |
| MK03 | 151K | 126K | 1,076,632 | 810,841 | 1,685 | 3,806 | 1,307 | 969 | 293 | 697 | 7,341 | 3,353 |
| MK04 | 59K | 7K | 958,717 | 398,324 | 8,123 | 5,180 | 3,101 | 543b | 105b | 257b | 1,056b | 687b |
| MK05 | 410K | 192K | 1,162,652 | 1,048,316 | 8,949 | 11,100 | 6,148 | 4,296 | 1,146 | 1,954 | 23,701 | 12,297 |
| MK08 | 50K | 25K | 916,527 | 898,976 | 3,236 | 2,115 | 1,579 | 864 | 436 | 1,609 | 3,390 | 3,110 |
| HD07 | 45K | 16K | 814,996 | 725,940 | 830b | 838b | 604b | 3,004 | 586 | 1,891 | 4,937 | 3,737 |
| HD09 | 171K | 38K | 1,332,467 | 535,370 | 14,079 | 14,124 | 7,304 | 2,167 | 501 | 954 | 9,021 | 4,616 |
| HD10 | 80K | 57K | 1,080,339 | 774,156 | 4,513 | 3,878 | 2,612 | 3,101 | 548 | 446 | 5,926 | 4,723 |
| HD13 | 89K | 21K | 933,799 | 401,302 | 6,262 | 7,732 | 3,224 | 2,552 | 1,223 | 1,265 | 7,467 | 4,585 |
Refers to the count of unique, error-corrected sequences which passed all quality control and filtering steps.
Excluded from analysis due to low yield of unique VH sequences.
MG patients do not exhibit a public H-CDR3 signature
Given the common antigen target in the MG subtypes (AChR-MG or MuSK-MG), we hypothesized that the subjects’ repertoires would contain evidence of convergent H-CDR3 sequences, which have been observed during infection and vaccination responses (55, 56). Clustering of identical length H-CDR3 amino acid sequences identified a small number of these convergent, or “public”, H-CDR3 clusters across subjects (Fig. S2A). The total abundance of H-CDR3 clusters public to AChR-MG and/or MuSK-MG subjects was a small fraction of the total H-CDR3 clusters (0.27%) (Fig. S2B). When considering H-CDR3 clusters exclusively public across HD and MG subjects, as an estimate of the expected public background, we observed a similarly low abundance of public clusters (0.22%) (Fig. S2C). Furthermore, in pairwise comparisons between subjects the average frequency of public H-CDR3 clusters was similar in MG (0.32% for AChR-MG and 0.30% for MuSK-MG) and HD (0.33%) subjects. Public sequences were dominated by short H-CDR3s, with an average length of 11.8 amino acids in public H-CDR3s that were observed exclusively in MG (Fig. S2B), and an average length of 10.8 amino acids for public H-CDR3s shared between MG and HD (Fig. S2C). We also investigated whether IGHV4-34 sequences of MG patients showed enrichment for the FWR1 hydrophobic patch characteristic of autoreactive 9G4+ antibodies in systemic lupus erythematosus (SLE) (57), but found no such signature (data not shown). Thus, we found no compelling evidence for enrichment of convergent H-CDR3s in the peripheral naïve and memory B cell populations derived from MG patients.
Clonality of the MG memory compartment is similar to that of healthy repertoires
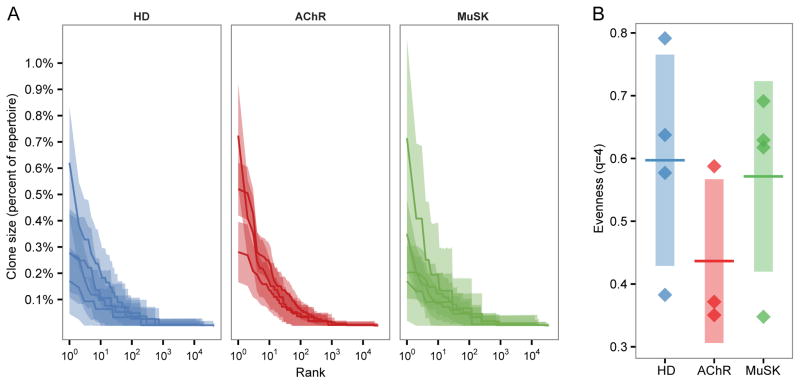
To test whether the B cell repertoires in MG patients were characterized by large clonal expansions, sequences were clustered into clonal groups, with each member of the group representing one unique variant (including variants that differed only by isotype). Relative clonal abundance distributions were calculated as previously described (38). Expanded clones were identified in all MG patients, but even the most abundant clones were less than 1% of the repertoire, which is consistent with previous observations of peripheral blood in the absence of an active immune response (58). Although a slight upward trend in clone sizes of AChR-MG was observed, this increase was, at most, 0.1% of the subjects’ repertoires (Fig. 1A). To assess the statistical significance of this trend, we compared the evenness of the HD and MG subjects. Evenness quantifies the extent to which an abundance distribution deviates from a uniform distribution, with a lower evenness indicating a less uniform distribution. As the shift in clonal abundance was most prevalent in the highly abundant clones, evenness scores were compared using the 4th order Hill diversity index (39) (q=4), which strongly up-weights the contribution of abundant clones to the evenness score (Fig. 1B). While a large decrease in evenness of the AChR-MG repertoires compared to HD was observed, implying greater clonal dominance, this shift was not statistically significant (AChR-MG vs HD t-test p=0.10; MuSK-MG vs HD t-test p=0.38). Overall, no significant evidence was found to support the idea that the peripheral memory compartment of MG patients is more oligoclonal than healthy donors.
Fig. 1. Memory compartment clonal abundance is comparable between healthy and MG repertoires.
(A) The rank-abundance distribution of memory compartment heavy chain clones with clone size (y-axis) as a percent of the repertoire against the size rank of the clone on a log10 scale (x-axis). Each dark line represents the estimated clonal abundance curve for a single subject, with shaded areas representing 95% confidence intervals derived via bootstrap (2,000 realizations). Subject status is denoted by HD, AChR (AChR-MG), and MuSK (MuSK-MG).
(B) Evenness at diversity order q=4 of the heavy chain memory compartment clone size distributions. Each point represents the estimated evenness score for a subject from the clonal abundance distributions in panel A. The vertical shading represents the standard deviation of the mean evenness scores and the horizontal bar represents the mean of the mean evenness scores. HD vs AChR-MG t-test p=0.100; HD vs MuSK-MG t-test p=0.382.
To assess ongoing recruitment of B cells into the memory compartment, we identified clones spanning the memory and naïve compartments. Between 0.3% and 3.9% of memory clones contained at least one member observed in the naïve compartment (Fig. S2D). The frequency of clones spanning the memory and naïve compartments was not significantly related to disease status, but was strongly dependent upon the total yield of unique sequences (Pearson R=0.78). We also observed a low percentage of clones that contained multiple isotypes (either IgM and IgG, IgM and IgA, or IgG and IgA) with between 0.5% and 3.0% of clones containing sequences with multiple isotypes (Fig. S2E). The frequency of clones containing multiple isotypes was also a highly sequencing depth-dependent effect (Pearson R=0.77). Overall, we found no evidence that circulating MG repertoires were biased due to active clonal expansion or transition of B cells from the naïve to memory compartments.
