Abstract
Data from multiple bacterial pathogens are consistent with regulator-encoding genes having higher mutation frequencies than the genome average. Such mutations drive both strain- and type- (e.g. serotype, haplotype) specific phenotypic heterogeneity, and may challenge public health due to the potential of variants to circumvent established treatment and/or preventative regimes. Here, using the human bacterial pathogen the group A Streptococcus (GAS; S. pyogenes) as a model organism, we review the types and regulatory-, phenotypic-, and disease-specific consequences of naturally occurring regulatory gene mutations. Strain-specific regulator mutations that will be discussed include examples that transform isolates into hyper-invasive forms by enhancing expression of immunomodulatory virulence factors, and examples that promote asymptomatic carriage of the organism. The discussion of serotype-specific regulator mutations focuses on serotype M3 GAS isolates, and how the identified rewiring of regulatory networks in this serotype may be contributing to a decades old epidemiological association of M3 isolates with particularly severe invasive infections. We conclude that mutation plays an outsized role in GAS pathogenesis and has clinical relevance. Given the phenotypic variability associated with regulatory gene mutations, the rapid examination of these genes in infecting isolates may inform with respect to potential patient complications and treatment options.
Graphical abstract
Abbreviated summary
The selection of mutations within regulator-encoding genes is a major driver of bacterial strain-and type-specific phenotypic heterogeneity. Here, we review the types and regulatory-, phenotypic-, and disease-specific consequences of naturally occurring regulatory gene mutations in the human bacterial pathogen the group A Streptococcus. We conclude that mutation plays an outsized role in group A Streptococcus pathogenesis and has clinical relevance.
Introduction
Bacterial pathogens commonly show intraspecies variation which has led to the creation of classification systems by which isolates can be distinguished (e.g. different clones, pulsed-field types, or serotypes). Importantly, at least in some instances it has been discovered that isolates from different classification groups have distinct virulence characteristics. For example, Staphylococcus aureus isolates from pulsed-field type USA300 show enhanced virulence relative to isolates from pulsed-field type USA200, and in part this is explained by differences in the Agr regulatory system which is a major regulator of virulence factor expression (Cheung et al., 2011, Uhlemann et al., 2014). Similarly, certain Salmonella enterica serotypes have the potential to become hyper-infectious following animal passage, although the molecular basis of this is unknown (Heithoff et al., 2012). Further, the human-specific pathogen group A Streptococcus (GAS, Streptococcus pyogenes) shows strain- and serotype-specific disease associations (Mitchell, 1962, Walker et al., 2007). Despite the public health challenges associated with variant emergence (Nicol & Wilkinson, 2008, Croucher et al., 2011), in most instances there is a dearth of information regarding the molecular mechanisms that drive this variation. Here, to summarize how the mutation-induced rewiring of regulatory networks can influence pathogen phenotypic heterogeneity, we will review how it drives strain- and serotype-specific phenotypic variation in GAS.
GAS cause human diseases that range from self-limiting pharyngitis, of which there are >600 million cases annually (Ralph & Carapetis, 2013), to the severely invasive necrotizing fasciitis, which has a mortality rate of between 25 and 50% (Olsen & Musser, 2010). The ability of this pathogen to cause such disease diversity is a consequence of its ability to express different virulence factor profiles, selected from dozens of secreted and cell-wall anchored proteins, in response to internal and external environmental cues (Hondorp & McIver, 2007, Shelburne et al., 2008). To this end, GAS utilize a combination of two-component regulatory systems, stand-alone transcription factors, and small non-coding regulatory RNAs (sRNAs) (Ribardo et al., 2004, McIver, 2009, Perez et al., 2009, Vega et al., 2016).
GAS strains are divided into serotypes based upon the sequence of the 5’ end of the emm gene, which encodes the classical GAS virulence factor the M protein (Cunningham, 2014). Importantly, epidemiological analyses spreading back more than five decades have identified non-random associations between certain serotypes and particular disease manifestations. For example, serotype M3 GAS isolates are associated with causing particularly severe invasive infections with a high mortality rate, while serotype M18 strains are associated with outbreaks of the post-GAS-infection sequela acute rheumatic fever (Mitchell, 1962, Beres et al., 2006). While the molecular mechanisms driving GAS-serotype disease-phenotype associations have yet to be fully elucidated, in some instances the data point to a critical role for regulatory rewiring through the serotype-specific mutation of regulator-encoding genes (Lynskey et al., 2013, Cao et al., 2014, Lynskey et al., 2015, Miller et al., 2015a, Miller et al., 2015b).
Strain-specific variation in GAS virulence has perhaps best been studied in relation to the selection of hyper-virulent derivatives during invasive infections (Engleberg et al., 2001, Sumby et al., 2006, Hollands et al., 2010, Li et al., 2014), the molecular basis of which is the mutation of regulator-encoding genes. The consequence of these mutations is an increased ability to evade the host immune response due to the enhanced expression, and/or the reduced degradation, of immunomodulatory virulence factors (Kwinn & Nizet, 2007). We will begin this review with a discussion of strain-specific regulatory gene mutations, and in particular mutations that lead to invasive disease hyper-virulence or enhanced carrier isolate status. We will subsequently discuss serotype-specific regulatory gene mutations, and in particular mutations that rewire regulation in serotype M3 isolates. In each case, we will review the function of the encoded proteins, the mutations that occur, and the transcriptional-, phenotypic-, and disease-specific consequences of these mutations. Given that regulatory genes have been identified as “hot-spots” for mutation in other bacteria (Yang et al., 2011, Lieberman et al., 2011), the information in this review may be applicable to variation observed for a broad swath of pathogens.
