Abstract
The BTK inhibitor ibrutinib is a highly effective, new targeted therapy for chronic lymphocytic leukemia (CLL) that thwarts leukemia cell survival, growth, and tissue homing. The effects of ibrutinib treatment on the T cell compartment, which is clonally expanded and thought to support the growth of the malignant B cells in CLL, are not fully characterized. Using next-generation sequencing technology we characterized the diversity of TCRβ chains in peripheral blood T cells from 15 CLL patients before and after one year of ibrutinib therapy. We noted elevated CD4+ and CD8+ T cell numbers and a restricted TCRβ repertoire in all pretreatment samples. After one year of ibrutinib therapy, elevated PB T cell numbers and T-cell related cytokine levels had normalized and T cell repertoire diversity significantly increased. Dominant TCRβ clones in pretreatment samples declined or became undetectable, and the number of productive unique clones significantly increased during ibrutinib therapy, with the emergence of large numbers of low-frequency TCRβ clones. Importantly, broader TCR repertoire diversity was associated with clinical efficacy and lower rates of infections during ibrutinib therapy. These data demonstrate that ibrutinib therapy increases diversification of the T cell compartment in CLL patients, which contributes to cellular immune reconstitution.
Keywords: Chronic lymphocytic leukemia, Ibrutinib, TCR repertoire, Next-generation sequencing
INTRODUCTION
Chronic lymphocytic leukemia (CLL), the most common leukemia in adults in Western societies, is characterized by the expansion of long-lived CD5+ mature monoclonal B lymphocytes in the blood and lymphatic tissues (1) .The expression and function of the B cell receptor (BCR) is central to disease pathogenesis and prognosis (2). Activation of BCR signaling occurs in the secondary lymphatic tissues, where CLL cells proliferate in areas called “pseudofollicles” or “proliferation centers” (3). In these areas, CLL cells are interspersed with T cells, which co-localize with Ki67+ proliferating CLL cells, suggesting that T cells provide help to the CLL cells for promoting their expansion (4–6). PB T cell numbers in untreated CLL patients are elevated and oligoclonal (4, 7), and emerging data indicate that these CLL T cells expand in an antigen-dependent fashion, in a process that resembles normal adaptive immune responses to antigen (8–10). Prior studies reported about T cell repertoire skewing and oligoclonality in CLL patients based on flow cytometric analysis, spectratyping (9, 11–13), and T cell receptor beta sequencing (10). TCRαβ diversity is generated by random rearrangements of V and J segments in the TCRα gene and V, D, and J segments in the TCRβ gene, concurrent with non-templated nucleotide insertions and deletions at the junctions (N-region); the resulting NDN region along with short segments of the flanking V and J genes comprise the complementarity-determining regions 3 (CDR3) of the TCR, which is primarily responsible for recognition of antigenic peptides (14). Prior studies in CLL established T cell dysfunction (anergy), which has been linked to increased susceptibility for infections (4, 15–17), and defective immunologic synapse formation between T cells and CLL cells or other antigen-presenting cells (APCs), resulting in impaired cytotoxicity against the malignant B cells (15, 16) and an “exhausted” T cell phenotype from chronic antigenic stimulation (4, 8, 17).
Ibrutinib is an orally bioavailable irreversible inhibitor of BTK (Bruton’s tyrosine kinase), a central BCR signaling molecule (2). In patients with CLL, ibrutinib is given continuously, and it characteristically causes rapid shrinkage of enlarged lymph nodes, along with a transient increase in PB CLL cells due to redistribution of tissue-resident CLL cells into the PB (18–20). With longer ibrutinib treatment, the vast majority of patients achieve durable remissions (19–21). We recently reported that T cell numbers in both CD4 and CD8 subsets normalized during ibrutinib-based therapy (19). How ibrutinib affects the T cell compartment, via direct or indirect mechanism, is unknown. Ibrutinib’s inhibition of IL-2 inducible kinase (ITK), a member of the TEC kinase family, which plays an important role in T-cell receptor (TCR) signaling, T cell polarization, adhesion, and migration, may argue for direct drug effects (22). Due to a conserved Cys in the kinase domain of ITK that is identical with BTK, ITK is also inhibited by ibrutinib and hence may affect T cell function, especially in Th2-polarized CD4+ T cells (23).
Given the importance of T cells in CLL pathophysiology and the effects of ibrutinib treatment on PB T cell numbers, we studied in detail the ibrutinib treatment-induced changes in the T cell compartment, utilizing high-throughput TCRβ chain sequencing to analyze the TCRβ repertoire in CLL patients before and after 12 months of ibrutinib therapy, along with cytokine profiling.
MATERIALS AND METHODS
Patient selection and clinical characteristics
This study was conducted using samples from patients treated on protocols that were reviewed and approved by the Institutional Review Board at MD Anderson Cancer Center (MDACC) in accordance with the Declaration of Helsinki; all patients provided informed consent for this study. Patients met clinical and immunophenotypic criteria for CLL and were treated at MDACC between February, 2012 and April, 2015 on ibrutinib monotherapy or ibrutinib plus rituximab (ClinicalTrials.gov, NCT01520519 and NCT02007044). Peripheral blood (PB) samples were collected before and after 3, 6 and 12 months on ibrutinib treatment. Plasma samples were collected after centrifugation and stored at −80°C until further use. PB mononuclear cells (PBMCs) were isolated via density gradient centrifugation over Ficoll-Paque (GE Healthcare, Pittsburgh, PA) and frozen in fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific, Waltham, MA) supplemented with 10% dimethylsulfoxide (DMSO; Sigma-Aldrich, St. Louis, MO) for storage in liquid nitrogen until further used. Clinical characteristics and laboratory data from the 29 patients analyzed in this study were collected using Clinic Station Version 3.4.4, an institutional electronic medical record system. Out of the samples from 29 patients, 15 patients’ samples underwent T cell repertoire analysis using purified CD3+ T cells (Table 1); serial samples from 14 patients were analyzed for changes in plasma cytokine levels. Additionally, T cell repertoire analyses from 5 age-matched healthy control samples (median 60 years, range 49 – 71 years; 3 females and 2 males) were obtained from the immunoSEQ platform database (Adaptive Biotechnologies, Seattle, WA). Baseline characteristics of the 29 patients are summarized in the Supplemental Table 1. Age-matched control plasma samples were purchased from ProteoGenex Inc. (Culver City, CA) for measurement of each cytokines in a cohort of age-matched healthy controls (median age: 62 years, range: 60 – 70, 5 females and 7 males).
Table 1.