The MG repertoire exhibits biases in V segment usage
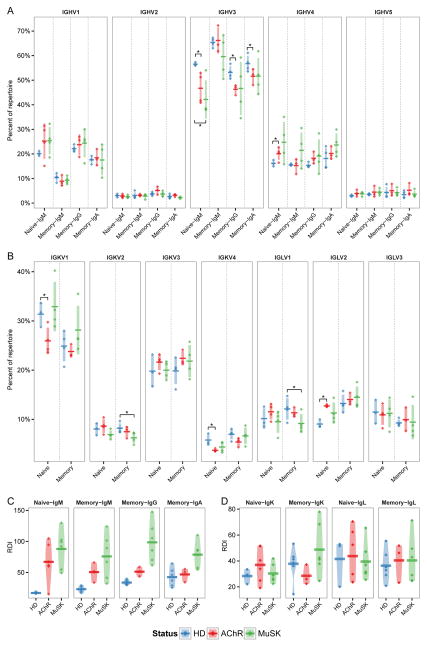
Previous studies have demonstrated V segment usage biases in several autoimmune diseases (57, 59). To determine whether similar V segment biases are also present in MG patients, we compared the relative abundance and inter-status variability of each V gene family between MG and HD subjects. In the naïve compartment of MG subjects, significant shifts were observed in the usage of IGHV3 and IGHV4 relative to HDs (ANOVA p=0.021) (Fig. 2A). Specifically, IGHV4 usage increased from 16.2% in HDs to 20.1% in AChR-MG (t-test p=0.027), while IGHV3 usage decreased from 56.5% in HDs to 46.7% in AChR-MG (t-test p=0.026) and 42.2% in MuSK-MG (t-test p=0.018). Although not statistically significant, an increase in IGHV4 was also observed in MuSK-MG (t-test p=0.067), as well as an increase in IGHV1 usage in both AChR-MG and MuSK-MG. Significant biases in V family usage of the light chains in the naïve compartment of AChR-MG were also observed (ANOVA p=0.016) (Fig. 2B). Specifically, IGLV2 usage increased from 9.0% in HD to 12.7% in AChR-MG (t-test p=0.006), IGKV1 usage decreased from 31.4% to 25.9% (t-test p=0.021), and IGKV4 decreased from 5.8% to 3.7% (t-test p=0.043). The same V family biases observed in naïve AChR-MG repertoires were apparent in the class-switched memory populations, particularly in the case of reduced IGHV3 usage for both IgG (p=0.006) and IgA (p=0.052). In the memory compartment we observed significant IGKV/IGLV shifts in MuSK-MG only, with a decrease in IGKV2 usage from 8.20% to 6.26% (p=0.023) and IGLV1 usage from 12.1% to 9.15% (p=0.048). Due to variation in the V segment genotypes of the patients, individual V gene usage was not comparable for all patients across all genes (data not shown). However, increases in IGHV1-69, IGHV4-34 and IGLV2-14 abundance comprised a large share of the naïve compartment biases observed in MG patients (data not shown). Overall, these results show that the MG B cell repertoire is abnormal, having biased V segment usage in both the VH and VL.
Fig. 2. V family usage is skewed in MG repertoires.
(A) Heavy chain V family usage for the naïve (Naïve-IgM) and memory (Memory-IgM, Memory-IgG and Memory-IgA) compartments. Usage is shown as a percent of the total unique IGHV sequences (y-axis) for HD, AChR-MG (AChR), and MuSK-MG (MuSK). Horizontal bars indicate the mean abundance over all subjects of a given status and vertical shading indicates +/− one standard deviation about the mean. Significance is denoted by: * (p <= 0.5).
(B) Light chain V family usage for the naïve and memory compartments. Usage is shown as a percent of the combined unique IGLV and IGLK sequences (y-axis). Significance is denoted by: * (p <= 0.5).
(C–D) Repertoire Dissimilarity Index (RDI) for naïve (Naïve-IgM, Naïve-IgK and Naïve-IgL) and memory (Memory-IgM, Memory-IgG, Memory-IgA, Memory-IgK, Memory-IgL) V families in (A) and (B). Each point indicates a pairwise RDI score (y-axis) for two subjects, with the mean score for each set of subjects marked by a horizontal bar and a density estimate indicated by the shaded region. RDI scores are grouped by within status comparisons for HD, AChR-MG (AChR), and MuSK-MG (MuSK).
The IGHV usage biases in the naïve compartment of AChR-MG and MuSK-MG subjects appeared to be associated with an increase in variability across subjects. To quantify this effect, we compared V family variability using the Repertoire Dissimilarity Index (RDI) (41). RDI scores the aggregate difference in gene usage between any pair of samples, providing a measure of how dissimilar two gene usage distribution are from one another. We observed a significant increase in IGHV RDI within both the naïve (ANOVA p=0.014) and memory (ANOVA p=6.7x10−8) compartments of AChR-MG and MuSK-MG repertoires, with the most pronounced difference in the naïve compartment (mean HD RDI=16.9, AChR-MG RDI=67.0, MuSK-MG RDI=87.9) (Fig. 2C). In contrast to IGHV usage, no significant differences in the variability of IGKV or IGLV usage across subjects was found in the naïve AChR-MG or MuSK-MG repertoires (Fig. 2D). Overall, these results show that the naïve MG repertoire is abnormal, having biased V segment usage in both the VH and VL, which are mirrored in the class-switched memory compartment. In addition, MG repertoires display considerably more individual variability within the naïve compartment compared to HD repertoires, suggesting B cell developmental abnormalities in MG may be partially patient-specific.
The MuSK-MG repertoire displays increased proximity of recombined IGLV-IGLJ genes
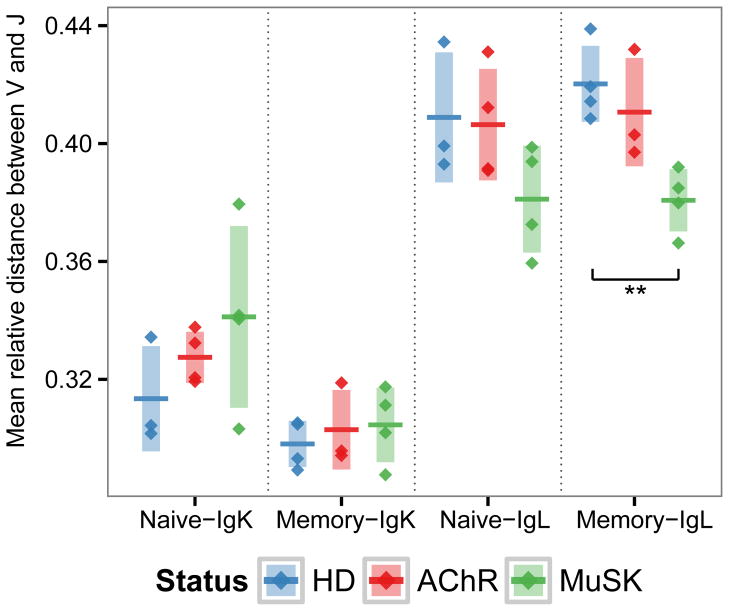
Light chain receptor editing is a primary mechanism by which self-reactive B cells are removed from the naïve B cell population (60, 61). In this process, a secondary V-J rearrangement may occur at either the original locus, the alternate chromosome, or through a kappa to lambda chain switch. Editing results in increased genomic distance between V-J, as secondary rearrangements at the same locus leads to utilization of V-J genes with greater intervening distance due to deletion of the original V and/or J genes. To investigate whether receptor editing was dysregulated in MG patients, we computed the relative genomic distance between the V gene and J gene for each unique VL sequence in AChR-MG and MuSK-MG repertoires and compared these to HD repertoires. While no significant differences were found in AChR-MG repertoires, IGLV-IGLJ pair distances in MuSK-MG were reduced within the memory and naïve compartments, although this was statistically significant only for memory (0.42 to 0.38 with t-test p=0.002 for memory, and 0.41 to 0.38 with t-test p=0.079 for naïve) (Fig. 3). While the shift in the naïve compartment of MuSK-MG was not statistically significant, we note that the mean IGLV-IGLJ distance in the naïve and memory compartments of MuSK-MG subjects were equivalent (0.38 naive vs. 0.38 memory). Similar to AChR-MG, no significant difference in IGKV-IGKJ distance in MuSK-MG subjects was observed. Thus, MuSK-MG, but not AChR-MG, subjects exhibit reduced IGLV-IGLJ genomic distance consistent with reduction in secondary rearrangement events at the IGL locus.
Fig. 3. Recombined lambda chains of MuSK-MG repertoires show reduced IGLV-IGLJ genomic distance.
The mean relative genomic distance between the V and J genes of kappa (IGK) and lambda (IGL) light chain sequences is shown for the naïve and memory compartments of HD, AChR-MG (AChR), and MuSK-MG (MuSK). Relative genomic distance is shown as a fractional rank position (y-axis) difference between the J gene and V gene. Significance is denoted by: * (p <= 0.5), ** (p <= 0.005).
