Abstract
Traditionally, vaccine development involves tradeoffs between immunogenicity and safety. Live-attenuated vaccines typically offer rapid and durable immunity but reduced safety, while the inability of inactivated vaccines to replicate enhances safety at the expense of immunogenicity, often necessitating multiple doses and boosters. To overcome these tradeoffs, we developed the insect-specific alphavirus, Eilat virus (EILV), as a vaccine platform. To address the chikungunya virus (CHIKV) pandemic, we used an EILV cDNA clone to design a chimeric virus containing the CHIKV structural proteins. The recombinant EILV/CHIKV virus was structurally identical at 10Å to wild-type CHIKV by single particle cryoelectron microscopy, mimicked the early stages of CHIKV replication in vertebrate cells from attachment and entry to viral RNA delivery, yet remained completely defective for productive replication, providing a high degree of safety. A single dose of EILV/CHIKV produced in mosquito cells elicited rapid (within 4 days) and long-lasting (>290 days) neutralizing antibodies that provided complete protection in two different mouse models. In nonhuman primates, EILV/CHIKV elicited rapid and robust immunity that protected against viremia and telemetrically-monitored fever. Our EILV platform represents the first structurally native application of an insect-specific virus in preclinical vaccine development and highlights the potential application of such viruses in vaccinology.
Traditionally, viral vaccine development begins with a pathogenic virus, followed by inactivation to render it replication-defective, or attenuation to maintain limited replication that stimulates rapid, robust and long-lived immunity. However, inherent in these attenuation approaches is the risk of disease, either from incomplete inactivation, underscored by the Cutter incident (1) that left 200 children paralyzed due to residual live polio virus, or from inadequate or unstable attenuation that is only recognized upon vaccination of large populations when rare vulnerable individuals develop disease. An example is the live-attenuated yellow fever vaccine 17D, which in rare cases produces life-threatening disease (2); this risk is particularly high for RNA viruses, which mutate frequently with the potential for reversion of attenuating mutations (3). Live-attenuated viruses have also been used as vectors to express heterologous viral proteins, but these backbones are still derived from pathogenic viruses and may carry the risk of disease, especially in persons immunocompromised by therapies for cancer, transplantation or other diseases.
Alternative vaccination approaches include immunization with DNA or RNA that expresses viral proteins, or in vitro-synthesized viral protein subunits, expressed individually or as assembled virus-like particles devoid of nucleic acids. These approaches offer a high degree of safety but typically require multiple doses and periodic boosters; some can also be expensive to manufacture. All of these traits are suboptimal for vaccines against diseases that occur in resource-poor endemic settings where outbreaks can be sudden and explosive, as highlighted by the recent expansions of chikungunya, Zika, and Ebola viruses (4,5).
To overcome these tradeoffs in vaccine design, we examined insect viruses that are abundant but largely ignored. Next generation sequencing of 70 different insect species revealed 112 novel viruses, previously unknown due to their lack of any disease association (6). Many of these viruses are descended from ancestors of virulent viral pathogens, such as Ebola virus, but typically are restricted to replication in insects only. They therefore offer a diverse resource for vaccine vector development because natural selection in the absence of vertebrate infection has resulted in host range restrictions that may provide a highly favorable safety profile. We also anticipated that the lack of chemical or physical inactivation would leave the CHIKV proteins present in a chimeric virus in native, functional form, thereby maintaining wild-type immunogenicity and antigenicity. Here, we report the first application of such an insect-specific virus in vaccine development.
Eilat virus (EILV) is an alphavirus isolated from mosquitoes in Israel (7). The genus Alphavirus (family Togaviridae) is comprised of small, icosahedral, enveloped viruses that package single-stranded, positive-sense RNA genomes ~11–12 kb in length. The genome encodes two open reading frames (ORFs): the nonstructural polyprotein, responsible for replicating the viral genome, is translated directly from the first ORF, while the structural polyprotein is translated from a second ORF located on subgenomic RNA transcripts driven by a subgenomic promoter (8). The majority of the 31 recognized alphavirus species are transmitted by mosquitoes and replicate in both invertebrates and many vertebrate taxa, allowing for their maintenance in enzootic cycles with occasional spillover into humans, sometimes causing severe disease during explosive outbreaks (9). EILV is the only described insect-specific alphavirus and is fundamentally and completely defective for vertebrate cell infection due to an inability to enter cells as well as to replicate its RNA genome, even after its artificial delivery into the cytoplasm (10). Phylogenetically, EILV groups within the mosquito-borne clade of alphaviruses that includes many important human pathogens such as chikungunya virus (CHIKV). The host-restricted characteristics of EILV coupled with the genetic tractability of alphaviruses (11,12) provide an opportunity to develop EILV as a novel vaccine platform for alphaviral diseases.
CHIKV, first associated with human disease in the 1950s in present-day Tanzania (13), has repeatedly emerged to cause widespread mosquito-borne epidemics with attack rates reaching 90% (14). Chikungunya fever (CHIKF) is characterized by severe, debilitating polyarthralgia with chronic joint pain that can persist for years in >60% of those affected (15). Although infection is rarely fatal, its disabling nature results in devastating economic impacts. For example, a La Reúnion Island outbreak resulted in an estimated loss of 55,000 life-years, and a health-care cost of US$50–55 million (14). Despite several vaccine candidates in pre-clinical and clinical development, none is licensed and no specific therapeutics are available for CHIKF.
We previously demonstrated the utility of an EILV/CHIKV chimeric virus as an antigen for serodiagnostics (16). Here, we further developed this chimera as a highly safe yet highly immunogenic vaccine for CHIKF, with protective efficacy demonstrated in multiple animal models after a single dose.
Results
EILV/CHIKV replicates to high titers in mosquito cells
We previously described an infectious cDNA clone of EILV/CHIKV (Fig. 1a) containing the structural polyprotein ORF of a 2014 CHIKV strain isolated from an infected patient from the British Virgin Islands (17). In vitro-transcribed RNA was electroporated into C7/10 mosquito cells, yielding a viral titer of 1×1010 plaque-forming units (PFU)/ml while passaged virus replicated to a mean peak titer of 1.4×109 PFU/ml by 36 hours post-infection (Fig. 1b). Interestingly, EILV/CHIKV replicated to higher titers and produced more overt cytopathic effects (CPE) than those previously reported for EILV (7), and formed plaques on mosquito cells similar in size and kinetics to those produced by EILV (Fig. 1c).
Figure 1.
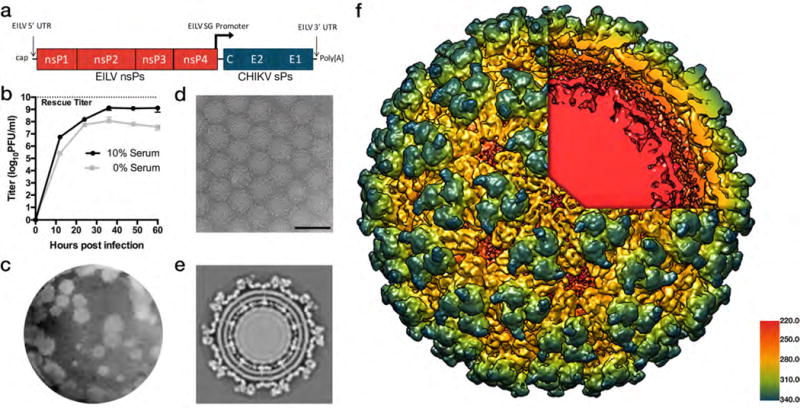
Characterization of EILV/CHIKV. (a) Genome organization of EILV/CHIKV chimeric virus containing the EILV 5′ and 3′ untranslated regions (UTRs) and subgenomic (SG) promoter as well as encoding genes for EILV nonstructural proteins (nsPs) and CHIKV structural proteins (sPs) (b) Replication kinetics of EILV/CHIKV on C7/10 cells in serum-containing or serum-free media. Monolayers were infected at a multiplicity of infection (measured in mosquito cells) of 0.1 PFU/cell and supernatants were collected at the indicated time points and titrated on C7/10 cell monolayers. Each data point represents the mean + s.d. titer of samples from triplicate infections and the experiment was performed twice. (c) EILV/CHIKV plaques 3 days after infection of C7/10 cells. (d) Cryo-electron micrograph (cryoEM) of EILV/CHIKV particles (scale bar = 100 nm). (e) Cross-section of EILV/CHIKV cryo-EM maps revealing transmembrane helices of E2/E1 glycoproteins interacting with capsid proteins. (f) A 9.85Å cryo-EM single-particle reconstruction of EILV/CHIKV, colored radially from E2/E1 heterodimer trimeric spikes in blue, green, and yellow through to the capsid proteins in red underlying the lipid bi-layer revealed through the cutaway. Particle reconstructions were identical to a previously published structure of CHIKV virus-like particles (Supplementary Fig.1).
Structural analysis and comparison with wild-type CHIKV
Single-particle cryo-electron microscopy (EM) reconstruction at 9.85Å resolution (gold-standard fourier shell correlation: FSC) revealed EILV/CHIKV virions with typical alphavirus structure, including trimeric spikes composed of heterodimers of the E1 and E2 envelope glycoproteins (Fig. 1d–f) embedded in the plasma membrane-derived lipid envelope and interacting with the capsid proteins (Fig. 1e–f). To evaluate whether the EILV/CHIKV structure differed from that of wild-type (wt) CHIKV, the previously published CHIKV cryo-EM structure (18)(European Molecular Data Base 5577) was low-pass filtered to 8Å, then aligned to the chimera and resampled onto a common grid before inspecting the maps and measuring the FSC. The cross-resolution (FSC=0.33) of the wt and EILV/CHIKV structures was 9.85Å, equal to the resolution of the EILV/CHIKV map, and no differences in structural protein conformation were observed (Supplementary Fig. 1).
