Abstract
Norovirus infections have a major impact on public health worldwide, yet there is a current dearth of norovirus-specific therapeutics and prophylactics. This report describes the discovery of a novel class of macrocyclic inhibitors of norovirus 3C-like protease, a cysteine protease that is essential for virus replication. SAR, structural, and biochemical studies were carried out to ascertain the effect of structure on pharmacological activity and permeability. Insights gained from these studies have laid a solid foundation for capitalizing on the therapeutic potential of the series of inhibitors described herein.
Graphical abstract
1. Introduction
Noroviruses belong to the family Caliciviridae and are classified into at least six genogroups (GI-GVI) [1–3]. Human noroviruses are the primary cause of non-bacterial acute gastroenteritis worldwide [4–5], and are associated with high morbidity and a heavy economic burden [6–8]. In the U.S. alone noroviruses account for >20 million cases annually and impact most severely the young and elderly, as well as immunocompromised individuals [9–12]. Combating norovirus infections presents a formidable challenge because of their robustness, high infectivity, ease of transmission via multiple routes [13–14], the current dearth of norovirus-specific therapeutics/prophylactics and vaccines [15–21], as well as a sub-optimal understanding of norovirus biology and pathophysiology [22–26].
Noroviruses are small, single-stranded, positive sense RNA viruses whose genome (7–8 kb) is covalently linked to a viral protein (VPg, virion protein, genome-linked) at the 5′ end and polyadenylated at the 3′ end [1,24]. The genome consists of three open reading frames (ORFs) that encode a 200 kDa polyprotein (ORF1), a major capsid protein VP1 (ORF2) and a small basic protein VP2 (ORF3) [1,24]. The polyprotein is processed by a virus-encoded protease to generate six mature non-structural proteins, including the viral protease (3C-like protease, 3CLpro or NS6pro) and the RNA dependent RNA polymerase (NS7pol). Co- and post-translational processing of the polyprotein by 3CLpro is essential for virus replication, consequently, 3CLpro is an attractive target for the discovery of anti-norovirus small molecule therapeutics and prophylactics [15–20].
Norovirus 3CLpro, including Norwalk virus (NV) 3CLpro, is a cysteine protease with a Cys-His-Glu catalytic triad, an extended binding site, and a chymotrypsin-like fold [27–31]. The protease displays a primary substrate specificity requirement for a P1 glutamine or glutamic acid residue, or equivalent [32]. We have recently described an array of transition state inhibitors and transition state mimics of NV 3CLpro [15]. In continuing our studies in this area, we describe herein the structure-guided design of a novel series of macrocyclic inhibitors of the protease, as well as pertinent structural, biochemical, and cell-based studies.
2. Results and Discussion
2.1. Inhibitor design rationale
Cocrystal structures of NV 3CLpro with peptidyl inhibitors reveal a network of backbone hydrogen bonds involving Ala158, Ala160, Gln110 and His30, as well as two critical hydrogen bonds between His157 and Thr134 and the carbonyl oxygen of the P1 Gln side chain [27,29,33]. The backbone hydrogen bonds mimic an antiparallel β-sheet and serve to correctly orient and position the inhibitor to the active site [34–35]. It was envisaged that the construction of a macrocyclic structure (Figure 1, structure (I)) capable of (a) maintaining the aforementioned favorable binding interactions and, (b) having a flexible diversity site that is well-suited to exploiting H-bonding and hydrophobic interactions with the S3-S4 subsites and modulating physicochemical properties, could potentially result in the identification of a molecule that displays high potency and drug-like characteristics [36–37]. Additional advantages frequently accrued by macrocyclization include higher pharmacological activity and selectivity, enhanced permeability, and improved stability [38–40]. The interaction of inhibitor (I) with the protease was further probed using different warheads, ring sizes, and P2 residues, a P1 alkoxyamide side chain replacement for the P1 Gln side chain that could potentially engage in intramolecular H-bonding, thereby attenuating cellular permeability [41–46] and, finally, structural variants focused on R1 which projects toward the S3-S4 subsites and affords opportunities for favorable binding interactions.
Fig. 1.
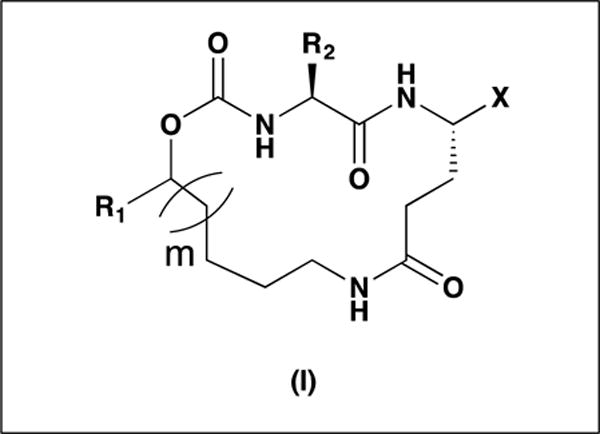
General structure of macrocyclic inhibitor (I)
2.2. Chemistry
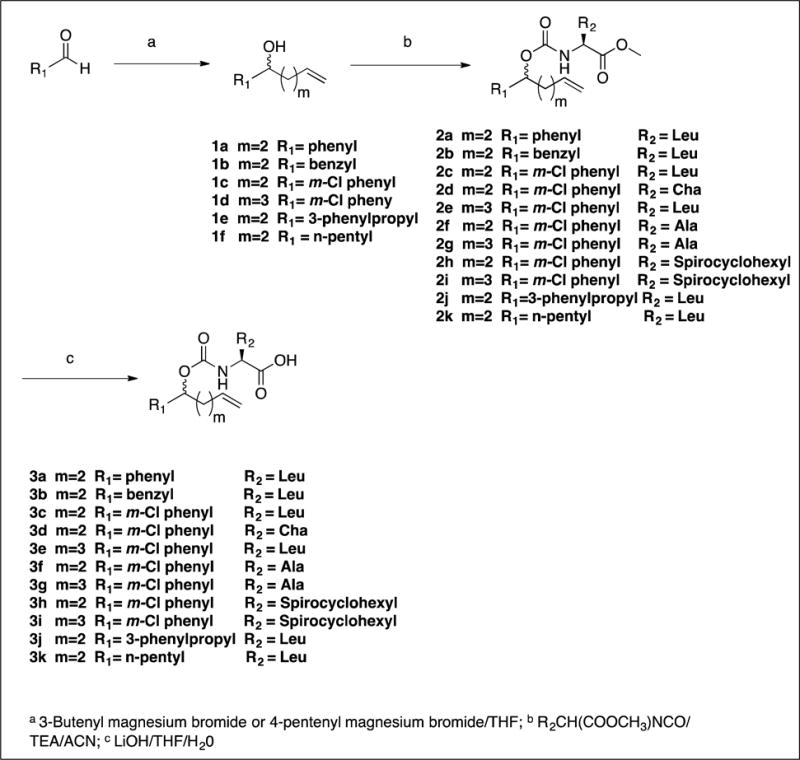
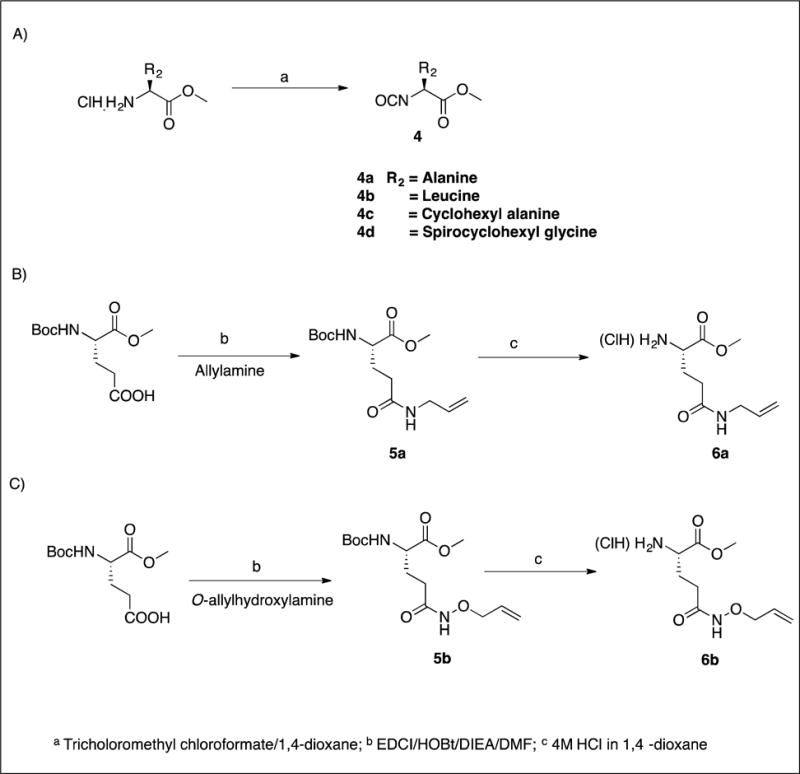
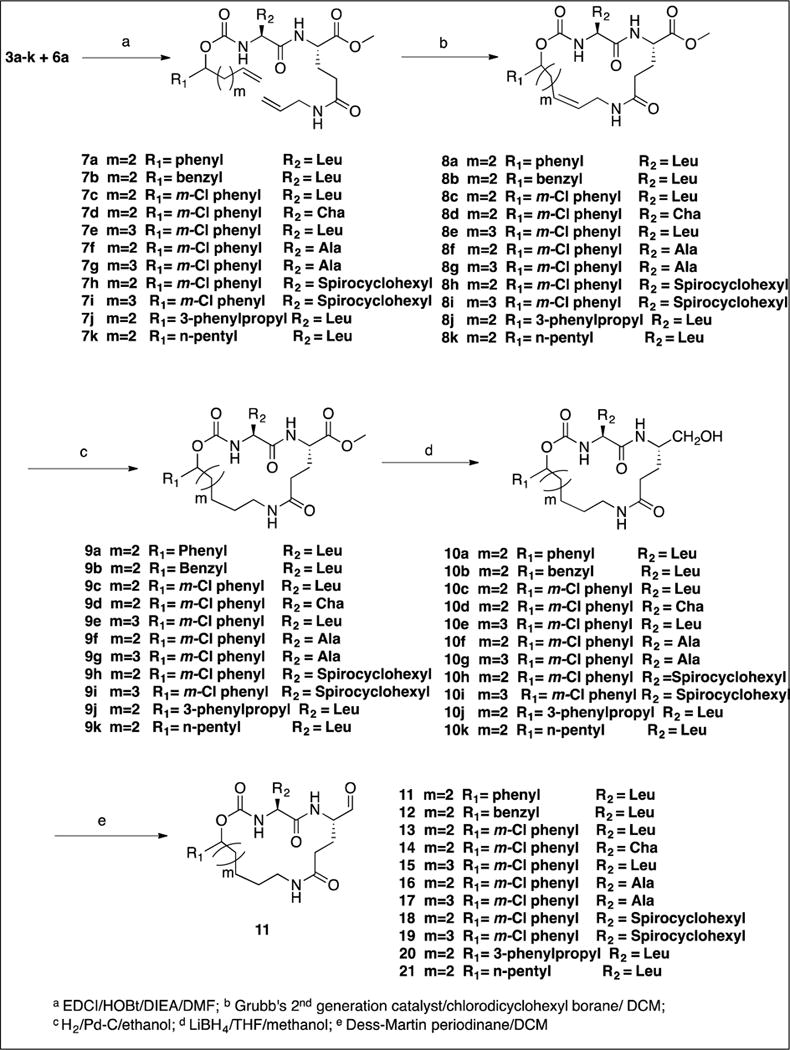
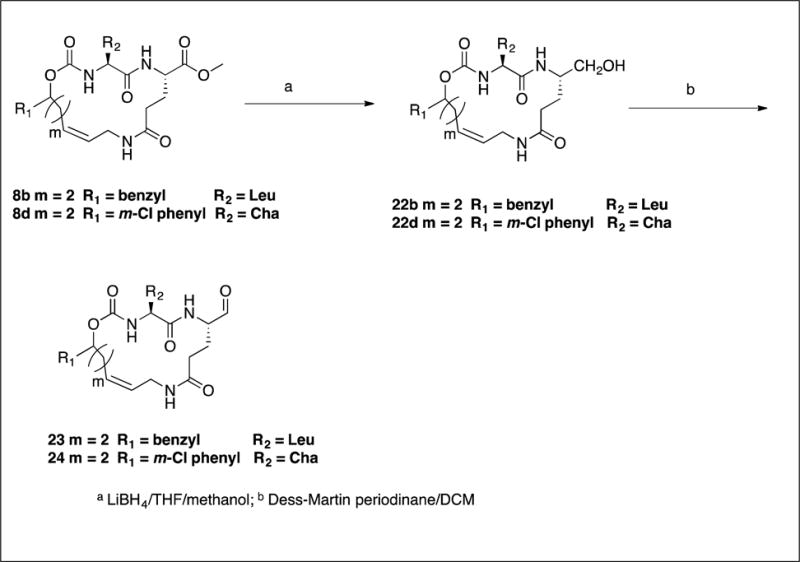
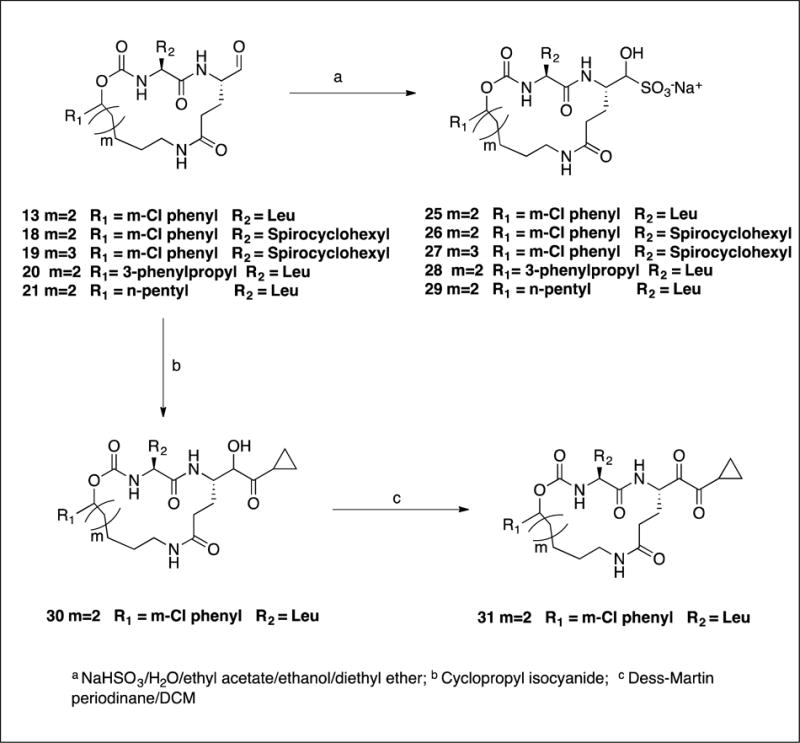
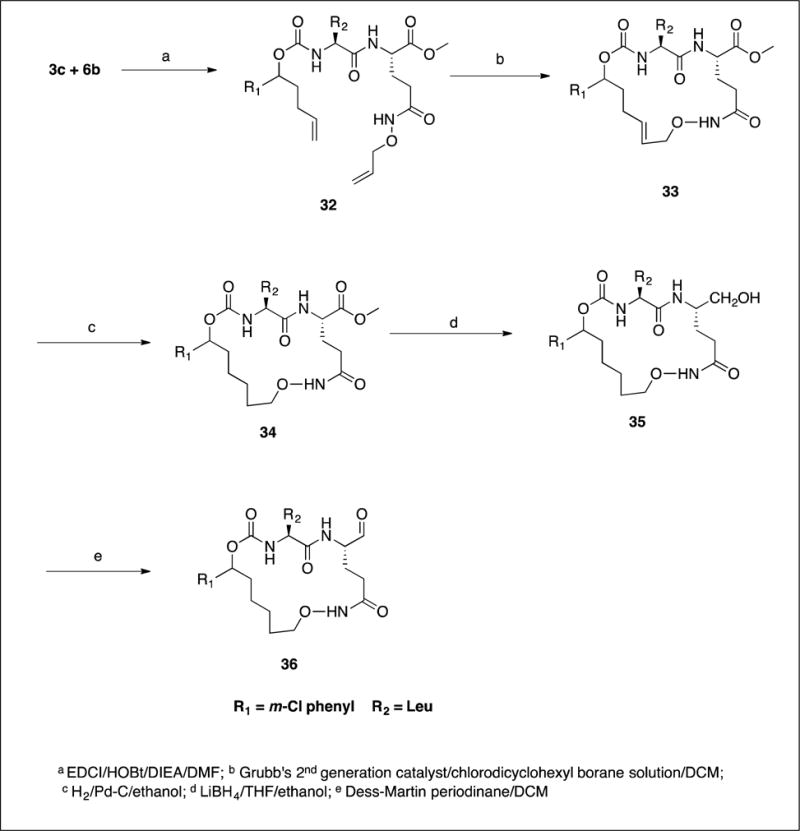
The synthesis of inhibitors 11–21 by coupling compounds 3a–k (Scheme 1) with compound 6a (Scheme 2B) to yield acyclic compounds 7a–k (Scheme 3). The reaction sequence outlined in Scheme 1 is flexible and permits ready manipulation of the ring size using appropriate alkenyl Grignard reagents. Furthermore, the nature of the P2 residue (R2) could be readily manipulated by reacting intermediates 1a–f (Scheme 1) with an appropriate amino acid ester isocyanate 4a–d (Scheme 2A). Ring closure using a metathesis reaction furnished compounds 8a–k which, upon sequential catalytic hydrogenation, reduction with lithium borohydride, and Dess-Martin periodinane oxidation yielded aldehydes 11–21. With the exception of compounds 14 and 36, these were obtained as diastereomeric mixtures. Aldehydes 23–24 having an unsaturated linker were synthesized in an analogous manner (Scheme 4). Aldehyde bisulfite adducts 25–29 and α-ketoamide 31 were synthesized as shown in Scheme 5. Finally, coupling of intermediate 3c with 6b (Scheme 2C) followed by further elaboration of the product yielded aldehyde 36 (Scheme 6).
Scheme 1.
Scheme 2.
Scheme 3.
Scheme 4.
Scheme 5.
Scheme 6.
2.3. Biochemical Studies
The inhibitory activity of the synthesized compounds against NV 3CLpro and their anti-norovirus activity in a cell-based replicon system, were evaluated as described in the experimental section [33,47]. The determined IC50 in enzyme assay, EC50 against NV in the replicon harboring cells (HG23 cells) or murine norovirus (MNV) in RAW264.7 cells, and CC50 values in HG23 cells are listed in Tables 1 and they are the average of at least two determinations.
Table 1.
Activity and cell toxicity of compounds 11–12, 20, 28, 13, 25, 31, 14a–b, 15–18, 26, 19, 27, 21, 29, and 36A–B (all values are μM).
| Compound | R1 | R2 | X | Ring Size | IC50 (μM) (NV) | EC50 (μM) (NV) | EC50 (μM) (MNV) | TC50 (EC50) |
|---|---|---|---|---|---|---|---|---|
| 11 | Phenyl | Leu | CHO | 17 | 1.3 | 7.7 | >20 | >100 |
|
| ||||||||
| 12 | Benzyl | Leu | CHO | 17 | 4.6 | 8.8 | >20 | >100 |
|
| ||||||||
| 20 | Propyl benzene | Leu | CHO | 17 | 4.2 | 7.5 | >20 | 71.5 |
| 28 | CH(OH)SO3Na | 6.8 | 4.6 | >20 | 69.2 | |||
|
| ||||||||
| 13 | m-Cl-phenyl | Leu | CHO | 17 | 1.5 | 1.8 | 13.2 | >100 |
| 25 | CH(OH)SO3Na | 1.1 | 1.6 | 15.1 | >100 | |||
| 31 | (C=O)(C=O) Cyclopropyl |
2.3 | 9.1 | 15.5 | 53.1 | |||
|
| ||||||||
| 14(major) | m-Cl-phenyl | Cha | CHO | 17 | 4.1 | 2.1 | 4.3 | 62.4 |
| 14(minor) | 6.5 | 2.5 | 6.5 | 45.3 | ||||
| 15 | m-Cl-phenyl | Leu | CHO | 18 | 6.3 | 5.4 | >20 | 67.4 |
|
| ||||||||
| 16 | m-Cl-phenyl | Ala | CHO | 17 | >100 | >20 | >20 | >100 |
| 17 | 18 | 62.3 | 9.5 | >20 | >100 | |||
|
| ||||||||
| 18 | m-Cl-phenyl | spirocyclohexyl | CHO | 17 | >100 | >20 | >20 | 62.4 |
| 26 | CH(OH)SO3Na | >100 | >20 | >20 | 48.5 | |||
| 19 | CHO | 18 | >100 | >20 | >20 | 53.3 | ||
| 27 | CH(OH)SO3Na | >100 | >20 | >20 | 46.6 | |||
|
| ||||||||
| 21 | n-pentyl | Leu | CHO | 17 | 3.2 | 3.1 | 12.1 | 72.7 |
| 29 | CH(OH)SO3Na | 5.1 | 3.5 | 14.6 | 78.5 | |||
|
| ||||||||
| 36A | m-Cl-phenyl | Leu | CHO | 18 | 30.5 | 3.6 | 15.5 | 68.3 |
| 36B | 46.7 | 5.1 | 13.4 | 73.8 | ||||
|
| ||||||||
| 23 | Benzyl | Leu | CHO | 17 | 2.5 | 3.1 | >20 | >100 |
|
| ||||||||
| 24 | m-Cl-phenyl | Cha | CHO | 17 | 1.3 | 1.5 | 3.1 | 42.3 |
The low cellular permeability and susceptibility to proteolytic degradation of peptide-based inhibitors provided the impetus behind the design of macrocyclic inhibitor (I). It is evident from the results shown in Table 1 that, with the exception of compounds 18, 26, 19, and 27 (Table 1) that were inactive, the rest of the compounds inhibited NV 3CLpro and displayed antiviral activity in cell based assays in the replicon harboring cells (NV) as well as RAW264.7 cells (MNV) with IC50 and EC50 values in the low micromolar range. The antiviral activities against NV and MNV in cell based assays demonstrate that the compounds have good permeability in different cell types (hepatoma cells [HG23] and macrophage-like cells [RAW264.7]). The aldehyde and aldehyde bisulfite adducts had comparable potency, however, replacement of the warhead with an α-ketoamide (Table 1, compounds 13 and 25 versus α-ketoamide 31) diminished activity. These observations are congruent with the results of previous studies with peptidyl and macrocyclic inhibitors of NV 3CLpro [48–49]. Replacement of the P2 Leu (R2) residue with cyclohexylalanine (Cha) had a minor effect on potency. This is contrast to the significant boost in potency observed in the dipeptidyl series of acyclic inhibitors having a P2 Cha [33]. As expected, a P2 Ala (R2) residue resulted in greatly diminished potency, a reflection of the strong preference of NV 3CLpro for a Cha or Leu residue at the P2 position [32]. Surprisingly, substitution of a spirocyclohexylglycine (1-aminocyclohexaneglycine) at P2 (R2) resulted in a dramatic loss of activity (Table 1, compounds 18, 26, 19, and 27). In those instances, where the diastereomers were separable, these displayed comparable potencies (Table 1, compounds 14A–B and 36A–B).