Mutational profiles in MG are similar to that of healthy repertoires
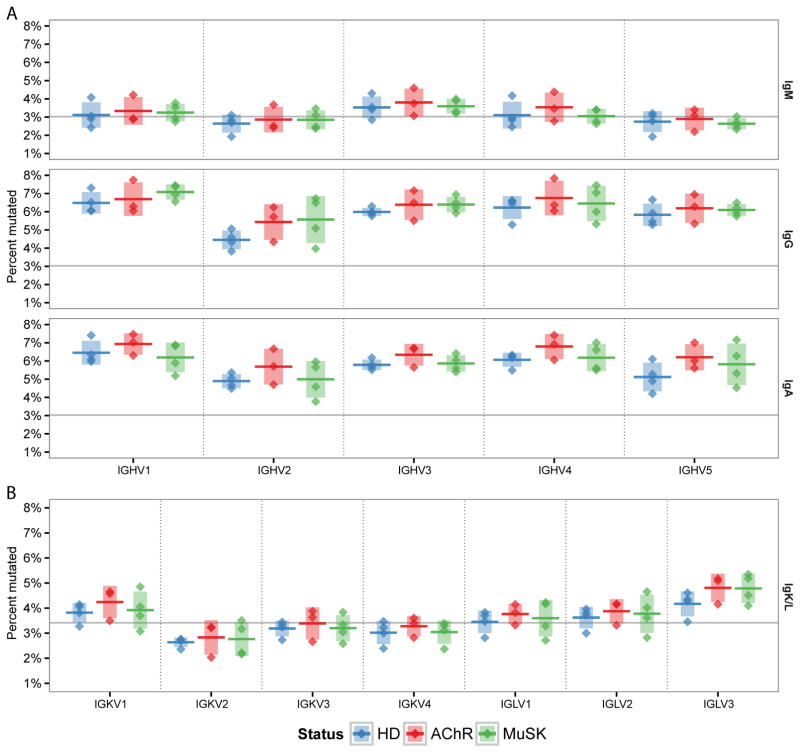
MG autoantibodies are class-switched and contain frequent somatic hypermutations (SHMs) (22, 62). To determine whether SHM was perturbed in MG, we analyzed somatic mutation patterns in the naïve and memory compartments of IGHV, IGLV and IGKV gene sequences. SHM exhibits well-known biases towards particular DNA motifs, leading to mutational hot-spots (such as WRC) and cold-spots (such as SYC), that are dependent upon the microsequence context (35). The intrinsic context biases of SHM, combined with differing sequence compositions of V gene families, leads to significant differences in the average mutational load across families (Fig. 4). In HD subjects, mutational loads ranged from 2.6% (IGHV2) to 3.5% (IGHV3) in IgM, 4.5% (IGHV2) to 6.5% (IGHV1) in IgG, 4.9% (IGHV2) to 6.5% (IGHV1) in IgA, and 2.6% (IGKV2) to 4.2% (IGLV3) in the VL. The mutational loads in MG subjects exhibited similar family-dependent mutational loads and, within each V family, there were no significant differences between AChR-MG, MuSK-MG and HD subjects (Fig. 4).
Fig. 4. Memory compartment mutational load is similar between healthy and MG repertoires.
Distribution of mean mutation frequency for heavy chain (A) and light chain (B) memory compartment sequences. Mutation frequency for each sequence was determined as the number of based changes from germline, excluding the N/P and D regions. Mean mutation frequency is shown for each chain, isotype and gene family. Horizontal bars indicate the mean of the mean mutation rates with the vertical shading indicating +/− one standard deviation about the mean of means.
To determine if the known SHM microsequence targeting biases were perturbed in MG subjects, and by implication the molecular mechanisms underlying SHM, we calculated SHM mutability profiles using a 5-mer context model (two nucleotides upstream and downstream of the mutated position) as previously described (35). Only synonymous mutations were used in the model to remove the influence of selection. We found that mutability profiles were highly consistent across all individuals (Pearson R 0.83 to 0.94, 0.67 to 0.95, and 0.58 to 0.93 for IGH, IGK, and IGL, respectively) (Figs. S3A, C, and E), and the AChR-MG, MuSK-MG and HD profiles all exhibited the well-known hot/cold-spot biases of SHM (Figs. S3B, D, and F). Furthermore, the full hierarchy of calculated mutabilities across DNA 5-mers was highly consistent across statuses (Pearson R 0.97 to 0.98, 0.91 to 0.96, and 0.97 to 0.98 for IGH, IGK, and IGL, respectively). No evidence was found that mutability profiles within a status groups (AChR-MG, MuSK-MG and HD) were more highly correlated to each other compared to another statuses (permutation test p-value > 0.1 for IGH, IGK, and IGL). Overall, both SHM targeting profiles and mutational loads in AChR-MG and MuSK-MG were highly consistent with those of HD subjects and previously reported models (35).
The AChR-MG repertoire exhibits decreased positive selection in CDRs
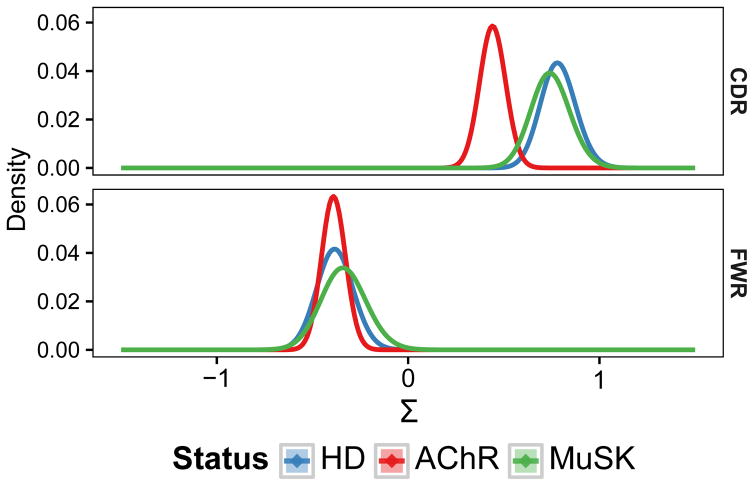
To determine if affinity maturation was perturbed in MG patients, we quantified selection strength (Σ) from the pattern of mutations in the V segment of expanded memory compartment lineages using BASELINe (47). This method compares the observed and expected frequency of non-synonymous mutations with positive Σ scores indicating an excess of non-synonymous mutations (positive selection) and negative Σ scores indicating fewer non-synonymous mutations than expected (negative selection). As the majority of positive selection events are shared among members of a B cell clone (and thus appear on the trunk of the lineage tree) (63), we quantified selection pressure on a per-lineage basis using the MRCA of each lineage as the representative sequence. The FWRs is critical to preserve the structure of the antibody receptor and, as expected, these regions showed significant evidence of negative selection in AChR-MG, MuSK-MG and HD subjects with no difference between status groups (Fig. 5). Furthermore, as expected, AChR-MG, MuSK-MG and HD subjects all exhibited significant positive selection in the CDRs, which generally contain antigen-contact residues. However, in this case, selection strength in AChR-MG was significantly reduced (Σ=0.44±0.13 in AChR-MG vs Σ=0.78±0.18 in HD) (Fig. 5). To determine whether the observed selection difference was due to a specific physicochemical property, we also used BASELINe to analyze the frequency of somatic mutations that led to changes in side chain hydrophobicity, charge, polarity, and volume classes. All of these properties individually displayed some degree of reduced positive selection in the CDRs of AChR-MG subjects, so that no single physicochemical property appeared primarily responsible for the decrease in selection pressure (Fig. S4). Taken together, these results suggest that while the underlying SHM process remains normal within MG subjects, the processes that guide antigen-driven selection (in the VH) are perturbed specifically in AChR-MG subjects.
Fig. 5. The AChR-MG memory compartment repertoire displays reduced selective pressure.
BASELINe PDFs are shown for HD, AChR-MG (AChR), and MuSK-MG (MuSK) repertoires, with density shown on the y-axis and the selection strength (Σ) shown on the x-axis. PDFs for each status were determined via convolution of the individual PDFs for subjects within each status group, resulting in a single aggregate PDF for each status.
