Abstract
Abscission is a cell separation process by which plants can shed organs such as fruits, leaves, or flowers. The process takes place in specific locations termed abscission zones. In fruit crops like citrus, fruit abscission represents a high percentage of annual yield losses. Thus, understanding the molecular regulation of abscission is of capital relevance to control production. To identify genes preferentially expressed within the citrus fruit abscission zone (AZ-C), we performed a comparative transcriptomics assay at the cell type resolution level between the AZ-C and adjacent fruit rind cells (non-abscising tissue) during ethylene-promoted abscission. Our strategy combined laser microdissection with microarray analysis. Cell wall modification-related gene families displayed prominent representation in the AZ-C. Phylogenetic analyses of such gene families revealed a link between phylogenetic proximity and expression pattern during abscission suggesting highly conserved roles for specific members of these families in abscission. Our transcriptomic data was validated with (and strongly supported by) a parallel approach consisting on anatomical, histochemical and biochemical analyses on the AZ-C during fruit abscission. Our work identifies genes potentially involved in organ abscission and provides relevant data for future biotechnology approaches aimed at controlling such crucial process for citrus yield.
Keywords: calyx abscission zone, cell wall modification, citrus fruit abscission, ethylene, lignin biosynthesis, phylogeny, transcriptomics
Introduction
Abscission is a cell separation process by which plants can shed their aerial organs. It takes place in groups of functionally specialized cells known as abscission zones (AZs), which are located at specific sites of organ detachment in the plant (Roberts et al., 2002; Estornell et al., 2013; Tucker and Kim, 2015).
Abscission is a fundamental process in plant biology that represented a highly beneficial evolutionary adaptation for plants: abscission allows for discarding senescent or physiologically damaged organs and for highly efficient seed dispersal. However, from an agricultural point of view, abscission has a tremendous impact on yield, leading to high yield losses in key crops like brassica or citrus. In this way, understanding abscission at the molecular level is of top relevance not only to understand a fundamental process for plant physiology but also to generate new, improved, highly productive crops.
Abscission related traits (i.e., reduced abscission of fruits or seeds) are among the main agronomic traits selected along plant domestication (Konishi et al., 2006; Pickersgill, 2007). A current example is the expansion of late-season varieties of sweet orange in the citrus market. In such varieties, the decline in the fruit retention force is delayed during the maturing period in comparison with early and mid-season varieties (Gallasch, 1996) that usually undergo pre-harvest fruit abscission (Spiegel-Roy and Goldschmidt, 1996). Thus, late-season varieties of sweet orange extend the fruit harvesting season benefiting growers and food industry.
Control of abscission is also relevant to facilitate mechanical harvesting, thus reducing collection costs. Mechanical fruit harvesting systems have been developed although they are still inefficient and cause tree damages (Li et al., 2011). On the other hand, several abscission-triggering compounds have been used to improve mechanical harvesting. In citrus, treatments with CMNP (5-chloro-3-methyl-4-nitro-1H-pyrazole) are used to promote fruit loosening and to facilitate and coordinate mechanical harvesting of fruits (Burns, 2002). In this regard, understanding the mechanisms underlying abscission is essential to control abscission and improve harvesting practices and productivity.
Studies on floral organ abscission in the model system Arabidopsis thaliana have provided a wealth of valuable information. However, the current information about the molecular mechanisms underlying abscission in crop species is rather scarce.
Most of the molecular studies of abscission in crops have mainly been focused on the characterization of individual or few genes. However, high-throughput approaches have recently been applied in AZ-containing tissues of tomato flowers (Meir et al., 2010) and apple (Zhu et al., 2011), mature olive (Gil-Amado and Gomez-Jimenez, 2013; Parra et al., 2013), melon (Corbacho et al., 2013), litchi (Li et al., 2015), and orange fruits (Cheng et al., 2015). In our previous studies (Agustí et al., 2008, 2009, 2012), global expression analyses provided a wide set of genes potentially involved in citrus leaf abscission. These datasets included a number of cell wall modification related genes as well as genes involved in signaling, transcription control, protein synthesis and degradation and vesicle transport.
Our current challenge is to identify key regulatory genes of citrus fruit abscission which is, indeed, an economically important process. In citrus, maturing fruits are shed through the abscission zone C (AZ-C), located at the boundary between the calyx button and the fruit rind (FR). In this region, different tissues converge and the isolation of exclusive AZ-C cells for molecular studies without any contamination of other cell-types is extremely complicated. In this study, we have taken advantage of the optimization of laser microdissection (LM) in citrus tissues (Agustí et al., 2009; Matas et al., 2010; Caruso et al., 2012) for the accurate sampling of fruit AZ-C cells. This strategy has allowed the precise quantification of the timing and magnitude of gene expression and associate metabolites involved in the process of ethylene-promoted abscission in the specific cells of the AZ-C. Moreover, phylogenetic analyses of the most representative gene families during abscission in citrus and different plant species have revealed a link between phylogenetic proximity and expression pattern during this process suggesting highly conserved functions for specific members of these families in abscission. Overall, this study, through the identification of potential abscission-related genes and the detailed spatio-temporal analysis of the anatomical and histochemical changes in the activated AZ-C, provides crucial information for future biotechnological approaches aimed at improving citrus yield.
Materials and methods
Plant material and in vitro treatments
We used fruits from two Citrus sinensis cultivars: a mid-season orange cultivar (cv. Washington Navel) that usually undergoes pre-harvest abscission and a late-season orange cultivar (cv. Ricalate Navel) with delayed abscission. Maturing fruits were harvested after color change from adult trees grown in a homogeneous experimental orchard under normal cultural practices at the Institut Valencià d'Investigacions Agràries (IVIA). Fruits were separated from the tree leaving 2 cm peduncles to isolate the AZ-C for further analyses. For abscission kinetics studies and tissue collection, Washington Navel fruits were incubated for 0, 24, 48, and 96 h in the presence or absence of ethylene (10 μL/L) in sealed 10 l containers at 22°C with a 16 h light period under fluorescent lighting. Ricalate Navel fruits were incubated for 0, 24, 48, 96, and 192 h in the presence of 1-aminocyclopropane-l-carboxylic acid (ACC; 0.1 mM) or water under the same temperature and light conditions. In this case, a 3 mL Pasteur pipette containing the ACC solution or water was fitted to the fruit peduncles.
Phloroglucinol staining
Phloroglucinol staining for lignin in fresh cut tissue portions (0.5 cm3) containing the AZ-C after 0, 24, and 48 h of ethylene or ACC treatment was performed according to Tadeo and Primo-Millo (1990). Samples were cut longitudinally to allow AZ-C staining and for further image acquisition. A saturated solution of phloroglucinol (Sigma-Aldrich) in 20% HCl was directly applied to samples. Observation was carried out with an Olympus SZ61 stereomicroscope (Olympus GmbH).
Cryoscanning electron microscopy (cryo-SEM)
Longitudinal sections as well as the proximal (peduncle) and distal (fruit) fracture plane of the ethylene-promoted AZ-C were observed using cryo-SEM. To examine longitudinal sections of the AZ-C, 1 cm portions of tissue were manually dissected with a razor blade. In the second case, the peduncle was forcibly separated from the fruit. Specimen mounting and AZ-C observation were carried out as previously described in Agustí et al. (2009). At least three samples containing the AZ-C after 24, 48, and 96 h of ethylene treatment were observed.
Periodic acid-schiff (PAS) staining
Tissue containing the AZ-C after 0, 24, and 48 h of ACC treatment was manually dissected using a razor blade in 0.5 cm3 portions. These samples were fixed overnight at 4°C in a 4% (w/v) paraformaldehyde-PBS solution. After fixation, samples were washed with PBS, dehydrated in a graded ethanol series and embedded in LR White (Electron Microscopy Sciences). Longitudinal sections of the calyx button area (1 μm thickness) were cut with a Leica RM2165 microtome and placed on glass slides. Slides were further stained with PAS (Sigma-Aldrich) and mounted with DPX Mountant (Fluka). Observations were performed on a Leica DMLA microscope (Leica Microsystems) and images were processed with Leica ASMLD Version 4.0 software.
Preparation of tissue containing the AZ-C for LM
Portions of tissue containing the AZ-C (0.5 cm3) were dissected from fruits after 0, 12, and 24 h of ethylene treatment for the transcriptomics assay, and after 0, 12, 24, and 36 h of ACC treatment for lignin intermediates quantification. Preparation of cryosections and microdissection were performed as previously described in Agustí et al. (2009). Cells from the AZ-C and the adjacent FR were selected from 30 to 40 cryosections and collected separately.
Phloroglucinol staining of cryosections
Cryosections of 14 μm of tissue containing the AZ-C after 48 h of ethylene treatment were processed as described in Agustí et al. (2009) and mounted on CryoJane® adhesive coated slides (Instrumedics) following the manufacturer's instructions. Slides were stored at −80°C until phloroglucinol staining. Staining for lignin was performed using a saturated solution of phloroglucinol (Sigma-Aldrich) in 20% HCl. Observation was carried out with an Olympus SZ61 stereomicroscope (Olympus GmbH).
RNA isolation, sample labeling, and microarray hybridization
Three independent biological replicates were collected for each cell type at 0, 12, and 24 h after ethylene treatment. For each independent sample, total RNA from ~40,000 pooled cells was extracted using the RNeasy Micro Kit (Qiagen) following the manufacturer's instructions. The RNA purity was assessed by measurements of OD260/OD280. Two RNA amplification rounds were performed utilizing the TargetAmp™ 2-Round Aminoallyl-aRNA Amplification Kit (EPICENTER) according to the manufacturer's instructions to synthesize the antisense cRNA. The quality of the amplified RNA was evaluated by OD260/OD280 measurements and agarose gel electrophoresis. Each sample was labeled with Cy5 and co-hybridized with Cy3-labeled antisense cRNA from a reference sample containing a mixture of equal amounts of RNA from all experimental samples (0, 6, 12, 24, 48, and 96 h of ethylene/air treatment). RNA labeling, microarray hybridization, and slide washes were performed as previously described in Cercos et al. (2006). Hybridized microarrays scanning, hybridization data acquisition, and microarray normalization and analysis were carried out as described in Agustí et al. (2009). A cDNA microarray including 21.081 putative genes of citrus was utilized (Martinez-Godoy et al., 2008). Gene expression differences were considered significant under a P-value lower than 0.05 and an M contrast cutoff value of +0.5 or −0.5. In this work, a time course experiment was designed for each cell type (AZ-C and FR), therefore, the expression level corresponds to M = log2 [AZ-Ct/AZ-C0] or M = log2 [FRt/FR0]. The raw microarray data as well as the protocols used to produce the data and the normalized data were deposited in the ArrayExpress database under the accession number E-MTAB-4538. Functional classification of the selected genes was performed using MIPS (Munich Information Center for Protein Sequences, http://www.helmholtzmuenchen.de/en/mips/) categorization. Amplified RNA was used for the validation of microarray hybridization data by semi-quantitative RT-PCR (sqRT-PCR) analysis (Figure S1).
sqRT-PCR analysis
sqRT-PCR analysis was carried out using the SuperScript II Reverse Transcriptase kit (Invitrogen, Carlsbad) following the manufacturer's instructions. After first-strand cDNA synthesis, PCR reactions were performed using the Biotools Taq DNA Polymerase (BIOTOOLS, B&M Labs). Size and intensity of expected bands were checked by 1% agarose gel electrophoresis. Citrus UBC gene (Ubiquitin-conjugating enzyme) was used as a reference to evaluate the amounts of mRNA in each sample. Primer sequences are available in Table S1.
In situ hybridization
RNA in situ hybridization with digoxigenin-labeled probes was performed as described (Gomez et al., 2011). Portions of tissue containing the AZ-C (0.5 cm3) were dissected from fruits after 24 h of ethylene treatment and immediately fixed at 4°C overnight in FAE (25% formaldehyde, 5% acetic acid, 50% ethanol), dehydrated, embedded in paraffin wax and sectioned to 8 μm. For CitCEL6 and CitPG20, RNA antisense and sense probes were generated with SP6 and T7 RNA polymerases, using as substrate a 1518 bp fragment of the CitCEL6 cDNA (1–1518 from ATG) or a 1110 bp fragment of the CitPG20 cDNA (217–1326 from ATG), amplified by PCR and cloned into the pGEM-T Easy vector (Promega).
Lignin intermediates quantification
Coumaric acid, caffeic acid, and ferulic acid were analyzed by UPLC coupled to tandem mass spectrometry (UPLC-MS/MS) as described by Argamasilla et al. (2014). Three independent samples containing ~40,000 pooled AZ-C cells were isolated by LM for each time point of ACC treatment (0, 12, 24, and 36 h) and collected in 60 μL of water.