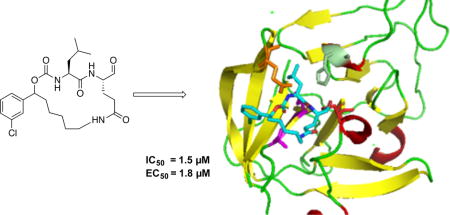
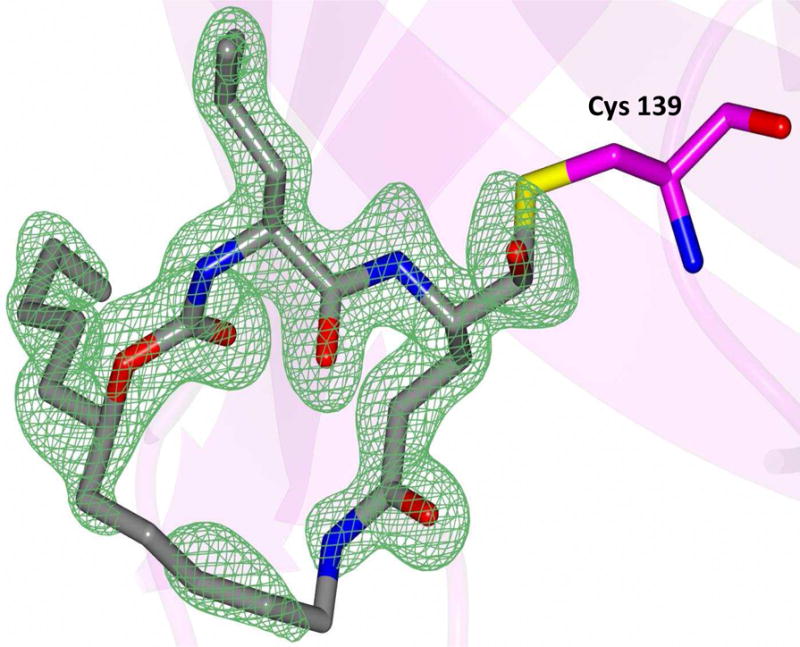
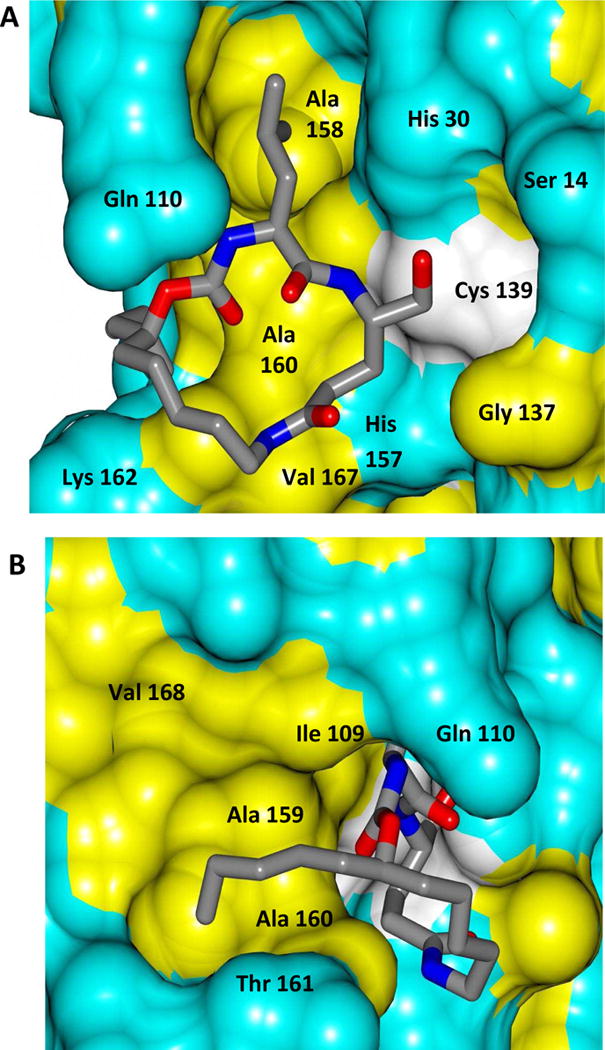
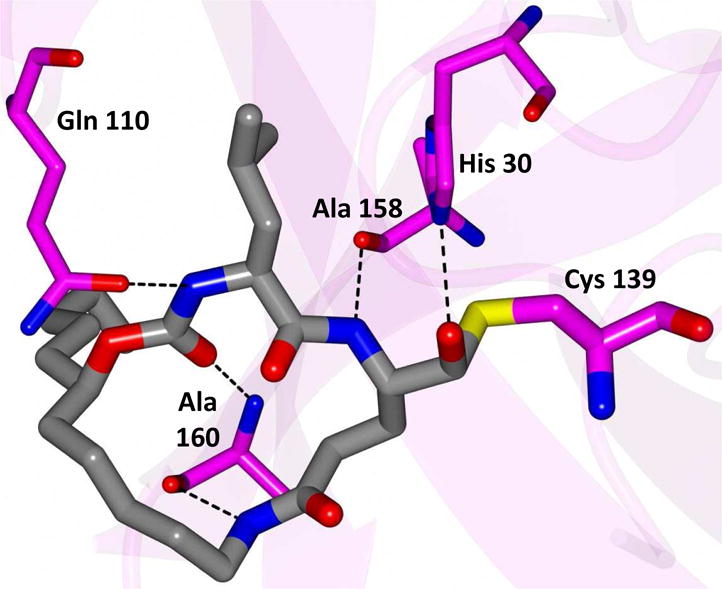
Delineation of the structural determinants impacting pharmacological activity, as well as demonstration of the mechanism of action, was made possible by determining a high resolution cocrystal structure of inhibitor 13 with NV 3CLpro. Examination of the active site revealed the presence of prominent difference electron density with inhibitor 13 covalently bound to Cys 139 and the entire inhibitor could be traced (Figure 2). The interactions between NV 3CLpro and 13 are shown in Figure 3 and electrostatic surface representations of the NV 3CLpro with the inhibitor nestled in the active site are shown in Figure 4. A network of H-bonds involving the backbone of inhibitor 13 and residues Gln110, Ala158, and Ala160 that serve to position the inhibitor correctly at the active site are clearly evident. Surprisingly, the His157 and Thr134 residues that are present in the vicinity of the Gln side chain and ordinarily engage in H-bonding interactions with the Gln side chain oxygen [33] are absent and seemingly displaced by Pro136, which forms a H-bond with the oxygen of the tetrahedral adduct (Figure 3). Thus, the loss of the pair of H-bonds between the Gln side chain oxygen and the His157 and Thr134 residues may account for the observed potency (vide infra). The inhibitor is covalently bonded to the active site Cys139 residue providing unequivocal demonstration of the mechanism of action of (I). The m-chlorophenyl group of inhibitor 13 is positioned within a hydrophobic cleft, as shown in Figure 5. Extending the position of the phenyl ring (Table 1, compounds 11–12, 20 and 28) resulted in a two to four-fold decrease in potency. Furthermore, one of the macrocycles having an unsaturated linker (compound 24, Table 1) was more potent than the corresponding macrocycle with a saturated linker (Table 1/compounds 14A–B) while the second one (Table 1/23) was ~2-fold more potent than compound 12 (Table 1). In order to enhance binding and permeability, the P1 Gln side chain was modified by introducing an additional H-bonding site. The structural modification did not have a major effect on potency (Table 1, compounds 36A–B).
Fig. 2.
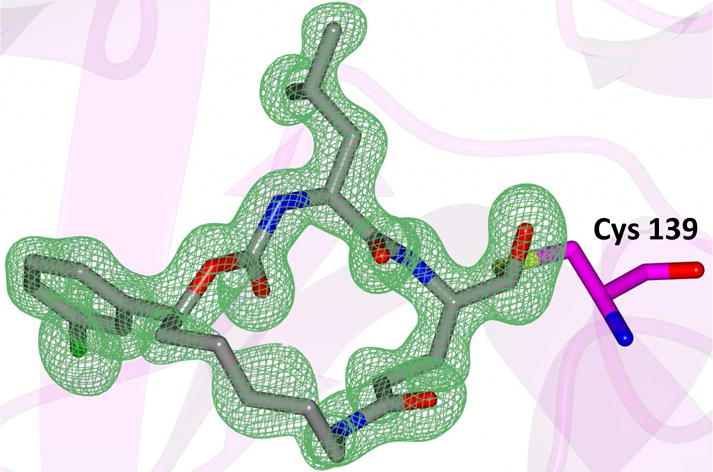
View of the Fo–Fc omit map for inhibitor 13 (green mesh) contoured at 3σ
Fig. 3.
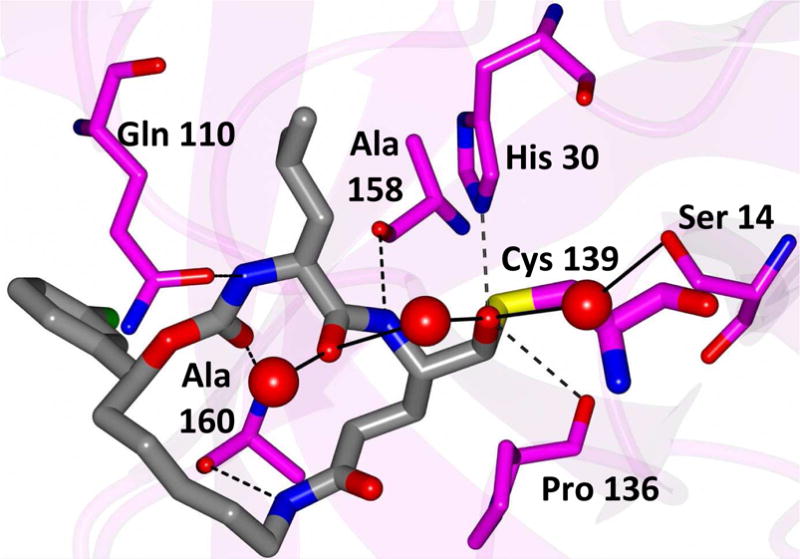
Hydrogen bond interactions (dashed lines) between NV 3CLpro and inhibitor 13. Contacts to water molecules are indicated by the solid lines
Fig. 4.
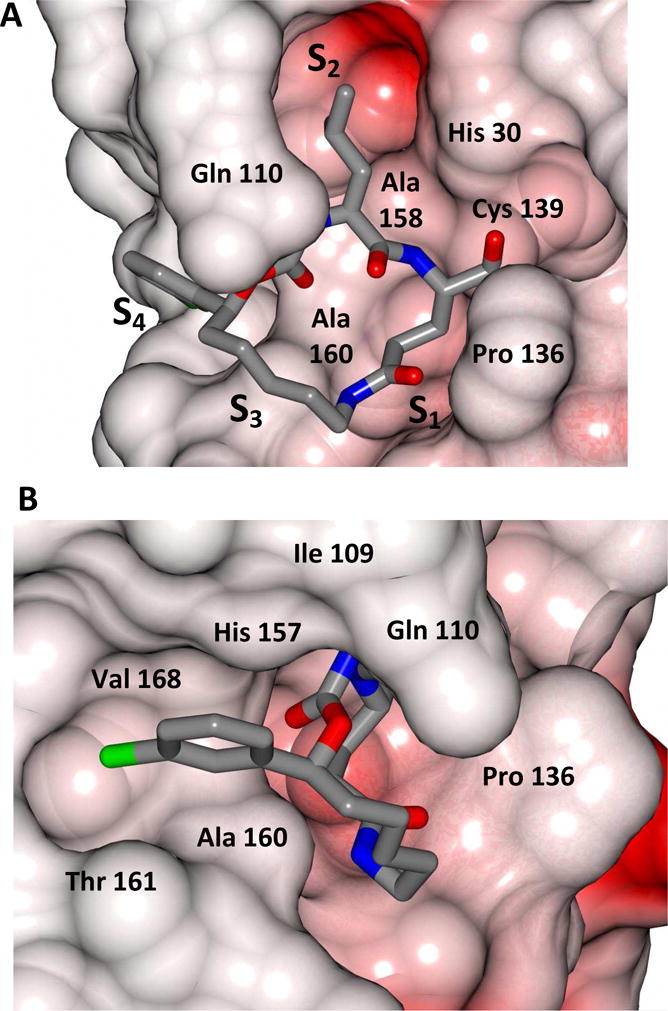
Two views showing the electrostatic surface representation NV 3CLpro binding site of inhibitor 13. A) View of the active site and B) the m-chlorophenyl group in the hydrophobic S4 pocket. View is rotated counterclockwise approximately 90° about the vertical axis relative to panel A.
Fig. 5.
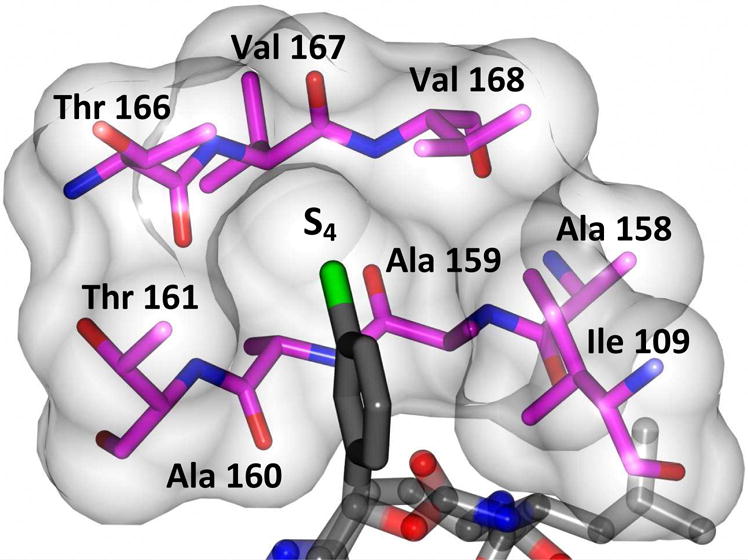
View of the m-chlorophenyl of inhibitor 13 which is positioned in a hydrophobic pocket of NV 3CLpro.
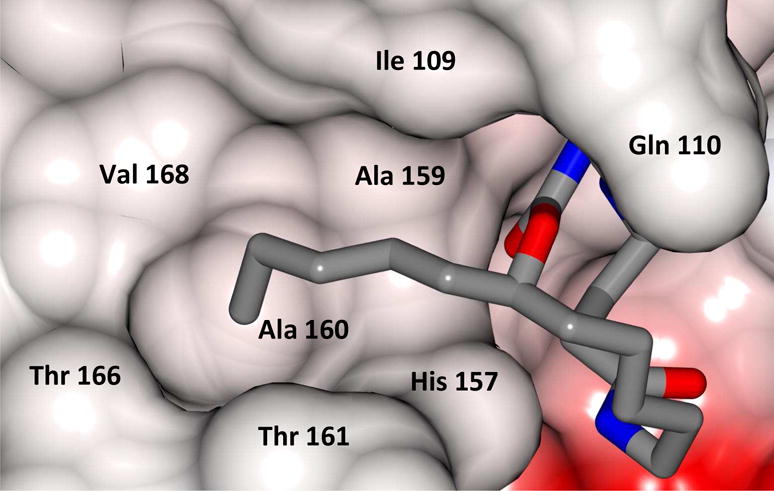
The m-chlorophenyl moiety occupies a predominantly hydrophobic pocket (Figure 5), consequently, this site was probed by synthesizing macrocycles having an n-pentyl chain. The resulting compounds were found to be ~ 2-fold less active (Table 1, compounds 21/29 and compounds 13/25). Inhibitor 21 is bound to the active site similarly to inhibitor 13 (Figures 6 and 7) and engages in similar H-bonding interactions (Figure 8), however, the Pro136 H-bond is missing. The latter, along with the higher entropic penalty associated with the alkyl chain, partially accounts for the lower affinity. The n-pentyl chain is clearly shown to engage in hydrophobic interactions (Figure 9). Modification of this portion of the inhibitor to facilitate the formation of new hydrogen bonds with the side chains of Thr161 or Thr168 could potentially enhance affinity (Figures 5 and 9).
Fig. 6.
Fo–Fc omit map of inhibitor 21 (green mesh) contoured at 3σ.
Fig. 7.
Surface representation of NV 3CL protease with bound inhibitor 21 site and with neighboring residues colored yellow (nonpolar), cyan (polar), and white (weakly polar). A) View of the inhibitor in the S1/S2 pocket and B) the S4 pocket. View is rotated counterclockwise approximately 90° about the vertical axis relative to panel A.
Fig. 8.
Hydrogen bond interactions (dashed lines) between inhibitor 21 and NV 3CL protease.
Fig. 9.
Electrostatic surface representation of NVPro showing the aliphatic chain of inhibitor 21 positioned in a hydrophobic S4 pocket.
3. Conclusions
Noroviruses are a leading cause of acute gastroenteritis in all age groups worldwide and have a significant impact on public health. There are currently no norovirus-specific therapeutics or prophylactics. In this report we describe the design, synthesis, and anti-norovirus activity of a novel class of macrocyclic inhibitors of NV 3CLpro. These studies provide new insights into the interaction of macrocyclic inhibitors with NV 3CLpro and demonstrate the nuanced interplay of structure, pharmacological activity, and cellular permeability. They also lay the ground work for conducting further studies related to the development of anti-norovirus therapeutics.
4. Experimental section
4.1. General
Reagents and dry solvents were purchased from various chemical suppliers (Aldrich, Acros Organics, ChemImpex, TCI America, Oakwood chemicals, Bachem, and Fisher) and were used as obtained. Silica gel (230–450 mesh) used for flash chromatography was purchased from Sorbent Technologies (Atlanta, GA). Thin layer chromatography was performed using Analtech silica gel plates. The 1H spectra were recorded in CDCl3 or DMSO-d6 on a Varian XL-400 NMR spectrometer and are reported relative to TMS (δH= 0.00 ppm). High resolution mass spectra (HRMS) were performed at the University of Kansas Mass Spectrometry lab using an LCT Premier mass spectrometer (Waters, Milford, MA) equipped with a time of flight mass analyzer and an electrospray ion source or a G6230B TOF MS (Agilent Technologies, Santa Clara, CA). Visualization was accomplished using UV light and/or iodine.
4.1.1. Synthesis of compounds 1a – f. General procedure
To a 250 mL round bottom flask kept under nitrogen atmosphere was added a 0.5 M solution (35 mmol) of the appropriate Grignard reagent (3-butenyl magnesium bromide or 4-pentenyl magnesium bromide) in THF and the solution was cooled to 0 – 5 °C. A solution of the appropriate aldehyde (35 mmol) in dry THF (20 mL) was added dropwise to the cooled Grignard solution over ~ 1 h. The reaction mixture was allowed to warm up to room temperature and stirred for 5 h under nitrogen. The disappearance of the aldehyde was monitored by TLC. The reaction mixture was cooled to 0 – 5°C and acidified to pH ~3.0 using 5% aqueous hydrochloric acid. The organic solvent was evaporated off and the residue was extracted with ethyl acetate (2 × 150 mL). The combined organic extracts were washed with brine (50 mL) dried over anhydrous Na2SO4, filtered, and concentrated. The crude product was purified by column chromatography to yield alcohols 1a–1f as oils.
4.1.1.1. 1-phenylpent-4-en-1-ol 1a
Oil (yield 80%); 1H NMR (400 MHz, CDCl3-d) δ ppm 1.68 – 1.99 (m, 2 H), 1.99 – 2.33 (m, 2 H), 4.58 – 4.74 (m, 1 H), 4.90 – 5.11 (m, 2 H), 5.73 – 5.94 (m, 1 H), 7.20 – 7.50 (m, 5 H). HRMS (ESI) calcd for C11H15O: [M+H]+: 163.1123 Found: 163.1126.
4.1.1.2. 1-phenylhex-5-en-2-ol 1b
Oil (yield 80%); 1H NMR (400 MHz, CDCl3-d) 1.53 – 1.69 (m, 2 H), 1,91 – 2.02 (m, 2 H), 2.70 – 2.93 (m, 2 H), 3.95 – 4.08 (m, 1 H), 4.88 – 5.09 (m, 3 H), 5.67 – 5.91 (m, 1 H), 7.23 – 7.34 (m, 5 H). HRMS (ESI) calcd for C12H17O: [M+H] + : 177.1279 Found: 177.1285.
