Abstract
Most knowledge on NK cells are based on studies of what are now known as conventional NK (cNK) cells in the mouse spleen or human peripheral blood. However, recent studies in mice indicate the presence of tissue-resident NK (trNK) cells in certain organs, such as the liver, that display different markers and transcription factor dependencies as compared to cNK cells. Here we provide evidence from cytometry by time-of-flight analysis and humanized mice indicating that human CD49e-negative NK cells are tissue-resident in the liver. Thus, these studies indicate that trNK cells are evolutionarily conserved in humans and mice, providing a foundation to explore their role in human disease.
Introduction
Natural killer (NK) cells are innate lymphocytes that are able to kill tumor and infected cells and produce cytokines (1). However, most studies of mouse NK cells have concentrated on NK cells isolated from the spleen. Recently, a population of non-circulating, tissue-resident NK (trNK) cells in the mouse liver was identified as being distinct from circulating and splenic conventional NK (cNK) cells by the mutually exclusive expression of CD49a and DX5, respectively (2, 3). Moreover, trNK cells (~30% of NK cells in the liver) tend to be CXCR6+ and require the transcription factor T-bet and not NFIL3, whereas cNK cells are CXCR6− and require NFIL3 (3–5). Thus, mouse trNK and cNK cells form distinct lineages.
Most studies of human NK cells have concentrated on NK cells derived from venous blood. They lack expression of CD3 and CD19 and are usually classified into 2 groups, CD56bright CD16+/− (CD56bright) or CD56dim CD16+ (CD56dim) (6). Human liver NK cells have not been as well studied. A recent report examining NK cells from liver biopsy specimens from humans with a variety of conditions identified CD49a+ CD56+ CD3− CD19− NK cells (7). However, these cells were variably present, averaging only ~2.3% of NK cells in positive samples, which comprised only 50% of donors. Thus, it is possible that other approaches may yield the identity of another NK cell population in the human liver that is tissue-resident.
Recent technological development of mass cytometry also known as cytometry by time-of-flight (CyTOF) provides another approach to study heterogeneous cell populations (8). CyTOF allows for single cell analysis with a higher number of markers than by conventional flow cytometry and can identify unique cell populations even when a specific marker for the given subset is not used. Thus, CyTOF provides a potential new approach to discover and define trNK cells in the human liver.
The liver is unique as it possesses two afferent blood vessels with the hepatic artery carrying oxygenated blood and the portal vein supplying nutrient-rich venous blood drained from the gastrointestinal tract and spleen (9). The two blood supplies mix in the liver sinusoid space, a highly fenestrated, low pressure vascular system that is an especially rich area for localization of innate immune cells, such as Kupffer cells (9) and liver trNK cells (2). Here we studied NK cells in the human liver, specifically cells remaining in the liver vasculature after copious perfusion of normal organs being prepared for transplantation. We used CyTOF to gain insights into a unique NK cell population present in the liver preps but not PBMC, prompting the identification and initial characterization of human trNK cells in the liver.
Materials and Methods
Human donors and cell preparation
After surgical dissection of liver and just prior to excision from the cadaveric donor, the aorta was cannulated and the liver was flushed with 5 liters of Custodiol histidine-tryptophan-ketoglutarate (HTK) solution (Essential Pharma, NJ, USA) which exited the superior vena cava. The liver was then excised and a further 700 mL of Custodiol HTK solution was used for a final flush that was collected (liver-post-excision flush, L-PxF) per the Midwest Transplant Services’ procurement protocol. The de-identified L-PxF perfusate was concentrated and the cell pellet resuspended in HBSS. L-PxF mononuclear cells and matched venous blood from the same donor were isolated by Ficoll-Hypaque gradient centrifugation. The Institutional Review Board (IRB) for Humans Research Protection at Washington University has determined that this study does not involve activities that are subject to IRB oversight, as the donors were deceased individuals.
CyTOF
Isolated mononuclear cells were stained with antibodies conjugated with transition element isotopes (Supplemental Table 1) and analyzed on a CyTOF 2 mass cytometer (Fluidigm, USA) in the Center for Human Immunology and Immunotherapy Programs (CHiiPs) as described previously (10). Antibodies were either purchased from Fluidigm or conjugated to metal isotopes in the Rheumatic Diseases Core Center. Data were processed and analyzed using Cytobank (http://wustl.cytobank.org).