Sequence identification, alignment, and phylogenetic analysis
Members of the different gene families associated with cell wall remodeling in citrus (based on CAZy classification; Carbohydrate-Active Enzymes; Cantarel et al., 2009; http://www.cazy.org/) were identified by TBLASTN search in the Citrus clementina haploid genome (version 0.9) database web browser (http://www.phytozome.net/search.php) using the consensus sequence for the catalytic domain of each family. Prediction of characteristic domains and conserved motifs was carried out through SMART (http://smart.embl-heidelberg.de/; Schultz et al., 1998; Letunic et al., 2009), PSORT (http://psort.hgc.jp/form.html), InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan/) and big-PI Plant Predictor (http://mendel.imp.ac.at/gpi/plant_server.html) servers. Phylogenetic trees are based on multiple alignments using the profile alignment function of ClustalW (www.ch.embnet.org/software/ClustalW-XXL.htm) and were generated with MEGA7 (Kumar et al., 2016) using the neighbor-joining algorithm with 1000 bootstrap replicates. Poisson correction for multiple substitutions was used and only values higher than 50% were considered.
Immunolocalization of pectic polyssacharides
The primary monoclonal antibodies (mAbs) used in this study and provided by Prof. Paul Knox (Centre for Plant Sciences, Faculty of Biological Sciences, University of Leeds, UK) were LM5 [anti-(1,4)-β-D-galactan; Jones et al., 1997], LM6 [anti-(1,5)-α-L-arabinan; Willats et al., 1998] and JIM5 [anti-partially methylesterified/de-esterified homogalacturonan; (Knox et al., 1990)]. The secondary antibody was fluorescein isothiocyanate (FITC)-conjugated anti-rat IgG (Sigma-Aldrich). Immunolocalization of pectic polysaccharides was performed on semi-thin sections of tissue containing the AZ-C (0.5 cm3) from fruits after 0, 24, and 48 h of ACC treatment. Immunolocalization of pectic polyssacharides, light microscopy and image acquisition were perfomed as described in Coimbra et al. (2007).
Results and discussion
Ethylene accelerates citrus fruit abscission
We performed a kinetics assay of citrus fruit abscission in response to abscission-accelerating treatments to determine the optimal sampling for the transcriptomic analysis. To that end, we carried out a comparison between orange (C. sinensis) fruits incubated with ethylene or its immediate metabolic precursor 1-aminocyclopropane-1-carboxylic acid (ACC) and fruits incubated with air or water (controls). We used maturing fruits from a mid-season orange cultivar (cv. Washington Navel) that usually undergoes pre-harvest abscission and from a late-season orange cultivar (cv. Ricalate Navel) with delayed abscission. We observed a faster decrease of fruit detachment force (FDF) in both Washington Navel and Ricalate Navel fruits treated with ethylene/ACC in comparison to air-/water-treated control fruits (Figure 1A). At 48 h after treatment, the FDF in fruits treated with ethylene or ACC was around 4 kgf. However, control fruits of Washington Navel and Ricalate Navel only showed values of FDF around 4 kgf at 96 and 192 h, respectively, a response that matches their pre-harvest abscission behavior. Thus, ethylene and ACC accelerated the abscission process in both varieties tested. This result strongly suggests that the natural delay in the schedule of FDF decline in fruits of the late-season variety Ricalate Navel in comparison with those of the mid-season variety Washington Navel was not related to any impairment in the response of well-developed tissues to ethylene (Zacarias et al., 1993). Based on these findings, we used induced AZ-C samples from both varieties for further analyses.
Figure 1.
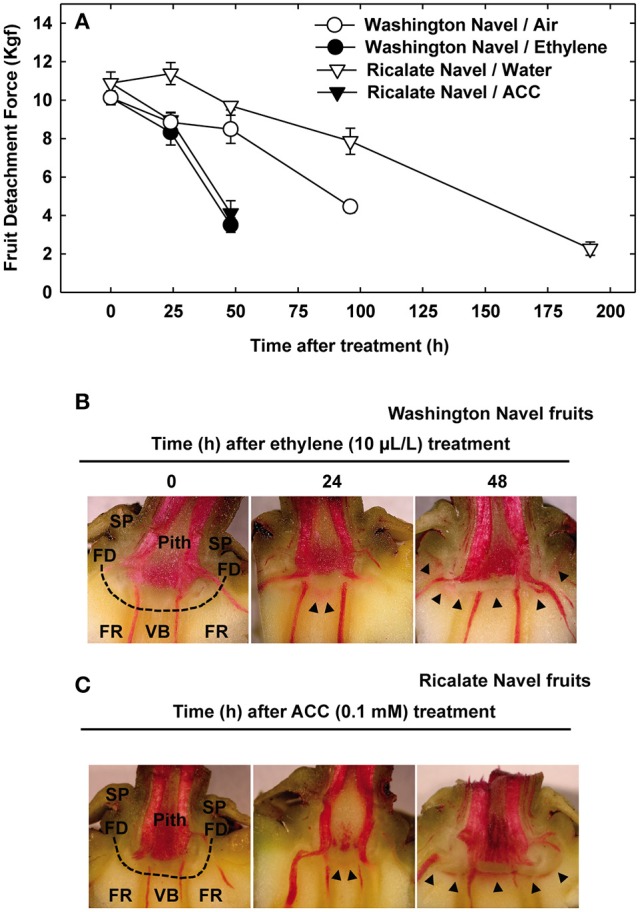
Effect of ethylene and 1-aminocyclopropane- 1-carboxylic acid (ACC) on citrus fruit abscission. (A) Abscission kinetics of Washington Navel fruits non-treated or treated with ethylene and Ricalate Navel fruits non-treated or treated with ACC. The results are means of 10 fruits ± SE. (B,C) Phloroglucinol staining for lignin in the AZ-C of Washington Navel fruits after ethylene treatment (B) and Ricalate Navel fruits after ACC treatment (C). Dashed line, abscission zone C;  , lignin deposition (phloroglucinol); FR, fruit rind; FD, floral disc; SP, sepals; VB, vascular bundles; P, parenchyma.
, lignin deposition (phloroglucinol); FR, fruit rind; FD, floral disc; SP, sepals; VB, vascular bundles; P, parenchyma.
Phloroglucinol staining reveals a positive correlation between abscission and lignin deposition
Phloroglucinol staining in receptacles of both Washington Navel and Ricalate Navel fruits revealed lignin deposition at the central core of the AZ-C, between the axial vascular bundles, 24 h after ethylene and ACC treatments (Figures 1B,C). Forty-eight hours after the treatments, lignin deposition spread out along the AZ-C, perfectly drawing the separation line between the calyx button and the FR. Accordingly, timing for lignin deposition positively correlated with abscission kinetics.
Morphological changes in activated AZ-C cells
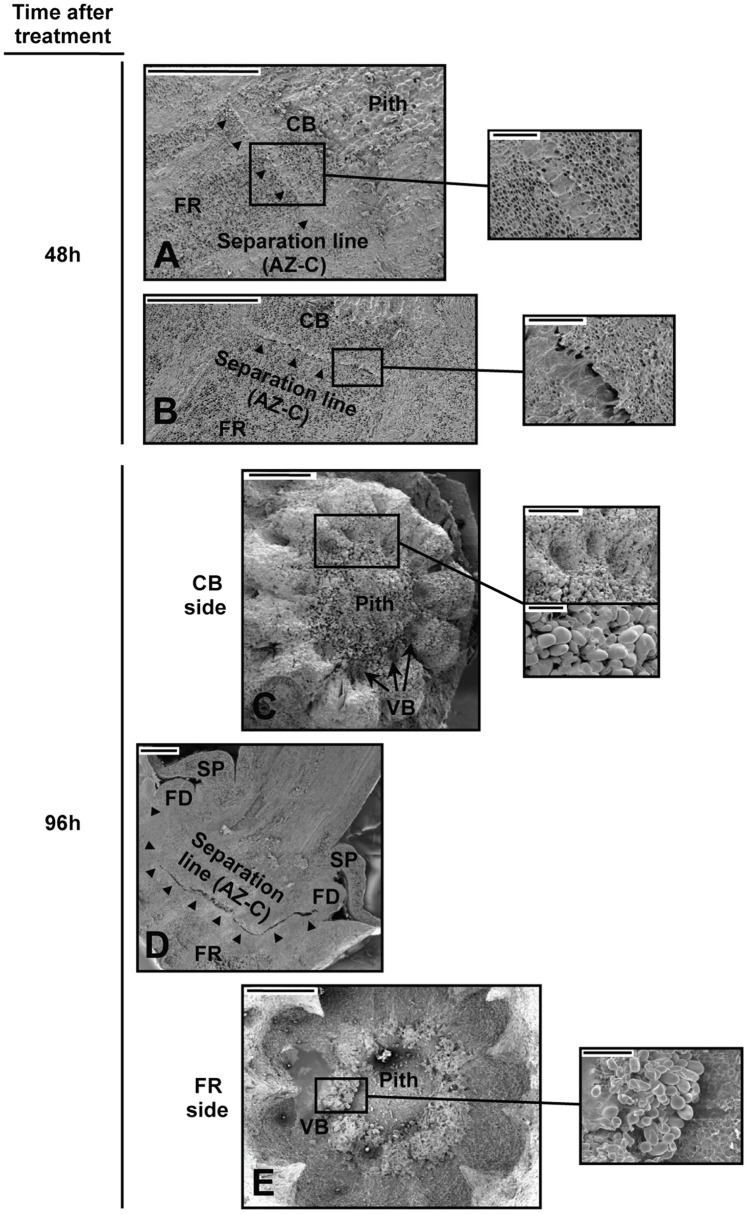
We used scanning electron microscopy (SEM) to examine changes in the cellular morphology of the AZ-C from Washington Navel fruits treated with ethylene (Figure 2). We observed the first cellular signs of activation of abscission by ethylene in the central core of the AZ-C at 48 h of treatment (Figures 2A,B). At that time, AZ-C samples could be split into two groups, one showing early stages of cell separation and the other one showing late events of cell separation. In the former group, the AZ-C was clearly distinguishable (Figure 2A), with accumulation of an amorphous material probably derived from the partial dissolution of the middle lamella and cell wall of the AZ-C cells. In the latter group of samples, cell separation was observed in the central core of the AZ-C (Figure 2B). A greater accumulation of amorphous material was observed, suggesting that cell wall and middle lamella degradation was complete after 48 h at the central region. At 96 h after ethylene treatment, cell separation extended from the central core to the periphery of the AZ-C (Figure 2D) and differential cell expansion was observed at proximal (calyx button) and distal (fruit) sides (Figures 2C,E). At the proximal side, parenchymatic pith cells underwent expansion (Figure 2C) while, at the distal side, expansion occurred in the cells of the axial vascular bundles (Figure 2E).
Figure 2.
Cellular morphology of the AZ-C. (A–E) Scanning electron micrographs of longitudinal sections (A,B,D) and the proximal (C; calyx button side) and distal (E; fruit rind side) fracture planes of the AZ-C from Washington Navel fruits after 48 h (A,B) and 96 h (C–E) of ethylene treatment. High magnification pictures show cells of separation layers. AZ-C, abscission zone C;  , separation line inside the AZ-C; CB, calyx button; FD, floral disc; FR, fruit rind; SP, sepal; VB, vascular bundles. Scale bars: 1 mm (A–E), 500 μm (A–C), 200 μm (E), 100 μm (C).
, separation line inside the AZ-C; CB, calyx button; FD, floral disc; FR, fruit rind; SP, sepal; VB, vascular bundles. Scale bars: 1 mm (A–E), 500 μm (A–C), 200 μm (E), 100 μm (C).
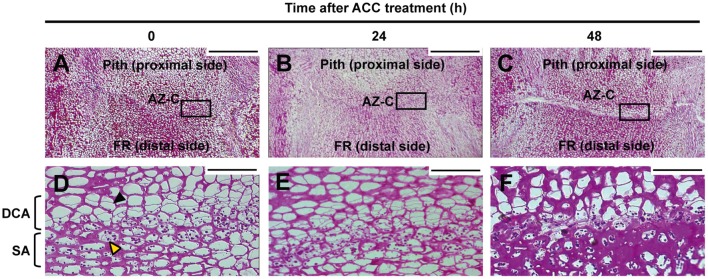
Two different cell areas form the AZ-C
Periodic acid-Schiff (PAS) staining was used to characterize anatomically the AZ-C after ACC treatment (Figure 3). This method detects insoluble carbohydrates and was used to distinguish the cells belonging to the AZ-C since these accumulate starch grains (Wilson and Hendershott, 1968; Huberman et al., 1983; Shiraishi and Yanagisawa, 1988; Goren, 1993). In addition to the starch-rich cell area (SA) previously identified by Wilson and Hendershott (1968) at the distal side of the AZ-C (FR side), we identified another cell area located at the proximal side of the AZ-C (pith side) and composed by recently divided cells based on the observation of thinner cell walls formed between cells (Divided Cells Area, DCA; Figure 3D). Then, the AZ-C was constituted by 10–15 cell layers distributed in cellular areas with two different cell morphologies and organellar composition (i.e., cells from the SA contain amyloplasts). The analysis of the AZ-C after 48 h of treatment suggests that cell wall degradation and cell degeneration occurred mainly at the layers of the SA in the fracture plane, adjacent to the mesocarp cells of the FR known as the albedo (see Figure 9). However, cell expansion occurred at the DCA (Figure 3F). These results together with kinetics (Figure 1A), phloroglucinol staining (Figures 1B,C) and SEM (Figure 2) data suggested that the activation of the fruit AZ-C by ethylene/ACC occurred early after treatment, probably prior to 24 h. The events related to cell wall loosening might start at 24 h, while cell separation seemed to begin at 48 h and to be completed at 96 h after treatment.