Strain-specific variation in regulatory activity
Mutation is a natural process that occurs at low frequency (calculated at ~10−9 mutations per generation in GAS) (Scott et al., 2012) during replication of the bacterial chromosome. In general, mutations that are beneficial to the organism are maintained within the population whereas detrimental mutations are lost. However, this distinction is not always clear cut. For example, as we will discuss, during invasive GAS infections there is the selection for gene mutations that result in an increased ability to resist neutrophil-mediated killing. Given the beneficial nature of resisting killing by neutrophils it would be expected that many, if not all, GAS isolates harbored these mutations. This is not the case however, as the same mutations that result in an increased ability to resist neutrophil-mediated killing also result in a decreased ability of GAS to colonize and proliferate in the upper respiratory tract. As upper respiratory tract infections represent the majority of infections caused by GAS, this reduces the likelihood that GAS strains containing the regulatory gene mutations will be transmitted to other hosts. Thus, this in part explains why some mutations that, at least under some conditions, would seemingly be beneficial are only present in a strain-specific manner.
Strain-specific regulatory gene mutations that enhance GAS immune evasion
Following whole genome sequencing of 87 invasive disease serotype M3 GAS isolates, Beres et al. identified 22 genes with increased nucleotide diversity relative to the genome as a whole (Beres et al., 2010). That some of these genes had higher levels of single nucleotide polymorphisms (SNPs) resulting in non-synonymous substitutions, rather than synonymous substitutions, is consistent with them undergoing diversifying selection (e.g. all 12 SNPs within the regulator of protease B [ropB, also known as rgg] gene result in non-synonymous changes) (Beres et al., 2010). The ropB gene, along with the control of virulence regulator and sensor (covR/S, also known as csrR/S) genes, which are also undergoing diversifying selection (Engleberg et al., 2001, Mayfield et al., 2014, Li et al., 2014), represented three of the five genes with the highest mutation rate. Why mutations in covR, covS, or ropB are positively selected for will be discussed in this section.
The CovR/S two-component regulatory system
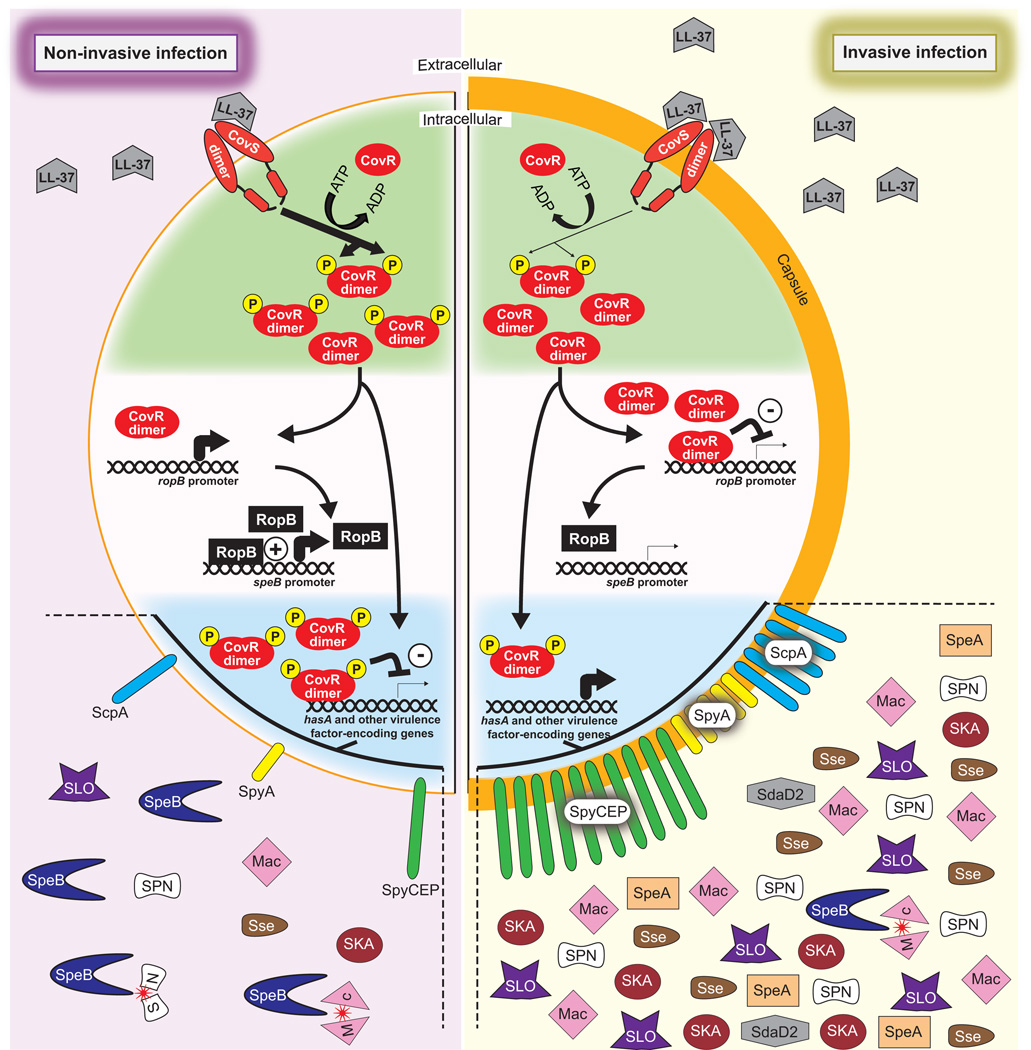
CovR/S form a two-component regulatory system, with CovS being a membrane-spanning sensor kinase that activates the cytoplasmically located CovR response regulator via phosphorylation, enhancing its ability to dimerize and bind DNA (Figure 1) (Graham et al., 2002, Gryllos et al., 2003, Gusa et al., 2006). The CovR/S system is an atypical two-component system as it primarily negatively regulates gene expression, with ~10% of GAS transcripts having reduced abundance due to CovR/S activity. Included within these repressed transcripts are almost two dozen that encode for key GAS virulence factors, a common characteristic of which is that they enhance the ability of GAS to evade the host immune response. For example, the capsule has anti-phagocytic properties (Wessels & Bronze, 1994) while SpyCEP reduces neutrophil migration and activation by degrading chemokine gradients (Zinkernagel et al., 2008). Given the function of CovR/S-repressed virulence factors, it is not surprising that this regulatory system is down-regulated during invasive GAS infections, enhancing virulence factor expression (Figure 1). Recent data indicates that the CovR/S system is down-regulated, at least in part, following binding of the human antimicrobial peptide LL-37 to the extracellular domain of CovS (Gryllos et al., 2008, Velarde et al., 2014). The interaction between LL-37 and CovS reduces CovS activity, which leads to a reduction in phosphorylated CovR.