Entry into vertebrate cells via endocytosis and genome translation
EILV was previously shown to be entry-deficient in vertebrate cells but EILV/CHIKV entry was not previously assessed. We therefore evaluated the ability of EILV/CHIKV to attach to vertebrate cells and assessed the effect of formalin-inactivation on this process. We purified EILV/CHIKV and live-attenuated CHIKV strain 181/clone25, a vaccine candidate developed by the U.S. Army (19). One-half of the EILV/CHIKV preparation was subjected to formalin-inactivation and the other mock-inactivated with diluent. These three preparations (181/clone25, live and inactivated EILV/CHIKV) were conjugated to Alexa Fluor 488 succinimidyl ester (AF488) as well as a mock conjugation to control for any nonspecific binding by the fluorophore. Unconjugated dye was removed by desalting column chromatography. NIH 3T3 cells were then exposed at a multiplicity of infection (MOI) of 100 PFU/cell to EILV/CHIKV-AF488, inactivated EILV/CHIKV-AF488, 181/clone25-AF488, or a PBS-AF488 control at 4°C. Mean fluorescence intensity (MFI) was quantified by flow cytometry and background fluorescence, determined by the PBS-AF488 control, was subtracted. While EILV/CHIKV- and 181/clone25-bound cells exhibited a MFI of 1987 and 2239, respectively, a significantly lower MFI (756) was detected from formalin-inactivated EILV/CHIKV-bound cells (Fig. 2a). The slightly higher MFI detected from 181/clone25-bound cells compared to live EILV/CHIKV may reflect the cell culture-adaptive Gly→Arg E2-82 substitution that increases binding to heparan sulfate and other glycosaminoglycans present on cultured cell surfaces (20,21).
Figure 2.
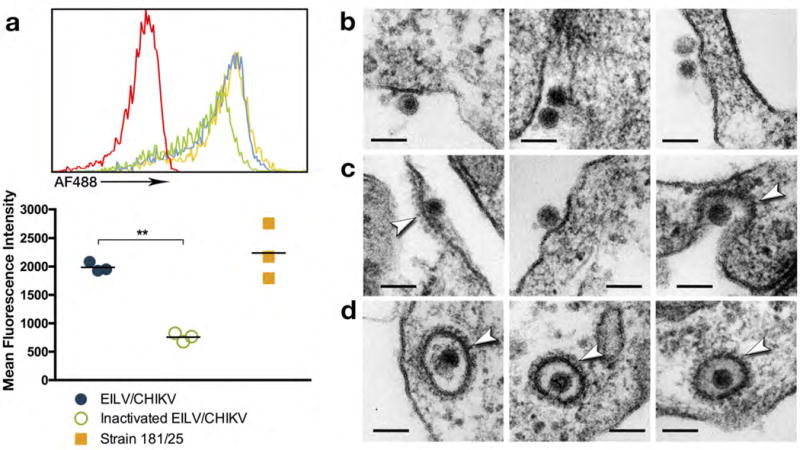
Binding and entry of EILV/CHIKV into vertebrate cells. (a) To study the effect of formalin-inactivation on binding, EILV/CHIKV, formalin-inactivated EILV/CHIKV, and live-attenuated strain 181/clone25 were conjugated to AF488 and dose-matched for binding to NIH 3T3 cells for 90 min at 4°C along with PBS-conjugation (red) to control for background fluorescence. Cells were then washed and three biological replicates per group were analyzed by flow cytometry (top graph) and background fluorescence was subtracted to calculate mean fluorescence intensity (bottom graph), two independent experiments were performed. Data reported as individual and mean values. (Statistical analysis: one-way ANOVA with Tukey’s multiple comparison test: ** = p < 0.0001). To assess downstream entry events, EILV/CHIKV was bound to Vero cells for 1hr at 4°C and monolayers were washed and then incubated at 37°C. (b) EILV/CHIKV particles were observed binding to the surface of Vero cells at time 0. (c) Fifteen seconds post-binding, clathrin-coated (arrows) pits formed around EILV/CHIKV particles. (d) Five-minutes post-binding, EILV/CHIKV particles were observed within clathrin-coated (arrows) vesicles (scale bar = 100 nm).
To visualize entry, we performed thin-section EM of Vero cells exposed to EILV/CHIKV, or formalin-inactivated EILV/CHIKV at a MOI of 1000 PFU/cell. Immediately after exposure to Vero cells, 70nm virions were observed binding to cell membranes (Fig. 2b) followed by the formation of clathrin-coated pit structures (22,23) within 15 seconds (Fig. 2c). At 5 minutes, EILV/CHIKV particles were associated with and observed within clathrin-coated vesicles, suggesting entry via clathrin-mediated endocytosis (Fig. 2d). In contrast, formalin-inactivated EILV/CHIKV was never detected entering cells and large aggregates of virus particles were visualized in close proximity to cell surfaces (Supplementary Fig. 2). This aggregation may also have contributed to an increase in MFI that was detected in the virus-binding experiment, suggesting that the specific cell surface binding of inactivated virus, per particle, is lower than we estimated (Fig. 2a).
Finally, to assess the ability of EILV/CHIKV to deliver genomic RNA and subsequent translation of this RNA by the host cell, we generated a recombinant virus containing GFP fused to nonstructural protein 3 of EILV (EILV/nsP3-GFP/CHIKV; Supplementary Fig. 3a). Vero or C7/10 cells (positive control) were infected at a MOI of 100 PFU/cell with EILV/nsP3-GFP/CHIKV, or mock-infected. Cells were then fixed, permeabilized, and stained at 8 HPI, and analyzed by fluorescence microscopy (Supplementary Fig. 3b–e). Similar to the C7/10 positive control, GFP foci were observed within the cytoplasm of Vero cells, indicating the localization of small nsP3-containing protein complexes previously described for replicating alphaviruses (24), while mock-infected controls were devoid of GFP foci.
EILV/CHIKV is replication-restricted and highly attenuated in vertebrates
We indirectly examined whether the translated non-structural proteins formed a functional replication complex by measuring translation of the subgenomic RNA, which is mediated by replicase-dependent transcription (8). We generated a recombinant construct containing nano-luciferase under the control of a second subgenomic promoter (EILV/nLUC/CHIKV)(Fig. 3a). 293T or C7/10 cells (positive control) were then infected at a MOI of 20 PFU/cell with EILV/nLUC/CHIKV and luciferase activity was measured over 8 hours. While infected C7/10 mosquito cells showed an exponential increase in luciferase activity, infected 293T cells exhibited only background levels (Fig. 3b).
Figure 3.
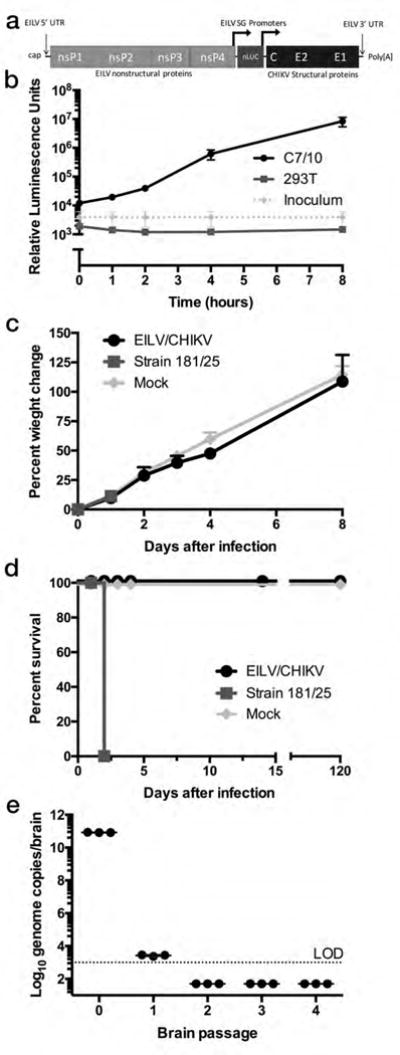
EILV/CHIKV is restricted from replication in vertebrate cells and does not cause disease in newborn IFN α/β receptor-knockout (IFNα/βR−/−) mice. (a) Genome organization of EILV/nLUC/CHIKV, generated by inserting nano luciferase (nLUC) directly after the subgenomic promoter of EILV followed by a duplication of this subgenomic promoter. (b) Luciferase assay of cells collected at 0, 1, 2, 4, and 8 hours after 293T or C7/10 cell infection in triplicate with EILV/nLUC/CHIKV at a multiplicity of infection of 20 PFU (measured in mosquito cells)/cell along with inoculum only at the same time points. Experiment was repeated three times and data were reported as mean + s.d.. (c, d) Newborn (7 day old, n=6/group) IFNα/βR−/− mice were infected intracranially with 8.7×108 PFU EILV/CHIKV, 1×104 PFU strain 181/clone25, or uninfected C7/10 cell supernatant and (c) weight change and (d) survival was recorded. Experiment was performed twice. (e) EILV/CHIKV was passaged in newborn (2–4 day old, n=3/passage) IFNα/βR−/− mouse brain and genomes were quantified in inoculum (passage 0) and following each passage, 2 days post-infection. All mice (n=8) survived the 5th passage (data not shown). Inoculum (passage 0) was reported as individual replicates as well as their mean value while remaining passages are reported as individual and mean values of 3 mouse replicates. (c–e) Experiment was performed once.
Next, we performed in vivo experiments to assess the host-restricted, replication-deficient phenotype of EILV/CHIKV. Previously, we reported on the safety of EILV/CHIKV as a diagnostic antigen by inoculating infant CD-1 mice intracranially and measuring viral loads in the brain, as well as survival (16). As a positive control for neurovirulence, we used the live-attenuated CHIKV strain 181/clone25 (19). Here we conducted a blinded analysis of histological brain sections collected during that study but not previously analyzed. At 7 days post-infection, 181/clone25-infected mice exhibited multifocal regions of neutrophilic inflammation and necrosis (microabcesses) within the neuronal parenchyma (Supplementary Fig. 4); one of 6 181/clone25-infected mice succumbed to illness, while none infected with EILV/CHIKV showed any signs of disease or histopathological lesions.
To more stringently examine the vertebrate cell replication deficiency and safety of EILV/CHIKV, we intracranially inoculated infant A129 IFN α/β receptor-knockout (IFNα/βR−/−) mice with 8.7 log10 PFU of EILV/CHIKV in C7/10 cell medium and compared weight change and survival to mice inoculated with a 10,000-fold lower dose of attenuated CHIKV strain 181/clone25 (Fig 3c–d), or C7/10 cell medium alone. All 6 of the mice died within 2 days of infection with CHIKV strain 181/clone25, while EILV/CHIKV-inoculated mice all survived at least 120 days and showed weight gains similar to those of mock-infected mice. To further confirm the host-restricted EILV/CHIKV phenotype, we performed 5 serial, intracranial, blind passages of EILV/CHIKV in 2–4 day-old IFNα/βR−/− mice (n=3 per passage), with 2-day incubations, starting with an 8.8 log10 PFU dose. When the 5th brain homogenate was inoculated into an additional infant A129 mouse cohort, all (n=8) survived. While we detected EILV/CHIKV RNA by qRT-PCR in passage 1 mouse brain homogenate, as well as infectious virus by plaque assay in 2 out of 3 mice (335 and 45 PFU), presumably due to the presence of residual inoculum virus, no evidence of replication was detected in subsequent passages (Fig. 3e).