Figure 3.
Anatomy of the AZ-C. Periodic acid-Schiff reactive (PAS) staining for insoluble carbohydrates of longitudinal sections of the AZ-C from Ricalate Navel fruits non-treated (A,D) and treated for 24 h (B,E) and 48 h (C,F) with ACC. Squares in (A–C) show the area magnified with the 40X objective. AZ-C, abscission zone C; FR, fruit rind; DCA, divided cells area; SA, starch-rich area;  , recently divided cell;
, recently divided cell;  , cell containing amyloplasts. Scale bars: 500 μm and 50 μm.
, cell containing amyloplasts. Scale bars: 500 μm and 50 μm.
Gene expression regulated by ethylene in AZ-C and FR cells
For gene expression analysis, we used the 20 K citrus cDNA microarray (Martinez-Godoy et al., 2008). We isolated cells from the central core of the AZ-C as well as from the FR located beneath the AZ-C through LM (Figure S2) to perform a time-course experiment (0, 12, and 24 h-ethylene) and compared data from each analysis. Results showed that ethylene differentially regulated 2280 genes exclusively in the AZ-C cells, 1742 genes exclusively in the FR cells, and 2001 genes were regulated by ethylene in both the AZ-C and the FR cells (Table S1).
All differentially regulated genes were grouped into functional categories according to the Munich Information Center for Protein Sequences (MIPS). The categories sugar, glucoside, polyol, and carboxylate metabolism and polysaccharide metabolism showed a higher percentage of regulation in the AZ-C compared with the FR (Table S2).
The set of genes discussed below was selected because of its prominent representation in the AZ-C or its particular biological interest. These gene families included those associated with cell wall metabolism and monolignol biosynthesis and polymerization (Table S3).
Many genes related to cell wall modification are regulated during fruit abscission
A high number of genes encoding cell wall modification enzymes were differentially regulated by ethylene exclusively in the fruit AZ-C. Our data suggested that this strong activation of cell wall metabolism occurred through both degradation and biosynthesis (Table 1) as we previously showed in the laminar abscission zone (LAZ) during leaf abscission (Agustí et al., 2008, 2009, 2012). Genes encoding enzymes that hydrolyze the cell wall and middle lamella would be responsible of the cell wall disassembly observed mainly in the SA at the distal side of the AZ-C (Figure 3F), which would lead to the effective organ separation. On the other hand, genes involved in the biosynthesis of new cell wall components would be related to the cell expansion observed mainly in the cell layers of the DCA located at the proximal side of the fracture plane (Figures 2C, 3F). The cell expansion might be associated with the generation of protective layers at the regions of the receptacle that remain exposed to the environment after abscission. Indeed, the recently divided cells at the proximal side of the AZ-C did not show cell degeneration (Figure 3F), suggesting that these cells might acquire such a function during the last step of organ abscission (Roberts et al., 2002; Estornell et al., 2013).
Table 1.
Relative gene expression values (AZ-Ct vs. AZ-C0) of genes involved in cell wall modification exclusively regulated by ethylene in AZ-C cells.
| Name | Contig/singleton ID | Microarray probe | Putative Ath orthologue | Relative expression [log2 (AZ-Ct/AZ-C0)] | |
|---|---|---|---|---|---|
| 12 h | 24 h | ||||
| ENDO-1,4-β-GLUCANASES|CELLULASES (GH9s) | |||||
| CitCEL3 | aCL1687Contig1 | IC0AAA38AD03 | AT2G32990 | – | 0.69 |
| CitCEL6* | aCL1347Contig1 | C21007H10 | AT4G02290 | – | 4.15 |
| CitCEL10 | aCL7597Contig1 | IC0AAA68DE06 | AT4G02290 | – | 0.60 |
| CitCEL17 | aCL1288Contig1 | C32011E04 | AT1G75680 | −0.88 | – |
| CitCEL22 | aC20010F01SK_c | C20010F01 | AT1G23210 | – | 0.72 |
| POLYGALACTURONASES (GH28s) | |||||
| CitPG6 | aCL2029Contig1 | C03009E03 | AT4G23820 | –1.58 | – |
| CitPG16 | aCL5261Contig1 | C01018A12 | AT3G61490 | 1.02 | – |
| CitPG20* | AT3G07970 | in-situ hybridization | |||
| CitPG41 | aC18008C05Rv_c | C18008C05 | AT3G57790 | 1.10 | 1.49 |
| aCL1063Contig1 | IC0AAA19CA02 | AT3G57790 | 0.88 | 0.95 | |
| CitPG42 | aIC0AAA85AB02RM1_c | IC0AAA85AB02 | AT3G48950 | – | 0.63 |
| CitPG43 | aCL675Contig4 | IC0AAA67DG09 | AT2G43870 | – | 2.34 |
| PECTATE-LYASES (PL1s) | |||||
| CitPL1 | aC03011D06SK_c | C03011D06 | AT5G63180 | −0.86 | −0.80 |
| CitPL5 | aIC0AAA15AF11RM1_c | IC0AAA15AF11 | AT1G67750 | 2.66 | |
| PECTIN-METHYLESTERASES (CE8s) | |||||
| CitPME8 | aC05807A09SK_c | C05807A09 | AT4G33220 | −3.13 | −2.57 |
| CitPME11 | aCL1691Contig1 | C08033H07 | AT1G11580 | −2.30 | – |
| CitPME13 | aCL4116Contig2 | C01011H09 | AT5G53370 | −2.15 | −2.90 |
| CitPME24 | aCL1451Contig1 | IC0AAA40DF03 | AT1G69940 | 0.71 | 0.71 |
| CitPME41 | aCL2379Contig1 | C32102B03 | AT5G09760 | – | 1.41 |
| PECTIN-ACETYLESTERASES (CE18s) | |||||
| CitPAE1 | aKN0AAP13YN19FM1_c | KN0AAP13YN19 | AT3G62060 | −0.72 | – |
| CitPAE4 | aCL67Contig4 | C08028G04 | AT4G19420 | 0.58 | 0.51 |
| CitPAE6 | aCL7344Contig1 | C02003B05 | AT5G26670 | −1.57 | – |
| aKN0AAI1DH10FM2_c | KN0AAI1DH10 | AT5G26670 | −2.07 | – | |
| β-GALACTOSIDASES (GH35s) | |||||
| CitGBAL16 | aC31805H10EF_c | C31805H10 | AT4G36360 | −1.59 | |
| aCL7104Contig1 | C02004B02 | AT4G36360 | −1.01 | ||
| β-GALACTOSIDASES (GH2s) | |||||
| CitGH22 | aCL4443Contig1 | C31401H10 | AT3G54440 | 0.71 | 0.95 |
| β-GLUCOSIDASES (GH1s) | |||||
| CitBGLU17 | aCL5744Contig1 | C31007D10 | AT2G44480 | −0.59 | −0.55 |
| CitBGLU24 | aCL1136Contig3 | IC0AAA1CB06 | AT3G06510 | – | 0.62 |
| β-MANNOSIDASES (GH5s) | |||||
| CitMAN4 | aC04002G09SK_c | C04002G09 | AT1G02310 | 1.41 | – |
| XYLOGLUCAN ENDOTRANSGLYCOSYLASES/HYDROLASES (GH16s) | |||||
| CitXTH16 | aC02023G10SK_c | C02023G10 | AT4G03210 | −1.55 | −2.33 |
| CitXTH24 | aIC0AAA99CH05RM1_c | IC0AAA99CH05 | AT1G32170 | 1.07 | 1.03 |
| CitXTH28 | aCL6772Contig1 | C01009B04 | AT4G37800 | – | −0.88 |
| α-XYLOSIDASES (GH31s) | |||||
| CitXYL4 | aCL6235Contig1 | C05075C11 | AT1G68560 | −0.98 | – |
| β-XYLOSIDASES (GH3s) | |||||
| CitBXL13 | aCL3345Contig1 | C02024D10 | AT1G78060 | −0.63 | – |
| CitBXL16 | aCL8110Contig1 | IC0AAA75AA10 | AT5G20950 | −0.99 | – |
| EXPANSINS | |||||
| CitEXP14 | aCL2131Contig1 | IC0AAA14BD04 | AT2G40610 | – | 3.01 |
| aIC0AAA87BH09RM1_c | IC0AAA87BH09 | AT2G40610 | 1.63 | 3.19 | |
| CitEXP15 | aKN0AAQ1YG09RM1_c | KN0AAQ1YG09 | AT4G17030 | 1.69 | 1.40 |
| CitEXP19 | aC02006G07SK_c | C02006G07 | AT1G20190 | – | 0.75 |
| CELLULOSE SYNTHASES/CELLULOSE SYNTHASE-LIKE PROTEINS | |||||
| CitCes1 | aC16012C03SK_c | C16012C03 | AT4G24010 | 1.82 | – |
| CitCes2 | aIC0AAA5DG11RM1_c | IC0AAA5DG11 | AT5G05170 | −1.74 | – |
| CitCsl3 | aCL5293Contig1 | C05070F01 | AT3G03050 | – | 0.68 |
| UDP-GLUCOSE 4-EPIMERASE | |||||
| CitUGE1 | aC31108G08EF_c | C31108G08 | AT4G10960 | – | 0.95 |
| CitUGE2 | aCL6604Contig1 | C03009D04 | AT1G12780 | – | 0.82 |
| MANNAN SYNTHASES | |||||
| CitManS1 | aCL3377Contig1 | IC0AAA16BH03 | AT5G22740 | – | 1.47 |
| CitManS2 | aCL3377Contig2 | IC0AAA99AD02 | AT5G22740 | – | 1.36 |
| GALACTOMANNAN GALACTOSYLTRANSFERASE (GMGT) | |||||
| CitGMGT1 | aC34004D03EF_c | IC0AAA58DH01 | AT2G22900 | – | 1.35 |
| RHAMNOSE BIOSYNTHETIC ENZYME | |||||
| CitRHM1 | aCL4478Contig1 | KN0AAI3AD11 | AT1G78570 | −0.52 | – |
| GLUCOSYLTRANSFERASES | |||||
| CitGTF1 | aCL3010Contig2 | IC0AAA58BE08 | AT1G77130 | 0.82 | – |
| CitGTF2 | aCL1592Contig1 | IC0AAA30DE02 | AT1G16570 | 0.58 | – |
| CitGTF3 | aCL3054Contig2 | KN0AAK3DE03 | AT3G50060 | – | 1.21 |
| CitGTF4 | aCL6931Contig1 | IC0AAA42BE09 | AT3G25140 | – | 0.65 |
| CitGTF5 | aCL5570Contig1 | C05056H08 | AT1G77990 | −1.42 | −1.51 |
| CitGTF6 | aCL6758Contig1 | C31701H10 | AT3G02100 | – | −1.23 |
| CitGTF7 | aKN0AAB3DB09ZM1_c | KN0AAB3DB09 | AT3G45400 | – | 0.60 |
| GDP-L-FUCOSE SYNTHASE | |||||
| CitGLFS1 | aCL790Contig1 | IC0AAA85AB07 | AT1G17890 | – | 0.73 |
| GALACTOSIDE 2-ALPHA-L-FUCOSYLTRANSFERASE | |||||
| CitGLFT1 | aIC0AAA69BA06RM1_c | IC0AAA69BA06 | AT1G74420 | – | 0.58 |
| CitGLFT2 | aCL5210Contig1 | C08029G10 | AT1G05575 | −0.87 | −1.24 |
| UDP-GLUCOSE DEHYDROGENASE | |||||
| CitUGD1 | aKN0AAP5YD20FM1_c | KN0AAP5YD20 | AT5G15490 | – | 0.51 |
| GLUCURONOSYL TRANSFERASE-LIKE PROTEIN | |||||
| CitGluT1 | aCL8573Contig1 | C02015B05 | AT3G55700 | – | −0.54 |
| UDP-GLUCURONATE DECARBOXYLASE | |||||
| CitUGluD1 | aCL1799Contig2 | C02011A11 | AT2G28760 | 1.67 | 1.57 |
| MANNOSYLTRANSFERASE-LIKE PROTEIN | |||||
| CitManT1 | aIC0AAA25BC01RM1_c | IC0AAA25BC01 | AT2G27100 | 0.77 | 0.60 |
Phylogenetic analyses reveal convergences between cell wall-remodeling proteins
Cell wall-remodeling enzymes are key players in abscission, but also in many other biological processes. Thus, the identification of abscission specific genes within these gene families acquires special relevance. However, this task becomes complicated due to the large size and high functional diversification of such gene families. To identify potential abscission specific cell wall-remodeling enzymes as well as reinforce the gene expression data, a phylogenetic analysis of gene families related to cell wall modification was carried out. For this purpose, genes encoding cell wall-remodeling proteins in the sequence of the C. clementina haploid genome (http://www.phytozome.net; Wu et al., 2014) were retrieved and compared to those of orange (C. sinensis; http://citrus.hzau.edu.cn/orange/) to identify all members of these citrus gene families (Table S3). We considered the phylogenetic relationships between cell wall-remodeling proteins of citrus and Arabidopsis along with proteins previously described as associated with abscission or dehiscence in other plant species.
