Abstract
Background
The neddylation pathway conjugates NEDD8 to cullin-RING ligases and controls the proteasomal degradation of specific proteins involved in essential cell processes. Pevonedistat (MLN4924) is a selective small molecule targeting the NEDD8-activating enzyme (NAE) and inhibits an early step in neddylation, resulting in DNA re-replication, cell cycle arrest and death. We investigated the anti-tumor potential of pevonedistat in preclinical models of melanoma.
Methods
Melanoma cell lines and patient-derived tumor xenografts (PDTX) treated with pevonedistat were assessed for viability/apoptosis and tumor growth, respectively, to identify sensitive/resistant models. Gene expression microarray and gene set enrichment analyses were performed in cell lines to determine the expression profiles and pathways of sensitivity/resistance. Pharmacodynamic changes in treated-PDTX were also characterized.
Results
Pevonedistat effectively inhibited cell viability (IC50 <0.3μM) and induced apoptosis in a subset of melanoma cell lines. Sensitive and resistant cell lines exhibited distinct gene expression profiles; sensitive models were enriched for genes involved in DNA repair, replication and cell cycle regulation, while immune response and cell adhesion pathways were upregulated in resistant models. Pevonedistat also reduced tumor growth in melanoma cell line xenografts and PDTX with variable responses. An accumulation of pevonedistat-NEDD8 adduct and CDT1 was observed in sensitive tumors consistent with its mechanism of action.
Conclusions
This study provided preclinical evidence that NAE inhibition by pevonedistat has anti-tumor activity in melanoma and supports the clinical benefits observed in recent Phase 1 trials of this drug in melanoma patients. Further investigations are warranted to develop rational combinations and determine predictive biomarkers of pevonedistat.
Keywords: neddylation, pevonedistat, MLN4924, protein degradation, melanoma
INTRODUCTION
Normal cellular functions such as cell growth, apoptosis and cell cycle require a highly regulated cascade of intracellular protein degradation through the ubiquitin-proteasome system (UPS) [1]. This proteolytic process is mediated by the ubiquitin-activating (E1) and ubiquitin-conjugating (E2) enzymes and ubiquitin ligases (E3), which ultimately result in the polyubiquitination of the target protein marked for degradation by the proteasome. The specificity of the degraded protein substrates is conferred by specific subunits of the E3 ligase [2].
Dysregulation of this multi-step degradation pathway and thus protein homeostasis may contribute to carcinogenesis [2]. In fact, the importance of UPS in cancer has been exploited in the development of the first proteasome inhibitor bortezomib (VELCADE), which has proven anti-tumor efficacy and is currently approved for the treatments of multiple myeloma and mantle cell lymphoma [3, 4]. However, as opposed to inhibiting general proteasomal degradation with its associated toxicities, there is an interest to more specifically target the degradation of proteins relevant to tumorigenesis [5]. The cullin-RING ligases (CRLs) are a family of E3 ubiquitin ligases that consist of a cullin and RING finger protein core with a substrate recognition model composed of adaptor and receptor proteins such as Skp-1 and F-box proteins [6]. CRLs have been shown to control the proteasomal degradation of a subset of biologically important proteins, including cell cycle regulators (e.g. cyclin D, p21), DNA repair proteins, and key signaling effectors such as β-catenin and Notch [7-9].
NEDD8 is an ubiquitin-like protein that modulates the activity of CRLs by covalently binding to their cullin subunits in a process called neddylation [2, 10-12]. Conjugation of NEDD8 to CRLs increases their activity, thus facilitating ubiquitination and proteasomal degradation of CRL-specific protein substrates [2]. NEDD8 itself is activated and conjugated to its target cullin by a step-wise process analogous to ubiquitination, requiring a corresponding E1 NEDD-activating enzyme (NAE), as well as NEDD8-specific E2 and E3 enzymes [6]. These and several other proteins play key roles in the neddylation pathway, and as such, may promote tumorigenesis when their normal function is impaired. In fact, various components of CRLs have been implicated in the development of cancer, including cullins and adaptor and receptor proteins [6]. For example, cullin-4A amplification has been observed in breast cancer [13, 14]. In addition, dysregulation of neddylation is involved in cholangiocarcinoma [15], prostate [16] and lung cancers [17].
Thus, NAE controls an early step in neddylation and represents a potential therapeutic target to modulate CRL activity and protein degradation [2]. Pevonedistat (MLN4924) is a synthesized derivative of N6-benzyl adenosine and a selective small molecule inhibitor of NAE [18]. The compound structurally resembles adenosine 5’-monophosphate (AMP), a product of the hydrolysis of adenosine 5’-triphsophate (ATP), which plays a key role in the enzymatic activity of NAE [18, 19]. The molecular mechanism of action and effects of pevonedistat were previously described by Soucy et al. [18], who provided the first proof-of-concept that targeting the neddylation pathway specifically impairs CRL-mediated protein degradation and has anti-tumor activity in preclinical model systems. Pevonedistat leads to the accumulation of known CRL substrates such as CDT1 and p27 in HCT-116 cell lines, which disrupts normal cell cycle progression [18, 20]. In particular, CDT1 is a component of the origin replication complex involved in the initiation of DNA replication [21], and is regulated by proteasomal degradation to avoid repeated firing of the replication origin. An increase in CDT1 levels results in a phenomenon of DNA re-replication leading to DNA damage [21].
Moreover, since the CRLs play a role in multiple DNA damage response and cell cycle pathways [22-24], it is likely that pevonedistat also exhibits additional mechanisms of action in the cell. In fact, a genome-wide siRNA screen in a melanoma cell line was conducted to investigate the genes or pathways that contribute to sensitivity to NAE inhibition [25]. This study identified 154 genes that modulated the anti-tumor activity of pevonedistat, half of which were involved in critical cell processes including cell cycle, apoptosis, DNA damage response, and the ubiquitin system (e.g. p53 pathway, BRCA1/2, base excision repair).
Pevonedistat has been demonstrated to promote apoptosis, senescence and autophagy in various cancer cell lines [18, 20, 26], while it also significantly inhibits tumor growth in colon and lung cancer cell line murine xenografts at tolerable doses [18]. Furthermore, the anti-tumor activity of pevonedistat has been confirmed in models of hematologic malignancies such as acute myeloid leukemia, myeloma and diffuse large B-cell lymphoma [27-29], as well as various solid tumors including cholangiocarcinoma [15], prostate [16], lung [17], ovarian [30], liver [31], head and neck cancer [32], and Ewing sarcoma [33]. Finally, pevonedistat also acts a radiosensitizer in pancreatic and breast cancer cells [34, 35]. In light of this preclinical evidence, several Phase 1 trials of pevonedistat have been completed in both hematologic [36, 37] and solid malignancies including melanoma [38, 39].
While targeted agents such as BRAF inhibitors for V600E-mutant tumors (e.g. vemurafenib, dabrafenib) and immunotherapies (e.g. ipilimumab, nivolumab) have improved outcomes in metastatic melanoma, the overall survival in this population remains limited [40]. This is an area in need of novel therapies. Intriguingly, in the Phase 1 trial of pevonedistat in advanced solid tumors [38], patients with melanoma were among those with the longest intervals of stable disease on study. Three of the 9 melanoma patients enrolled were treated with 7 or more cycles of pevonedistat, and one maintained stable disease for 6 months [38]. Subsequently, pevonedistat was evaluated in a melanoma-specific Phase 1 trial of 37 patients with 2 different dosing schedules (schedule A: on days 1, 4, 8, 11 every 21 days, at 50-278mg/m2; schedule B: on days 1, 8, 15 every 21 days, at 157mg/m2) [39]. Similar to the previous Phase 1 trial, there was a significant rate of stable disease in melanoma patients (48%), while 1 patient achieved a partial response [39].
The results of these recent Phase 1 clinical trials indicate that NAE inhibition may have potential anti-tumor effects in melanoma. However, the significance of the neddylation pathway of protein degradation and the biological effects of pevonedistat specifically in melanoma have not been comprehensively investigated in the preclinical setting. This is the first study to characterize the preclinical anti-tumor activity of pevonedistat, as well as its PD effects and predictors of response in a large number of melanoma models including multiple patient-derived tumor xenografts (PDTX).
MATERIALS AND METHODS
Reagents
Pevonedistat (MW 443.5) was supplied by Millennium Pharmaceuticals, Inc. in lyophilized form. Pevonedistat was solubilized in 100% DMSO for in vitro experiments and in 10% HPbCD for in vivo experiments.