Clinical characteristics of CLL patients with TCR repertoire analysis (n = 15)
| Characteristic | N (frequency, %) or median (range) |
|---|---|
| Sex | |
| Female | 7 (46.7) |
| Male | 8 (53.3) |
| Age (years) | 65 (48 – 75) |
| Rai stage | |
| 0 | 0 (0.0) |
| I | 3 (20.0) |
| II | 3 (20.0) |
| III | 2 (13.3) |
| IV | 7 (46.7) |
| Cytogenetic abnormalities (FISH) | |
| Del 17p | 6 (40.0) |
| Del 11q | 4 (26.6) |
| Trisomy 12 | 1 (6.7) |
| Diploid | 1 (6.7) |
| Del 13q | 3 (20.0) |
| Survival status | |
| Alive | 15 (100.0) |
| Dead | 0 (0.0) |
| IgVH | |
| Mutated | 1 (6.7) |
| Unmutated | 13 (86.7) |
| Unknown | 1 (6.7) |
| CD38 | |
| Positive | 5 (33.3) |
| Negative | 10 (66.7) |
| ZAP-70 | |
| Positive | 9 (60.0) |
| Negative | 4 (26.6) |
| Unknown | 2 (13.3) |
| Disease status | |
| CR | 3 (20.0) |
| PR | 12 (80.0) |
| Relapsed/refractory | 12 (80.0) |
| Number of previous treatments | 2 (1 – 4) |
| Time from last treatment to ibrutinib therapy | 3 (1.5 – 13) |
| Previous purine analog | 10 (66.7) |
| Previous alkylating agent | 11 (73.3) |
| Previous anti-CD20 monoclonal antibody | 11 (73.3) |
| Previous lenalidomide | 0 (0) |
CR, complete remission; PR, partial remission.
Cytokine detection using multiplex-bead array assay
We used the MILLIPLEX® map Human High Sensitivity T Cell Panel (EMD Millipore, Billerica, MA) to measure plasma levels of 21 cytokines before and after 3, 6, and 12 months of ibrutinib therapy. The kit includes specific components for quantification of human IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12p70, IL-13, IL-17A, IL-21, IL-23, TNF-α, γ-IFN, GM-CSF, CCL3 (MIP-1α), CCL4 (MIP-1β), CXCL11 (ITAC), CCL20 (MIP-3α), and fractalkine. The assay was performed according to manufacture’s instructions. The resulting raw data were acquired using a Luminex 200 plate reader and analyzed with the associated Bio-Plex® Manager 4.0 software (Bio-Rad, Hercules, CA).
Analysis of TCRβ repertoire diversity by next-generation sequencing
To analyze changes in TCR repertoire during ibrutinib therapy, we applied next-generation sequencing technology to characterize TCRβ CDR3 sequences before and after 12 months of ibrutinib monotherapy (n = 10) or ibrutinib plus rituximab therapy (n = 5). To obtain purified T cell DNA templates, PBMC were thawed, and CD3+ T cells were stained with a mouse monoclonal antibody anti human CD3 conjugated to allophycocyanin (APC), clone UCHT1 (BD Biosciences) and then purified by fluorescence activated cell sorting (FACS) using a FACSAria II (BD Biosciences, San Jose, CA); the gating strategy comprised a first gate on live cells, followed by a second and third gate on single cells (FSC-A vs FSC-H and SSC-A vs SSC-H, respectively). The purity of sorted CD3+ population was above 98% for all the samples. Then, genomic DNA was extracted from purified CD3+ T cells using the QIAamp DNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions, quantification and purity assessment was performed using a 1000 NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE). Input template DNA amount for next generation sequencing was 1.2 µg per sample. Bias-controlled multiplexed PCR amplification, high-throughput sequencing, and TCRβ CDR3 region analysis via a bioinformatics pipeline were performed using the immunoSEQ platform (Adaptive Biotechnologies, Seattle, WA) (24, 25). The total sequences per sample were composed of productive sequences (“in frame”) and non-productive sequences (including “out of frame” and “has stop”, which indicates a stop codon within the CDR3 region that was generated during VDJ recombination). Incomplete and out-of-frame rearrangements were filtered out. All productive unique sequences (clonotypes) per sample were ranked in 4 groups classified according to their frequencies (based on the ratio of read counts per clone to input cell numbers) to analyze the features of every TCRβ clonotype. Group “4” refers to the 1000 most frequent clones (top 1000) within each sample; “3” refers to clones with frequency ranging from 0.0005% up to the top 1000; “2” refers to clones with frequency ranging from singleton up to 0.0005%; and “1” denotes singleton clones that were from nearly single cells (with read counts less than 10). Next, to longitudinally observe how the frequency changed for each patient in terms of distribution of clones among groups 1–4 from pretreatment to 1 year after ibrutinib therapy, all TCRβ productive unique clones from each patient were assumed as a whole, and each clone was assigned to one of 3 classes: “new”, “undetected”, and “persistent”. “New” indicates that the clones appeared after ibrutinib therapy and were not present before treatment; “undetected” refers to clones that were present before treatment and then disappeared after therapy; and “persistent” indicates the clones that existed both before and after treatment.
Statistics
All statistical analyses were performed using GraphPad Prism version 6.00 for Mac (GraphPad Software, La Jolla, CA, 2013). The results were expressed as mean ± standard error of the mean (SEM) or median and range as appropriate. Comparisons of proportions and variables between different groups were performed by the Mann-Whitney test, Wilcoxon matched-paired signed rank test, and paired or unpaired t test, as appropriate. Pearson correlation was used to analyze the correlation between univariates. Using a 2-sided analysis, P ≤ 0.05 was considered statistical significance.
RESULTS
Peripheral blood T cell counts decline and normalize during ibrutinib therapy
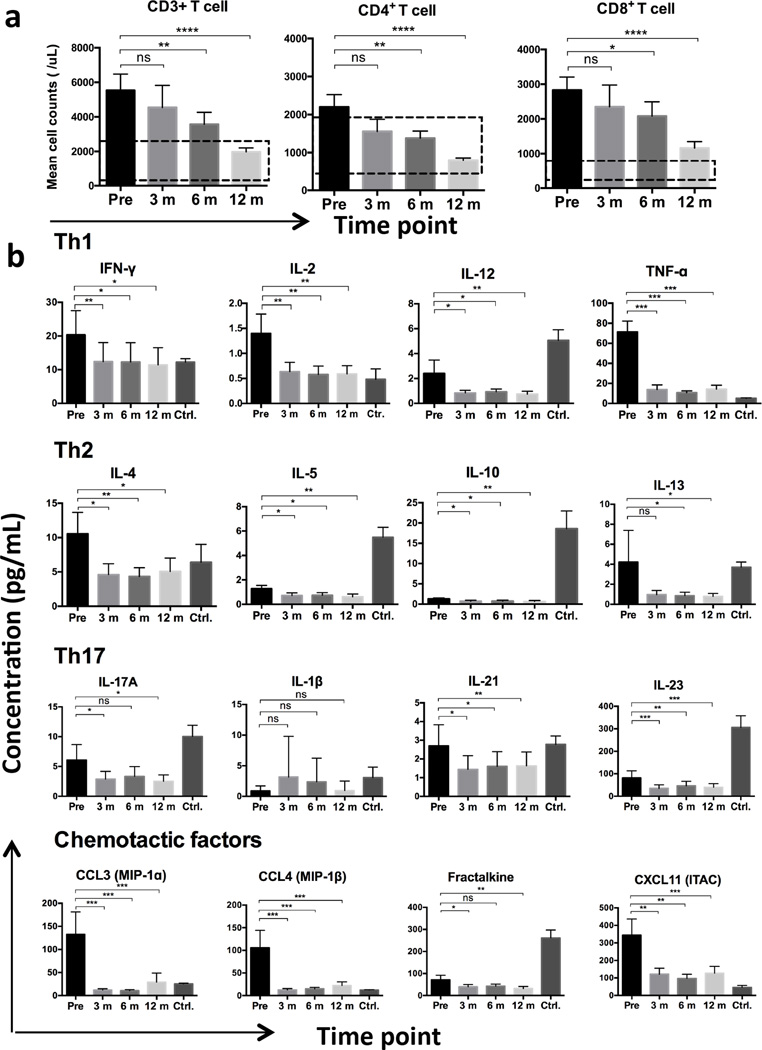
Prior to ibrutinib treatment we noted elevated counts of CD3+ lymphocyte (mean ± SEM, 5458 ± 971/µL, n = 26), which dropped to 4531 ± 1401/µL at 3 months on ibrutinib therapy (n = 11), to 3486 ± 713/µL after 6 months (n = 25), and further significantly declined to 1876 ± 209/µL after 12 months (n = 27, P = 0.0009; Figure 1a). Accordingly, CD4+ and CD8+ T cell counts synchronously decreased on ibrutinib therapy over time (r = 0.981, P = 0.0189; Supplemental Figure 1a). The mean CD4+ T cell count decreased from 2209 ± 337/µL prior to treatment (n = 27) to 1601 ± 346/µL after 3 months (n = 11), to 1378 ± 194/µL after 6 months (n = 25, P = 0.0157), and to 788 ± 59/µL after 12 months on ibrutinib therapy (n = 27, P < 0.0001; Figure 1a). CD8+ T cell counts decreased from 2711 ± 372/µL prior to therapy (n = 27) to 2224 ± 674/µL after 3 months (n = 11), to 1994 ± 421/µL after 6 months (n = 25), and to 1068 ± 169/µL after 12 months (n = 27, P < 0.0001; Figure 1a). We noted significant negative correlations between the CD3+, CD4+, and CD8+ T cell counts and duration of ibrutinib therapy (CD3+ T cells: r = −0.998, P = 0.0016; CD4+ T cells: r = −0.980, P = 0.0199; and CD8+ T cells: r = −0.995, P = 0.0055; Supplemental Figure 1a). The declines in T cell counts were accompanied by a reduction in CD19+ CLL cell counts (Supplemental Figure 1b).