Immunization of C57BL/6 mice induces robust humoral and cellular responses
An immunocompetent C57BL/6 mouse model was used to assess immunogenicity of EILV/CHIKV compared to dose-matched, formalin-inactivated EILV/CHIKV or a dose of live-attenuated strain 181/clone25 used in clinical trials (25). Four-week-old mice were vaccinated subcutaneously (SC) with a single 8.8 log10 PFU dose of live or inactivated EILV/CHIKV, or with a 5.5 log10 PFU dose of strain 181/clone25 used in prior studies (25), or mock-vaccinated with PBS. Rapid seroconversion was observed at 4 days post-vaccination (DPV) in 4 out of 5 mice vaccinated with live EILV/CHIKV, with 80% plaque-reduction neutralizing test (PRNT80) titers of 1:20–1:80, measured on Vero cells, while all other cohorts remained seronegative (<1:20). After 4 DPV, all groups showed an upward trend in PRNT80 titers and at 28 DPV there was a significant difference (p < 0.0001) between live and inactivated EILV/CHIKV, or strain 181/clone25 (Fig. 4a). One mouse vaccinated with 181/clone25 failed to seroconvert. Three mice per group were sacrificed at 4 or 8 DPV and splenocytes were isolated, stimulated with a peptide pool of CHIKV CD8 T-cell epitopes corresponding to proteins E1, E2, and C, as previously described (26), stained for CD3, CD8, and IFN-γ, and analyzed by flow cytometry (Fig. 4b & Supplementary Fig. 5). At 4 DPV, compared to mock-vaccinated mice, live EILV/CHIKV-vaccinated mice exhibited higher mean numbers of antigen-specific IFN-γ-producing CD8+ T cells (21.5×104 versus 7.6×104 cells, p < 0.0001, Fig. 4b). At 8 DPV, EILV/CHIKV-vaccinated mice exhibited further elevation in numbers of activated CD8+ T-cells, following peptide stimulation, compared to mock-vaccinated animals (33.7×104 versus 5.9×104 cells, p < 0.0001, Fig. 4b).
Figure 4.
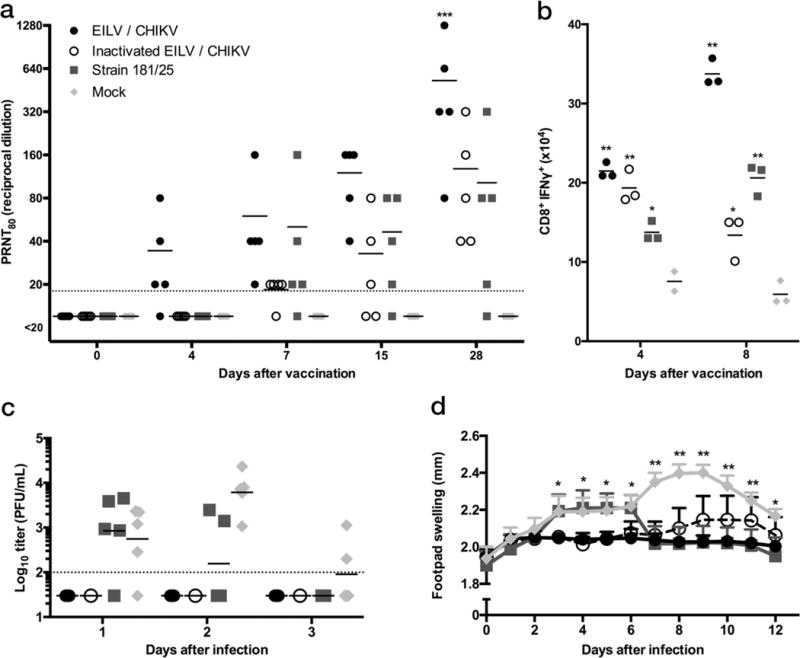
Single-dose of EILV/CHIKV is immunogenic and efficacious in a non-lethal, immunocompetent model of CHIKV infection. C57BL/6 mice (n=5/group) were vaccinated with EILV/CHIKV, formalin-inactivated EILV/CHIKV, strain 181/clone25, or mock vaccinated, and (a) 80% plaque-reduction neutralizing antibody titers (PRNT80) for serum collected 4, 8, 15, and 28 days post-vaccination (DPV) were determined. Dashed line indicates the lower limit of detection (1:20). (b) Splenocytes were harvested at 4 and 8 DPV (independent experiment, n=3/group/time point), stimulated with a peptide pool containing CHIKV murine CD8 T-cell epitopes (26), stained for CD3, CD8, and IFN-γ, and analyzed by flow cytometry (representative flow plots located in Supplementary Fig. 5). (c,d) All mice were challenged intradermally with 1×106 PFU of CHIKV-99659 in the rear footpad at 30 DPV and (c) viremia was determined at 1, 2, and 3 days post-challenge from 3 animals per group and (d) footpad height was measured for all 5 animals per group represented as mean + s.d.. (a–c) Individual data points as well as mean values reported. (Statistical analysis: two-way ANOVA with Tukey’s multiple comparison test, (a) EILV/CHIKV compared to all groups p < 0.0001, (b) compared to mock ** p< 0.0001, * p < 0.001, (d) compared to EILV/CHIKV or strain 181/clone25 * p < 0.05, ** p < 0.0001). Experiment performed twice.
Due to a lack of characterized CHIKV CD4 T-cell epitopes, a similar experiment was performed and splenocytes were harvested at 4, 8, and 28 DPV, stimulated with PMA and ionomycin, stained for CD3, CD4, CD8, and IFN-γ, and analyzed by flow cytometry (Supplementary Fig. 6). At 4 DPV, live EILV/CHIKV-vaccinated mice exhibited higher percentages of CD4+ (3.9% versus 1.6%) and CD8+ T cells (8.1% versus 4.7%) producing IFN-γ compared to sham- and inactivated EILV/CHIKV-vaccinated animals (p < 0.0001, Fig. 4b–c). Immune responses in live EILV/CHIKV-vaccinated mice remained stronger than observed in the other groups at later time points (8, 28 DPV). Formalin-inactivation of EILV/CHIKV delayed the activated phenotype for both CD4+ and CD8+ T-cells until 28 DPV, compared to live EILV/CHIKV (p < 0.0001). Strain 181/clone25-vaccinated mice demonstrated an enhancement of activated CD8+ T-cells, peaking at 8 DPV and CD4+ T-cells at 28 DPV (p < 0.001), compared to sham-vaccinated mice.
At day 30 post-vaccination, all mice were challenged with 6 log10 PFU of wt CHIKV strain 99659, a 2014 human isolate from the British Virgin Islands. All mice vaccinated with live or inactivated EILV/CHIKV were completely protected from viremia, while mock-vaccinated mice exhibited viremia with a mean peak titer of 2.7 log10 PFU/ml; 181/clone25-vaccinated mice had a mean viremia titer of 2.2 log10 PFU/ml at 2 days post-challenge (DPC). At 3 DPC, mock-vaccinated mice had a mean titer of 2 log10 PFU/ml whereas no viremia could be detected in any EILV/CHIKV- or 181/clone25-vaccinated mice (Fig. 4d). All mice vaccinated with live EILV/CHIKV were completely protected from footpad swelling, while the duration of swelling was shortened in mice vaccinated with 181/clone25. In contrast, inactivated EILV/CHIKV initially protected mice from swelling, but by 7 DPC a slight but insignificant increase in footpad thickness was observed (Fig. 4e).
Single-dose, long-term efficacy in IFNα/βR−/− mice
Long-term efficacy was evaluated in A129 IFNα/βR−/− mice because this model provides a lethal end-point upon CHIKV challenge. Six-week-old mice were vaccinated SC with a single 8.5 log10 PFU dose of live or inactivated EILV/CHIKV, or with 5.5 log10 PFU of strain 181/clone25, or mock-vaccinated with PBS. By day 28, live EILV/CHIKV induced PRNT80 titers between 1:160–1:1280, similar to those in C57BL/6 mice at 28 DPV, with a slight decline to 1:40–1:1280 at 289 DPV (Fig. 5a). In contrast, strain 181/clone25 induced >1:1280 PRNT80 titers at day 28, which were sustained through 289 DPV. In the C57BL/6 model, 181/clone25 induced PRNT80 titers of only <1:20–1:320 at 28 DPV, significantly lower (p<0.0001) than those following EILV/CHIKV vaccination (Fig 4a). In IFNα/βR−/− mice, inactivated EILV/CHIKV induced similar PRNT titers to those in C57BL/6 mice at 28 DPV and significantly lower (p<0.05) than those induced by EILV/CHIKV in both animal models, followed by a slow decline to <1:20–1:160 at 289 DPV (Fig. 5a). Following challenge at 292 DPV with 1,000 PFU of CHIKV-99659, live EILV/CHIKV- and 181/clone25-vaccinated mice were fully protected from weight loss, footpad swelling, viremia, and death, compared to mock-vaccinated mice (p<0.05), which lost on average 12% of their weight, exhibited a mean peak viremia of 6.2 log10 PFU at 4 DPC, and were moribund between 6–7 DPC (Fig. 5b–e). While inactivated EILV/CHIKV protected mice from death and viremia, they experienced a mean 8% weight loss and prolonged footpad swelling. Blinded histopathological analysis of pelvic limb sections showed that, upon challenge, mock-vaccinated animals exhibited full thickness subcutaneous edema of the distal digit with associated perivascular scant mixed inflammation (Supplementary Fig. 7). Inactivated EILV/CHIKV-vaccinated mice had a locally extensive neutrophilic cellulitis with scattered myocyte degeneration and regeneration. Neither 181/clone 25- nor live EILV/CHIKV-vaccinated mice had significant histopathological findings.
Figure 5.