Cell lines and cell culture
Thirty-five authenticated human melanoma cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). All cells were grown in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% fetal bovine serum, 1% non-essential amino acids, 1% penicillin/streptomycin, and 1% Normocin (anti-mycoplasma agent). Cells were incubated at 37°C in a humidified incubator with 5% CO2. All in vitro treatments with the compound were conducted with the use of complete growth media.
Cell viability assay
Cell viability was quantitated using CellTiter-Glo® Luminescent Cell Viability Assay (Promega Corporation, Madison, WI, US) according to manufacturer instructions. Subconfluent cells in culture were collected and transferred to 96-well white-walled flat bottom plates at 1,500-5,000 viable cells in 100μl suspension per well. Cells were incubated overnight at 37°C, and then exposed to increasing concentrations of pevonedistat up to 3μM for 72 hrs. Then, 100μl of CellTiter-Glo® reagent was mixed in each well and incubated for 10 min, followed by luminescence detection using a plate reader (BiotekSynergy2, Winooski, VT, USA). Cell viability curves were derived from the raw luminescent data and used to determined IC50 values.
Gene microarray (RNA expression)
From the cell viability assay, cell lines were called sensitive or resistant to pevonedistat based on the cutoffs of IC50 <0.3μM and >1μM, respectively. Eleven sensitive (SKMEL1, HMCB, C32, A101D, 451LU, 1205LU, A2058, HS695T, G361, HS294T and SH4) and 6 resistant (HT144, HS394T, MEWO, WM75, COLO829 and SKMEL24) cell lines were selected for gene expression analysis using the Cancer Cell Line Encyclopedia (CCLE, GSM36133) and in-house gene expression profiling by the Affymetrix HG-U133 Plus 2.0 microarray platform. In addition, gene expression of 4 cell line xenografts in the pevonedistat-sensitive (HMCB, A2058, HS294T and SH4) and in the MLN-resistant (HT144, HS934T, MEWO and SKMEL24) groups were profiled by the Affymetrix HuGene 1.0 ST microarray. Sample preparation and processing procedures were performed as described in the Affymetrix GeneChip® Expression Analysis Manual (Affymetrix Inc., Santa Clara, CA, USA). Gene expression profiles were normalized by the Robust Multiarray Average method using the Affymetrix Power Tools (APT) for each platform. Multiple probe sets representing the same gene were collapsed using the maximum expression values, and common genes across the 2 platforms were identified for further study.
Gene set enrichment analysis (GSEA)
GSEA of the microarray data was done using the GSEA software version 2.0.6 obtained from the Broad Institute (http://www.broad.mit.edu/gsea) [52]. Gene set permutations were done 1,000 times for each analysis. We used the nominal P-value and normalized enrichment score to sort the pathways enriched in the sensitive and resistant phenotypes, where the pathways were defined by the gene sets from the Kyoto Encyclopedia of Genes and Genomes (KEGG) and BioCARTA databases.
Apoptosis assay
Apoptosis was measured using the Caspase-Glo® 3/7 assay (Promega Corporation, Madison, WI, USA). One hundred microliters of cell suspension containing 1,500-5,000 cells per well were plated in a 96-well, white-walled tissue culture plate and allowed to adhere overnight. Media was removed, 100μl of media without and with pevonedistat at indicated concentrations was added, and cells were incubated for 24 hrs; this was found to be the optimal duration based on multiple experiments using various other time points. Following exposure to pevonedistat, 100μl of Caspase-Glo® 3/7 reagent was added, cells were incubated in the dark for 1 hr, and the plate was read on a plate reader (BiotekSynergy2, Winooski, VT, USA) using luminescence as the method of detection of apoptosis measured by relative light units and normalized against the untreated (control) cells. An increase in luminescence indicated more apoptotic cells.
Cell line xenografts and patient-derived tumor xenografts (PDTX)
Female 5–6-week-old athymic nude mice were purchased from Harlan Laboratories (Indianapolis, IN, USA). Animals were allowed to acclimate for at least 7 days before any handling. All studies were conducted at the University of Colorado Anschutz Medical Campus in accordance with the National Institutes of Health guidelines for the care and use of laboratory animals, and animals were housed in a facility accredited by the American Association for Accreditation of Laboratory Animal Care (3-5 mice per cage).
For cell line xenografts, melanoma cells were harvested in logarithmic growth phase and resuspended in a 1:1 mixture of serum-free RPMI 1640 and Matrigel (BD Biosciences, San Jose, CA, USA). Five to ten million cells were injected subcutaneously into the flank of each mouse using a 23-gauge needle. Mice were monitored daily for signs of toxicity and were weighed twice weekly. Tumor size was evaluated twice per week by caliper measurements using the following formula: tumor volume = length × width2 × 0.52. When tumors reached 100–300mm3, mice were treated with vehicle or pevonedistat 90mg/kg subcutaneously twice daily until the end of study. Duration of treatment varied between xenograft models and was determined by the observed response and maximal tumor size allowed (1,500mm3). Tumor volume was monitored and expressed as percent of volume on Day 1 for each treatment group at the time points evaluated. At least 5 xenografts per treatment group (2 tumors per mouse) were assessed to ensure statistical power and the results were averaged. The effect of pevonedistat on tumor growth was calculated as %TGI at the end of study, defined as %TGI = (Tx/Cx) × 100, where Tx = average tumor volume of treated mice on Day X (i.e. end of study) and Cx = average tumor volume of control mice on Day X. TGI <50% represents significant growth inhibition.
For PDTX, surgical specimens from patients undergoing resection of a primary or metastatic tumor at the University of Colorado Hospital were cut into 3-5mm pieces and implanted subcutaneously with a 10-gauge trochar into the flanks of mice for each patient sample. After subsequent growth and passage in mice (to 3-6 generations), tumors were excised and expanded into cohorts for study treatment. Treatment with pevonedistat, monitoring of mice and measurement of tumor were conducted as described above.
Immunohistochemistry (IHC)
Tumor tissues were harvested from PDTX, formalin-fixed and paraffin-embedded (FFPE). IHC assays were performed on 5-micron FFPE tumor sections using the Ventana XT® auto-stainer (Ventana Medical Systems, Tucson, AZ, USA). Antigen retrieval consisted of incubation with Ventana's CC1 antigen retrieval solution for 20 min. Primary antibody to pevonedistat-NEDD8 adduct (Millennium Pharmaceuticals, Inc., Cambridge, MA, USA) was incubated with tissue for 1 hr at 37°C, labeled with biotinylated goat anti-rabbit IgG secondary antibody (Vector Laboratories, Burlingame, CA, USA) for 32 min at room temperature, and detected with horseradish peroxidase (HRP) DAB Map system (Ventana Medical Systems) followed by hematoxylin and bluing reagent counter-stain (Ventana Medical Systems). Primary antibody to CDT1 (Millennium Pharmaceuticals, Inc.) was incubated with tissue for 1 hr at room temperature, labeled with UltraMap anti-mouse HRP multimer secondary antibodies (Ventana Medical Systems) and detected with the ChromoMap DAB Kit (Ventana Medical Systems), followed by hematoxylin and bluing reagent counter-stain. Images were captured at 20x magnification using the ScanScope XT whole-slide imaging system (Aperio Technologies, Vista, CA, USA).
Statistical analyses
Statistical analyses were performed using GraphPad Prism Software (La Jolla, CA, USA). For comparisons of 2 groups, the unpaired t-test was performed. For correlations, a Spearman correlation was used. P-values <0.05 were considered statistically significant.