Figure 1. T cell counts and the levels of related plasma cytokines significantly decreased during ibrutinib therapy.
(a) T cell subsets decreased over time during ibrutinib therapy. (b) Levels of plasma Th1-, Th2-, and Th17-type cytokines and chemotactic factors significantly declined when compared to pretreatment levels. The bars represent mean values, and dotted lines indicate the normal ranges. “Pre”, “3 m”, “6 m”, and “12 m” refer to pretreatment, 3 month, 6 month, and 12 month on ibrutinib therapy, respectively. Ctrl. Indicates age-matched control group. (Asterisks represent statistical significance; *P < 0.05, ** P < 0.01, ***P < 0.005, **** P < 0.0001. ns, not significant.)
Plasma Th1, Th2, and Th17-type cytokine levels decreased during ibrutinib treatment
The majority of the plasma cytokine and chemokine concentrations, elevated at pretreatment in CLL patients compared to healthy volunteers as reported previously (26), were significantly reduced after 3 months of ibrutinib therapy levels. IL-6 and IL-8 were the only exceptions, with a moderate increase at 3 months and subsequent decrease (supplemental Fig 1c). Th1, Th2, and Th17-type cytokines remained low after 3, 6, 9, and 12 months of continuous treatment (Figure 1b). As IFN-γ and IL-4 are the important differentiation factors for Th1 and Th2 T cells respectively (23), the IFN-γ to IL-4 ratio can be used as an approximation of the Th1/Th2 balance. The mean IFN-γ/IL-4 ratio increased from 2.48 ± 0.83 at baseline to 2.94 ± 1.23 after 12 months of therapy (n = 14, P = 0.708), suggesting that IFN-γ-producing Th1 cells become more prevalent during ibrutinib treatment (23). Consistent with previous studies (19, 27), plasma CCL3 and CCL4 levels were significantly decreased 3 months after ibrutinib therapy, and remained low during follow-up of 12 months (Figure 1b).
TCRβ repertoire diversity increased during ibrutinib therapy
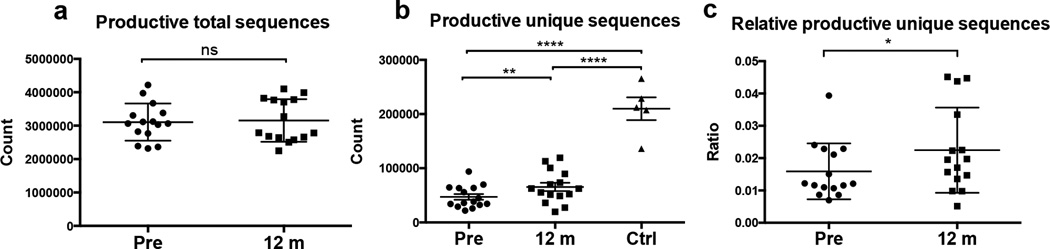
TCRβ sequences were generated with equal amounts of input template DNA that were extracted from matched numbers of purified CD3+ T cells in pretreatment samples and samples collected after 1 year of ibrutinib therapy. Consequently, we did not observe any significant difference in total productive sequence counts between pre- and post-treatment samples (P = 0.792; Figure 2a). In contrast, we noted a significant increase in total unique TCRβ sequences from 58783 ± 6505 (n = 15) per sample before treatment to 80945 ± 9211 (n = 15) after 1 year of ibrutinib therapy (P = 0.0055; Supplemental Figure 1c). More importantly, absolute productive unique sequence counts, an indicator for richness of clones, also significantly increased from 47227 ± 5258 (n = 15) per sample before treatment to 65482 ± 7633 (n = 15) after 1 year of ibrutinib therapy (P = 0.0056; Figure 2b), with a median increase of 76.1% (range 0.53% – 115.46%; Supplemental Figure 1d and 1e). However, these counts were still significantly lower than those in age-matched healthy controls (P < 0.0001), suggesting that it may take more than one year before the process of TCR diversity recovery is complete. Furthermore, to quantify TCR diversity we analyzed the relative productive unique sequences (i.e. ratio of productive unique sequences to productive total sequences), and confirmed that TCRβ repertoire diversity significantly increased 1 year after ibrutinib therapy relative to pretreatment (P = 0.0215; Figure 2c).
Figure 2. TCRβ repertoire diversity increased after one year of ibrutinib therapy.
(a) There was no significant difference in numbers of productive total TCRβ sequences between samples taken before and after 1 year of ibrutinib therapy. In contrast, (b) the number of absolute productive unique sequences, representing the richness of the clones, and (c) relative productive unique sequences significantly increased after 1 year of ibrutinib therapy. “Pre” and “12 m” refer to pretreatment and 12 months on ibrutinib therapy, respectively. (*P < 0.05, ** P < 0.01, **** P < 0.0001; ns, not significant.)