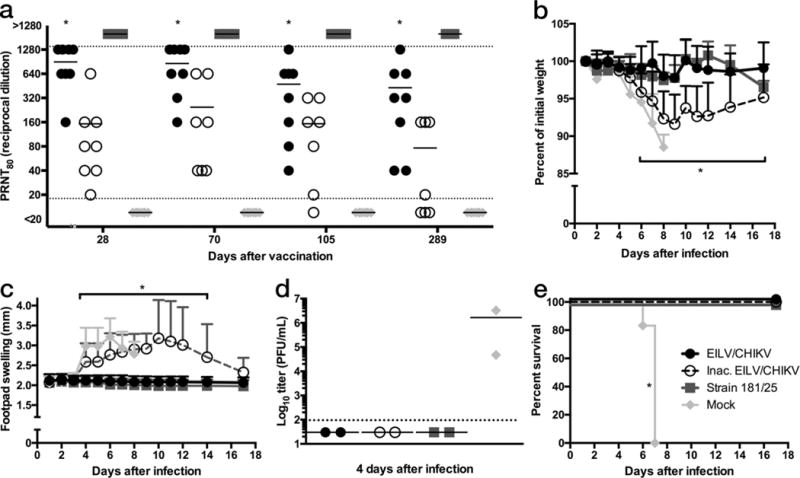
A single-dose of EILV/CHIKV is immunogenic and provides long-term protection in a lethal, immunocompromised model of CHIKV-infection. IFNα/βR−/− mice (n=8/group) were vaccinated with EILV/CHIKV, formalin-inactivated EILV/CHIKV, live-attenuated strain 181/clone25, or mock vaccinated and (a) 80% plaque-reduction neutralizing antibody titers (PRNT80) were determined at 28, 70, 105, and 289 days post vaccination (DPV) for all 8 animals and each data point and mean value was plotted. (b–e) Four days after challenge with 1×103 PFU of CHIKV-99650 ID in the rear footpad, three mice per group were sacrificed and blood and pelvic limbs were harvested for (d) viremia and histopathological analyses (Supplementary Fig. 7), respectively. The remaining five mice per group were utilized for analysis of (b) weight change, (c) footpad swelling, and (e) survival at the indicated time points with (b,c) represented as mean + s.d.. (d) Blood could not be collected from one mouse so the remaining two are plotted as individual data points as well as the mean value(Statistical analysis: two-way ANOVA with Tukey’s multiple comparison test, (a) EILV/CHIKV compared to all groups * p < 0.05, (b–c) formalin-inactivated EILV/CHIKV and mock * p < 0.05, Fisher’s exact test of final survival proportions, (e) compared to mock * p < 0.05). Experiment performed once.
Single-dose of EILV/CHIKV protects NHPs against CHIKV challenge
Because CHIKV infection of nonhuman primates (NHPs), especially cynomolgus macaques, closely mimics human CHIKF (27,28), we tested the efficacy of EILV/CHIKV as a single-dose vaccine in this model. Five male macaques 3–5 years of age and seronegative for anti-alphavirus immunity by hemagglutination inhibition (data not shown) were divided into two cohorts to achieve a similar weight distribution (3.0 – 5.8 kg), and each was implanted with a telemetric device to measure temperature at one-minute intervals throughout the study. Three animals were vaccinated intramuscularly in the right quadriceps with 8.1 log10 PFU of live EILV/CHIKV and 2 macaques were mock-vaccinated with uninfected C7/10 mosquito cell supernatant to assess potential hypersensitivity to mosquito cell proteins potentially present in vaccine preparations. Following the injections, no swelling or redness was detected at the injection sites and all body temperatures remained normal. Neutralizing antibody titers were determined on 4, 7, 14, and 28 DPV. By 4 DPV, all three vaccinated subjects seroconverted with PRNT80 titers of 1:20, after which antibody titers increased with the oldest animal peaking at 1:80 while the other 2 macaques developed titers of 1:320 and 1:640 at 28 DPV (Fig. 6a).
Figure 6.
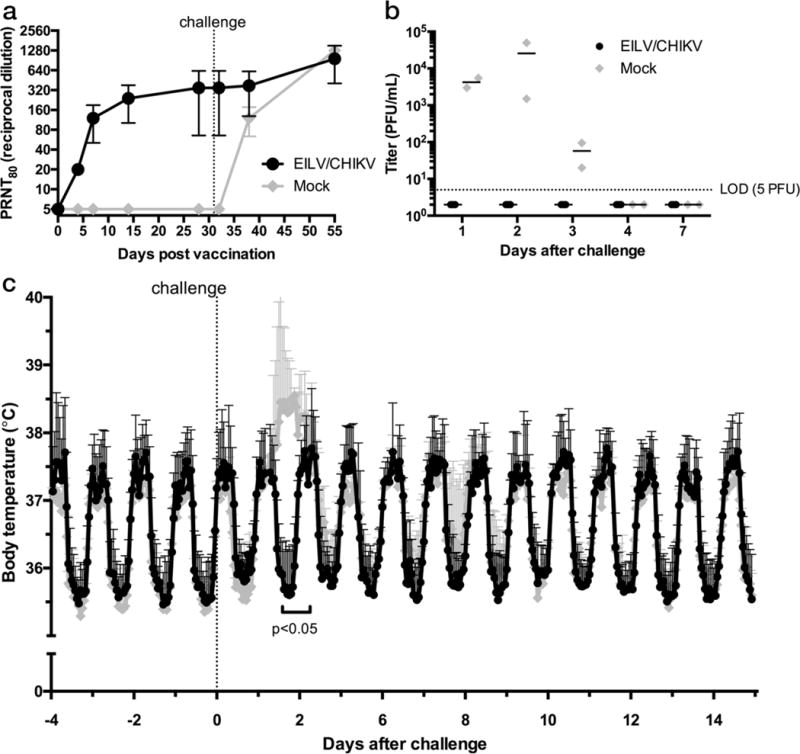
Nonhuman primates are protected against CHIKV-infection after a single-dose of EILV/CHIKV. Cynomolgus macaques, implanted with telemetrically-monitored temperature sensors, were vaccinated with 1.3×108 PFU of EILV/CHIKV (n=3) or mock-vaccinated with uninfected C7/10 cell supernatant (n=2) intramuscularly and (a) 80% plaque-reduction neutralizing antibody titers (PRNT80) were determined at 4, 7, 14, 28, 32, 38, and 51 DPV. Each data point represents mean + s.d. (b,c) All animals were challenged SC with 105 PFU of the virulent La Reunion strain of CHIKV at 30 DPV and (b) viremia titers were measured and reported as individual values as well as mean values, and (c) body temperature-measurements were recorded every minute and reported as hourly means + s.d. Experiment performed once. (Statistical analysis: (b) Multiple two-sided T-tests without assuming consistent s.d. * p < 0.05)
At 31 DPV, all animals were challenged SC with 5 log10 PFU of CHIKV strain La Réunion, shown previously to induce viremia, fever, and hypothermia in cynomolgus macaques (28), and monitored twice daily for signs of disease with continued telemetric monitoring. Approximately 1 DPC, both mock-vaccinated subjects experienced fever while the 3 vaccinated subjects continued to exhibit typical diurnal temperature fluctuations similar to baseline readings (Fig. 6c). Fevers in the mock-vaccinated subjects peaked at 2 DPC with a maximum mean temperature difference of 2.6°C compared to the 3 vaccinated subjects, coinciding with a peak mean viremia of 4.4 log10 PFU/ml in the mock-vaccinated animals (Fig. 6b). All 3 vaccinated subjects exhibited normal, baseline diurnal body temperatures throughout the study and viremia remained undetectable (limit of detection 5 PFU/ml; Fig. 6b,c). By 20 DPC, an anamnestic immune response was observed in EILV/CHIKV-vaccinated NHPs with a 2–4 fold increase in neutralizing titers (Fig. 6a).
To assess T-cell responses, frozen PBMCs from (7, 14, and 51 DPV), were thawed, stimulated with PMA and ionomycin (due to a lack of characterized CHIKV T-cell epitopes in NHPs), stained for CD3, CD4, CD8, and IFN-γ, and analyzed by flow cytometry (Supplementary Fig. 8). In contrast to the C57BL/6 mouse study, only a small, insignificant difference in the percentage of IFN-γ CD4 double-positive cells, compared to mock-vaccinated animals, was observed at 7 (0.83% versus 0.55%) and 14 DPV (0.73% versus 0.3 %), this response increased at 20 days after challenge to 2.4% compared to 2.2% in the mock-vaccinated animals (Supplementary Fig. 8b). Interestingly, while an enhancement of activated CD8 T-cells was not detected in the EILV/CHIKV-vaccinated NHPs (Supplementary Fig. 8c), the total number of CD8+ T-cells was elevated compared to mock-vaccinated NHPs (Supplementary Fig. 8d).
Finally, to test the ability of EILV/CHIKV-induced antibodies to cross-neutralize other CHIKV strains, sera from the 3 vaccinated NHPs collected 28 DPV were tested against 3 strains representing the Asian, West African, and Indian Ocean lineages, which span the maximum genetic and antigenic diversity within the CHIKV species (Table 1). All three CHIKV strains were neutralized, with PRNT80 titers reduced only 2–8-fold compared to homotypic titers (Supplementary Table 1).
Discussion
Live-attenuated vaccines typically offer rapid and durable immunity but can be reactogenic, as exemplified by 5 Phase II human volunteers who developed arthralgia after vaccination with CHIKV vaccine strain 181/clone25 (25). While the inability of inactivated vaccines to replicate offers higher safety, immunogenicity tends to be lower than that of live-attenuated formulations, often necessitating multiple doses and periodic boosters. Also, genetic instability of live RNA viruses can result in reversion to a pathogenic phenotype.
We overcame the tradeoffs of safety versus immunogenicity using the host-restricted EILV as a genetic backbone for CHIKV chimeras. The replication-defective nature of EILV/CHIKV in vertebrate cells, despite its ability to replicate to exceptionally high titers in insect cells, provides attractive safety and cost features. Our repeated inability to detect EILV/CHIKV genomes after the first murine brain passage indicated the absence of genome replication and lack of potential for the acquisition of replication-competence. Previously, utilizing a Sindbis virus (SINV) chimera containing the EILV 3′ untranslated genome region (UTR), we demonstrated that the EILV 3′ UTR is able to initiate minus strand transcription in vertebrate cells. However, following electroporation of the wt EILV genome into vertebrate cells, minus-strand replication could only be detected by RT-PCR if SINV nsPs were provided in trans (10). Here we demonstrated that the EILV nsP ORF is translated in vertebrate cells (Supplemental Fig. 3b); however, translation of the subgenomic RNA could not be detected (Fig. 3b), indicating that EILV nsPs are unable to mediate transcription needed for mutation and adaptation. Therefore, the EILV/CHIKV block in vertebrate cell replication appears to reflect incompatibilities between EILV nsPs and vertebrate host-factors.