4.1.1.3. 1-(3-chlorophenyl) pent-4-en-1-ol 1c
Oil, yield (80%), 1H NMR (400 MHz, CDCl3) δ ppm 1.71 – 1.96 (m, 2 H), 2.04 – 2.24 (m, 2 H), 4.69 (s, 1 H), 4.97 – 5.13 (m, 2 H), 5.77 – 5.93 (m, 1 H), 7.17 – 7.50 (m, 4 H). HRMS (ESI) calcd for C11H14ClO: [M+H]+: 197.0733 Found: 197.0735.
4.1.1.4. 1-(3-chlorophenyl) hex-5-en-1-ol 1d
Oil (yield 75%); 1H NMR (400 MHz, CDCl3-d) δ ppm 1.26 – 1.62 (m, 1 H), 1.59 – 1.85 (m, 1 H), 1.83 – 1.89 (m, 2 H), 2.02 – 2.13 (m, 2 H), 4.61 – 4.71 (m, 1 H), 4.95 – 5.08 (m, 3 H), 5.69 – 5.86 (m, 1 H), 7.16 – 7.30 (m, 3 H), 7.35 (s, 1 H). HRMS (ESI) calcd for C12H16ClO: [M+H]+: 211.0890 Found: 211.0894.
4.11.5. 1-phenyloct-7-en-4-ol 1e
Oil (yield 55%);1H NMR (400 MHz, CDCl3-d) δ ppm 1.41 – 1.59 (m, 4 H), 1.61 – 1.86 (m, 2 H), 2.04 – 2.26 (m, 2 H), 2.57 – 2.71 (t, 2 H), 3.59 – 3.70 (m, 1 H), 4.91 – 5.10 (m, 3 H), 5.76 – 5.89 (m, 1 H), 7.14 – 7.20 (d, 2 H), 7.23 – 7.30 (t, 3 H). HRMS (ESI) calcd for C14H21O: [M+H]+: 205.1592 Found: 205.1598.
4.1.1.6. Dec-1-en-5-ol 1f
Oil (yield 93%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.82 – 0.91(t, 2 H), 1.21 – 1.33 (m, 4 H), 1.35 – 1.44 (m, 2 H), 1.47 – 1.61 (m, 2 H), 2.05 – 2.25 (m, 2 H),3.55 – 3.63 (m, 1 H), 4.90 – 5.07 (m, 3 H), 5.76 – 5.89 (m, 1 H). HRMS (ESI) calcd for C10H21O: [M+H] +: 157.1592 Found: 157.1954.
4.1.2. Synthesis of carbamates 2a – k. General procedure
A solution of compound 1 (35 mmol) in dry acetonitrile (60 mL) was treated with trimethylamine (7.1 g; 70 mmol) followed by an appropriate amino acid methyl ester isocyanate 4 (35 mmol). The resulting reaction mixture was refluxed for 3 h and then allowed to cool to room temperature. The disappearance of the alcohol was monitored by TLC. The solvent was evaporated and the residue was taken up in ethyl acetate (250 mL) and the organic layer was washed with 5% aqueous HCl (2 × 50 mL) and saturated NaCl (50 mL). The organic layer was dried over anhydrous sulfate, filtered, and concentrated to yield an oily product. Purification by flash chromatography yielded esters 2a-k as colorless oils.
4.1.2.1. Methyl (((1-phenylpent-4-en-1-yl) oxy) carbonyl)-L-leucinate 2a
Oil (yield 80%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.68 – 1.05 (d, 6 H), 1.40 – 1.73 (m, 1 H), 1.73 – 1.95 (m, 2 H), 1.95 – 2.21 (m, 4 H), 3.64 – 3.86 (s,3 H), 4.45 – 4.53 (m, 1 H), 4.86 – 5.20 (m, 2 H), 5.54 – 5.69 (m, 1 H), 5.69 – 5.92 (m, 1 H), 7.17 – 7.35 (m, 5 H), 7.40 – 7.50 (d, 1 H). HRMS (ESI) calcd for C19H28NO4:[M+H] +: 334.2018 Found: 334.2021.
4.1.2.2. Methyl (((1-phenylhex-5-en-2-yl) oxy) carbonyl)-L-leucinate 2b
Oil (yield 80%); 1H NMR (400 MHz, CDCl3) δ ppm 0.81 – 0.97 (d, 6 H), 1.53 – 1.69 (m, 5 H), 1,91 – 2.02 (m, 2 H), 2.70 – 2.93 (m, 2 H), 3.71 – 3.78 (s, 3 H), 4.18 – 4.28 (m, 1 H), 4.88 – 5.09 (m, 3 H), 5.67 – 5.91 (m, 1 H), 6.20 – 6 60 (br, 1 H), 7.13 – 7.43 (m, 5 H), 7.71 – 7.80 (d, 1 H). HRMS (ESI) calcd for C20H30NO4: [M+H] +: 348.2175 Found: 348.2180.
4.1.2.3. Methyl (((1-(3-chlorophenyl) pent-4-en-1-yl) oxy) carbonyl)-L-leucinate 2c
Oil (yield 80%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.84 – 1.06 (d, 6 H), 1.41 – 1.72 (m, 1 H), 1.73 – 2.24 (m, 5 H), 2.42 – 2.57 (m, 2 H),3.62 – 3.77 (d, 3 H), 4.23 – 4.42 (m, 1 H), 4.60 – 4.74 (m, 1 H), 4.92 – 5.24 (m, 1 H), 5.54 – 5.68 (m, 1 H), 5.72 – 5.98 (m, 1 H), 7.15 – 7.32 (m, 3 H), 7.41 (s, 1 H). HRMS (ESI) calcd for C19H27ClNO4: [M+H] +: 368.1629 Found: 368.1633.
4.1.2.4. Methyl (2S)-2-((((1-(3-chlorophenyl) pent-4-en-1-yl) oxy) carbonyl) amino)-3-cyclohexylpropanoate 2d
Oil (yield 75%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.73 – 0.93 (m, 4 H), 1.03 – 1.31 (m, 2H), 1.31 – 1.55 (m, 2 H),1.55 – 1.75 (m, 2 H), 1.74 – 1.89 (m, 4 H), 2.00 – 2.21 (m, 2 H), 3.52 – 3.61 (s, 3H), 4.44 – 4.50 (m, 1 H), 4.95 – 5.18 (m, 3 H), 5.67 – 5.89 (m, 2 H), 7.14 – 7.35 (m, 4 H), 7.44 – 7.49 (s, 1 H). HRMS (ESI) calcd for C22H31ClNO4: [M+H] +: 408.1942 Found: 408.1947.
4.1.2.5. Methyl (((1-(3-chlorophenyl) hex-5-en-1-yl) oxy) carbonyl)-L-leucinate 2e
Oil (yield 80%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.87 – 0.92 (d, 6 H), 1.30 – 1.62 (m, 3 H), 1.61 – 1.84 (m, 2 H), 1.97 – 2.16 (m, 4 H), 3.61 – 3.70 (s, 3 H), 4.60 – 4.70 (m, 1 H), 4.89 – 5.06 (m, 2 H), 5.66 – 5.87 (m, 2 H), 7.13 – 7.38 (m, 4 H), 7.40 – 7.45 (s, 1 H). HRMS (ESI) calcd for C20H29ClNO4: [M+H] +: 382.1785 Found: 382.1789.
4.1.2.6. Methyl (((1-(3-chlorophenyl) pent-4-en-1-yl) oxy) carbonyl)-L-alaninate 2f
Oil (yield 80%); 1H NMR (400 MHz, CDCl3-d) δ ppm 1.42 (d, J=7.03 Hz, 3 H), 1.75 – 1.92 (m, 2 H), 1.90 – 2.14 (m, 2 H), 3.74 (br d, J=16.01 Hz, 3 H), 4.27 – 4.39 (m, 1 H), 4.94 – 5.09 (m, 2 H), 5.24 – 5.44 (d, 1 H), 5.55 – 5.68 (m, 1 H), 5.72 – 5.87 (m, 1 H), 7.26 (dd, J=4.10, 0.98 Hz, 4 H). HRMS (ESI) calcd for C16H21ClNO4: [M+H] +: 326.1159 Found: 326.1162.
4.1.2.7. Methyl (((1-(3-chlorophenyl) hex-5-en-1-yl) oxy) carbonyl)-L-alaninate 2g
Oil (yield 80%); 1H NMR (400 MHz, CDCl3-d) δ ppm 1.27 – 1.56 (m, 5 H), 1.66 – 2.14 (m, 4 H), 3.71 (br d, J=16.01 Hz, 3 H), 4.24 – 4.39 (m, 1 H), 4.89 – 5.06 (m, 2 H), 5.29 – 5.49 (m, 1 H), 5.54 – 5.83 (m, 2 H), 7.10 – 7.36 (m, 4 H). 8.10 – 8.15 (br s, 1 H). HRMS (ESI) calcd for C17H23ClNO4: [M+H] +: 340.1316 Found: 340.1322.
4.1.2.8. Methyl 1-((((1-(3-chlorophenyl) pent-4-en-1-yl) oxy) carbonyl) amino) cyclohexane-1-carboxylate 2h
Oil (yield 70%); 1H NMR (400 MHz, CDCl3-d) δ ppm 1.18 – 1.66 (m, 6H), 1.67 – 2.21 (m, 6 H), 2.23 – 2.40 (m, 2 H), 3.59 (s, 3 H), 4.53 – 4.73 (m, 1 H), 4.93 – 5.13 (m, 2 H), 5.48 – 5.64 (m, 1 H), 5.71 – 5.93 (m, 1 H), 7.24 (m, 4 H). HRMS (ESI) calcd for C20H27ClNO4: [M+H] +: 380.1629 Found: 380.1635.
4.1.2.9. Methyl 1-((((1-(3-chlorophenyl) hex-5-en-1-yl) oxy) carbonyl) amino) cyclohexane-1-carboxylate 2i
Oil, yield (65%); 1H NMR (400 MHz, CDCl3-d) δ ppm 1.30 – 2.16 (m, 16 H), 3.57 – 3.65 (s, 3 H), 4.62 – 4.71 (m, 1 H), 4.88 – 5.08 (m, 2 H), 5.48 – 5.63 (m, 1 H), 5.67 – 5.87 (m, 1 H), 7.11 – 7.30 (m, 3 H), 7.32 – 7.39 (s, 1 H). HRMS (ESI) calcd for C21H29ClNO4: [M+H] +: 394.1785 Found: 394.1790.
4.1.2.10. Methyl (((1-phenyloct-7-en-4-yl) oxy) carbonyl)-L-leucinate 2j
Oil (yield 88%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.83 – 0.96 (d, 6 H), 1.37 – 1.92 (m, 7 H), 1.95 – 2.07 (m, 3 H), 2.49 – 2.65 (t, 2 H), 3.63 – 3.73 (s, 3 H), 4.28 – 4.48 (m, 1 H), 4.71 – 4.81 (m, 1 H), 4.88 – 5.08 (m, 2 H), 5.48 – 5.63 (m, 1 H), 5.67 – 5.87 (m, 1 H), 7.08 – 7.18 (m, 2 H), 7.18 – 7.28 (m, 3 H). HRMS (ESI) calcd for C22H34NO4: [M+H] +: 376.2488 Found: 376.2452.
4.1.2.11. Methyl ((dec-1-en-5-yloxy) carbonyl)-L-leucinate 2k
Oil (yield 91%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.83 – 0.90 (t, 2 H),0.90 – 0.98 (d, 6 H), 1.20 – 1.35 (m, 6 H), 1.45 – 1.70 (m, 4 H), 1.98 – 2.15 (m, 4 H), 3.69 – 3.76 (s, 3 H), 4.30 – 4.43 (m, 1 H), 4.70 – 4.80 (m, 1 H), 4.90 – 5.05 (m, 2 H), 5.70 – 5.80 (m, 1 H), 7.80 – 7.89 (d, 1 H). HRMS (ESI) calcd for C18H34NO4: [M+H] +: 328.2488 Found: 328.2454.
4.1.3. Synthesis of acids (3a–g and 3j–k). General procedure A
A solution of ester 2 (10 mmol) in tetrahydrofuran (15 mL) was treated with 1M aqueous LiOH (20 mL). The reaction mixture was stirred for 3 h at room temperature while monitoring the disappearance of the ester by TLC. Most of the solvent was evaporated off and the solution was acidified to pH ~3 using 5% hydrochloric acid (10 mL). The aqueous layer was extracted with ethyl acetate (2 × 100 mL) and the combined organic layer was washed with brine (50 mL). The organic layer was dried over anhydrous sodium sulfate, filtered, and concentrated to yield compounds 3a–g and 3j–k as colorless oils.
4.1.3.1. (((1-phenylpent-4-en-1-yl) oxy) carbonyl)-L-leucine 3a
Oil (yield 92%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.68 – 1.05 (d, 6 H), 1.40 – 1.73 (m, 1 H), 1.73 – 1.95 (m, 2 H), 1.95 – 2.21 (m, 4 H), 4.45 – 4.53 (m, 1 H), 4.86 – 5.20 (m, 2 H), 5.54 – 5.69 (m, 1 H), 5.69 – 5.92 (m, 1 H), 7.17 – 7.35 (m, 5 H), 7.40 – 7.50 (d, 1 H). HRMS (ESI) calcd for C18H26 NO4: [M+H] +: 320.1862 Found: 320.1865.
4.1.3.2. (((1-phenylhex-5-en-2-yl) oxy) carbonyl)-L-leucine 3b
Oil (yield 91%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.81 – 0.97 (d, 6 H), 1.53 – 1.69 (m, 5 H), 1,91 – 2.02 (m, 2 H), 2.70 – 2.93 (m, 2 H), 4.18 – 4.28 (m, 1 H), 4.88 – 5.09 (m, 3 H), 5.67 – 5.91 (m, 1 H), 6.20 – 6 60 (br., 1 H), 7.13 – 7.43 (m, 5 H), 7.71 – 7.80 (d, 1 H). HRMS (ESI) calcd for C19H28NO4: [M+H] +: 334.2018 Found: 334.2022.
4.1.3.3. (((1-(3-chlorophenyl) pent-4-en-1-yl) oxy) carbonyl)-L-leucine 3c
Oil (yield 92%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.84 – 1.06 (d, 6 H), 1.41 – 1.72 (m, 1 H), 1.73 – 2.24 (m, 5 H), 2.42 – 2.57 (m, 2 H), 4.23 – 4.42 (m, 1 H), 4.60 – 4.74 (m, 1 H), 4.92 – 5.24 (m, 1 H), 5.54 – 5.68 (m, 1 H), 5.72 – 5.98 (m, 1 H), 7.15 – 7.32 (m, 3 H), 7.41 (s, 1 H). HRMS (ESI) calcd for C18H25ClNO4: [M+H] +: 354.1472 Found: 354.1475.
4.1.3.4. (2S)-2-((((1-(3-chlorophenyl) pent-4-en-1-yl) oxy) carbonyl) amino)-3-cyclohexylpropanoic acid 3d
Oil (yield 90%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.77 – 1.30 (m, 7 H), 1.58 – 2.21 (m, 8 H), 4.25 – 4.44 (m, 2 H), 4.65 – 4.73 (m, 1 H), 4.95 – 5.22 (m, 2 H), 5.53 – 5.71 (m, 1 H), 5.71 – 5.88 (m, 1 H), 7.14 – 7.41 (m, 4 H), 7.44 – 7.49 (s, 1 H). HRMS (ESI) calcd for C21H29ClNO4: [M+H] +: 394.1785 Found: 394.1791.
4.1.3.5. (((1-(3-chlorophenyl) hex-5-en-1-yl) oxy) carbonyl)-L-leucine 3e
Oil (yield 95%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.85 – 1.00 (d, 6 H), 1.41 – 1.78 (m, 5 H), 1.95 – 2.17 (m, 4 H), 4.29 – 4.48 (m, 1 H), 4.63 – 4.73 (m, 1 H), 4.84 – 5.01 (m, 1 H), 5.18 – 5.30 (m, 2 H), 5.72 – 5.85 (m, 2 H), 7.14 – 7.40 (m, 4 H). HRMS (ESI) calcd for C19H27ClNO4: [M+H] +: 368.1629 Found: 368.1637.
4.1.3.6. (((1-(3-chlorophenyl) pent-4-en-1-yl) oxy) carbonyl)-L-alanine 3f
Oil (yield 92%); 1H NMR (400 MHz, CDCl3-d) δ ppm 1.34 – 1.47 (d, 3 H), 1.75 – 1.91 (m, 2 H), 1.91 – 2.11 (m, 2 H), 4.28 – 4.46 (m, 1 H), 4.97 – 5.13 (m, 2 H), 5.35 – 5.46 (m, 1 H), 5.57 – 5.66 (m, 1 H), 5.73 – 5.87 (m, 1 H), 7.28 (br d, J=5.47 Hz, 3 H), 7.31 – 7.42 (s, 1 H). HRMS (ESI) calcd for C15H19ClNO4: [M+H] +: 312.1003 Found: 312.1010.
4.1.3.7. (((1-(3-chlorophenyl) hex-5-en-1-yl) oxy) carbonyl)-L-alanine 3g
Oil (yield 92%); 1H NMR (400 MHz, CDCl3-d) δ ppm 1.27 – 1.56 (m, 5 H), 1.66 – 2.14 (m, 4 H), 4.24 – 4.39 (m, 1 H), 4.89 – 5.06 (m, 2 H), 5.29 – 5.49 (m, 1 H), 5.54 – 5.83 (m, 2 H), 7.10 – 7.36 (m, 4 H). 8.10 – 8.15 (br s, 1 H). HRMS (ESI) calcd for C16H21ClNO4: [M+H] +: 326.1159 Found: 326.1168.
4.1.3.8. (((1-phenyloct-7-en-4-yl) oxy) carbonyl)-L-leucine 3j
Oil (yield 94%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.83 – 0.96 (d, 6 H), 1.37 – 1.92 (m, 7 H), 1.95 – 2.07 (m, 3 H), 2.49 – 2.65 (t, 2 H), 4.28 – 4.48 (m, 1 H), 4.71 – 4.81 (m, 1 H), 4.88 – 5.08 (m, 2 H), 5.48 – 5.63 (m, 1 H), 5.67 – 5.87 (m, 1 H), 7.08 – 7.18 (m, 2 H), 7.18 – 7.28 (m, 3 H), 8.10 – 8.16 (d, 1 H). HRMS (ESI) calcd for C21H32NO4: [M+H] +: 362.2331 Found 362.2333.
4.1.3.9. ((Dec-1-en-5-yloxy) carbonyl)-L-leucine 3k
Oil (yield 91%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.83 – 0.90 (t, 2 H),0.90 – 0.98 (d, 6 H), 1.20 – 1.35 (m, 6 H), 1.45 – 1.70 (m, 4 H), 1.98 – 2.15 (m, 4 H), 4.30 – 4.43 (m, 1 H), 4.70 – 4.80 (m, 1 H), 4.90 – 5.05 (m, 2 H), 5.70 – 5.80 (m, 1 H), 7.80 – 7.89 (d, 1 H). HRMS (ESI) calcd for C17H32NO4: [M+H] +: 314.2331 Found 314.2339.
4.1.3. Synthesis of acids 3h–i. General procedure B
A solution of ester 2a or 2i (10 mmol) in tetrahydrofuran (10 mL) and methanol (10 mL) was treated with 2M aqueous LiOH (20 mL) and the reaction mixture was heated to 60°C for 12 h with stirring. The disappearance of the ester was monitored by TLC. The reaction mixture was cooled to room temperature, the solvent was removed under reduced pressure, and the solution was acidified to pH ~3 using 5% hydrochloric acid (~20 mL). The aqueous layer was extracted with ethyl acetate (2 × 100 mL) and the combined extracts were washed with brine (50 mL). The organic layer was dried over anhydrous Na2SO4, filtered, and concentrated to yield compounds 3h–i as oils.
4.1.3.10. 1-((((1-(3-chlorophenyl) pent-4-en-1-yl) oxy) carbonyl) amino) cyclohexane-1-carboxylic acid 3h
Oil (yield 90%); 1H NMR (400 MHz, CDCl3) δ ppm 1.18 – 1.66 (m, 6H), 1.67 – 2.21 (m, 6 H), 2.23 – 2.40 (m, 2 H), 4.69 (dd, J=7.62, 5.27 Hz, 1 H), 4.94 – 5.09 (m, 2 H), 5.57 – 5.62 (m, 1 H), 5.72 – 5.90 (m, 1 H), 7.14 – 7.17 (m, 1 H), 7.15 – 7.31 (m, 4 H). HRMS (ESI) calcd for C19H25ClNO4: [M+H] +: 366.1472 Found 366.1478.
4.1.3.11. 1-((((1-(3-chlorophenyl) hex-5-en-1-yl) oxy) carbonyl) amino) cyclohexane-1-carboxylic acid 3i
Oil (yield 95%); 1H NMR (400 MHz, CDCl3) δ ppm 1.30 – 2.16 (m, 16 H), 4.62 – 4.71 (m, 1 H), 4.88 – 5.08 (m, 2 H), 5.48 – 5.63 (m, 1 H), 5.67 – 5.87 (m, 1 H), 7.11 – 7.30 (m, 3 H), 7.32 – 7.39 (s, 1 H). HRMS (ESI) calcd for C20H27ClNO4: [M+H] +: 380.1629 Found 380.1637.
4.1.4. Synthesis of amino acid methyl ester isocyanates 4a–d. General procedure
The appropriate amino acid methyl ester hydrochloride (55 mmol) was placed in a dry 500-mL RB flask and then dried overnight on the vacuum pump. The flask was flushed with nitrogen and dry dioxane (150 mL) was added followed by trichloromethyl chloroformate (16.26 g, 82.5 mmol). After refluxing for 12 h, the solvent was removed on the rotary evaporator and the residue was vacuum distilled to yield pure isocyanates 4a–d as colorless oils.