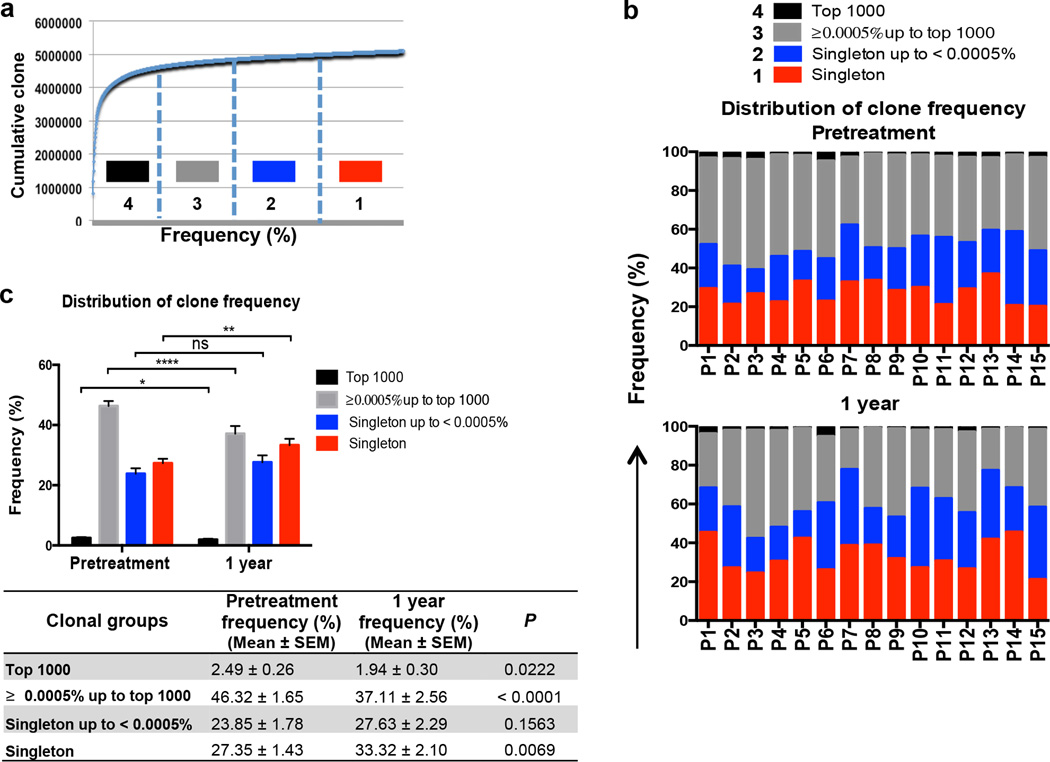
To further characterize TCRβ repertoire restoration during ibrutinib therapy, clonotypes within the TCR libraries from each sample were ranked in 4 groups according to frequency (Figure 3a). We found that there was significantly greater diversity in T cell repertoire after 1 year of ibrutinib therapy, because the fraction of “top 1000” clones declined from 2.49% ± 0.26% (n = 15) in pretreatment samples to 1.94% ± 0.30% (P = 0.0222) and the fraction of “singleton” clones significantly increased from 27.35% ± 1.43% (n = 15) to 33.32% ± 2.10% (P = 0.0069; Figure 3b and 3c). The diversity in the CDR3 regions of the TCRβ chains is generated by non-templated nucleotide insertions at the Vβ-Dβ and Dβ-Jβ junctions. Accordingly, we noted more nucleotide insertions in the CDR3 regions in clones with low-frequency TCRβ sequences than in clones with high-frequency sequences (Supplemental Figure 2a), together with greater CDR3 length (Supplemental Figure 2b). These results, collectively, indicate that patients acquire more low-frequency clones with greater diversification of TCR repertoire during ibrutinib therapy.
Figure 3. High-frequency TCRβ clones diminished and low-frequency T cell clones increased during ibrutinib therapy.
(a) Graphic representation of the model used for assignment of clones according to their frequency. (b and c) TCRβ repertoire diversity significantly increased after 1 year of ibrutinib therapy compared to pretreatment samples, as the fraction of high-frequency clones significantly declined and the fractions of “singleton” clones significantly increased. (*P < 0.05, ** P < 0.01, **** P < 0.0001; ns, not significant.)
Correlation between TCRβ repertoire diversity and clinical characteristics
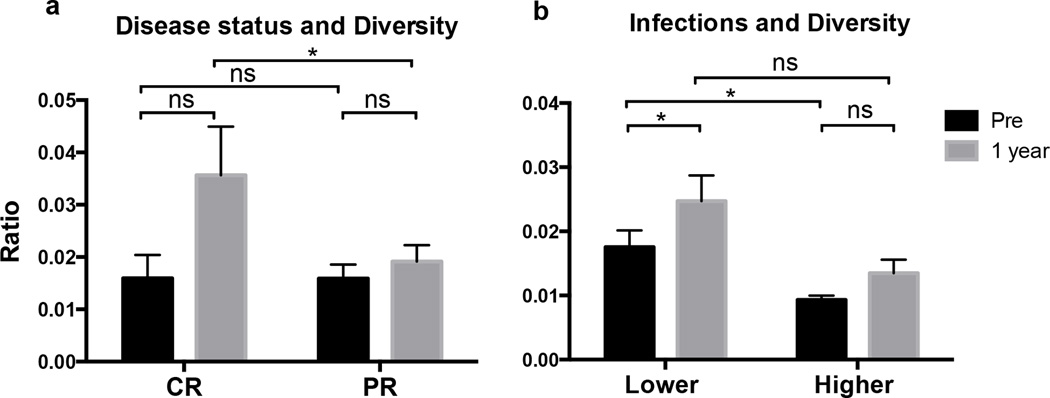
For analysis of correlations between TCRβ repertoire diversity and established prognostic markers, individual TCRβ repertoire diversity was stratified based on patients ZAP-70 and CD38 expression, prior treatment, cytogenetic abnormalities, disease duration, Rai stage, age, β2 microglobulin levels (β2M), and ALCs at pretreatment and 1 year. Before ibrutinib therapy, there was no significant difference in TCRβ repertoire diversity between patients with low- or high-risk prognostic markers (Supplemental Figure 3a). We also did not find any significant impact of ZAP-70, CD38, disease duration, disease status, or prior treatment (ibrutinib vs. ibrutinib plus rituximab) on TCRβ repertoire diversification (Supplemental Figure 3a and 3b). However, we noted that patients who achieved complete remission (CR) and who had no prior treatment had broader TCR repertoire diversification after 1 year of ibrutinib therapy than patients who achieved partial remissions (PR) and had received prior treatments (both P = 0.0482, Figure 4a and Supplemental Figure 3a). Significantly higher TCR repertoire diversification during ibrutinib therapy also was noted in younger patients (age < 65 years, P = 0.0298), patients with lower β2M level (< 4.5 mg/L, P = 0.0303), lower Rai stages (stage I or II, P = 0.0107), and lower pretreatment CD3+ and CD8+ T cell counts (P = 0.0099 and P = 0.029, respectively; Supplemental Figure 3a). However, these correlations are based on relatively small numbers of patients and therefore need to be interpreted with caution and should be validated in a larger cohort.
Figure 4. Broader TCR repertoire diversification was associated with clinical response and lower infection rates during ibrutinib therapy.