The AChR-MG memory repertoire exhibits abnormal CDR3 physicochemical properties
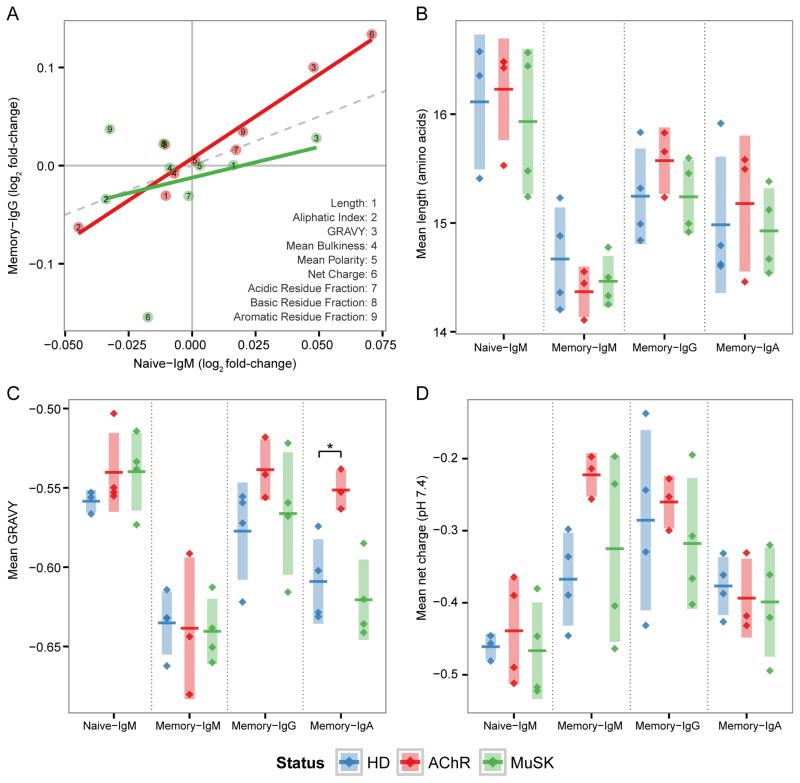
The H-CDR3 encodes the majority of B cell repertoire diversity, and contributes significantly to antigen specificity (64). Previous investigations into autoimmune B cell repertoires have observed an increase in the hydrophobicity (65), positive charge (57) or length (66) of the H-CDR3. To determine if such repertoire-scale biases in H-CDR3 properties were present in MG patients, we analyzed nine amino acid physicochemical properties: the length of the H-CDR3 region, the mean net charge, the grand average of hydrophobicity (GRAVY), mean side chain bulkiness, mean polarity, the aliphatic index, and the abundance of acidic, basic and aromatic residues. Consistent with previous studies (67, 68), significant differences were found between the naïve IgM, memory IgM and class-switched memory populations in HD subjects. Specifically, naïve B cells expressed H-CDR3s that were longer (ANOVA p=0.003) (Fig. 6B), more negatively charged (ANOVA p=0.188) (Fig. 6C), and had higher hydrophobicity (ANOVA p=0.020) (Fig. 6D). H-CDR3 properties of MuSK-MG subjects were similar to those of HDs. In contrast, AChR-MG subjects displayed significant abnormalities in the H-CDR3 GRAVY scores (ANOVA p=0.012). Specifically, the mean GRAVY score of the memory IgA repertoire increased 1.1-fold in AChR-MG compared to HD (from −0.609 to −0.552; t-test p=0.008) (Fig. 6D). Although not statistically significant, we note that the mean net charge and GRAVY scores in the AChR-MG memory IgM repertoire both displayed large fold increases compared to HDs (Fig. 6C). Thus, AChR-MG subjects exhibit repertoire-scale biases in H-CDR3 physicochemical properties across multiple B cell compartments.
Fig. 6. The AChR-MG memory compartment repertoire displays abnormal H-CDR3 physicochemical properties.
(A) Log2 fold-change of nine H-CDR3 physicochemical properties for AChR-MG (AChR) and MuSK-MG (MuSK), with respect to scores in HD. Shown are the mean scores for each property for both naïve (Naïve-IgM) and memory IgG (Memory-IgG) repertoires along with a linear fit for each status; correlation is significant only for AChR-MG (R2=0.93; p=3.06x10−5). (B) H-CDR3 mean length in amino acid residues, (C) GRAVY index, and (D) mean net charge at pH 7.4 for each subject is shown as a single point for both the naive (Naïve-IgM) and memory (Memory-IgM, Memory-IgG and Memory-IgA) compartments. Horizontal bars indicated the mean of the mean property scores with the vertical shading indicating +/− one standard deviation about the mean of means. Significance is denoted by: * (p <= 0.5).
Given the B cell tolerance checkpoint defects we previously identified in MG (54), we sought to assess the extent to which the observed H-CDR3 differences in the memory compartment of AChR-MG repertoires were driven by the naïve repertoire. We found that the physicochemical property biases observed in the class-switched memory compartment (AChR-MG vs. HD) were significantly correlated with changes that were already apparent in the naïve IgM compartment (R2=0.93; p=3.06x10−5). In general, the biases within the AChR-MG naïve compartment were further amplified within the class-switched memory compartment, with the largest fold-change biases displayed by net charge, GRAVY score, and the aliphatic index (Fig. 6A). While we also observed a correlation between naive IgM and memory IgG biases in MuSK-MG, this effect was small and not statistically significant (R2=0.08; p=0.469). Taken together these results show that AChR-MG, but not MuSK-MG repertoires, display aberrant H-CDR3 physicochemical properties, and that these biases are likely initiated in the naïve compartment.
Discussion
MG is a prototypical B cell-mediated autoimmune disease and holds historical significance as the first neurological disease shown to be mediated by autoantibodies. The study of MG has driven discoveries in other autoimmune diseases (69), and it remains both clinically and scientifically relevant. Understanding the shape of the B cell repertoire in MG may lead to more effective therapeutics, and may further serve to elucidate potential pathogenic mechanisms in other B cell-mediated autoimmune diseases. Through the use of high-throughput BCR sequencing we have, for the first time, described the naïve and memory B cell repertoire of both AChR-MG and MuSK-MG patients in high-resolution. A recent study by our group demonstrated that both AChR-MG and MuSK-MG fail to properly enforce B cell tolerance (54). Given that tolerance checkpoints counter-select a considerable fraction of developing B cells (61), one may reason that distinctive BCR repertoire characteristics would be conspicuous in a naïve B cell compartment that is shaped without proper tolerance regulation. We have uncovered defects that materialize in the absence of properly regulated tolerance checkpoints, revealed by marked differences from healthy controls in the naïve B cell repertoires of both AChR-MG and MuSK-MG subjects. Furthermore, we reasoned that an abnormally formed naïve compartment would produce similar abnormalities in the memory compartment which it supplies. Our test of this hypothesis, shown in this study, indicates that deformations in the naïve B cell compartment of AChR-MG and MuSK-MG are transferred to the memory compartment.
The naïve repertoire and defective B cell counter selection
We observed significant differences in IGHV gene family usage in MG patients, including an increase in IGHV4 family genes (Fig. 2A). Increases in IGHV4 usage have been previously implicated in several other autoimmune diseases, including SLE, multiple sclerosis and Wiskott-Aldrich syndrome (59, 70–72). A mechanistic explanation for the relationship between IGHV4-34 and SLE has been partially defined (57), but no clear explanation for the abundance of IGHV4 family genes in other autoimmune disease is known. In the case of MG, the biases observed in the class-switched memory population are already apparent in the naïve compartment. It is possible that the IGHV4 biases found in other autoimmune diseases similarly originate within the naïve compartment. Previous studies have observed an increase in IGKV4 usage in SLE, celiac disease and type 1 diabetes (73), but we did not observe this hallmark in MG light chain repertoires. In fact, the naïve repertoires of both AChR-MG and MuSK-MG subjects showed a decrease in IGKV4 usage compared to healthy controls.
Healthy naïve repertoires show highly consistent IGHV usage in naïve IgM+ cells in both our data and previous reports (41). In contrast, we found that naïve MG repertoires were highly variable between individuals (Fig. 2C). The increased variability in naïve MG repertoires implies a difference in selection – either the introduction of otherwise absent selection or the lack of appropriate selection. We also note that both H-CDR3 charge and hydrophobicity exhibited low variability in naïve HD repertoires, and that these properties showed higher variance in naïve MG repertoires, further implicating selective forces as the root of the disorderliness in naïve MG repertoires, given that nucleotide diversity in the V segment contributes only partially to H-CDR3 physicochemical character.
To evade the development of an immune response against self, central and peripheral tolerance mechanisms counter-select B cells during their development (61, 66). Deficiencies in the integrity of B cell tolerance mechanisms can be demonstrated in a number of autoimmune diseases, including SLE, rheumatoid arthritis, multiple sclerosis (74–76) and recently MG (54). Our sequencing data, derived from naïve IgM+ B cells, indicates that both AChR-MG and MuSK-MG subjects display characteristics in their naïve B cell repertoire that are conspicuously different from those observed in HDs. We suggest that these findings are consistent with, and may be reflective of, the reported inadequate counter-selection during B cell development in MG (54).
The memory compartment and BCR selection abnormalities
We examined the MG memory B cell repertoire for abnormalities in the abundance or character of B cell clones relative to the HDs. No statistically significant difference in clonal abundance was observed. The analysis of convergent H-CDR3 sequences specific to MG subjects revealed no evidence of public H-CDR3 sequences that could be considered a signature for either AChR-MG or MuSK-MG subjects. Mutational load and SHM hot- and cold-spot targeting preferences of MG repertoires were examined; neither mutational loads nor SHM targeting preferences were perturbed with respect to HDs.