Figure 1. Differential regulatory activity of the CovR/S two-component system between invasive and non-invasive infections.
Shown is a schematic of a GAS cell, with the left half of the cell showing CovR/S-mediated regulation that occurs during non-invasive infections, and the right half of the cell showing CovR/S-mediated regulation that occurs during invasive infections. The upper (green) section highlights the interactions between CovR and CovS. Binding of the human antimicrobial peptide LL-37 by CovS, which occurs at higher frequency during invasive infections, diminishes CovS kinase activity and hence CovR phosphorylation. The middle (white) section highlights the hypothesized regulation of SpeB protease expression, with non-phosphorylated CovR being able to bind, only when at a sufficiently high concentration as is the case during invasive infections (or in a covS mutant), to the ropB promoter and repressing transcription. RopB is a transcription factor that is required for high level SpeB expression. The lower (blue) section highlights the consequences of CovR phosphorylation status with regard to the expression of multiple immunomodulatory virulence factors. The non-exhaustive list of CovR/S-repressed virulence factors shown are Mac-1-like protein (Mac), streptokinase (SKA), Streptolysin O (SLO), S. pyogenes NADase (SPN), secreted streptococcal esterase (Sse), Streptococcal pyogenic exotoxin A (SpeA), cysteine protease (SpeB), C5a peptidase (ScpA), chemokine protease (SpyCEP), S. pyogenes ADP-ribosylating toxin (SpyA), and group A streptococcal DNase D2 (SdaD2). The ability of SpeB to cleave and inactivate GAS virulence factors is also shown (red stars; see Figure 2 for more detail about SpeB activity).
The importance of reducing CovR/S activity during invasive infections is perhaps best exemplified by the fact that covR/S mutant strains are commonly selected for, and can be recovered from, patients with invasive infections (Ikebe et al., 2010). That covR/S mutants are derived from parental strains with wild-type covR/S genes is evident from a study in which mice that were infected via the subcutaneous injection of wild-type GAS developed, in a time-dependent manner, abscesses containing a mixture of parental and covR/S mutant derivatives (Sumby et al., 2006). Relative to the parental strains, covR/S mutants show increased lethality in murine models of bacteremia and soft tissue infection, and increased resistance to neutrophil-mediated killing. However, it should be stressed that not all invasive disease GAS isolates harbor covR or covS mutations, rather invasive infections appear to be caused by a mixture of parental and mutant GAS strains. We hypothesize that the large number of secreted virulence factors produced by the cov mutants have a positive bystander effect on neighboring parental GAS cells, enhancing the probability of parental cells disseminating to other hosts (Trevino et al., 2009).
Interestingly, mutations occur at a higher frequency in covS than covR, even after adjusting for gene size (Friaes et al., 2015). This is believed to be a consequence of the fact that covS and covR null mutant strains are not identical. For example, covS mutants produce undetectable levels of the secreted protease SpeB, while covR mutants produce high levels, two-fold higher than parental strains (Trevino et al., 2009). We hypothesize that this is due to non-phosphorylated CovR being the form that is capable of binding to the ropB promoter and inhibiting its transcription, and subsequently preventing SpeB production. Thus, in a covS mutant strain non-phosphorylated CovR is highly abundant shutting off SpeB expression, while in a covR mutant SpeB expression is not repressed (Figure 1). This hypothesis awaits experimental validation.