(a) Patients who achieved CR had greater TCR repertoire diversification than patients who achieved PR after one year of ibrutinib therapy. (b) Broader TCR repertoire diversity was associated with lower rates of infections during the first 6 months of ibrutinib therapy, and TCR repertoire diversity significantly increased after 1 year of ibrutinib therapy in patients with lower rates of infection. (*P < 0.05, ns, not significant.)
Next, we explored the impact of TCR repertoire diversity on the risk of infection in CLL patients. Twelve out of 15 patients (80%) developed 27 cases of infections during a median follow-up time of 25 months (range 22 – 26). Infections were more frequent during the first 6 months with an average rate of 16.7 infections per 100 patient-months compared with 4.2 thereafter. Respiratory tract infections were the most common (51.9%), followed by skin (14.8%) and genitourinary (11.1%) infections. We noted a significant correlation between broader TCR repertoire diversity and lower rates of infections during the first 6 months of ibrutinib therapy (P = 0.0425, Figure 4b); accordingly, TCR repertoire diversity significantly increased after 1 year in patients with lower rates of infections (P = 0.0308, Figure 4b).
The change and redistribution of TCRβ clones after ibrutinib therapy
Sixteen and twenty-two Vβ-Jβ segment usages, with more than 20000 reads, were found before and after 1 year of ibrutinib therapy, respectively. Of the top 16 Vβ-Jβ segment usages before treatment, 6 usages (i.e. Vβ 06-05/Jβ 01-02, Vβ 02-01/Jβ 01-01, Vβ 06-05/Jβ 01-01, Vβ 06-01/Jβ 01-05, Vβ 05-06/Jβ 01-03, and Vβ 07-09/Jβ 02-07) had further increased at 1 year after ibrutinib therapy. These findings suggest that usages of Vβ-Jβ subfamilies, skewed before treatment, were improved and increased after 1 year of ibrutinib therapy, and in the meantime antigenic stimulations (by unknown antigens) promotes the persistence or overexpression of certain Vβ-Jβ subfamilies during therapy.
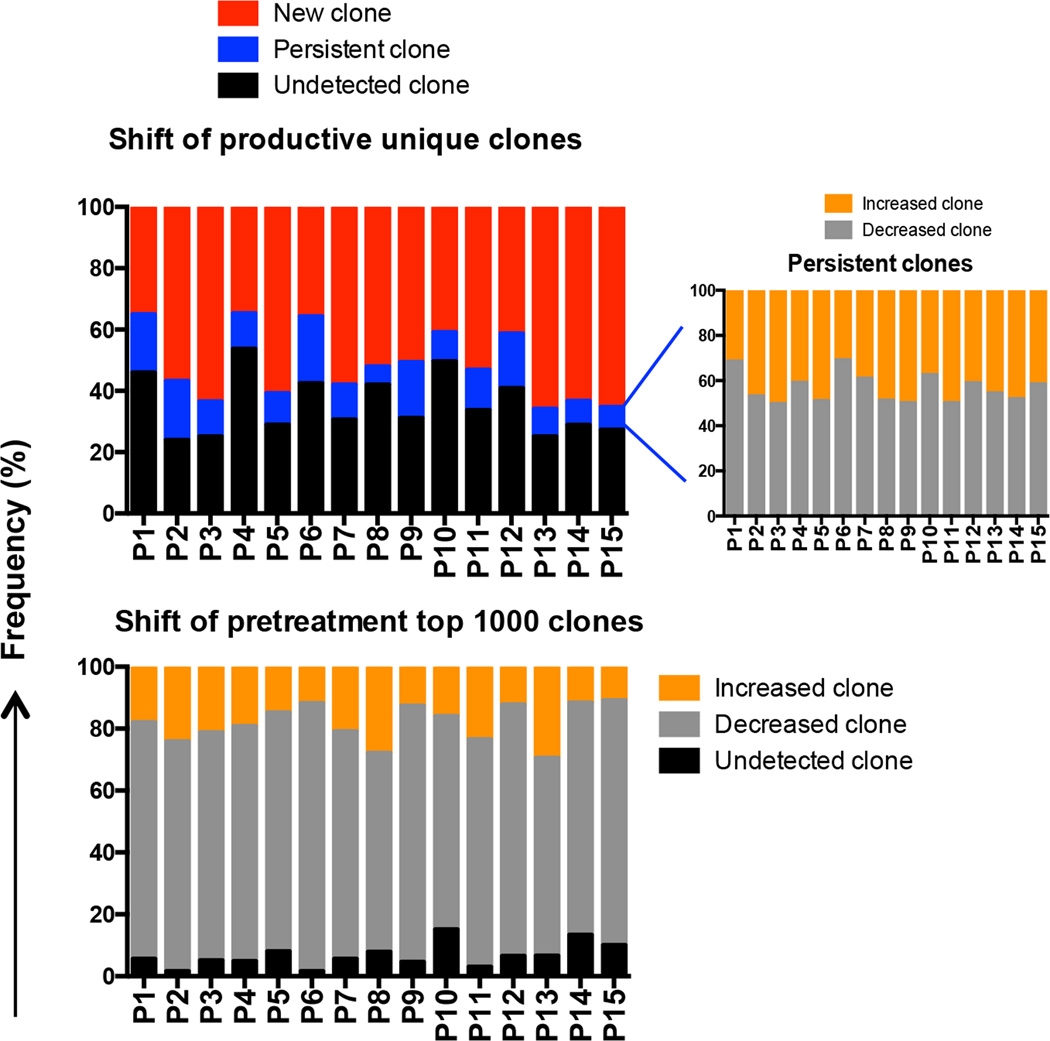
To track each productive unique clone in CLL patients from pretreatment to 1 year of ibrutinib therapy, we categorized all productive unique clones from each patient into one of the following categories: a) new, b) undetected, or c) persistent T cell clones. We detected a large number of new clones that emerged during 1 year of ibrutinib therapy (median 53.16%, range 34.67% – 65.91%) (Figure 5). Interestingly, the majority of these new clones (median 61.95%, range 41.57% – 81.07%) were small-size clones with less than 0.0005% frequency (groups “2” and “1”; Supplemental Figure 2c), suggesting that these represent newly generated T cell clones rather than being derived from T cells present before treatment. In contrast, a median 31.14% of pretreatment clones (range 23.96% – 53.69%) were undetectable after 1 year of ibrutinib therapy (Figure 5). These were mainly high-frequency clones (greater than 0.0005% frequency, groups “4” and “3”; Supplemental Figure 2d). Moreover, a median 75% of the top 1000 pretreatment clones (range 63.9% – 86.6%) declined in frequency, and 5.60% (range 1.60% – 15.10%) became undetectable. Increases were seen only in 18% of these clones (range 10.90% – 29.50%; Figure 5). The fraction of persistent T cell clones after 1 year of ibrutinib therapy was 11.43% (range 5.98% – 21.89%), primarily derived from high-frequency clones; of these, a median 45.48% of clones (range 30.79% – 50.23%) had increased in frequency and 54.52% (range 49.77% – 69.21%) had decreased (Figure 5). These results, collectively, indicate that the vast majority of dominant TCRβ clones that were present before treatment were reduced or vanished during ibrutinib therapy, while at the same time a large number of new low-frequency T cell clones, presumably naïve T cells, emerged.