4.1.5. Synthesis of compounds 5a–b. General procedure
To a solution of Boc-L-glutamic acid α-methyl ester (80 mmol) in dry DMF (200 mL) were added EDCI (19.93 g; 104 mmol), HOBt (15.92 g; 104 mmol) and the reaction mixture was stirred for 30 min at room temperature. Following the sequential addition of allylamine/O-allylhydroxylamine hydrochloride (120 mmol) and DIEA (20.68 g; 160 mmol), the reaction mixture was stirred for 16 h at room temperature. Completion of the reaction was monitored by TLC. The solvent was removed and the residue was partitioned between ethyl acetate (400 mL) and 5% aqueous HCl (100 mL). The layers were separated and the organic layer was further washed with saturated aqueous NaHCO3 (2 × 100 mL), followed by saturated NaCl (100 mL). The organic layer was dried over anhydrous sodium sulfate, filtered and concentrated to yield a yellow-colored oily product. Purification by flash chromatography yielded a white solid.
4.1.5.1. Methyl N5-allyl-N2-(tert-butoxycarbonyl)-L-glutaminate 5a
White solid (yield 85%); m.p. 69–70°C; 1H NMR (400 MHz, CDCl3-d) δ ppm 1.42 – 1.50 (s, 9 H), 1.90 – 2.00 (m, 1 H), 2.16 – 2.28 (m. 1 H), 3.74 – 3.80 (s, 3 H), 3.91 – 4.00 (t, 2 H), 4.34 – 4.41 (m, 1 H), 5.09 – 5.22 (m, 2 H), 5.30 – 5.40 (br. s, 1 H), 5.78 – 5.94 (m, 1 H), 6.10 – 6.20 (br. s, 1 H). HRMS (ESI) calcd for C14H25N2O5: [M+H] + : 301.1763 Found 301.1771.
4.1.5.2. Methyl N5-(allyloxy)-N2-(tert-butoxycarbonyl)-L-glutaminate 5b
Sticky oil (yield 54%); 1H NMR (400 MHz, CDCl3-d) δ ppm 1.41 – 1.50 (s, 9 H), 1.78 – 1.96 (br. s, 1 H), 2.12 – 2.28 (m. 4 H), 3.72 – 3.80 (s, 3 H), 4.24 – 4.37 (m, 2 H), 4.44 – 4.51 (m, 1 H), 5.29 – 5.50 (m, 2 H), 5.92 – 6.07 (m, 1 H), 9.29 – 9.42 (br. s, 1 H). HRMS (ESI) calcd for C14H25N2O6: [M+H] +: 317.1713 Found 317.1718.
4.1.6. Synthesis of compounds 6a–b. General procedure
To a solution of compound 5 (43 mmol) in dry DCM (30 mL) was added a solution of 4 M HCl in dioxane (130 mL) with stirring. The reaction mixture was stirred for 3 h at room temperature. The disappearance of the starting material was monitored by TLC. The solvent was evaporated under reduced pressure and compound 6 was used in the next step without further purification.
4.1.7. Synthesis of acyclic compounds 7a–k and 32
General procedure
To a solution of compound 3 (15 mmol) in dry DMF (40 mL) was added EDCI (3.74 g, 19.5 mmol, 1.30 eq), HOBt (2.97 g, 19.5 mmol, 1.30 eq) and the mixture was stirred for 30 min at room temperature. In a separate flask, a solution of compound 6a (3.55 g, 15 mmol) in DMF (20 mL) cooled to 0–5°C was treated with diisopropyl ethylamine (DIEA) (7.75 g, 60 mmol, 4 eq), stirred for 30 min, and then added to the reaction mixture containing acid 3. The reaction mixture was stirred for 16 h while monitoring the reaction by TLC. The solvent was removed and the residue was partitioned between ethyl acetate (300 mL) and 10% citric acid (50 mL). The layers were separated and the organic layer was further washed with saturated aqueous NaHCO3 (2 × 50 mL), followed by brine (50 mL). The organic layer was dried over anhydrous sodium sulfate, filtered and concentrated to yield a yellow-colored oily product. Purification by flash chromatography yielded esters 7a–k.
4.1.7.1. Methyl N5-allyl-N2-((((1-phenylpent-4-en-1-yl) oxy) carbonyl)-L-leucyl)-L-glutaminate 7a
Oil (yield 62%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.68 – 1.05 (d, 6 H), 1.40 – 1.73 (m, 3 H), 1.73 – 1.95 (m, 2 H), 1.95 – 2.21 (m, 6 H), 3.64 – 3.76 (s,3 H), 4.45 – 4.53 (m, 4 H), 4.86 – 5.20 (m, 4 H), 5.54 – 5.69 (m, 2 H), 5.69 – 5.92 (m, 2 H), 7.17 – 7.35 (m, 5 H), 7.40 – 7.50 (d, 1 H), 7.90 – 7. 80 (d, 1 H). HRMS (ESI) calcd for C27H40N3O6: [M+H] +: 502.2917 Found 502.2921.
4.1.7.2. Methyl N5-allyl-N2-((((1-phenylhex-5-en-2-yl) oxy) carbonyl)-L-leucyl)-L-glutaminate 7b
Oil (yield 60%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.81 – 0.97 (d, 6 H), 1.53 – 1.69 (m, 5 H), 1, 91 – 2.02 (m, 6 H), 2.70 – 2.93 (m, 2 H), 3.73 (s, 3 H), 3.80 – 3.94 (t, 2 H), 4.40 – 4.62 (m, 2 H), 4.88 – 5.04 (m, 2 H), 5.06 – 5.26 (m, 2 H), 5.68 – 5.87 (m, 3 H), 6.20 – 6.41 (br. d, 1 H), 6.91 – 7.10 (d, 1 H), 7.14 – 7.36 (m, 5 H), 7.85 – 7.92 (d, 1 H). HRMS (ESI) calcd for C28H42N3O6: [M+H] +: 516.3074 Found 516.3080.
4.1.7.3. Methyl N5-allyl-N2-((((1-(3-chlorophenyl) pent-4-en-1-yl) oxy) carbonyl)-L-leucyl)-L-glutaminate 7c
Oil (yield 65%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.74 – 1.02 (d, 6 H), 1.43 – 2.13 (m, 4 H), 2.13 – 2.38 (m, 6 H), 3.60 – 3.71 (s, 3 H), 4.07 – 4.24 (m, 1 H), 4.41 – 4.61 (m, 2 H), 4.91 – 5.25 (m, 1 H), 5.25 – 5.42 (m, 2 H), 5.50 – 5.68 (m, 2 H), 5.70 – 5.92 (m, 2 H), 6.07 – 6.26 (m, 2 H), 6.30 – 6.39 (m, 1 H), 6.96 – 7.34 (m, 4 H), 7.7 – 7.8 (d, 1 H), 8.1 – 8.2 (d, 1 H). HRMS (ESI) calcd for C27H39ClN3O6: [M+H] +: 536.2527 Found 536.2534.
4.1.7.4. Methyl N5-allyl-N2-((2S)-2-((((1-(3-chlorophenyl) pent-4-en-1-yl) oxy) carbonyl) amino)-3-cyclohexylpropanoyl) glutaminate 7d
Oil (yield70%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.75 – 1.59 (m, 4 H), 1.55 – 1.73 (m, 4 H), 1.74 – 1.89 (m, 4 H), 1.89 – 2.14 (m, 5 H), 2.14 – 2.31 (m, 4 H), 3.65 – 3.76 (d, 3 H), 3.76 – 4.02 (m, 1 H), 4.07 – 4.29 (m, 1 H), 4.40 – 4.61 (m, 2 H), 4.93 – 5.38 (m, 2 H), 5.43 – 5.64 (m, 2 H), 5.69 – 5.90 (m, 3 H), 6.10 – 6.20 (d, 1 H), 6.35 – 6.44 (d, 1 H), 6.86 – 7.05 (d, 1 H), 7.12 – 7.37 (m, 4 H). HRMS (ESI) calcd for C30H43ClN3O6: [M+H] +: 576.2840 Found 576.2848.
4.1.7.5. Methyl N5-allyl-N2-((((1-(3-chlorophenyl) hex-5-en-1-yl) oxy) carbonyl)-L-leucyl)-L-glutaminate 7e
Oil (yield 75%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.82 – 1.03 (d, 6 H), 1.20 – 2.32 (m, 13 H), 3.73 (br d, J=12.89 Hz, 3 H), 4.03 – 4.20 (m, 2 H), 4.38 – 4.59 (m, 2 H), 4.89 – 5.03 (m, 2 H), 5.08 – 5.32 (m, 2 H), 5.48 – 5.67 (m, 1 H), 5.68 – 5.94 (m, 1 H), 5.98 – 6.12 (m, 1 H), 6.21 – 6.34 (br s, 1 H), 6.86 – 7.03 (br s, 2 H), 7.13 – 7.37 (m, 4 H). HRMS (ESI) calcd for C28H41ClN3O6: [M+H] +: 550.2684 Found 550.2691.
4.1.7.6. Methyl N5-allyl-N2-((((1-(3-chlorophenyl) pent-4-en-1-yl) oxy) carbonyl)-L-alanyl)-L-glutaminate 7f
Oil (yield 80%); 1H NMR (400 MHz, CDCl3-d) δ ppm 1.39 (d, J=7.03 Hz, 3 H), 1.75 – 1.91 (m, 4 H), 1.91 – 2.11 (m, 4 H), 3.66 – 3.71 (s, 3 H), 4.17 – 4.27 (d, 2 H), 4.41 – 4.61 (m, 2 H), 4.92 – 5.25 (m, 4 H), 5.47 – 5.65 (m, 1 H), 5.71 – 5.88 (m, 2 H), 6.15 – 6.20 (br s, 1 H), 6.40 – 6.45 (br s, 1 H), 7.12 – 7.32 (m, 4 H), 7.50 – 7.60 (d, 1 H). HRMS (ESI) calcd for C24H33ClN3O6: [M+H] +: 494.2058 Found 494.2067.
4.1.7.7. Methyl N5-allyl-N2-((((1-(3-chlorophenyl) hex-5-en-1-yl) oxy) carbonyl)-L-alanyl)-L-glutaminate 7g
Oil (yield 80%); 1H NMR (400 MHz, CDCl3-d) δ ppm 1.30 – 1.39 (m, 2 H), 1.39 – 1.42 (d, J=7.03 Hz, 3 H), 1.75 – 1.91 (m, 4 H), 1.91 – 2.11 (m, 4 H), 3.66 – 3.71 (s, 3 H), 4.17 – 4.27 (d, 2 H), 4.41 – 4.61 (m, 2 H), 4.92 – 5.25 (m, 4 H), 5.47 – 5.65 (m, 1 H), 5.71 – 5.88 (m, 2 H), 6.15 – 6.20 (br s, 1 H), 6.40 – 6.45 (br s, 1 H), 7.12 – 7.32 (m, 4 H), 7.50 – 7.60 (d, 1 H). HRMS (ESI) calcd for C25H35ClN3O6: [M+H] +: 508.2214 Found 508.2218.
4.1.7.8. Methyl N5-allyl-N2-(1-((((1-(3-chlorophenyl) pent-4-en-1-yl) oxy) carbonyl) amino) cyclohexane-1-carbonyl)-L-glutaminate 7h
Oil (yield 55%); 1H NMR (400 MHz, CDCl3) δ ppm 1.20 – 1.48 (m, 5 H), 1.49 – 1.74 (m, 5 H), 1.75 – 1.90 (m, 4 H), 1.90 – 2.35 (m, 4 H), 3.71 (s, 3 H), 3.76 – 3.91 (m, 2 H), 4.41 – 4.61 (m, 1 H), 4.95 – 5.26 (m, 4 H), 5.72 – 5.91 (m, 3 H), 6.29 – 6.45 (br. d, 1 H), 6.57 – 6.71 (br. s, 1 H), 6.95 – 7.06 (d, 1 H), 7.15 – 7.35 (m, 4 H). HRMS (ESI) calcd for C28H39ClN3O6: [M+H] +: 548.2527 Found 548.2531.
4.1.7.9. Methyl N5-allyl-N2-(1-((((1-(3-chlorophenyl) hex-5-en-1-yl) oxy) carbonyl) amino) cyclohexane-1-carbonyl)-L-glutaminate 7i
Oil (yield 57%); 1H NMR (400 MHz, CDCl3-d) δ ppm 1.16 – 1.53 (m, 7 H), 1.54 – 1.74 (m, 7 H), 1.75 – 2.34 (m, 6 H), 3.71 (s, 3 H), 3.73 – 3.90 (m, 2 H), 4.39 – 4.63 (m, 1 H), 4.90 – 5.27 (m, 4 H), 5.41 – 5.61 (m, 1 H), 5.65 – 5.91 (m, 2 H), 6.29 – 6.44 (br. s, 1 H), 6.60 – 6.66 (d, 1 H), 6.89 – 7.03 (d, 1 H), 7.13 – 7.18 (m, 3 H), 7.21 – 7.31 (s, 1 H). HRMS (ESI) calcd for C29H41ClN3O6: [M+H] +: 562.2684 Found 562.2685.
4.1.7.10. Methyl N5-allyl-N2-((((1-phenyloct-7-en-4-yl) oxy) carbonyl)-L-leucyl)-L-glutaminate 7j
White solid (yield 62%); m.p 99 – 100° C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.90 – 0.99 (d, 6 H), 1.46 – 1.74 (m, 7 H), 1.75 – 1.82 (t, 2 H),1.90 – 2.12 (m, 4 H), 2.14 – 2.35 (m, 2 H), 2.52 – 2.60 (t, 2 H), 3.70 – 3.75 (s, 3 H), 3.80 – 3.90 (d, 2 H), 4.10 – 4.20 (m, 1 H), 4.48 – 4.59 (m, 1 H), 4.71 – 4.84 (m, 1 H), 4.90 – 5.23 (m, 4 H), 5.70 – 5.90 (m, 2 H), 6.20 – 6.30 (br. s, 1 H), 6.30 – 6.40 (br. s, 1 H), 6.93 – 6.70 (d, 1 H), 7.11 – 7.20 (d, 2 H), 7.22 – 7.30 (m, 3 H). HRMS (ESI) calcd for C30H46N3O6: [M+H] +: 544.3387 Found 544.3390.
4.1.7.11. Methyl N5-allyl-N2-(((dec-1-en-5-yloxy) carbonyl)-L-leucyl)-L-glutaminate 7k
White solid (yield 70%); m.p 103 – 104°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.83 – 0.91 (t, 2 H),0.91 – 0.99 (d, 6 H), 1.20 – 1.35 (m, 6 H), 1.45 – 1.70 (m, 4 H), 1.98 – 2.12 (m, 4 H), 2.14 – 2.35 (m, 2 H), 2.52 – 2.60 (t, 2 H), 3.72 – 3.76 (s, 3 H), 3.80 – 3.90 (d, 2 H), 4.10 – 4.20 (m, 1 H), 4.50 – 4.60 (m, 1 H), 4.66 – 4.78 (m, 1 H), 4.91 – 5.24 (m, 4 H), 5.71 – 5.90 (m, 2 H), 6.35 – 6.50 (br. s, 2 H), 6.91 – 7.00 (d, 1 H). HRMS (ESI) calcd for C26H46N3O6: [M+H] +: 496.3387 Found 496.3388.
4.1.7.12. Methyl N5-(allyloxy)-N2-((((1-(3-chlorophenyl) pent-4-en-1-yl)oxy)carbonyl)-L-leucyl)-L-glutaminate 32
Oil (yield 52%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.83 – 1.00 (dd, 6 H), 1.23 – 1.29 (t, 2 H), 1.43 – 2.15 (m, 5 H), 2.14 – 2.35 (m, 4 H), 3.69 – 3.80 (d, 3 H), 4.03 – 4. 22 (m, 2 H), 4.32 – 4.48 (m, 2 H), 4.93 – 5.06 (m, 2 H), 5.23 – 5.44 (m, 2 H), 5.50 – 5.61 (m, 1 H), 5.73 – 5.84 (m, 1 H), 5.87 – 6.05 (m, 1 H), 6.76 – 6. 89 (br. s, 1 H), 7.13 – 7.20 (s, 1 H), 7.23 – 7.33 (m, 3 H), 9.22 – 9.29 (d, 1 H), 9.43 – 9.51 (d, 1 H). HRMS (ESI) calcd for C27H39ClN3O7: [M+H] +: 552.2477 Found 552.2479.
4.1.8. Synthesis of compounds 8a–k and 33. General procedure for ring closing metathesis.
A solution of acyclic diene 7 (1.21 mmol) in dry DCM (1.5 L) was degassed for 30 min using nitrogen. Chloro dicyclohexyl borane (1 M in hexane) (1.21 mL; 1.21 mmol) and Grubb’s 2nd generation catalyst (104 mg; 10 mol%) were added and degassing was continued for 10 min. The reaction mixture was heated to 45 °C and stirred for 30 min under a nitrogen atmosphere. An additional portion of Grubb’s 2nd generation catalyst (52 mg; 5 mol%) was added and the solution stirred for 16 h at 45 °C under a nitrogen atmosphere. The reaction was quenched by adding activated charcoal (600 mg) and stirring the reaction for 18 h at room temperature. The reaction mixture was filtered through a Celite bed and the solvent was removed on the rotary evaporator. The crude residue was purified by flash chromatography to give macrocyclic esters 8a–k as off-white solids.
4.1.8.1. Methyl (4S,7S, E)-4-isobutyl-2,5,10-trioxo-17-phenyl-1-oxa-3,6,11-triazacycloheptadec-13-ene-7-carboxylate 8a
Off-white solid (yield 65%); m.p 79 – 80°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.68 – 1.05 (d, 6 H), 1.40 – 1.73 (m, 3 H), 1.73 – 1.95 (m, 2 H), 1.95 – 2.21 (m, 6 H), 3.64 – 3.76 (s, 3 H), 3.80 – 3.89 (m, 2 H), 4.45 – 4.53 (m, 2 H), 5.51 – 5.74 (m, 2 H), 5.76 – 5.90 (m, 1 H), 7.17 – 7.35 (m, 5 H), 7.40 – 7.50 (d, 1 H), 7.90 – 7. 80 (d, 1 H), 8.25 – 8.35 (d, 1 H). HRMS (ESI) calcd for C25H36N3O6: [M+H] +: 474.2604 Found: 474.2608.
4.1.8.2. Methyl (4S,7S, E)-17-benzyl-4-isobutyl-2,5,10-trioxo-1-oxa-3,6,11-triazacycloheptadec-13-ene-7-carboxylate 8b
Off-white solid (yield 68%); m.p 70 – 71°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.81 – 0.97 (d, 6 H), 1.53 – 1.69 (m, 4 H), 1,91 – 2.02 (m, 5 H), 2.70 – 2.93 (m, 2 H), 3.73 – 3.80 (s, 3 H), 3.80 – 3.94 (m, 2 H), 4.40 – 4.62 (m, 2 H), 4.88 – 5.04 (m, 1 H), 5.06 – 5.26 (m, 2 H), 5.68 – 5.87 (m, 2 H), 6.20 – 6.41 (br. d, 1 H), 6.91 – 7.10 (d, 1 H), 7.14 – 7.36 (m, 5 H), 7.85 – 7.92 (d, 1 H). HRMS (ESI) calcd for C26H38N3O6: [M+H] +: 488.2761 Found: 488.2767.
4.1.8.3. Methyl (4S,7S, E)-17-(3-chlorophenyl)-4-isobutyl-2,5,10-trioxo-1-oxa-3,6,11-triazacycloheptadec-13-ene-7-carboxylate 8c
Off-white solid (yield 70%); m.p 84 – 85 °C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.74 – 0.99 (d, 6 H), 1.42 – 1.83 (m, 3 H), 2.11 – 2.43 (m, 6 H),2.47 – 2.64 (m, 2 H), 3.28 – 3.40 (m, 2 H), 3.69 – 3.81 (s, 3 H), 3.81 – 3.96 (t, 2 H), 4.62 – 4.77 (m, 1 H), 5.24 – 5.35 (m, 1 H), 5.54 – 5.75 (m, 1 H), 5.91 – 6.03 (m, 1 H), 6.14 – 6.24 (d, 1 H), 6.96 – 7.34 (m, 4 H), 8.02 – 8.11 (d, 1 H), 8.60 – 8.65 (d, 1 H). HRMS (ESI) calcd for C25H35ClN3O6: [M+H] +: 508.2214 Found: 508.2219.
4.1.8.4. Methyl (4S,7S, E)-17-(3-chlorophenyl)-4-(cyclohexylmethyl)-2,5,10-trioxo-1-oxa-3,6,11-triazacycloheptadec-13-ene-7-carboxylate 8d
Off-white solid (yield 69%); m.p (A: 74 – 75°C), (B: 177 – 178°C).1H NMR (400 MHz, CDCl3-d) δ ppm 0.66 – 0.93 (m, 11 H), 0.95 – 2.32 (m, 10 H), 3.61 – 3.74 (s, 3 H), 3.74 – 3.86 (d, 2 H), 4.00 – 4.20 (m, 1 H), 4.29 – 4.51 (m, 1 H), 4.84 – 5.01 (m, 1 H), 5.42 – 5.83 (m, 2 H), 5.94 – 6.11 (m, 1 H), 6.22 – 6.31 (d, 1 H), 6.75 – 6.92 (d, 1 H), 7.04 – 7.27 (m, 4 H). HRMS (ESI) calcd for C28H38ClN3NaO6: [M+Na] +: 570.2347 Found: 570.2346.