Conventional flow cytometry
Isolated mononuclear cells were stained with fluorescently labeled antibodies for cell surface expression of CD56 (NCAM16.2), CD62L (DREG-56), CXCR6 (13B1E5), CD45 (HI30), CD49e (IIA1), CD49a (SR84), CD3 (UCHT1), CD19 (HIB19), CD49a (TS2/7), CD94 (DX22), CD69 (FN50), CD16 (3G8), and CD27 (L128) (eBioscience, CA, USA). Dead cells were excluded with the fixable viability dye eFluor 506 (eBioscience). Intracellular transcription factor staining was performed using the FoxP3 staining kit according to manufacturer’s instructions for expression of Tbet (eBio4B10) and EOMES (WD1928) (eBioscience). IFN-γ, TNF-α and CD107a expression production was measured by intracellular cytokine staining of isolated mononuclear cells 6h post stimulation with PMA (20 ng/mL) and ionomycin (1 ug/mL). Brefeldin A (cytokines) or monensin (CD107a)was added 1h after stimulation. For CD107a staining, anti-CD107a (eBioH4A3, eBioscience) was added during culture. Cells were fixed and permeabilized with the BD cytofix/cytoperm kit according to manufacturer’s instructions and stained with IFN-γ (4S.B3) and TNF-α (Mab11). Stained cells were analyzed on a FACSCanto or LSR Fortessa X20 (BD Biosciences, USA) and data was analyzed with FlowJo (version 10.0.7, Tree Star, OR, USA). Statistical analysis was conducted in GraphPad Prism 6.01 (GraphPad Software, CA, USA).
NSG mice
Humanized NOD-SCID-gamma (NSG) mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and housed according to institutional guidelines (Protocol # 20160002) approved by the animal studies committee. Liver mononuclear cells were isolated on Percoll gradient as previously described (3). Collected blood was lysed in red blood cell lysis buffer before staining with fluorescent antibodies as above.
Results
CyTOF analysis reveals CD49e as a discriminating marker
To evaluate the possibility that there are trNK cells in the human liver, we took advantage of previous studies indicating that mouse trNK cells reside in the liver sinusoids and are enriched when the excised liver is flushed with saline (2). Herein, we collected samples from organ donor livers being prepared for cadaveric transplantation that were extensively flushed in situ then excised for a final flush. Paired PBMC samples were also collected from each donor. Our analysis revealed that NK cells do not differentially express CD49a in PBMC versus L-PxF (Supplemental Fig 1). Thus, unlike previously reported (7), we were unable to discern a liver-specific population of NK cells based on CD49a expression in L-PxF samples in our cohort.
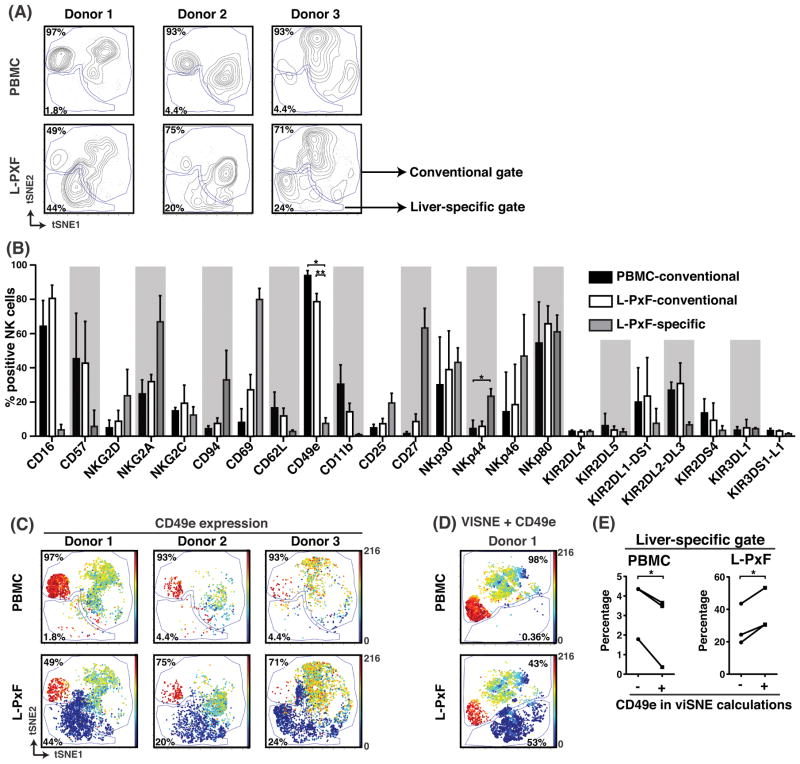
To further evaluate the potential presence of human liver trNK cells, we utilized CyTOF analysis of 40 parameters, including 30 markers (Supplemental Table 1), an expansion of our previously published CyTOF panel (10) to include other markers used to identify human NK cell subsets (11). Due to the multidimensionality of the single cell data, we visualized our CyTOF results comparing NK cells in matched PBMC and L-PxF samples by applying the viSNE algorithm (12) on gated CD56+ CD3− CD19− NK cells. Clustering on the NK-associated markers CD11b, CD16, CD45, CD56, and CD57 revealed a population of NK cells present in both PBMC and L-PxF samples (conventional) and another population present only in L-PxF (liver-specific) samples from three donors (Fig 1A). To quantify and further analyze these NK populations, manual gates were drawn to encompass the conventional and liver-specific cells that represent a substantial portion of NK cells in the L-PxF, ranging from 19.64 – 43.53% (mean 29.19%). Thus, our CyTOF analysis suggested the presence of a liver-specific NK cell population.
Figure 1.
Multidimensional analysis of venous blood (PBMC) or liver post-excision flush (L-PxF) isolated NK cells identifies a unique liver-specific subset lacking expression of CD49e. (A) viSNE plots of NK cells from 3 donors were generated by clustering with the markers CD11b, CD16, CD45, CD56, and CD57. Gates were manually drawn in the contour plots based on the distribution of cells from the PBMC fraction. Numbers indicate percentage of cells in respective gates. (B) Average percentage of indicated marker expression derived from the gates in (A). Bars indicate SD, * = p<0.05, ** = p<0.01, Student t test after Bonferroni correction. (C) viSNE plots showing CD49e expression for the 3 donors in (A). Color intensity indicates CD49e expression intensity. (D) viSNE plots with CD49e expression from donor 1 that includes CD49e in the list of clustering markers. (E) Summary of cell percentages in liver-specific gates from PBMC and L-PxF when CD49e was excluded (-) or included (+) from clustering. * = p<0.05, Student paired t test.
To identify potential markers that could distinguish liver-specific from conventional NK cells, we analyzed the individual expression of additional surface molecules on each subset (Fig 1B). We observed increased CD69, CD94, CD27, NKG2D, and NKG2A expression on NK cells in the liver-specific gate as compared to those in the conventional gate in both PBMC and L-PxF samples, although the differences were not statistically significant, likely due to the low sample number. However, two markers, NKp44 and CD49e, were differentially expressed in a statistically significant manner. While NKp44 was found on a subpopulation in the liver-specific gate, particularly striking was the expression of CD49e. Essentially all NK cells in the conventional gate in either PBMC or L-PxF samples expressed CD49e, while CD49e expression was almost completely absent on NK cells in the liver-specific gate, confirmed by viSNE graphs when CD49e expression was overlaid (Fig 1C). Furthermore, by including CD49e in our viSNE clustering calculations, the liver-specific cell cluster was better separated from conventional NK cells (Fig 1D) as compared to when CD49e was not used in the viSNE algorithm (Fig 1C, Donor 1) because the percentage of cells in the liver-specific gate was reduced in the PBMC fraction whilst it increased in the L-PxF fraction (Fig 1E). Thus, CyTOF analysis strongly suggested that the lack of CD49e expression specifically marks a liver-specific NK cell subset.