Figure 5. Longitudinal evaluation of shift of T cell clones from pretreatment to 1 year after ibrutinib therapy.
The dominant clones present before treatment decreased in frequency or vanished, while a large numbers of new clones emerged 1 year after ibrutinib therapy.
DISCUSSION
This deep-sequencing analysis of the TCRβ repertoire in CLL patients undergoing ibrutinib therapy revealed a significant diversification of the TCRβ repertoire during ibrutinib therapy. At the same time, elevated numbers of PB T cells in pretreatment samples, affecting the CD4 and CD8 subsets, normalized. More specifically, we noted a significant increase in numbers of productive unique TCRβ clones after 1 year of ibrutinib therapy when compared to the matched pretreatment samples. Furthermore, this increase in TCRβ productive unique sequences and emergence of large numbers of new T cell clones with low frequency, along with a major reduction or deletion of the majority of clonally expanded high-frequency T cell clones that were present before therapy, corroborate the recovery of the T cell repertoire diversification during ibrutinib therapy in CLL patients.
T cell immune dysfunction is a common feature in CLL that has been linked to increased susceptibility to infectious and autoimmune complications (15, 16, 28). Studies of T cell repertoire and function in CLL patients established a clonal expansion of T cells with increased numbers of CD4+ and CD8+ T cells, elevated levels of T-cell related cytokines, and restrictive usages of specific Vβ-Jβ subfamilies driven by antigen selection, resulting in a skewed TCR repertoire (4, 9–12, 17, 29). The synchronous decline in blood CLL and T cell numbers and the associated decrease in plasma chemokine and cytokine levels during ibrutinib therapy (Fig. 1) support the concept of co-evolution and interdependence between T cells and CLL cells (4, 23, 27). Regarding the decreases in plasma cytokine levels during ibrutinib therapy, more direct effects of ibrutinib have to be considered. Through inhibition of BTK and off-target enzymes, such as ITK, ibrutinib can directly alter the activity of T cells (23), NK cells (30, 31), and monocytes/macrophages (27) resulting in reduced cytokine secretion by these cells. The functional role of these clonally expanded T cells in CLL patients remains controversial; both pro-tumoral and tumor-suppressive activities have been discussed (32–35). In addition to CLL-promoting CD4+ cells (32, 33), other studies characterized cytotoxic T lymphocytes (CTLs) that recognize CLL specific idiotype peptides or other tumor-associated antigens (34, 35) and identified leukemia-associated mono/oligoclonal T cells within the circulating T cell compartment (7). However, these tumor-associated-antigen specific T cell clones apparently cannot effectively eliminate the malignant B cells, which may be related to T cell exhaustion.
Notably, ibrutinib therapy in CLL results in a decline and normalization of elevated CD4+ and CD8+ T cell numbers and inhibition of T cell related chemokine and cytokine production (19, 23, 27), also alter the composition of T cell subsets by exerting a Th1-selective pressure and inhibiting differentiation of the Th17 T cell subset that was skewed in CLL as described previously (23, 27, 36), indicating a normalized T cell state. More importantly, the TCRβ repertoire diversification was reconstituted after one year of ibrutinib therapy, which is reminiscent of changes in the T cell compartment after allogeneic hematopoietic stem cell transplantation (allo-HSCT), where also large numbers of new TCRβ sequences and diverse T cell clonotypes were seen, with a simultaneous decline or deletion of TCRβ clones that were dominant before allo-HSCT (37–39). These findings suggest that effective ibrutinib therapy, over time, reduces TCR repertoire skewing and T cell clonal expansion, and patients acquire a more normal TCR repertoire. Moreover, these findings are also interesting in the context of clinical findings showing a major decrease in infectious complications in CLL patients treated with ibrutinib, once remissions were achieved (22, 28, 40). Interestingly, we also noted that the rate of infection was relatively low in patients with broader TCR repertoire diversity, especially during the first 6 months of ibrutinib treatment, when infections were more frequent. Moreover, patients that achieved CRs had higher TCR repertoire diversity than PR patients after one year of ibrutinib therapy, which indicates that deeper remission allows for better recovery of the TCR repertoire diversity.
Additionally, we found broader TCR repertoire diversification during ibrutinib therapy in patients with low-risk prognostic makers, for example, in younger patients, and those with low β2M level, and lower Rai stages. Therefore, the changes in the TCR repertoire diversity during ibrutinib therapy reflect, to some extent, treatment response and patient characteristics and may have an impact on longer-term outcome. However, given the relatively small number of cases analyzed in this study, these findings need to be corroborated in a larger cohort of patients.
In summary, our data demonstrate that ibrutinib therapy promotes the recovery of TCR repertoire diversity in CLL patients. The emergence of a large number of new TCRβ clones along with the increased diversity of the TCR repertoire indicate that CLL patients acquire a broader TCR Vβ repertoire after 1 year of ibrutinib therapy. These findings strengthen the concept of co-evolution of CLL cells with T cells, and provide novel insight into ibrutinib’s effects on the T cell compartment in CLL patients.
Supplementary Material
Acknowledgments
J.A.B. and S.O.B. received research funding from Pharmacyclics.
The authors are grateful to Sunita Patterson for outstanding manuscript edits and Benjamin Hayes for assistance with sample and data collection.
This work was supported by a Leukemia & Lymphoma Society Scholar Award in Clinical Research (to J.A.B.), a Cancer Center Support Grant (NCI Grant P30 CA016672), MD Anderson’s Moon Shot Program in CLL, and the National Natural Science Foundation of China (Grant # 81000921)
Footnotes
CONFLICT OF INTEREST
The remaining authors declared no competing financial interests.
AUTHOR CONTRIBUTIONS
Q.Y collected the clinical information, analyzed the data and results, designed the figures, and wrote the paper; M.S. selected the patient samples, performed isolation of CD3+ cells, DNA extraction and performed cytokine arrays and corrected the manuscript. H.R., E.Y., and M.V. performed TCR repertoire analyses, assisted with the data interpretation, and reviewed the paper. M.J.K., A.F., Z.E., H.K., N.J., W.G.W., and S.O.B. contributed to the clinical patient management and sample collection. J.A.B. designed and supervised the study, and wrote the paper.
