Abstract
Cytokinins are important signals that participate in different plant processes, and are well known for their strong influence in plant development. With the years, knowledge has been built about their effects, chemical nature, metabolism, and signaling mechanisms. However, one aspect about cytokinins that has been lagging behind is cytokinin transport. Recent reports are providing more information about how cytokinins are transported and how their transport is connected to their effects in development. This review provides a general overview of what is known about cytokinin transport, with a focus on the latest reports.
Keywords: cytokinin transport, cytokinin uptake, cytokinin export, ENT, PUP, ABCG, cytokinin signaling
For a long time, the molecular mechanisms of cytokinin action were poorly understood, because information about the physiological role of cytokinins came from external application experiments, and did not involve the use of plants with altered levels of or responses to cytokinin. It took over 40 years to identify the first genes involved in cytokinin signaling. In 1996, using an activation tagging strategy, Kakimoto isolated Arabidopsis thaliana mutants exhibiting cytokinin responses that were independent of cytokinin application. In this screen, CYTOKININ INDEPENDENT 1 (CKI1) was identified (Kakimoto, 1996). Though unlikely a cytokinin receptor (Yamada, 2001), CKI1, encoding an integral membrane protein similar to bacterial two-component His kinases, provided the first hint about cytokinin perception and signaling. The first authentic cytokinin receptor, CYTOKININ RESPONSE 1 (CRE1), was identified by Inoue et al. (2001) by screening for Arabidopsis mutants whose hypocotyl explants failed to form shoots in medium with high cytokinin/auxin ratio, and it was found to sense cytokinins in heterologous systems (Inoue et al., 2001; Suzuki et al., 2001; reviewed in Heyl et al., 2012).
Further research on cytokinin receptors yielded insights into the biochemical properties and functions of these sensor histidine kinases. Their nature suggested that other downstream elements of a two-component signaling pathway (TCS) could participate in the signal transduction, such as Arabidopsis histidine-containing phosphotransfer proteins (AHPs) and Arabidopsis response regulators (ARRs) (summarized in Kieber and Schaller, 2010; Hwang et al., 2012).
Cytokinin-overproducing plants obtained by activation tagging helped to identify an endogenous plant isopentenyl transferase (IPT) that produced active CKs in planta (Zubko et al., 2002). Moreover, additional knowledge about the biological function of cytokinins was obtained from cytokinin-deficient plants, such as transgenic tobacco plants overexpressing genes that encode catabolic cytokinin oxidase (CKX) enzymes from Arabidopsis (Werner et al., 2003).
Further genetic and molecular analyses boosted significant progress in unraveling cytokinin metabolism, perception, and signal transduction (summarized in the following reviews: Sakakibara, 2006; Hirose et al., 2008; Del Bianco et al., 2013; El-Showk et al., 2013; Kieber and Schaller, 2014; Zürcher and Müller, 2016).
However, less knowledge was available about the molecular mechanisms involved in cytokinin transport. Mainly, three kinds of cytokinin transporters have been reported to date: purine permeases (PUP), equilibrative nucleoside transporters (ENT), and G subfamily ATP-binding cassette (ABCG) transporters (reviewed in Kudo et al., 2010; Girke et al., 2014; Borghi et al., 2015; summarized in Table 1).
Table 1.
Cytokinin transporter families.
| Transporter family | Evidence of cytokinin transport activity | Members with experimental evidence for cytokinin transport | Assayed cytokinin∗, K (when available), system | Expression pattern | Subcellular localization | Reference | |
|---|---|---|---|---|---|---|---|
| PUP | Cytokinin uptake (zeatin, kinetin, isopentenyladenine benzyladenine, and some ribosides) | AtPUP1 | Ki = 20–35 μM (kinetin and zeatin vs. adenine transport), Km = 40 μM (trans-zeatin), in yeast | Leaves, stems, flowers, siliques, hydathodes, stigmata | Gillissen et al., 2000; Bürkle et al., 2003 | Review: Girke et al., 2014 | |
| AtPUP2 | Adenine transport inhibition by iPA, trans and cis-zeatin, kinetin, BAP and tZR, in yeast | Flowers, roots, leaf vasculature (phloem), pollen | Bürkle et al., 2003 | ||||
| AtPUP14 | trans-zeatin, iPA, BA, in plant-derived protoplasts and microsomes | Many organs and stages | Plasma membrane | Zürcher et al., 2016 | |||
| OsPUP7 | Higher iP and iPR levels in the mutant plant | Stigma, style, stamens, hull vein, vascular bundle of roots, culms and leaves | Qi and Xiong, 2013 | ||||
| ENT | Cytokinin uptake (ribosides, inactive transport forms) | AtENT8 AtENT3 | Reduced uptake efficiency of iPR, in mutant hypocotyls | ENT8: SAM, hypocotyl, petioles, stems, flowers, siliques | Sun et al., 2005 | Review: Girke et al., 2014 | |
| AtENT6 | Km = 17 μM iPR, Km = 630 μM tZR, in yeast | Root, leaf, and flower vasculature, and stomata | Plasma membrane | Wormit et al., 2004; Hirose et al., 2008 | |||
| OsENT2 | Km = 32 μM iPR, Km = 660 μM tZR, in yeast | Vasculature (grains, roots, stems, and leaves), and roots (stele, lateral roots) | Hirose et al., 2005 | ||||
| ABCG | Cytokinin export (trans-zeatin type) | AtABCG14 | Reduced trans-zeatin root-to-shoot translocation in mutant plants | Mainly roots (phloem, procambium and pericycle) | Plasma membrane | Ko et al., 2014; Zhang et al., 2014 Reviews: Borghi et al., 2015; Hwang et al., 2016 | |
∗More biochemical data for transport of other molecules, such as adenine, is available for these transporters, but this table only contains data related to cytokinin transport. iPA or iP, isopentenyladenine; iPR, isopentenyladenosine; BAP or BA, benzylaminopurine or benzyladenine; tZR, trans-zeatin riboside.