RESULTS
Pevonedistat inhibits cell viability in a subset of melanoma cell lines
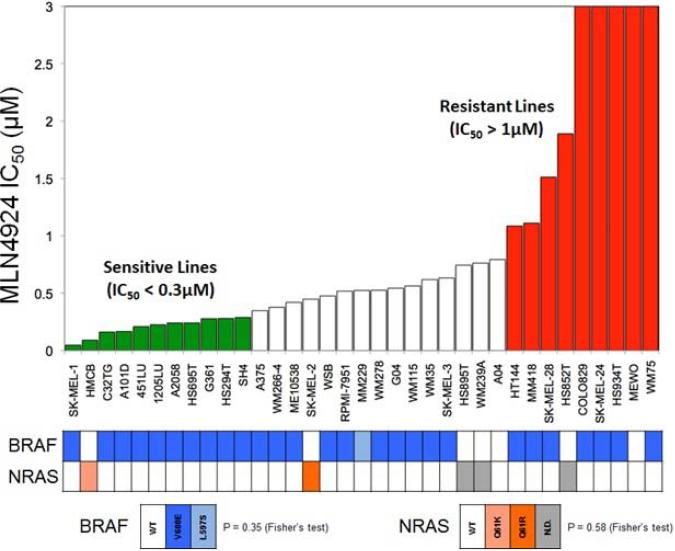
In order to assess the ability of pevonedistat to inhibit cell viability in vitro, a panel of 35 melanoma cell lines was exposed to increasing concentrations of pevonedistat up to 3μM and analyzed by the CellTiter-Glo® cell viability assay to determine the IC50. A wide range of sensitivity was observed across the cell lines (Figure 1). Cell lines with IC50 >1μM were defined as relatively resistant, and based on the distribution of IC50s of the remainder cell lines, those with IC50 <0.3μM were defined as relatively sensitive. Among relatively sensitive cell lines, SKMEL-1 was the most responsive to the growth-inhibitory effects of pevonedistat. Other cell lines had intermediate sensitivities (IC50 0.3-1μM).
Figure 1.
Anti-proliferative effects of pevonedistat in melanoma cell lines as determined by the cell viability assay (CellTiter-Glo®). Cells were treated with pevonedistat at increasing concentrations for 72 hrs followed by cell viability assay. IC50 (μM) values are shown (3 independent experiments per cell line). Cell lines are defined as relatively sensitive (IC50 <0.3μM, in green) or resistant (IC50 >1μM, in red). The profile of BRAF and NRAS mutations for each cell line is indicated below.
The variability in IC50s suggests that there may be a specific genetic profile that predicts response to pevonedistat in a subset of melanoma cells. Since there was only one sensitive cell line (HMCB) with wild-type BRAF and all cell lines tested had wild-type KIT and PIK3CA, it was not possible to assess whether the anti-proliferative effects of the drug were affected by mutations in these genes, particularly BRAF.
Sensitive and resistant cell lines are characterized by differential gene expression
Next, we assessed the gene expression profiles of several melanoma cell lines by conventional gene array to see if we could identify an inherent genetic signature associated with responsiveness to pevonedistat, using the most sensitive (IC50 <0.3μM; n=11) and most resistant (IC50 >1μM; n=6) cell lines as the dataset. This analysis identified the genes that were most upregulated in the sensitive and resistant models (Table 1, Supplementary Figure 1).
Table 1.
The 20 most upregulated genes in pevonedistat-sensitive and -resistant melanoma cell lines by gene expression microarray
| Sensitive cells | Resistant cells |
|---|---|
| PASD1 | IL13RA2 |
| TEX15 | EDIL3 |
| PLS1 | ABCA6 |
| RAPGEF5 | IFI44L |
| CDCA7 | XAF1 |
| WNK3 | CCDC80 |
| KIF5C | GJA1 |
| ANKRD7 | CTSK |
| FAM70A | SEMA3A |
| FRMD3 | LPAR1 |
| CTSL2 | RGS4 |
| MAP7D2 | MEOX2 |
| PIP5K1B | DOK5 |
| FAM38B | NPTX1 |
| SCRN1 | IFIT2 |
| CXORF48 | ABI3BP |
| LYN | CFH |
| F11R | RBMS3 |
| BAIAP2L1 | OLFML3 |
| SHISA2 | FAT4 |
Furthermore, GSEA was performed by mapping the gene expression data against the KEGG and BioCARTA pathway databases, yielding the pathways relevant in pevonedistat-sensitive and -resistant cell lines (Supplementary Table 1). The common core genes enriched in the most sensitive and resistant pathways identified are shown in Supplementary Table 2. For pevonedistat-sensitive cells, the common core genes enriched in the KEGG and BioCARTA databases were found to be involved in cell cycle, DNA repair, p53 signaling and spliceosome (Supplementary Figure 2A and 2B). Conversely, the pevonedistat-resistant models demonstrated increased expression of genes involved in immune response and cell adhesion (Supplementary Figure 2C and 2D).
These results indicate that the sensitivity of melanoma cell lines to inhibition of neddylation by pevonedistat might be associated with a distinct profile of gene expression and cellular pathways.
Pevonedistat exhibits variable apoptotic effects in melanoma cell lines
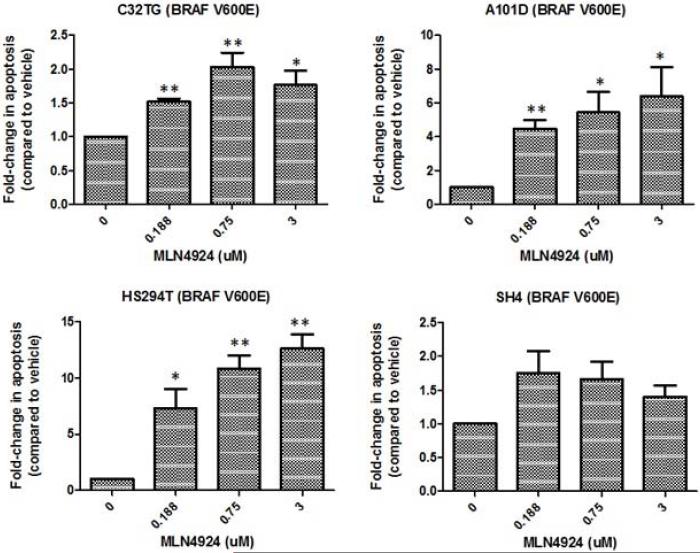
Pevonedistat is known to cause DNA damage and apoptosis. Therefore, we investigated the impact of pevonedistat on the fold-change in apoptosis compared to vehicle in sensitive melanoma cell lines (selected based on their IC50s from the cell viability assay) using the Caspase-Glo® 3/7 assay (Figure 2). Pevonedistat significantly increased apoptosis in all cell lines tested compared to vehicle, and a dose-dependent effect was observed in most of the cell lines examined. However, the degree of pro-apoptotic effect was variable across cell lines. In HS294T cells in which the greatest induction was observed, apoptosis was increased by up to 7.3-fold at 0.188μM, 10.8-fold at 0.75μM, and 12.6-fold at 3μM.
Figure 2.
Pevonedistat induces apoptosis in melanoma cell lines. The most sensitive cell lines based on their IC50s from the cell viability assay were selected for study. Cell lines were treated with various concentrations of pevonedistat for 24 hrs and evaluated for level of apoptosis using the Caspase-Glo® 3/7 assay. Results of representative cell lines are shown, expressed as fold-increase in apoptosis compared to vehicle control. Treated groups were compared to the untreated group (vehicle) by unpaired T-test, with statistically significant differences indicated by asterisks (* if p<0.05, ** if p<0.005).