Despite the lack of any significant bias in clonal structure, mutational loads, or SHM targeting preferences, we did observe several abnormalities in the memory compartments of AChR-MG and MuSK-MG subjects related to clonal selection. BASELINe (47) analysis of mutation patterns identified reduced selection pressure in the H-CDR1 and H-CDR2 of AChR-MG subjects. AChR-MG repertoires were also characterized by an increase in mean hydrophobicity of the H-CDR3. Interestingly, the biases observed in the AChR-MG H-CDR3 appear to originate in the naïve compartment, and are subsequently amplified in the IgG memory repertoire. In contrast, MuSK-MG repertoires showed no significant differences in H-CDR1 and H-CDR2 selection pressure or H-CDR3 properties compared to HDs. Furthermore, class-switched MuSK-MG repertoires exhibited the same characteristic shifts in H-CDR3 length, net change and hydrophobicity compared to naïve cells that have been demonstrated in previous studies of healthy repertoires (67, 68). Together these results show that the peripheral AChR-MG memory compartment exhibits reduced selection pressure on the VH sequence.
A primary mechanism by which self-reactive B cells are removed from the naïve B cell population is through light chain receptor editing (60, 61), wherein a secondary V-J rearrangement may occur at either the original locus, the alternate chromosome, or through a kappa to lambda chain switch. Previous work has estimated that 20–50% of naive B cells have undergone receptor editing through lambda chain replacement (61). In both SLE and rheumatoid arthritis, subjects have been shown to exhibit elevated levels of auto-reactive naïve B cells, and these disease have also shown biases in V to J gene proximity in the light chain genes utilized (61). When we examined the proximity of rearranged IGLV and IGLJ genes used, we found that both the naïve and memory repertoire of MuSK-MG subjects used more proximal IGLV-IGLJ pairs. While an increase in IGLV-IGLJ proximity is not a direct indicator of a deficiency in secondary receptor rearrangement, it is an explanation consistent with the observed tolerance defect in MuSK-MG (54).
We have shown that there are significant abnormalities in the memory compartments of both AChR-MG and MuSK-MG patients. However, these abnormalities are already apparent in the naïve compartment of MG subjects - a pattern consistent with defective counter-selection during B cell development. Additionally, we have shown that MG memory compartment biases are not consistent between the two MG subtypes. These results suggest fundamental differences in the underlying mechanisms leading to AChR-MG and MuSK-MG autoimmunity, and may underlie the divergent disease characteristics, including IgG subclass usage and response to therapy, especially B cell depletion (9).
Therapeutics and MG B cell repertoire
Biologics have been increasingly used to treat autoimmune disease. In the treatment of MG, biologics, such as anti-CD20, which directly affect depletion of the B cell compartment, are promisingly effective in a subset of patients (9, 10, 77). However, recent studies demonstrate that B cell depletion therapy does not correct tolerance checkpoint defects, which persist after such treatment (78). CD20-mediated B cell depletion targets the naïve and memory repertoire for deletion, but it leaves antibody (and presumably autoantibody) producing cells intact. Our study, which describes a deformed naive B cell repertoire, raises questions regarding the durability of such treatment and further suggests the potential need for chronic administration. Recent application of autologous stem cell transplantation has shown promise in MG treatment (79), although the treatment is restricted to a very small subgroup of patients. Nonetheless, it may be that immune ablation is required for lasting disease remission, given the abnormalities that are established in the naïve B cell compartment.
Repertoire analysis, coupled with clinical diagnostics, may provide useful insight into the immunological changes associated with a number of biologic treatment approaches, and may serve as a biomarker for tracking or predicting the response to particular therapeutic approaches. Both the results and approach of this study could be applied to first identify abnormalities in the repertoire that associate with clinical parameters, and then used to monitor the repopulated B cell compartment along with its associated clinical presentation. In this regard, repertoire sequencing may provide a valuable benchmark for tracking changes in the B cell repertoire in patients with MG that could adjudicate re-treatment decisions prior to the onset of potentially harmful clinical relapses.
Limitations and future directions
The number of total sequences in our study was large enough (> 0.5 million) to allow conclusions to be drawn from the data; however, the limited set of AChR-MG and MuSK-MG subjects may restrict the generalizability of our findings. Given that this study was the first to employ high-throughput, next-generation sequencing to investigate the repertoire of human B cells in AChR and MuSK MG, it was specifically designed to determine whether broad repertoire abnormalities are commonly present in the disease. Extended interpretation meant to associate the repertoire with minor disease subclasses, disease severity/variability, and the influence of treatment was not built into the experimental design. Further study with larger cohorts of clinically heterogeneous MG subjects, including longitudinally acquired samples, will be necessary to accurately associate the abnormal repertoire shape with clinical status and demographics and the response to treatment.
This study was also not designed to directly associate the abnormalities in the repertoire with production of MG specific autoantibodies, although all of the MG patients studied expressed such autoantibodies. The number of circulating autoantibody-producing cells present in MG or any autoantibody mediated disease is likely to be small. This is highlighted by a very limited number of studies that describe the identification of B cells with particular specificity. An example of such includes a post-vaccination study demonstrating that specific memory B cells represent as few as 0.01%–0.11% (80) of the compartment. Furthermore, the specific B cell compartment in which these cells reside in MG patients is not completely understood. They may be long-lived plasma cells inhabiting the bone marrow or thymus (as shown in some cases of MG), circulating plasmablasts, or a combination of contributions from these compartments. These MG-specific sequences are thus likely to constitute a minute fraction of the sequencing data collected and analyzed herein. Consequently, our data present a generalized broader distortion of the whole MG B cell repertoire, likely reflecting underlying immune dysregulation in AChR-MG and MuSK-MG. This leaves open the question as to how the distorted MG repertoire contributes to pathogenic autoantibody production. A number of approaches can be used to address this question, such as single cell VH:VL pairing coupled with sorting of specific B cells via labeled antigen. However, these technologies still require considerable development to overcome throughput limitations and were accordingly outside the scope of this study. Approaches currently in development will likely enable high-throughput autoantibody characterization at the repertoire scale.
We restricted our investigative scope to consideration of only the major naïve (CD20+CD19+CD27−) and memory (CD20+CD19+CD27+) B cell populations. We recognize that a more complex sorting strategy, which includes additional B cell subpopulations may reveal further details about the nature and origin of MG B cell repertoire abnormalities. In particular, our results show many MG repertoire abnormalities are likely initiated in the naïve compartment. As such, separation of the peripheral naïve compartment into new emigrant (CD19+CD10+CD27−IgM+) and mature naïve (CD19+CD10−CD27− IgM+) populations may reveal how AChR-MG and MuSK-MG abnormalities reflect central or peripheral tolerance checkpoints defects, and whether these defects occur at the same developmental stage in both disease subtypes. Similarly, investigation of the plasmablast (CD27+CD20−CD38+), double negative (CD27−IgM−) and unswitched memory populations may prove informative. However, the low abundance of these sub-populations likely limited their impact on the repertoire scale perturbations we observed in our data sets.
Conclusions
We have shown that patients with AChR-MG or MuSK-MG are unable to generate or sustain the B cell repertoire found in healthy individuals. Despite the commonality of having a deformed repertoire, AChR-MG and MuSK-MG each exhibit unique abnormalities, which implies fundamental differences in the underlying mechanisms leading to AChR-MG and MuSK-MG autoimmunity. These abnormalities likely originate in the naïve compartment due, in part, to defective tolerance checkpoints, which are then subsequently transferred to the memory compartment.
Supplementary Material
Acknowledgments
The authors thank Dr. Kaya Bilguvar and Christopher Castaldi from the Yale Center for Genome Analysis for expert assistance with executing the sequencing on the next-generation platform and Dr. Aditya Kumar for technical expertise with the flow cytometry. The authors also thank the Yale Center for Research Computing (funded by NIH grants RR19895 and RR029676-01) for use of their computing resources.
Funding: Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) through a grant to K.C.O., under award number R01AI114780, by the NIH under award number R01AI104739 to S.H.K, by the National Library of Medicine (NLM) under award number T15 LM07056 to J.A.V.H, and by the Gruber Science Fellowship awarded to J.Q.Z. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Author contributions: The study was directed by the co-principal investigators S.H.K and K.C.O. It was designed and initiated by J.A.V.H., S.H.K, F.V. and K.C.O. The next-generation BCR sequencing was performed using an approach designed by F.V. and executed by T.J.G and T.J.B. Experiments were performed and data were collected by J.A.V.H., P.S., T.J.G. and T.J.B. The sequencing data were processed, analyzed and interpreted by J.A.V.H., J.Q.Z., L.C., and S.H.K. using code authored by J.A.V.H., C.R.B. and S.H.K. The manuscript was written by J.A.V.H., S.H.K. and K.C.O and edited through contributions by all of the authors. R.J.N., R.J.B. and M.M.D. and E.C. recruited study-appropriate subjects, collected clinical specimens along with providing interpreted clinical data.
