Abstract
Besides donor T cells, natural killer (NK) cells are considered to have a major role in preventing relapse after allogeneic hematopoietic stem cell transplantation (HSCT). After T-cell-depleted haploidentical HSCT, a strong NK alloreactivity has been described. These effects have been attributed to killer-cell immunoglobulin-like receptors (KIR). Abundant reports suggest a major role of KIR not only on outcome after haploidentical HSCT but also in the unrelated donor setting. In this review, we give a brief overview of the mechanism of NK cell activation, nomenclature of KIR haplotypes, human leukocyte antigen (HLA) groups, and distinct models for prediction of NK cell alloreactivity. It can be concluded that KIR-ligand mismatch seems to provoke adverse effects in unrelated donor HSCT with reduced overall survival and increased risk for high-grade acute graft-versus-host disease. The presence of activating KIR, as seen in KIR haplotype B, as well as the patient’s HLA C1/x haplotype might reduce relapse in myeloid malignancies.
Keywords: NK-cell, killer-cell immunoglobulin-like receptor, unrelated, stem cell, transplantation, HSCT, haplotype
Introduction
Natural killer (NK) cells are considered to contribute important immune effects against leukemia [graft-versus-leukemia (GVL) effect] after allogeneic hematopoietic stem cell transplantation (HSCT). Alloreactive NK cells are considered rather save concerning the development of graft-versus-host disease (GVHD) (1–5), although a high number of activating killer-cell immunoglobulin-like receptors (KIR) (6) or extensive NK cell stimulation (7) might promote GVHD, maybe due to remaining T-cells in the graft. Shah et al. (7) found an association between infusion of activated NK cells and occurrence of acute GVHD (aGVHD): Children with ultrahigh-risk sarcoma received T-cell-repleted grafts from matched unrelated donors (URDs) or matched sibling donors with subsequent infusion of IL-15 and 4-1BBL preactivated NK cells. Five of nine patients developed aGVHD. Those effects were attributed to NK cell-mediated T-cell activation (7). The biology of NK cells is complex, but activation by human leukocyte antigen (HLA) via the group of KIR is considered to be a relevant mechanism of activation. Within this review, we will provide a summary of concepts of KIR-mediated NK cell activation and an overview of GVL effects in haploidentical (haplo), but especially in URD HSCT.
Biology and Activation of NK Cells
Natural killer cells were named after their ability to kill infected or tumor cells without the need for prior antigen contact (8–10). They are defined by surface expression of CD56 and lack of CD3 (11). Unlike T cells, NK-cell receptors do not undergo rearrangement. In a process called licensing, NK cells with inhibitory receptors for present HLA class I (HLA-I) molecules (indicating “self”) are positively selected and stimulated for proliferation, leading to a licensed and self-tolerant subset. Missing inhibitory receptors against HLA-I do not lead to depletion but to a second subset of unlicensed but self-tolerant NK cells (12). Activation of NK cells might be initiated by antigen contact, but it is executed only after integration of abundant activating and inhibitory signals (13, 14). Today, several NK-cell receptors are known. Besides KIR, other NK-cell receptors that have been shown to have the potential to positively influence outcome after allogeneic HSCT are natural cytotoxicity receptors (15–17) as well as activating NKG2D (18) and DNAM-1 (19, 20) that bind to MICA/B and ULBPs or CD112/CD155, respectively. Both can be induced by DNA damage (21) and seem to play a role in negative regulation of T-cell responses (22) and acute myeloid leukemia (AML)/myelodysplastic syndrome immune evasion (15, 23).
KIR and HLA
Killer-cell immunoglobulin-like receptors belong to type-I transmembrane proteins of the immunoglobulin-like receptor superfamily and recognize classical HLA-I molecules (14). The 15 KIR genes and 2 pseudogenes are located on chromosome 19q13.4. According to the number of extracellular immunoglobulin-like domains (D), the receptors are named KIR2D and KIR3D (24, 25). On the cytoplasmic side, they have either long (L) inhibitory or short (S) activating domains (14). Inhibitory KIR bind to the highly polymorphic regions of HLA-I molecules: HLA-A, B, and C (26), while the ligands for activating KIR are poorly defined (14, 27).
To facilitate description of KIR-ligands, HLA-C phenotypes can be grouped into HLA-C group 1 and 2 according to their respective KIR-binding motif. HLA-C group 1 contains all ligands with serine at residue 77 and asparagine at residue 80 of the α1 helix (HLA-Casn80), binding KIR2DL2/3 and 2DS2. Members of this group are HLA-C*01/*03/*07/*08/*12/*14/*16. HLA-C group 2 (HLA-Clys80) has asparagine at residue 77 and lysine at residue 80 and contains HLA-C*02/*04/*05/*06/*15/*17/*18. They are ligands for KIR2DL1 and KIR2DS1 (28–31).
KIR3DL1 binds HLA-Bw4, and KIR3DL2 and 2DS2 bind HLA-A3 and A11 (14, 18, 32–38). Despite its structure, KIR2DL4 exhibits activating capacities and might bind soluble HLA-G (39–45). The KIR phenotype of an individual is his or her distinct set of inhibitory or activating KIR with an underlying distinct genotype (27, 46, 47). All genotypes can be summarized to a set of distinct haplotypes, which again result in the superordinated KIR haplotypes A or B (27, 46). KIR haplotype B is defined as the presence of KIR2DL5, 2DS1/2/3/5, or 3DS1, which have to be absent in KIR haplotype A (48). KIR2DS4 is the only activating KIR in haplotype A (46). KIR haplotype B/x (B/B or B/A) is found in about 30% of the Caucasian population (49). A more detailed analysis includes the information, whether the individual KIR is coded in the centromeric (Cen) or telomeric (Tel) gene motif of the KIR locus, resulting in Cen-A/A, Cen-B/x, and the respective Tel haplotypes (49–52). Thus, each individual expresses a certain KIR haplotype and a distinct HLA-C haplotype (C1/C1, C1/C2, or C2/C2). For prediction of alloreactive NK cell effects, the presence of HLA-C1, C2, and Bw4, as well as their respective KIR, are investigated (53). KIR2DL4 stimulation by HLA-G is considered to induce tolerance at the maternal–fetal barrier as well as IFN-gamma release of NK cells but not cytotoxicity (39, 43). KIR3DL2 and 2DS2 stimulation by HLA-A3 and A11 is also not in the primary focus of altering NK cell alloreactivity. KIR3DL2 has been identified as a surface marker in cutaneous T-cell lymphoma (54–56). For KIR2DS2, a reduced survival after URD-HSCT is suspected due to higher incidence of GVHD (57).
Model Situations Predicting NK Cell Alloreactivity
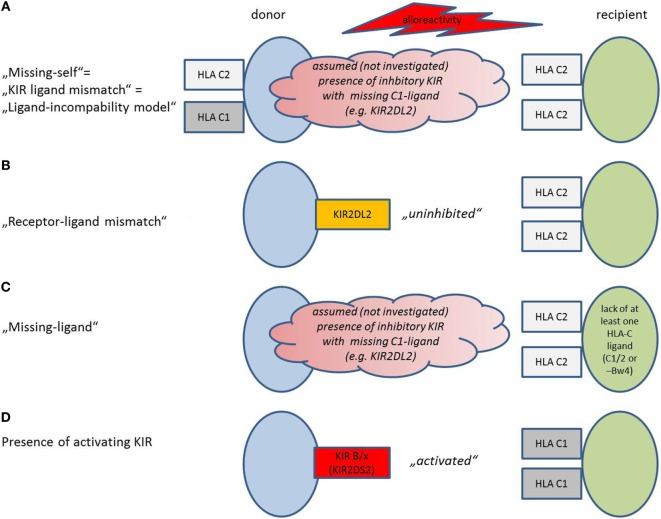
Different definitions of a mismatch between the donor’s NK cells and the recipient’s HLA exist, depending on the method that was chosen for KIR and HLA (HLA-C1, C2, and Bw4) evaluation (Figure 1).
Figure 1.