Pevonedistat inhibits tumor growth in a subset of melanoma cell line and patient-derived tumor xenografts
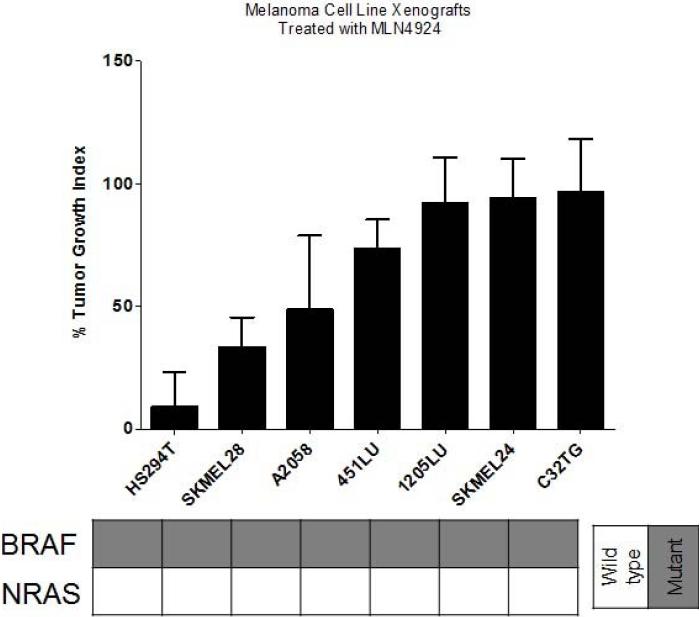
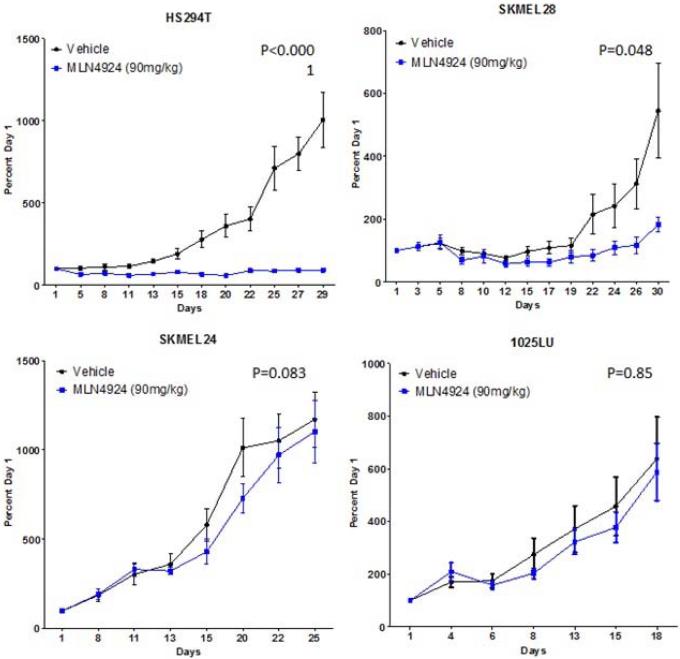
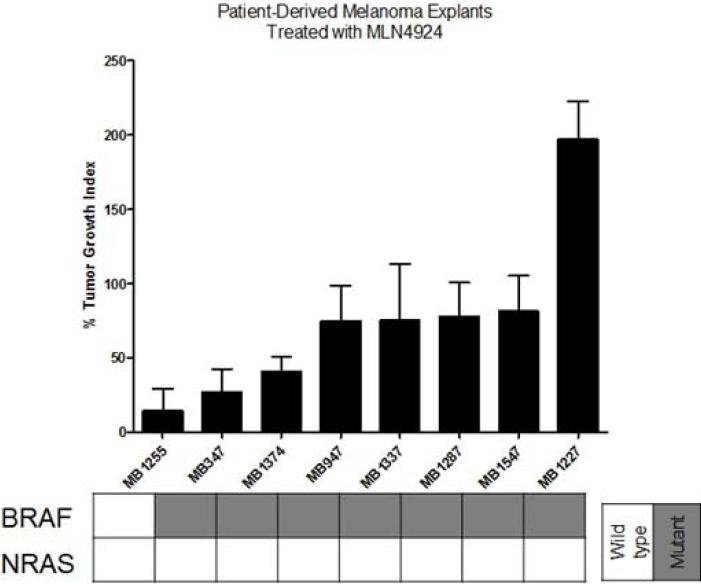
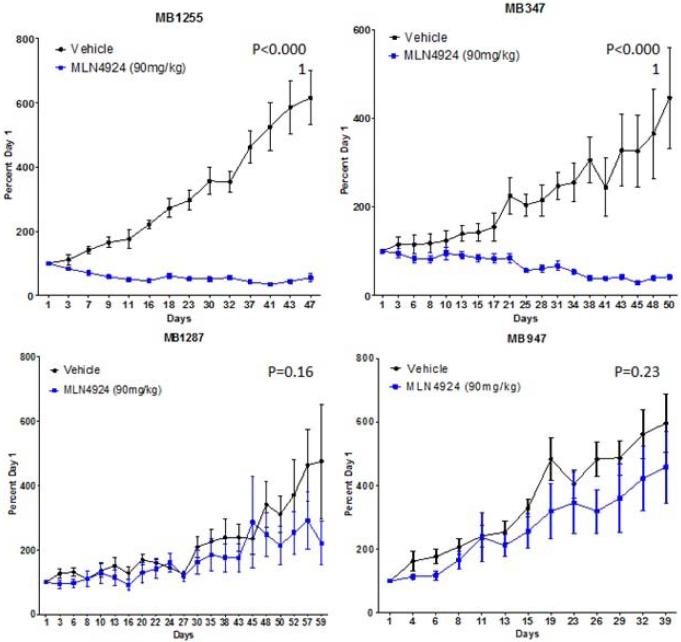
To assess the tumor activity of pevonedistat in vivo, subcutaneous murine xenografts were generated by injecting selected melanoma cell lines into nude mice. Relatively sensitive (n=5) and resistant (n=2) cell lines were selected based on their IC50s from the cell viability assay. The xenografts were treated with vehicle or pevonedistat 90mg/kg subcutaneously twice daily (at least 5 mice per treatment group (2 tumors per mouse) to ensure statistical power), and the tumor volume (expressed as percent of volume on Day 1) was followed twice weekly and averaged for each group; the percent tumor growth index (%TGI) was calculated at the end of study for each group to assess the growth inhibitory effects of pevonedistat (Figure 3a). The xenografts were treated with pevonedistat for 30 days or until mice had to be sacrificed for excess tumor volume (>1,500mm3).
Figure 3.
Effects of pevonedistat on in vivo tumor growth in melanoma xenografts. (a) The most sensitive (n=5) and resistant (n=2) cell lines were selected based on their IC50s from the cell viability assay to generate subcutaneous cell line xenografts. Mice were treated with vehicle or pevonedistat (90mg/kg subcutaneously twice daily), with at least 5 mice per treatment group. Tumor volume (mm3) was measured twice weekly and expressed as percent of volume on Day 1, which was averaged for each treatment group in the growth curves shown. The average percent tumor growth index (TGI) was determined for each xenograft model at the end of study, where TGI <50% indicated significant growth inhibition. (b) Melanoma patient-derived tumor xenografts (PDTX) (n=8) were generated, treated with vehicle or pevonedistat, and assessed for tumor volume and %TGI as described. Growth curves and %TGIs for representative xenografts are shown.
The HS294T xenograft was most sensitive and the SK-MEL-24 xenograft was among the most resistant, which correlated with their in vitro responses to pevonedistat (i.e. cell viability assay). However, for most of the other cell line models, in vitro sensitivity (IC50 <0.3μM) did not consistently correlate with in vivo sensitivity (TGI <50%). In particular, the SK-MEL-28 model, which showed in vitro resistance to pevonedistat, demonstrated a large reduction in tumor volume in the cell line xenografts treated with pevonedistat compared to vehicle. Furthermore, the effect of pevonedistat on tumor growth was evaluated in 8 melanoma PDTX (Figure 3b). There was significant tumor growth inhibition in 3 PDTX models (MB1255, MB347, MB1374). On the other hand, there was significant tumor progression in the treated MB1227 PDTX.
Thus, pevonedistat demonstrated anti-tumor activity in a subset of melanoma cell line xenografts and PDTX. In addition, there were no significant toxicities or loss of body weight observed in the mice treated with pevonedistat.
Pevonedistat produces pharmacodynamic changes in melanoma PDTX that correlate with anti-tumor activity
Pevonedistat inhibits NAE by forming a pevonedistat-NEDD8 adduct and preventing the activation and transfer of NEDD8 to cullin proteins [41], which is essential for the function of CRLs and their degradation of protein substrates. In fact, the mechanism of action and downstream molecular effects of MLN4824 were previously reported by Soucy et al. [18]. Treatment with pevonedistat was shown to decrease levels of NEDD8-cullin and increase the CRL substrate CDT1, a protein important for initiating DNA origins of replication and promoting the DNA re-replication phenotype characteristic of pevonedistat-treated cells [42]. Accumulation of CDT1 is hypothesized to induce DNA damage activating CHK1 and other pro-apoptotic mediators [18].