References
- 1.Berrih-Aknin S. Myasthenia Gravis: paradox versus paradigm in autoimmunity. Journal of autoimmunity. 2014;52:1–28. doi: 10.1016/j.jaut.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Phillips LH., 2nd The epidemiology of myasthenia gravis. Ann N Y Acad Sci. 2003;998:407–412. doi: 10.1196/annals.1254.053. [DOI] [PubMed] [Google Scholar]
- 3.Santos E, Coutinho E, Moreira I, Silva AM, Lopes D, Costa H, Silveira F, Nadais G, Morais H, Martins J, Branco MC, Veiga A, Silva RS, Ferreira A, Sousa F, Freijo M, Matos I, Andre R, Negrao L, Fraga C, Santos M, Sampaio M, Lopes C, Leite MI, Goncalves G. Epidemiology of myasthenia gravis in Northern Portugal: Frequency estimates and clinical epidemiological distribution of cases. Muscle Nerve. 2016;54:413–421. doi: 10.1002/mus.25068. [DOI] [PubMed] [Google Scholar]
- 4.Cetin H, Fulop G, Zach H, Auff E, Zimprich F. Epidemiology of myasthenia gravis in Austria: rising prevalence in an ageing society. Wien Klin Wochenschr. 2012;124:763–768. doi: 10.1007/s00508-012-0258-2. [DOI] [PubMed] [Google Scholar]
- 5.Hoch W, McConville J, Helms S, Newsom-Davis J, Melms A, Vincent A. Autoantibodies to the receptor tyrosine kinase MuSK in patients with myasthenia gravis without acetylcholine receptor antibodies. Nat Med. 2001;7:365–368. doi: 10.1038/85520. [DOI] [PubMed] [Google Scholar]
- 6.Higuchi O, Hamuro J, Motomura M, Yamanashi Y. Autoantibodies to lowdensity lipoprotein receptor-related protein 4 in myasthenia gravis. Annals of Neurology. 2011;69:418–422. doi: 10.1002/ana.22312. [DOI] [PubMed] [Google Scholar]
- 7.Zisimopoulou P, Evangelakou P, Tzartos J, Lazaridis K, Zouvelou V, Mantegazza R, Antozzi C, Andreetta F, Evoli A, Deymeer F, Saruhan-Direskeneli G, Durmus H, Brenner T, Vaknin A, Berrih-Aknin S, Frenkian Cuvelier M, Stojkovic T, DeBaets M, Losen M, Martinez-Martinez P, Kleopa KA, Zamba-Papanicolaou E, Kyriakides T, Kostera-Pruszczyk A, Szczudlik P, Szyluk B, Lavrnic D, Basta I, Peric S, Tallaksen C, Maniaol A, Tzartos SJ. A comprehensive analysis of the epidemiology and clinical characteristics of anti-LRP4 in myasthenia gravis. J Autoimmun. 2014;52:139–145. doi: 10.1016/j.jaut.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Querol L, Illa I. Myasthenia gravis and the neuromuscular junction. Curr Opin Neurol. 2013;26:459–465. doi: 10.1097/WCO.0b013e328364c079. [DOI] [PubMed] [Google Scholar]
- 9.Diaz-Manera J, Martinez-Hernandez E, Querol L, Klooster R, Rojas-Garcia R, Suarez-Calvet X, Munoz-Blanco JL, Mazia C, Straasheijm KR, Gallardo E, Juarez C, Verschuuren JJ, Illa I. Long-lasting treatment effect of rituximab in MuSK myasthenia. Neurology. 2012;78:189–193. doi: 10.1212/WNL.0b013e3182407982. [DOI] [PubMed] [Google Scholar]
- 10.Nowak RJ, Dicapua DB, Zebardast N, Goldstein JM. Response of patients with refractory myasthenia gravis to rituximab: a retrospective study. Ther Adv Neurol Disord. 2011;4:259–266. doi: 10.1177/1756285611411503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leite MI, Jacob S, Viegas S, Cossins J, Clover L, Morgan BP, Beeson D, Willcox N, Vincent A. IgG1 antibodies to acetylcholine receptors in ‘seronegative’ myasthenia gravis. Brain. 2008;131:1940–1952. doi: 10.1093/brain/awn092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodgaard A, Nielsen FC, Djurup R, Somnier F, Gammeltoft S. Acetylcholine receptor antibody in myasthenia gravis: predominance of IgG subclasses 1 and 3. Clinical and experimental immunology. 1987;67:82–88. [PMC free article] [PubMed] [Google Scholar]
- 13.Niks EH, van Leeuwen Y, Leite MI, Dekker FW, Wintzen AR, Wirtz PW, Vincent A, van Tol MJ, Jol-van der Zijde CM, Verschuuren JJ. Clinical fluctuations in MuSK myasthenia gravis are related to antigen-specific IgG4 instead of IgG1. J Neuroimmunol. 2008;195:151–156. doi: 10.1016/j.jneuroim.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 14.Koneczny I, Cossins J, Waters P, Beeson D, Vincent A. MuSK myasthenia gravis IgG4 disrupts the interaction of LRP4 with MuSK but both IgG4 and IgG1-3 can disperse preformed agrin-independent AChR clusters. PLoS One. 2013;8:e80695. doi: 10.1371/journal.pone.0080695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viegas S, Jacobson L, Waters P, Cossins J, Jacob S, Leite MI, Webster R, Vincent A. Passive and active immunization models of MuSK-Ab positive myasthenia: electrophysiological evidence for pre and postsynaptic defects. Exp Neurol. 2012;234:506–512. doi: 10.1016/j.expneurol.2012.01.025. [DOI] [PubMed] [Google Scholar]
- 16.Lindstrom JM, Engel AG, Seybold ME, Lennon VA, Lambert EH. Pathological mechanisms in experimental autoimmune myasthenia gravis. II. Passive transfer of experimental autoimmune myasthenia gravis in rats with anti-acetylcholine recepotr antibodies. J Exp Med. 1976;144:739–753. doi: 10.1084/jem.144.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oda K, Korenaga S, Ito Y. Myasthenia gravis: passive transfer to mice of antibody to human and mouse acetylcholine receptor. Neurology. 1981;31:282–287. doi: 10.1212/wnl.31.3.282. [DOI] [PubMed] [Google Scholar]
- 18.Sterz R, Hohlfeld R, Rajki K, Kaul M, Heininger K, Peper K, Toyka KV. Effector mechanisms in myasthenia gravis: end-plate function after passive transfer of IgG, Fab, and F(ab′)2 hybrid molecules. Muscle Nerve. 1986;9:306–312. doi: 10.1002/mus.880090404. [DOI] [PubMed] [Google Scholar]
- 19.Marx A, Pfister F, Schalke B, Saruhan-Direskeneli G, Melms A, Strobel P. The different roles of the thymus in the pathogenesis of the various myasthenia gravis subtypes. Autoimmun Rev. 2013;12:875–884. doi: 10.1016/j.autrev.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Wolfe GI, Kaminski HJ, Aban IB, Minisman G, Kuo HC, Marx A, Strobel P, Mazia C, Oger J, Cea JG, Heckmann JM, Evoli A, Nix W, Ciafaloni E, Antonini G, Witoonpanich R, King JO, Beydoun SR, Chalk CH, Barboi AC, Amato AA, Shaibani AI, Katirji B, Lecky BR, Buckley C, Vincent A, Dias-Tosta E, Yoshikawa H, Waddington-Cruz M, Pulley MT, Rivner MH, Kostera-Pruszczyk A, Pascuzzi RM, Jackson CE, Garcia Ramos GS, Verschuuren JJ, Massey JM, Kissel JT, Werneck LC, Benatar M, Barohn RJ, Tandan R, Mozaffar T, Conwit R, Odenkirchen J, Sonett JR, Jaretzki A, 3rd, Newsom-Davis J, Cutter GR MS Group. Randomized Trial of Thymectomy in Myasthenia Gravis. N Engl J Med. 2016;375:511–522. doi: 10.1056/NEJMoa1602489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leprince C, Cohen-Kaminsky S, Berrih-Aknin S, Vernet-Der Garabedian B, Treton D, Galanaud P, Richard Y. Thymic B cells from myasthenia gravis patients are activated B cells. Phenotypic and functional analysis. J Immunol. 1990;145:2115–2122. [PubMed] [Google Scholar]
- 22.Sims GP, Shiono H, Willcox N, Stott DI. Somatic hypermutation and selection of B cells in thymic germinal centers responding to acetylcholine receptor in myasthenia gravis. J Immunol. 2001;167:1935–1944. doi: 10.4049/jimmunol.167.4.1935. [DOI] [PubMed] [Google Scholar]
- 23.Vrolix K, Fraussen J, Losen M, Stevens J, Lazaridis K, Molenaar PC, Somers V, Bracho MA, Le Panse R, Stinissen P, Berrih-Aknin S, Maessen JG, Van Garsse L, Buurman WA, Tzartos SJ, De Baets MH, Martinez-Martinez P. Clonal heterogeneity of thymic B cells from early-onset myasthenia gravis patients with antibodies against the acetylcholine receptor. J Autoimmun. 2014;52:101–112. doi: 10.1016/j.jaut.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Cardona A, Pritsch O, Dumas G, Bach JF, Dighiero G. Evidence for an antigendriven selection process in human autoantibodies against acetylcholine receptor. Mol Immunol. 1995;32:1215–1223. doi: 10.1016/0161-5890(95)00101-8. [DOI] [PubMed] [Google Scholar]