In contrast to formalin-inactivated virus, EILV/CHIKV is exceptionally immunogenic. Using three different animal models, we demonstrated rapid, robust, and durable immunogenicity (Figs. 4a, b, 5a, 6a), following a single SC or IM dose. Compared to dose-matched, formalin-inactivated EILV/CHIKV, significant differences in murine neutralizing antibodies and activated CD4+ and CD8+ T-cells were detected as well as differences in the longevity of neutralizing responses (Figs. 4, 5, Supplementary Fig. 6). Structurally, EILV/CHIKV was identical to CHIKV (Fig. 1 & Supplementary Fig. 1), corroborating our previous findings of its antigenic authenticity (16). However, unlike wt EILV, EILV/CHIKV chimeras attach and enter vertebrate cells via clathrin-mediated endocytosis (Fig. 3) to deliver genomic RNA that is subsequently translated but nonfunctional for replication (Supplementary Fig. 3).
There is no well-supported mechanism to explain the reduced immunogenicity of formalin-inactivated versus live alphavirus vaccines (29–32). As the endocytic-pathway intersects with antigen presentation on major histocompatibility complex (MHC) I (33) and II molecules (34), the efficiency of antigen presentation via this pathway is improved up to 10,000-fold compared phagocytosis (35,36). Formalin-inactivation abolishes MHC I presentation of measles virus (37), entry of porcine reproductive and respiratory syndrome virus (38), binding, entry, and viral RNA infectivity of polio virus (39), and modifies the antigenic structures of Japanese encephalitis virus (40).
We demonstrated that EILV/CHIKV activates the endocytic pathway like vertebrate-infectious alphaviruses (22), while inactivation significantly reduces binding to vertebrate cells (Fig. 3d), presumably by interfering with receptor-mediated entry. We also demonstrated that the EILV/CHIKV genome is translated upon delivery into vertebrate cells (Supplementary Fig. 3), but the role of this delivery and/or translation in enhancing immunogenicity requires further investigation. The improved infectivity of antigen presenting cells such as dendritic cells by alphaviruses derived from mosquito cells such as EILV/CHIKV compared to mammalian cells (41), due to mannose-enriched glycosylation, suggests an additional immunogenicity mechanism.
Finally, we showed that a single dose of EILV/CHIKV protected two mouse models from all measures of disease up to 292 DPV. Formalin-inactivated EILV/CHIKV, despite inducing measurable immune responses and protecting IFNα/βR−/− mice from a fatal outcome, reduced but did not eliminate disease in this model. For a NHP model, we utilized cynomolgus macaques, which develop human-like CHIKF (27) and have proven valuable to assess CHIKF vaccines (28,30,42). In contrast to replication-deficient vaccines that require multiple doses to protect NHPs (42), EILV/CHIKV provided complete protection after a single dose in our pilot study. Rapid, single-dose immunogenicity, coupled with a high degree of safety, is desirable to vaccine travelers to CHIKF-endemic countries as well as to control explosive outbreaks when prompt immune responses can halt transmission.
In summary, we introduce EILV as an affordable and efficient alphavirus vaccine platform. Due to their host-restricted nature, EILV-based chimeras do not require inactivation, maximally preserving native antigens to permit receptor-mediated endocytosis and improved antigen presentation. Due to their fundamental host restriction, EILV chimeras combine the safety of inactivated vaccines with the immunogenicity and ease-of-production of live-attenuated vaccines.
We have also generated EILV chimeras for eastern and Venezuelan equine encephalitis viruses, and both replicate efficiently in mosquito cells. Further work is needed to determine if this approach can be extended to other families of viruses. Over 28 insect-specific flaviviruses have been characterized (43) and their application in vaccine development for flavivirus pathogens should be explored, especially considering the recent expansion of Zika virus and congenital Zika syndrome (44).
Online Methods
Cell cultures
C7/10 cells (American Type Culture Collection, Rockville, MD), derived from A. albopictus mosquitoes, were maintained at 29°C with 5% CO2 in Dulbecco’s minimal essential medium (DMEM) containing 10% (V/V) heat-inactivated fetal bovine serum (FBS), sodium pyruvate (1 mM), penicillin (100 U/mL), streptomycin (100 μg/mL), and 1% (v/v) tryptose phosphate broth (Sigma, St. Louis, MO). Vero, NIH 3T3, and 293T cells (ATCC, Manassas, VA) were propagated at 37°C with 5% CO2 in DMEM containing 10% (V/V) heat-inactivated FBS, penicillin (100U/mL), and sreptomycin (100 μg/mL). Our lab utilizes C7/10 and Vero cells for virus isolations and samples are routinely analyzed by deep sequencing. By this method, no contaminating viral sequences were detected. Vero cells were also tested and found to be negative for mycoplasma contamination.
Plasmid constructs
The original plasmid encoding the cDNA of EILV (pEILV) was previously described (7,10). EILV/CHIKV (Fig. 1a) was generated by replacing the structural protein open reading frame (str ORF) with that of CHIKV-99659, a human isolate from the British Virgin Islands (17). CHIKV-99659 was isolated on Vero cells, passaged twice on C7/10 cells and cDNA was generated from trizol-extracted total RNA. Two overlapping fusion RT-PCR fragments containing the EILV subgenomic promoter from an XbaI site to Bsu36I in CHIKV str ORF and from NcoI in CHIKV str ORF to NotI in the EILV 3′ UTR were joined with a third RT-PCR fragment from Bsu36I to NcoI in CHIKV str ORF by restriction digest and ligation reactions and sub-cloned into a shuttle vector. Finally, the assembled fragment between XbaI and NotI sites was cloned back into pEILV by restriction digest and ligation reactions, to complete the EILV/CHIKV construct. Additional constructs are described in the figure legends. All full-length cDNA clones were Sanger-sequenced using ABI PRISM Big Dye (Applied Biosystems, Foster City, CA).
Recombinant virus rescue
Recombinant viruses were rescued as previously described (7). Briefly, plasmids encoding the full-length viral genomes were purified by CsCl-gradient ultracentrifugation, linearized by digestion with NotI, and purified by phenol-chloroform extraction. RNA was then transcribed and capped using an mMessage mMachine SP6 kit (Ambion, Waltham, MA), analyzed by agarose gel-electrophoresis, and electroporated into C7/10 cells without further purification. After 24 hours incubation at 29°C, cell supernatant was replaced with fresh complete medium. At 48 hours after electroporation, supernatant was harvested, clarified by centrifugation, and stored at −80°C.
Virus replication, concentration, and purification
EILV/CHIKV was amplified by infection of C7/10 cells at a multiplicity (MOI) of 0.1 PFU/cell and at 48 hours post-infection (HPI), supernatants were harvested and clarified by centrifugation for 10 min at 3,000×g. To concentrate virus for vaccine studies, clarified supernatants were mixed with polyethylene glycol (PEG) 8000 and NaCl to 7% and 2.3% (wt/vol), respectively, incubated overnight at 4°C, and the precipitate was pelleted by centrifugation for 30 min at 3,100×g. Pellets were then resuspended in PBS and titrated by C7/10 plaque assay as described below. To further purify PEG-precipitated virus for virus-cell binding assays and cryo-electron microscopic analysis, resuspended virus was overlaid onto a 20–70% continuous sucrose gradient and separated from contaminants by ultracentrifugation for 1.5 hrs at 210,000xg. The virus band was collected and applied to a 100-kDa Amicon filter (Millipore, Billerica, MA). Sucrose was then removed by 5 washes with PBS.
To prepare dose-matched, formalin-inactivated EILV/CHIKV, PEG-precipitated EILV/CHIKV was split in half. One preparation was formalin-inactivated as previously described (29), while the other was mock-inactivated. Briefly, virus was passed through a membrane filter to remove debris and warmed to 37°C. Virus was inactivated by the addition of 10% formaldehyde solution (Sigma, St. Louis, MO) with stirring to a final concentration of 0.1% (v/v), or mock-inactivated by the addition of an equivalent volume of PBS. Virus suspensions were incubated with mixing for 3 days at 37°C followed by 4 days at 4°C with vigorous mixing twice daily. Formaldehyde was neutralized by the addition of 3.5% (w/v) sodium metabisulfite (Sigma, St. Louis, MO) to a final concentration of 0.035% (w/v). Inactivation was confirmed by cytopathic effect (CPE) assay on C7/10 cells.
Virus and neutralizing-antibody titers
Plaque assays on mosquito cells were performed as previously described (7). Eighty percent plaque-reduction neutralization tests (PRNT80) were performed on Vero cells as previously described (45) using strain 181/clone25 as the control virus. NHP sera were further tested against additional strains of CHIKV as noted in the figure legends. To quantify EILV/CHIKV genome copies in mouse brain tissue, viral RNA was extracted from brain homogenate using Qiagen viral RNA mini kit according to manufacturer’s instructions (Qiagen, Venlo, Netherlands). RNA was then reverse-transcribed and cDNA was subsequently amplified and quantified by qRT-PCR using TaqMan RNA-to-CT 1-step kit (Life Technologies, Carlsbad, CA) with forward and reverse primers: TGGAGCTTCTGTCTGTCACC, and ACGTACGGAGACGGGATAAC, respectively, and probe: 56-FAM-TCGCTTGATTAATCACGTGCGAG-3BHQ_1, according to manufacturer’s instructions, using the QuantStudio 6 Flex Real-Time PCR system (Thermo Fisher Scientific, Waltham, MA). An RNA standard, used to generate an absolute standard curve, was generated by transcribing RNA from the EILV/CHIKV plasmid using SP6 polymerase as described above. Transcribed RNA was treated with DNase to remove DNA template, purified by phenol-chloroform extraction, and quantified by measurement of UV absorption at 260nm using the DeNovix DS-11 spectrophotometer (DeNovix Inc, Wilmington, DE).