The first PUP transporter was identified through a complementation assay using an adenine uptake deficient yeast mutant transformed with an Arabidopsis cDNA library. Competition assays indicated that, besides adenine, PUP1 could also transport the cytokinins kinetin and zeatin, and at a lower extent their ribosides (Gillissen et al., 2000). In this way, a new membrane protein family of 21 members was identified (Schwacke et al., 2003). Further analyses of the transport function of two other members of the family, PUP2 and PUP3, showed that PUP2 could also transport cytokinins (trans- and cis-zeatin, kinetin, benzyladenine, isopentenyladenine, and to a lesser extent trans-zeatin riboside), while no transport evidence could be obtained for PUP3 (Bürkle et al., 2003). Moreover PUP1. PUP2 and PUP3 presented very different expression patterns (in hydathodes and stigmatic papillae, vasculature and pollen, respectively), giving a hint about their physiological roles in the plant (Bürkle et al., 2003). Later studies on cytokinin uptake kinetics in Arabidopsis cell cultures and seedlings, suggested the existence of low and high-affinity transport systems. Furthermore, an exploration of expression data of more than 12 PUP genes revealed that they were expressed in diverse tissues and levels (Cedzich et al., 2008). Hildreth et al. (2011) have proposed that PUPs are only present in vascular plants, where they can transport also other molecules (besides cytokinins) such as caffeine and nicotine (Gillissen et al., 2000; Bürkle et al., 2003; Hildreth et al., 2011; Jelesko, 2012; Qi and Xiong, 2013; reviewed in Girke et al., 2014).
More evidence for PUP function in cytokinin transport has been obtained from studies in rice. Qi and Xiong (2013) identified 12 OsPUP members, which also present varied expression patterns in different tissues and developmental stages. They characterized OsPUP7, exploring its expression and making use of an available insertional mutant (Qi and Xiong, 2013). OsPUP7 was found to be expressed mainly in the pistils, stamens, and the vasculature of different organs, and to be induced by the addition of abscisic acid and kinetin. Because isopentenyladenine and isopentenyladenosine levels were altered in the mutant, the authors proposed that OsPUP7 is also able to transport cytokinins and derivatives. However, levels of trans-zeatin and trans-zeatin riboside were not affected, leading the authors to suggest that this transporter has a selective transport activity. The insertional mutant presented clear alterations in organ size and flowering time, and had increased sensitivity to abiotic stresses (osmotic and saline stress) and hormone treatments (abscisic acid and benzyladenine). However, how the function of these transporters was affecting the cytokinin signaling response of cells during plant development was unknown.