4.1.8.5. Methyl (4S,7S, E)-18-(3-chlorophenyl)-4-isobutyl-2,5,10-trioxo-1-oxa-3,6,11-triazacyclooctadec-13-ene-7-carboxylate 8e
Off-white solid (yield 70%); m.p 63 – 64°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.82 – 1.03 (d, 6 H), 1.20 – 2.32 (m, 7 H), 2.35 – 2.49 (m, 6 H), 3.76 (s, 3 H), 4.01 – 4.25 (m, 2 H), 4.49 – 4.58 (m, 2 H), 5.48 – 5.86 (m, 3 H),6.32 – 6.38 (d, 1 H), 6.89 – 6.93 (d, 1 H), 7.13 – 7.44 (m, 4 H), 8.05 – 8.11 (br s, 1 H). HRMS (ESI) calcd for C26H37ClN3O6: [M+H] +: 522.2371 Found: 522.2378.
4.1.8.6. Methyl (4S,7S, E)-17-(3-chlorophenyl)-4-methyl-2,5,10-trioxo-1-oxa-3,6,11-triazacycloheptadec-13-ene-7-carboxylate 8f
Off-white solid (yield 70%); m.p 105 – 106°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 1.32 – 1.50 (d, 3 H), 1.55 – 2.44 (m, 4 H), 2.44 – 2.75 (m, 4 H), 3.66 – 3.70 (s, 3 H), 4.05 – 4.10 (d, 2 H), 4.50 – 4.61 (m, 1 H), 4.66 – 4.82 (m, 1 H), 5.46 – 5.73 (m, 2 H), 5.89 – 6.01 (m, 1 H), 6.20 – 6.25 (d, 1 H),6.91 – 6.95 (br s, 1 H), 7.07 – 7.42 (m, 4 H), 8.50 – 8.54 (br s, 1 H). HRMS (ESI) calcd for C22H29ClN3O6: [M+H] +: 466.1745 Found: 466.1751.
4.1.8.7. Methyl (4S,7S, E)-18-(3-chlorophenyl)-4-methyl-2,5,10-trioxo-1-oxa-3,6,11-triazacyclooctadec-13-ene-7-carboxylate 8g
Off-white solid (yield 70%); m.p 74 – 75°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 1.20 – 1.30 (m, 2 H), 1.37 (br d, J=7.03 Hz, 3 H), 1.56 – 2.49 (m, 8 H), 3.76 (s, 3 H), 3.81 – 3.90 (m, 2 H), 4.54 – 4.86 (m, 2 H), 5.48 – 5.80 (m, 3 H), 6.14 – 6.32 (br d, 1 H), 6.90 – 7.02 (d, 1 H), 7.06 – 7.38 (m, 4 H), 7.70 – 7.75 (br s, 1 H). HRMS (ESI) calcd for C23H31ClN3O6: [M+H] +: 480.1901 Found: 480.1907.
4.1.8.8. Methyl (20S, E)-10-(3-chlorophenyl)-8,17,22-trioxo-9-oxa-7,16,21-triazaspiro [5.16] docos-13-ene-20-carboxylate 8h
Off-white solid (yield 70%); m.p 105 – 106°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 1.20 – 1.26 (m, 4 H), 1.35 – 1.76 (m, 4 H), 1.77 – 1.90 (m, 5 H), 2.26 – 2.49 (m, 6 H), 3.69 – 3.72 (s, 3 H), 3.381 – 3.88 (m, 2 H), 4.35 – 4.49 (m, 1 H), 5.46 – 5.70 (m, 2 H), 5.91 – 6.04 (m, 1 H), 6.08 – 6.15 (d, 1 H), 7.14 – 7.38 (m, 4 H), 7.87 – 7.96 (d, 1 H). HRMS (ESI) calcd for C26H35ClN3O6: [M+H] +: 520.2214 Found: 520.2222.
4.1.8.9. Methyl (21S, E)-10-(3-chlorophenyl)-8,18,23-trioxo-9-oxa-7,17,22-triazaspiro [5.17] tricos-14-ene-21-carboxylate 8i
Off-white solid (yield 72%); m.p 87 – 88°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 1.20 – 1.26 (m, 4 H), 1.35 – 1.55 (m, 6 H), 1.55 – 1.74 (m, 5 H), 2.05 – 2.36 (m, 4 H), 2.37 – 2.55 (m, 2 H), 3.76 (d, J=2.34 Hz, 3 H), 3.80 – 3.87 (m, 2 H), 4.53 – 4.74 (m, 1 H), 5.59 – 5.75 (m, 2 H), 6.38 – 6.51 (d, 1 H), 6.51 – 6.61 (d, 1 H), 6.83 – 6.91 (d, 1 H), 7.13 – 7.39 (m, 4 H). HRMS (ESI) calcd for C27H37ClN3O6: [M+H] +: 534.2371 Found: 534.2381.
4.1.8.10. Methyl (4S, 7S, Z)-4-isobutyl-2,5,10-trioxo-17-(3-phenylpropyl)-1-oxa-3,6,11-triazacycloheptadec-13-ene-7-carboxylate 8j
Off-white solid (yield 68%); m.p 72 – 73°C.1H NMR (400 MHz, CDCl3-d) δ ppm 0.86 – 1.02 (d, 6 H), 1.46 – 1.86 (m, 7 H), 2.04 – 2.36 (m, 8 H), 2.52 – 2.60 (t, 2 H), 3.70 – 3.77 (s, 3 H), 3.82 – 3.93 (d, 2 H), 4.13 – 4.23 (m, 1 H), 4.63 – 4.80 (m, 1 H), 5.00 – 5.08 (t, 1 H), 5.33 – 5.63 (m, 2 H), 5.73 – 5.83 (d, 1H), 6.00 – 6.06 (d, 1 H), 6.93 – 6.70 (d, 1 H), 7.10 – 7.20 (d, 2 H), 7.22 – 7.32 (m, 3 H). HRMS (ESI) calcd for C28H42N3O6: [M+H] +: 516.3074 Found: 516.3079.
4.1.8.11. Methyl (4S, 7S, Z)-4-isobutyl-2,5,10-trioxo-17-pentyl-1-oxa-3,6,11-triazacycloheptadec-13-ene-7-carboxylate 8k
Off-white solid (yield 54%); m.p 64 – 65°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.85 – 0.92 (t, 2 H),0.93 – 1.04 (d, 6 H), 1.22 – 1.37 (m, 6 H), 1.43 – 1.92 (m, 6 H), 2.12 – 2.40 (m, 4 H), 2.52 – 2.64 (m, 2 H), 3.70 – 3.78 (s, 3 H), 3.80 – 3.90 (d, 2 H), 4.15 – 4.25 (m, 1 H), 4.50 – 4.60 (m, 1 H), 4.62 – 4.78 (m, 2 H), 4.91 – 5.24 (m, 1 H), 5.42 – 5.50 (m, 2 H), 5.84 – 5.89 (d, 1 H), 6.03 – 6.09 (d, 1 H), 6.31 – 6.37 (br. s, 1 H). HRMS (ESI) calcd for C24H42N3O6: [M+H] +: 468.3074 Found: 468.3081.
4.1.8.12. Methyl (6S, 9S, E)-13-(3-chlorophenyl)-9-isobutyl-3,8,11-trioxo-1,12-dioxa-2,7,10-triazacyclooctadec-16-ene-6-carboxylate 33
Off-white solid (yield 52%); m.p 160 – 161°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.84 – 0.99 (d, 6 H), 1.23 – 1.29 (t, 2 H), 1.49 – 2.57 (t, 8 H), 3.72 – 3.79 (s, 3 H), 3.82 – 3.90 (d, 1 H), 4.03 – 4. 17 (d, 2 H), 4.24 – 4.43 (m, 1 H), 4.59 – 4.74 (m, 1 H), 5.18 – 5.24 (d, 1 H), 5.52 – 5.67 (m, 2 H), 5.68 – 5.96 (m, 1 H), 6. 24 – 6. 32 (d, 1 H), 7.13 – 7.21 (s, 1 H), 7.25 – 7.36 (m, 3 H), 9.24 – 9.30 (d, 1 H). HRMS (ESI) calcd for C25H35ClN3O7: [M+H] +: 524.2164 Found: 524.2171.
4.1.9. Synthesis of compounds 9a–k and 34. General procedure for the hydrogenation reaction
To a solution of the appropriate olefin 8 (1.0 mmol) in anhydrous ethanol (10 mL) was added palladium on carbon (10% Pd-C, 2.0 eq) and the mixture was stirred under a hydrogen atmosphere (a balloon was used) at room temperature and atmospheric pressure for 18 h. The mixture was filtered through Celite and the filtrate was concentrated in vacuo to yield a crude product which was purified by flash chromatography to yield compounds 9a–k as white solids.
4.1.9.1. Methyl (4S, 7S)-4-isobutyl-2,5,10-trioxo-17-phenyl-1-oxa-3,6,11-triazacycloheptadecane-7-carboxylate 9a
White solid (yield 92%); m.p 85 – 86°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.68 – 1.05 (d, 6 H), 1.40 – 1.73 (m, 5 H), 1.73 – 1.95 (m, 4 H), 1.95 – 2.21 (m, 6 H), 3.20 – 3.29 (t, 2 H), 3.64 – 3.76 (s, 3 H), 4.45 – 4.53 (m, 2 H), 5.76 – 5.90 (m, 1 H), 7.17 – 7.35 (m, 5 H), 7.40 – 7.50 (d, 1 H), 8.25 – 8.35 (d, 1 H), 8. 72 – 8.80 (d, 1 H). HRMS (ESI) calcd for C25H38N3O6: [M+H] +: 476.2761 Found: 476.2768.
4.1.9.2. Methyl (4S, 7S)-17-benzyl-4-isobutyl-2,5,10-trioxo-1-oxa-3,6,11-triazacycloheptadecane-7-carboxylate 9b
White solid (yield 95%); m.p 190 – 191°C. 1H NMR (400 MHz, CDCl3) δ ppm 0.81 – 0.97 (d, 6 H), 1.53 – 1.69 (m, 6 H), 1,91 – 2.02 (m, 5H), 2.20 – 2.31 (4 H), 2.70 – 2.93 (m, 2 H), 3.21 – 3.31 (t, 2 H), 3.73 – 3.80 (s, 3 H), 4.40 – 4.62 (m, 2 H), 4.88 – 5.04 (m, 1 H), 6.20 – 6.41 (br. d, 1 H), 6.91 – 7.10 (d, 1 H), 7.14 – 7.36 (m, 5 H), 7.85 – 7.92 (d, 1 H). HRMS (ESI) calcd for C26H40N3O6: [M+H] +: 490.2917 Found: 490.2921.
4.1.9.3. Methyl (4S, 7S)-17-(3-chlorophenyl)-4-isobutyl-2,5,10-trioxo-1-oxa-3,6,11-triazacycloheptadecane-7-carboxylate 9c
White solid (yield 95%); m.p 149 – 150°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.74 – 0.99 (d, 6 H), 1.42 – 1.83 (m, 3 H), 2.11 – 2.43 (m, 6 H), 2.47 – 2.64 (m, 2 H), 3.28 – 3.40 (m, 2 H), 3.69 – 3.81 (s, 3 H), 3.81 – 3.96 (t, 2 H), 4.50 – 5.60 (m, 1 H), 4.62 – 4.77 (m, 1 H), 5.91 – 6.03 (m, 1 H), 6.14 – 6.24 (d, 1 H), 6.96 – 7.34 (m, 4 H), 8.02 – 8.11 (d, 1 H), 8.60 – 8.65 (d, 1 H). HRMS (ESI) calcd for C25H37ClN3O6: [M+H] +: 510.2371 Found: 510.2378.
4.1.9.4. Methyl (4S, 7S)-17-(3-chlorophenyl)-4-(cyclohexylmethyl)-2,5,10-trioxo-1-oxa-3,6,11-triazacycloheptadecane-7-carboxylate 9d
White solid (yield 95%); m.p (A: 71 – 72°C, B: 161 – 162°C).1H NMR (400 MHz, CDCl3-d) δ 0.66 – 0.93 (m, 12 H), 0.95 – 2.32 (m, 10 H), 3.18 – 3.21 (t, 2 H), 3.63 – 3.81 (s, 3 H), 3.96 – 4.10 (t, 2 H), 4.00 – 4.20 (m, 1 H), 4.29 – 4.51 (m, 1 H), 4.84 – 5.01 (m, 1 H), 5.94 – 6.11 (m, 1 H), 6.22 – 6.31 (d, 1 H), 6.75 – 6.92 (d, 1 H), 7. 12 – 7.27 (m, 4 H), 8.25 – 8.40 (br. s, 1 H), 8.82 – 8.90 (d, 1 H). HRMS (ESI) calcd for C28H41ClN3O6: [M+H] +: 550.2684 Found: 550.2689.
4.1.9.5. Methyl (4S, 7S)-18-(3-chlorophenyl)-4-isobutyl-2,5,10-trioxo-1-oxa-3,6,11-triazacyclooctadecane-7-carboxylate 9e
White solid (yield 94%); m.p 122 – 123°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.74 – 0.99 (d, 6 H), 1.42 – 1.83 (m, 5 H), 2.11 – 2.43 (m, 6 H),2.47 – 2.64 (m, 2 H), 3.28 – 3.40 (m, 2 H), 3.69 – 3.81 (s, 3 H), 3.81 – 3.96 (t, 2 H), 4.50 – 5.60 (m, 1 H), 4.62 – 4.77 (m, 1 H), 5.91 – 6.03 (m, 1 H), 6.14 – 6.24 (d, 1 H), 6.96 – 7.34 (m, 4 H), 8.02 – 8.11 (d, 1 H), 8.60 – 8.65 (d, 1 H). HRMS (ESI) calcd for C26H39ClN3O6: [M+H] +: 525.2527 Found: 524.2533.
4.1.9.6. Methyl (4S, 7S)-17-(3-chlorophenyl)-4-methyl-2,5,10-trioxo-1-oxa-3,6,11-triazacycloheptadecane-7-carboxylate 9f
White solid (yield 93%); m.p 131 – 132°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.82 – 0.99 (m, 4 H), 1.40 (d, J=6.64 Hz, 3 H), 1.52 – 1.84 (m, 2 H), 1.82 – 2.06 (m, 2 H), 2.09 – 2.50 (m, 4 H), 3.12 – 3.31 (m, 2 H), 3.70 – 3.81 (s, 3 H), 4.65 – 4.77 (m, 1 H), 5.18 – 5.26 (m, 1 H), 5.84 – 5.93 (m, 1 H), 6.17 – 6.33 (br s, 1 H), 6.52 – 6.63 (d, 1 H), 7.10 – 7.30 (m, 4 H), 7.30 – 7.44 (m, 1 H). HRMS (ESI) calcd for C22H31ClN3O6: [M+H] +: 468.1901 Found: 468.1911.
4.1.9.7. Methyl (4S, 7S)-18-(3-chlorophenyl)-4-methyl-2,5,10-trioxo-1-oxa-3,6,11-triazacyclooctadecane-7-carboxylate 9g
White solid (yield 93%); m.p 100 – 101°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.75 – 0.96 (m, 4 H), 1.37 – 1.58 (d, m, 5 H), 1.65 – 1.81 (m, 2 H), 1.81 – 2.10 (m, 2 H), 2.10 – 2.54 (m, 4 H), 3.21 – 3.34 (t, 2 H), 3.75 (s, 3 H), 4.17 – 4.28 (m, 1 H), 4.56 – 4.70 (m, 1 H), 5.25 – 5.40 (br s, 1 H), 5.47 – 5.82 (m, 1 H), 6.14 – 6.29 (br s, 1 H), 7.01 – 7.51 (m, 4 H). 7.90 – 8.00 (br s, 1 H). HRMS (ESI) calcd for C23H33ClN3O6: [M+H] +: 482.2058 Found: 482.2069.
4.1.9.8. Methyl (20S)-10-(3-chlorophenyl)-8,17,22-trioxo-9-oxa-7,16,21-triazaspiro [5.16] docosane-20-carboxylate 9h
White solid (yield 95%); m.p 105 – 106°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.84 – 0.97 (m, 2 H), 1.09 – 1.40 (m, 7 H), 1.40 – 2.65 (m, 13 H), 3.10 – 3.28 (m, 2 H), 3.67 – 3.70 (s, 3 H), 4.31 – 4.44 (m, 1 H), 5.81 – 5.90 (m, 1 H), 6.47 – 6.62 (t, 1 H), 7.11 – 7.39 (m, 4 H), 8.26 – 8. 30 (br. S, 1 H), 8.64 – 8.79 (br. s, 1 H). HRMS (ESI) calcd for C26H37ClN3O6: [M+H] +: 522.2371 Found: 522.2389.
4.1.9.9. Methyl (21S)-10-(3-chlorophenyl)-8,18,23-trioxo-9-oxa-7,17,22-triazaspiro [5.17] tricosane-21-carboxylate 9i
White solid (yield 95%); m.p 91 – 92°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.83 – 0.91 (m, 2 H), 1.12 – 2.11 (m, 14 H), 2.18 – 2.40 (m, 8 H), 3.28 – 3.38 (m, 2 H), 3.76 (s, 3 H), 4.55 – 4.69 (m, 1 H), 5.52 – 5.61 (m, 1 H), 6.29 – 6.37 (br. s, 1 H), 6.44 – 6.59 (br. s, 1 H), 6.63 – 6.73 (br. s, 1 H), 7.13 – 7.39 (m, 4 H). HRMS (ESI) calcd for C27H39ClN3O6: [M+H] +: 536.2527 Found: 536.2543.
4.1.9.10. Methyl (4S, 7S)-4-isobutyl-2,5,10-trioxo-17-(3-phenylpropyl)-1-oxa-3,6,11-triazacycloheptadecane-7-carboxylate 9j
White solid (yield 97%); m.p 200 – 201°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.85 – 1.01 (d, 6 H), 1.15 – 1.81 (m, 10 H), 2.09 – 2.19 (m, 9 H), 2.54 – 2.67 (t, 2 H), 3.02 – 3.14 (t, 2 H), 3.68 – 3.77 (s, 3 H), 3.90 – 3.98 (d, 1 H), 4.66 – 4.75 (m, 1 H), 4.90 – 5.00 (m, 2 H), 6.07 – 6.16 (d, 1 H), 6.17 – 6.24 (d, 1 H), 7.10 – 7.20 (d, 2 H), 7.24 – 7.34 (m, 3 H). HRMS (ESI) calcd for C28H44N3O6: [M+H] +: 518.3230 Found: 518.3253.
4.1.9.11. Methyl (4S, 7S)-4-isobutyl-2,5,10-trioxo-17-pentyl-1-oxa-3,6,11-triazacycloheptadecane-7-carboxylate 9k
White solid (yield 96%); m.p 203 – 204°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.83 – 0.90 (t, 2 H),0.91 – 1.01 (d, 6 H), 1.20 – 1.34 (m, 8 H), 1.38 – 1.84 (m, 9 H), 2.24 – 2.51 (m, 4 H), 3.02 – 3.17 (br m, 1 H), 3.44 – 3.55 (br. s, 1 H), 3.69 – 3.78 (s, 3 H), 3.90 – 4.00 (d, 1 H), 4.65 – 4.75 (m, 2 H), 4.86 – 4.98 (d, 2 H), 5.84 – 5.89 (d, 1 H), 6.09 – 6.14 (d, 1 H), 6.16 – 6.24 (d, 1 H), 8.03 – 8.13 (br. s, 1 H). HRMS (ESI) calcd for C24H44N3O6: [M+H] +: 470.3230 Found: 470.3271.
4.1.9.12. Methyl (6S, 9S)-13-(3-chlorophenyl)-9-isobutyl-3,8,11-trioxo-1,12-dioxa-2,7,10-triazacyclooctadecane-6-carboxylate 34
White solid (yield 92%); m.p 73 – 73°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.87 – 1.00 (d, 6 H), 1.16 – 1.34 (m, 3 H), 1.36 – 1.78 (m, 9 H), 1.83 – 2.05 (t, 2 H), 2.14 – 2.30 (m, 2 H), 3.74 – 3.82 (s, 3 H), 3.83 – 4.16 (t, 2 H), 4.61 – 4.76 (m, 1 H), 5.10 – 5.19 (m, 1 H), 5.71 – 5.83 (m, 1 H), 7.15 – 7.23 (s, 1 H), 7.23 – 7.39 (m, 3 H), 9.12 – 9.19 (d, 1 H), 9.76 – 9.80 (d, 1 H). HRMS (ESI) calcd for C25H37ClN3O7: [M+H] +: 526.2320 Found: 526.2332.
4.1.10. Synthesis of alcohols 10a–k and 35. General procedure
To a solution of an appropriate macrocyclic ester 9 (5 mmol) in anhydrous THF (30 mL) was added dropwise a solution of lithium borohydride in THF (2M in THF, 7.5 mL, 15 mmol) and the reaction mixture was stirred for 30 min at room temperature. Absolute ethyl alcohol (15 mL) was added and the reaction mixture was stirred at room temperature overnight. The reaction mixture was then acidified by adding 1.0 M aqueous potassium bisulfate until the pH of the solution was ~3. Removal of the solvent left a residue which was taken up in ethyl acetate (150 mL). The organic layer was washed with brine (25 mL), dried over anhydrous sodium sulfate, filtered, and concentrated to yield compounds 10a–k as white solids.
4.1.10.1. (4S, 7S)-7-(Hydroxymethyl)-4-isobutyl-17-phenyl-1-oxa-3,6,11-triazacycloheptadecane-2,5,10-trione 10a
White solid (yield 93%); m.p 144 – 145°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.68 – 1.05 (d, 6 H), 1.40 – 1.73 (m, 5 H), 1.73 – 1.95 (m, 4 H), 1.95 – 2.21 (m, 6 H), 3.20 – 3.29 (t, 2 H), 3.35 – 3.46 (m, 2 H), 4.45 – 4.53 (m, 2 H), 5.76 – 5.90 (m, 1 H), 7.17 – 7.35 (m, 5 H), 7.40 – 7.50 (d, 1 H), 8.25 – 8.35 (d, 1 H), 8. 72 – 8.80 (d, 1 H). HRMS (ESI) calcd for C24H38N3O5: [M+H] +: 448.2811 Found: 448.2834.