Phenotype of CD49e− CD3− CD19− NK cells
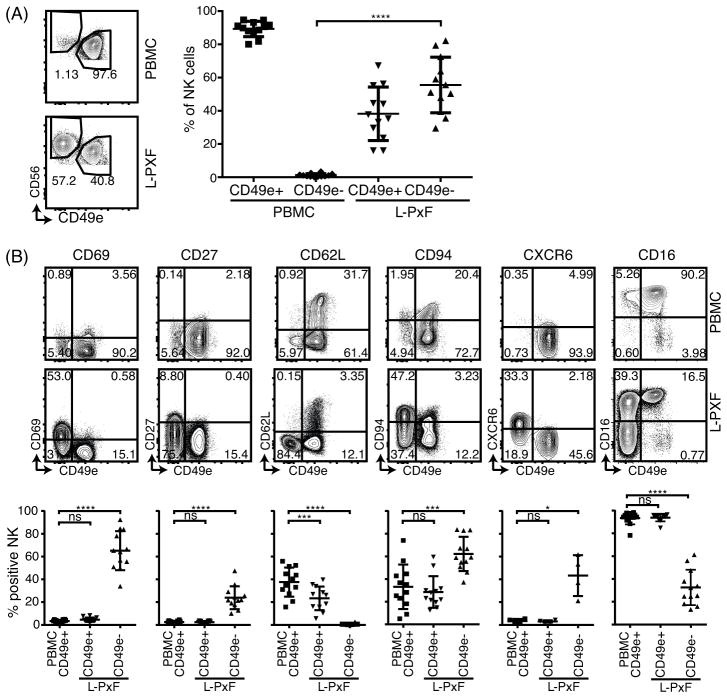
We validated lack of CD49e expression as a marker for liver NK cells in 14 healthy donors using traditional flow cytometry. Consistent with CyTOF data, we found that a relatively large subpopulation (55.53% ± 16.77) of CD56bright CD3− CD19− cells were CD49e− in L-PxF samples (Fig 2A). In contrast, this subpopulation was virtually absent in the PBMCs from each donor (1.23% ± 1.01). The liver CD49e− NK cells had a significantly higher expression of CD69 (65.18% ± 17.09 vs 3.43% ± 1.11), CD94 (62.15% ± 15.12 vs 33.16% ± 19.63) and CD27 (23.89% ± 10.05 vs 2.50% ± 1.09) but lower expression of CD62L (0.60% ± 0.69 vs 37.60% ± 12.72) when compared to NK cells in PBMCs, respectively (Fig 2B). Furthermore, CD49e− NK cells had elevated expression of CXCR6 (43.30% ± 17.97 vs 3.41% ± 1.77) similar to CD56bright liver NK cells previously reported (1). Of note, a subpopulation of the CD49e− NK cells from L-PxF also lacked the expression of CD16 (32.55% ± 15.52 vs 93.33% ± 5.47) when compared to NK cells isolated from venous blood respectively, indicating heterogeneity of CD16 expression within the CD49e− NK cell population.
Figure 2.
Differential cell surface marker expression on CD49e+ and CD49e− NK cells. (A) Expression of CD49e and CD56 on isolated single cell viable CD3− CD19− lymphocytes from healthy donor PBMC and L-PxF samples. Summary of percentage of NK cells from the 2 subsets of CD49e+ and CD49e− cells from venous blood and L-PxF is shown on the right. **** = p<0.0001, Student t test. Bars indicate mean ± SD. (B) Representative scatter plots (upper panel) and summary graph (lower panel) of indicated cell surface markers of NK cells from PBMC and L-PxF. * = p<0.05, ** = p<0.01, *** = p<0.001, **** = p<0.0001, ns = not significant, Student t test. Bars indicate mean ± SD
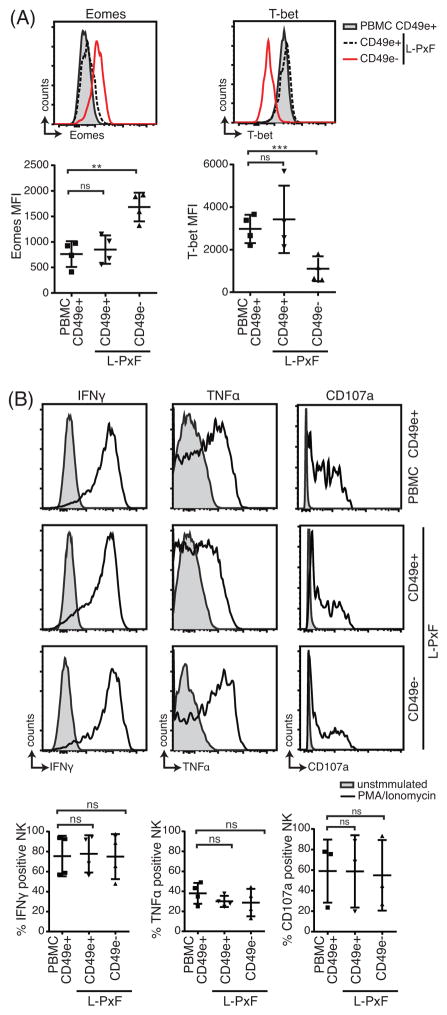
Surprisingly, we found that human CD49e− liver NK cells had significantly elevated expression of Eomes (1683 MFI (median fluorescence intensity) ± 281 vs 762 MFI ± 253) compared to CD49e+ NK cells (Fig 3A). In contrast, there was lower expression of T-bet (1102 MFI ± 581) compared to CD49e+ NK cells (2977 MFI ± 666) (Fig 4A). Upon stimulation with PMA and ionomycin, all NK cell populations produced similar amounts of IFNγ and TNFα (Fig 3B). Though there was wide variation, all stimulated NK cell populations also displayed CD107a, suggesting that they all have the capacity to perform cytotoxicity.