Recently, a report by Zürcher et al. (2016) shed light on these questions. To identify regulatory steps in the cytokinin signaling response pathway, they used the Arabidopsis embryo at the heart stage as a model. When embryos were treated with the cytokinin benzyladenine, the signal of the cytokinin-sensitive two-component sensor TCSn:GFP (Zürcher et al., 2013) expanded from the provascular tissue to other tissues. However, no response was detected in the prospective cotyledons. The authors noticed that the lack of TCSn:GFP response in the prospective cotyledons was most likely not due to a lack of cytokinin receptors or downstream signaling elements. The Arabidopsis histidine kinase 4 (AHK4/CRE1) cytokinin receptor is expressed there. Moreover, downstream signaling appeared to be functional as overexpression of the cytokinin-independent constitutively active CKI1 kinase, which acts upstream of the AHPs (Deng et al., 2010), caused ubiquitous TCSn:GFP activation. Therefore, the authors hypothesized that cytokinin transporters could be responsible for the lack of TCSn:GFP activation in the prospective cotyledons. To test their hypothesis, they surveyed the expression patterns of PUP transporter genes to find one expressed in this tissue. Among them, they found PUP14 to be highly expressed in many stages and tissues, including the prospective cotyledons suggesting that it could be negatively affecting the cytokinin response in this tissue. Indeed, induction of PUP14 silencing (using an artificial microRNA, amirPUP14) caused ectopic cytokinin signaling in the prospective cotyledons, indicating that PUP14 could restrict cytokinin signaling. Later in development, these tissues with ectopic signaling presented morphological defects (also observed in the nascent root meristem). Furthermore, constitutive PUP14 ectopic expression had deleterious effects, and its induction caused a reduction in cytokinin signaling. Uptake experiments using labeled trans-zeatin confirmed that PUP14, which was localized at the plasma membrane, could transport this cytokinin. Moreover, adenine and bioactive cytokinins (isopentenyladenine and benzyladenine) could inhibit trans-zeatin transport in competition assays. In contrast, neither the transport form trans-zeatin riboside, auxin, nor allantoin, inhibited uptake by PUP14. The authors reasoned that uptake of bioactive cytokinins by PUP14, together with the effects of PUP14 down or up-regulation in the expression TCSn:GFP reporter line, could suggest that this transporter could be depleting cytokinins from the apoplast, making them unavailable to be sensed by plasma membrane-localized receptors. To test this, they compared the effect of PUP14 uptake, to the effects of CKXs (cytokinin-degrading enzymes) targeted for secretion to the apoplast or localized in the cytoplasm, in the cytokinin signaling response. They found that both enzymatic clearing of external cytokinins, and cytokinin uptake had a similar effect on the cytokinin signaling response, suggesting that apoplastic and not cytoplasmic cytokinins initiate signaling as reported by the TCSn:GFP line.
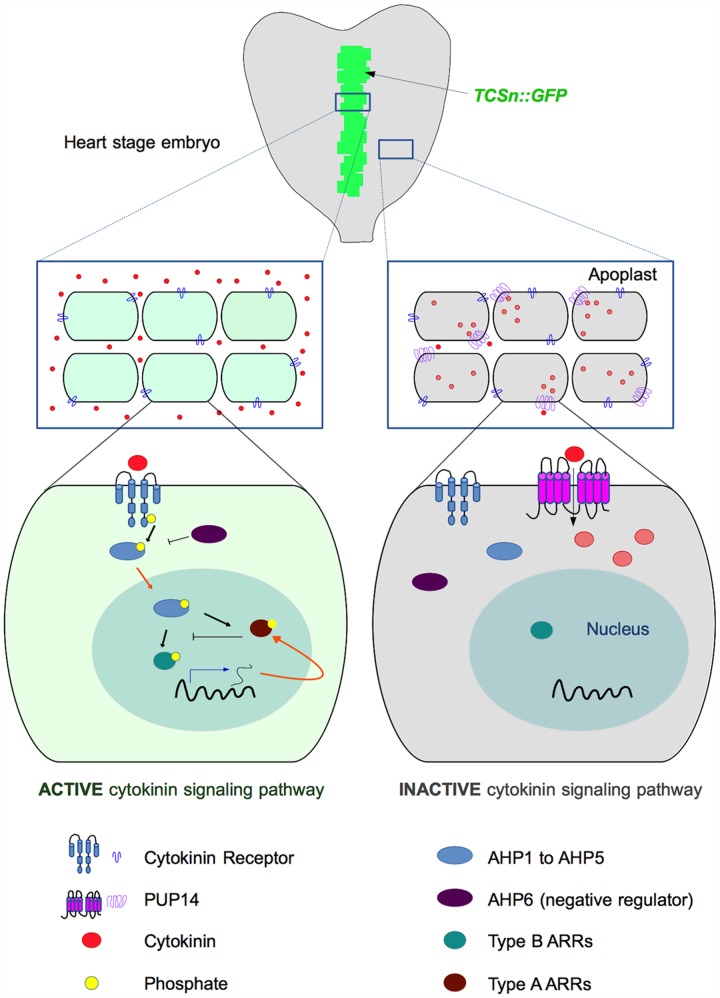
Therefore, the authors proposed a model in which PUP14-mediated uptake of cytokinins depletes them from the apoplast, constraining, and therefore contributing to define the topography of cytokinin signaling responses that guide morphogenesis in a developing plant (the model proposed is depicted in Figure 1).
FIGURE 1.