REFERENCES
- 1.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. The New England journal of medicine. 2005;352:804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 2.Burger JA, Buggy JJ. Bruton tyrosine kinase inhibitor ibrutinib (PCI-32765) Leukemia & lymphoma. 2013;54:2385–2391. doi: 10.3109/10428194.2013.777837. [DOI] [PubMed] [Google Scholar]
- 3.Messmer BT, Messmer D, Allen SL, Kolitz JE, Kudalkar P, Cesar D, Murphy EJ, Koduru P, Ferrarini M, Zupo S, Cutrona G, Damle RN, Wasil T, Rai KR, Hellerstein MK, Chiorazzi N. In vivo measurements document the dynamic cellular kinetics of chronic lymphocytic leukemia B cells. The Journal of clinical investigation. 2005;115:755–764. doi: 10.1172/JCI23409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagnara D, Kaufman MS, Calissano C, Marsilio S, Patten PE, Simone R, Chum P, Yan XJ, Allen SL, Kolitz JE, Baskar S, Rader C, Mellstedt H, Rabbani H, Lee A, Gregersen PK, Rai KR, Chiorazzi N. A novel adoptive transfer model of chronic lymphocytic leukemia suggests a key role for T lymphocytes in the disease. Blood. 2011;117:5463–5472. doi: 10.1182/blood-2010-12-324210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pascutti MF, Jak M, Tromp JM, Derks IA, Remmerswaal EB, Thijssen R, van Attekum MH, van Bochove GG, Luijks DM, Pals ST, van Lier RA, Kater AP, van Oers MH, Eldering E. IL-21 and CD40L signals from autologous T cells can induce antigen-independent proliferation of CLL cells. Blood. 2013;122:3010–3019. doi: 10.1182/blood-2012-11-467670. [DOI] [PubMed] [Google Scholar]
- 6.Patten PE, Buggins AG, Richards J, Wotherspoon A, Salisbury J, Mufti GJ, Hamblin TJ, Devereux S. CD38 expression in chronic lymphocytic leukemia is regulated by the tumor microenvironment. Blood. 2008;111:5173–5181. doi: 10.1182/blood-2007-08-108605. [DOI] [PubMed] [Google Scholar]
- 7.Rezvany MR, Jeddi-Tehrani M, Wigzell H, Osterborg A, Mellstedt H. Leukemia-associated monoclonal and oligoclonal TCR-BV use in patients with B-cell chronic lymphocytic leukemia. Blood. 2003;101:1063–1070. doi: 10.1182/blood-2002-03-0746. [DOI] [PubMed] [Google Scholar]
- 8.Christopoulos P, Pfeifer D, Bartholome K, Follo M, Timmer J, Fisch P, Veelken H. Definition and characterization of the systemic T-cell dysregulation in untreated indolent B-cell lymphoma and very early CLL. Blood. 2011;117:3836–3846. doi: 10.1182/blood-2010-07-299321. [DOI] [PubMed] [Google Scholar]
- 9.Goolsby CL, Kuchnio M, Finn WG, Peterson L. Expansions of clonal and oligoclonal T cells in B-cell chronic lymphocytic leukemia are primarily restricted to the CD3(+)CD8(+) T-cell population. Cytometry. 2000;42:188–195. [PubMed] [Google Scholar]
- 10.Vardi A, Agathangelidis A, Stalika E, Karypidou M, Siorenta A, Anagnostopoulos A, Rosenquist R, Hadzidimitriou A, Ghia P, Sutton LA, Stamatopoulos K. Antigen Selection Shapes the T-cell Repertoire in Chronic Lymphocytic Leukemia. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22:167–174. doi: 10.1158/1078-0432.CCR-14-3017. [DOI] [PubMed] [Google Scholar]
- 11.Farace F, Orlanducci F, Dietrich PY, Gaudin C, Angevin E, Courtier MH, Bayle C, Hercend T, Triebel F. T cell repertoire in patients with B chronic lymphocytic leukemia. Evidence for multiple in vivo T cell clonal expansions. Journal of immunology. 1994;153:4281–4290. [PubMed] [Google Scholar]
- 12.Rezvany MR, Jeddi-Tehrani M, Osterborg A, Kimby E, Wigzell H, Mellstedt H. Oligoclonal TCRBV gene usage in B-cell chronic lymphocytic leukemia: major perturbations are preferentially seen within the CD4 T-cell subset. Blood. 1999;94:1063–1069. [PubMed] [Google Scholar]
- 13.Zaborsky N, Holler C, Geisberger R, Asslaber D, Gassner FJ, Egger V, Pinon-Hofbauer J, Kocher T, Hartmann TN, Greil R, Egle A. B-cell receptor usage correlates with the sensitivity to CD40 stimulation and the occurrence of CD4+ T-cell clonality in chronic lymphocytic leukemia. Haematologica. 2015;100:e307–e310. doi: 10.3324/haematol.2015.124719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton L, Wilson IA. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science. 1996;274:209–219. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- 15.Ramsay AG, Johnson AJ, Lee AM, Gorgun G, Le Dieu R, Blum W, Byrd JC, Gribben JG. Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug. The Journal of clinical investigation. 2008;118:2427–2437. doi: 10.1172/JCI35017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramsay AG, Clear AJ, Fatah R, Gribben JG. Multiple inhibitory ligands induce impaired T-cell immunologic synapse function in chronic lymphocytic leukemia that can be blocked with lenalidomide: establishing a reversible immune evasion mechanism in human cancer. Blood. 2012;120:1412–1421. doi: 10.1182/blood-2012-02-411678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riches JC, Davies JK, McClanahan F, Fatah R, Iqbal S, Agrawal S, Ramsay AG, Gribben JG. T cells from CLL patients exhibit features of T-cell exhaustion but retain capacity for cytokine production. Blood. 2013;121:1612–1621. doi: 10.1182/blood-2012-09-457531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burger JA, Montserrat E. Coming full circle: 70 years of chronic lymphocytic leukemia cell redistribution, from glucocorticoids to inhibitors of B-cell receptor signaling. Blood. 2013;121:1501–1509. doi: 10.1182/blood-2012-08-452607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burger JA, Keating MJ, Wierda WG, Hartmann E, Hoellenriegel J, Rosin NY, de Weerdt I, Jeyakumar G, Ferrajoli A, Cardenas-Turanzas M, Lerner S, Jorgensen JL, Nogueras-Gonzalez GM, Zacharian G, Huang X, Kantarjian H, Garg N, Rosenwald A, O'Brien S. Safety and activity of ibrutinib plus rituximab for patients with high-risk chronic lymphocytic leukaemia: a single-arm, phase 2 study. The Lancet. Oncology. 2014;15:1090–1099. doi: 10.1016/S1470-2045(14)70335-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herman SE, Niemann CU, Farooqui M, Jones J, Mustafa RZ, Lipsky A, Saba N, Martyr S, Soto S, Valdez J, Gyamfi JA, Maric I, Calvo KR, Pedersen LB, Geisler CH, Liu D, Marti GE, Aue G, Wiestner A. Ibrutinib-induced lymphocytosis in patients with chronic lymphocytic leukemia: correlative analyses from a phase II study. Leukemia. 2014;28:2188–2196. doi: 10.1038/leu.2014.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byrd JC, Furman RR, Coutre SE, Burger JA, Blum KA, Coleman M, Wierda WG, Jones JA, Zhao W, Heerema NA, Johnson AJ, Shaw Y, Bilotti E, Zhou C, James DF, O'Brien S. Three-year follow-up of treatment-naive and previously treated patients with CLL and SLL receiving single-agent ibrutinib. Blood. 2015;125:2497–2506. doi: 10.1182/blood-2014-10-606038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berg LJ, Finkelstein LD, Lucas JA, Schwartzberg PL. Tec family kinases in T lymphocyte development and function. Annual review of immunology. 2005;23:549–600. doi: 10.1146/annurev.immunol.22.012703.104743. [DOI] [PubMed] [Google Scholar]