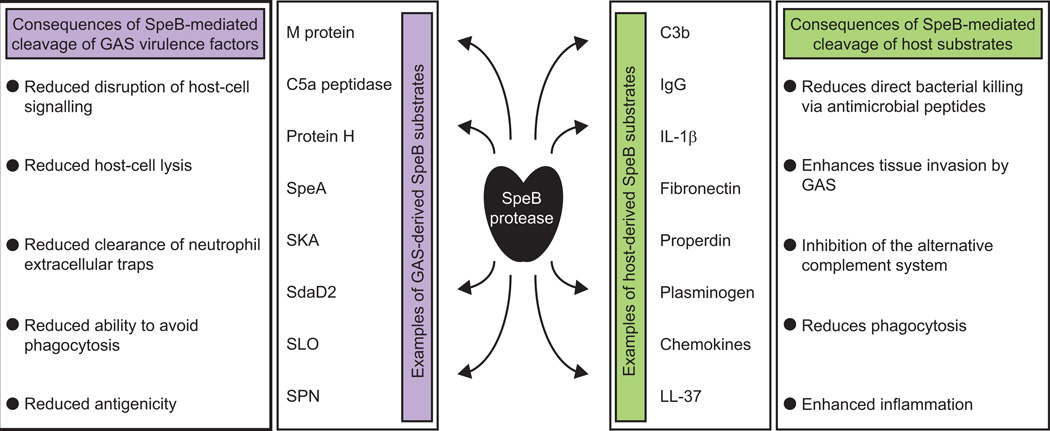
The differential regulation of SpeB expression is significant as SpeB targets a wide range of substrates for degradation, from host substrates such as anti-microbial peptides and chemokines, to many of the virulence factors that are part of the CovR/S regulon (Figure 2) (Carroll & Musser, 2011, Nelson et al., 2011). That disrupting CovS activity is more advantageous than disrupting CovR activity is also evident from the analysis of covR mutant strains. While some naturally occurring covR mutants harbor null mutations, completely abrogating CovR function, others maintain repressor activity but are unable to interact with CovS, resulting in a regulatory pattern equivalent to a covS mutant (Trevino et al., 2009).
Figure 2. SpeB targets an array of host and GAS proteins for degradation.
Shown are examples of host (green) and GAS (purple) proteins that are targeted for degradation by SpeB, as well as some of the consequences of this protease activity.
Studies investigating the frequency of covR/S mutations identified between 30 and 50% of invasive GAS isolates, and between 2 and 5% of non-invasive isolates, harbor mutations (Ikebe et al., 2010, Shea et al., 2011). To help explain this infection-specific difference in covR/S mutation frequency, competition assays were performed between covS mutant and parental strains during growth in human saliva, an ex vivo model of an upper respiratory tract infection. It was identified that covS mutant strains are significantly outcompeted by parental strains during human saliva growth (Trevino et al., 2009). Thus, it was concluded that the CovR/S system is critical to the ability of GAS to transition between infection types, with CovR/S activity contributing to the ability of GAS to cause non-invasive infections, and a reduction in CovR/S activity contributing to the ability of GAS to cause invasive infections. This was further supported by comparisons between covS mutant and parental strains in murine skin adherence and upper respiratory tract models of infection, with the covS mutants being attenuated in their ability to colonize (Hollands et al., 2010, Alam et al., 2013). Thus, while covR/S mutants are positively selected for during invasive infections due to their greater resistance to neutrophil-mediated killing, these mutant strains are not maintained in the population at high frequency due to their attenuated ability to cause non-invasive infections. Consequently, covR/S mutant strains that are recovered from invasive infections are, in most cases, thought to have arisen as a result of de novo mutations. In summary, the strain-specific mutation of covR/S has clinically relevant consequences, with these mutant strains enhancing the severity of invasive infections cause by this pathogen. However, as will be covered below, covR/S mutation is only one mechanism by which GAS may enhance virulence factor expression.
The regulator of protease B (RopB)
As mentioned above, a contributing factor behind the enhanced ability of covS mutant strains to cause severe invasive infections is thought to be the reduction in the expression of the SpeB protease, and therefore an enhancement in the abundance of secreted and cell-wall anchored virulence factors. If this were the case, mutation of the speB gene, or of a positive regulator of SpeB expression, could feasibly be other mechanisms by which GAS could achieve a so-called “invasive phenotype”. Consistent with this is the finding that the most polymorphic GAS gene identified in several population-based studies is ropB (Beres et al., 2010, Friaes et al., 2015), given that RopB is the major positive regulator of SpeB expression (Lyon et al., 1998). During the transition from exponential to stationary phase, RopB binds to the speB promoter and dramatically upregulates transcription (Neely et al., 2003) (Figure 1). However, unlike the situation regarding covR/S mutations, there is currently a controversy as to which conditions promote the selection of ropB mutants (Kansal et al., 2000, Ikebe et al., 2010, Olsen et al., 2015). Given that the GAS reservoir is the human upper respiratory tract, and that SpeB expression contributes to upper respiratory tract infection (Shelburne et al., 2005), we cannot envision a scenario whereby ropB mutants would be selected for during pharyngeal infections. However, the substrate specificity of SpeB, which includes as many host immune system proteins as it does GAS anti-immune system virulence factors (Figure 2), does not provide an obvious case for the loss of SpeB expression being advantageous during invasive infections. We favor the view that GAS encounters multiple micro-environments during the course of an invasive infection, and that while SpeB may promote GAS virulence in some of these environments, there is at least one in which SpeB expression is strongly selected against. Further research is required to identify where and when ropB mutants arise.