Proposed mechanism of cytokinin-signaling restriction by PUP14 (based on models published by Bürkle et al., 2003; Zürcher et al., 2016). Top: Diagram depicting the region of cytokinin response (indicated in green) revealed by the TCSn:GFP marker in a heart-stage embryo. Zürcher et al. (2016) proposed that PUP14 import of apoplastic cytokinins to the cytosol causes their depletion from the apoplast and makes them unavailable for the membrane-bound AHK receptors that initiate the signaling cascade that activates the TCS response. In the middle and bottom diagrams, cytokinin responsive cells are light green, while non-responsive cells are represented in gray. In the responsive cells (left), cytokinins are perceived by the AHK receptors, which autophosphorylate and subsequently transfer the phosphate (black arrows represent phosphorylation) to Arabidopsis histidine-containing phosphotransfer proteins (AHP1 to AHP5). Then, the AHPs enter the nucleus (orange arrows represent intracellular traffic) and phosphorylate type B Arabidopsis response regulators (ARRs), which activate transcription (blue arrow) of CK response genes, reported by the TCSn:GFP marker line and including type A ARRs (negative regulators of cytokinin signaling, as indicated by blocked lines). In the right part of the diagram, the expression of PUP14 promotes the depletion of cytokinins from the apoplast and the cytokinin response (reported by the TCSn:GFP marker) is not activated, even when exogenous cytokinins are applied. Cytokinins are represented by red circles, and internalized cytokinins are represented by light red circles. For simplicity, only one TCS element or PUP14 transporter per cell is depicted.
With the proposed model, new questions emerge. For example, it would be interesting to assess the ratio of transporters vs. receptors in the cell, or at least the tissue, under study to provide a better understanding of their competing interactions with cytokinins, especially considering that high affinities have been reported for the cytokinin receptors (in the low nM range, Yamada, 2001; Romanov et al., 2005, 2006; Lomin et al., 2015, reviewed in Heyl et al., 2012). Affinities for cytokinins have been measured for only two PUPs (PUP1 and 2, K-values in the μM range, Gillissen et al., 2000; Bürkle et al., 2003) using different methods, and they do not seem to be comparable to the reported receptor affinities. Therefore, to better understand the physiological role of PUP14 and further support the proposed model, it would be very informative to measure its affinity for cytokinins and compare it to the affinities of the receptors under the same conditions. In this comparison, conditions such as pH should be considered. In the case of cytokinin receptors, changes in their sensitivity depending on pH have been reported (Romanov et al., 2006; Lomin et al., 2015), and this is a relevant factor to consider, since different cellular and extracellular compartments have different pHs (Lomax et al., 1985; Li et al., 2005). Previous studies have reported cytokinin receptor localization, and cytokinin sensing, mostly at the endoplasmic reticulum (Caesar et al., 2011; Lomin et al., 2011; Wulfetange et al., 2011). In this regard, we are still missing key information that can reconcile the data that indicates that cytokinins are sensed at the plasma membrane, as in the proposed model for PUP14 function, with the data that indicates that cytokinins are sensed at the endoplasmic reticulum, as reported in the works cited above. Here, further research in cytokinin transport may also contribute to unravel this enigma, answering questions regarding how cytokinins reach each of these locations inside and outside the cell, and the advantages of a sophisticated system of cytokinin depletion over the downregulation of receptors in a group of cells.
The second family of cytokinin transporters, the ENTs, were found initially as putative plant nucleoside transporters with similarity to the human transporters hENT1 and hENT2, and reported to localize at the plasma membrane (Li and Wang, 2000). Complementation of a yeast mutant unable to synthesize adenine, with Arabidopsis cDNA, demonstrated that AtENT1 could transport adenosine (Möhlmann et al., 2001; reviewed in Girke et al., 2014), while its silencing affected pollen germination (Bernard et al., 2011). FURTHER characterization of the nucleoside transport capabilities and expression of other AtENTs followed (Li et al., 2003; Wormit et al., 2004; Traub et al., 2007). The finding of an AtENT8 mutation as a suppressor of the altered phenotypes of a cytokinin overproducing plant, together with the characterization of AtENT6 and the rice OsENT2 in yeast, provided evidence of a role for ENTs in cytokinin transport (Hirose et al., 2005, 2008; Sun et al., 2005). For AtENT3, a loss-of-function mutant presented reduced cytokinin uptake (Sun et al., 2005), but cytokinin transport activity was not detected when the transporter was expressed in yeast (Hirose et al., 2008). Until now, the data indicates that the cytokinin species transported by a subset of the ENTs are the riboside forms of isopentenyladenine and trans-zeatin (isopentenyladenine riboside and trans-zeatin riboside) (Hirose et al., 2005, 2008; Sun et al., 2005; reviewed in Girke et al., 2014). These forms are not active cytokinins, and are considered to be transport forms (reviewed in Hirose et al., 2008; Lomin et al., 2015 reported in planta binding assays to receptors showing strongly reduced affinity of riboside forms). Also, as reported for PUPs, ENTs can transport many non-cytokinin molecules, opening the question of whether only some members are involved in cytokinin transport, and whether they may play broader roles, not specific to cytokinin homeostasis.