- 25.Farrar J, Portolano S, Willcox N, Vincent A, Jacobson L, Newsom-Davis J, Rapoport B, McLachlan SM. Diverse Fab specific for acetylcholine receptor epitopes from a myasthenia gravis thymus combinatorial library. Int Immunol. 1997;9:1311–1318. doi: 10.1093/intimm/9.9.1311. [DOI] [PubMed] [Google Scholar]
- 26.Zuckerman NS, Hazanov H, Barak M, Edelman H, Hess S, Shcolnik H, Dunn-Walters D, Mehr R. Somatic hypermutation and antigen-driven selection of B cells are altered in autoimmune diseases. Journal of autoimmunity. 2010;35:325–335. doi: 10.1016/j.jaut.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Hill ME, Shiono H, Newsom-Davis J, Willcox N. The myasthenia gravis thymus: a rare source of human autoantibody-secreting plasma cells for testing potential therapeutics. J Neuroimmunol. 2008;201–202:50–56. doi: 10.1016/j.jneuroim.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 28.Scadding GK, Vincent A, Newsom-Davis J, Henry K. Acetylcholine receptor antibody synthesis by thymic lymphocytes: correlation with thymic histology. Neurology. 1981;31:935–943. doi: 10.1212/wnl.31.8.935. [DOI] [PubMed] [Google Scholar]
- 29.Leite MI, Strobel P, Jones M, Micklem K, Moritz R, Gold R, Niks EH, Berrih-Aknin S, Scaravilli F, Canelhas A, Marx A, Newsom-Davis J, Willcox N, Vincent A. Fewer thymic changes in MuSK antibody-positive than in MuSK antibody-negative MG. Ann Neurol. 2005;57:444–448. doi: 10.1002/ana.20386. [DOI] [PubMed] [Google Scholar]
- 30.Glanville J, Zhai W, Berka J, Telman D, Huerta G, Mehta GR, Ni I, Mei L, Sundar PD, Day GMR, Cox D, Rajpal A, Pons J. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20216–20221. doi: 10.1073/pnas.0909775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vander Heiden JA, Yaari G, Uduman M, Stern JN, O’Connor KC, Hafler DA, Vigneault F, Kleinstein SH. pRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics. 2014;30:1930–1932. doi: 10.1093/bioinformatics/btu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alamyar E, Duroux P, Lefranc MP, Giudicelli V. IMGT(®) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods in molecular biology. 2012;882:569–604. doi: 10.1007/978-1-61779-842-9_32. [DOI] [PubMed] [Google Scholar]
- 33.Gupta NT, Vander Heiden JA, Uduman M, Gadala-Maria D, Yaari G, Kleinstein SH. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015;31:3356–3358. doi: 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Team, R. D. C. Vienna Austria R Foundation for Statistical Computing. R Foundation for Statistical Computing; 2010. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 35.Yaari G, Vander Heiden JA, Uduman M, Gadala-Maria D, Gupta N, Stern JN, O’Connor KC, Hafler DA, Laserson U, Vigneault F, Kleinstein SH. Models of somatic hypermutation targeting and substitution based on synonymous mutations from high-throughput immunoglobulin sequencing data. Front Immunol. 2013;4:358. doi: 10.3389/fimmu.2013.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glanville J, Kuo TC, von Büdingen HC, Guey L, Berka J, Sundar PD, Huerta G, Mehta GR, Oksenberg JR, Hauser SL, Cox DR, Rajpal A, Pons J. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20066–20071. doi: 10.1073/pnas.1107498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chao A. Nonparametric Estimation of the Number of Classes in a Population. Scandinavian Journal of Statistics. 1984;11:265–270. [Google Scholar]
- 38.Chao A, Hsieh TC, Chazdon RL, Colwell RK, Gotelli NJ. Unveiling the species-rank abundance distribution by generalizing the Good-Turing sample coverage theory. Ecology. 2015;96:1189–1201. doi: 10.1890/14-0550.1. [DOI] [PubMed] [Google Scholar]
- 39.Hill M. Diversity and evenness: a unifying notation and its consequences. Ecology. 1973;54:427–432. [Google Scholar]
- 40.Gadala-Maria D, Yaari G, Uduman M, Kleinstein SH. Automated analysis of high-throughput B-cell sequencing data reveals a high frequency of novel immunoglobulin V gene segment alleles. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1417683112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubelt F, Bolen CR, McGuire HM, Heiden JA, Gadala-Maria D, Levin M, MEG, Mamedov MR, Swan GE, Dekker CL, Cowell LG, Kleinstein SH, Davis MM. Individual heritable differences result in unique cell lymphocyte receptor repertoires of naive and antigen-experienced cells. Nat Commun. 2016;7:11112. doi: 10.1038/ncomms11112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lefranc MP. Nomenclature of the human immunoglobulin heavy (IGH) genes. Exp Clin Immunogenet. 2001;18:100–116. doi: 10.1159/000049189. [DOI] [PubMed] [Google Scholar]
- 43.Watson CT, Steinberg KM, Graves TA, Warren RL, Malig M, Schein J, Wilson RK, Holt RA, Eichler EE, Breden F. Sequencing of the human IG light chain loci from a hydatidiform mole BAC library reveals locus-specific signatures of genetic diversity. Genes Immun. 2015;16:24–34. doi: 10.1038/gene.2014.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watson CT, Steinberg KM, Huddleston J, Warren RL, Malig M, Schein J, Willsey aJ, Joy JB, Scott JK, Graves Ta, Wilson RK, Holt RA, Eichler EE, Breden F. Complete Haplotype Sequence of the Human Immunoglobulin Heavy-Chain Variable, Diversity, and Joining Genes and Characterization of Allelic and Copy-Number Variation. The American Journal of Human Genetics. 2013;92:530–546. doi: 10.1016/j.ajhg.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Felsenstein J. PHYLIP - Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 46.Stern JN, Yaari G, Vander Heiden JA, Church G, Donahue WF, Hintzen RQ, Huttner AJ, Laman JD, Nagra RM, Nylander A, Pitt D, Ramanan S, Siddiqui BA, Vigneault F, Kleinstein SH, Hafler DA, O’Connor KC. B cells populating the multiple sclerosis brain mature in the draining cervical lymph nodes. Sci Transl Med. 2014;6:248ra107. doi: 10.1126/scitranslmed.3008879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yaari G, Uduman M, Kleinstein SH. Quantifying selection in high-throughput Immunoglobulin sequencing data sets. Nucleic Acids Res. 2012;40:e134. doi: 10.1093/nar/gks457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 49.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 50.Moore DS. Amino acid and peptide net charges: A simple calculational procedure. Biochemical Education. 1985;13:10–11. [Google Scholar]
- 51.Zimmerman JM, Eliezer N, Simha R. The characterization of amino acid sequences in proteins by statistical methods. J Theor Biol. 1968;21:170–201. doi: 10.1016/0022-5193(68)90069-6. [DOI] [PubMed] [Google Scholar]
- 52.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 53.Ikai A. Thermostability and Aliphatic Index of Globular Proteins. J Biochem. 1980;1898:1895–1898. [PubMed] [Google Scholar]