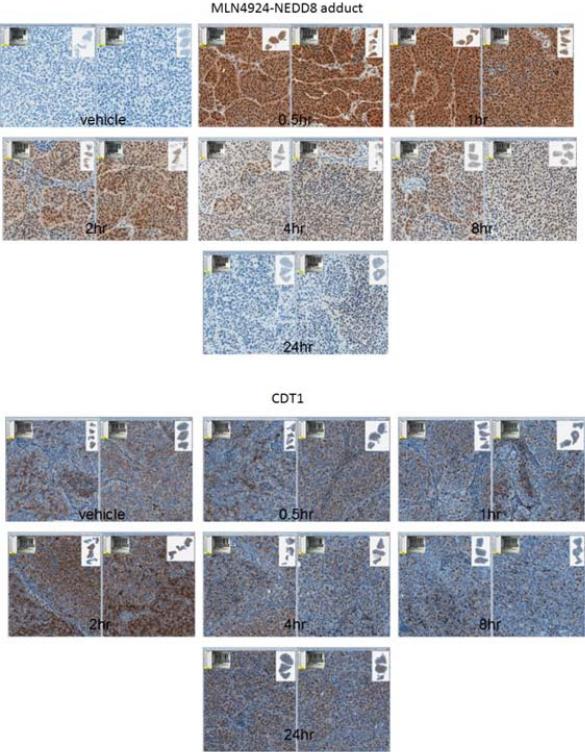
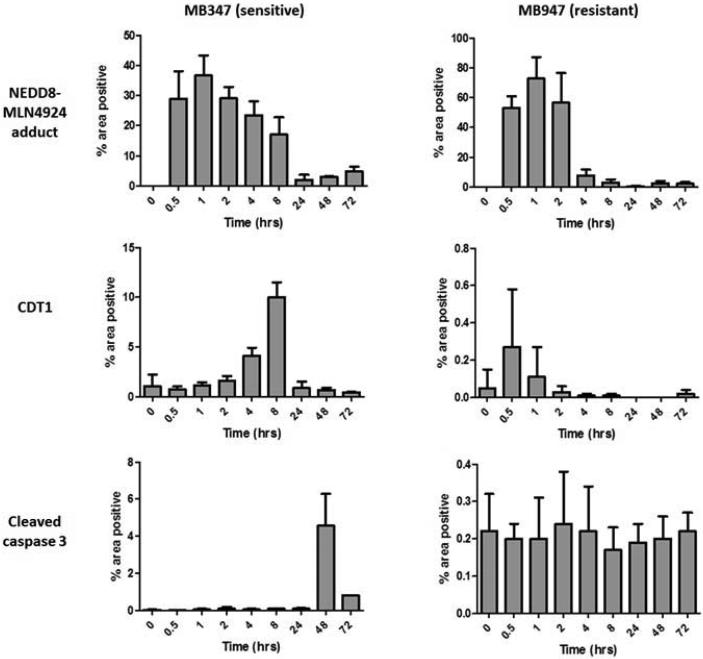
The PD effects of pevonedistat were evaluated in sensitive and resistant PDTX models of melanoma. PDTX were treated with a single dose of pevonedistat, and tumor tissues were harvested at various time points and analyzed by IHC for the pevonedistat-NEDD8 adduct, CDT1 and cleaved caspase 3 (Figure 4). Exposure to pevonedistat induced a significant and rapid increase in pevonedistat-NEDD8 adduct by 30 min in all models (Figure 4a). However, there was a faster depletion of the adduct in the resistant (MB947) compared to the sensitive (MB347) PDTX (Figure 4b); in the latter, pevonedistat-NEDD8 remained elevated for at least 8 hrs after treatment. Moreover, there was a rise in CDT1 levels in MB347, but not in MB947. Although there was low expression of cleaved caspase 3 in most tumors, this increased to significantly higher levels in one PDTX with tumor regression (MB347) (Figure 4b).
Figure 4.
Pevonedistat binds to its intended target and demonstrates expected pharmacodynamic (PD) effects in melanoma PDTX models. Sensitive and resistant PDTX were administered a single dose of pevonedistat, and tumor specimens were harvested at various time points from 30 min to 24 hrs. (a) Tumor sections were analyzed by immunohistochemistry (IHC) for NEDD8-pevonedistat adduct and CDT1, a protein substrate of cullin-RING ligases (CRLs). Representative results from a sensitive xenograft (MB1374) are shown. NEDD8-pevonedistat adduct increased rapidly at 30 min, followed by CDT1 accumulation up to 2 hrs after pevonedistat administration due to inhibition of CRL activity. (b) Levels of NEDD8-pevonedistat adduct, CDT1 and cleaved caspase 3 were quantified by the percent area of positive IHC staining at serial time points up to 72 hrs. At least 3 tumors were analyzed per time point (mean and standard deviation shown). Results from a sensitive (MB347) and a resistant (MB947) PDTX are shown.
These findings suggest that pevonedistat inhibits CRL activity and degradation of its protein substrates, leading to persistently elevated levels of pevonedistat-NEDD8 adduct and CDT1 in sensitive melanoma PDTX.
DISCUSSION
Disruption of protein homeostasis plays an important role in tumorigenesis, and the neddylation pathway is particularly relevant for the degradation of proteins needed for essential cell functions such as cell cycle and apoptosis [7]. Pevonedistat (MLN4924) is an investigational agent that specifically targets NAE, prevents NEDD8 conjugation and activation of CRLs, thereby disrupting the degradation of CRL-specific substrates. Unlike bortezomib, a proteasome inhibitor that targets general protein degradation, pevonedistat acts at an earlier step in this process by interfering with the neddylation pathway, and is thus able to inhibit the degradation of a more specific subset of proteins.
In this study, we utilized a large panel of melanoma cell lines and xenografts, including multiple PDTX, and demonstrated preclinical evidence of the anti-tumor effects of pevonedistat in a subgroup of these models. The findings support the neddylation pathway as a relevant and novel druggable target in this disease. The PD changes induced by pevonedistat in these melanoma models were consistent with its proposed mechanism of action, which was previously described [18]. Moreover, sensitivity and resistance to pevonedistat were associated with different profiles of gene expression and cellular pathways.
Consistent with the preclinical activity of pevonedistat in melanoma demonstrated in our study, the recently published Phase 1 trials of this agent showed clinical benefit in patients with melanoma on the basis of their high rate and durability of disease control [38, 39]. Particularly, in the melanoma-specific Phase 1 study, 48% of patients had stable disease, lasting ≥6.5 months in 4 patients [39]. Interestingly, all of these 4 patients had wild-type BRAF status. Moreover, the drug demonstrated an acceptable toxicity profile in previous trials. Therefore, the promising results of this preclinical study complement the recent early clinical trials of pevonedistat to provide the rationale for further development of this therapeutic agent in melanoma.
There was variable sensitivity to pevonedistat across melanoma cell lines and PDTX, which is commonly observed with targeted agents. This was similarly seen in the clinical setting in the fact that some of the pevonedistat-treated melanoma patients in the Phase 1 trials had stable disease, while others did not gain clinical benefit [38, 39]. Predictive markers that allow the selection of treatment-sensitive tumors will be needed to maximize the benefits of this NAE inhibitor. Response to targeted therapies is frequently modulated by aberrations in other interacting pathways, and global gene expression analysis may identify expression profiles that distinguish sensitive and resistant models. Our gene expression analysis yielded a set of genes that were differentially regulated in sensitive and resistant melanoma cell lines. GSEA using the KEGG and BioCARTA databases, which cover different spectrums of cellular pathways, showed that genes involved in cell cycle regulation, DNA repair, p53 signaling and spliceosome were more enriched in sensitive melanoma cell lines. In contrast, genes involved in immune response and cell adhesion were enriched in resistant cells. These results provide insights into the potential molecular differences between sensitivity and resistance, which need to be further validated. To our knowledge, this is the first study to utilize these genomic approaches to explore the landscape of global gene expression and cellular pathways associated with pevonedistat sensitivity and resistance in preclinical melanoma models. Furthermore, this is the first characterization of potential pathways of intrinsic resistance to NAE inhibition implicating the role of the immune system. In support of this, a recent study demonstrating that pevonedistat inhibits IFN-β production corroborates with the observed enrichment of immune-related genes in resistant melanoma cells [43]. In addition, other mechanisms of resistance to pevonedistat, which were not explored in this paper but should be considered particularly for cases of acquired resistance, include mutations in components of the neddylation machinery such as UBA3 (i.e. catalytic subunit of NAE) [44, 45].