Figure 3.
Differential expression of transcription factors but not functional profile of CD49e+ and CD49e− NK cells. (A) Expression and MFI summary of T-bet and Eomes of PBMC and L-PxF derived NK cells. ** = p<0.01, *** = p<0.001, ns = not significant, Student t test. (B) Representative intracellular cytokine staining for IFNγ and TNFα production by NK cells from PBMC and L-PxF after 6 hours stimulation with PMA/ionomycin. Panels also show CD107a expression after stimulation. Summary graphs of intracellular cytokine production are shown at the bottom. ns = not significant. Bars indicate mean ± SD.
Figure 4.
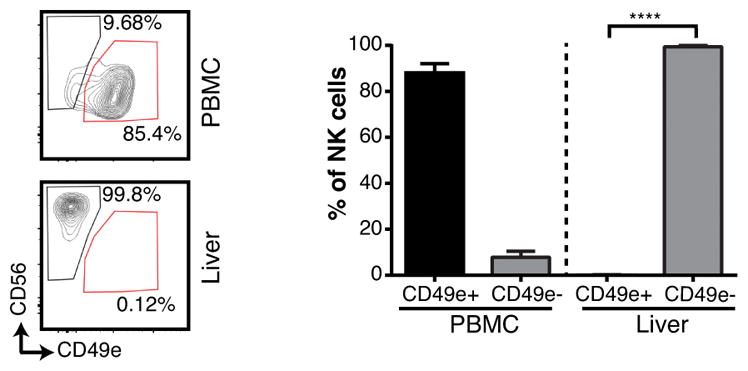
Human CD49e− NK cells are preferentially found in the liver of humanized NSG mice. Representative contour plots and summary graphs of humanized-NSG mice NK cells isolated from venous blood (PBMC) and liver homogenate (n= 3). Single cell viable hCD45+, CD3−, CD19−, lymphocytes were gated on before determining the expression of CD56 and CD49e. **** = p<0.0001, Student t test. Bars indicate SD.
CD49e− NK cells are present in the livers of humanized-NSG mice
To further determine if CD49e− NK cells specifically reside in the liver as compared to the circulation, we analyzed NOD-SCID-gamma common chain knockout (NSG) mice reconstituted with human CD34+ cord blood cells that establish a human immune system with functional NK cells (13). The vast majority of liver NK cells were CD49e−, and they were selectively present in liver as compared to blood (95% vs 0.5%, p<0.01) (Fig 4). Interestingly, relatively few CD49e+ NK cells were found in the liver whereas these cells were present in peripheral blood. Nonetheless, these data indicate that CD49e− NK cells selectively reside in the liver, consistent with tissue-residency.
Discussion
Here we exploited the localization of mouse trNK cells in the liver sinusoids, specifically in post-excision flush samples, to enrich for trNK cells derived from human livers. By CyTOF analysis, we found that lack of CD49e expression discriminates NK cells found in L-PxF samples but not in PBMCs. The CD49e− NK cells have phenotypic differences, characteristic for tissue resident cells, from CD49e+ NK cells in both PBMC and L-PxF samples. Studies published while the current manuscript was in preparation provide data consistent with our findings (14–16). However, the current studies markedly advance our understanding since we provide extensive CyTOF analysis and also show that human CD49e− NK cells selectively reside in the liver of humanized NSG mice. Taken together, the data effectively establish that human CD49e− NK cells are trNK cells in the human liver.