Current evidence indicates that both PUP and ENT transporters participate in cytokinin uptake. But are there transporters that have been shown to export cytokinins?
Different cytokinin species are translocated between different plant organs. trans-zeatin types are transported via the xylem from roots to shoots and isopentenyladenine types via the phloem systemically or from shoots to roots (reviewed in Kudo et al., 2010; Zürcher and Müller, 2016). Therefore, the existence of cytokinin exporters was suspected, but remained an enigma until 2014. Then, two groups independently reported that the Arabidopsis ABCG14 transporter functions as a cytokinin exporter that loads cytokinin in the root xylem (Ko et al., 2014; Zhang et al., 2014). In one case, using a genetic approach to characterize the ABCG proteins, Zhang et al. (2014) found that knock-out mutations in ABCG14 led to defects in the long-distance translocation and distribution of the root-synthesized trans-zeatin type cytokinins, evidenced by their overaccumulation in the root and reduced content in the shoot. These defects caused morphological alterations such as small inflorescences and rosettes, slender stems with altered vasculature, and short primary roots. Efflux pump activity was demonstrated by employing radiolabeled trans-zeatin species in in planta feeding and detached leaf assays (Zhang et al., 2014). In the other case, to elucidate the molecular mechanism underlying the translocation of cytokinins, Ko et al. (2014) searched for genes that were highly expressed in root phloem companion cells, and co-expressed with the cytokinin synthesis adenylate isopentenyltransferase 3 (IPT3) gene and with genes induced by cytokinin treatments. In this way they found ABCG14, and confirmed that this gene was also induced in cytokinin-treated seedlings. The developmental defects observed in the abcg14 insertional knock-out mutant resembled those of cytokinin biosynthetic mutants, and were rescued with exogenous trans-zeatin, but not with isopentenyladenine, applications. As Ko et al. (2014) and Zhang et al. (2014) also tested transport capabilities and observed increased accumulation of trans-zeatin type cytokinins in the root and reduced content in the shoot. Moreover, they also found that ABCG14 is expressed in the vasculature, and that the protein it encodes is localized at the plasma membrane. Interestingly, like the application of exogenous trans-zeatin, the grafting of abcg14 shoots to wild type roots could rescue defective shoot growth, suggesting that the translocation of cytokinins from roots to shoots, mediated by ABCG14, is necessary for proper shoot growth and development (Ko et al., 2014).
The ABCGs belong to a subfamily of ATP-binding cassette (ABC) transporters that participate in various biological processes, including the transport of other hormones (reviewed by Borghi et al., 2015). Interestingly, plants have many more ABC transporters than other organisms, and ABCG transporters are particularly abundant in plants (reviewed by Hwang et al., 2016). These transporters have been clustered in six groups (Rajsz et al., 2016), and although the activity of many has been characterized, the biological and biochemical functions of many ABCG proteins remain undetermined and it is possible that other proteins of this subfamily can also export cytokinin (reviewed in Borghi et al., 2015; Hwang et al., 2016).
In summary, the work on PUP, ENT and ABCG transporters has provided more pieces to put together the puzzle of cytokinin transport and to improve the current understanding of cytokinin action.
Still, there are many questions that remain open about these matters. For cytokinin transport, some of the many possible questions that come to mind are: Which roles do the different members of these families play in the local, paracrine and systemic effects of cytokinin? Do other members of each transporter family have the same biochemical functions as the members that have been studied? For example, human ENTs can facilitate nucleoside transport in a bidirectional fashion, and some have been associated with nuclear or endoplasmic reticulum membranes (reviewed in Young et al., 2008). Do members of this family in plants function or localize in similar ways? Moreover, phloem cytokinin (isopentenyladenine type) transport from shoot to root has an important function in root vascular development and nodulation control (Bishopp et al., 2011; Sasaki et al., 2014; brief summary: Muraro et al., 2011). Do members of one or more of these families participate in this translocation? Are there more cytokinin transporters that remain to be discovered?
It will be very interesting to further explore the localization, transport capabilities and role of all the members of these transporter families in cytokinin movement, their impact on plant development and other processes, and integrate the new knowledge in the current models of cytokinin homeostasis and action.
Author Contributions
All the authors revised published information, discussed and prepared the table, and drafted the manuscript. DD-R and YD-M prepared the figure, and NM-M prepared the table and edited the final versions of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the reviewers and Stefan de Folter for critically reading the manuscript.
Footnotes
Funding. Work in the Marsch-Martínez lab is supported by the National Council of Science and Technology of Mexico (CONACyT, grant CB2015-255069), and YD-M and DD-R by CONACyT fellowships (262981 and 254467, respectively).