Glycoside hydrolases and pectate lyases
Cellulases
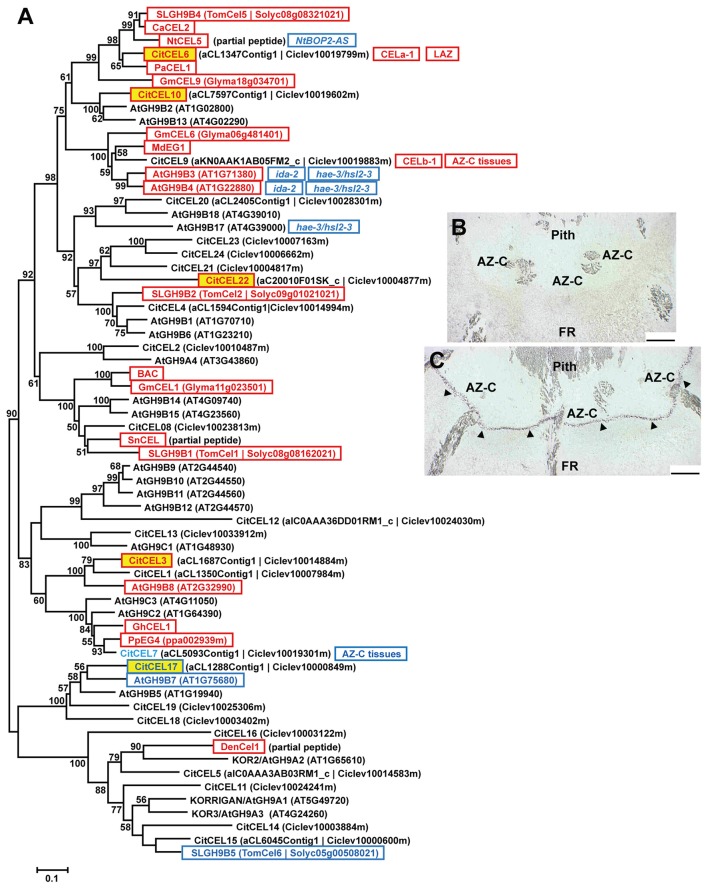
Cellulases/endo-1,4-β-glucanases (CELs; EC 3.2.1.4) belong to the glycoside hydrolase family 9 (GH9; CAZy; http://www.cazy.org; Henrissat, 1991; Lombard et al., 2014) and are classified in Groups A, B and C based on their particular protein domains (Libertini et al., 2004; Urbanowicz et al., 2007). Figure 4A shows the phylogenetic analysis of the citrus and Arabidopsis GH9 families and the deduced proteins of this family previously related to the abscission process in other plant species. Interestingly, three CELs up-regulated exclusively in the AZ-C (CitCEL6, CitCEL10, and CitCEL22) fell in the same subclade with other CELs up-regulated during abscission in other plant species (Figure 4A, Figure S4). CitCEL6 and CitCEL10 were closely related to CELs induced during abscission in avocado (PaCEL1; Tonutti et al., 1995), tomato (SLGH9B4; Brummell et al., 1999), pepper (CaCEL2; Ferrarese et al., 1995), tobacco (NtCEL5; Wu et al., 2012), and soybean (GmCEL09; Tucker et al., 2007), while CitCEL22 was more closely related to the tomato CEL SLGH9B2 (Brummell et al., 1999). CitCEL6 was previously reported as up-regulated in the citrus laminar abscission zone (LAZ) and the AZ-C (CsCEL-a1; Burns et al., 1998; Agustí et al., 2008, 2009, 2012). In addition, in-situ hybridizations on longitudinal AZ-C sections for CitCEL6 showed specific signal in activated AZ-C cells (Figures 4B,C), supporting a potential role of this CEL in cell wall degradation during the abscission process. However, CitCEL9 did not show a significant change in gene expression although it has been reported as up-regulated in both the LAZ and the fruit AZ-C (CsCEL-b1; Burns et al., 1998; Cheng et al., 2015). This gene showed a very close phylogenetic relationship to AtGH9B3 and AtGH9B4, which were down-regulated in ida-2 and hae hsl2 Arabidopsis mutant plants (Liu et al., 2013; Niederhuth et al., 2013). This could be due to the different methods used for AZ-C cells isolation and gene expression analyses. CitCEL3 was also up-regulated exclusively in the AZ-C (Figure 4A). It was located in a different subclade, closely related to AtGH9B8 that was up-regulated during stamen abscission in Arabidopsis (Lashbrook and Cai, 2008). On the other hand, CitCEL17, down-regulated exclusively in the AZ-C, was closely related to AtGH9B7 that was also down-regulated during stamen abscission in Arabidopsis (Lashbrook and Cai, 2008).
Figure 4.
Phylogenetic relationships between cellulases/endo-1,4-β-glucanases (CELs) and gene expression changes in response to ethylene inAZ-C and FR cells. (A) CELs annotated in the Citrus clementina haploid genome (Wu et al., 2014), regulated by ethylene in AZ-C cells and/or FR cells of Washington Navel maturing fruits and previously described as related to the abscission process in other plant species are shown. Phylogenetic trees are based on multiple alignments of proteins using the profile alignment function of ClustalW (http://www.ch.embnet.org/software/ClustalW-XXL.html) and were generated with MEGA7 (Kumar et al., 2016) using the neighbor-joining algorithm with 1000 bootstrap replicates. Only bootstrap supports higher than 50% were considered and are shown in the nodes. Sequences are color-coded as follows: , up-regulated exclusively in AZ-C cells; , down-regulated exclusively in AZ-C cells; , represented in the 20 K citrus microarray (Martinez-Godoy et al., 2008) but without hybridization results. and , up- and down-regulated, respectively, in Arabidopsis stamen-AZ cells (Lashbrook and Cai, 2008); , and , up- and down-regulated, respectively, during AZ activation in other plant species. CEL-a1 and CEL-b1, cellulases previously identified and characterized in citrus AZs (Burns et al., 1998). , up-regulated in the citrus laminar AZ (LAZ) cells during ethylene- and water stress-promoted leaf abscission (Agustí et al., 2008, 2012); , up-regulated in citrus AZ-C and surrounding tissues (Cheng et al., 2015). Transcripts of NtCEL5 were down-regulated in the corolla base of tobacco plants over-expressing an antisense-oriented sequence of NtBOP2 (; Wu et al., 2012). Transcripts of AtGH9B3 and AtGH9B4 were down-regulated in receptacles of mutant plants (Liu et al., 2013) and double mutant plants (Niederhuth et al., 2013) while transcripts of AtGH9B17 were down-regulated only in receptacles of hae-2/hsl2-3 double mutant plants (Niederhuth et al., 2013). (B,C)
CitCEL6 expression by in situ hybridization in the AZ-C of Ricalate Navel fruits after 24 h of ACC treatment (B, sense probe; C, anti-sense probe). Hybridization is indicated by the presence of a dark purple precipitate ( ). AZ-C, abscission zone C; FR, fruit rind. Scale bars: 500 μm.
). AZ-C, abscission zone C; FR, fruit rind. Scale bars: 500 μm.
Polygalacturonases
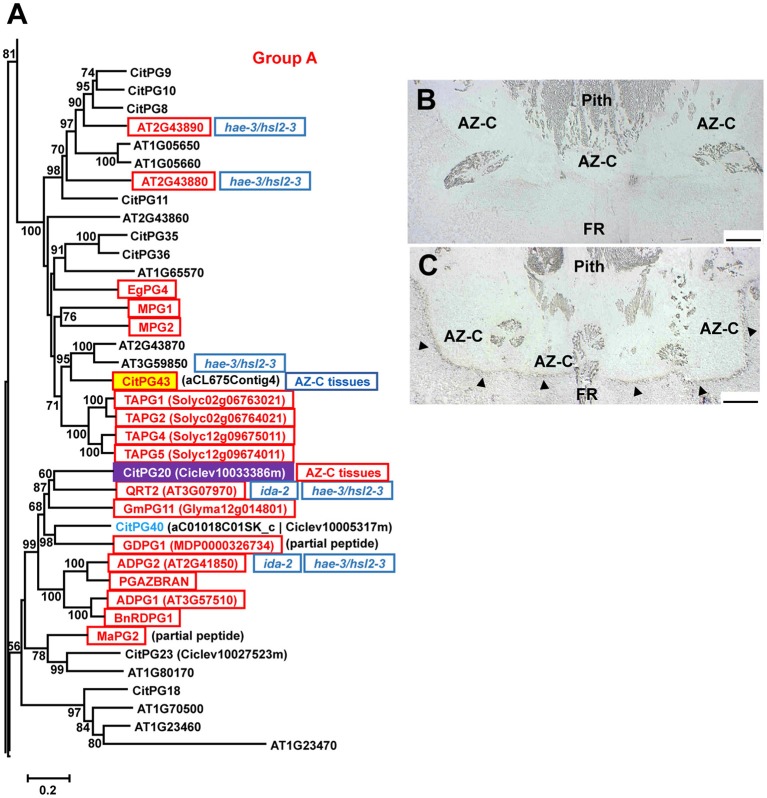
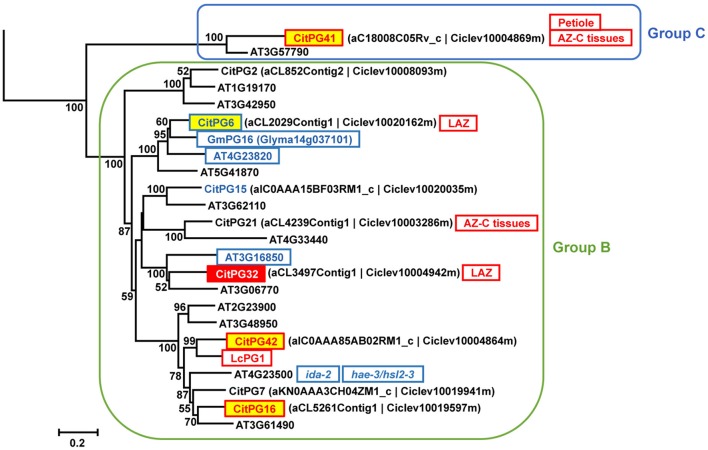
Polygalacturonases (PGs; EC 3.2.1.15) belong to the glycoside hydrolase family 28 (GH28) and are classified in three different groups (Groups A, B, and C) based on phylogenetic analyses (Kim et al., 2006). The large PG gene family of citrus (Figures 5A, 6, Figure S4) was highly represented in the AZ-C transcriptome. Regarding Group A, to which most of the PGs belong, CitPG43 was up-regulated exclusively in the AZ-C cells (Figure 5A) and CitPG20 was detected specifically in the abscission-activated AZ-C cells by in-situ hybridization (Figures 5B,C). Another three PGs, CitPG16, and CitPG42, belonging to Group B, and the only PG belonging to Group C, CitPG41, were also up-regulated exclusively in AZ-C cells during ethylene-promoted fruit abscission (Figure 6). On the other hand, a PG belonging to Group B, CitPG6, was down-regulated exclusively in the AZ-C (Figure 6) although it has been reported to be predominantly expressed in the LAZ cells of citrus leaves during ethylene-promoted abscission (Agustí et al., 2009).
Figure 5.