- 54.Lee JY, Stathopoulos P, Gupta S, Bannock JM, Barohn RJ, Cotzomi E, Dimachkie MM, Jacobson L, Lee CS, Morbach H, Querol L, Shan JL, Vander Heiden JA, Waters P, Vincent A, Nowak RJ, O’Connor KC. Compromised fidelity of B-cell tolerance checkpoints in AChR and MuSK myasthenia gravis. Annals of Clinical and Translational Neurology. 2016;3:443–454. doi: 10.1002/acn3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parameswaran P, Liu Y, Roskin KM, Jackson KK, Dixit VP, Lee JY, Artiles KL, Zompi S, Vargas MJ, Simen BB, Hanczaruk B, McGowan KR, Tariq MA, Pourmand N, Koller D, Balmaseda A, Boyd SD, Harris E, Fire AZ. Convergent antibody signatures in human dengue. Cell Host Microbe. 2013;13:691–700. doi: 10.1016/j.chom.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jackson KJ, Liu Y, Roskin KM, Glanville J, Hoh RA, Seo K, Marshall EL, Gurley TC, Moody MA, Haynes BF, Walter EB, Liao HX, Albrecht RA, Garcia-Sastre A, Chaparro-Riggers J, Rajpal A, Pons J, Simen BB, Hanczaruk B, Dekker CL, Laserson J, Koller D, Davis MM, Fire AZ, Boyd SD. Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell Host Microbe. 2014;16:105–114. doi: 10.1016/j.chom.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Richardson C, Chida AS, Adlowitz D, Silver L, Fox E, Jenks SA, Palmer E, Wang Y, Heimburg-Molinaro J, Li QZ, Mohan C, Cummings R, Tipton C, Sanz I. Molecular basis of 9G4 B cell autoreactivity in human systemic lupus erythematosus. J Immunol. 2013;191:4926–4939. doi: 10.4049/jimmunol.1202263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laserson U, Vigneault F, Gadala-Maria D, Yaari G, Uduman M, Vander Heiden JA, Kelton W, Taek Jung S, Liu Y, Laserson J, Chari R, Lee JH, Bachelet I, Hickey B, Lieberman-Aiden E, Hanczaruk B, Simen BB, Egholm M, Koller D, Georgiou G, Kleinstein SH, Church GM. High-resolution antibody dynamics of vaccine-induced immune responses. Proc Natl Acad Sci U S A. 2014;111:4928–4933. doi: 10.1073/pnas.1323862111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tipton CM, Fucile CF, Darce J, Chida A, Ichikawa T, Gregoretti I, Schieferl S, Hom J, Jenks S, Feldman RJ, Mehr R, Wei C, Lee FE, Cheung WC, Rosenberg AF, Sanz I. Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat Immunol. 2015;16:755–765. doi: 10.1038/ni.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Halverson R, Torres RM, Pelanda R. Receptor editing is the main mechanism of B cell tolerance toward membrane antigens. Nat Immunol. 2004;5:645–650. doi: 10.1038/ni1076. [DOI] [PubMed] [Google Scholar]
- 61.Meffre E, Wardemann H. B-cell tolerance checkpoints in health and autoimmunity. Curr Opin Immunol. 2008;20:632–638. doi: 10.1016/j.coi.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 62.Graus YF, de Baets MH, Parren PW, Berrih-Aknin S, Wokke J, van Breda Vriesman PJ, Burton DR. Human anti-nicotinic acetylcholine receptor recombinant Fab fragments isolated from thymus-derived phage display libraries from myasthenia gravis patients reflect predominant specificities in serum and block the action of pathogenic serum antibodies. J Immunol. 1997;158:1919–1929. [PubMed] [Google Scholar]
- 63.Yaari G, Benichou JI, Vander Heiden JA, Kleinstein SH, Louzoun Y. The mutation patterns in B-cell immunoglobulin receptors reflect the influence of selection acting at multiple time-scales. Philos Trans R Soc Lond B Biol Sci. 2015:370. doi: 10.1098/rstb.2014.0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu JL, Davis MM. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity. 2000;13:37–45. doi: 10.1016/s1074-7613(00)00006-6. [DOI] [PubMed] [Google Scholar]
- 65.Meffre E, Schaefer A, Wardemann H, Wilson P, Davis E, Nussenzweig MC. Surrogate Light Chain Expressing Human Peripheral B Cells Produce Self-reactive Antibodies. The Journal of experimental medicine. 2004;199:145–150. doi: 10.1084/jem.20031550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wardemann H. Predominant Autoantibody Production by Early Human B Cell Precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 67.Wu YC, Kipling D, Dunn-Walters DK. The relationship between CD27 negative and positive B cell populations in human peripheral blood. Front Immunol. 2011;2:81. doi: 10.3389/fimmu.2011.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu YC, Kipling D, Leong HS, Martin V, Ademokun AA, Dunn-Walters DK. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood. 2010;116:1070–1078. doi: 10.1182/blood-2010-03-275859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vincent A. Unravelling the pathogenesis of myasthenia gravis. Nat Rev Immunol. 2002;2:797–804. doi: 10.1038/nri916. [DOI] [PubMed] [Google Scholar]
- 70.Castiello MC, Bosticardo M, Pala F, Catucci M, Chamberlain N, van Zelm MC, Driessen GJ, Pac M, Bernatowska E, Scaramuzza S, Aiuti A, Sauer AV, Traggiai E, Meffre E, Villa A, van der Burg M. Wiskott-Aldrich Syndrome protein deficiency perturbs the homeostasis of B-cell compartment in humans. J Autoimmun. 2014;50:42–50. doi: 10.1016/j.jaut.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Palanichamy A, Apeltsin L, Kuo TC, Sirota M, Wang S, Pitts SJ, Sundar PD, Telman D, Zhao LZ, Derstine M, Abounasr A, Hauser SL, von Budingen HC. Immunoglobulin class-switched B cells form an active immune axis between CNS and periphery in multiple sclerosis. Sci Transl Med. 2014;6:248ra106. doi: 10.1126/scitranslmed.3008930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cameron EM, Spencer S, Lazarini J, Harp CT, Ward ES, Burgoon M, Owens GP, Racke MK, Bennett JL, Frohman EM, Monson NL. Potential of a unique antibody gene signature to predict conversion to clinically definite multiple sclerosis. J Neuroimmunol. 2009;213:123–130. doi: 10.1016/j.jneuroim.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hehle V, Fraser LD, Tahir R, Kipling D, Wu YC, Lutalo PM, Cason J, Choong L, D’Cruz DP, Cope AP, Dunn-Walters DK, Spencer J. Immunoglobulin kappa variable region gene selection during early human B cell development in health and systemic lupus erythematosus. Mol Immunol. 2015;65:215–223. doi: 10.1016/j.molimm.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 74.Samuels J, Ng YS, Coupillaud C, Paget D, Meffre E. Impaired early B cell tolerance in patients with rheumatoid arthritis. The Journal of experimental medicine. 2005;201:1659–1667. doi: 10.1084/jem.20042321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yurasov S, Wardemann H, Hammersen J, Tsuiji M, Meffre E, Pascual V, Nussenzweig MC. Defective B cell tolerance checkpoints in systemic lupus erythematosus. J Exp Med. 2005;201:703–711. doi: 10.1084/jem.20042251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kinnunen T, Chamberlain N, Morbach H, Cantaert T, Lynch M, Preston-Hurlburt P, Herold KC, Hafler DA, O’Connor KC, Meffre E. Specific peripheral B cell tolerance defects in patients with multiple sclerosis. J Clin Invest. 2013;123:2737–2741. doi: 10.1172/JCI68775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Keung B, Robeson KR, DiCapua DB, Rosen JB, O’Connor KC, Goldstein JM, Nowak RJ. Long-term benefit of rituximab in MuSK autoantibody myasthenia gravis patients. J Neurol Neurosurg Psychiatry. 2013;84:1407–1409. doi: 10.1136/jnnp-2012-303664. [DOI] [PubMed] [Google Scholar]
- 78.Chamberlain N, Massad C, Oe T, Cantaert T, Herold KC, Meffre E. Rituximab does not reset defective early B cell tolerance checkpoints. J Clin Invest. 2016;126:282–287. doi: 10.1172/JCI83840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bryant A, Atkins H, Pringle CE, Allan D, Anstee G, Bence-Bruckler I, Hamelin L, Hodgins M, Hopkins H, Huebsch L, McDiarmid S, Sabloff M, Sheppard D, Tay J, Bredeson C. Myasthenia Gravis Treated With Autologous Hematopoietic Stem Cell Transplantation. JAMA Neurol. 2016 doi: 10.1001/jamaneurol.2016.0113. [DOI] [PubMed] [Google Scholar]
- 80.Franz B, May KF, Jr, Dranoff G, Wucherpfennig K. Ex vivo characterization and isolation of rare memory B cells with antigen tetramers. Blood. 2011;118:348–357. doi: 10.1182/blood-2011-03-341917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.