References
- Amasino R. (2005). 1955: Kinetin arrives. The 50th anniversary of a new plant hormone. Plant Physiol. 138 1177–1184. 10.1104/pp.104.900160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard C., Traub M., Kunz H., Hach S., Trentmann O., Möhlmann T. (2011). Equilibrative nucleoside transporter 1 (ENT1) is critical for pollen germination and vegetative growth in Arabidopsis. J. Exp. Bot. 62 4627–4637. 10.1093/jxb/err183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopp A., Lehesranta S., Vatén A., Help H., El-Showk S., Scheres B., et al. (2011). Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr. Biol. 21 927–932. 10.1016/j.cub.2011.04.049 [DOI] [PubMed] [Google Scholar]
- Borghi L., Kang J., Ko D., Lee Y., Martinoia E. (2015). The role of ABCG-type ABC transporters in phytohormone transport. Biochem. Soc. Trans. 43 924–930. 10.1042/bst20150106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürkle L., Cedzich A., Döpke C., Stransky H., Okumoto S., Gillissen B., et al. (2003). Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. Plant J. 34 13–26. 10.1046/j.1365-313x.2003.01700.x [DOI] [PubMed] [Google Scholar]
- Caesar K., Thamm A., Witthoft J., Elgass K., Huppenberger P., Grefen C., et al. (2011). Evidence for the localization of the Arabidopsis cytokinin receptors AHK3 and AHK4 in the endoplasmic reticulum. J. Exp. Bot. 62 5571–5580. 10.1093/jxb/err238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedzich A., Stransky H., Schulz B., Frommer W. B. (2008). Characterization of cytokinin and adenine transport in Arabidopsis cell cultures. Plant Physiol. 148 1857–1867. 10.1104/pp.108.128454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bianco M., Giustini L., Sabatini S. (2013). Spatiotemporal changes in the role of cytokinin during root development. New Phytol. 199 324–338. 10.1111/nph.12338 [DOI] [PubMed] [Google Scholar]
- Deng Y., Dong H., Mu J., Ren B., Zheng B., Ji Z., et al. (2010). Arabidopsis histidine kinase CKI1 acts upstream of histidine phosphotransfer proteins to regulate female gametophyte development and vegetative growth. Plant Cell 22 1232–1248. 10.1105/tpc.108.065128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Showk S., Ruonala R., Helariutta Y. (2013). Crossing paths: cytokinin signalling and crosstalk. Development 140 1373–1383. 10.1242/dev.086371 [DOI] [PubMed] [Google Scholar]
- Gillissen B., Burkle L., Andre B., Kuhn C., Rentsch D., Brandl B., et al. (2000). A new family of high-affinity transporters for adenine, cytosine, and purine derivatives in Arabidopsis. Plant Cell 12 291–300. 10.2307/3870929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke C., Daumann M., Niopek-Witz S., Möhlmann T., Kerr I. (2014). Nucleobase and nucleoside transport and integration into plant metabolism. Front. Plant Sci. 5:443 10.3389/fpls.2014.00443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A., Riefler M., Romanov G. A., Schmülling T. (2012). Properties, functions and evolution of cytokinin receptors. Eur. J. Cell Biol. 91 246–256. 10.1016/j.ejcb.2011.02.009 [DOI] [PubMed] [Google Scholar]
- Hildreth S. B., Gehman E. A., Yang H., Lu R.-H., Ritesh K. C., Harich K. C., et al. (2011). Tobacco nicotine uptake permease (NUP1) affects alkaloid metabolism. Proc. Natl. Acad. Sci. U.S.A 108 18179–18184. 10.1073/pnas.1108620108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N., Makita N., Yamaya T., Sakakibara H. (2005). Functional characterization and expression analysis of a gene, OsENT2, encoding an equilibrative nucleoside transporter in rice suggest a function in cytokinin transport. Plant Physiol. 138 196–206. 10.1104/pp.105.060137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N., Takei K., Kuroha T., Kamada-Nobusada T., Hayashi H., Sakakibara H. (2008). Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 59 75–83. 10.1093/jxb/erm157 [DOI] [PubMed] [Google Scholar]
- Hwang I., Sheen J., Müller B. (2012). Cytokinin signaling networks. Annu. Rev. Plant Biol. 63 353–380. 10.1146/annurev-arplant-042811-105503 [DOI] [PubMed] [Google Scholar]
- Hwang J., Song W., Hong D., Ko D., Yamaoka Y., Jang S., et al. (2016). Plant ABC transporters enable many unique aspects of a terrestrial plant’s lifestyle. Mol. Plant 9 338–355. 10.1016/j.molp.2016.02.003 [DOI] [PubMed] [Google Scholar]