Cryo-electron microscopy and reconstruction
EILV/CHIKV virions in Tris/EDTA/NaCl (TEN) buffer (0.05 M Tris-HCl [pH 7.4], 0.1 M NaCl, 0.001 M EDTA) were diluted with an equal volume of TEN buffer to reduce particle concentration. Three μL of the diluted virion solution was applied to a Quantifoil grid (mesh size 200) and blotted with an FEI Vitrobot Mark IV for 2 s at 100% relative humidity and room temperature before plunge freezing. Preliminary imaging on a JEM-2010F cryomicroscope was used for initial model generation in EMAN2 (46). Final imaging was performed in a JEM-3200FSC cryomicroscope with Omega-type energy filter. 348 micrographs were recorded at 1.96Å/pix on a DE-20 direct electron detector. 60 electrons per Å2 were captured. Damage compensation and motion correction were performed as previously described (47).
The dataset was split in half and kept independent through all processing until the generation of the final map, except that handedness was corrected manually by reference to published alphavirus structures (48). Reconstruction was performed by multi-pathway simulated annealing (49). Particles were excluded if orientation solutions from multiple simulated annealing initializations did not agree to within 2°. Of the 21,024 particles boxed, 13,455 were retained by this criterion in the final reconstruction. A radial structure factor was applied and the map was low-pass filtered to 8Å.
Thin-section electron microscopy
Vero cell monolayers were prepared and infected with EILV/CHIKV or formalin-inactivated EILV/CHIKV for 1hr at 4°C. Following cold-binding to cells at a MOI of 1000 PFU/cell and fixation for 0 or 15 seconds, or 5 minutes after incubation at 37°C, the cells were processed for thin-section EM as previously described (22). Infected Vero (3 time points including 0, 15 secs, and 5 mins) cell monolayers were fixed in 2.5% formaldehyde/0.1% glutaraldehyde in 0.05 M cacodylate buffer pH 7.3 containing 0.03% trinitrophenol and 0.03% CaCl2 for at least 1 hour at room temperature. Monolayers were then washed in 0.1 M cacodylate buffer, scraped and pelleted by centrifugation. Pellets were post-fixed in 1% OsO4, 0.1 M cacodylate buffer, pH 7.3, washed with distilled water, en bloc stained with 2% aqueous uranyl acetate for 20 minutes at 60°C, then dehydrated in graded series of ethanol and embedded in Poly/Bed 812 (Polysciences, Warrington, PA). Ultrathin sections were cut on Leica EM UC7 ultramicrotome (Leica Microsystems, Buffalo grove, IL), stained with lead citrate and examined on a Philips (Eindhoven, Netherlands) 201 electron microscope at 60 KV.
Virus-fluorophore conjugation and binding assay
Viruses were conjugated to Alexa Fluor 488 (AF488) succinimidyl ester (Thermo Fisher Scientific, Waltham, MA) as previously described (50). Viruses were diluted to 1×109PFU/ml (as determined by mosquito cell plaque assay) and AF488 in DMSO was added to a final concentration of 100 uM and suspensions were incubated for 1 hr at room temp with occasional mixing. Unconjugated dye was then removed by desalting-column chromatography (GE Healthcare Bio-Sciences, Pittsburgh, PA) according to manufacturer’s instructions and conjugated virus was stored at −80°C before the binding assay was performed.
Quantification of virus-bound cells was performed as previously described with modifications (51). Briefly, NIH 3T3 cells were detached non-enzymatically using CellStripper (Corning), pelleted, and washed once with binding buffer (50 mM HEPES, 100 mM NaCl, 1 mg/ml BSA, pH 7.4), and 2×105 cells per sample were equilibrated to 4°C. Cells were then pelleted and groups of triplicate samples were resuspended in binding buffer containing EILV/CHIKV, formalin-inactivated EILV/CHIKV, or strain 181/clone25 at a MOI of 100 PFU/cell followed by shaking incubation at 4°C for 90 min. Unbound virus was then washed from pelleted cells with 1ml of binding buffer and virus-bound cells were pelleted and resuspended in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) followed by incubation at 4°C for 30 min. Samples were then washed with 1ml binding buffer, pelleted, and resuspended in 300 μl binding buffer for analysis by flow cytometry using a C6 Flow Cytometer (Accuri cytometers, Ann Arbor, MI)
Fluorescence microscopy
Vero or C7/10 cells were seeded onto glass coverslips in 6-well plates (Corning, Tewksbury, MA) and infected with EILV/nsP3-GFP/CHIKV at a MOI of 100, followed by incubation at 29°C. Eight hours after infection, cells were washed and fixed in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA). Cells were then washed and permeabilized in PBS (0.5% Triton X-100) and stained with phalloidin DyLight 550 conjugate and DAPI (ThermoFisher Scientific, Waltham, MA). Coverslips were then washed and mounted onto glass slides with MOWIOL (Sigma, St. Louis, MO) and imaged on an Olympus BX61 microscope (Olympus Life Sciences, Waltham, MA).
Luciferase assay
C7/10 or 293T cell monolayers were cooled to 4°C and infected in triplicate in 6-well costar plates at a MOI of 20 with EILV/nLUC/CHIKV. After 1hr incubation at 4°C, monolayers were washed once with cold PBS and C7/10 cells or 293T cells were incubated in complete media at 29°C or 37°C, respectively. At 0, 1, 2, 4, and 8 hours post-infection, cells were harvested, pelleted, resuspended in PBS, and frozen at −80°C until all samples were collected. Luciferase activity was determined by Nano-Glo Luciferase Assay (Promega, Madison, WI) according to manufacturer’s instructions. Samples were thawed and equal volume of luciferase substrate-containing buffer was added to the cells and mixed. Luminescence was then measured on a SpectraMax luminometer (Molecular Devices, Sunnyvale, CA).
Flow cytometry
Larger volumes of murine blood were collected at 7 and 14 DPV and peripheral blood mononuclear cells (PBMCs) were isolated and frozen at −80°C until all samples could be processed at the same time for flow cytometry. To measure intracellular IFN-γ production in murine CD8 T-cells in response to CHIKV antigen, splenocytes were isolated and stimulated for 6 hours with previously described pool of peptides (26) corresponding to CD8 T-cell epitopes located in E1 (HSMTNAVTI), E2 (IILYYYELY), and C (ACLVGDKVM) (10 ug/ml/peptide, Sigma, St. Louis, MO), in a Golgi-plug (BD Biosciences, San Jose, CA) containing medium. To measure non-specific intracellular IFN-γ production in CD4 and CD8 murine or NHP T-cells, splenocytes or PBMCs were isolated from mice or NHPs, respectively, and stimulated with 50 ng/ml PMA (Sigma, St. Louis, MO) and 500 ng/ml ionomycin (Sigma, St. Louis, MO) for 4 h at 37°C in a Golgi-plug containing medium. Following peptide or mitogen stimulation, cells were harvested, stained with antibodies for CD3, CD4, or CD8, fixed and permeabilized with BD Cytofix/Cytoperm (BD Biosciences, San Jose, CA) before adding PE-conjugated anti-IFN-γ, or control PE-conjugated rat IgG1 (eBiosciences, San Diego, CA) for murine splenocytes, or PE-conjugated mouse IgG1 for NHP PBMCs (BD Biosciences, San Jose, CA). Cells were then washed and analyzed by using a C6 Flow Cytometer (Accuri cytometers, Ann Arbor, MI).
Animal studies
All animal studies were approved by the UTMB Institutional Animal Care and Use Committee. The facility where animal studies were conducted is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International and follows guidelines set forth by the Guide for the Care and Use of Laboratory Animals, National Research Council, 2011. All sample sizes were determined based on power analysis assuming LD100 challenge doses and subsequent survival rates of 0.99 versus 0.01 with α=0.05 using a one-sided Fisher’s exact test. Mice were non-specifically and blindly distributed into their respective groups but sample collection and analyses were not blinded except for the histological tissue sections which were blindly analyzed. No exclusion criteria were established prior to beginning the studies but we failed to collect blood from one mouse during an experiment as described in the figure legends. Cynomolgus macaques were ranked by weight and alternately distributed into two groups to achieve an approximately similar distribution of weights between groups and sample collection and analyses were blinded. No data was excluded from analyses.
Mouse safety studies
Seven-day old male and female A129 IFN α/β receptor knockout mice (n=6/group), obtained from colonies maintained under specific pathogen-free conditions at University of Texas Medical Branch (UTMB; Galveston, TX), were infected intracranially (IC) with 8.7 log10 PFU of EILV/CHIKV, 4 log10 PFU of strain 181/clone25, or PEG-precipitated C7/10 cell-supernatant as a negative control in a 10 μl volume and monitored daily for weight change and survival.
To assess the potential of EILV/CHIKV to acquire a replication-competent phenotype in vivo, we infected three 2- to 4-day old male and female IFNα/βR−/− mice IC with 8.8 log10 PFU of EILV/CHIKV or 1 mouse with PBS in a 10 μl volume and sacrificed all 4 mice on day 2 post-infection. We then harvested and homogenized the brain in a 500 μl volume of PBS and re-infected a new cohort of 2- to 3-day old IFNα/βR−/− mice. This was repeated for 5 passages. Upon infection of the 5th cohort, mice were monitored for weight-change and survival for 21 days.
Mouse immunogenicity and efficacy studies
To assess immunogenicity and efficacy in an immunocompetent mouse model of CHIKV-infection, five 4-week old female C57BL/6 mice (Charles River, Wilmington, MA) per group were vaccinated subcutaneously (SC) with 8.8 log10 PFU of EILV/CHIKV, or with dose-matched formalin-inactivated EILV/CHKV, or with 5.5 log10 PFU of live-attenuated CHIKV strain 181/clone25, or with PBS in a 100 μl volume. Mice were bled at 4, 7, 15, and 28 DPV to assess neutralizing antibody titers and, in a separate experiment, 3 mice per group were sacrificed at 4, 8, and 28 DPV so spleen could be harvested for analysis of T-cell activation. Mice were then challenged intradermally (ID) in the rear footpad on day 30 with 6 log10 PFU of CHIKV-99659 in a 10 μl volume, monitored daily for signs of illness, weight loss, and footpad swelling, and were bled at 1, 2, and 3 DPC to assess viremia. Animals were euthanized on day 12 post-challenge.