Interestingly, a genomic siRNA screen of one melanoma cell line (A375) found that the knockdown of several genes with functions in cell cycle and DNA damage response sensitized cells to pevonedistat [25]. Although similar pathways were implicated, the specific genes that were most upregulated in sensitive melanoma cells in our study did not significantly overlap with the list of genes associated with greatest modulation of pevonedistat effects from the siRNA screen. Given the complexity of pathways that govern important cell processes such as cell cycle and DNA repair, which are regulated by both activating and inhibitory signaling proteins, it is not surprising that sensitivity to pevonedistat might require overexpression of some genes and repression of others. Moreover, these two strategies for analyzing genes that interact with pevonedistat were intrinsically different techniques. In the siRNA screen, only one gene in a single cell line was examined at any one time, and knockdown of a specific gene would likely induce changes in the network of interconnected signaling pathways that would inevitably affect drug sensitivity. On the other hand, our study used gene expression analysis and GSEA, in which a large panel of genes was interrogated simultaneously over multiple cell lines; this might be a more comprehensive approach to assess the genomic profile predictive of pevonedistat response but also leads to more complex results. Nevertheless, NAE inhibition certainly appears to interact with DNA repair, p53 signaling, and cell cycle checkpoints, although the precise changes induced by pevonedistat on these pathways have not yet been elucidated. While pevonedistat likely has many effects in the cell other than stabilization of CDT1 [25], sensitivity to this agent was nevertheless reduced with siRNA-silencing of CDT1, thus implying that the induction of DNA re-replication through CDT1 accumulation is an important mechanism of action.
BRAF mutation occurs in 40-60% of patients with melanoma and is the key predictive biomarker for BRAF inhibitors [46]. Due to the limited number of melanoma cell lines and PDTX with wild-type BRAF, we were unable to assess whether the effects of pevonedistat were modulated by BRAF status. Interestingly, the only BRAF wild-type PDTX in this study (MB1255) was also the most sensitive one to NAE inhibition, and the patients with the longest durations of stable disease in the Phase 1 study of pevonedistat in melanoma all had BRAF wild-type tumors [39]. However, further studies in BRAF wild-type melanoma models are necessary before any definite conclusion can be drawn.
Although a significant rate of stable disease was achieved, tumor response was limited in pevonedistat-treated melanoma patients in the Phase 1 clinical trial (e.g. only 1 partial response observed). The ability of pevonedistat to shrink tumors might be improved by combining it with other agents. In fact, the gene expression profiles of sensitive tumors provide insights into rational combination partners with this agent. The identification of DNA repair, replication and cell cycle pathways upregulated in pevonedistat-sensitive cells makes biological sense in light of the downstream effects of this NAE inhibitor. CDT1, which facilitates the initiation of DNA replication origins and is a CRL-specific degradation substrate, accumulates in pevonedistat-treated cells, thus inducing a DNA re-replication and S-phase defective phenotype that leads to DNA damage and apoptosis [18, 25, 42]. As such, pevonedistat activity might be enhanced by gene aberrations or drugs that impair DNA repair or promote apoptosis. Indeed, p53, BRCA1/2, transcription-coupled and base excision repair, as well as genes mediating DNA replication were shown by siRNA screen to be particularly important for synthetic lethality with pevonedistat in melanoma cells [25].
Evidence exists for the combination of pevonedistat with certain known therapeutic agents. Synergy was observed between pevonedistat and DNA-damaging drugs (e.g. mitomycin C, cisplatin, gemcitabine, cytarabine) in vitro [47]. In fact, pevonedistat and mitomycin C synergy in cell lines was dependent on genes responsible for DNA repair (e.g. ATR, BRCA1/2, transcription-coupled repair). Further, the additive effects of the combination of pevonedistat and platinum agents were clearly demonstrated in ovarian cancer models, including platinum-resistant models [48, 49]. Three currently accruing trials are investigating pevonedistat in combination with azacitadine in acute myelogenous leukemia (NCT01814826), with docetaxel, gemcitabine or carboplatin/paclitaxel in solid tumors (NCT01862328), and with azacitadine in high risk myelodysplastic syndromes, chronic myelomonocytic leukemia and low blast acute myelogenous leukemia in a randomized Phase 2 trial (vs. azacitadine alone in the comparator arm) (NCT02610777). Future studies should focus on rational combinations of pevonedistat to maximize its anti-tumor potential, particularly with novel agents targeting DNA damage or repair mechanisms.
Sensitivity to pevonedistat was clearly associated with PD effects in melanoma PDTX indicating target engagement and inhibition of the neddylation pathway. Our analysis revealed the following time course of molecular events: a rapid rise in pevonedistat-NEDD8 adduct within 30 minutes due to NAE inhibition, followed by the accumulation of the CRL-specific substrate CDT1 consistent with reported literature [18, 34, 42, 50]. These PD alterations confirmed that pevonedistat (given at 90mg/kg subcutaneously twice daily in our murine xenografts) bound its intended target in vivo and resulted in downstream molecular effects as expected based on its mechanism of action [18, 25]. These same PD markers, particularly pevonedistat-NEDD8 and CDT1 levels measured by IHC in patient tumor specimens, have been incorporated into clinical trials of this agent. Indeed, in the Phase 1 studies of pevonedistat in solid tumors and melanoma [38, 39], there was a significant increase in the protein expressions of both pevonedistat-NEDD8 adduct and CDT1 in post-dose tumor biopsies (after second dose of cycle 1) compared to baseline. In addition, gene transcripts regulated by NAE as detected by RT-PCR were increased in whole blood samples post-treatment, even at the lowest pevonedistat dose evaluated (50mg/m2) [39]. Thus, the consistency of PD changes in the preclinical and clinical settings not only strengthens the fact that this novel inhibitor engages its intended target and the proposed mechanism of action, but also establishes the validity of these PD markers for use in future clinical trials. While induction of apoptosis is believed to be a phenotypic endpoint of NAE inhibition, we observed low levels of cleaved caspase 3 during most of the time points after treatment with pevonedistat in the sensitive PDTX (i.e. MB347). Previous studies have demonstrated other non-apoptotic anti-tumor effects of pevonedistat, including senescence and autophagy, which might be relevant in this case [20, 26].
Pevonedistat had potent activity in sensitive melanoma cell lines with IC50s <0.3μM. This is superior to many targeted agents and is comparable to the potency of the BRAF inhibitor vemurafenib in BRAF-mutant cells, currently used for the standard treatment of BRAF V600E melanoma. For instance, the most sensitive melanoma cell line SK-MEL-1 in our panel demonstrated IC50 for pevonedistat <0.1μM, while vemurafenib has IC50 0.10μM and sorafenib (inhibitor of RAF and other kinases) has IC50 2.83μM in these cells [51]. Further, a given cell line may also have different sensitivities to different agents. SK-MEL-28 was relatively resistant to pevonedistat (IC50 1.5μM), but is highly sensitive to vemurafenib (IC50 0.04μM) [51]. This supports the concept that pevonedistat targets a unique pathway independent of the MAPK signaling cascade, and that the activities of pevonedistat and MAPK inhibitors are likely governed by distinct genetic predictors.
Both in vitro and in vivo preclinical models of melanoma were used in this study to evaluate the efficacy of pevonedistat. We found that the in vitro sensitivity of melanoma cell lines as determined by the cell viability assay did not consistently correlate with in vivo tumor response when the same cell lines were implanted into xenografts. In vivo models are more likely to recapitulate the true tumor microenvironment, where interactions between tumor cells, stroma and humoral factors (e.g. growth factors, cytokines) can occur and might partly account for the discrepancy observed in cell lines. It is interesting that cell adhesion pathways were preferentially upregulated in resistant melanoma cells in the GSEA. This implies that the tumor microenvironment might play a role in modulating the response to pevonedistat, although the mechanism of how signalling through cell adhesion molecules may interact with the neddylation pathway is unknown. Nevertheless, the observed discrepancy in sensitivity highlights the importance of including in vivo models in the preclinical evaluation of novel agents, while in vitro models provide a good starting point. Interestingly, in the PDTX studies, there was paradoxical doubling of tumor volume with pevonedistat treatment in the MB1227 xenograft. This might be related to a negative feedback interaction between neddylation and other oncogenic pathways, which might be activated when neddylation is repressed.