Phylogenetic relationships between polygalacturonases (PGs) of the Group A (GH28) and gene expression changes in response to ethylene in AZ-C and FR cells. PGs of the Group A located at the Citrus clementina haploid genome, regulated by ethylene in AZ-C cells and/or FR cells of Washington Navel maturing fruits and previously described as related to the abscission process in Arabidopsis (Lashbrook and Cai, 2008), tomato (Kalaitzis et al., 1995; Li et al., 2015; Hong and Tucker, 1998), apple (Atkinson et al., 2002; Li and Yuan, 2008), oilseed rape (Petersen et al., 1996; Sander et al., 2001; Gonzalez-Carranza et al., 2002), melon (Hadfield et al., 1998), oil palm (Roongsattham et al., 2012), and banana (Mbéguié-a-Mbéguié et al., 2009) are shown. Phylogenetic trees are based on multiple alignments of proteins using the profile alignment function of ClustalW (http://www.ch.embnet.org/software/ClustalW-XXL.html) and were generated with MEGA7 (Kumar et al., 2016) using the neighbor-joining algorithm with 1000 bootstrap replicates. Only bootstrap supports higher than 50% were considered and are shown in the nodes. Sequences are color-coded as follows: , up-regulated exclusively in AZ-C cells; , localization of transcripts in the citrus fruit AZ-C by in situ hybridization; , represented in the 20 K citrus microarray (Martinez-Godoy et al., 2008) but without hybridization results. , up-regulated in citrus AZ-C and surrounding tissues (Cheng et al., 2015). , up-regulated in Arabidopsis stamen-AZ cells (Lashbrook and Cai, 2008); , up-regulated during AZ activation in other plant species. Transcripts of ADPG2 (AT2G41850) and QRT2 (AT3G07970) were down-regulated in receptacles of mutant plants (Liu et al., 2013) and hae-2/hsl2-3 double mutant plants (Niederhuth et al., 2013) while transcripts of AT2G43880, AT2G43890 and AT3G59850 were down-regulated only in receptacles of double mutant plants (Niederhuth et al., 2013). (B,C)
CitPG20 expression by in situ hybridization in the AZ-C of Ricalate Navel fruits after 24 h of ACC treatment (B, sense probe; C, anti-sense probe). Hybridization is indicated by the presence of a dark purple precipitate ( ). AZ-C, abscission zone C; FR, fruit rind. Scale bars: 500 μm.
). AZ-C, abscission zone C; FR, fruit rind. Scale bars: 500 μm.
Figure 6.
Phylogenetic relationships between PGs from Groups B and C (GH28) and gene expression changes in response to ethylene in AZ-C and FR cells. PGs of the Groups B and C located at the Citrus clementina haploid genome, regulated by ethylene in AZ-C cells and/or FR cells of Washington Navel maturing fruits and previously described as related to the abscission process in Arabidopsis (Lashbrook and Cai, 2008), soybean (Tucker et al., 2007), and lychee (Peng et al., 2013). Phylogenetic trees are based on multiple alignments of proteins using the profile alignment function of ClustalW (http://www.ch.embnet.org/software/ClustalW-XXL.html) and were generated with MEGA7 (Kumar et al., 2016) using the neighbor-joining algorithm with 1000 bootstrap replicates. Only bootstrap supports higher than 50% were considered and are shown in the nodes. Sequences are color-coded as follows: , up-regulated exclusively in AZ-C cells; , down-regulated by exclusively in AZ-C cells; , up-regulated in both AZ-C and fruit rind (FR) cells; , down-regulated exclusively in FR cells. , down-regulated in Arabidopsis stamen-AZ cells (Lashbrook and Cai, 2008); , up-regulated during AZ activation in other plant species. , preferentially expressed in the citrus laminar AZ (LAZ) cells during ethylene-promoted leaf abscission (Agustí et al., 2009); Petiole, preferentially expressed in the citrus leaf petiolar cells during ethylene-promoted leaf abscission (Agustí et al., 2009); , up-regulated in citrus AZ-C and surrounding tissues (Cheng et al., 2015). Transcripts of AT4G23500 were down-regulated in receptacles of mutant plants (Liu et al., 2013) and double mutant plants (Niederhuth et al., 2013).
Many PGs were shown to be involved in organ abscission in different plant species. Moreover, several of the citrus PGs up-regulated in AZ-C cells during fruit abscission showed close phylogenetic relationship with some of the most actively expressed abscission-associated PGs. This was the case of CitPG43 and CitPG20 (Figure 5A). CitPG43 was grouped in the same subclade as different abscission-related PGs from tomato (TAPG1, TAPG2, TAPG4, and TAPG5; Kalaitzis et al., 1995, 1997; Hong and Tucker, 1998), ripe melon (MPG1 and MPG2; Hadfield et al., 1998), and oil palm fruit (EgPG4; Roongsattham et al., 2012). On the other hand, CitPG20 was grouped in the same subclade as several PGs associated with abscission or dehiscence in Arabidopsis (ADPG1, ADPG2, and QRT2; Gonzalez-Carranza et al., 2002, 2007; Ogawa et al., 2009), rape (PGAZBRAN and RDPG1; Petersen et al., 1996; Sander et al., 2001; Gonzalez-Carranza et al., 2002), apple (GDPG1; Atkinson et al., 2002), soybean (GmPG11; Tucker et al., 2007), and banana (MaPG2; Mbéguié-a-Mbéguié et al., 2009). Both CitPG20 and CitPG43 have recently been reported to be up-regulated by ethephon in citrus fruit AZ-C-enriched tissues (Cheng et al., 2015). In relation to the Group B of PGs up-regulated exclusively in the AZ-C, CitPG16 and CitPG42, that were closely related phylogenetically, were also closely related to PGs involved in lychee fruitlet abscission (LcPG1; Peng et al., 2013) and floral organ abscission (Liu et al., 2013; Niederhuth et al., 2013). Regarding CitPG6, it was grouped in the same subclade as GmPG16 (Figure 6) that was down-regulated during abscission in soybean (GmPG16; Tucker et al., 2007). Therefore, despite PGs comprise a large gene family in plants, it has been possible to highlight a strong phylogenetic connection to PGs that are actively regulated during organ abscission.
Pectate lyases
Pectate lyases (PLs; EC 4.2.2.2) belong to pectate lyase family 1 (PL1; CAZy; http://www.cazy.org; Henrissat, 1991; Lombard et al., 2014) and are classified in five different groups (Groups I, II, III, IV, and V; Sun and Van Nocker, 2010). Group I of PLs contained all enzymes previously reported to be induced during organ abscission in Arabidopsis (AtPLL18, AtPLL19, AtPLL22, AtPLL23, AtPLL24, and AtPLL25; Lashbrook and Cai, 2008; Niederhuth et al., 2013), soybean (GmPL01 and GmPL02; Tucker et al., 2007), banana (MaPEL1; Mbéguié-a-Mbéguié et al., 2009), and rose (RbPEL1; Singh et al., 2011; Figure S5). In agreement with that, the expression of CitPL5 was up-regulated exclusively in the AZ-C during ethylene-promoted fruit abscission. CitPL19 was up-regulated in both the AZ-C and the FR and was closely related to PLs that were also up-regulated in the AZ of rose (RbPEL1; Singh et al., 2011) and Arabidopsis (AtPLL25 and AtPLL26; Lashbrook and Cai, 2008; Sun and Van Nocker, 2010). Furthermore, CitPL19 was previously reported to be up-regulated exclusively in the citrus LAZ (Agustí et al., 2008, 2009), suggesting a role in fruit and leaf abscission but also in cell wall degradation events taking place in the FR. Three citrus PLs (CitPL5, CitPL7, and CitPL19) have recently been reported to be up-regulated by ethephon in fruit AZ-C-enriched tissues (Cheng et al., 2015; Figure S5). On the other hand, CitPL1, also belonging to Group I-PLs, was down-regulated exclusively in the AZ-C.
Other cell wall modifying enzymes
In addition to the gene families mentioned above, several cell wall modification-related families included genes regulated by ethylene in the AZ-C and/or the FR such as pectin-methylesterases (PMEs; family CE8), pectin-acetylesterases (PAEs; family CE13), β-galactosidases (β-GALs and members of the GH2 gene family), β-glucosidases (β-GLUs; family GH1), endo-β-mannosidases (β-MANs; family GH5), xyloglucan endotransglucosylases/hydrolases (XTHs; family GH16), β-xylosidases (β-XYLs; family GH3), α-xylosidases (α-XYLs; family GH31), and expansins (EXPs; Table 1, Tables S2, S4). Phylogenetic relationships between citrus, Arabidopsis and abscission-related genes from different species are shown in Figures S3–S6.
In summary, this study revealed a link between phylogenetic proximity and expression pattern between cell wall remodeling enzymes of citrus and those previously reported to be involved in organ abscission in other plant species, suggesting that different plant species use common genes to control similar processes. Therefore, those genes exclusively up-regulated in the AZ-C and closely related to genes that were also up-regulated during abscission in other plant species (CitCEL3, CitCEL6, CitCEL10, CitCEL22, CitPG43, CitPG16, CitPG20, CitPG42, CitPL5, CitXTH24, and CitEXP14) might have a molecular function during citrus fruit abscission (Table 1, Figures 4–6, Figures S3–S6). Furthermore, phylogenetic analysis also provided insights into the functional divergence between members of these gene families. More importantly, our results provided a set of genes belonging to groups, subfamilies, or families that have not yet been described as involved in abscission. These include, the Group 1 of PMEs and the family CE13 (Figure S3), the Group C of PGs (Figure 6), the family GH2 of β-GALs, the family GH1 (Figure S4), and the subfamily EXLB of EXPs (Figure S6).
Pectic polysaccharides change their spatial distribution in the cell walls of AZ-C cells during abscission
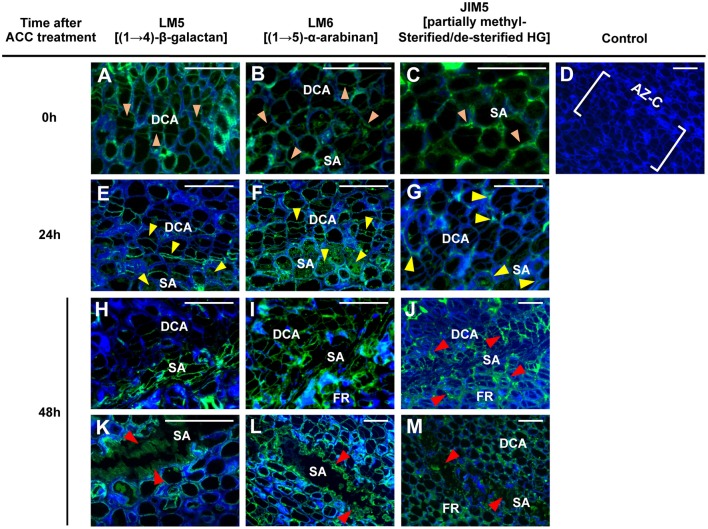
Having identified a set of cell wall-related genes that were up-regulated during fruit abscission and to test whether such gene expression changes corresponded to enzymatic activity, we next attempted to identify alterations in pectic polysaccharides arrangements at the AZ-C cell walls. To examine such changes, we carried out an immunolocalization of pectic polysaccharides assay during ACC-promoted abscission. At 0 h, the epitopes (1 → 4)-β-D-galactan, (1 → 5)-α-L-arabinan, and partially methylesterified/de-esterified homogalacturonan (HG) recognized by the mAbs LM5 and LM6 and JIM5, respectively, were homogeneously distributed along cell walls of the AZ-C (Figures 7A–C). An increase in the (1 → 4)-β-D-galactan (LM5) labeling intensity in the starch-rich cell area of the AZ-C was observed at 24 h after ACC treatment (Figure 7E) whereas (1 → 5)-α-L-arabinan (LM6) and partially methylesterified/de-esterified HGs (JIM5) labeling highlighted cell wall changes in the divided cell area (Figures 7F,G). At a more advanced stage of the process (48 h), the labeling intensity detected for the three mAbs was higher at the cell layers closer to the separation line (Figures 7H–J), probably due to the accumulation of residual compounds of these pectic polysaccharides from the dissolution of the middle lamella and cell walls. Furthermore, an amorphous material with high fluorescence was observed at the regions where cell separation was completed (Figures 7K–M), in accordance with findings observed in Figure 2B and also with the first anatomical and histological description of citrus fruit abscission reported by Wilson and Hendershott (1968).
Figure 7.
Immunolocalization of pectic polysaccharides in the AZ-C. Longitudinal sections of tissue containing the AZ-C of Ricalate Navel maturing fruits were incubated with the monoclonal antibodies (mAb) LM5 (A,E,H,K), LM6 (B,F,I,L), and JIM5 (C,G,J,M) to detect (1,4)-β-D-galactans, (1,5)-α-L-arabinans and partially methylesterified/de-esterified HGs, respectively, after 0 (A, B and C), 24 (E,F,G) and 48 h (H–M) of ACC treatment. Control did not show immunofluorescence (D). Scale bars: 5 μm. Key labeling: walls at the two AZ-C cell areas (divided cells area [DCA] and starch-rich area [SA]) and at the fruit rind (FR) cell layers just below the SA showing fluorescence due to each of the mAbs after 0 ( ), 24 (
), 24 ( ) or 48 (
) or 48 ( ) h of ACC treatment. Micrographs represent the merger of images from pectic epitopes detection by mAbs (green) and from cellulose detection by calcofluor white (blue).
) h of ACC treatment. Micrographs represent the merger of images from pectic epitopes detection by mAbs (green) and from cellulose detection by calcofluor white (blue).