Strain-specific regulatory mutations enhance the carrier isolate status of GAS
Many bacterial pathogens have the ability to asymptomatically colonize a host, although the molecular mechanisms that drive asymptomatic carriage, relative to symptomatic infection, are for the most part poorly defined. GAS is the prototype for studying asymptomatic carriage with, depending on the study, GAS carriage rates in the upper respiratory tracts of children ranging from 5 to 15% (Shaikh et al., 2010). Animal models of infection comparing carrier and non-carrier GAS isolates identified that carrier isolates are attenuated in their ability to cause invasive infections (Krause et al., 1962). Thus, there appears to be a genetic factor that distinguishes carrier GAS isolates from non-carrier isolates. Recent genome sequence comparisons of carrier and non-carrier GAS isolates have identified two regulatory networks whose disruption appear to contribute to the development of carrier isolates:
The multigene regulator in GAS (Mga)
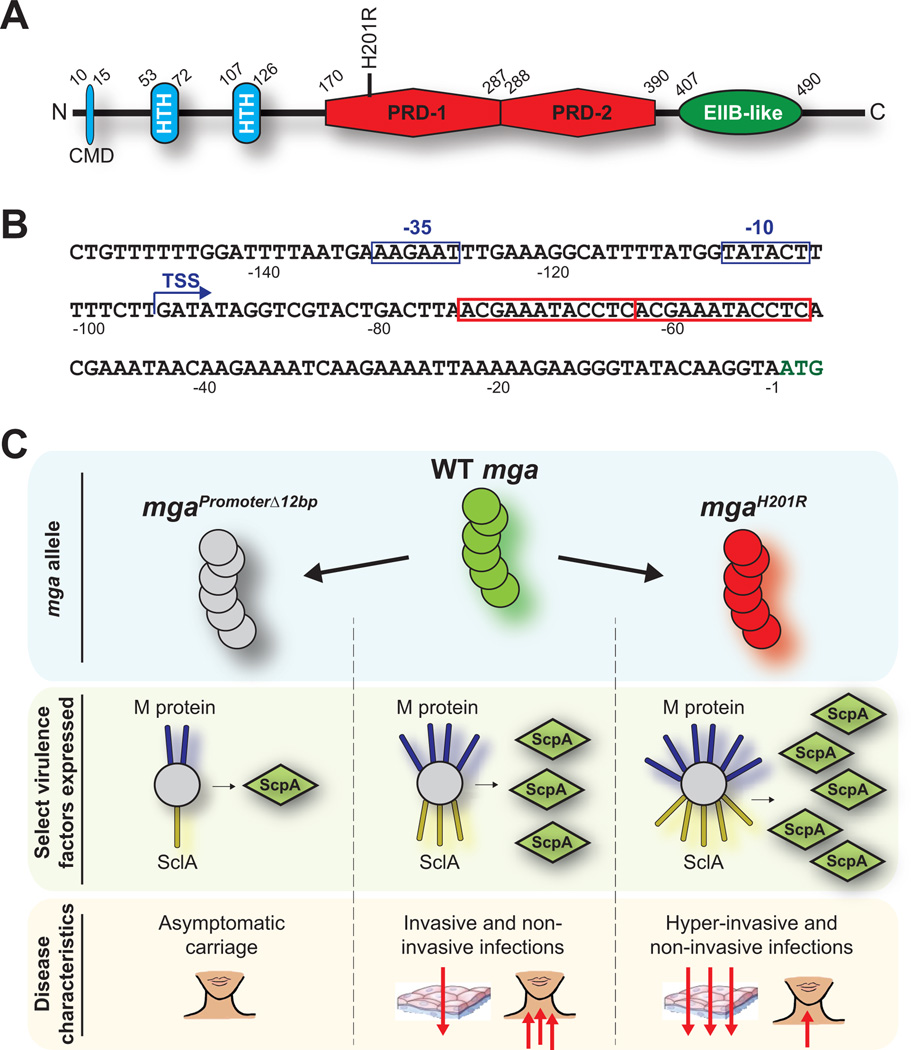
Mga is the founding member of the Mga-like family of proteins that are present within multiple Gram-positive pathogens (e.g. the AtxA anthrax toxin regulator of Bacillus anthracis) (Hondorp & McIver, 2007, Hammerstrom et al., 2015). That Mga contributes to GAS virulence was discovered more than 30 years ago (Spanier et al., 1984). Mga positively regulates the expression of several classical GAS virulence factors, including the M protein and the C5a peptidase. In addition to regulating virulence factor expression, Mga regulates, and is also regulated by, carbohydrate metabolism (Ribardo & McIver, 2006). The regulation of Mga activity by carbohydrate metabolism occurs through the presence of two phosphotransferase system (PTS) regulatory domains (PRDs) located within Mga (Figure 3A). PTSs regulate carbohydrate transport and metabolism, and in part, this is achieved by the phosphorylation of PRD-containing proteins (Deutscher et al., 2014). The importance of the PRDs to Mga activity will be further discussed later in the review, when we cover a strain-specific mga mutation that results in an alteration within one of these domains.
Figure 3. Distinct mga mutations lead to altered disease characteristics.
(A) Domain structure of the Mga protein. The amino acids that boarder each domain are numbered. The identified domains are: conserved Mga domain (CMD), helix-turn-helix DNA-binding motif (HTH), phosphotransferase system regulatory domain (PRD), and a domain important in oligomerization (EIIB-like). The location of the H201R mutation is highlighted. (B) Nucleotide sequence of the mga promoter region. Nucleotides are numbered relative to the A of the ATG start codon (green nucleotides). The transcriptional start site (TSS) of the major mga promoter is shown with a bent arrow, and the −10 and −35 promoter sequences are highlighted with blue boxes. In red boxes are the two 12 bp direct repeat sequences present upstream of mga, one of which is deleted in select carrier GAS isolates. (C) Regulatory and disease-specific consequences of select mga mutations. The number of red arrows highlight the severity of the infections.