This study was the first comprehensive preclinical evaluation to characterize the activity of the NAE inhibitor pevonedistat specifically in melanoma, by utilizing a large set of in vitro and in vivo models, including several PDTX, as well as global gene expression and enrichment analyses. Our results provided evidence for the anti-tumor effects of pevonedistat in a subset of melanoma models, and showed that the neddylation pathway is a potential anti-cancer target in this malignancy. We determined that genes and pathways involved in DNA repair, replication and cell cycle regulation appear to be important determinants of sensitivity to pevonedistat and may provide rational targets for combination therapy. These findings are particularly significant and timely, as they firmly substantiate the results of the recently published Phase 1 trials of pevonedistat that demonstrated its clinical activity in a subgroup of melanoma patients. Taken together, the preclinical and clinical data strongly support further development of pevonedistat in melanoma. Future studies will also be needed to identify predictors of response and optimal combination partners for clinical evaluation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the PETT lab members for critical comments
Funding
This work was supported by a commercial research grant (A.C. Tan, T.M. Pitts, J.J. Tentler and S.G. Eckhardt) and by grants from the University of Colorado Cancer Center and Millennium Pharmaceuticals, Inc.
Footnotes
Conflicts of interest
S.J. Blakemore, P.G. Smith, A. McDonald and A. Berger were employed by Millennium Pharmaceuticals Inc. at the time of conducting this research.
COMPLIANCE WITH ETHICAL STANDARDS
Conflict of Interest
Kit Man Wong declares that she has no conflict of interest. Lindsey N. Micel declares that she has no conflict of interest. Heather M. Selby declares that she has no conflict of interest. Aik Choon Tan declares that he has no conflict of interest. Todd M. Pitts declares that he has no conflict of interest. Stacey M. Bagby declares that she has no conflict of interest. Anna Spreafico declares that she has no conflict of interest. Peter J. Klauck declares that he has no conflict of interest. Stephen J. Blakemore was employed by Millennium Pharmaceuticals Inc. at the time of conducting this research. Peter F. Smith was employed by Millennium Pharmaceuticals Inc. at the time of conducting this research. Alice McDonald was employed by Millennium Pharmaceuticals Inc. at the time of conducting this research. Allison Berger was employed by Millennium Pharmaceuticals Inc. at the time of conducting this research. John J. Tentler declares that he has no conflict of interest. S. Gail Eckhardt declares that she has no conflict of interest.
Ethical Approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All animal studies described in this study were conducted at the University of Colorado Anschutz Medical Campus in accordance with the National Institutes of Health guidelines for the care and use of laboratory animals, and animals were housed in a facility accredited by the American Association for Accreditation of Laboratory Animal Care (3-5 mice per cage).
Informed Consent
Not applicable (study does not involve human subjects)
REFERENCES
- 1.Schwartz AL, Ciechanover A. Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annual Review of Pharmacology & Toxicology. 2009;49:73. doi: 10.1146/annurev.pharmtox.051208.165340. [DOI] [PubMed] [Google Scholar]
- 2.Nawrocki ST, Griffin P, Kelly KR, Carew JS. MLN4924: a novel first-in-class inhibitor of NEDD8-activating enzyme for cancer therapy. Expert opinion on investigational drugs. 2012;21(10):1563. doi: 10.1517/13543784.2012.707192. [DOI] [PubMed] [Google Scholar]
- 3.Richardson PGSPSMWIDSEAFTHJLB-YDLSGH. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. New England Journal of Medicine. 2005;352(24):2487. doi: 10.1056/NEJMoa043445. [DOI] [PubMed] [Google Scholar]
- 4.Kane RC, Dagher R, Farrell A, Ko CW, Sridhara R, Justice R, Pazdur R. Bortezomib for the treatment of mantle cell lymphoma. Clinical Cancer Research. 2007;13(18 Pt 1):5291. doi: 10.1158/1078-0432.CCR-07-0871. [DOI] [PubMed] [Google Scholar]
- 5.Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nature ReviewsDrug Discovery. 2006;5(7):596. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka T, Nakatani T, Kamitani T. Inhibition of NEDD8-conjugation pathway by novel molecules: potential approaches to anticancer therapy. Molecular Oncology. 2012;6(3):267. doi: 10.1016/j.molonc.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147(2):459. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guardavaccaro D, Pagano M. Oncogenic aberrations of cullin-dependent ubiquitin ligases. Oncogene. 2004;23(11):2037. doi: 10.1038/sj.onc.1207413. [DOI] [PubMed] [Google Scholar]
- 9.Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nature cell biology. 2004;6(10):1003. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- 10.Chiba T, Tanaka K. Cullin-based ubiquitin ligase and its control by NEDD8-conjugating system. Current Protein & Peptide Science. 2004;5(3):177. doi: 10.2174/1389203043379783. [DOI] [PubMed] [Google Scholar]
- 11.Read MA, Brownell JE, Gladysheva TB, Hottelet M, Parent LA, Coggins MB, Pierce JW, Podust VN, Luo RS, Chau V, et al. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Molecular & Cellular Biology. 2000;20(7):2326. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Podust VN, Brownell JE, Gladysheva TB, Luo RS, Wang C, Coggins MB, Pierce JW, Lightcap ES, Chau V. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(9):4579. doi: 10.1073/pnas.090465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen LC, Manjeshwar S, Lu Y, Moore D, Ljung BM, Kuo WL, Dairkee SH, Wernick M, Collins C, Smith HS. The human homologue for the Caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer research. 1998;58(16):3677. [PubMed] [Google Scholar]
- 14.Melchor L, Saucedo-Cuevas LP, Munoz-Repeto I, Rodriguez-Pinilla SM, Honrado E, Campoverde A, Palacios J, Nathanson KL, Garcia MJ, Benitez J. Comprehensive characterization of the DNA amplification at 13q34 in human breast cancer reveals TFDP1 and CUL4A as likely candidate target genes. Breast Cancer Research. 2009;11(6):R86. doi: 10.1186/bcr2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Q, Yu GY, Shi JY, Li LH, Zhang WJ, Wang ZC, Yang LX, Duan M, Zhao H, Wang XY, et al. Neddylation pathway is up-regulated in human intrahepatic cholangiocarcinoma and serves as a potential therapeutic target. Oncotarget. 2014;5(17):7820. doi: 10.18632/oncotarget.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Li L, Liang Y, Li C, Zhao H, Ye D, Sun M, Jeong LS, Feng Y, Fu S, et al. Targeting the neddylation pathway to suppress the growth of prostate cancer cells: therapeutic implication for the men's cancer. BioMed Research International. 2014;2014:974309. doi: 10.1155/2014/974309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, Wang M, Yu G, Chen P, Li H, Wei D, Zhu J, Xie L, Jia H, Shi J, et al. Overactivated neddylation pathway as a therapeutic target in lung cancer. Journal of the National Cancer Institute. 2014;106(6):ju083. doi: 10.1093/jnci/dju083. [DOI] [PubMed] [Google Scholar]
- 18.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458(7239):732. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 19.Bohnsack RN, Haas AL. Conservation in the mechanism of Nedd8 activation by the human AppBp1-Uba3 heterodimer. Journal of Biological Chemistry. 2003;278(29):26823. doi: 10.1074/jbc.M303177200. [DOI] [PubMed] [Google Scholar]