- Inoue T., Higuchi M., Hashimoto Y., Seki M., Kobayashi M., Kato T., et al. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409 1060–1063. 10.1038/35059117 [DOI] [PubMed] [Google Scholar]
- Jelesko J. (2012). An expanding role for purine uptake permease-like transporters in plant secondary metabolism. Front. Plant Sci. 3:78 10.3389/fpls.2012.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto T. (1996). CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 274 982–985. 10.1126/science.274.5289.982 [DOI] [PubMed] [Google Scholar]
- Kieber J., Schaller G. (2010). The perception of cytokinin: a story 50 years in the making. Plant Physiol. 154 487–492. 10.1104/pp.110.161596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber J., Schaller G. (2014). Cytokinins. Arabidopsis Book 12:e0168 10.1199/tab.0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko D., Kang J., Kiba T., Park J., Kojima M., Do J., et al. (2014). Arabidopsis ABCG14 is essential for the root-to-shoot translocation of cytokinin. Proc. Natl. Acad. Sci. U.S.A. 111 7150–7155. 10.1073/pnas.1321519111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo T., Kiba T., Sakakibara H. (2010). Metabolism and long-distance translocation of cytokinins. J. Integr. Plant Biol. 52 53–60. 10.1111/j.1744-7909.2010.00898.x [DOI] [PubMed] [Google Scholar]
- Li G., Liu K., Baldwin S., Wang D. (2003). Equilibrative nucleoside transporters of Arabidopsis thaliana: cDNA cloning, expression pattern, and analysis of transport activities. J. Biol. Chem. 278 35732–35742. 10.1074/jbc.m304768200 [DOI] [PubMed] [Google Scholar]
- Li J., Wang D. (2000). Cloning and in vitro expression of the cDNA encoding a putative nucleoside transporter from Arabidopsis thaliana. Plant Sci. 157 23–32. 10.1016/s0168-9452(00)00261-2 [DOI] [PubMed] [Google Scholar]
- Li J., Yang H., Peer W., Richter G., Blakeslee J., Bandyopadhyay A., et al. (2005). Arabidopsis H-PPase AVP1 regulates auxin-mediated organ development. Science 310 121–125. 10.1126/science.1115711 [DOI] [PubMed] [Google Scholar]
- Lomax T., Mehlhorn R., Briggs W. (1985). Active auxin uptake by zucchini membrane vesicles: quantitation using ESR volume and pH determinations. Proc. Natl. Acad. Sci. U.S.A. 82 6541–6545. 10.1073/pnas.82.19.6541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomin S., Krivosheev D., Steklov M., Arkhipov D., Osolodkin D., Schmulling T., et al. (2015). Plant membrane assays with cytokinin receptors underpin the unique role of free cytokinin bases as biologically active ligands. J. Exp. Bot. 66 1851–1863. 10.1093/jxb/eru522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomin S., Yonekura-Sakakibara K., Romanov G., Sakakibara H. (2011). Ligand-binding properties and subcellular localization of maize cytokinin receptors. J. Exp. Bot. 62 5149–5159. 10.1093/jxb/err220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C., Skoog F., Okumura F., Von Saltza M., Strong F. (1955a). Structure and synthesis of kinetin. J. Am. Chem. Soc. 77 2662–2663. 10.1021/ja01614a108 [DOI] [Google Scholar]
- Miller C., Skoog F., Von Saltza M., Strong F. (1955b). Kinetin, a cell division factor from deoxyribonucleic acid. J. Am. Chem. Soc. 77 1392–1392. 10.1021/ja01610a105 [DOI] [Google Scholar]
- Möhlmann T., Mezher Z., Schwerdtfeger G., Neuhaus H. (2001). Characterisation of a concentrative type of adenosine transporter from Arabidopsis thaliana (ENT1, At). FEBS Lett. 509 370–374. 10.1016/s0014-5793(01)03195-7 [DOI] [PubMed] [Google Scholar]
- Muraro D., Wilson M., Bennett M. (2011). Root development: cytokinin transport matters, too! Curr. Biol. 21 R423–R425. 10.1016/j.cub.2011.04.048 [DOI] [PubMed] [Google Scholar]
- Osugi A., Sakakibara H. (2015). Q&A: how do plants respond to cytokinins and what is their importance? BMC Biol. 13:102 10.1186/s12915-015-0214-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z., Xiong L. (2013). Characterization of a purine permease family gene OsPUP 7 involved in growth and development control in rice. J. Integr. Plant Biol. 55 1119–1135. 10.1111/jipb.12101 [DOI] [PubMed] [Google Scholar]
- Rajsz A., Warzybok A., Migocka M. (2016). Genes encoding cucumber full-size ABCG proteins show different responses to plant growth regulators and sclareolide. Plant Mol. Biol. Reporter 34 720–736. 10.1007/s11105-015-0956-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov G., Lomin S., Schmulling T. (2006). Biochemical characteristics and ligand-binding properties of Arabidopsis cytokinin receptor AHK3 compared to CRE1/AHK4 as revealed by a direct binding assay. J. Exp. Bot. 57 4051–4058. 10.1093/jxb/erl179 [DOI] [PubMed] [Google Scholar]