Changes in the pectic polysaccharides distribution in the AZ-C cell walls enabled us to correlate evidences of enzymatic activity with gene expression results. In particular, based on immunodetection of partially methylesterified/de-esterified HG and expression results, we propose that PMEs CitPME24 and CitPME41 and PAE CitPAE4 may act on de-esterification of HGs in the AZ-C cell walls (Table 1, Figure S3). The activity of PMEs and PAEs is thought to facilitate the subsequent action of pectin hydrolases (Chen and Mart, 1996). Thus, the PGs CitPG43, CitPG16, CitPG20, CitPG41, and CitPG42, and the PLs CitPL5, and CitPL19 may potentially hydrolyze the HGs highly accessible after CitPME24, CitPME41, and CitPAE4 activity (Table 1, Figures 5, 6, Figures S3–S5). Finally, the only α-L-arabinofuranosidase identified in citrus (CitASD1) did not show significant changes in gene expression based on the statistical cutoff mentioned in materials and methods (Figure S4). In Arabidopsis, it has been reported that AtBXL1 and AtBXL3 acted as bifunctional α-L-arabinofuranosidase/β-D-xylosidases (Minic et al., 2004; Arsovski et al., 2009). Therefore, changes observed in (1, 5)-α-L-arabinans at the AZ-C might be due to the dual activity of β-XYLs such as CitBXL11 and CitBXL19, which were in the same clade as AtBXL1 and AtBXL3 (Figure S4).
A set of genes involved in lignin biosynthesis and polymerization are regulated in the AZ-C cells
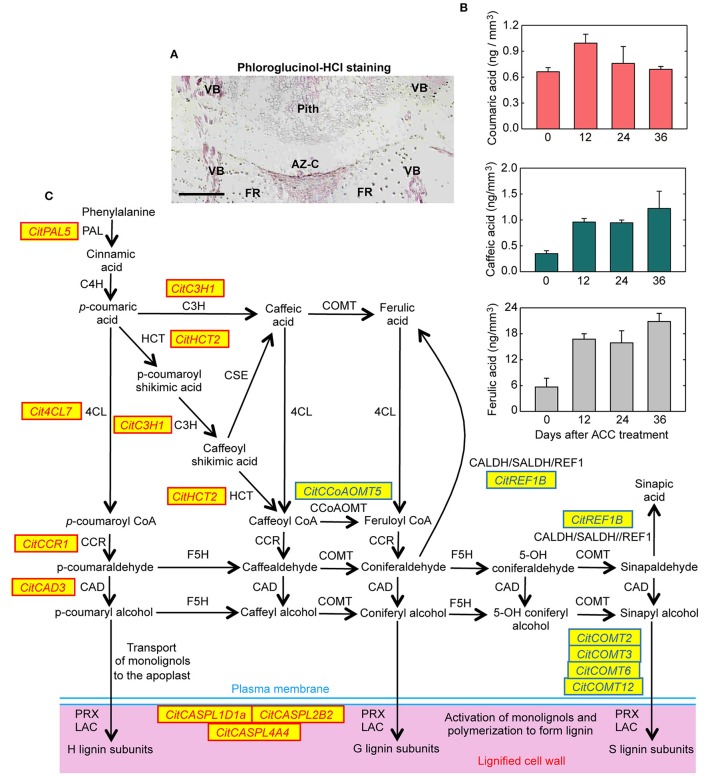
Significant expressed genes belonging to different gene families involved the monolignol biosynthesis pathway were up-regulated by ethylene exclusively in the AZ-C (Table 2, Figure 8, Figure S7). These included a phenylalanine ammonia-lyase (CitPAL5), a p-coumarate 3-hydroxylase (CitC3H1), a hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase (CitHCT2), a 4-coumarate-CoA ligase-like protein (Cit4CL7), a cinnamoyl-CoA reductase-like protein (CitCCR1), and a cinnamyl alcohol dehydrogenase (CitCAD3). In addition, three genes encoding proteins involved in the oxidative coupling of monolignols and belonging to the CASPARIAN STRIP MEMBRANE DOMAIN PROTEIN family (CitCASPL1D1a, CitCASPL2B2, and CitCASPL4A4) were also up-regulated by ethylene exclusively in the AZ-C (Table 2, Figure 8, Figure S7). These results correlated with the observation of lignin deposition in the AZ-C after 24 and 48 h of ethylene/ACC treatment (Figures 1B,C) and the increase in the level of lignin intermediates detected by UPLC-MS/MS in AZ-C cells (Figure 8B). A significant increase of coumaric acid was observed at 12 h after ACC treatment. In addition, levels of caffeic acid and ferulic acid, compounds that are synthesized from coumaric acid, increased at 12 h and were maintained up to 36 h. Taken together, these data mainly reflected the activation of the H lignin pathway which results from the incorporation of p-hydroxyphenyl (H) units into the lignin polymer (Figure 8C). However, G lignin (lignin with guaiacyl units) biosynthesis would be also possible in activated AZ-C cells since an increase in CitC3H1 and CitHCT2 expression and caffeic and ferulic acids levels also occurred in AZ-C cells despite any member of the CCoAOMTs gene family were up-regulated (Figure 8). Regarding S lignin biosynthesis, four caffeic acid O-methyltransferases (CitCOMT2, CitCOMT3, CitCOMT6, and CitCOMT12) were down-regulated exclusively in AZ-C cells (Table 2, Figure 8C). CitCOMT3 was closely related in sequence to AtCOMT1 (Figure S7), a member of the Arabidopsis COMT gene family with 5-hydroxyconiferaldehyde O-methyltransferase activity that has been implicated directly in S lignin synthesis (Nakatsubo et al., 2008). These results suggest that the S lignin pathway might be inactive in AZ-C cells during fruit abscission. In woody dicotyledons, such as citrus, lignin is polymerized from mostly G and S lignin subunits (Sarkanen and Ludwig, 1971). However, lignin composition can differ among cell types (Nakashima et al., 2008; Ruel et al., 2009) and expression data suggested that cell walls of AZ-C cells might be mainly enriched in H lignin and probably also in G lignin subunits.
Table 2.
Relative gene expression values (AZ-Ct vs. AZ-C0) of genes involved in lignin biosynthesis exclusively regulated by ethylene in AZ-C cells.
| Name | Contig/Singleton ID | Microarray probe | Putative Ath orthologue | Relative expression [log2 (AZ-Ct/AZ-C0)] | |
|---|---|---|---|---|---|
| 12 h | 24 h | ||||
| PHENYLALANINE AMMONIA-LYASES (PALs) | |||||
| CitPAL5 | aC02002A11SK_c | C02002A11 | AT2G37040 | 2.33 | 1.22 |
| 4-COUMARATE:CoA LIGASES (4CLs) | |||||
| Cit4CL7 | aC31504D07EF_c | C31504D07 | AT4G19010 | – | 1.00 |
| p-COUMARATE 3-HYDROXYLASES (C3Hs) | |||||
| CitC3H1 | aC32104F09EF_c | C32104F09 | AT2G40890 | 1.03 | 0.65 |
| HYDROXYCINNAMOYL CoA:SHIKIMATE/QUINATE HYDROXYCINNAMOYLTRANSFERASES (HCTs) | |||||
| CitHCT2 | aIC0AAA81CG08RM1_c | IC0AAA81CG08 | AT5G48930 | 0.58 | 0.56 |
| CAFFEOYL-CoA 3-O-METHYLTRANSFERASES (CCoAOMTs) | |||||
| CitCCoAOMT5 | aCL18Contig10 | C31100A08 | AT4G34050 | – | −0.78 |
| CINNAMOYL-CoA REDUCTASES (CCRs) | |||||
| CitCCR1 | aCL8119Contig1 | C34205C03 | AT2G23910 | 0.94 | 1.39 |
| CONIFERALDEHYDE/SINAPALDEHYDE DEHYDROGENASES (CALDH/SALDH//REF1) | |||||
| CitREF1B | aCL1370Contig1 | C01019E09 | AT3G24503 | – | −0.97 |
| CAFFEIC ACID O-METHYLTRANSFERASES (COMTs) | |||||
| CitCOMT2 | aCL4061Contig1 | C03007H04 | AT5G54160 | −0.75 | – |
| CitCOMT3 | aCL3343Contig1 | C05133H01 | AT5G54160 | – | −0.87 |
| CitCOMT6 | aCL4936Contig1 | C02003G01 | AT5G54160 | −1.41 | – |
| CitCOMT12 | aC08039B05SK_c | C08039B05 | AT5G54160 | – | −0.56 |
| CINNAMYL ALCOHOL DEHYDROGENASES (CADs) | |||||
| CitCAD3 | aCL33Contig1 | C18001A01 | AT4G37970 | 0.90 | – |
| CASPARIAN STRIP MEMBRANE DOMAIN PROTEINS (CASPs) | |||||
| CitCASPL1D1a | aCL6688Contig1 | C04010H11 | AT4G15610 | 0.74 | – |
| CitCASPL2B2 | aCL5933Contig1 | C32104D12 | AT2G35760 | 0.82 | – |
| CitCASPL4A4 | aC08028B10SK_c | C08028B10 | AT4G11655 | 0.93 | 0.61 |
–, No significant regulation. Putative gene identifications are based on sequence homology with Arabidopsis thaliana. Additional data are shown in Figure S7.
Figure 8.
Lignin biosynthesis and deposition in the abscission zone area during citrus fruit abscission. (A) Tissue localization of lignin through phloroglucinol-HCl staining in longitudinal sections of the AZ-C from Washington Navel fruits treated for 48 h with ethylene. Lignin is deposited at the central core of the AZ-C, at the separation line, and spreads to the adjacent cells of the fruit rind through the distal side of the AZ-C. Scale bar: 500 μm. Key labeling: AZ-C, abscission zone C; FR, fruit rind; VB, vascular bundles. (B) Lignin biosynthesis intermediates were quantified through UPLC-MS/MS in AZ-C cells at 0, 12, 24, and 36 h after ACC treatment. Data are expressed as ng of coumaric acid, caffeic acid and ferulic acid per mm3 of microdissected tissue. The results are means of three independent samples containing ~40,000 pooled AZ-C cells ± SE. (C) Genes belonging to the general phenylpropanoid and monolignol biosynthesis pathways and lignin polymerization up- or down-regulated exclusively in the fruit AZ-C cells during ethylene-promoted citrus fruit abscission. Enzymes and proteins associated with monolignol biosynthesis and polymerization are: phenylalanine ammonia lyase (PAL), trans-cinnamate 4-hydroxylase (C4H), 4-coumarate:CoA ligase (4CL), hydroxycinnamoyl-CoA:shikimate/quinate hydroxycinnamoyl transferase (HCT), coniferaldehyde dehydrogenase/sinapaldehyde dehydrogenase (CALDH/SALDH), caffeoyl shikimate esterase (CSE), p-coumarate 3-hydroxylase (C3H), caffeoyl-CoA 3-O-methyltransferase (CCoAOMT), cinnamoyl-CoA reductase (CCR), ferulate 5-hydroxylase (F5H), caffeic acid O-methyltransferase (COMT), cinnamyl alcohol dehydrogenase (CAD), Casparian strip membrane domain protein-like (CASPL), laccase (LAC) and peroxidase (PRX).
The role of lignin deposition has been associated with the generation of protective layers at the tissues remaining in the plant during the last step of the abscission process (Addicot, 1982; Agustí et al., 2008; Van Nocker, 2009). However, it has been suggested that lignification could also facilitate the mechanical cell wall breakage during cell separation processes (Sexton, 1979; Liljegren et al., 2000). In the AZ-C, lignin deposition only occurred at the distal side of the AZ-C (Figure 8A). This differential deposition of lignin strongly suggests that this polymer mainly acts by generating a tension in the fracture plane to facilitate cell wall breakage during citrus fruit abscission rather than forming protective layers.
Conclusion
In this work, the isolation of specialized cell types through LM, combined with the global transcriptional analysis of ethylene-promoted AZ-C cells and the comparison with adjacent FR cells, enabled us to identify an AZ-C-exclusive gene set potentially involved in citrus fruit abscission. This set of genes includes those related to cell wall remodeling as well as lignin biosynthesis and polymerization. The combined function of these genes would enable cell wall modifications necessary for organ detachment (Figure 9). These results, together with the anatomical and morphological analysis of the AZ-C, the determination of changes in pectic polysaccharides distribution and the deposition of extracellular polymers observed in the activated AZ-C, lead to the most comprehensive characterization of citrus fruit abscission performed to date. In addition, our work shows the first classification in citrus of gene families involved in cell wall modification, which are crucial in abscission, and reveals a robust nexus between phylogenetic proximity and expression pattern during abscission in citrus and other plant species, not previously described. Therefore, this study strongly suggests that different plant species use common genes to control the abscission process. The dataset provided in this study is a highly valuable resource for guiding future functional analyses in order to answer specific abscission-related questions. In particular, those cell wall-related genes, which are evolutionarily conserved in citrus and other plant species with similar expression pattern during abscission, would represent major candidate genes for further biotechnological approaches.