Model situations that provoke natural killer (NK) cell alloreactivity. Models are depicted as used in the present review, adopted and modified from Symons and Fuchs (53). Details concerning the activation mechanism are provided in the text. (A) Missing-self model, also described as “killer-cell immunoglobulin-like receptors (KIR)-ligand mismatch” or “ligand-incompability model”: Potential alloreactivity in the graft-versus-host direction is predicted by investigation of human leukocyte antigen (HLA) on donor and recipient. An HLA for inhibitory KIR that is present in the donor lacks in the recipient. The presence of the respective inhibitory KIR in the donor is assumed but not verified. (B) Receptor–ligand mismatch: NK cells become activated in the graft-versus-host direction, if they have an inhibitory KIR, for which the HLA ligand in the recipient is missing. Thus, the NK cells are “uninhibited.” Other than in (A), KIR on donor cells and HLA on recipient cells are investigated, not “assumed.” (C) Missing ligand: If the presence of the respective inhibitory KIR is not evaluated, but assumed in a model where at least one HLA-ligand is missing (HLA-C1/2 or Bw4). Other than in (A), only HLA on recipient cells but not on donor cells are evaluated. (C) The presence of activating KIR predicts alloreactivity in the presence of the respective activating ligand. KIR haplotype B/x contains more activating KIR than KIR haplotype A/A.
Missing-Self/KIR-Ligand Mismatch (Figure 1A)
Alloreactivity was initially thought to be only dependent on lack of inhibitory HLA-I molecules in the recipient that are present in the donor (“missing-self” or “KIR-ligand mismatch” or “ligand-incompability model”) (53, 58–60). For evaluation of KIR-ligand mismatch, donor and recipient are screened for expression of HLA: NK cells from a HLA C1/C1 donor will be alloreactive against a C2/C2 recipient. If a recipient expresses HLA-C1, C2, and Bw4, he will be resistant toward NK cell killing, as seen in one-third of the population (61). It is assumed, but not verified that the respective KIR, necessary for alloreactivity, is present in the donor.
Receptor–Ligand Mismatch (Figure 1B)
The receptor–ligand model states that donor NK cells become activated in the graft-versus-host direction; if they have inhibitory KIR, for which the HLA ligands in the recipient are missing, the NK cells become “uninhibited” (4). Thus, in addition to the HLA status of the recipient, confirmatory KIR genotyping of the patient is required. Other than in the first model, KIR on donor cells and HLA on recipient cells are investigated, not “assumed.” This model can be considered as an improvement of the “missing ligand model.”
Missing Ligand Model (Figure 1C)
Here, only the recipients’ HLA are genotyped, and missing HLA-C1, C2, or Bw4 for inhibitory KIR predict an alloreactivity of the graft; the presence of the respective KIR that would bind to the missing HLA is only assumed (53).
Presence of Activating KIR (Figure 1D)
Some theories emphasize that to achieve NK cell alloreactivity, “un-inhibiton” of NK cells by missing inhibitory HLA ligands might not be sufficient. Activation requires additional stimulation of activating KIR in the graft (62). In this model, alloreactivity can be predicted by measurement of activating KIR on donor cells. Some studies increase the predictive validity by detection of the respective activating ligands on donor cells.
Therefore, we determine mismatches on the donor and recipient side between ligand–ligand, receptor–ligand, and receptor–receptor or identify activating KIR in the donor (49, 53). HLA and KIR can be investigated by genotyping, phenotyping, or functional NK cell assays to predict alloreactivity. We would suggest to follow the well-described methods of Ruggeri et al. for genotyping and phenotyping (61).
All approaches were initially tested in the haploidentical setting: The Perugia group suggests donor and recipient HLA typing to identify mismatch (2), followed by confirmatory donor KIR typing to verify a mismatch between donor KIR and recipient HLA ligand (“KIR-ligand mismatch” combined with a “receptor–ligand” concept) (61). They found ligand incompatibility between donor and recipient in haploidentical HSCT to be associated with increased GVL effects and lower relapse in acute leukemia (2, 4). KIR-ligand mismatch can be prevalent either in the graft-versus-host direction when the donor’s KIR ligand is not shared by the recipient or in the host-versus-graft direction when the recipient’s KIR ligand is not present in the donor. The St. Jude group rather focuses on receptor–ligand mismatch in the haplo setting (63), while the researchers from Minnesota implemented their strategy for the URD setting by selecting KIR B/x donors for HLA-C1-positive recipients for improved alloreactivity (50, 64, 65).
Evidence of NK Cell-Mediated GVL Effects
Lessons Learned from the Haploidentical HSCT Setting
Much knowledge concerning NK cell-mediated alloreactivity has been collected due to the implementation of haploidentical HSCT. To reduce the risk of GVHD, T-cell depletion was performed before graft infusion at the cost of graft rejection (66). These effects could be partially overcome by infusion of high numbers of stem cells (67). Ruggeri et al. were the first to show NK cell-mediated alloreactivity in the T-cell-depleted haploidentical graft (5). Facilitated engraftment as well as tumor lysis by NK cells occurred by donor grafts that were KIR-ligand incompatible in the graft-versus-host direction without occurrence of GVHD. Since then, many other groups have investigated the beneficial effect of alloreactive NK cells in the haploidentical HSCT (2, 68) and have refined criteria for potential donor choice (61). The results are promising for AML (4, 68, 69), while lymphoid malignancies have been shown to be resistant in some (2, 69) but not all cases (63, 70) for KIR-mediated NK cell effects. The present status of NK cell-mediated effects in haploidentical HSCT has been reviewed elsewhere (53, 71, 72).
Results in the Unrelated-Donor HSCT Setting
After the identification of beneficial NK cell-mediated alloreactivity in haploidentical HSCT, efforts were made to adopt the findings for transplantations with URD (Table 1). Even though many patients already have the opportunity to receive a graft from an HLA-matched donor, donor choice by KIR repertoire is useful. Since HLA and KIR are inherited separately, approximately 75% of HLA-identical sibling donors and almost 100% of matched URDs will show KIR disparities and might therefore be a potential source for alloreactive NK cells (73, 74).
Table 1.
Studies on NK cell alloreactivity for unrelated donors.
| Reference | N | Median age (years)a | Disease (n) | Tx (n) | Model | Conditioning and graft source | Immunosuppression | Main results |
|---|---|---|---|---|---|---|---|---|
| Davies et al. (75) | 175 | 17 | CML, AML, ALL, MDS, others | MMUD (175) | KIR-L MMb | Myeloablative
|
TCD or CSA ± MTX | Adverse KIR-L MM in myeloid malignancies: Lower OS at 1 and 5 years (P < 0.01). No difference between KIR-L M/MM in any endpoint for total cohort |
| Schaffer et al. (76) | 104 | 29 | Diverse | MUD (62)/MMUD (42) | KIR-L MM | Myeloablative
|
MTX + CSA, ATG | Adverse KIR-L MM: Reduced OS and RFS |
| Giebel et al. (85) | 130 | 18–20.5 | Diverse | MUD (61)/MMUD (49) | KIR-L MM | Myeloablative
|
CSA, MTX, ATG | Beneficial KIR-L MM: Higher OS and RFS (P = 0.0007; 4.5 years). No influence of HLA-MM in the patients without KIR-L MM |
| Bornhauser et al. (77) | 118 | 42–44 | AML, CML, MDS | MUD (54)/MMUD (64) | KIR-L MM | Myeloablative
|
ATG (118) | Adverse KIR-L MM: Higher relapse for KIR-L MM (P = 0.02), but no difference in survival after KIR-L MM, MUD, and MMUD transplantation |
| Schaffer et al. (78) | 190 | 35–39 | Diverse | MUD (94)/MMUD (96) | KIR-L MM | Myeloablative (168)RIC (22)
|
CSA based (179) or TCD (11) plus ATG (all) | Adverse KIR-L MM: Higher infections, leading to increased TRM and reduced OS (P = 0.01), but no increase of relapse or GVHD |
| Venstrom et al. (96) | 1,277 | 40.5–41.7 | AML | MUD (664)/MMUD (613) | Missing ligand | Myeloablative (1,069)RIC/NMA (189)
|
Diverse, no ATG | Adverse absence of C1: HLAC2/C2 recipients have higher relapse than HLAC1/x recipients (P = 0.05) |
| Receptor–ligand KIR genes | Beneficial KIR2DS1 from C1/x donor associated with lower relapse compared to absence of KIR2DS1 (P = 0.003) and lower mortality (P = 0.04) w/o higher high-grade aGVHD or TRM | |||||||
| Beneficial KIR3DS1 associated with lower mortality (P = 0.01) by lower TRM and aGVHD | ||||||||
| No predictive effects in ALL patients (separate cohort) | ||||||||