While characterizing a collection of carrier GAS isolates, it was discovered that two epidemiologically unassociated isolates both harbored the same 12 bp deletion within the mga promoter region (Flores et al., 2013). The deletion removed one of two adjacent 12 bp direct repeats (Figure 3B), and likely occurred as a consequence of slipped-strand mispairing during DNA replication. Analysis of the consequences of this deletion determined that it significantly reduced the transcription of both mga and Mga-regulated genes (e.g. a >20-fold decrease in scpA mRNA abundance), and reduced GAS virulence in animal models of necrotizing fasciitis (Figure 3C) (Flores et al., 2013). It has been hypothesized, but not yet proven, that the observed 12 bp deletion is selected for among carrier strains as the decreased levels of extracellular protein expression, and in particular of the M protein, lowers the immunogenicity of these isolates during oropharyngeal colonization.
The LiaFSR three-component regulatory system
Where studied (e.g. Streptococcus agalactiae and Enterococcus faecalis), the Lia regulatory system has been identified as controlling species-specific responses to cell wall-active antibiotics and antimicrobial peptides (Klinzing et al., 2013, Reyes et al., 2015). A recent GAS study discovered that a non-synonymous mutation in the liaS gene, a putative sensor kinase, was one of three SNPs that distinguished GAS isolates recovered from a single individual during acute pharyngitis and subsequent asymptomatic carriage (Flores et al., 2015). Through allelic exchange it was determined that the liaSR135G mutation enhanced characteristics associated with carriage (e.g. murine nasopharyngeal colonization), at the expense of characteristics associated with invasive infections (e.g. virulence in a murine necrotizing fasciitis model). While a full understanding of why the liaSR135G mutation results in the phenotypes observed remains to be determined, this mutation alters the transcription of multiple GAS genes, including mga, which is down-regulated. Thus, the data support the notion that the disruption of regulatory systems is an important contributor to the transition of GAS from disease to carriage states (Flores et al., 2015). This may be a widespread phenomenon as similar findings have also been observed following comparison of disease and carrier isolates of pathogens such as S. aureus and Neisseria meningitidis (Schoen et al., 2008, Young et al., 2012).
A strain-specific mutation in mga enhances invasive GAS infections
A whole-genome sequencing study of serotype M59 GAS isolates identified a higher than expected number of SNPs in mga (Sanson et al., 2015b). The most commonly occurring SNP, which arose independently at least five times, resulted in a H201R amino acid substitution. The H201 amino acid resides within the first of two PRD domains present within Mga (Figure 3A). To determine whether the H201R substitution altered Mga regulatory activity, comparisons were performed between parental and H201R isogenic mutant strains. The mutant strain had significantly increased expression of mga and Mga-regulated genes, altered regulation that resulted in the mutant strain having enhanced virulence in non-human primate and murine models of invasive GAS infection (Figure 3C) (Sanson et al., 2015b). Recently, it was identified that the phosphorylation of two histidine residues located within the first Mga PRD domain, H207 and H273, reduces Mga activity (Hondorp et al., 2013, Sanson et al., 2015a). H201 is located on the same α-helix as H207, and is also in close proximity to H273 (Sanson et al., 2015a). Thus, the data are consistent with the hypothesis that the H201R substitution enhances Mga activity by preventing the inhibitory phosphorylation of H207 and/or H273.
Serotype-specific variation in regulatory activity
As mentioned, epidemiological analyses have identified associations between certain GAS serotypes and disease phenotypes (Colman et al., 1993). This implies that isolates of these serotypes are distinct, in gene content or gene regulation, from those of other serotypes and that these differences promote the ability of isolates of these serotypes to preferentially cause individual diseases. If so, the study of these serotypes may shed light on the virulence factors and regulatory patterns required for GAS to cause these diseases, information that may be useful with regard to the development of novel therapeutic or preventive regimes. While serotypes show variation in the assortment of bacteriophage and pathogenicity islands integrated into their genomes (Green et al., 2005, Zhu et al., 2015), none are unique to a single serotype. Therefore, we favor the hypothesis that it is the alternate regulation of core chromosomally-encoded virulence factors that is at the crux of GAS-serotype disease-phenotype associations.
Rewiring of regulatory networks in serotype M3 GAS isolates
Serotype M3 GAS isolates are associated with causing particularly severe invasive infections with a high mortality rate (Beres et al., 2006). Investigations to gain insight into why isolates of this serotype are hyper-virulent during invasive infections have discovered that M3 isolates, since at least the 1920’s, harbor mutations within at least three regulatory genes, rocA, fasC, and rivR (Cao et al., 2014, Lynskey et al., 2015, Miller et al., 2015a). The type and consequences of mutations in these genes will be discussed in this section, including how they contribute to the observed invasive disease hyper-virulence of M3 isolates.
The regulator of Cov (RocA) protein
The rocA gene was discovered from a transposon mutagenesis screen in which rocA disruption enhanced the abundance of covR/S mRNA (Biswas & Scott, 2003). Given that RocA shares sequence similarity to membrane-spanning sensor kinases, it is likely that this protein enhances covR mRNA levels indirectly. Indeed, while a full understanding of RocA activity is lacking, recent data are consistent with RocA being a pseudokinase that functions through the CovR/S system, and that it does this by indirectly enhancing the phosphorylation status of CovR (Miller et al., 2015a) (Sumby, unpublished).