4.1.10.2. (4S, 7S)-17-Benzyl-7-(hydroxymethyl)-4-isobutyl-1-oxa-3,6,11-triazacycloheptadecane-2,5,10-trione 10b
White solid (yield 96%); m.p 214 – 215°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.81 – 0.97 (d, 6 H), 1.53 – 1.69 (m, 6 H), 1,91 – 2.02 (m, 5H), 2.20 – 2.31 (4 H), 2.70 – 2.93 (m, 2 H), 3.21 – 3.31 (t, 2 H), 3.52 – 3.60 (m, 2 H), 4.40 – 4.62 (m, 3 H), 4.88 – 5.04 (m, 1 H), 6.20 – 6.41 (br. d, 1 H), 6.91 – 7.10 (d, 1 H), 7.14 – 7.36 (m, 5 H), 7.85 – 7.92 (d, 1 H). HRMS (ESI) calcd for C25H40N3O5: [M+H] +: 462.2968 Found: 462.2984.
4.1.10.3. (4S, 7S)-17-(3-Chlorophenyl)-7-(hydroxymethyl)-4-isobutyl-1-oxa-3,6,11-triazacycloheptadecane-2,5,10-trione 10c
White solid (yield 95%); m.p 98 – 99°C. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.74 – 0.99 (d, 6 H), 1.42 – 1.83 (m, 3 H), 2.11 – 2.43 (m, 6 H), 2.47 – 2.64 (m, 2 H), 3.28 – 3.40 (m, 2 H), 3.4 – 3.5 (m, 2H), 3.81 – 3.96 (t, 2 H), 4.50 – 5.60 (m, 1 H), 4.62 – 4.77 (m, 1 H), 5.91 – 6.03 (m, 1 H), 6.14 – 6.24 (d, 1 H), 6.96 – 7.34 (m, 4 H), 8.02 – 8.11 (d, 1 H), 8.60 – 8.65 (d, 1 H). HRMS (ESI) calcd for C24H37ClN3O5: [M+H] +: 482.2422 Found: 482.2444.
4.1.10.4. (4S, 7S)-17-(3-Chlorophenyl)-4-(cyclohexylmethyl)-7-(hydroxymethyl)-1-oxa-3,6,11-triazacycloheptadecane-2,5,10-trione 10d
White solid (yield 93%); m.p (A: 71 – 72°C, B: 161 – 162°C). 1H NMR (400 MHz, CDCl3-d) δ 0.66 – 0.93 (m, 12 H), 0.95 – 2.32 (m, 10 H), 3.18 – 3.21 (t, 2 H), 3.96 – 4.10 (t, 2 H), 4.00 – 4.20 (m, 1 H), 4.29 – 4.51 (m, 1 H), 4.84 – 5.01 (m, 1 H), 5.94 – 6.11 (m, 1 H), 6.22 – 6.31 (d, 1 H), 6.75 – 6.92 (d, 1 H),7. 12 – 7.27 (m, 4 H), 8.25 – 8.40 (br s, 1 H), 8.82 – 8.90 (d, 1 H). HRMS (ESI) calcd for C27H41ClN3O5: [M+H] +: 522.2735 Found: 522.2744.
4.1.10.5. (4S, 7S)-18-(3-Chlorophenyl)-7-(hydroxymethyl)-4-isobutyl-1-oxa-3,6,11-triazacyclooctadecane-2,5,10-trione 10e
White solid (yield 95%); m.p 71 – 72°C. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.79 – 0.94 (d, 6 H), 1.15 – 1.30 (m, 4 H), 1.31 – 1.41 (m, 4 H), 1.61 – 1.81 (m, 5 H), 2.00 – 2.10 (m, 4 H), 3.53 – 3.77 (m, 5 H), 4.55 – 4.74 (m, 2 H), 5.75 – 5.82 (m, 1 H), 6.48 – 6.54 (t, 1 H), 7.21 – 7.46 (m, 4 H), 7.54 – 7.63 (d, 1 H), 7.73 – 7.83 (d, 1 H). HRMS (ESI) calcd for C25H39ClN3O5: [M+H] +: 496.2578 Found: 496.2612.
4.1.10.6. (4S, 7S)-17-(3-Chlorophenyl)-7-(hydroxymethyl)-4-methyl-1-oxa-3,6,11-triazacycloheptadecane-2,5,10-trione 10f
White solid (yield 82%); m.p 154 – 155°C.1H NMR (400 MHz, DMSO-d6) δ ppm 0.75 – 0.91 (m, 2 H), 1.13 – 1.25 (m, 2 H), 1.39 – 1.41 (d, 3 H), 1.44 – 1.57 (m, 2 H), 1.57 – 1.71 (m, 2 H), 1.79 – 1.95 (m, 2 H), 2.11 – 2.32 (m, 2 H), 3.23 – 3.38 (m, 4 H), 3.72 – 3.90 (m, 1 H), 4.64 – 4.72 (m, 2 H), 5.82 – 5.89 (m, 1 H), 6.86 – 6.97 (br. s, 1 H), 7.22 – 7.45 (m, 4 H), 7.57 – 7.68 (d, 1 H), 7.72 – 7.83 (d, 1 H). HRMS (ESI) calcd for C21H31ClN3O5: [M+H] +: 440.1952 Found: 440.1983.
4.1.10.7. (4S, 7S)-18-(3-Chlorophenyl)-7-(hydroxymethyl)-4-methyl-1-oxa-3,6,11-triazacyclooctadecane-2,5,10-trione 10g
White solid (yield 88%); m.p 192 – 193°C. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.75 – 0.96 (m, 4 H), 1.37 – 1.58 (d, m, 5 H), 1.65 – 1.81 (m, 2 H), 1.81 – 2.10 (m, 2 H), 2.10 – 2.54 (m, 4 H), 3.21 – 3.34 (t, 2 H), 3.45 – 3.50 (m, 3 H), 4.17 – 4.28 (m, 1 H), 4.56 – 4.70 (m, 1 H), 5.25 – 5.40 (br s, 1 H), 5.47 – 5.82 (m, 1 H), 6.14 – 6.29 (br s, 1 H), 7.01 – 7.51 (m, 4 H). 7.90 – 8.00 (br s, 1 H). HRMS (ESI) calcd for C22H33ClN3O5: [M+H] +: 454.2109 Found: 454.2477.
4.1.10.8. (20S)-10-(3-Chlorophenyl)-20-(hydroxymethyl)-9-oxa-7,16,21-triazaspiro [5.16] docosane-8,17,22-trione 10h
White solid (yield 90%); m.p 107 – 108°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.82 – 0.92 (m, 2 H), 1.12 – 1.54 (m, 9 H), 1.54 – 1.99 (m, 10 H), 2.16 – 2.43 (m, 6 H), 2.96 – 3.40 (m, 2 H), 3.44 – 3.70 (m, 2 H), 5.21 – 5.24 (s, 1 H), 5.54 – 5.65 (m, 1 H), 6.54 – 6.60 (br. s, 1 H), 6.71 – 6.84 (d, 1 H), 7.01 – 7.09 (d, 1 H), 7.11 – 7.38 (m, 4 H). HRMS (ESI) calcd for C25H37ClN3O5: [M+H] +: 494.2422 Found: 494.2440.
4.1.10.9. (21S)-10-(3-Chlorophenyl)-21-(hydroxymethyl)-9-oxa-7,17,22-triazaspiro [5.17] tricosane-8,18,23-trione 10i
White solid (yield 94%); m.p 92 – 93°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.84 – 0.92 (m, 2 H), 1.14 – 1.51 (m, 7 H), 1.51 – 1.99 (m, 10 H), 2.14 – 2.47 (m, 6 H), 2.94 – 3.41 (m, 2 H), 3.45 – 3.71 (m, 2 H), 5.19 – 5.21 (s, 1 H), 5.53 – 5.65 (m, 1 H), 6.49 – 6.60 (br. s, 1 H), 6.76 – 6.86 (d, 1 H), 6.94 – 7.04 (d, 1 H), 7.11 – 7.38 (m, 4 H). HRMS (ESI) calcd for C26H39ClN3O5: [M+H] +: 508.2578 Found: 508.2601.
4.1.10.10. (4S, 7S)-7-(Hydroxymethyl)-4-isobutyl-17-(3-phenylpropyl)-1-oxa-3,6,11-triazacycloheptadecane-2,5,10-trione 10j
White solid (yield 98%); m.p 216 – 217°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.85 – 1.01 (d, 6 H), 1.15 – 1.81 (m, 10 H), 2.09 – 2.19 (m, 9 H), 2.54 – 2.67 (t, 2 H), 3.02 – 3.14 (t, 2 H), 3.53 – 3.74 (m, 2 H), 3.90 – 3.98 (d, 1 H), 4.66 – 4.75 (m, 1 H), 4.90 – 5.00 (m, 2 H), 6.07 – 6.16 (d, 1 H), 6.17 – 6.24 (d, 1 H), 7.10 – 7.20 (d, 2 H), 7.24 – 7.34 (m, 3 H). HRMS (ESI) calcd for C27H44N3O5: [M+H] +: 490.3281 Found: 490.3312.
4.1.10.11. (4S, 7S)-7-(Hydroxymethyl)-4-isobutyl-17-pentyl-1-oxa-3,6,11-triazacycloheptadecane-2,5,10-trione 10k
White solid (yield 98%); m.p 230°C (d). 1H NMR (400 MHz, CDCl3-d) δ ppm 0.85 – 0.93 (t, 2 H), 0.93 – 1.01 (d, 6 H), 1.20 – 1.34 (m, 8 H), 1.38 – 1.84 (m, 9 H), 2.24 – 2.51 (m, 4 H), 3.02 – 3.17 (br m, 1 H), 3.44 – 3.55 (br. s, 1 H), 3.54 – 3.73 (m, 2 H), 3.90 – 4.00 (d, 1 H), 4.65 – 4.75 (m, 2 H), 4.86 – 4.98 (d, 2 H), 5.00 – 5.06 (d, 1 H), 6.09 – 6.14 (d, 1 H), 6.29 – 6.39 (d, 1 H), 8.03 – 8.13 (br. s, 1 H). HRMS (ESI) calcd for C23H34N3O5: [M+H] +: 442.3281 Found: 442.3319.
4.1.10.12. (6S, 9S)-13-(3-Chlorophenyl)-6-(hydroxymethyl)-9-isobutyl-1,12-dioxa-2,7,10-triazacyclooctadecane-3,8,11-trione 35
White solid (yield 97%); m.p 138 – 139°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.83 – 0.97 (d, 6 H), 1.21 – 1.34 (m, 3 H), 1.41 – 1.99 (m, 9 H), 2.01 – 2.10 (t, 2 H), 2.17 – 2.30 (m, 2 H), 3.54 – 3.72 (t, 2 H), 3.80 – 4.16 (m, 2 H), 5.23 – 5.31 (m, 1 H), 5.47 – 5.57 (m, 1 H), 5.69 – 5.79 (m, 1 H), 6.57 – 6.63 (d, 1 H), 6.94 – 6.99 (d, 1 H), 7.15 – 7.20 (s, 1 H), 7.23 – 7.34 (m, 3 H), 9.33 – 9.39 (d, 1 H). HRMS (ESI) calcd for C24H37ClN3O6: [M+H] +: 498.2371 Found: 498.2388.
4.1.11. Synthesis of macrocyclic aldehydes 11–21 and 36. General procedure
An appropriate alcohol (0.6 mmol) was dissolved in anhydrous dichloromethane (20 mL) under a nitrogen atmosphere and cooled to 0° C. Dess-Martin periodinane (1.2 mmol, 2.0 eq.) was added to the reaction mixture with stirring. The ice bath was removed and the reaction mixture was stirred at room temperature for 3 h (monitoring by TLC indicated complete disappearance of the starting material). A solution of 40 mM sodium thiosulfate in saturated aqueous NaHCO3 (50 mL) was added and the solution was stirred for another 15 min. The aqueous layer was removed and the organic layer was washed with saturated sodium bicarbonate (25 mL), water (2 × 25 mL) and brine (25 mL). The organic layer was dried over anhydrous sodium sulfate, filtered and concentrated. The yellow residue was purified by flash chromatography (silica gel/methylene chloride/ethyl acetate/methanol) to yield the aldehyde.
4.1.11.1. (4S, 7S)-4-Isobutyl-2,5,10-trioxo-17-phenyl-1-oxa-3,6,11-triazacycloheptadecane-7-carbaldehyde 11
White solid (yield 60%); m.p 130 – 131°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.68 – 1.05 (d, 6 H), 1.40 – 1.73 (m, 5 H), 1.73 – 1.95 (m, 4 H), 1.95 – 2.21 (m, 6 H), 3.20 – 3.29 (t, 2 H), 4.45 – 4.53 (m, 2 H), 5.76 – 5.90 (m, 1 H), 7.17 – 7.35 (m, 5 H), 7.40 – 7.50 (d, 1 H), 8.25 – 8.35 (d, 1 H), 8. 72 – 8.80 (d, 1 H), 9.55 – 9.64 (m, 1 H). HRMS (ESI) calcd for C24H36N3O5: [M+H] +: 446.2655 Found: 446.2658.
4.1.11.2. (4S, 7S)-17-Benzyl-4-isobutyl-2,5,10-trioxo-1-oxa-3,6,11-triazacycloheptadecane-7-carbaldehyde 12
White solid (yield 55%); m.p 82 – 83°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.81 – 0.97 (d, 6 H), 1.53 – 1.69 (m, 6 H), 1,91 – 2.02 (m, 5H), 2.20 – 2.31 (4 H), 2.70 – 2.93 (m, 2 H), 3.21 – 3.31 (t, 2 H), 4.40 – 4.62 (m, 2 H), 4.88 – 5.04 (m, 1 H), 6.20 – 6.41 (br. d, 1 H), 6.91 – 7.10 (d, 1 H), 7.14 – 7.36 (m, 5 H), 7.85 – 7.92 (d, 1 H), 9.53 – 9.62 (s, 1 H). HRMS (ESI) calcd for C25H38N3O5: [M+H] +: 460.2811 Found: 460.2813.
4.1.11.3. (4S, 7S)-17-(3-Chlorophenyl)-4-isobutyl-2,5,10-trioxo-1-oxa-3,6,11-triazacycloheptadecane-7-carbaldehyde 13
White solid (yield 75%); m.p 90 – 91°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.74 – 0.99 (d, 6 H), 1.42 – 1.83 (m, 5 H), 2.11 – 2.43 (m, 6 H),2.47 – 2.64 (m, 2 H), 3.28 – 3.40 (t, 2 H), 3.4 – 3.5 (m, 2H), 4.50 – 5.60 (m, 1 H), 4.62 – 4.77 (m, 1 H), 5.91 – 6.03 (m, 1 H), 6.14 – 6.24 (d, 1 H), 6.96 – 7.34 (m, 4 H), 8.02 – 8.11 (d, 1 H), 8.60 – 8.65 (d, 1 H), 9.54 – 9.61 (s, 1 H). HRMS (ESI) calcd for C24H34ClN3NaO5: [M+Na] +: 502.2085 Found: 502.2071.
4.1.11.4. (4S, 7S)-17-(3-Chlorophenyl)-4-(cyclohexylmethyl)-2,5,10-trioxo-1-oxa-3,6,11-triazacycloheptadecane-7-carbaldehyde 14A
White solid (yield 70%); m.p 71 – 72°C, 1H NMR (400 MHz, CDCl3-d) δ 0.66 – 0.93 (m, 12 H), 0.95 – 2.32 (m, 10 H), 3.18 – 3.21 (t, 2 H), 3.96 – 4.10 (t, 2 H), 4.29 – 4.51 (m, 1 H), 4.84 – 5.01 (m, 1 H), 5.94 – 6.11 (m, 1 H), 6.75 – 6.92 (d, 1 H), 7. 12 – 7.27 (m, 4 H), 8.25 – 8.40 (br s, 1 H), 8.82 – 8.90 (d, 1 H), 9.48 – 9.58 (s, 1 H). HRMS (ESI) calcd for C27H39ClN3O5: [M+H] +: 520.2578 Found: 520.2771.
4.1.11.5. (4S, 7S)-17-(3-Chlorophenyl)-4-(cyclohexylmethyl)-2,5,10-trioxo-1-oxa-3,6,11-triazacycloheptadecane-7-carbaldehyde 14B
White solid (yield 70%); m.p 75 – 76°C. 1H NMR (400 MHz, CDCl3-d) δ 0.66 – 0.93 (m, 12 H), 0.95 – 2.32 (m, 10 H), 3.18 – 3.21 (t, 2 H), 3.96 – 4.10 (t, 2 H), 4.29 – 4.51 (m, 1 H), 4.84 – 5.01 (m, 1 H), 5.94 – 6.11 (m, 1 H), 6.75 – 6.92 (d, 1 H), 7. 12 – 7.27 (m, 4 H), 8.25 – 8.40 (br s, 1 H), 8.82 – 8.90 (d, 1 H), 9.48 – 9.58 (s, 1 H). HRMS (ESI) calcd for C27H38ClN3NaO5: [M+Na] +: 542.2398 Found: 542.2506.
4.1.11.6. (4S, 7S)-18-(3-Chlorophenyl)-4-isobutyl-2,5,10-trioxo-1-oxa-3,6,11-triazacyclooctadecane-7-carbaldehyde 15
White solid (yield 68%); m.p 69 – 70°C. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.79 – 0.94 (d, 6 H), 1.15 – 1.30 (m, 4 H), 1.31 – 1.41 (m, 4 H), 1.61 – 1.81 (m, 5 H), 2.00 – 2.10 (m, 4 H), 3.18 – 3.27 (t, 2 H), 4.55 – 4.74 (m, 2 H), 5.75 – 5.82 (m, 1 H), 6.48 – 6.54 (t, 1 H), 7.21 – 7.46 (m, 4 H), 7.54 – 7.63 (d, 1 H), 8.53 – 8.59 (d, 1 H), 9.57 – 9.63 (s, 1 H). HRMS (ESI) calcd for C25H37ClN3O5: [M+H] +: 494.2422 Found: 494.2446.
4.1.11.7. (4S, 7S)-17-(3-Chlorophenyl)-4-methyl-2,5,10-trioxo-1-oxa-3,6,11-triazacycloheptadecane-7-carbaldehyde 16
White solid (yield 70%); m.p 84 – 85°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.79 – 0.91 (m, 2 H), 1.13 – 1.36 (m, 2 H), 1.44 – 1.48 (d, 3 H), 1.76 – 2.01 (m, 4 H), 2.19 – 2.70 (m, 4 H), 3.27 – 3.43 (m, 2 H), 4.33 – 4.40 (m, 1 H), 4.63 – 4.71 (m, 1 H), 5.81 – 5.93 (m, 1 H), 6.06 – 6.16 (m, 1 H), 6.57 – 6.65 (d, 1 H), 6.92 – 7.01 (d, 1 H), 7.10 – 7.40 (m, 4 H), 9.56 – 9.64 (s, 1 H). HRMS (ESI) calcd for C21H29ClN3O5: [M+H] + : 438.1796 Found: 438.2153.
4.1.11.8. (4S, 7S)-18-(3-Chlorophenyl)-4-methyl-2,5,10-trioxo-1-oxa-3,6,11-triazacyclooctadecane-7-carbaldehyde 17
White solid (yield 64%); m.p 123 – 124°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.83 – 0.91 (m, 2 H), 1.17 – 1.38 (m, 4 H), 1.42 – 1.47 (d, 3 H), 1.80 – 2.00 (m, 4 H), 2.21 – 2.65 (m, 4 H), 3.30 – 3.41 (m, 2 H), 4.35 – 4.43 (m, 1 H), 4.61 – 4.71 (m, 1 H), 5.83 – 5.93 (m, 1 H), 6.06 – 6.14 (m, 1 H), 6.67 – 6.75 (d, 1 H), 6.81 – 6.91 (d, 1 H), 7.11 – 7.35 (m, 4 H), 9.61 – 9.71 (s, 1 H). HRMS (ESI) calcd for C22H31ClN3O5: [M+H] +: 452.1952 Found: 452.2315.
4.1.11.9. (20S)-10-(3-Chlorophenyl)-8,17,22-trioxo-9-oxa-7,16,21-triazaspiro[5.16]docosane-20-carbaldehyde 18
White solid (yield 63%); m.p 110 – 111°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.91 – 1.41 (m, 9 H), 1.51 – 1.70 (m, 5 H), 2.12 – 2.16 (m, 2 H), 2.20 – 2.50 (m, 4 H), 2.87 – 3.03 (m, 2 H), 4.50 – 4.60 (m, 1 H), 5.02 – 5.10 (m, 1 H), 5.68 – 5.85 (m, 1 H), 6.30 – 6.41 (d, 1 H), 7.18 – 7.34 (m, 4 H), 7.69 – 7.74 (d, 1 H), 8.23 – 8.30 (d, 1 H), 9.58 – 9.65 (s, 1 H). HRMS (ESI) calcd for C25H35ClN3O5: [M+H] +: 492.2265 Found: 492.2293.