| De Santis et al. (80) | 104 | 24 | Diverse | MMUD (104) | KIR-L MM | Myeloablative
|
No ATG | Adverse: KIR-L MM (HVG): Increased graft rejection |
| BM: CSA, MTX (59), T-cell depletion (9) | Adverse KIR-L MM (GVH): Increased aGVHD grade 3–4 | |||||||
| PB: No CSA (39) | Adverse KIR-L MM (GVH or HVG): Increased TRM, decrease RFS | |||||||
| Beneficial high number of donor KIR: Lower GVHD and improved survival | ||||||||
| Giebel et al. (57) | 111 | 18.5–21 | Diverse | MUD (90)/MMUD (21) | Missing ligand | Myloablative
|
CSA, MTX, ATG | Adverse absence of C1: C2/C patients have lower OS and DFS, due to higher relapse |
| Sun et al. (97) | 65 | 45–46 | AML | MUD (39)/MMUD (26) | Receptor–receptor | Diverse | CSA + MTX (65) | Prediction of incidence of aGVHD possible: Activating KIR in the donor that lack in recipient and the lack of inhibitory KIR in the donor that are present in the recipient predict increased aGVHD |
| No ATG or TCD | Indifferent results for KIR-L MM, missing ligand, number of activating KIR | |||||||
| Giebel et al. (98) | 25 | 27 | ALL, AML, MDS, CML, NHL | MUD (23)/MMUD (2) | KIR genes | Myeloablative
|
CSA, MTX, ATG | Adverse presence of KIR2DS1: Reduced OS and DFS due to increased GVHD and relapse |
| Indifferent presence of KIR2DS1 | ||||||||
| Kröger et al. (79) | 142 | 33 | AML, MDS, CMML, CML, ALL | MUD (103)/MMUD (39) | KIR haplotype | Myeloablative
|
ATG, CSA, MTX | Adverse KIR B/x: Higher relapse than KIR A/A (P = 0.03), but only in AML/MDS/CML/CMML, not ALL, resulting in lower OS |
| KIR-L MM | Adverse KIR-L MM: Higher TRM, lower OS, no increase of GVHD | |||||||
| Adverse KIR3DS1, 2DS1, 2DS5 in UVA, only 2DS5 in MVA, all resulting in higher relapse | ||||||||
| Farag et al. (83) | 1,571 | 59–68 | AML, MDS, CML | MMUD KIR-L MM GVH (137) | KIR-L MM | Myeloablative
|
± T-cell depletion | Indifferent KIR-L MM: For KIR-L MM (GVH/HVG) as well as KIR-L M but HLA MM at HLA B ± C versus HLA- and KIR-L M grafts: Same rates of increased aGVHD grade 3–4, TRM, treatment failure, and overall mortality compared to HLA- and KIR-L matched grafts |
| MMUD KIR-L MM HVG (170) | ||||||||
| MMUD KIR-L M (260) | ||||||||
| MUD (1,004) | ||||||||
| Hsu et al. (60) | 1,770 | 34.5–35 | AML, MDS, CML, ALL | MMUD (1,190)/MUD (580) | Missing ligand | Myeloablative
|
T-cell replete grafts | Beneficial: missing ligand in MMUD (defined as homozygosity of recipient HLA-B or C epitopes) resulting in lower relapse (P = 0.004), but not for MUD |
| KIR-L MM | Absence of HLA-C2 or Bw4 associated with reduced relapse, no survival benefit | |||||||
| Indifferent KIR-L MM model in subgroup of 428 patients: no difference in relapse (but also not with applied missing-ligand model in same subgroup P = 0.07) | ||||||||
| Miller et al. (93) | 2,062 | – | AML, CML, MDS | MMUD/MUD | Missing ligand | - | ± ATG or TCD | Beneficial absence of one ligand in early stage AML or MDS: reduced relapse, independent from HLA match (C1/C2/Bw4) |
| Adverse absence of ≥1 ligand in CML: Increased late-onset high-grade acute GVHD | ||||||||
| Willemze et al. (82) | 218 | 12.8–15 | AML, ALL | MUD (42)/MMUD (176) | KIR-L MM | RIC (202)Myeloablative (6)
|
CSA based (174) Other (44)± ATG (196) |
Beneficial KIR-L MM: Improved DFS, OS, and decrease relapse |
| Gagne et al. (84) | 264 | 24.5 | Diverse | MUD (164)/MMUD (100) | KIR-L MM Missing ligand Receptor–ligand Receptor–receptor |
Myeloablative
|
Unmanipulated BM |
Indifferent KIR-L MM Adverse missing-ligand: Decreased survival but only in C1-deficient recipients, in myeloid malignancies Adverse receptor–ligand mismatch: KIR3DL1 as well as KIR3DL1/3DS1 mismatch (GVH: D+ R–, absence of recipient HLA-Bw4) from a HLA-Bw4-negative donor is correlated with low OS in HLA-identical and high relapse in MMUD HSCT |
| Ludajic et al. (94) | 124 | 42 | Diverse | MUD | Missing ligand | Myeloablative (90)RIC (34)
|
CSA-based (124) ± ATG (30) | Adverse absence of HLA-C2 in recipients of KIR2DL1-positive grafts or KIR A/A grafts: Increased aGVHD |
| Beneficial absence of HLA-C2 in recipients of KIR2DS2-positive grafts: Decreased aGVHD | ||||||||
| Cooley et al. (64) | 448 | 33–34 | AML | MUD (209)/MMUD (239) | KIR haplotype KIR-L MM |
Myeloablative
|
T-cell replete MMUD grafts | Beneficial KIR B/x in KIR-L M HSCT: Compared to KIR A/A higher RFS in KIR-L M (MUD and MMUD) but not in KIR-L MM (MMUD) |
| Beneficial survival rates for KIR2DL2 and 2DS2 positive grafts | ||||||||
| Cooley et al. (50) | 1,409 | 19/39 | ALL, AML | MUD (687) MMUD (722) | KIR haplotype | Myeloablative
|
T-cell replete MMUD grafts | Beneficial KIR B/x: Higher RFS in AML but not ALL |
| Cen-B motifs improve outcome without increased aGVHD/cGVHD or TRM | ||||||||
| Venstrom et al. (99) | 1,087 | 35.3–37.5 | AML, MDS, CML, ALL | MUD (670)/MMUD (417) | KIR genes KIR haplotype |
Myloablative
|
CSA (751) | Beneficial presence of KIR3DS1: Same rate of relapse but reduced TRM and aGVHD, resulting in lower mortality in AML and MDS. Beneficial effects increase with copy numbers of donor KIR3DS1 |
| No CSA (120) | Beneficial effect of KIR B/x (including KIR3DS1) similar but weaker | |||||||
| TCD (216) | ||||||||
| Kröger et al. (100) | 118 | 51 | MM | Unrelated (81) | KIR haplotype | Myeloablative (12)RIC (106)
|
ATG (110) | Beneficial KIR B/x B in MUD: MUD but not MMUD haplotype B/x reaches lower 1-year relapse than haplotype AA (P = 0.005), resulting in higher 5-year DFS (P = 0.009). |
| Related (37) | ||||||||
| Venstrom et al. (96) | 1,277 | 40.5–41.7 | AML | MUD (664)/MMUD (613) | Missing ligand Receptor–ligand KIR genes |
Myloablative (1,069)
|
CSA (346) | Adverse absence of C1 and beneficial KIRSDS1: Reduced risk of relapse, if the allograft was derived from an HLA-C1/x donor |
| Tac (428) | Beneficial presence of KIR3DS1: Not lower relapse but reduced TRM and aGVHD, resulting in lower mortality in AML | |||||||
| TCD (348) | ||||||||
| Cooley et al. (65) | 1,532 | Adults and children | AML | MUD (856)/MMUD (676) | KIR haplotype | Myeloablative | T-cell replete MMUD grafts | Beneficial KIR B/x, adverse absence of C1: Relapse protection improved by high KIR-B content in recipients HLA-C1/x but not C2/C2 (significant only in MMUD, not MUD). No effect of donor HLA |
| KIR gene content | ||||||||
| Missing-ligand | ||||||||
| Sobecks et al. (95) | 909 | 56–57 | AML, MDS | MUD (712)/MMUD (197) | Missing ligand | RIC
|
Diverse ± ATG (317) | Adverse KIR2DS1 educated in a C2/C2 donor: Higher GVHD and TRM without reduced relapse (AML) |
| Adverse ≥1 missing ligand or absence of HLA-C2: Higher aGVHD (AML) | ||||||||
| Indifferent KIR centromeric gene content or donor activating KIR | ||||||||
| Faridi et al. (49) | 281 | 50 | AML, ALL | MSD (153)/MUD (128) | Comparison of different models | Myeloablative
|
ATG, CSA, MTX | Adverse KIR-KIR mismatch: Increased cGVHD in HLA C1/x recipients |
| Beneficial ≥1 missing ligand: Reduced relapse without improved OS | ||||||||
| Indifferent results for KIR B | ||||||||
| Bachanova et al. (101) | 614 | 48–52 | NHL | MUD (396)/MMUD (218) | KIR haplotype | Myeloablative (253)RIC (361)
|
Diverse | Beneficial KIR B/x in MUD HSCT: Lower relapse after 5 years compared to KIR A/A donors (P = 0.5) with improved progression-free survival (P = 0.007) |
| Rocha et al. (81) | 461 | Adults and children | AML | MMUD (461) | KIR-L MM | Myeloablative
|
With (145) or w/o (35) in vivo T-cell depletion in 3–5/8 MM cohort | Adverse: KIR-L MM (HVG): At 3–5/8 HLA-MM level: Higher mortality (P = 0.008) and NRM (P = 0.008), no difference in relapse or GVHD in KIR-L MM versus KIR-L M (HVG) for AML and ALL |
| 3–5/8 HLA-MM (212) | Indifferent KIR-L MM (GVH): No differences in GVH direction or higher HLA-matched subgroup or complete patient cohort | |||||||
ALL, acute lymphoid leukemia; AML, acute myeloid leukemia; ATG, antithymocyte globulin (rabbit ATG, Campath or OTK3); BM, bone marrow; Cen, centromeric; CML, chronic myeloid leukemia; CMML, chronic myelomonocytic leukemia; CSA, cyclosporine A; GVH, graft-versus-host direction; GVHD, graft-versus-host disease (a, acute; c, chronic); HD, Hodgkin’s disease; HLA, human leukocyte antigen; HVG, host-versus-graft direction; KIR, killer-cell immunoglobulin-like receptors; KIR-L M, KIR-ligand match; KIR-L MM, KIR-ligand mismatch; M, matched; MDS, myelodysplastic syndrome; MM, multiple myeloma or mismatch; MSD, matched sibling donor; MMUD, mismatched unrelated donor, defined as HLA 10/10 – 8/8 or not further specified; MTX, methotrexate; MUD, matched unrelated donor; MVA, multivariate analysis; NMA, non-myeloablative; n.s., not significant; Tel, telomeric; PB, peripheral blood stem cells; RIC, reduced intensity conditioning; TCD, T-cell depletion; Tx, transplantation details; UVA, univariate analysis.