- 20.Lin JJ, Milhollen MA, Smith PG, Narayanan U, Dutta A. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer research. 2010;70(24):10310. doi: 10.1158/0008-5472.CAN-10-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Truong LN, Wu X. Prevention of DNA re-replication in eukaryotic cells. Journal of molecular cell biology. 2011;3(1):13. doi: 10.1093/jmcb/mjq052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li JM, Jin J. CRL Ubiquitin Ligases and DNA Damage Response. Frontiers in oncology. 2012;2:29. doi: 10.3389/fonc.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hannah J, Zhou P. Regulation of DNA damage response pathways by the cullin-RING ubiquitin ligases. DNA repair. 2009;8(4):536. doi: 10.1016/j.dnarep.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abbas T, Dutta A. CRL4Cdt2: master coordinator of cell cycle progression and genome stability. Cell cycle (Georgetown, Tex) 2011;10(2):241. doi: 10.4161/cc.10.2.14530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blank JL, Liu XJ, Cosmopoulos K, Bouck DC, Garcia K, Bernard H, Tayber O, Hather G, Liu R, Narayanan U, et al. Novel DNA damage checkpoints mediating cell death induced by the NEDD8-activating enzyme inhibitor MLN4924. Cancer research. 2013;73(1):225. doi: 10.1158/0008-5472.CAN-12-1729. [DOI] [PubMed] [Google Scholar]
- 26.Luo Z, Pan Y, Jeong LS, Liu J, Jia L. Inactivation of the Cullin (CUL)-RING E3 ligase by the NEDD8-activating enzyme inhibitor MLN4924 triggers protective autophagy in cancer cells. Autophagy. 2012;8(11):1677. doi: 10.4161/auto.21484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, Oberheu K, Padmanabhan S, O'Dwyer M, Nawrocki ST, et al. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115(18):3796. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
- 28.McMillin DW, Jacobs HM, Delmore JE, Buon L, Hunter ZR, Monrose V, Yu J, Smith PG, Richardson PG, Anderson KC, et al. Molecular and cellular effects of NEDD8-activating enzyme inhibition in myeloma. Molecular Cancer Therapeutics. 2012;11(4):942. doi: 10.1158/1535-7163.MCT-11-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E, Langston SP, et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood. 2010;116(9):1515. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- 30.Pan WW, Zhou JJ, Yu C, Xu Y, Guo LJ, Zhang HY, Zhou D, Song FZ, Fan HY. Ubiquitin E3 ligase CRL4(CDT2/DCAF2) as a potential chemotherapeutic target for ovarian surface epithelial cancer. Journal of Biological Chemistry. 2013;288(41):29680. doi: 10.1074/jbc.M113.495069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo Z, Yu G, Lee HW, Li L, Wang L, Yang D, Pan Y, Ding C, Qian J, Wu L, et al. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer research. 2012;72(13):3360. doi: 10.1158/0008-5472.CAN-12-0388. [DOI] [PubMed] [Google Scholar]
- 32.Zhao L, Yue P, Lonial S, Khuri FR, Sun SY. The NEDD8-activating enzyme inhibitor, MLN4924, cooperates with TRAIL to augment apoptosis through facilitating c-FLIP degradation in head and neck cancer cells. Molecular Cancer Therapeutics. 2011;10(12):2415. doi: 10.1158/1535-7163.MCT-11-0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackintosh C, Garcia-Dominguez DJ, Ordonez JL, Ginel-Picardo A, Smith PG, Sacristan MP, de Alava E. WEE1 accumulation and deregulation of S-phase proteins mediate MLN4924 potent inhibitory effect on Ewing sarcoma cells. Oncogene. 2013;32(11):1441. doi: 10.1038/onc.2012.153. [DOI] [PubMed] [Google Scholar]
- 34.Wei D, Li H, Yu J, Sebolt JT, Zhao L, Lawrence TS, Smith PG, Morgan MA, Sun Y. Radiosensitization of human pancreatic cancer cells by MLN4924, an investigational NEDD8-activating enzyme inhibitor. Cancer research. 2012;72(1):282. doi: 10.1158/0008-5472.CAN-11-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang D, Tan M, Wang G, Sun Y. The p21-dependent radiosensitization of human breast cancer cells by MLN4924, an investigational inhibitor of NEDD8 activating enzyme. PLoS ONE [Electronic Resource] 2012;7(3):e34079. doi: 10.1371/journal.pone.0034079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah JJ, Jakubowiak AJ, O'Connor OA, Orlowski RZ, Harvey RD, Smith MR, Lebovic D, Diefenbach C, Kelly K, Hua Z, et al. Phase I Study of the Novel Investigational NEDD8-Activating Enzyme Inhibitor Pevonedistat (MLN4924) in Patients with Relapsed/Refractory Multiple Myeloma or Lymphoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015 doi: 10.1158/1078-0432.CCR-15-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swords RT, Erba HP, DeAngelo DJ, Bixby DL, Altman JK, Maris M, Hua Z, Blakemore SJ, Faessel H, Sedarati F, et al. Pevonedistat (MLN4924), a First-in-Class NEDD8-activating enzyme inhibitor, in patients with acute myeloid leukaemia and myelodysplastic syndromes: a phase 1 study. Br J Haematol. 2015;169(4):534–543. doi: 10.1111/bjh.13323. [DOI] [PubMed] [Google Scholar]
- 38.Sarantopoulos J, Shapiro GI, Cohen RB, Clark JW, Kauh JS, Weiss GJ, Cleary JM, Mahalingam D, Pickard MD, Faessel HM, et al. Phase I Study of the Investigational NEDD8-Activating Enzyme Inhibitor Pevonedistat (TAK-924/MLN4924) in Patients with Advanced Solid Tumors. Clin Cancer Res. 2016;22(4):847–857. doi: 10.1158/1078-0432.CCR-15-1338. [DOI] [PubMed] [Google Scholar]
- 39.Bhatia S, Pavlick AC, Boasberg P, Thompson JA, Mulligan G, Pickard MD, Faessel H, Dezube BJ, Hamid O. A phase I study of the investigational NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in patients with metastatic melanoma. Invest New Drugs. 2016 doi: 10.1007/s10637-016-0348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olszanski AJ. Current and future roles of targeted therapy and immunotherapy in advanced melanoma. Journal of Managed Care Pharmacy. 2014;20(4):346. doi: 10.18553/jmcp.2014.20.4.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Molecular cell. 2010;37(1):102. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 42.Milhollen MA, Narayanan U, Soucy TA, Veiby PO, Smith PG, Amidon B. Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer research. 2011;71(8):3042. doi: 10.1158/0008-5472.CAN-10-2122. [DOI] [PubMed] [Google Scholar]
- 43.Song H, Huai W, Yu Z, Wang W, Zhao J, Zhang L, Zhao W. MLN4924, a First-in-Class NEDD8-Activating Enzyme Inhibitor, Attenuates IFN-β Production. J Immunol. 2016 doi: 10.4049/jimmunol.1501752. [DOI] [PubMed] [Google Scholar]
- 44.Xu GW, Toth JI, da Silva SR, Paiva SL, Lukkarila JL, Hurren R, Maclean N, Sukhai MA, Bhattacharjee RN, Goard CA, et al. Mutations in UBA3 confer resistance to the NEDD8-activating enzyme inhibitor MLN4924 in human leukemic cells. PLoS ONE [Electronic Resource] 2014;9(4):e93530. doi: 10.1371/journal.pone.0093530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Milhollen MA, Thomas MP, Narayanan U, Traore T, Riceberg J, Amidon BS, Bence NF, Bolen JB, Brownell J, Dick LR, et al. Treatment-emergent mutations in NAEbeta confer resistance to the NEDD8-activating enzyme inhibitor MLN4924. Cancer Cell. 2012;21(3):388. doi: 10.1016/j.ccr.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 46.Long GV, Menzies AM, Nagrial AM, Haydu LE, Hamilton AL, Mann GJ, Hughes TM, Thompson JF, Scolyer RA, Kefford RF. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. Journal of Clinical Oncology. 2011;29(10):1239. doi: 10.1200/JCO.2010.32.4327. [DOI] [PubMed] [Google Scholar]
- 47.Garcia K, Blank JL, Bouck DC, Liu XJ, Sappal DS, Hather G, Cosmopoulos K, Thomas MP, Kuranda M, Pickard MD, et al. Nedd8-activating enzyme inhibitor MLN4924 provides synergy with mitomycin C through interactions with ATR, BRCA1/BRCA2, and chromatin dynamics pathways. Molecular Cancer Therapeutics. 2014;13(6):1625. doi: 10.1158/1535-7163.MCT-13-0634. [DOI] [PubMed] [Google Scholar]
- 48.Jazaeri AA, Shibata E, Park J, Bryant JL, Conaway MR, Modesitt SC, Smith PG, Milhollen MA, Berger AJ, Dutta A. Overcoming platinum resistance in preclinical models of ovarian cancer using the neddylation inhibitor MLN4924. Molecular Cancer Therapeutics. 2013;12(10):1958. doi: 10.1158/1535-7163.MCT-12-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nawrocki ST, Kelly KR, Smith PG, Espitia CM, Possemato A, Beausoleil SA, Milhollen M, Blakemore S, Thomas M, Berger A, et al. Disrupting protein NEDDylation with MLN4924 is a novel strategy to target cisplatin resistance in ovarian cancer. Clinical Cancer Research. 2013;19(13):3577. doi: 10.1158/1078-0432.CCR-12-3212. [DOI] [PubMed] [Google Scholar]
- 50.Jia L, Li H, Sun Y. Induction of p21-dependent senescence by an NAE inhibitor, MLN4924, as a mechanism of growth suppression. Neoplasia (New York) 2011;13(6):561. doi: 10.1593/neo.11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Genomics of Drug Sensitivity in Cancer. [ http://www.cancerrxgene.org/]
- 52.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.