Long-term immunogenicity and efficacy was assessed in an immunocompromised mouse model of CHIKV-infection. IFNα/βR−/− mice were bred at the University of Texas Medical Branch (UTMB). Eight male or female IFNα/βR−/− mice per group, 6-weeks old, were vaccinated SC with 8.5 log10 PFU of EILV/CHIKV, or with dose-matched formalin-inactivated EILV/CHIKV, or with 5.5 log10 PFU of strain 181/clone25, or with PBS in a 100 μl volume. Mice were bled at 28, 70, 105, and 289 DPV to assess neutralizing antibody titers, and challenged ID in the rear footpad on day 292 with 3 log10 PFU of CHIKV-99659 in a 10 μl volume. Animals were monitored daily for signs of disease, weight loss, footpad swelling, and were euthanized by CO2 exposure if they became moribund. Three mice from each group were sacrificed on day 5 post-challenge, blood was collected for viremia titers, and rear-legs were harvested for histology. Seventeen days post challenge, surviving mice were euthanized.
Tissues were fixed in 10% neutral buffered formalin (RICCA Chemical Company, Arlington, TX) and bone was decalcified overnight in fixative/decalcifier (VWR International, Radnar, PA) and processed for histology as previously described (52).
Nonhuman primate immunogenicity and efficacy studies
Five colony-bred, male Chinese-origin cynomolgus macaques (Macaca fascicularis), 3–5 years old and weighing 3–6 kg were obtained from Shin Nippon Biomedical Laboratories (Alice, TX). All subjects tested negative for infection with simian immunodeficiency virus, simian type D retrovirus, simian T-lymphotropic virus, and fecal swabs were negative for shigella, and campylobacter. Serum was also screened by hemagglutination inhibition and found to be negative for antibodies against CHIKV, Semliki Forest, and Sindbis viruses (data not shown). All blood collections, vaccinations, and infections were performed under anesthesia by intramuscular (IM) injection of ketamine.
One-week prior to the start of the study, subjects were bled via the femoral vein to obtain baseline serology, and TA-D70 telemetry devices (Data Sciences International, St. Paul, MN) that record body temperature and activity, were surgically implanted inter-muscularly in the dorsum of all 5 subjects. Hair was clipped along the dorsum and the surgical site was prepared for aseptic placement of the telemetry device. A 2–4 cm incision was made in the skin and an inter-muscular pocket was made for the telemetry device. The device was anchored to the underlying tissue with suture and the subcutaneous tissue and skin were closed with suture. Analgesics were provided during and after the surgical procedure along with a topical antibiotic. Animals were housed individually in cages specifically designed to be mounted with receivers matched to each TA-D70 device and configured to transmit the signals to a data acquisition base station running Ponemah. Primary (temperature) and secondary (activity) sample rates were set to 20 and 10 samples/second, respectively, and data collection was started on the day following surgery and was collected every minute, 24 hours per day and moving averages of temperature and activity were calculated in 60 minute intervals and reported as an hourly average.
On the day of vaccination, animals were anesthetized and their right thighs were shaved. EILV/CHIKV, 8.1 log10 PFU (n=3), or C7/10 cell supernatant from mock-infected cells (n=2) was administered IM into the right quadriceps of each animal in a 1ml volume and the injection site was monitored for any adverse reactions. Animals were then monitored twice daily for any clinical signs of disease until challenge. On days 4, 7, 14, and 28 post-vaccination, animals were anesthetized and blood was collected in EDTA, heparin, or serum Vacutainers (Beckton Dickinson, Franklin Lakes, NJ). Sera were collected and anamnestic neutralizing antibody responses were determined at 1, 7, and 20 DPC, while viremia titers were determined at 1–4 and 7 DPC. PBMCs from 20 DPC were also isolated and frozen at −80°C.
Thirty-one days after vaccination, all subjects were challenged SC with 5 log10 PFU of CHIKV-LR in a 100 μl volume and monitored three times daily for clinical signs of disease and bled on days 1, 2, 3, 4, 7 post-challenge to assess viremia, and antibody titers. At the end of the study, 20 days after challenge, all subjects were bled for final measurements and transferred to another protocol.
For T-cell assays, approximately 10 ml of blood was collected at 7, 14, and 51 DPV and PBMCs were isolated by density gradient centrifugation in 90% ficol-hypaque (Sigma), counted, and frozen at −80°C in freezing media (90% FBS, 10% DMSO). Prior to flow cytometry assays, PBMCs were thawed and re-counted to assess viability. Approximately 50% of cells from 7 and 14 DPV did not survive the freeze-thaw while only ~10% loss was noted for the 51 DPV samples.
Statistics
Statistical analysis was performed using GraphPad Prism software (version 6.0h) and RStudio (version 0.99.491). Data distribution and variance was evaluated for normality and similarity, respectively, by qqplot and boxplot analyses in RStudio, as sample sizes were too small for traditional normality tests. One-way and two-way ANOVAs with Tukey’s multiple comparison tests as well as multiple T- and Fisher exact tests were used as specified in the figure legends.
Data availability
Cryo-em micrographs and reconstructions are deposited at emdatabank.org: accession number EMD-3406.
Supplementary Material
Acknowledgments
We thank C. Klages and W. Lawrence for their technical assistance with the nonhuman primate study, V. Popov and F. Murphy for providing electron microscopy training and discussion, N. Dobias and the UTMB research histology core for assistance with tissue preparations, O. Escaffre for confocal microscopy assistance, and C. Mire for critically reading the manuscript
Funding: Support for this work came from NIH grants AI120942 (to SCW), Welch Foundation grant (to WC), and a grant from the UTMB Technology Commercialization Program. JHE is supported by a UTMB McLaughlin Fellowship. Electron microscopy was supported by NIH grant P41GM103832 (to WC).
Footnotes
Author contributions: JHE, FN, IF, TW, WC, and SCW conceived the project and designed experiments; JHE, FN, DYK, HL, AJA, SLR, GL, and JTK performed experiments; JHE, AJA, KF, FN, IF, TW, WC, JTK and SCW did data analysis; JHE and SCW wrote the paper.
Competing interests: SCW, FN, and JHE have a patent (US 9,422,529) entitled “Alphavirus Compositions and Methods of Use”, which includes technology reported in this manuscript. The views expressed in this article are those of the authors and do not reflect the official policy or position of the U.S. Department of Defense, or the Department of the Army.
References
- 1.Offit PA. The cutter incident: How america’s first polio vaccine led to the growing vaccine crisis. Yale University Press; 2005. [Google Scholar]
- 2.Whittembury A, Ramirez G, Hernandez H, Ropero AM, Waterman S, Ticona M, et al. Viscerotropic disease following yellow fever vaccination in Peru. Vaccine. 2009;27(43):5974–81. doi: 10.1016/j.vaccine.2009.07.082. [DOI] [PubMed] [Google Scholar]
- 3.Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. Rapid evolution of RNA genomes. Science. 1982 Mar 26;215(4540):1577–85. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 4.Weaver SC, Lecuit M. Chikungunya virus and the global spread of a mosquito-borne disease. N Engl J Med. 2015 Mar 26;372(13):1231–9. doi: 10.1056/NEJMra1406035. [DOI] [PubMed] [Google Scholar]
- 5.Weaver SC, Costa F, Garcia-Blanco MA, Ko AI, Ribeiro GS, Saade G, et al. Zika virus: History, emergence, biology, and prospects for control. Antiviral Res. 2016 Mar 18;130:69–80. doi: 10.1016/j.antiviral.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li CX, Shi M, Tian JH, Lin XD, Kang YJ, Chen LJ, et al. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife. 2015;4 doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nasar F, Palacios G, Gorchakov RV, Guzman H, Da Rosa AP, Savji N, et al. Eilat virus, a unique alphavirus with host range restricted to insects by RNA replication. Proc Natl Acad Sci U S A. 2012 Sep 4;109(36):14622–7. doi: 10.1073/pnas.1204787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strauss JH, Strauss EG. The alphaviruses: Gene expression, replication, and evolution. Microbiol Rev. 1994 Sep;58(3):491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powers AM, Brault AC, Shirako Y, Strauss EG, Kang W, Strauss JH, Weaver SC. Evolutionary relationships and systematics of the alphaviruses. J Virol. 2001 Nov;75(21):10118–31. doi: 10.1128/JVI.75.21.10118-10131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nasar F, Gorchakov RV, Tesh RB, Weaver SC. Eilat virus host range restriction is present at multiple levels of virus life cycle. J Virol. 2014 Nov 12; doi: 10.1128/JVI.01856-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhn RJ, Griffin DE, Owen KE, Niesters HG, Strauss JH. Chimeric sindbis-ross river viruses to study interactions between alphavirus nonstructural and structural regions. J Virol. 1996 Nov;70(11):7900–9. doi: 10.1128/jvi.70.11.7900-7909.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paessler S, Fayzulin RZ, Anishchenko M, Greene IP, Weaver SC, Frolov I. Recombinant sindbis/venezuelan equine encephalitis virus is highly attenuated and immunogenic. J Virol. 2003 Sep;77(17):9278–86. doi: 10.1128/JVI.77.17.9278-9286.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lumsden WH. An epidemic of virus disease in southern province, tanganyika territory, in 1952–53: II. General description and epidemiology. Trans R Soc Trop Med Hyg. 1955 Jan;49(1):33–57. doi: 10.1016/0035-9203(55)90081-x. [DOI] [PubMed] [Google Scholar]