aMedian age was not always declared for the total cohort but only the investigated subgroups. In this case, age-age does not describe a range but the youngest and oldest median age stated.
bKIR-L MM in the GVH direction unless stated otherwise.
KIR-Ligand Mismatch Seems to Induce Adverse Effects in URD HSCT
Davies et al. (75) were the first to perform a retrospective analysis of patients with HLA mismatched URD HSCT, comparing KIR-ligand mismatch. In the analysis, no difference in any of the primary endpoints was achieved. Concerning the subgroup of myeloid malignancies, KIR-ligand mismatch resulted in worse OS at 5 years [13 versus 38%, P < 0.01, no use of antithymocyte globulin (ATG)], which was even more surprising. Others confirmed worse outcome for KIR-ligand mismatch in URD HSCT after conditioning with ATG (76–79) or without ATG (80), accompanied with higher infections in the early posttransplant period (78) or increased graft rejection, TRM, and GVHD (80). A recent study confirmed higher mortality and higher TRM without difference in relapse in 3–5/8 HLA-mismatched KIR-ligand mismatched (in the host-versus-graft direction) unrelated cord blood transplantations for AML and acute lymphoid leukemia (ALL) compared to KIR-matched cord blood, while no difference was found for mismatch in the graft-versus-host direction or in a higher HLA-matched subgroup or the complete patient cohort (81). The authors did suggest to not using KIR-ligand mismatch as a criterion for cord blood selection. An earlier Eurocord study (82) detected favorable outcome for KIR-ligand mismatched transplantations in AML and ALL but used lower HLA-resolution techniques.
No difference in mortality after either KIR-ligand mismatched or HLA-mismatched but KIR-ligand matched donor–recipient pairs was detected by a comprehensive study of CIBMT, EBMT, and the Dutch transplant registry (83), investigating the results of 1,571 patients with myeloid malignancies with or without T-cell depletion. KIR-ligand mismatch was associated with significantly higher high-grade aGVHD, just as HLA mismatch at HLA-C and/or B. No predictive effects of KIR-ligand mismatch on outcome after T-cell-repleted unrelated HSCT were detected in a retrospective multicenter study in France (84). Here, different models of NK cell alloreactivity were compared in a very heterogeneous cohort of patients. These investigations were partially designed as a response to the positive results in haploidentical HSCT and in a previous study by Giebel et al. (85) with different results: KIR-ligand mismatch in patients with myeloid malignancies achieved significant higher OS and RFS as well as lower TRM and relapse compared to HLA mismatch with KIR ligand match or compared to matched URD HSCT with the use of pretransplant ATG. The differing results could be only partially attributed to the use or sparing of ATG (85): Although toxic (86) or immunosuppressive (87, 88) on NK cells, ATG has been shown to accelerate NK-cell and B-cell reconstitution in some (89) but not all investigations (90, 91). It has also been shown to decelerate the recovery of CD4+ and CD8+ T cells (89, 91) while sparing effector-memory T cells and T-regulatory cells (91). The results indicated that knowledge from haploidentical cannot be transferred to unrelated HSCT without further adaptations (75). Grafts for haploidentical HSCT were mainly highly T-cell depleted and performed with high stem-cell doses as well as no or low immunosuppression, resulting in fast NK cell but slow T-cell reconstitution with low T-cell numbers and eradication of antigen-presenting cells by alloreactive NK clones (2, 67, 92). Therefore, the immunological environment during engraftment in haploidentical HSCT is much different from URD-HSCT.
Missing-Ligand Model and Presence of Activating KIR Are Predictive for Outcome
Later, Hsu et al. (60) identified not only KIR-ligand mismatch but also missing KIR ligands as protective against relapse in HLA mismatched but not in matched URD HSCT. These effects were seen in myeloid and lymphoid malignancies and supported by later investigations by other authors (93). In the study by Hsu et al. (60), the absence of HLA-C2 or HLA-Bw4 KIR ligands was associated with lower relapse. Other authors confirmed the impact of HLA-C2: Absence of HLA-C2 in recipients of KIR2DL1-positive grafts resulted in higher incidence of aGVHD after myeloablative (94) as well as reduced intensity (95) conditioning. The absence of C1 epitopes, as seen for C2/C2 recipients, has been claimed responsible for poorer outcome (57, 65, 96). In search for favorable KIR in URD HSCT, Sun et al. (97) prospectively analyzed outcome of URD AML patients without in vivo T-cell depletion by ATG. According to the presence or absence of activating or inhibitory KIR in donor and recipient, they calculated a new predictive algorithm for GVHD, in which an inhibitory KIR in the donor that lacks in the patients has a negative value vice versa a positive value. On the other hand, they could not find other models such as KIR-ligand, missing-ligand, or high numbers of activating KIR to be predictive for aGVHD (97). In general, among the activating receptors, the presence of KIR2DS2 has been shown to be associated with lower OS and DFS as well as higher incidence of GVHD, resulting in high TRM (98). The alloreactivity of KIR2DS1 educated in a C2/C2 donor results in higher GVHD and TRM without reduced relapse (95). KIRSDS1 has been claimed responsible for reduced risk of relapse, if the allograft was derived from an HLA-C1/x donor (96), but did not show any beneficial effects in other investigations (98). The presence of KIR3DS1 was not associated with lower relapse but reduced TRM and aGVHD, resulting in lower mortality in AML patients (96, 99). KIR3DL1 and KIR3DL1/3DS1 mismatch in the GVH direction (donor positive, recipient negative, absence of recipient HLA-Bw4) from a HLA-Bw4-negative donor is correlated with low OS in HLA-identical and high relapse in HLA-mismatched URD HSCT (84). There are several other investigations apart from the environment of URD HSCT, which might be even more conflicting and difficult to transfer. Our early investigations showed the low-alloreactive KIR haplotype A to be associated with lower relapse after HSCT for leukemia (79), while in a later analysis, KIR haplotype B was associated with improved PFS and OS in patients with multiple myeloma (100). Cooley et al. (50, 64, 65) systematically investigated the influence of the KIR haplotype B. In summary, a high number of KIR haplotype B defining receptors, especially of those coded in the centromeric regions, showed beneficial effects on survival of HLA C1/x AML recipients after ATG-free HSCT without increased GVHD and without benefit of KIR-ligand mismatch. No positive influence of haplotype B was seen in recent investigations for leukemia (49) but in HLA-matched URD-HSCT of non-Hogdkin lymphoma patients, where KIR B/x grafts led to significant lower relapse after 5 years compared to KIR A/A donors (P = 0.5) (101). The role of KIR genotypes in matched unrelated and sibling HSCT has recently also been investigated by Faridi et al. (49). Their aim was to compare the predictive value of KIR-ligand mismatch (61) versus the “missing-ligand” hypothesis (63) or the advantage of a specific KIR haplotype (50, 64, 65). They found KIR–KIR match to be associated with lower cGVHD for HLA C1/x recipients as well as lower RFS. One or more missing ligand in the unrelated recipient for donor KIR resulted in reduced relapse (21.6 versus 63.6%, P = 0.001) and higher RFS without improved OS. None of the tested hypotheses had influence on OS, and no effect of donor KIR haplotype was detected.