- 23.Dubovsky JA, Beckwith KA, Natarajan G, Woyach JA, Jaglowski S, Zhong Y, Hessler JD, Liu TM, Chang BY, Larkin KM, Stefanovski MR, Chappell DL, Frissora FW, Smith LL, Smucker KA, Flynn JM, Jones JA, Andritsos LA, Maddocks K, Lehman AM, Furman R, Sharman J, Mishra A, Caligiuri MA, Satoskar AR, Buggy JJ, Muthusamy N, Johnson AJ, Byrd JC. Ibrutinib is an irreversible molecular inhibitor of ITK driving a Th1-selective pressure in T lymphocytes. Blood. 2013;122:2539–2549. doi: 10.1182/blood-2013-06-507947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH, Carlson CS. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, Parsons JM, Steen MS, LaMadrid-Herrmannsfeldt MA, Williamson DW, Livingston RJ, Wu D, Wood BL, Rieder MJ, Robins H. Using synthetic templates to design an unbiased multiplex PCR assay. Nature communications. 2013;4:2680. doi: 10.1038/ncomms3680. [DOI] [PubMed] [Google Scholar]
- 26.Yan XJ, Dozmorov I, Li W, Yancopoulos S, Sison C, Centola M, Jain P, Allen SL, Kolitz JE, Rai KR, Chiorazzi N, Sherry B. Identification of outcome-correlated cytokine clusters in chronic lymphocytic leukemia. Blood. 2011;118:5201–5210. doi: 10.1182/blood-2011-03-342436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niemann CU, Herman SE, Maric I, Gomez-Rodriguez J, Biancotto A, Chang BY, Martyr S, Stetler-Stevenson M, Yuan CM, Calvo KR, Braylan RC, Valdez J, Lee YS, Wong DH, Jones J, Sun C, Marti GE, Farooqui MZ, Wiestner A. Disruption of in vivo Chronic Lymphocytic Leukemia Tumor-Microenvironment Interactions by Ibrutinib - Findings from an Investigator-Initiated Phase II Study. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22:1572–1582. doi: 10.1158/1078-0432.CCR-15-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riches JC, Gribben JG. Understanding the immunodeficiency in chronic lymphocytic leukemia: potential clinical implications. Hematology/oncology clinics of North America. 2013;27:207–235. doi: 10.1016/j.hoc.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 29.O'Brien S, Furman RR, Coutre SE, Sharman JP, Burger JA, Blum KA, Grant B, Richards DA, Coleman M, Wierda WG, Jones JA, Zhao W, Heerema NA, Johnson AJ, Izumi R, Hamdy A, Chang BY, Graef T, Clow F, Buggy JJ, James DF, Byrd JC. Ibrutinib as initial therapy for elderly patients with chronic lymphocytic leukaemia or small lymphocytic lymphoma: an open-label, multicentre, phase 1b/2 trial. The Lancet. Oncology. 2014;15:48–58. doi: 10.1016/S1470-2045(13)70513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohrt HE, Sagiv-Barfi I, Rafiq S, Herman SE, Butchar JP, Cheney C, Zhang X, Buggy JJ, Muthusamy N, Levy R, Johnson AJ, Byrd JC. Ibrutinib antagonizes rituximab-dependent NK cell-mediated cytotoxicity. Blood. 2014;123:1957–1960. doi: 10.1182/blood-2014-01-547869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Da Roit F, Engelberts PJ, Taylor RP, Breij EC, Gritti G, Rambaldi A, Introna M, Parren PW, Beurskens FJ, Golay J. Ibrutinib interferes with the cell-mediated anti-tumor activities of therapeutic CD20 antibodies: implications for combination therapy. Haematologica. 2015;100:77–86. doi: 10.3324/haematol.2014.107011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Os A, Burgler S, Ribes AP, Funderud A, Wang D, Thompson KM, Tjonnfjord GE, Bogen B, Munthe LA. Chronic lymphocytic leukemia cells are activated and proliferate in response to specific T helper cells. Cell reports. 2013;4:566–577. doi: 10.1016/j.celrep.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 33.Burgler S, Gimeno A, Parente-Ribes A, Wang D, Os A, Devereux S, Jebsen P, Bogen B, Tjonnfjord GE, Munthe LA. Chronic lymphocytic leukemia cells express CD38 in response to Th1 cell-derived IFN-gamma by a T-bet-dependent mechanism. Journal of immunology. 2015;194:827–835. doi: 10.4049/jimmunol.1401350. [DOI] [PubMed] [Google Scholar]
- 34.Trojan A, Schultze JL, Witzens M, Vonderheide RH, Ladetto M, Donovan JW, Gribben JG. Immunoglobulin framework-derived peptides function as cytotoxic T-cell epitopes commonly expressed in B-cell malignancies. Nature medicine. 2000;6:667–672. doi: 10.1038/76243. [DOI] [PubMed] [Google Scholar]
- 35.Rajasagi M, Shukla SA, Fritsch EF, Keskin DB, DeLuca D, Carmona E, Zhang W, Sougnez C, Cibulskis K, Sidney J, Stevenson K, Ritz J, Neuberg D, Brusic V, Gabriel S, Lander ES, Getz G, Hacohen N, Wu CJ. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood. 2014;124:453–462. doi: 10.1182/blood-2014-04-567933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lad DP, Varma S, Varma N, Sachdeva MU, Bose P, Malhotra P. Regulatory T-cell and T-helper 17 balance in chronic lymphocytic leukemia progression and autoimmune cytopenias. Leukemia & lymphoma. 2015;56:2424–2428. doi: 10.3109/10428194.2014.986479. [DOI] [PubMed] [Google Scholar]
- 37.van Heijst JW, Ceberio I, Lipuma LB, Samilo DW, Wasilewski GD, Gonzales AM, Nieves JL, van den Brink MR, Perales MA, Pamer EG. Quantitative assessment of T cell repertoire recovery after hematopoietic stem cell transplantation. Nature medicine. 2013;19:372–377. doi: 10.1038/nm.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muraro PA, Robins H, Malhotra S, Howell M, Phippard D, Desmarais C, de Paula Alves Sousa A, Griffith LM, Lim N, Nash RA, Turka LA. T cell repertoire following autologous stem cell transplantation for multiple sclerosis. The Journal of clinical investigation. 2014;124:1168–1172. doi: 10.1172/JCI71691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burkhardt UE, Hainz U, Stevenson K, Goldstein NR, Pasek M, Naito M, Wu D, Ho VT, Alonso A, Hammond NN, Wong J, Sievers QL, Brusic A, McDonough SM, Zeng W, Perrin A, Brown JR, Canning CM, Koreth J, Cutler C, Armand P, Neuberg D, Lee JS, Antin JH, Mulligan RC, Sasada T, Ritz J, Soiffer RJ, Dranoff G, Alyea EP, Wu CJ. Autologous CLL cell vaccination early after transplant induces leukemia-specific T cells. The Journal of clinical investigation. 2013;123:3756–3765. doi: 10.1172/JCI69098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun C, Tian X, Lee YS, Gunti S, Lipsky A, Herman SE, Salem D, Stetler-Stevenson M, Yuan C, Kardava L, Moir S, Maric I, Valdez J, Soto S, Marti GE, Farooqui MZ, Notkins AL, Wiestner A, Aue G. Partial reconstitution of humoral immunity and fewer infections in patients with chronic lymphocytic leukemia treated with ibrutinib. Blood. 2015;126:2213–2219. doi: 10.1182/blood-2015-04-639203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.