4.1.11.10. (21S)-10-(3-Chlorophenyl)-8,18,23-trioxo-9-oxa-7,17,22-triazaspiro [5.17] tricosane-21-carbaldehyde 19
White solid (yield 65%); m.p 87 – 88°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.90 – 1.36 (m, 9 H), 1.53 – 1.72 (m, 7 H), 2.10 – 2.14 (m, 2 H), 2.14 – 2.54 (m, 4 H), 2.86 – 3.02 (m, 2 H), 4.42 – 4.60 (m, 1 H), 5.00 – 5.11 (m, 1 H), 5.66 – 5.81 (m, 1 H), 6.15 – 6.23 (d, 1 H), 7.19 – 7.36 (m, 4 H), 7.89 – 7.94 (d, 1 H), 8.10 – 8.16 (d, 1 H), 9.55 – 9.62 (s, 1 H). HRMS (ESI) calcd for C26H37ClN3O5: [M+H] +: 506.2422 Found: 506.2440.
4.1.11.11. (4S, 7S)-4-Isobutyl-2,5,10-trioxo-17-(3-phenylpropyl)-1-oxa-3,6,11-triazacycloheptadecane-7-carbaldehyde 20
White solid (yield 60%); m.p 163 – 164°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.86 – 1.03 (d, 6 H), 1.15 – 1.81 (m, 10 H), 2.09 – 2.19 (m, 9 H), 2.54 – 2.67 (t, 2 H), 3.02 – 3.14 (t, 2 H), 3.95 – 4.05 (d, 1 H), 4.65 – 4.73 (m, 1 H), 4.90 – 5.00 (d, 2 H), 6.09 – 6.17 (d, 1 H), 6.36 – 6.44 (d, 1 H), 7.12 – 7.22 (d, 2 H), 7.24 – 7.32 (m, 3 H), 9.56 – 9.60 (s, 1 H). HRMS (ESI) calcd for C27H41N3O5: [M+H] +: 487.3046 Found: 487.3051.
4.1.11.12. (4S, 7S)-4-Isobutyl-2,5,10-trioxo-17-pentyl-1-oxa-3,6,11-triazacycloheptadecane-7-carbaldehyde 21
White solid (yield 61%), m.p 172 – 173°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.83 – 0.91 (t, 2 H),0.92 – 1.01 (d, 6 H), 1.20 – 1.34 (m, 8 H), 1.38 – 1.84 (m, 9 H), 2.24 – 2.51 (m, 4 H), 3.15 – 3.27 (m, 1 H), 3.39 – 3.49 (m, 1 H), 3.97 – 4.07 (m, 1 H), 4.63 – 4.73 (m, 2 H), 4.85 – 4.96 (m, 2 H), 4.96 – 5.03 (d, 1 H), 6.14 – 6.22 (d, 1 H), 6.49 – 6.56 (d, 1 H), 9.56 – 9.63 (s, 1 H). HRMS (ESI) calcd for C23H42N3O5: [M+H] +: 440.3124 Found: 440.3163.
4.1.11.13. (6S, 9S)-13-(3-Chlorophenyl)-9-isobutyl-3,8,11-trioxo-1,12-dioxa-2,7,10-triazacyclooctadecane-6-carbaldehyde 36A
White solid (yield (30%); m.p 62 – 63°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.83 – 1.00 (d, 6 H), 1.21 – 1.29 (m, 4 H), 1.44 – 1.96 (m, 7 H), 2.36 – 2.77 (m, 4 H), 4.00 – 4.22 (t, 2 H), 5.08 – 5.11 (m, 1 H), 5.45 – 5.60 (m, 1 H), 6.14 – 6.22 (d, 1 H), 6.49 – 6.56 (d, 1 H), 7.12 – 7.20 (s, 1 H),7.23 – 7.33 (m, 3 H),7.39 – 7.44 (d, 1 H), 9.59 – 9.63 (d, 1 H), 9.72 – 9.77 (s, 1 H). HRMS (ESI) calcd for C24H34ClN3O6: [M] +: 495.2136 Found: 495.2279.
4.1.11.14. (6S, 9S)-13-(3-Chlorophenyl)-9-isobutyl-3,8,11-trioxo-1,12-dioxa-2,7,10-triazacyclooctadecane-6-carbaldehyde 36B
White solid (yield 50%); m.p 78 – 79°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.81 – 0.99 (d, 6 H), 1.23 – 1.31 (m, 4 H), 1.44 – 1.94 (m, 7 H), 2.36 – 2.70 (m, 4 H), 3.58 – 3.64 (t, 2 H), 4.00 – 4.16 (m, 2 H), 4.25 – 4.34 (m, 1 H), 5.50 – 5.63 (m, 1 H), 6.60 – 6.70 (d, 1 H), 7.12 – 7.22 (s, 1 H),7.23 – 7.34 (m, 3 H),7.85 – 7.91 (d, 1 H), 8.22 – 8.27 (d, 1 H), 9.72 – 9.76 (s, 1 H). HRMS (ESI) calcd for C24H34ClN3O6: [M] +: 495.2136 Found: 495.2283.
4.1.12. Synthesis of alcohols 22b and 22d. General procedure
To a solution of representative ester (5 mmol) in anhydrous THF (30 mL) was added lithium borohydride (2M in THF, 7.5 mL, 15 mmol) dropwise, followed by absolute ethyl alcohol (15 mL), and the reaction mixture was stirred at room temperature overnight. The reaction mixture was then acidified by adding 5% HCl and the pH adjusted to ~2. Removal of the solvent left a residue which was taken up in ethyl acetate (100 mL). The organic layer was washed with brine (25 mL), dried over anhydrous sodium sulfate, filtered, and concentrated to yield compounds 22b and 22d as white solids.
4.1.12.1. (4S, 7S, E)-17-Benzyl-7-(hydroxymethyl)-4-isobutyl-1-oxa-3,6,11-triazacycloheptadec-13-ene-2,5,10-trione 22b
Sticky solid (yield 83%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.81 – 0.97 (d, 6 H), 1.53 – 1.69 (m, 4 H), 1,91 – 2.02 (m, 5 H), 2.70 – 2.93 (m, 2 H), 3.53 – 3.64 (m, 2 H), 3.80 – 3.94 (m, 2 H), 4.40 – 4.62 (m, 2 H), 4.88 – 5.04 (m, 1 H), 5.06 – 5.26 (m, 2 H), 5.68 – 5.87 (m, 2 H), 6.20 – 6.41 (br. d, 1 H), 6.91 – 7.10 (d, 1 H), 7.14 – 7.36 (m, 5 H), 7.85 – 7.92 (d, 1 H). HRMS (ESI) calcd for C25H38N3O5: [M+H] +: 460.2811 Found: 460.2831.
4.1.12.2. (4S, 7S, E)-17-(3-Chlorophenyl)-4-(cyclohexylmethyl)-7-(hydroxymethyl)-1-oxa-3,6,11-triazacycloheptadec-13-ene-2,5,10-trione 22d
Sticky solid (yield 80%); 1H NMR (400 MHz, CDCl3-d) δ ppm 0.66 – 0.93 (m, 11 H), 0.95 – 2.32 (m, 10 H), 3.51 – 3.61 (m, 2 H), 3.74 – 3.86 (d, 2 H), 4.00 – 4.20 (m, 1 H), 4.29 – 4.51 (m, 1 H), 4.84 – 5.01 (m, 1 H), 5.42 – 5.83 (m, 2 H), 5.94 – 6.11 (m, 1 H), 6.22 – 6.31 (d, 1 H), 6.75 – 6.92 (d, 1 H), 7.04 – 7.27 (m, 4 H). HRMS (ESI) calcd for C27H38ClN3O5: [M+H] +: 519.2500 Found: 519.2561.
4.1.13. Synthesis of aldehydes 23 and 24. General procedure
Representative alcohol (0.6 mmol) was dissolved in anhydrous dichloromethane (20 mL) under a nitrogen atmosphere and cooled to 0°C. Dess-Martin periodinane (0.75 g, 1.78 mmol, 3.0 eq.) was added to the reaction mixture with stirring. The ice bath was removed and the reaction mixture was stirred at room temperature for 3 h (monitoring by TLC indicated complete disappearance of the starting material). A solution of 40 mM sodium thiosulfate in saturated aqueous NaHCO3 (50 mL) was added and the solution was stirred for another 15 min. The aqueous layer was removed and the organic layer was washed with sodium bicarbonate (25 mL), water (2 × 25 mL) and brine (25 mL). The organic layer was dried over anhydrous sodium sulfate, filtered and concentrated. The yellow residue was purified by flash chromatography (silica gel/methylene chloride/ethyl acetate/methanol) to yield the desired aldehyde.
4.1.13.1 (4S, 7S, E)-17-Benzyl-4-isobutyl-2,5,10-trioxo-1-oxa-3,6,11-triazacycloheptadec-13-ene-7-carbaldehyde 23
White solid (yield 53%); m.p 88 – 89°C. 1H NMR (400 MHz, CDCl3-d) δ ppm 0.83 – 0.98 (d, 6 H), 1.55 – 1.70 (m, 4 H), 1,93 – 2.04 (m, 5 H), 2.71 – 2.93 (m, 2 H), 3.81 – 3.95 (m, 2 H), 4.41 – 4.61 (m, 2 H), 4.90 – 5.05 (m, 1 H), 5.08 – 5.27 (m, 2 H), 5.69 – 5.89 (m, 2 H), 6.20 – 6.37 (br. d, 1 H), 6.95 – 7.11 (d, 1 H), 7.14 – 7.36 (m, 5 H), 7.85 – 7.92 (d, 1 H), 9.54 – 9.67 (s, 1 H). HRMS (ESI) calcd for C25H36N3O5: [M+H] +: 458.2655 Found: 458.2666.
4.1.13.2. (4S, 7S, E)-17-(3-Chlorophenyl)-4-(cyclohexylmethyl)-2,5,10-trioxo-1-oxa-3,6,11-triazacycloheptadec-13-ene-7-carbaldehyde 24
White solid (yield 59%); m.p 83 – 84°C.1H NMR (400 MHz, CDCl3-d) δ ppm 0.68 – 0.95 (m, 11 H), 0.99 – 2.34 (m, 10 H), 3.74 – 3.86 (d, 2 H), 4.00 – 4.20 (m, 1 H), 4.29 – 4.51 (m, 1 H), 4.84 – 5.01 (m, 1 H), 5.42 – 5.83 (m, 2 H), 5.94 – 6.11 (m, 1 H), 6.22 – 6.31 (d, 1 H), 6.75 – 6.92 (d, 1 H), 7.04 – 7.27 (m, 4 H), 9.56 – 9.63 (s, 1 H). HRMS (ESI) calcd for C27H37ClN3O5: [M+H] +: 518.2422 Found: 518.2430.
4.1.14. Synthesis of aldehyde bisulfite salts 25–29. General procedure
To a solution of aldehydes 13, 18, 19, or 21 (0.52 mmol) in dry ethyl acetate (3.5 mL) was added absolute ethanol (1.8 mL) with stirring, followed by a solution of sodium bisulfite (55 mg; 0.52 mmol) in water (0.5 mL). The reaction mixture was stirred for 3 h at 50 °C. The reaction mixture was allowed to cool to room temperature and then vacuum filtered. The solid was thoroughly washed with absolute ethanol and the filtrate was dried over anhydrous sodium sulfate, filtered, and concentrated to yield yellowish oil. The oily product was treated with ethyl ether (2 × 10 mL) to form white solid. The white solid was stirred with ethyl ether (5 mL) and ethyl acetate (2.5 mL) for 5 minutes. Careful removal of the solvent using a pipette left compound as a white solid.
4.1.14.1. Sodium ((4S, 7S)-17-(3-chlorophenyl)-4-isobutyl-2,5,10-trioxo-1-oxa-3,6,11-triazacycloheptadecan-7-yl) (hydroxy) methanesulfonate 25
White solid (yield 77%); m.p 145 – 146°C. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.81 – 0.97 (d, 6 H), 1.42 – 1.83 (m, 5 H), 2.11 – 2.43 (m, 6 H), 2.47 – 2.64 (m, 2 H), 3.28 – 3.40 (t, 2 H), 3.40 – 3.50 (m, 2H), 4.50 – 4.60 (m, 1 H), 4.62 – 4.77 (m, 1 H), 5.23 – 5.31 (br. s, 1 H), 5.62 – 5.71 (m, 1 H), 5.91 – 6.03 (m, 1 H), 6.14 – 6.24 (d, 1 H), 6.96 – 7.34 (m, 4 H), 8.02 – 8.11 (d, 1 H), 8.60 – 8.65 (d, 1 H). HRMS (ESI) calcd for C24H35ClN3O8S−: [M] −: 560.1839 Found: 560.1925.
4.1.14.2. Sodium ((20S)-10-(3-chlorophenyl)-8,17,22-trioxo-9-oxa-7,16,21-triazaspiro [5.16] docosan-20-yl) (hydroxy) methanesulfonate 26
White solid (yield 75%); m.p 148 – 149°C. 1H NMR (400 MHz, DMSO-d6) δ ppm 1.07 – 1.23 (m, 7 H), 1.23 – 1.98 (m, 9 H), 2.09 – 2.27 (m, 6 H), 3.42 – 3.56 (m, 3 H), 5.12 – 5.18 (br. s, 1 H), 5.43 – 5.66 (m, 1 H), 5.78 – 5.82 (m, 1 H), 6.97 – 7.08 (d, 1 H), 7.08 – 7.19 (d, 1 H), 7.24 – 7.46 (m, 4 H), 8.11 – 8.17 (d, 1H). HRMS (ESI) calcd for C25H35ClN3O8S−: [M] −: 572.1839 Found: 572.1849.
4.1.14.3. Sodium ((21S)-10-(3-chlorophenyl)-8,18,23-trioxo-9-oxa-7,17,22-triazaspiro[5.17] tricosan-21-yl) (hydroxy) methanesulfonate 27
White solid (yield 65%); m.p 145 – 146°C. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.90 – 1.36 (m, 11 H), 1.53 – 1.72 (m, 7 H), 2.10 – 2.14 (m, 2 H), 2.14 – 2.54 (m, 4 H), 2.86 – 3.02 (m, 2 H), 4.42 – 4.60 (m, 1 H), 5.00 – 5.11 (m, 1 H), 5.13 – 5.20 (br. s, 1 H), 5.66 – 5.81 (m, 1 H), 6.15 – 6.23 (d, 1 H), 7.19 – 7.36 (m, 4 H), 7.89 – 7.94 (d, 1 H), 8.10 – 8.16 (d, 1 H). HRMS (ESI) calcd for C26H37ClN3O8S−: [M] −: 586.1995 Found: 586.2064.
4.1.14.4. Sodium hydroxyl ((4S, 7S)-4-isobutyl-2,5,10-trioxo-17-(3-phenylpropyl)-1-oxa-3,6,11-triazacycloheptadecan-7-yl) methanesulfonate 28
White solid (yield 70%); m.p 173 – 174°C. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.90 – 1.03 (d, 6 H), 1.15 – 1.81 (m, 10 H), 2.09 – 2.19 (m, 9 H), 2.53 – 2.64 (t, 2 H), 3.08 – 3.19 (t, 2 H), 3.50 – 3.65 m, 1 H), 4.65 – 4.73 (m, 1 H), 4.90 – 5.00 (d, 2 H), 5.21 – 5.30 (s, 1 H), 5.68 – 5.70 (d, 1 H), 6.36 – 6.44 (d, 1 H), 7.12 – 7.22 (d, 2 H), 7.24 – 7.32 (m, 3 H), 8.32 – 8.40 (d, 1 H). HRMS (ESI) calcd for C26H37ClN3O8S−: [M] −: 586.1995 Found: 586.2064.
4.1.14.5. Sodium hydroxyl ((4S, 7S)-4-isobutyl-2,5,10-trioxo-17-pentyl-1-oxa-3,6,11-triazacycloheptadecan-7-yl) methanesulfonate 29
White solid (yield 75%); m.p 143 – 144°C. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.85 – 0.92 (t, 2 H), 0.93 – 1.04 (d, 6 H), 1.21 – 1.35 (m, 8 H), 1.40 – 1.85 (m, 9 H), 2.22 – 2.50 (m, 4 H), 3.15 – 3.27 (m, 1 H), 3.39 – 3.49 (m, 1 H), 3.97 – 4.07 (m, 1 H), 4.63 – 4.73 (m, 2 H), 4.85 – 4.96 (m, 2 H), 4.96 – 5.03 (d, 1 H), 5.21 – 5.30 (br. s, 1 H), 5.67 – 5.71 (d, 1 H), 6.14 – 6.22 (d, 1 H), 6.49 – 6.56 (d, 1 H), 8.12 – 8.17 (d, 1 H). HRMS (ESI) calcd for C23H42ClN3O8S−: [M] −: 520.2698 Found: 520.2715.
4.1.15. Synthesis of compound 30.
A solution of aldehyde 13 (1.25 mmol) in ethyl acetate (10 mL) kept at 0° C was treated with acetic acid (90 mg; 1.44 mmol) followed by cyclopropyl isocyanide (92 mg; 1.375 mmol), and the reaction mixture was stirred at room temperature for 18 h. The solution was concentrated in vacuo and the residue was dissolved in methanol (10 mL) and treated with a solution of K2CO3 (0.4 g; 2.95 mmol) in water (7 mL). The reaction mixture was stirred at room temperature for 2 h. Methanol was evaporated off and the aqueous layer was extracted with ethyl acetate (3 × 20 mL). The combined organic layers were washed with 5% HCl (2 × 20 mL) and brine (25 mL). The organic layer was dried over anhydrous sodium sulfate and the solvent was removed to yield compound 30 as a off-white solid. This was used in the next step without further purification.
4.1.15.1. 2-((4S, 7S)-17-(3-Chlorophenyl)-4-isobutyl-2,5,10-trioxo-1-oxa-3,6,11-triazacycloheptadecan-7-yl)-N-cyclopropyl-2-hydroxyacetamide 30
Off-white solid (yield (57%); m.p 189 – 190°C. 1H NMR (400 MHz, DMSO-d6) δ ppm 0.37 – 0.48 (m, 2 H), 0.51 – 0.63 (m, 2 H), 0.73 – 0.96 (d, 6 H), 1.06 – 1.86 (m, 11 H), 2.11 – 2.33 (m, 4 H), 2.60 – 2.75 (m, 1 H), 3.18 – 3.23 (t, 2 H), 3.97 – 4.16 (m, 1 H), 4.48 – 4.59 (m, 1 H), 4.87 – 4.94 (m, 1 H), 5.72 – 5.87 (m, 1 H), 6.26 – 6.44 (s, 1 H), 6.82 – 6.99 (d, 1 H), 6.99 – 7.08 (d, 1 H), 7.22 – 7.51 (m, 4 H), 7.60 – 7.65 (m, 1 H), 7.65 – 7.80 (d, 1 H). HRMS (ESI) calcd for C28H42ClN4O6: [M+H] +: 565.2793 Found: 565.2811.
4.1.16. Synthesis of α-ketoamide 31
To a solution of compound 30 (0.62 mmol) in anhydrous dichloromethane (20 mL) cooled to 0° C and kept under a nitrogen atmosphere was added Dess-Martin periodinane reagent (0.53 g, 1.24 mmol, 2.0 eq) with stirring. The ice bath was removed and the reaction mixture was stirred at room temperature for 3 h. The reaction was monitored by TLC until the starting material disappeared. A solution of 10 % aqueous sodium thiosulfate (20 mL) was added and the solution was stirred for 15 min. The solution was poured into a separatory funnel and the aqueous layer was removed. The organic layer was washed with 10 % aqueous sodium thiosulfate (20 mL), followed by saturated aqueous sodium bicarbonate (2 × 20 mL), water (2 × 20 mL) and brine (20 mL). The organic layer was dried over anhydrous sodium sulfate, filtered and concentrated leaving a yellow solid which was purified by flash chromatography (silica gel/methylene chloride/ethyl acetate/methanol) to yield 31 as a white solid.
4.1.16.1. 2-((4S, 7S)-17-(3-Chlorophenyl)-4-isobutyl-2,5,10-trioxo-1-oxa-3,6,11-triazacycloheptadecan-7-yl)-N-cyclopropyl-2-oxoacetamide 31
White solid (yield 50%); m.p 210°C (d). 1H NMR (400 MHz, DMSO-d6) δ ppm 0.34 – 0.44 (m, 2 H), 0.49 – 0.60 (m, 2 H), 0.83 – 0.96 (d, 6 H), 1.12 – 1.84 (m, 11 H), 2.10 – 2.30 (m, 4 H), 2.63 – 2.75 (m, 1 H), 3.18 – 3.23 (t, 2 H), 4.00 – 4.16 (m, 1 H), 4.49 – 4.57 (m, 1 H), 4.86 – 4.92 (m, 1 H), 5.75 – 5.87 (m, 1 H), 6.80 – 6.90 (d, 1 H), 6.98 – 7.08 (d, 1 H), 7.21 – 7.50 (m, 4 H), 7.62 – 7.68 (m, 1 H), 7.70 – 7.80 (d, 1 H). HRMS (ESI) calcd for C28H40ClN4O6: [M+H] +: 563.2636 Found: 563.2660.
4.2. Biochemical studies. FRET protease assays
The FRET protease assay 3CLpro was performed by preparing stock solutions of the substrate (Edans-DFHLQ/GP-Dabcyl) and inhibitor in DMSO and diluting into assay buffer which was comprised of 20mM HEPES buffer, pH 8, containing NaCl (200 mM), 0.4 mM EDTA, glycerol (60%), and 6 mM dithiothreitol (DTT). The protease was mixed with serial dilutions of each compound up to 100 μM or with DMSO in 25 μL of assay buffer and incubated at 37°C for 30 min, followed by the addition of 25 μL of assay buffer containing substrate. Fluorescence readings were obtained using an excitation wavelength of 360 nm and an emission wavelength of 460 nm on a fluorescence microplate reader (FLx800; Biotec, Winoosk, VT) 1 h following the addition of substrate. Relative fluorescence units (RFU) were determined by subtracting background values (substrate-containing well without protease) from the raw fluorescence values, as described previously [33,47–49]. The dose-dependent FRET inhibition curves were fitted with a variable slope by using GraphPad Prism software (GraphPad, La Jolla, CA) in order to determine the IC50 values of the inhibitors.