Future Directions
During the past years, improvement in understanding NK cell alloreactivity has been made by wisely modeled analyzes (49, 60, 84). Despite clinical relevance (102–105), we still know too little about the NK cell education after HSCT (95). The interplay of NK cells and T-cells after HSCT is still subject of further investigation (105), and as we now know about KIR expression on T-cells (106), we need to be precise in our technical methods. To overcome the problem of heterogeneity, we would suggest beginning with a simple multicenter prospective trial in adult patients with AML in first molecular complete remission, testing the hypothesis that the number of activating KIR in the unmanipulated graft improves overall survival without increasing GVHD. KIR and HLA of donor and recipient should be measured by high-resolution genotyping and phenotyping. Every patient should receive the same conditioning and first-line immune suppression.
Conclusion
Due to heterogeneity of the conducted studies, a general recommendation cannot be made. In matched URD-HSCT, a donor with high numbers of activating KIR can be chosen to optimize patient’s chances for survival.
Author Contributions
SH and NK contributed equally to the manuscript writing.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Asai O, Longo DL, Tian ZG, Hornung RL, Taub DD, Ruscetti FW, et al. Suppression of graft-versus-host disease and amplification of graft-versus-tumor effects by activated natural killer cells after allogeneic bone marrow transplantation. J Clin Invest (1998) 101(9):1835–42. 10.1172/JCI1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science (2002) 295(5562):2097–100. 10.1126/science.1068440 [DOI] [PubMed] [Google Scholar]
- 3.Olson JA, Leveson-Gower DB, Gill S, Baker J, Beilhack A, Negrin RS. NK cells mediate reduction of GVHD by inhibiting activated, alloreactive T cells while retaining GVT effects. Blood (2010) 115(21):4293–301. 10.1182/blood-2009-05-222190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruggeri L, Mancusi A, Capanni M, Urbani E, Carotti A, Aloisi T, et al. Donor natural killer cell allorecognition of missing self in haploidentical hematopoietic transplantation for acute myeloid leukemia: challenging its predictive value. Blood (2007) 110(1):433–40. 10.1182/blood-2006-07-038687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruggeri L, Capanni M, Casucci M, Volpi I, Tosti A, Perruccio K, et al. Role of natural killer cell alloreactivity in HLA-mismatched hematopoietic stem cell transplantation. Blood (1999) 94(1):333–9. [PubMed] [Google Scholar]
- 6.Bishara A, De Santis D, Witt CC, Brautbar C, Christiansen FT, Or R, et al. The beneficial role of inhibitory KIR genes of HLA class I NK epitopes in haploidentically mismatched stem cell allografts may be masked by residual donor-alloreactive T cells causing GVHD. Tissue Antigens (2004) 63(3):204–11. 10.1111/j.0001-2815.2004.00182.x [DOI] [PubMed] [Google Scholar]
- 7.Shah NN, Baird K, Delbrook CP, Fleisher TA, Kohler ME, Rampertaap S, et al. Acute GVHD in patients receiving IL-15/4-1BBL activated NK cells following T-cell-depleted stem cell transplantation. Blood (2015) 125(5):784–92. 10.1182/blood-2014-07-592881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herberman RB, Nunn ME, Holden HT, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic and allogeneic tumors. II. Characterization of effector cells. Int J Cancer (1975) 16(2):230–9. 10.1002/ijc.2910160205 [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Penarrubia P, Koster FT, Kelley RO, McDowell TD, Bankhurst AD. Antibacterial activity of human natural killer cells. J Exp Med (1989) 169(1):99–113. 10.1084/jem.169.1.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biron CA, Brossay L. NK cells and NKT cells in innate defense against viral infections. Curr Opin Immunol (2001) 13(4):458–64. 10.1016/S0952-7915(00)00241-7 [DOI] [PubMed] [Google Scholar]
- 11.Robertson MJ, Ritz J. Biology and clinical relevance of human natural killer cells. Blood (1990) 76(12):2421–38. [PubMed] [Google Scholar]
- 12.Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature (2005) 436(7051):709–13. 10.1038/nature03847 [DOI] [PubMed] [Google Scholar]
- 13.Leiden JM, Gottesdiener KM, Quertermous T, Coury L, Bray RA, Gottschalk L, et al. T-cell receptor gene rearrangement and expression in human natural killer cells: natural killer activity is not dependent on the rearrangement and expression of T-cell receptor alpha, beta, or gamma genes. Immunogenetics (1988) 27(4):231–8. 10.1007/BF00376117 [DOI] [PubMed] [Google Scholar]
- 14.Lanier LL. NK cell recognition. Annu Rev Immunol (2005) 23:225–74. 10.1146/annurev.immunol.23.021704.115526 [DOI] [PubMed] [Google Scholar]
- 15.Sanchez-Correa B, Gayoso I, Bergua JM, Casado JG, Morgado S, Solana R, et al. Decreased expression of DNAM-1 on NK cells from acute myeloid leukemia patients. Immunol Cell Biol (2012) 90(1):109–15. 10.1038/icb.2011.15 [DOI] [PubMed] [Google Scholar]
- 16.Sivori S, Carlomagno S, Pesce S, Moretta A, Vitale M, Marcenaro E. TLR/NCR/KIR: which one to use and when? Front Immunol (2014) 5:105. 10.3389/fimmu.2014.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghasemzadeh M, Hosseini E, Schwarer AP, Pourfathollah AA. NK cell maturation to CD56(dim) subset associated with high levels of NCRs overrides the inhibitory effect of NKG2A and recovers impaired NK cell cytolytic potential after allogeneic hematopoietic stem cell transplantation. Leuk Res (2016) 43:58–65. 10.1016/j.leukres.2015.12.002 [DOI] [PubMed] [Google Scholar]
- 18.Farag SS, Caligiuri MA. Human natural killer cell development and biology. Blood Rev (2006) 20(3):123–37. 10.1016/j.blre.2005.10.001 [DOI] [PubMed] [Google Scholar]
- 19.Kearney CJ, Ramsbottom KM, Voskoboinik I, Darcy PK, Oliaro J. Loss of DNAM-1 ligand expression by acute myeloid leukemia cells renders them resistant to NK cell killing. Oncoimmunology (2016) 5(8):e1196308. 10.1080/2162402X.2016.1196308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruse V, Hamann C, Monecke S, Cyganek L, Elsner L, Hubscher D, et al. Human induced pluripotent stem cells are targets for allogeneic and autologous natural killer (NK) cells and killing is partly mediated by the activating NK receptor DNAM-1. PLoS One (2015) 10(5):e0125544. 10.1371/journal.pone.0125544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerboni C, Fionda C, Soriani A, Zingoni A, Doria M, Cippitelli M, et al. The DNA damage response: a common pathway in the regulation of NKG2D and DNAM-1 ligand expression in normal, infected, and cancer cells. Front Immunol (2014) 4:508. 10.3389/fimmu.2013.00508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zingoni A, Ardolino M, Santoni A, Cerboni C. NKG2D and DNAM-1 activating receptors and their ligands in NK-T cell interactions: role in the NK cell-mediated negative regulation of T cell responses. Front Immunol (2012) 3:408. 10.3389/fimmu.2012.00408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlsten M, Baumann BC, Simonsson M, Jadersten M, Forsblom AM, Hammarstedt C, et al. Reduced DNAM-1 expression on bone marrow NK cells associated with impaired killing of CD34+ blasts in myelodysplastic syndrome. Leukemia (2010) 24(9):1607–16. 10.1038/leu.2010.149 [DOI] [PubMed] [Google Scholar]
- 24.Colonna M, Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science (1995) 268(5209):405–8. 10.1126/science.7716543 [DOI] [PubMed] [Google Scholar]