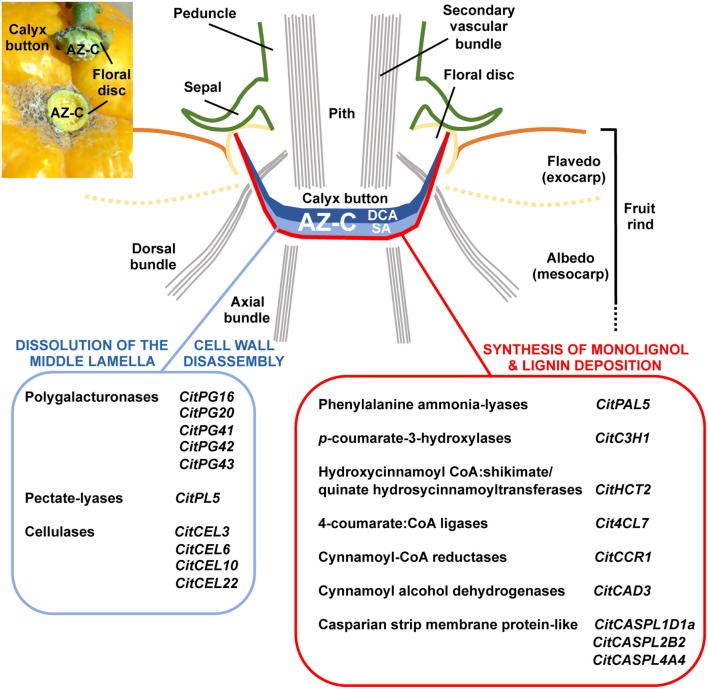
Figure 9.
Specific cellular and molecular events involved in the dissolution of the middle lamella, the disassembly of cell walls and the synthesis and deposition of lignin in the AZ-C during ethylene-promoted abscission. The AZ-C consists of two different cell areas, the Divided Cells Area (DCA) and the Starch-rich Area (SA). The early cellular and molecular events associated with citrus fruit abscission occur in the central core of the AZ-C between the axial vascular bundles and spread up to the calyx button periphery reaching then the floral disc. The final outcome of this cell separation process is the shedding of the fruit remaining the calyx button attached to the tree as shown in the inset of the upper-left corner of the figure. Two parallel cellular events involving cell wall dissolution and synthesis and deposition of lignin occurred specifically in the SA of the AZ-C cells during abscission. These cellular events are potentially promoted by the tissue-specific expression of particular members of several gene families that have been clearly involved in those metabolic pathways.
Author contributions
PM, JA, MT, and FT conceived the survey and designed the experiments; PM performed laser microdissection of citrus tissues; VA and AG performed metabolite profiling analyses; PM, JA, and CD performed microarray experiments and analyses; MC, SC, and FT performed immunolocalization experiments; LE, MG, MP, and FT performed in situ hybridization experiments; PM, JA, MT, and FT wrote the article with contributions of all the authors; all authors discussed the results and edited the article.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank E. Blázquez, I. Sanchís, A. Boix, and M. A. Argomániz for the help on all of the assays and experiments performed both in the laboratory and in the field. This study was financially supported by the Spanish Instituto Nacional de Investigaciones Agrarias (grant RTA2008-00065-00-00 to FT [including a PhD fellowship for PM] and RTA2014-00071-C06-01 to MT), the Spanish Ministerio de Economia e Innovación (grants PSE-060000-2009-8 and IPT-010000-2010-43 to MT and BIO2011-26302 to MP) and the Spanish Ministerio de Industria (grant AGL2011-30240 to MT). VA and CD were recipients of a “Juan de la Cierva” and an INIA/CCAA postdoctoral contract, respectively.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00126/full#supplementary-material
Specific primers used for sqRT-PCR.
Genes regulated by ethylene exclusively in AZ-C or FR cells, and in both AZ-C and FR cells.
Functional categories related to polysaccharide metabolism and subcellular localization regulated in the AZ-C and the FR.
Identification of citrus genes belonging to different families of cell wall remodeling enzymes, and monolignol biosynthesis and polymerization.
sqRT-PCR-based relative expression in the AZ-C and the FR.
LM isolation of AZ-C and FR cells.
Phylogenetic relationships between Carbohydrate esterases.
Phylogenetic relationships between Glycoside hydrolases.
Phylogenetic relationships between Pectate lyases.
Phylogenetic relationships between Expansins.
Phylogenetic relationships between members of gene families associated with monolignol biosynthesis and polymerization.
References
- Addicot F. T. (1982). Abscission. University of California Press, Berkeley. [Google Scholar]
- Agustí J., Gimeno J., Merelo P., Serrano R., Cercos M., Conesa A., et al. (2012). Early gene expression events in the laminar abscission zone of abscission-promoted citrus leaves after a cycle of water stress/rehydration: involvement of CitbHLH1. J. Exp. Bot. 63, 6079–6091. 10.1093/jxb/ers270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agustí J., Merelo P., Cercos M., Tadeo F. R., Talon M. (2008). Ethylene-induced differential gene expression during abscission of citrus leaves. J. Exp. Bot. 59, 2717–2733. 10.1093/jxb/ern138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agustí J., Merelo P., Cercos M., Tadeo F. R., Talon M. (2009). Comparative transcriptional survey between laser-microdissected cells from laminar abscission zone and petiolar cortical tissue during ethylene-promoted abscission in citrus leaves. BMC Plant Biol. 9:127. 10.1186/1471-2229-9-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argamasilla R., Gómez-Cadenas A., Arbona V. (2014). Metabolic and regulatory responses in citrus rootstocks in response to adverse environmental conditions. J. Plant Growth Regul. 33, 169–180. 10.1007/s00344-013-9359-z [DOI] [Google Scholar]
- Arsovski A. A., Popma T. M., Haughn G. W., Carpita N. C., McCann M. C., Western T. L. (2009). AtBXL1 encodes a bifunctional beta-D-xylosidase/alpha-L-arabinofuranosidase required for pectic arabinan modification in Arabidopsis mucilage secretory cells. Plant Physiol. 150, 1219–1234. 10.1104/pp.109.138388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson R. G., Schroder R., Hallett I. C., Cohen D., Macrae E. A. (2002). Overexpression of polygalacturonase in transgenic apple trees leads to a range of novel phenotypes involving changes in cell adhesion. Plant Physiol. 129, 122–133. 10.1104/pp.010986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell D. A., Hall B. D., Bennett A. B. (1999). Antisense suppression of tomato endo-1,4-beta-glucanase Cel2 mRNA accumulation increases the force required to break fruit abscission zones but does not affect fruit softening. Plant Mol. Biol. 40, 615–622. 10.1023/A:1006269031452 [DOI] [PubMed] [Google Scholar]
- Burns J. K. (2002). Using molecular biology tools to identify abscission materials for citrus. HortScience 37, 459–464. [Google Scholar]
- Burns J. K., Lewandowski D. J., Nairn C. J., Brown G. E. (1998). Endo-1,4-β-glucanase gene expression and cell wall hydrolase activities during abscission in Valencia orange. Physiol. Plant. 102, 217–225. 10.1034/j.1399-3054.1998.1020209.x [DOI] [Google Scholar]
- Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238. 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso M., Merelo P., Distefano G., La Malfa S., Lo Piero A. R., Tadeo F. R., et al. (2012). Comparative transcriptome analysis of stylar canal cells identifies novel candidate genes implicated in the self-incompatibility response of Citrus clementina. BMC Plant Biol. 12:20. 10.1186/1471-2229-12-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cercos M., Soler G., Iglesias D. J., Gadea J., Forment J., Talon M. (2006). Global analysis of gene expression during development and ripening of citrus fruit flesh. A proposed mechanism for citric Acid utilization. Plant Mol. Biol. 62, 513–527. 10.1007/s11103-006-9037-7 [DOI] [PubMed] [Google Scholar]
- Chen E. M. W., Mart A. J. (1996). Nature of sites hydrolyzable by endopolygalacturonase in partially-esterified homogalacturonans. Carbohydr. Polym. 29, 129–136. 10.1016/0144-8617(96)00005-7 [DOI] [Google Scholar]
- Cheng C., Zhang L., Yang X., Zhong G. (2015). Profiling gene expression in citrus fruit calyx abscission zone (AZ-C) treated with ethylene. Mol. Genet. Genomics 290, 1991–2006. 10.1007/s00438-015-1054-2 [DOI] [PubMed] [Google Scholar]
- Coimbra S., Almeida J., Junqueira V., Costa M. L., Pereira L. G. (2007). Arabinogalactan proteins as molecular markers in Arabidopsis thaliana sexual reproduction. J. Exp. Bot. 58, 4027–4035. 10.1093/jxb/erm259 [DOI] [PubMed] [Google Scholar]
- Corbacho J., Romojaro F., Pech J. C., Latche A., Gomez-Jimenez M. C. (2013). Transcriptomic events involved in melon mature-fruit abscission comprise the sequential induction of cell-wall degrading genes coupled to a stimulation of endo and exocytosis. PLoS ONE 8:e58363. 10.1371/journal.pone.0058363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estornell L. H., Agusti J., Merelo P., Talon M., Tadeo F. R. (2013). Elucidating mechanisms underlying organ abscission. Plant Sci. 199–200, 48–60. 10.1016/j.plantsci.2012.10.008 [DOI] [PubMed] [Google Scholar]
- Ferrarese L., Trainotti L., Moretto P., Polverino De Laureto P., Rascio N., Casadoro G. (1995). Differential ethylene-inducible expression of cellulase in pepper plants. Plant Mol. Biol. 29, 735–747. 10.1007/BF00041164 [DOI] [PubMed] [Google Scholar]
- Gallasch P. (1996). Evaluating new selections of late hanging navel orange, in International Citrus Congress (Sun City: International Society of Citriculture; ). [Google Scholar]
- Gil-Amado J. A., Gomez-Jimenez M.C. (2013). Transcriptome analysis of mature fruit abscission control in olive. Plant Cell Physiol. 54, 244–269. 10.1093/pcp/pcs179 [DOI] [PubMed] [Google Scholar]
- Gomez M. D., Urbez C., Perez-Amador M. A., Carbonell J. (2011). Characterization of constricted fruit (ctf) mutant uncovers a role for AtMYB117/LOF1 in ovule and fruit development in Arabidopsis thaliana. PLoS ONE 6:e18760. 10.1371/journal.pone.0018760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Carranza Z. H., Elliott K. A., Roberts J. A. (2007). Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J. Exp. Bot. 58, 3719–3730. 10.1093/jxb/erm222 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Carranza Z. H., Whitelaw C. A., Swarup R., Roberts J. A. (2002). Temporal and spatial expression of a polygalacturonase during leaf and flower abscission in oilseed rape and Arabidopsis. Plant Physiol. 128, 534–543. 10.1104/pp.010610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren R. (1993). Anatomical, physiological, and hormonal aspects of abscission in citrus. Hort. Rev. 15:182 10.1002/9780470650547.ch4 [DOI] [Google Scholar]
- Hadfield K. A., Rose J. K. C., Yaver D. S., Berka R. M., Bennett A. B. (1998). Polygalacturonase gene expression in ripe melon fruit supports a role for polygalacturonase in ripening-associated pectin disassembly. Plant Physiol. 117, 363–373. 10.1104/pp.117.2.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B. (1991). A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316. 10.1042/bj2800309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. B., Tucker M. L. (1998). Genomic organization of six tomato polygalacturonases and 5′ upstream sequence identity with tap1 and win2 genes. Mol. Gen. Genet. 258, 479–487. 10.1007/s004380050758 [DOI] [PubMed] [Google Scholar]
- Huberman M., Goren R., Zamski E. (1983). Anatomical aspects of hormonal regulation of abscission in citrus – The shoot-peduncle abscission zone in the non-abscising stage. Physiol. Plant. 59, 445–454. 10.1111/j.1399-3054.1983.tb04228.x [DOI] [Google Scholar]
- Jones L., Seymour G. B., Knox J. P. (1997). Localization of pectic galactan in tomato cell walls using a monoclonal antibody specific to (1[−>]4)-[beta]-D-galactan. Plant Physiol. 113, 1405–1412. 10.1104/pp.113.4.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaitzis P., Koehler S. M., Tucker M. L. (1995). Cloning of a tomato polygalacturonase expressed in abscission. Plant Mol. Biol. 28, 647–656. 10.1007/BF00021190 [DOI] [PubMed] [Google Scholar]
- Kalaitzis P., Solomos T., Tucker M. L. (1997). Three different polygalacturonases are expressed in tomato leaf and flower abscission, each with a different temporal expression pattern. Plant Physiol. 113, 1303–1308. 10.1104/pp.113.4.1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Shiu S.-H., Thoma S., Li W.-H., Patterson S. E. (2006). Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol. 7:R87. 10.1186/gb-2006-7-9-r87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox J. P., Linstead P. J., King J., Cooper C., Roberts K. (1990). Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta 181, 512–521. 10.1007/BF00193004 [DOI] [PubMed] [Google Scholar]