Whereas CD49a is a reliable marker of naïve trNK cells in mouse liver (2), here we found that the main human trNK cell population is marked by absence of CD49e expression as opposed to expression of CD49a, which was reported by others studying liver biopsy specimens (7). While we have not yet identified a mutually exclusive marker set for human NK cells, analogous to CD49a and DX5 (CD49b) in mice, the human CD49e− NK cell subset appears to be similar in this regard to DX5− NK cells in mice (2) in that liver trNK cells in both species selectively lack expression of an integrin expressed by all cNK cells in their respective species. {CD49 molecules are α chains heterodimerizing with the β1 chain to form integrins whose expression may be affected by activation and that confer overlapping functions (17).} However, to our surprise, human CD49e− NK cells appear to selectively express Eomesodermin rather than T-bet, whereas mouse CD49a+ DX5− NK cells express these T-box transcription factors in a reciprocal fashion and require T-bet for development (3). Furthermore, others have shown that human liver NK cells selectively express Eomes rather than T-bet (15, 16, 18). Thus, these data and studies highlight the challenges in directly translating findings from the mouse to humans when relying solely on expression of specific molecules and are consistent with differences in T-bet and Eomes expression in other immune cells in mice and humans (19).
The differences in molecule expression by mouse and liver trNK cells is reminiscent of the convergent evolution of Ly49 in mice and KIRs in humans, whereby each specific species evolved different genetic solutions for critical functions (20). Here, it is important to note not that the molecules selectively expressed by trNK and cNK cells in mouse and human are different. Rather the most important finding here is that both species have substantial numbers of trNK cells in the liver, raising the possibility of trNK cells will be found in other human organs as illustrated in mice (3).
Yet, the functional significance of liver trNK cells remains somewhat unclear. Here we show that their capacities to produce cytokines and degranulate upon stimulation with PMA and ionomycin are similar to cNK cells. While others have found that the liver NK cells have some functional differences (15, 16), as also noted with mouse liver trNK versus cNK cells (3), the primary physiological function of liver trNK cells relative to cNK cells remains to be fully determined. They could be important in immune responses attributed to cNK cells, such as tumor killing, control of viral infections, etc. However, it is likely that liver trNK cells may be especially important due to their location. More data are available in rodents, indicating that trNK cells reside in the liver sinusoids where they should be better positioned to deal with intravascular tumors and pathogens as well as interact with Kupffer cells (21). Since we identified CD49e− NK cells in L-PxF samples, it is very likely that human trNK cells are also selectively in the sinusoids. Regardless, the presence of liver trNK cells in humans strongly suggest that they have evolutionary significance, and their identification lays the foundation for studying liver trNK cells in human disease.
Supplementary Material
Acknowledgments
This work was supported by the Howard Hughes Medical Institute, Barnes-Jewish Hospital Foundation, Center for Human Immunology and Immunotherapy Programs, and NIH grant R01-AI106561 to W.M.Y.
We thank our colleagues at Midwest Transplant Services for their help in procuring samples for analysis.
Abbreviations
- cNK
conventional NK
- CyTOF
cytometry by time-of-flight
- KIR
killer Ig-like receptor
- L-PxF
liver post-excision flush
- MFI
mean fluorescence intensity
- NSG
NOD-Scid-gamma common chain
- trNK
tissue-resident NK
References
- 1.Yokoyama WM. Chapter 17. Natural killer cells. In: Paul WE, editor. Fundamental immunology. 7. Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 395–431. [Google Scholar]