- 25.Wagtmann N, Biassoni R, Cantoni C, Verdiani S, Malnati MS, Vitale M, et al. Molecular clones of the p58 NK cell receptor reveal immunoglobulin-related molecules with diversity in both the extra- and intracellular domains. Immunity (1995) 2(5):439–49. 10.1016/1074-7613(95)90025-X [DOI] [PubMed] [Google Scholar]
- 26.Borges L, Cosman D. LIRs/ILTs/MIRs, inhibitory and stimulatory Ig-superfamily receptors expressed in myeloid and lymphoid cells. Cytokine Growth Factor Rev (2000) 11(3):209–17. 10.1016/S1359-6101(00)00007-1 [DOI] [PubMed] [Google Scholar]
- 27.Hsu KC, Chida S, Geraghty DE, Dupont B. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol Rev (2002) 190:40–52. 10.1034/j.1600-065X.2002.19004.x [DOI] [PubMed] [Google Scholar]
- 28.Mandelboim O, Reyburn HT, Vales-Gomez M, Pazmany L, Colonna M, Borsellino G, et al. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J Exp Med (1996) 184(3):913–22. 10.1084/jem.184.3.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyington JC, Sun PD. A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol Immunol (2002) 38(14):1007–21. 10.1016/S0161-5890(02)00030-5 [DOI] [PubMed] [Google Scholar]
- 30.Igarashi T, Wynberg J, Srinivasan R, Becknell B, McCoy JP, Jr, Takahashi Y, et al. Enhanced cytotoxicity of allogeneic NK cells with killer immunoglobulin-like receptor ligand incompatibility against melanoma and renal cell carcinoma cells. Blood (2004) 104(1):170–7. 10.1182/blood-2003-12-4438 [DOI] [PubMed] [Google Scholar]
- 31.Faridi RM, Agrawal S. Killer immunoglobulin-like receptors (KIRs) and HLA-C allorecognition patterns implicative of dominant activation of natural killer cells contribute to recurrent miscarriages. Hum Reprod (2011) 26(2):491–7. 10.1093/humrep/deq341 [DOI] [PubMed] [Google Scholar]
- 32.Colonna M, Navarro F, Bellon T, Llano M, Garcia P, Samaridis J, et al. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J Exp Med (1997) 186(11):1809–18. 10.1084/jem.186.11.1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samaridis J, Colonna M. Cloning of novel immunoglobulin superfamily receptors expressed on human myeloid and lymphoid cells: structural evidence for new stimulatory and inhibitory pathways. Eur J Immunol (1997) 27(3):660–5. 10.1002/eji.1830270313 [DOI] [PubMed] [Google Scholar]
- 34.Fehniger TA, Shah MH, Turner MJ, VanDeusen JB, Whitman SP, Cooper MA, et al. Differential cytokine and chemokine gene expression by human NK cells following activation with IL-18 or IL-15 in combination with IL-12: implications for the innate immune response. J Immunol (1999) 162(8):4511–20. [PubMed] [Google Scholar]
- 35.Vivier E, Anfossi N. Inhibitory NK-cell receptors on T cells: witness of the past, actors of the future. Nat Rev Immunol (2004) 4(3):190–8. 10.1038/nri1306 [DOI] [PubMed] [Google Scholar]
- 36.Fuchs A, Colonna M. The role of NK cell recognition of nectin and nectin-like proteins in tumor immunosurveillance. Semin Cancer Biol (2006) 16(5):359–66. 10.1016/j.semcancer.2006.07.002 [DOI] [PubMed] [Google Scholar]
- 37.Guo H, Qian X. Clinical applications of adoptive natural killer cell immunotherapy for cancer: current status and future prospects. Onkologie (2010) 33(7):389–95. 10.1159/000315698 [DOI] [PubMed] [Google Scholar]
- 38.Orr MT, Lanier LL. Natural killer cell education and tolerance. Cell (2010) 142(6):847–56. 10.1016/j.cell.2010.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajagopalan S, Fu J, Long EO. Cutting edge: induction of IFN-gamma production but not cytotoxicity by the killer cell Ig-like receptor KIR2DL4 (CD158d) in resting NK cells. J Immunol (2001) 167(4):1877–81. 10.4049/jimmunol.167.4.1877 [DOI] [PubMed] [Google Scholar]
- 40.Kikuchi-Maki A, Catina TL, Campbell KS. Cutting edge: KIR2DL4 transduces signals into human NK cells through association with the Fc receptor gamma protein. J Immunol (2005) 174(7):3859–63. 10.4049/jimmunol.174.7.3859 [DOI] [PubMed] [Google Scholar]
- 41.Cantoni C, Verdiani S, Falco M, Pessino A, Cilli M, Conte R, et al. p49, a putative HLA class I-specific inhibitory NK receptor belonging to the immunoglobulin superfamily. Eur J Immunol (1998) 28(6):1980–90. [DOI] [PubMed] [Google Scholar]
- 42.Allan DS, Colonna M, Lanier LL, Churakova TD, Abrams JS, Ellis SA, et al. Tetrameric complexes of human histocompatibility leukocyte antigen (HLA)-G bind to peripheral blood myelomonocytic cells. J Exp Med (1999) 189(7):1149–56. 10.1084/jem.189.7.1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ponte M, Cantoni C, Biassoni R, Tradori-Cappai A, Bentivoglio G, Vitale C, et al. Inhibitory receptors sensing HLA-G1 molecules in pregnancy: decidua-associated natural killer cells express LIR-1 and CD94/NKG2A and acquire p49, an HLA-G1-specific receptor. Proc Natl Acad Sci U S A (1999) 96(10):5674–9. 10.1073/pnas.96.10.5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajagopalan S, Long EO. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J Exp Med (1999) 189(7):1093–100. 10.1084/jem.189.7.1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyson JE, Erskine R, Whitman MC, Chiu M, Lau JM, Koopman LA, et al. Disulfide bond-mediated dimerization of HLA-G on the cell surface. Proc Natl Acad Sci U S A (2002) 99(25):16180–5. 10.1073/pnas.212643199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, et al. Human diversity in killer cell inhibitory receptor genes. Immunity (1997) 7(6):753–63. 10.1016/S1074-7613(00)80394-5 [DOI] [PubMed] [Google Scholar]
- 47.Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, et al. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol (2002) 168(5):2307–15. 10.4049/jimmunol.168.5.2307 [DOI] [PubMed] [Google Scholar]
- 48.Marsh SG, Parham P, Dupont B, Geraghty DE, Trowsdale J, Middleton D, et al. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Tissue Antigens (2003) 62(1):79–86. 10.1034/j.1399-0039.2003.00072.x [DOI] [PubMed] [Google Scholar]
- 49.Faridi RM, Kemp TJ, Dharmani-Khan P, Lewis V, Tripathi G, Rajalingam R, et al. Donor-recipient matching for KIR genotypes reduces chronic GVHD and missing inhibitory KIR ligands protect against relapse after myeloablative, HLA matched hematopoietic cell transplantation. PLoS One (2016) 11(6):e0158242. 10.1371/journal.pone.0158242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Le CT, et al. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood (2010) 116(14):2411–9. 10.1182/blood-2010-05-283051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yawata M, Yawata N, Abi-Rached L, Parham P. Variation within the human killer cell immunoglobulin-like receptor (KIR) gene family. Crit Rev Immunol (2002) 22(5–6):463–82. 10.1615/CritRevImmunol.v22.i5-6.70 [DOI] [PubMed] [Google Scholar]
- 52.Martin AM, Kulski JK, Gaudieri S, Witt CS, Freitas EM, Trowsdale J, et al. Comparative genomic analysis, diversity and evolution of two KIR haplotypes A and B. Gene (2004) 335:121–31. 10.1016/j.gene.2004.03.018 [DOI] [PubMed] [Google Scholar]
- 53.Symons HJ, Fuchs EJ. Hematopoietic SCT from partially HLA-mismatched (HLA-haploidentical) related donors. Bone Marrow Transplant (2008) 42(6):365–77. 10.1038/bmt.2008.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Musette P, Michel L, Jean-Louis F, Bagot M, Bensussan A. Polymorphic expression of CD158k/p140/KIR3DL2 in Sezary patients. Blood (2003) 101(3):1203. 10.1182/blood-2002-09-2915 [DOI] [PubMed] [Google Scholar]
- 55.Bagot M, Moretta A, Sivori S, Biassoni R, Cantoni C, Bottino C, et al. CD4(+) cutaneous T-cell lymphoma cells express the p140-killer cell immunoglobulin-like receptor. Blood (2001) 97(5):1388–91. 10.1182/blood.V97.5.1388 [DOI] [PubMed] [Google Scholar]