- Romanov G., Spíchal L., Lomin S., Strnad M., Schmülling T. (2005). A live cell hormone-binding assay on transgenic bacteria expressing a eukaryotic receptor protein. Anal. Biochem. 347 129–134. 10.1016/j.ab.2005.09.012 [DOI] [PubMed] [Google Scholar]
- Sakakibara H. (2006). Cytokinins: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57 431–449. 10.1146/annurev.arplant.57.032905.105231 [DOI] [PubMed] [Google Scholar]
- Sasaki T., Suzaki T., Soyano T., Kojima M., Sakakibara H., Kawaguchi M. (2014). Shoot-derived cytokinins systemically regulate root nodulation. Nat. Commun. 5:4983 10.1038/ncomms5983 [DOI] [PubMed] [Google Scholar]
- Schwacke R., Schneider A., Van Der Graaff E., Fischer K., Catoni E., Desimone M., et al. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131 16–26. 10.1104/pp.011577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Hirose N., Wang X., Wen P., Xue L., Sakakibara H., et al. (2005). Arabidopsis SOI33/AtENT8 gene encodes a putative equilibrative nucleoside transporter that is involved in cytokinin transport in planta. J. Integr. Plant Biol. 47 588–603. 10.1111/j.1744-7909.2005.00104.x [DOI] [Google Scholar]
- Suzuki T., Miwa K., Ishikawa K., Yamada H., Aiba H., Mizuno T. (2001). The Arabidopsis sensor his-kinase, AHK4, can respond to cytokinins. Plant Cell Physiol. 42 107–113. 10.1093/pcp/pce037 [DOI] [PubMed] [Google Scholar]
- Traub M., Flörchinger M., Piecuch J., Kunz H., Weise-Steinmetz A., Deitmer J., et al. (2007). The fluorouridine insensitive 1 (fur1) mutant is defective in equilibrative nucleoside transporter 3 (ENT3), and thus represents an important pyrimidine nucleoside uptake system in Arabidopsis thaliana. Plant J. 49 855–864. 10.1111/j.1365-313x.2006.02998.x [DOI] [PubMed] [Google Scholar]
- Werner T., Motyka V., Laucou V., Smets R., Van Onckelen H., Schmülling T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 2532–2550. 10.1105/tpc.014928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormit A., Traub M., Flörchinger M., Neuhaus H., Möhlmann T. (2004). Characterization of three novel members of the Arabidopsis thaliana equilibrative nucleoside transporter (ENT) family. Biochem. J. 383 19–26. 10.1042/bj20040389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulfetange K., Lomin S., Romanov G., Stolz A., Heyl A., Schmulling T. (2011). The cytokinin receptors of Arabidopsis are located mainly to the endoplasmic reticulum. Plant Physiol. 156 1808–1818. 10.1104/pp.111.180539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H. (2001). The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 42 1017–1023. 10.1093/pcp/pce127 [DOI] [PubMed] [Google Scholar]
- Young J., Yao S., Sun L., Cass C., Baldwin S. (2008). Human equilibrative nucleoside transporter (ENT) family of nucleoside and nucleobase transporter proteins. Xenobiotica 38 995–1021. 10.1080/00498250801927427 [DOI] [PubMed] [Google Scholar]
- Zhang K., Novak O., Wei Z., Gou M., Zhang X., Yu Y., et al. (2014). Arabidopsis ABCG14 protein controls the acropetal translocation of root-synthesized cytokinins. Nat. Commun. 5:3274 10.1038/ncomms4274 [DOI] [PubMed] [Google Scholar]
- Zubko E., Adams C., Macháèková I., Malbeck J., Scollan C., Meyer P. (2002). Activation tagging identifies a gene from Petunia hybrid responsible for the production of active cytokinins in plants. Plant J. 29 797–808. 10.1046/j.1365-313x.2002.01256.x [DOI] [PubMed] [Google Scholar]
- Zürcher E., Liu J., Donato M. D., Geisler M., Muller B. (2016). Plant development regulated by cytokinin sinks. Science 353 1027–1030. 10.1126/science.aaf7254 [DOI] [PubMed] [Google Scholar]
- Zürcher E., Müller B. (2016). “Cytokinin synthesis, signaling, and function - advances and new insights,” in International Review of Cell and Molecular Biology Vol. 324 ed. Jeon K. W. (Amsterdam: Elsevier Inc; ), 1–38. [DOI] [PubMed] [Google Scholar]
- Zürcher E., Tavor-Deslex D., Lituiev D., Enkerli K., Tarr P. T., Muller B. (2013). A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 161 1066–1075. 10.1104/pp.112.211763 [DOI] [PMC free article] [PubMed] [Google Scholar]