- Konishi S., Izawa T., Lin S. Y., Ebana K., Fukuta Y., Sasaki T., et al. (2006). An SNP caused loss of seed shattering during rice domestication. Science 312, 1392–1396. 10.1126/science.1126410 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrook C. C., Cai S. (2008). Cell wall remodeling in Arabidopsis stamen abscission zones: temporal aspects of control inferred from transcriptional profiling. Plant Signal. Behav. 3, 733–736. 10.4161/psb.3.9.6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. (2009). SMART 6: recent updates and new developments. Nucleic Acids Res. 37, D229–D232. 10.1093/nar/gkn808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Lee S.-H., Hsu H.-Y. (2011). Review on fruit harvesting method for potential use of automatic fruit harvesting systems. Proc. Eng. 23, 351–366. 10.1016/j.proeng.2011.11.2514 [DOI] [Google Scholar]
- Li C., Wang Y., Ying P., Ma W., Li J. (2015). Genome-wide digital transcript analysis of putative fruitlet abscission related genes regulated by ethephon in litchi. Front. Plant Sci. 6:502. 10.3389/fpls.2015.00502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. G., Yuan R. C. (2008). NAA and ethylene regulate expression of genes related to ethylene biosynthesis, perception, and cell wall degradation during fruit abscission and ripening in ‘Delicious’ apples. J. Plant Growth. Regul. 27, 283–295. 10.1007/s00344-008-9055-6 [DOI] [Google Scholar]
- Libertini E., Li Y., McQueen-Mason S. J. (2004). Phylogenetic analysis of the plant endo-beta-1,4-glucanase gene family. J. Mol. Evol. 58, 506–515. 10.1007/s00239-003-2571-x [DOI] [PubMed] [Google Scholar]
- Liljegren S. J., Ditta G. S., Eshed Y., Savidge B., Bowman J. L., Yanofsky M. F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. 10.1038/35008089 [DOI] [PubMed] [Google Scholar]
- Liu B., Butenko M. A., Shi C. L., Bolivar J. L., Winge P., Stenvik G. E., et al. (2013). NEVERSHED and INFLORESCENCE DEFICIENT IN ABSCISSION are differentially required for cell expansion and cell separation during floral organ abscission in Arabidopsis thaliana. J. Exp. Bot. 64, 5345–5357. 10.1093/jxb/ert232 [DOI] [PubMed] [Google Scholar]
- Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Godoy M. A., Mauri N., Juarez J., Marques M. C., Santiago J., Forment J., et al. (2008). A genome-wide 20 K citrus microarray for gene expression analysis. BMC Genomics 9:318. 10.1186/1471-2164-9-318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matas A. J., Agustí J., Tadeo F. R., Talón M., Rose J. K. C. (2010). Tissue-specific transcriptome profiling of the citrus fruit epidermis and subepidermis using laser capture microdissection. J. Exp. Bot. 61, 3321–3330. 10.1093/jxb/erq153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbéguié-a-Mbéguié D., Hubert O., Baurens F. C., Matsumoto T., Chillet M., Fils-Lycaon B., et al. (2009). Expression patterns of cell wall-modifying genes from banana during fruit ripening and in relationship with finger drop. J. Exp. Bot. 60, 2021–2034. 10.1093/jxb/erp079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meir S., Philosoph-Hadas S., Sundaresan S., Selvaraj K. S. V., Burd S., Ophir R., et al. (2010). Microarray analysis of the abscission-related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiol. 154, 1929–1956. 10.1104/pp.110.160697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minic Z., Rihouey C., Do C. T., Lerouge P., Jouanin L. (2004). Purification and characterization of enzymes exhibiting β-D-xylosidase activities in stem tissues of arabidopsis. Plant Physiol. 135, 867–878. 10.1104/pp.104.041269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima J., Chen F., Jackson L., Shadle G., Dixon R. A. (2008). Multi-site genetic modification of monolignol biosynthesis in alfalfa (Medicago sativa): effects on lignin composition in specific cell types. New Phytol. 179, 738–750. 10.1111/j.1469-8137.2008.02502.x [DOI] [PubMed] [Google Scholar]
- Nakatsubo T., Kitamura Y., Sakakibara N., Mizutani M., Hattori T., Sakurai N., et al. (2008). At5g54160 gene encodes Arabidopsis thaliana 5-hydroxyconiferaldehyde O-methyltransferase. J. Wood Sci. 54, 312–317. 10.1007/s10086-008-0958-4 [DOI] [Google Scholar]
- Niederhuth C. E., Patharkar O. R., Walker J. C. (2013). Transcriptional profiling of the Arabidopsis abscission mutant hae hsl2 by RNA-Seq. BMC Genomics 14:37. 10.1186/1471-2164-14-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Kay P., Wilson S., Swain S. M. (2009). ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are Polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21, 216–233. 10.1105/tpc.108.063768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra R., Paredes M., Sanchez-Calle I., Gomez-Jimenez M. C. (2013). Comparative transcriptional profiling analysis of olive ripe-fruit pericarp and abscission zone tissues shows expression differences and distinct patterns of transcriptional regulation. BMC Genomics 14:866. 10.1186/1471-2164-14-866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Wu J., Lu W., Li J. (2013). A polygalacturonase gene clustered into clade E involved in lychee fruitlet abscission. Sci. Hortic. 150, 244–250. 10.1016/j.scienta.2012.10.029 [DOI] [Google Scholar]
- Petersen M., Sander L., Child R., Van Onckelen H., Ulvskov P., Borkhardt B. (1996). Isolation and characterisation of a pod dehiscence zone-specific polygalacturonase from Brassica napus. Plant Mol. Biol. 31, 517–527. 10.1007/BF00042225 [DOI] [PubMed] [Google Scholar]
- Pickersgill B. (2007). Domestication of plants in the Americas: insights from mendelian and molecular genetics. Ann. Bot. 100, 925–940. 10.1093/aob/mcm193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. A., Elliott K. A., Gonzalez-Carranza Z. H. (2002). Abscission, dehiscence, and other cell separation processes. Annu. Rev. Plant Biol. 53, 131–158. 10.1146/annurev.arplant.53.092701.180236 [DOI] [PubMed] [Google Scholar]
- Roongsattham P., Morcillo F., Jantasuriyarat C., Pizot M., Moussu S., Jayaweera D., et al. (2012). Temporal and spatial expression of polygalacturonase gene family members reveals divergent regulation during fleshy fruit ripening and abscission in the monocot species oil palm. BMC Plant Biol. 12:150. 10.1186/1471-2229-12-150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel K., Berrio-Sierra J., Derikvand M. M., Pollet B., Thevenin J., Lapierre C., et al. (2009). Impact of CCR1 silencing on the assembly of lignified secondary walls in Arabidopsis thaliana. New Phytol. 184, 99–113. 10.1111/j.1469-8137.2009.02951.x [DOI] [PubMed] [Google Scholar]
- Sander L., Child R., Ulvskov P., Albrechtsen M., Borkhardt B. (2001). Analysis of a dehiscence zone endo-polygalacturonase in oilseed rape (Brassica napus) and Arabidopsis thaliana: evidence for roles in cell separation in dehiscence and abscission zones, and in stylar tissues during pollen tube growth. Plant Mol. Biol. 46, 469–479. 10.1023/A:1010619002833 [DOI] [PubMed] [Google Scholar]
- Sarkanen K. V., Ludwig C. H. (1971). Lignins: Occurrence, Formation, Structure and Reactions. New York, NY: Wiley-Interscience. [Google Scholar]
- Schultz J., Milpetz F., Bork P., Ponting C. P. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U.S.A. 95, 5857–5864. 10.1073/pnas.95.11.5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton R. (1979). Spatial and temporal aspects of cell separation in the foliar abscission zones of Impatiens sultani Hook. Protoplasma 99, 53–66. 10.1007/BF01274069 [DOI] [Google Scholar]
- Shiraishi M., Yanagisawa T. (1988). Anatomical and histochemical studies of starch distribution during abscission in filaments and petals of Wase Satsuma mandarin. J. Jpn. Soc. Hortic. Sci. 56, 365–374. 10.2503/jjshs.56.365 [DOI] [Google Scholar]
- Singh A. P., Pandey S. P., Rajluxmi, Pandey S., Nath P., Sane A. P. (2011). Transcriptional activation of a pectate lyase gene, RbPel1, during petal abscission in rose. Postharvest Biol. Technol. 60, 143–148. 10.1016/j.postharvbio.2010.12.014 [DOI] [Google Scholar]
- Spiegel-Roy P., Goldschmidt E. E. (1996). The Biology of Citrus. Cambridge: Cambridge University Press. [Google Scholar]
- Sun L., Van Nocker S. (2010). Analysis of promoter activity of members of the PECTATE LYASE-LIKE (PLL) gene family in cell separation in Arabidopsis. BMC Plant Biol. 10:152. 10.1186/1471-2229-10-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadeo F. R., Primo-Millo E. (1990). Peroxidase activity changes and lignin deposition during the senescence process in citrus stigmas and styles. Plant Sci. 68, 47–56. 10.1016/0168-9452(90)90151-D [DOI] [Google Scholar]
- Tonutti P., Cass L. G., Christoffersen R. E. (1995). The expression of cellulase gene family members during induced avocado fruit abscission and ripening. Plant Cell Environ. 18, 709–713. 10.1111/j.1365-3040.1995.tb00573.x [DOI] [Google Scholar]
- Tucker M. L., Kim J. (2015). Abscission research: what we know and what we still need to study. Stewart Postharvest Rev. 2:1 10.2212/spr.2015.2.1 [DOI] [Google Scholar]
- Tucker M. L., Burke A., Murphy C. A., Thai V. K., Ehrenfried M. L. (2007). Gene expression profiles for cell wall-modifying proteins associated with soybean cyst nematode infection, petiole abscission, root tips, flowers, apical buds, and leaves. J. Exp. Bot. 58, 3395–3406. 10.1093/jxb/erm188 [DOI] [PubMed] [Google Scholar]
- Urbanowicz B. R., Bennett A. B., Del Campillo E., Catala C., Hayashi T., Henrissat B., et al. (2007). Structural organization and a standardized nomenclature for plant endo-1,4-beta-glucanases (cellulases) of glycosyl hydrolase family 9. Plant Physiol. 144, 1693–1696. 10.1104/pp.107.102574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nocker S. (2009). Development of the abscission zone. Stewart Postharvest Rev. 5, 1–6. 10.2212/spr.2009.1.5 [DOI] [Google Scholar]
- Willats W. G. T., Marcus S. E., Knox J. P. (1998). Generation of a monoclonal antibody specific to (1 → 5)-α-l-arabinan. Carbohydr. Res. 308, 149–152. 10.1016/S0008-6215(98)00070-6 [DOI] [PubMed] [Google Scholar]
- Wilson W. C., Hendershott C. (1968). Anatomical and histochemical studies of abscission of oranges. Proc. Am. Soc. Hortic. Sci. 92, 203–210. [Google Scholar]
- Wu G. A., Prochnik S., Jenkins J., Salse J., Hellsten U., Murat F., et al. (2014). Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 32, 656–662. 10.1038/nbt.2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.-M., Yu Y., Han L.-B., Li C.-L., Wang H.-Y., Zhong N.-Q., et al. (2012). The tobacco BLADE-ON-PETIOLE2 gene mediates differentiation of the corolla abscission zone by controlling longitudinal cell expansion. Plant Physiol. 159, 835–850. 10.1104/pp.112.193482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacarias L., Tadeo F. R., Bono R., Primo-Millo E. (1993). Abscission studies in a new mutant of navel oranges, in Cellular and Molecular Aspects of the Plant Hormone Ethylene: Proceedings of the International Symposium on Cellular and Molecular Aspects of Biosynthesis and Action of the Plant Hormone Ethylene, Agen, France, August 31–September 4, 1992, eds Pech J. C., Latché A., Balagué C. (Dordrecht: Springer; ), 284–290. [Google Scholar]
- Zhu H., Dardick C., Beers E., Callanhan A., Xia R., Yuan R. (2011). Transcriptomics of shading-induced and NAA-induced abscission in apple (Malus domestica) reveals a shared pathway involving reduced photosynthesis, alterations in carbohydrate transport and signaling and hormone crosstalk. BMC Plant Biol. 11:138. 10.1186/1471-2229-11-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Specific primers used for sqRT-PCR.
Genes regulated by ethylene exclusively in AZ-C or FR cells, and in both AZ-C and FR cells.
Functional categories related to polysaccharide metabolism and subcellular localization regulated in the AZ-C and the FR.
Identification of citrus genes belonging to different families of cell wall remodeling enzymes, and monolignol biosynthesis and polymerization.
sqRT-PCR-based relative expression in the AZ-C and the FR.
LM isolation of AZ-C and FR cells.
Phylogenetic relationships between Carbohydrate esterases.
Phylogenetic relationships between Glycoside hydrolases.
Phylogenetic relationships between Pectate lyases.
Phylogenetic relationships between Expansins.
Phylogenetic relationships between members of gene families associated with monolignol biosynthesis and polymerization.