- 56.Battistella M, Janin A, Jean-Louis F, Collomb C, Leboeuf C, Sicard H, et al. KIR3DL2 (CD158k) is a potential therapeutic target in primary cutaneous anaplastic large-cell lymphoma. Br J Dermatol (2016) 175(2):325–33. 10.1111/bjd.14626 [DOI] [PubMed] [Google Scholar]
- 57.Giebel S, Locatelli F, Wojnar J, Velardi A, Mina T, Giorgiani G, et al. Homozygosity for human leucocyte antigen-C ligands of KIR2DL1 is associated with increased risk of relapse after human leucocyte antigen-C-matched unrelated donor haematopoietic stem cell transplantation. Br J Haematol (2005) 131(4):483–6. 10.1111/j.1365-2141.2005.05797.x [DOI] [PubMed] [Google Scholar]
- 58.Karre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature (1986) 319(6055):675–8. 10.1038/319675a0 [DOI] [PubMed] [Google Scholar]
- 59.Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today (1990) 11(7):237–44. 10.1016/0167-5699(90)90097-S [DOI] [PubMed] [Google Scholar]
- 60.Hsu KC, Gooley T, Malkki M, Pinto-Agnello C, Dupont B, Bignon JD, et al. KIR ligands and prediction of relapse after unrelated donor hematopoietic cell transplantation for hematologic malignancy. Biol Blood Marrow Transplant (2006) 12(8):828–36. 10.1016/j.bbmt.2006.04.008 [DOI] [PubMed] [Google Scholar]
- 61.Ruggeri L, Mancusi A, Urbani E, Velardi A. Identifying NK alloreactive donors for haploidentical hematopoietic stem cell transplantation. Methods Mol Biol (2016) 1393:141–5. 10.1007/978-1-4939-3338-9_14 [DOI] [PubMed] [Google Scholar]
- 62.Chewning JH, Gudme CN, Hsu KC, Selvakumar A, Dupont B. KIR2DS1-positive NK cells mediate alloresponse against the C2 HLA-KIR ligand group in vitro. J Immunol (2007) 179(2):854–68. 10.4049/jimmunol.179.2.854 [DOI] [PubMed] [Google Scholar]
- 63.Leung W, Iyengar R, Turner V, Lang P, Bader P, Conn P, et al. Determinants of antileukemia effects of allogeneic NK cells. J Immunol (2004) 172(1):644–50. 10.4049/jimmunol.172.1.644 [DOI] [PubMed] [Google Scholar]
- 64.Cooley S, Trachtenberg E, Bergemann TL, Saeteurn K, Klein J, Le CT, et al. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood (2009) 113(3):726–32. 10.1182/blood-2008-07-171926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Marsh SG, et al. Donor killer cell Ig-like receptor B haplotypes, recipient HLA-C1, and HLA-C mismatch enhance the clinical benefit of unrelated transplantation for acute myelogenous leukemia. J Immunol (2014) 192(10):4592–600. 10.4049/jimmunol.1302517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reisner Y, Kapoor N, Kirkpatrick D, Pollack MS, Cunningham-Rundles S, Dupont B, et al. Transplantation for severe combined immunodeficiency with HLA-A,B,D,DR incompatible parental marrow cells fractionated by soybean agglutinin and sheep red blood cells. Blood (1983) 61(2):341–8. [PubMed] [Google Scholar]
- 67.Aversa F, Tabilio A, Terenzi A, Velardi A, Falzetti F, Giannoni C, et al. Successful engraftment of T-cell-depleted haploidentical “three-loci” incompatible transplants in leukemia patients by addition of recombinant human granulocyte colony-stimulating factor-mobilized peripheral blood progenitor cells to bone marrow inoculum. Blood (1994) 84(11):3948–55. [PubMed] [Google Scholar]
- 68.Bachanova V, Cooley S, Defor TE, Verneris MR, Zhang B, McKenna DH, et al. Clearance of acute myeloid leukemia by haploidentical natural killer cells is improved using IL-2 diphtheria toxin fusion protein. Blood (2014) 123(25):3855–63. 10.1182/blood-2013-10-532531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stringaris K, Adams S, Uribe M, Eniafe R, Wu CO, Savani BN, et al. Donor KIR Genes 2DL5A, 2DS1 and 3DS1 are associated with a reduced rate of leukemia relapse after HLA-identical sibling stem cell transplantation for acute myeloid leukemia but not other hematologic malignancies. Biol Blood Marrow Transplant (2010) 16(9):1257–64. 10.1016/j.bbmt.2010.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pende D, Marcenaro S, Falco M, Martini S, Bernardo ME, Montagna D, et al. Anti-leukemia activity of alloreactive NK cells in KIR ligand-mismatched haploidentical HSCT for pediatric patients: evaluation of the functional role of activating KIR and redefinition of inhibitory KIR specificity. Blood (2009) 113(13):3119–29. 10.1182/blood-2008-06-164103 [DOI] [PubMed] [Google Scholar]
- 71.Moretta A, Pende D, Locatelli F, Moretta L. Activating and inhibitory killer immunoglobulin-like receptors (KIR) in haploidentical haemopoietic stem cell transplantation to cure high-risk leukaemias. Clin Exp Immunol (2009) 157(3):325–31. 10.1111/j.1365-2249.2009.03983.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rezvani K, Rouce RH. The application of natural killer cell immunotherapy for the treatment of cancer. Front Immunol (2015) 6:578. 10.3389/fimmu.2015.00578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shilling HG, Young N, Guethlein LA, Cheng NW, Gardiner CM, Tyan D, et al. Genetic control of human NK cell repertoire. J Immunol (2002) 169(1):239–47. 10.4049/jimmunol.169.1.239 [DOI] [PubMed] [Google Scholar]
- 74.Shilling HG, McQueen KL, Cheng NW, Shizuru JA, Negrin RS, Parham P. Reconstitution of NK cell receptor repertoire following HLA-matched hematopoietic cell transplantation. Blood (2003) 101(9):3730–40. 10.1182/blood-2002-08-2568 [DOI] [PubMed] [Google Scholar]
- 75.Davies SM, Ruggieri L, DeFor T, Wagner JE, Weisdorf DJ, Miller JS, et al. Evaluation of KIR ligand incompatibility in mismatched unrelated donor hematopoietic transplants. Killer immunoglobulin-like receptor. Blood (2002) 100(10):3825–7. 10.1182/blood-2002-04-1197 [DOI] [PubMed] [Google Scholar]
- 76.Schaffer M, Aldener-Cannava A, Remberger M, Ringden O, Olerup O. Roles of HLA-B, HLA-C and HLA-DPA1 incompatibilities in the outcome of unrelated stem-cell transplantation. Tissue Antigens (2003) 62(3):243–50. 10.1034/j.1399-0039.2003.00089.x [DOI] [PubMed] [Google Scholar]
- 77.Bornhauser M, Schwerdtfeger R, Martin H, Frank KH, Theuser C, Ehninger G. Role of KIR ligand incompatibility in hematopoietic stem cell transplantation using unrelated donors. Blood (2004) 103(7):2860–1; author reply 2. 10.1182/blood-2003-11-3893 [DOI] [PubMed] [Google Scholar]
- 78.Schaffer M, Malmberg KJ, Ringden O, Ljunggren HG, Remberger M. Increased infection-related mortality in KIR-ligand-mismatched unrelated allogeneic hematopoietic stem-cell transplantation. Transplantation (2004) 78(7):1081–5. 10.1097/01.TP.0000137103.19717.86 [DOI] [PubMed] [Google Scholar]
- 79.Kröger N, Binder T, Zabelina T, Wolschke C, Schieder H, Renges H, et al. Low number of donor activating killer immunoglobulin-like receptors (KIR) genes but not KIR-ligand mismatch prevents relapse and improves disease-free survival in leukemia patients after in vivo T-cell depleted unrelated stem cell transplantation. Transplantation (2006) 82(8):1024–30. 10.1097/01.tp.0000235859.24513.43 [DOI] [PubMed] [Google Scholar]
- 80.De Santis D, Bishara A, Witt CS, Nagler A, Brautbar C, Slavin S, et al. Natural killer cell HLA-C epitopes and killer cell immunoglobulin-like receptors both influence outcome of mismatched unrelated donor bone marrow transplants. Tissue Antigens (2005) 65(6):519–28. 10.1111/j.1399-0039.2005.00396.x [DOI] [PubMed] [Google Scholar]