- 14.Soumahoro MK, Boelle PY, Gaüzere BA, Atsou K, Pelat C, Lambert B, et al. The chikungunya epidemic on la réunion island in 2005–2006: A cost-of-illness study. PLoS Negl Trop Dis. 2011 Jun;5(6):e1197. doi: 10.1371/journal.pntd.0001197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schilte C, Staikovsky F, Couderc T, Madec Y, Carpentier F, Kassab S, Michault A. Chikungunya Virus-associated Long-term Arthralgia: A 36-month Prospective Longitudinal Study. PLoS Neglected Tropical Diseases. 2013;7(3) doi: 10.1371/journal.pntd.0002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erasmus JH, Needham J, Raychaudhuri S, Diamond MS, Beasley DW, Morkowski S, et al. Utilization of an eilat virus-based chimera for serological detection of chikungunya infection. PLoS Negl Trop Dis. 2015 Oct;9(10):e0004119. doi: 10.1371/journal.pntd.0004119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanciotti Robert S, Valadere Anne Marie. Transcontinental Movement of Asian Genotype Chikungunya Virus. Emerging infectious diseases. 2014;20(8) doi: 10.3201/eid2008.14026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun S, Xiang Y, Akahata W, Holdaway H, Pal P, Zhang X, et al. Structural analyses at pseudo atomic resolution of chikungunya virus and antibodies show mechanisms of neutralization. Elife. 2013;2:e00435. doi: 10.7554/eLife.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levitt NH, Ramsburg HH, Hasty SE, Repik PM, Cole FE, Lupton HW. Development of an attenuated strain of chikungunya virus for use in vaccine production. Vaccine. 1986 Sep;4(3):157–62. doi: 10.1016/0264-410x(86)90003-4. [DOI] [PubMed] [Google Scholar]
- 20.Gardner CL, Hritz J, Sun C, Vanlandingham DL, Song TY, Ghedin E, et al. Deliberate attenuation of chikungunya virus by adaptation to heparan sulfate-dependent infectivity: A model for rational arboviral vaccine design. PLoS Negl Trop Dis. 2014 Feb;8(2):e2719. doi: 10.1371/journal.pntd.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorchakov R, Wang E, Leal G, Forrester NL, Plante K, Rossi SL, et al. Attenuation of chikungunya virus vaccine strain 181/clone 25 is determined by two amino acid substitutions in the E2 envelope glycoprotein. J Virol. 2012 Jun;86(11):6084–96. doi: 10.1128/JVI.06449-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helenius A, Kartenbeck J, Simons K, Fries E. On the entry of semliki forest virus into BHK-21 cells. J Cell Biol. 1980 Feb;84(2):404–20. doi: 10.1083/jcb.84.2.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eyden B. Organelles in tumor diagnosis: An ultrastructural atlas. Igaku-Shoin Medical Publishers. 1996 [Google Scholar]
- 24.Frolova E, Gorchakov R, Garmashova N, Atasheva S, Vergara LA, Frolov I. Formation of nsp3-specific protein complexes during sindbis virus replication. J Virol. 2006 Apr;80(8):4122–34. doi: 10.1128/JVI.80.8.4122-4134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edelman R, Tacket CO, Wasserman SS, Bodison SA, Perry JG, Mangiafico JA. Phase II safety and immunogenicity study of live chikungunya virus vaccine TSI-GSD-218. Am J Trop Med Hyg. 2000 Jun;62(6):681–5. doi: 10.4269/ajtmh.2000.62.681. [DOI] [PubMed] [Google Scholar]
- 26.Muthumani Karuppiah, Lankaraman Katthikbabu M, Laddy Dominick J, Sundaram Senthil G, Chung Christopher W, Sako Eric, Wu Ling, et al. Immunogenicity of Novel Consensus-based DNA Vaccines Against Chikungunya Virus. Vaccine. 2008;26(40) doi: 10.1016/j.vaccine.2008.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labadie K, Larcher T, Joubert C, Mannioui A, Delache B, Brochard P, et al. Chikungunya disease in nonhuman primates involves long-term viral persistence in macrophages. J Clin Invest. 2010 Mar;120(3):894–906. doi: 10.1172/JCI40104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy CJ, Adams AP, Wang E, Plante K, Gorchakov R, Seymour RL, et al. Chikungunya vaccine candidate is highly attenuated and protects nonhuman primates against telemetrically monitored disease following a single dose. J Infect Dis. 2014 Jun 15;209(12):1891–9. doi: 10.1093/infdis/jiu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White A, Berman S, Lowenthal JP. Comparative immunogenicities of chikungunya vaccines propagated in monkey kidney monolayers and chick embryo suspension cultures. Appl Microbiol. 1972 May;23(5):951–2. doi: 10.1128/am.23.5.951-952.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakao E, Hotta S. Immunogenicity of purified, inactivated chikungunya virus in monkeys. Bull World Health Organ. 1973 May;48(5):559–62. [PMC free article] [PubMed] [Google Scholar]
- 31.Tiwari M, Parida M, Santhosh SR, Khan M, Dash PK, Rao PV. Assessment of immunogenic potential of vero adapted formalin inactivated vaccine derived from novel ECSA genotype of chikungunya virus. Vaccine. 2009 Apr 21;27(18):2513–22. doi: 10.1016/j.vaccine.2009.02.062. [DOI] [PubMed] [Google Scholar]
- 32.Kumar M, Sudeep AB, Arankalle VA. Evaluation of recombinant E2 protein-based and whole-virus inactivated candidate vaccines against chikungunya virus. Vaccine. 2012 Sep 21;30(43):6142–9. doi: 10.1016/j.vaccine.2012.07.072. [DOI] [PubMed] [Google Scholar]
- 33.Burgdorf S, Lukacs-Kornek V, Kurts C. The mannose receptor mediates uptake of soluble but not of cell-associated antigen for cross-presentation. J Immunol. 2006 Jun 1;176(11):6770–6. doi: 10.4049/jimmunol.176.11.6770. [DOI] [PubMed] [Google Scholar]
- 34.Mellman I, Steinman RM. Dendritic cells: Specialized and regulated antigen processing machines. Cell. 2001 Aug 10;106(3):255–8. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 35.Engering AJ, Cella M, Fluitsma DM, Hoefsmit EC, Lanzavecchia A, Pieters J. Mannose receptor mediated antigen uptake and presentation in human dendritic cells. Adv Exp Med Biol. 1997;417:183–7. doi: 10.1007/978-1-4757-9966-8_31. [DOI] [PubMed] [Google Scholar]
- 36.Arnold-Schild D, Hanau D, Spehner D, Schmid C, Rammensee HG, de la Salle H, Schild H. Cutting edge: Receptor-mediated endocytosis of heat shock proteins by professional antigen-presenting cells. J Immunol. 1999 Apr 1;162(7):3757–60. [PubMed] [Google Scholar]
- 37.Cardoso AI, Beauverger P, Gerlier D, Wild TF, Rabourdin-Combe C. Formaldehyde inactivation of measles virus abolishes cd46-dependent presentation of nucleoprotein to murine class i-restricted ctls but not to class ii-restricted helper T cells. Virology. 1995 Sep 10;212(1):255–8. doi: 10.1006/viro.1995.1479. [DOI] [PubMed] [Google Scholar]
- 38.Delrue I, Delputte PL, Nauwynck HJ. Assessing the functionality of viral entry-associated domains of porcine reproductive and respiratory syndrome virus during inactivation procedures, a potential tool to optimize inactivated vaccines. Vet Res. 2009;40(6):62. doi: 10.1051/vetres/2009047. [DOI] [PubMed] [Google Scholar]
- 39.Wilton T, Dunn G, Eastwood D, Minor PD, Martin J. Effect of formaldehyde inactivation on poliovirus. J Virol. 2014 Oct;88(20):11955–64. doi: 10.1128/JVI.01809-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan YC, Chiu HC, Chen LK, Chang GJ, Chiou SS. Formalin inactivation of japanese encephalitis virus vaccine alters the antigenicity and immunogenicity of a neutralization epitope in envelope protein domain III. PLoS Negl Trop Dis. 2015 Oct;9(10):e0004167. doi: 10.1371/journal.pntd.0004167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klimstra WB, Nangle EM, Smith MS, Yurochko AD, Ryman KD. DC-SIGN and L-SIGN can act as attachment receptors for alphaviruses and distinguish between mosquito cell- and mammalian cell-derived viruses. J Virol. 2003 Nov;77(22):12022–32. doi: 10.1128/JVI.77.22.12022-12032.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akahata W, Yang ZY, Andersen H, Sun S, Holdaway HA, Kong WP, et al. A virus-like particle vaccine for epidemic chikungunya virus protects nonhuman primates against infection. Nat Med. 2010 Mar;16(3):334–8. doi: 10.1038/nm.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolling BG, Weaver SC, Tesh RB, Vasilakis N. Insect-Specific virus discovery: Significance for the arbovirus community. Viruses. 2015 Sep;7(9):4911–28. doi: 10.3390/v7092851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.França GVA, Schuler-Faccini L, Oliveira WK, Henriques CMP, Carmo EH, Pedi VD, Victora CG. Congenital Zika virus syndrome in Brazil: A case series of the first 1501 livebirths with complete investigation. The Lancet. 2016:891–897. doi: 10.1016/S0140-6736(16)30902-3. [DOI] [PubMed] [Google Scholar]
- 45.Beaty BJ, Calisher CH, Shope RE. Arboviruses. In: Schmidt NJ, Emmons RW, editors. Diagnostic procedures for viral, rickettsial and chlamydial infections. Washington DC: American Public Health Association; 1989. pp. 797–855. [Google Scholar]
- 46.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007 Jan;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Bammes BE, Chen D-H, Jin L, Bilhorn RB. Visualizing and correcting dynamic specimen processes in TEM using a direct detection device. Microscopy and Microanalysis. 2013;19(S2):1320–1. [Google Scholar]
- 48.Zhang R, Hryc CF, Cong Y, Liu X, Jakana J, Gorchakov R, et al. 4.4 Å cryo-em structure of an enveloped alphavirus venezuelan equine encephalitis virus. EMBO J. 2011 Sep 14;30(18):3854–63. doi: 10.1038/emboj.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X, Jiang W, Jakana J, Chiu W. Averaging tens to hundreds of icosahedral particle images to resolve protein secondary structure elements using a multi-path simulated annealing optimization algorithm. J Struct Biol. 2007 Oct;160(1):11–27. doi: 10.1016/j.jsb.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang S, Tan HC, Ooi EE. Visualizing dengue virus through alexa fluor labeling. J Vis Exp. 2011;(53):e3168. doi: 10.3791/3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kroschewski H, Allison SL, Heinz FX, Mandl CW. Role of heparan sulfate for attachment and entry of tick-borne encephalitis virus. Virology. 2003 Mar 30;308(1):92–100. doi: 10.1016/s0042-6822(02)00097-1. [DOI] [PubMed] [Google Scholar]
- 52.Plante K, Wang E, Partidos CD, Weger J, Gorchakov R, Tsetsarkin K, et al. Novel chikungunya vaccine candidate with an ires-based attenuation and host range alteration mechanism. PLoS Pathog. 2011 Jul;7(7):e1002142. doi: 10.1371/journal.ppat.1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-em micrographs and reconstructions are deposited at emdatabank.org: accession number EMD-3406.