Analysis of more than 450 serotype M3 GAS isolates, which were recovered between the 1920s and 2010, identified that they all harbor a frameshift mutation within rocA (Lynskey et al., 2015, Miller et al., 2015a). This mutation, a 1 bp deletion from within a homopolymeric tract (Figure 4A), truncates the protein within the c-terminal ATPase domain (Figure 4B). RNAseq analysis comparing a parental M3 isolate with a derivative in which a functional rocA allele had been introduced identified 56 mRNA transcripts that were differentially regulated (Miller et al., 2015a). Importantly, a third of the differentially regulated genes encode for virulence factors, including the hyaluronic acid capsule (120-fold higher mRNA levels in the parental strain) and the chemokine protease SpyCEP (90-fold higher mRNA levels in the parental strain). Thus, the natural rocA mutation of serotype M3 GAS isolates is the molecular basis behind a significant upregulation in virulence factor expression by isolates of this serotype (see pink-shaded section of Figure 5). Virulence assays identified that a parental M3 isolate could survive the bactericidal properties of human blood, and cause mortality in a murine bacteremia model of infection, at higher levels than a rocA complemented derivative (Miller et al., 2015a). Thus, the rocA mutation contributes to the invasive disease hyper-virulence of serotype M3 GAS.
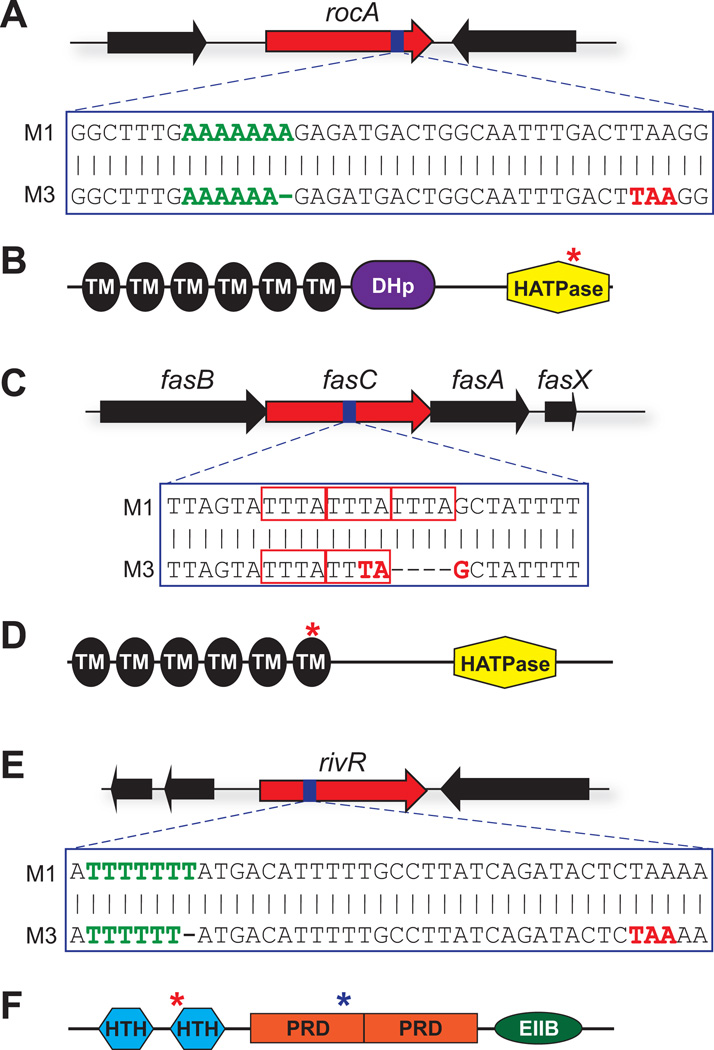
Figure 4. Serotype M3 GAS isolates harbor mutations in the rocA, fasC, and rivR regulator-encoding genes.
(A) M3 isolates have a 1 bp deletion in rocA. Shown is a schematic of the rocA gene, with the region of the gene that contains the 1 bp deletion in serotype M3 isolates being highlighted with blue shading. The nucleotide sequence of this region is shown in a comparison between M1 and M3 isolates. The green nucleotides highlight the homopolymeric tract which contains the M3 GAS deletion, while the red nucleotides highlight the premature start codon which is introduced into the M3 GAS rocA gene due to the deletion. (B) Domain analysis of the RocA protein. Identified domains are: transmembrane domains (TM; black), a dimerization and histidine phosphotransfer domain (DHp; purple), and a histidine-kinase-like catalytic domain (HATPase; yellow). The red asterisk highlights the location of the truncation that occurs in serotype M3 isolates. (C) M3 isolates have a 4 bp deletion in fasC. The red rectangles highlight the three tetra-nucleotide repeat sequences, one of which is deleted in serotype M3 GAS isolates. (D) Domain analysis of the FasC protein. The red asterisk highlights the location of the truncation that occurs in serotype M3 isolates. (E) M3 isolates have a 1 bp deletion in rivR. (F) Domain analysis of the RivR protein. RivR is a member of the Mga-like family of transcriptional regulators, and similar to Mga has helix-turn-helix DNA-binding motifs (HTH), phosphotransferase system regulatory domains (PRD), and a domain important in oligomerization (EIIB-like). The red asterisk highlights the location of the truncation that occurs in serotype M3 isolates. The blue asterisk highlights the location of the L242P amino acid substitution that occurs in M3 isolates.