- 2.Peng H, Jiang X, Chen Y, Sojka DK, Wei H, Gao X, Sun R, Yokoyama WM, Tian Z. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. The Journal of clinical investigation. 2013;123:1444–1456. doi: 10.1172/JCI66381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, Zhong C, Chase JM, Rothman PB, Yu J, Riley JK, Zhu J, Tian Z, Yokoyama WM. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, Charrier E, Bienvenu J, Henry T, Debien E, Hasan UA, Marvel J, Yoh K, Takahashi S, Prinz I, de Bernard S, Buffat L, Walzer T. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med. 2014;211:563–577. doi: 10.1084/jem.20131560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seillet C, Huntington ND, Gangatirkar P, Axelsson E, Minnich M, Brady HJ, Busslinger M, Smyth MJ, Belz GT, Carotta S. Differential requirement for Nfil3 during NK cell development. J Immunol. 2014;192:2667–2676. doi: 10.4049/jimmunol.1302605. [DOI] [PubMed] [Google Scholar]
- 6.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marquardt N, Beziat V, Nystrom S, Hengst J, Ivarsson MA, Kekalainen E, Johansson H, Mjosberg J, Westgren M, Lankisch TO, Wedemeyer H, Ellis EC, Ljunggren HG, Michaelsson J, Bjorkstrom NK. Cutting edge: identification and characterization of human intrahepatic CD49a+ NK cells. J Immunol. 2015;194:2467–2471. doi: 10.4049/jimmunol.1402756. [DOI] [PubMed] [Google Scholar]
- 8.Bendall SC, Simonds EF, Qiu P, Amirel AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, Balderas RS, Plevritis SK, Sachs K, Pe’er D, Tanner SD, Nolan GP. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crispe IN. The liver as a lymphoid organ. Annu Rev Immunol. 2009;27:147–163. doi: 10.1146/annurev.immunol.021908.132629. [DOI] [PubMed] [Google Scholar]
- 10.Miner JJ, Aw Yeang HX, Fox JM, Taffner S, Malkova ON, Oh ST, Kim AH, Diamond MS, Lenschow DJ, Yokoyama WM. Brief report: chikungunya viral arthritis in the United States: a mimic of seronegative rheumatoid arthritis. Arthritis & rheumatology. 2015;67:1214–1220. doi: 10.1002/art.39027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, Dekker CL, Mackey S, Maecker H, Swan GE, Davis MM, Norman PJ, Guethlein LA, Desai M, Parham P, Blish CA. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med. 2013;5:208ra145. doi: 10.1126/scitranslmed.3006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amirel AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, Pe’er D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strowig T, Chijioke O, Carrega P, Arrey F, Meixlsperger S, Ramer PC, Ferlazzo G, Munz C. Human NK cells of mice with reconstituted human immune system components require preactivation to acquire functional competence. Blood. 2010;116:4158–4167. doi: 10.1182/blood-2010-02-270678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hudspeth K, Donadon M, Cimino M, Pontarini E, Tentorio P, Preti M, Hong M, Bertoletti A, Bicciato S, Invernizzi P, Lugli E, Torzilli G, Gershwin ME, Mavilio D. Human liver-resident CD56(bright)/CD16(neg) NK cells are retained within hepatic sinusoids via the engagement of CCR5 and CXCR6 pathways. J Autoimmun. 2016;66:40–50. doi: 10.1016/j.jaut.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmon C, Robinson MW, Fahey R, Whelan S, Houlihan DD, Geoghegan J, O’Farrelly C. Tissue-resident Eomes(hi) T-bet(lo) CD56(bright) NK cells with reduced proinflammatory potential are enriched in the adult human liver. Eur J Immunol. 2016;46:2111–2120. doi: 10.1002/eji.201646559. [DOI] [PubMed] [Google Scholar]
- 16.Stegmann KA, Robertson F, Hansi N, Gill U, Pallant C, Christophides T, Pallett LJ, Peppa D, Dunn C, Fusai G, Male V, Davidson BR, Kennedy P, Maini MK. CXCR6 marks a novel subset of T-bet(lo)Eomes(hi) natural killer cells residing in human liver. Scientific reports. 2016;6:26157. doi: 10.1038/srep26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abram CL, Lowell CA. The ins and outs of leukocyte integrin signaling. Annu Rev Immunol. 2009;27:339–362. doi: 10.1146/annurev.immunol.021908.132554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marotel M, Hasan U, Viel S, Marcais A, Walzer T. Back to the drawing board: Understanding the complexity of hepatic innate lymphoid cells. Eur J Immunol. 2016;46:2095–2098. doi: 10.1002/eji.201646584. [DOI] [PubMed] [Google Scholar]
- 19.Knox JJ, Cosma GL, Betts MR, McLane LM. Characterization of T-bet and eomes in peripheral human immune cells. Front Immunol. 2014;5:1–12. doi: 10.3389/fimmu.2014.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelley J, Walter L, Trowsdale J. Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet. 2005;1:129–139. doi: 10.1371/journal.pgen.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng H, Wisse E, Tian Z. Liver natural killer cells: subsets and roles in liver immunity. Cell Mol Immunol. 2015;13:328–336. doi: 10.1038/cmi.2015.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.