- 81.Rocha V, Ruggeri A, Spellman S, Wang T, Sobecks R, Locatelli F, et al. Killer cell immunoglobulin-like receptor-ligand matching and outcomes after unrelated cord blood transplantation in acute myeloid leukemia. Biol Blood Marrow Transplant (2016) 22(7):1284–9. 10.1016/j.bbmt.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Willemze R, Rodrigues CA, Labopin M, Sanz G, Michel G, Socie G, et al. KIR-ligand incompatibility in the graft-versus-host direction improves outcomes after umbilical cord blood transplantation for acute leukemia. Leukemia (2009) 23(3):492–500. 10.1038/leu.2008.365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Farag SS, Bacigalupo A, Eapen M, Hurley C, Dupont B, Caligiuri MA, et al. The effect of KIR ligand incompatibility on the outcome of unrelated donor transplantation: a report from the center for international blood and marrow transplant research, the European blood and marrow transplant registry, and the Dutch registry. Biol Blood Marrow Transplant (2006) 12(8):876–84. 10.1016/j.bbmt.2006.05.007 [DOI] [PubMed] [Google Scholar]
- 84.Gagne K, Busson M, Bignon JD, Balere-Appert ML, Loiseau P, Dormoy A, et al. Donor KIR3DL1/3DS1 gene and recipient Bw4 KIR ligand as prognostic markers for outcome in unrelated hematopoietic stem cell transplantation. Biol Blood Marrow Transplant (2009) 15(11):1366–75. 10.1016/j.bbmt.2009.06.015 [DOI] [PubMed] [Google Scholar]
- 85.Giebel S, Locatelli F, Lamparelli T, Velardi A, Davies S, Frumento G, et al. Survival advantage with KIR ligand incompatibility in hematopoietic stem cell transplantation from unrelated donors. Blood (2003) 102(3):814–9. 10.1182/blood-2003-01-0091 [DOI] [PubMed] [Google Scholar]
- 86.Penack O, Fischer L, Gentilini C, Nogai A, Muessig A, Rieger K, et al. The type of ATG matters – natural killer cells are influenced differentially by Thymoglobulin, Lymphoglobulin and ATG-Fresenius. Transpl Immunol (2007) 18(2):85–7. 10.1016/j.trim.2007.05.001 [DOI] [PubMed] [Google Scholar]
- 87.Neudorf S, Jones M. The effects of antithymocyte globulin on natural killer cells. Exp Hematol (1988) 16(10):831–5. [PubMed] [Google Scholar]
- 88.Stauch D, Dernier A, Sarmiento Marchese E, Kunert K, Volk HD, Pratschke J, et al. Targeting of natural killer cells by rabbit antithymocyte globulin and campath-1H: similar effects independent of specificity. PLoS One (2009) 4(3):e4709. 10.1371/journal.pone.0004709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bosch M, Dhadda M, Hoegh-Petersen M, Liu Y, Hagel LM, Podgorny P, et al. Immune reconstitution after anti-thymocyte globulin-conditioned hematopoietic cell transplantation. Cytotherapy (2012) 14(10):1258–75. 10.3109/14653249.2012.715243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Giebel S, Dziaczkowska J, Wojnar J, Krawczyk-Kulis M, Markiewicz M, Kruzel T, et al. The impact of immunosuppressive therapy on an early quantitative NK cell reconstitution after allogeneic haematopoietic cell transplantation. Ann Transplant (2005) 10(2):29–33. [PubMed] [Google Scholar]
- 91.Servais S, Menten-Dedoyart C, Beguin Y, Seidel L, Gothot A, Daulne C, et al. Impact of pre-transplant anti-t cell globulin (ATG) on immune recovery after myeloablative allogeneic peripheral blood stem cell transplantation. PLoS One (2015) 10(6):e0130026. 10.1371/journal.pone.0130026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aversa F, Tabilio A, Velardi A, Cunningham I, Terenzi A, Falzetti F, et al. Treatment of high-risk acute leukemia with T-cell-depleted stem cells from related donors with one fully mismatched HLA haplotype. N Engl J Med (1998) 339(17):1186–93. 10.1056/NEJM199810223391702 [DOI] [PubMed] [Google Scholar]
- 93.Miller JS, Cooley S, Parham P, Farag SS, Verneris MR, McQueen KL, et al. Missing KIR ligands are associated with less relapse and increased graft-versus-host disease (GVHD) following unrelated donor allogeneic HCT. Blood (2007) 109(11):5058–61. 10.1182/blood-2007-01-065383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ludajic K, Balavarca Y, Bickeboller H, Rosenmayr A, Fae I, Fischer GF, et al. KIR genes and KIR ligands affect occurrence of acute GVHD after unrelated, 12/12 HLA matched, hematopoietic stem cell transplantation. Bone Marrow Transplant (2009) 44(2):97–103. 10.1038/bmt.2008.432 [DOI] [PubMed] [Google Scholar]
- 95.Sobecks RM, Wang T, Askar M, Gallagher MM, Haagenson M, Spellman S, et al. Impact of KIR and HLA genotypes on outcomes after reduced-intensity conditioning hematopoietic cell transplantation. Biol Blood Marrow Transplant (2015) 21(9):1589–96. 10.1016/j.bbmt.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Venstrom JM, Pittari G, Gooley TA, Chewning JH, Spellman S, Haagenson M, et al. HLA-C-dependent prevention of leukemia relapse by donor activating KIR2DS1. N Engl J Med (2012) 367(9):805–16. 10.1056/NEJMoa1200503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun JY, Gaidulis L, Dagis A, Palmer J, Rodriguez R, Miller MM, et al. Killer Ig-like receptor (KIR) compatibility plays a role in the prevalence of acute GVHD in unrelated hematopoietic cell transplants for AML. Bone Marrow Transplant (2005) 36(6):525–30. 10.1038/sj.bmt.1705089 [DOI] [PubMed] [Google Scholar]
- 98.Giebel S, Nowak I, Wojnar J, Markiewicz M, Dziaczkowska J, Wylezol I, et al. Impact of activating killer immunoglobulin-like receptor genotype on outcome of unrelated donor-hematopoietic cell transplantation. Transplant Proc (2006) 38(1):287–91. 10.1016/j.transproceed.2005.11.091 [DOI] [PubMed] [Google Scholar]
- 99.Venstrom JM, Gooley TA, Spellman S, Pring J, Malkki M, Dupont B, et al. Donor activating KIR3DS1 is associated with decreased acute GVHD in unrelated allogeneic hematopoietic stem cell transplantation. Blood (2010) 115(15):3162–5. 10.1182/blood-2009-08-236943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kröger N, Zabelina T, Berger J, Duske H, Klyuchnikov E, Binder T, et al. Donor KIR haplotype B improves progression-free and overall survival after allogeneic hematopoietic stem cell transplantation for multiple myeloma. Leukemia (2011) 25(10):1657–61. 10.1038/leu.2011.138 [DOI] [PubMed] [Google Scholar]
- 101.Bachanova V, Weisdorf DJ, Wang T, Marsh SG, Trachtenberg E, Haagenson MD, et al. Donor KIR B genotype improves progression-free survival of non-Hodgkin lymphoma patients receiving unrelated donor transplantation. Biol Blood Marrow Transplant (2016) 22(9):1602–7. 10.1016/j.bbmt.2016.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nowak J, Koscinska K, Mika-Witkowska R, Rogatko-Koros M, Mizia S, Jaskula E, et al. Donor NK cell licensing in control of malignancy in hematopoietic stem cell transplant recipients. Am J Hematol (2014) 89(10):E176–83. 10.1002/ajh.23802 [DOI] [PubMed] [Google Scholar]
- 103.Foley B, Felices M, Cichocki F, Cooley S, Verneris MR, Miller JS. The biology of NK cells and their receptors affects clinical outcomes after hematopoietic cell transplantation (HCT). Immunol Rev (2014) 258(1):45–63. 10.1111/imr.12157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nowak J, Koscinska K, Mika-Witkowska R, Rogatko-Koros M, Mizia S, Jaskula E, et al. Role of donor activating KIR-HLA ligand-mediated NK cell education status in control of malignancy in hematopoietic cell transplant recipients. Biol Blood Marrow Transplant (2015) 21(5):829–39. 10.1016/j.bbmt.2015.01.018 [DOI] [PubMed] [Google Scholar]
- 105.Foley B, Cooley S, Verneris MR, Curtsinger J, Luo X, Waller EK, et al. NK cell education after allogeneic transplantation: dissociation between recovery of cytokine-producing and cytotoxic functions. Blood (2011) 118(10):2784–92. 10.1182/blood-2011-04-347070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Horowitz A, Guethlein LA, Nemat-Gorgani N, Norman PJ, Cooley S, Miller JS, et al. Regulation of adaptive NK cells and CD8 T cells by HLA-C correlates with allogeneic hematopoietic cell transplantation and with cytomegalovirus reactivation. J Immunol (2015) 195(9):4524–36. 10.4049/jimmunol.1401990 [DOI] [PMC free article] [PubMed] [Google Scholar]