4.3. Cell-based inhibition assays
The effects of each inhibitor on virus replication were examined against NV or MNV in the NV replicon harboring cells (HG23 cells) or RAW264.7 cells, respectively [33]. Briefly, confluent and semi-confluent HG23 cells were incubated with medium containing DMSO (<0.1%) or each compound (up to 20 μM) for 48 h for NV. After the incubation, total RNA was extracted and viral genome was quantitated with real-time quantitative RT-PCR (qRT-PCR). For MNV, confluent RAW264.7 cells were inoculated with MNV at a multiplicity of infection (MOI) of 0.1 for 1 h, and the inoculum was replaced with medium containing DMSO (0.1%) or each compound (up to 20 μM). The virus-infected cells were further incubated for 24 h, and the replication of virus was measured by the 50% tissue culture infective dose (TCID50) method. The EC50 values were determined by GraphPadPrism software [33].
4.4. Nonspecific cytotoxic effects
The cytotoxic dose for 50% cell death (CC50) for each compound was determined for HG23 cells used in this study. Confluent cells grown in 96-well plates were treated with various concentrations (1 to 100 μM) of each compound for 48 h. Cell cytotoxicity was measured by a CytoTox 96 nonradioactive cytotoxicity assay kit (Promega, Madison, WI) and crystal violet staining. The in vitro therapeutic index was calculated by dividing the CC50 by the EC50.
4.5. X-ray crystallographic studies. Crystallization and Data Collection
Purified NV 3CL pro in 100 mM NaCl, 50 mM PBS pH 7.2, 1 mM DTT at a concentration of 10 mg/mL was used for preparation of the NV 3CLpro:inhibitor complexes. A 100 mM stock solution of the inhibitors was prepared in DMSO and the NV 3CLpro:inhibitor complex was prepared by mixing 9 μL of inhibitor (3 mM) with 291 μL (0.49 mM) of NV 3CLpro and incubating on ice for 1 h. The buffer was exchanged to 100 mM NaCl, 20 mM Tris pH 8.0 in a Vivaspin-20 (MWCO=5kDa, Vivaproducts, Inc.) concentrator and the sample was concentrated to 10.0 mg/mL for crystallization screening. All crystallization experiments were conducted Compact Jr. (Rigaku Reagents) sitting drop vapor diffusion plates at 20 °C using equal volumes of protein and crystallization solution equilibrated against 75 μL of the latter. Crystals of the inhibitor 13 complex (NV 3CLpro:13), that displayed a prismatic morphology, were obtained from the Index HT screen (Hampton Research) condition H12 (30% (w/v) PEG 2000 MME, 150 potassium bromide) in 1–2 days. Crystals of the complex with inhibitor 21 (NV 3CLpro:21) were also obtained from the Index HT G5 condition. Samples were transferred to a fresh drop composed of 80% crystallization solution and 20% PEG 400 and stored in liquid nitrogen. X-ray diffraction data were collected at the Advanced Photon Source IMCA-CAT beamline 17-ID using a Dectris Pilatus 6M pixel array detector.
4.6. Structure Solution and Refinement
Intensities were integrated using XDS [50–51] via Autoproc [52] and the Laue class analysis and data scaling were performed with Aimless [53] which suggested that the highest probability Laue class was 2/m and space group C2. The Matthew’s coefficient [54] suggested that there was a single molecule in the asymmetric unit (Vm=1.8 Å3/Da, % solvent=32%). Structure solution was conducted by molecular replacement with Phaser [55] using a previously determined structure of inhibitor bound NV 3CLpro (PDB: 3UR9) [33] as the search model. Structure refinement using and manual model building were conducted with Phenix [56] and Coot [57] respectively. Anisotropic atomic displacement parameters were refined for all atoms except solvent molecules for the NV 3CLpro:13 structure. Disordered side chains were truncated to the point for which electron density could be observed. Structure validation was conducted with Molprobity [58] and figures were prepared using the CCP4MG package [59].
4.7. Accession Codes
Coordinates and structure factors were deposited to the Worldwide Protein Databank (wwPDB) with the accession codes 5TG1 (NV 3CLpro:13) and 5TG2 (NV 3CLpro:21).
Table 2.
Crystallographic data for NV 3CLpro:inhibitor 13 and 21 structures
| NV 3CLpro:13 | NV 3CLpro:21 | |
|---|---|---|
| Data Collection | ||
| Unit-cell parameters (Å, °) | a=64.40, b=36.49, c=61.47, β=112.5 | a=67.02, b=36.70, c=61.72, β =110.2 |
| Space group | C2 | C2 |
| Resolution (Å)1 | 31.83–1.40 (1.42–1.40) | 32.79–1.75 (1.78–1.75) |
| Wavelength (Å) | 1.0000 | 1.0000 |
| Temperature (K) | 100 | 100 |
| Observed reflections | 83,610 | 46,353 |
| Unique reflections | 25,218 | 14,350 |
| <I/σ(I)>1 | 13.4 (1.9) | 9.5 (1.9) |
| Completeness (%)1 | 96.8 (95.3) | 99.6 (94.6) |
| Multiplicity1 | 3.3 (3.2) | 3.2 (2.4) |
| Rmerge (%)1, 2 | 4.8 (69.1) | 8.2 (46.4) |
| Rmeas (%)1, 4 | 5.8 (82.9) | 9.8 (58.9) |
| Rpim (%)1, 4 | 3.1 (45.4) | 5.4 (35.6) |
| CC1/21, 5 | 0.998 (0.704) | 0.996 (0.780) |
| Refinement | ||
| Resolution (Å) 1 | 31.83–1.40 | 32.79–1.75 |
| Reflections (working/test)1 | 24,030/1,183 | 13,624/710 |
| Rfactor/Rfree (%)1,3 | 14.3/19.2 | 17.6/22.7 |
| No. of atoms (Protein/Ligand/Water) | 1,253/33/128 | 1,196/31/83 |
| Model Quality | ||
| R.m.s deviations | ||
| Bond lengths (Å) | 0.009 | 0.01 |
| Bond angles (°) | 1.035 | 0.998 |
| Average B-factor (Å2) | ||
| All Atoms | 19.2 | 20.4 |
| Protein | 17.9 | 19.7 |
| Ligand | 20.9 | 22 |
| Water | 31.4 | 29.9 |
| Coordinate error(maximum likelihood) (Å) | 0.15 | 0.19 |
| Ramachandran Plot | ||
| Most favored (%) | 98.8 | 98.1 |
| Additionally allowed (%) | 1.2 | 1.9 |
Values in parenthesis are for the highest resolution shell.
Rmerge = ΣhklΣi |Ii(hkl) − <I(hkl)>|/ΣhklΣi Ii(hkl), where Ii(hkl) is the intensity measured for the ith reflection and <I(hkl)> is the average intensity of all reflections with indices hkl.
Rfactor = Σhkl ||Fobs (hkl) | − |Fcalc (hkl) ||/Σhkl |Fobs (hkl)|; Rfree is calculated in an identical manner using 5% of randomly selected reflections that were not included in the refinement.
A novel series of macrocyclic inhibitors of norovirus 3CL protease inhibitors is reported.
High resolution crystal structures of enzyme-inhibitor complexes have confirmed the mechanism of action.
The structural determinants involved in binding have been delineated.
The inhibitor design rationale was validated by x-ray crystallographic studies.
Acknowledgments
The generous financial support of this work by the National Institutes of Health (AI109039) is gratefully acknowledged. Use of the University of Kansas Protein Structure Laboratory was supported by a grant from the National Institute of General Medical Sciences (P30GM110761) from the National Institutes of Health. Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02-06CH11357.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Green KY. Caliciviridae: The Noroviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Vol. 1. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 949–979. [Google Scholar]
- 2.Lee BE, Pang XL. New strains of norovirus and the mystery of viral gastroenteritis epidemics. Can Med Assoc. 2013;185:1381–1382. doi: 10.1503/cmaj.130426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parra GL, Green KY. Genome of emerging norovirus GII.17, United States, 2014. Emerg Infect Dis. 2015;21:1477–1479. doi: 10.3201/eid2108.150652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koo HL, Ajami N, Atmar RL, DuPont HL. Noroviruses: the leading cause of gastroenteritis worldwide. Discov Med. 2010;10:61–70. [PMC free article] [PubMed] [Google Scholar]
- 5.Hall AJ, Lopman BA, Payne DC, Patel MM, Gastanaduy PA, Vinje J, Parashar UD. Norovirus disease in the United States. Emerg Infect Dis. 2013;19:1198–1205. doi: 10.3201/eid1908.130465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartsch SM, Lopman BA, Ozawa S, Hall AJ, Lee BY. Global economic burden of norovirus gastroenteritis. PLoS One. 2016;11(4):e0151219. doi: 10.1371/journal.pone.0151219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belliott G, Lopman BA, Ambert-Balay K, Pothier P. The burden of norovirus gastroenteritis: an important foodborne and healthcare-related infection. Clin Microbiol Infect. 2014;20:724–730. doi: 10.1111/1469-0691.12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Institutes of Health/U.S. National Library of Medicine/Medline Plus. Norovirus, a Costly Bug. https://www.nih.gov/medlineplus/news/fullstory_158512.html.
- 9.Pringle K, Lopman B, Vega E, Vinje J, Parashar UD, Hall AJ. Noroviruses: epidemiology, immunity and prospects for prevention. Future Microbiol. 2015;10:53–67. doi: 10.2217/fmb.14.102. [DOI] [PubMed] [Google Scholar]
- 10.Bok K, Green KY. Norovirus gastroenteritis in immunocompromised patients. N Engl J Med. 2012;367:2126–2132. doi: 10.1056/NEJMra1207742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robilotti E, Deresinski S, Pinsky BA. Noroviruses. Clin Microbiol Rev. 2015;28:134–164. doi: 10.1128/CMR.00075-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanata CF, Fischer-Walker CL, Olascoaga AC, Torres CX, Aryee MKJ, Black RE. Global causes of diarrheal disease mortality in children <5 years of age: a systematic review. PLoS One. 2013;8:e72788. doi: 10.1371/journal.pone.0072788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall AJ. Noroviruses: the perfect human pathogen. J Infect Dis. 2012;205:1622–1624. doi: 10.1093/infdis/jis251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore MD, Goulter RM, Jaykus LA. Human norovirus as a foodborne pathogen: challenges and developments. Ann Rev Food Sci Technol. 2015;6:411–433. doi: 10.1146/annurev-food-022814-015643. [DOI] [PubMed] [Google Scholar]
- 15.Kim Y, Galasiti Kankanamalage AC, Chang K-O, Groutas WC. Recent advances in the discovery of norovirus therapeutics. J Med Chem. 2015;58:9438–9450. doi: 10.1021/acs.jmedchem.5b00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galasiti Kankanamalage AC, Weerawarna PM, Kim Y, Chang KO, Groutas WC. Anti-norovirus therapeutics: a patent review (2010–2015) Expert Opin Ther Pat. 2016;26:297–308. doi: 10.1517/13543776.2016.1153065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venkataraman Prasad BV, Shanker S, Muhaxhiri Z, Deng L, Choi J-M, Estes MK, Song Y, Palzkill T, Atmar RL. Antiviral targets of human noroviruses. Curr Opin Virol. 2016;18:117–125. doi: 10.1016/j.coviro.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weerasekara S, Prior AM, Hua DH. Current tools for norovirus drug discovery. Expert Opin Drug Discov. 2016;11:529–541. doi: 10.1080/17460441.2016.1178231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kocher J, Yuan L. Norovirus vaccines and potential anti-norovirus drugs: recent advances and future perspectives. Future Virol. 2015;10:899–913. doi: 10.2217/fvl.15.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rocha-Pereira J, Neyts J, Jochmans D. Norovirus: targets and tools in antiviral drug discovery. Biochem Pharmacol. 2014;91:1–11. doi: 10.1016/j.bcp.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atmar RL, Baehner F, Cramer JP, Song E, Borkowski A, Mendelman PM. Rapid responses to 2 virus-like particle norovirus vaccine candidate formulations in healthy adults: a randomized controlled trial. J Infect Dis. 2016;214:845–853. doi: 10.1093/infdis/jiw259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karst SM, Tibbets SA. Recent advances in understanding norovirus pathogenesis. J Med Virol. 2016;88:1837–1843. doi: 10.1002/jmv.24559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karst SM, Wobus CE. A working model of how noroviruses infect the intestine. PLoS Pathog. 2015 Feb 27;11(2):e1004626. doi: 10.1371/journal.ppat.1004626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thorne LG, Goodfellow IG. Norovirus gene expression and replication. J Gen Virol. 2014;95:278–291. doi: 10.1099/vir.0.059634-0. [DOI] [PubMed] [Google Scholar]
- 25.Karst SM. The influence of commensal bacteria on infection with enteric viruses. Nat Rev Microbiol. 2016;14:197–204. doi: 10.1038/nrmicro.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orchard RC, Wilen CB, Doench JG, Baldridge MT, McCune BT, Lee YCJ, Lee S, Pruett-Miller SM, Nelson CA, Fremont CA, Virgin HW. Discovery of a proteinaceous cellular receptor for a norovirus. Science. 2016;10:1126. doi: 10.1126/science.aaf1220. science.aaf1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hussey RJ, Coates L, Gill RS, Erskine PT, Coker SF, Mitchell E, Cooper JB, Broadbridge R, Clarke IN, Lambden PR, Shoolingin-Jordan PM. Structural study of norovirus 3C specificity: binding of a designed active site-directed peptide inhibitor. Biochemistry. 2011;50:240–249. doi: 10.1021/bi1008497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardy ME, Crone TJ, Brower JE, Ettayebi K. Substrate specificity of the Norwalk virus 3C-like proteinase. Virus Res. 2002;89:29–39. doi: 10.1016/s0168-1702(02)00114-4. [DOI] [PubMed] [Google Scholar]
- 29.Muhaxhiri Z, Deng L, Shanker S, Sankaran B, Estes MK, Palzkill T, Song Y, Venkataram Prasad BV. Structural basis of substrate specificity and protease inhibition in Norwalk virus. J Virol. 2013;87:4281–4292. doi: 10.1128/JVI.02869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi D, Hiromasa Y, Kim Y, Anbanandam A, Yao X, Chang KO, Prakash O. Structural and dynamics characterization of norovirus protease. Protein Sci. 2013;22:347–357. doi: 10.1002/pro.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allaire M, Chernaia MM, Malcolm BA, James MN. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases. Nature. 1994;369:72–76. doi: 10.1038/369072a0. [DOI] [PubMed] [Google Scholar]
- 32.Nomenclature used is that of; Schechter L, Berger A. Biochem Biophys Res Comm. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]; where S1, S2, S3, …. Sn and S1′, S2′, S3′, …. Sn′ correspond to the enzyme subsites on the N-terminus and C-terminus side, respectively, of the scissile bond. Each subsite accommodates a corresponding amino acid residue side chain designated P1, P2, P3,…Pn and P1′, P2′, P3′,…..Pn′ of a substrate or inhibitor. P1 is the primary substrate specificity residue and P1-P1′ is the scissile bond.
- 33.Galasiti Kankanamalage AC, Kim Y, Weerawarna PM, Uy RAZ, Damalanka VC, Mandadapu SR, Alliston KR, Mehzabeen N, Battaille KP, Lovell S, Chang K-O, Groutas WC. Structure-guided design and optimization of dipeptidyl inhibitors of norovirus 3CL protease. Structure-activity relationships and biochemical, x-ray crystallographic, cell-based and in vivo studies. J Med Chem. 2015;58:3144–3155. doi: 10.1021/jm5019934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tyndall JD, Fairlie DP. Proteases universally recognize beta strands in their active sites. Chem Rev. 2005;105:973–999. doi: 10.1021/cr040669e. [DOI] [PubMed] [Google Scholar]
- 35.Tyndall JD, Fairlie DP. Macrocycles mimic the extended peptide conformation recognized by aspartic, serine, cysteine, and metallo proteases. Current Med Chem. 2001;8:893–907. doi: 10.2174/0929867013372715. [DOI] [PubMed] [Google Scholar]
- 36.Leeson P, Young RJ. Molecular property design: does everyone get it? Med Chem Lett. 2015;6:722–725. doi: 10.1021/acsmedchemlett.5b00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meanwell N. Improving drug candidates by design: a focus on physicochemical properties as a means of improving compound disposition and safety. Chem Res Toxicol. 2011;24:1420–1456. doi: 10.1021/tx200211v. [DOI] [PubMed] [Google Scholar]
- 38.Bhat A, Roberts LR, Dwyer JJ. Lead discovery and optimization strategies for peptide macrocycles. Eur J Med Chem. 2015;94:471–479. doi: 10.1016/j.ejmech.2014.07.083. [DOI] [PubMed] [Google Scholar]
- 39.Marsault E, Peterson ML. Macrocycles are great cycles: applications, opportunities, and challenges of synthetic macrocycles. J Med Chem. 2011;54:1961–2004. doi: 10.1021/jm1012374. [DOI] [PubMed] [Google Scholar]
- 40.Giordanetto F, Kihlberg J. Macrocyclic drugs and clinical candidates: what can medicinal chemists learn from their properties? J Med Chem. 2014;57:278–295. doi: 10.1021/jm400887j. [DOI] [PubMed] [Google Scholar]
- 41.Doak BC, Zheng J, Dobritzsch D, Kihlberg J. How beyond the rule of 5 drugs and clinical candidates bind to their targets. J Med Chem. 2016;59:2312–2327. doi: 10.1021/acs.jmedchem.5b01286. [DOI] [PubMed] [Google Scholar]
- 42.Matsson P, Doak BC, Over B, Kihlberg J. Cell permeability beyond the rule of 5. Adv. Drug Deliv Rev. 2016;101:42–61. doi: 10.1016/j.addr.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 43.Doak BC, Zheng J, Dobritzsch D, Kihlberg J. How beyond the rule of 5 drugs and clinical candidates bind to their targets. J Med Chem. 2016;59:2312–2327. doi: 10.1021/acs.jmedchem.5b01286. [DOI] [PubMed] [Google Scholar]
- 44.Alex A, Millan DS, Perez M, Wakenhut F, Whitlock GA. Intramolecular hydrogen bonding to improve membrane permeability and absorption in beyond the rule of five chemical space. Med Chem Commun. 2011;2:669–674. [Google Scholar]
- 45.Bockus AT, Lexa KW, Pye CR, Kalgutkar AS, Gardner JW, Hund KCR, Hewitt WM, Schwochert JA, Glassey E, Price DA, Mathiowetz AM, Liras S, Jacobson MP, Lokey RS. Probing the physicochemical boundaries of cell permeability and oral bioavailability in lipophilic macrocycles inspired by natural products. J Med Chem. 2015;58:4581–4589. doi: 10.1021/acs.jmedchem.5b00128. [DOI] [PubMed] [Google Scholar]
- 46.Thansandote P, Harris RM, Dexter HL, Simpson GL, Pal S, Upton RJ, Valko K. Improving the passive permeability of macrocyclic peptides: balancing permeability with other physicochemical properties. Bioorg Med Chem. 2015;23:322–327. doi: 10.1016/j.bmc.2014.11.034. [DOI] [PubMed] [Google Scholar]
- 47.Kim Y, Lovell S, Tiew KC, Mandadapu SR, Alliston KR, Battaille KP, Groutas WC, Chang KO. Broad spectrum antivirals against 3Cl or 3C-like proteases of picornaviruses, noroviruses and coronaviruses. J Virol. 2012;6:11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Damalanka VC, Kim Y, Alliston KR, Weerawarna PM, Galasiti Kankanamalage AC, Lushington GH, Mehzabeen N, Battaille KP, Lovell S, Chang K-O, Groutas WC. Oxadiaxole-based vell-permeable macrocyclic transition state inhibitors of norovirus 3CL protease. J Med Chem. 2016;59:1899–1913. doi: 10.1021/acs.jmedchem.5b01464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weerawarna PM, Kim Y, Galasiti Kankanamalage AC, Damalanka VC, Lushington GH, Alliston KR, Mehzabeen N, Battaille KP, Lovell S, Chang K-O, Groutas WC. Structure-based design and synthesis of triazole-based macrocyclic inhibitors of norovirus protease: structural, biochemical, spectroscopic and antiviral studies. Eur J Med Chem. 2016;25:300–318. doi: 10.1016/j.ejmech.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kabsch W. Automatic Indexing of Rotation Diffraction Patterns. J Appl Crystallogr. 1988;21:67–72. [Google Scholar]
- 51.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, Bricogne G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evans PR. An Introduction to Data Reduction: Space-group Determination, Scaling and Intensity Statistics. Acta Crystallogr D Biol Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 55.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn M, Storoni LC, Read RJ. Phaser Crystallographic Software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams PD, Afonine PV, Bunkoczi G, Chen VB, David IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a Comprehensive Python-based System for Macromolecular Structure Solution. Acta Crystallogr Sect D: Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and Development of Coot. Acta Crystallogr Sect D: Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: All-atom Structure Validation for Macromolecular Crystallography. Acta Crystallogr Sect D: Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowlan K, Emlsey P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 Molecular Graphics Project. Acta Crystallogr Sect D: Biol Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 60.Evans P. Scaling and Assessment of Data Quality. Acta Crystallogr Sect D: Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 61.Diederichs K, Karplus PA. Improved R-factors for Diffraction Data Analysis in Macromolecular Crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- 62.Weiss MS. Global Indicators of X-ray Data Quality. J Appl Crystallogr. 2001;34:130–135. [Google Scholar]
- 63.Karplus PA, Diederichs K. Linking Crystallographic Model and Data Quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evans P. Biochemistry. Resolving Some Old Problems in Protein Crystallography. Science. 2012;336:986–987. doi: 10.1126/science.1222162. [DOI] [PubMed] [Google Scholar]