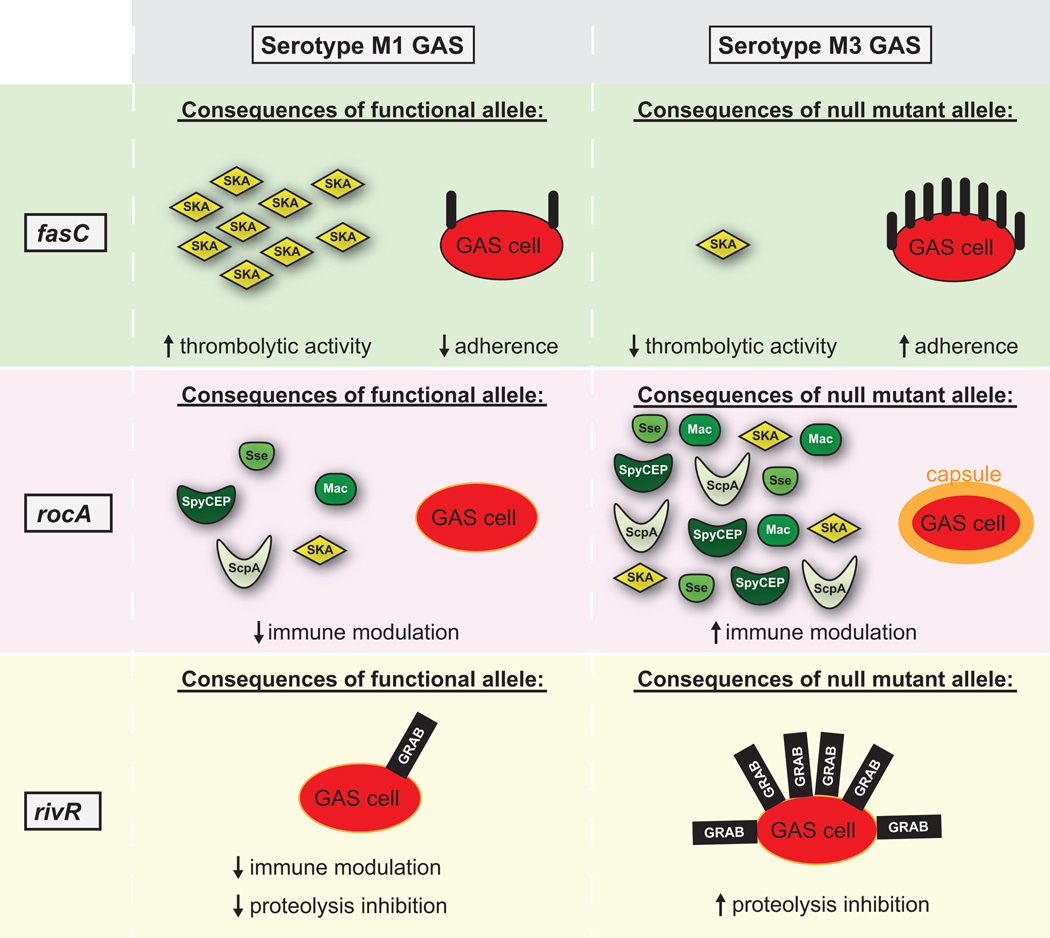
Figure 5. Serotype M3 GAS isolates have unique virulence characteristics as a consequence of the fasC, rocA, and rivR gene mutations.
Shown is a comparison of virulence characteristics, and virulence factor expression, that occurs in serotype M1 and M3 GAS isolates due to the presence/absence of functioning FasC (green-shaded regions), RocA (pink-shaded regions) and RivR (beige-shaded regions) proteins. The Fas system: A functioning Fas system leads to the production of the sRNA FasX. FasX enhances the stability of mRNA encoding the thrombolytic agent streptokinase, and reduces the translation of mRNAs encoding adhesins. Due to the inactivation of the fasC gene, serotype M3 isolates do not have this regulatory activity. RocA: In serotype M1 isolates RocA enhances the activity of the CovR/S system which represses the expression of multiple immunomodulatory virulence factors. Due to inactivation of the rocA gene, serotype M3 isolates do not have this regulatory activity. RivR: In serotype M1 isolates RivR represses transcription of genes encoding for the immunomodulatory capsule and cell-surface protease inhibitor GRAB. Due to inactivation of the rivR gene, serotype M3 isolates do not have this regulatory activity (although, for unknown reasons, a functional RivR has no regulatory effect on the capsule in the M3 background).
The fibronectin/fibrinogen-binding / hemolytic activity / streptokinase regulatory (Fas) system
The Fas system is encoded by a four gene locus, fasBCAX (Figure 4C), with FasB and FasC having sequence similarity to membrane-spanning sensor kinases, FasA having sequence similarity to response regulators, and FasX being a small regulatory RNA (sRNA). While FasBCA have not been studied in detail, all three are required for significant production of FasX, which is the effector molecule of this system (Kreikemeyer et al., 2001). FasX is hypothesized to mediate the transition of GAS from the colonization to the dissemination stages of infection. This hypothesis is based upon the findings that FasX positively regulates streptokinase expression, a virulence factor that promotes blood clot degradation and tissue barrier destruction, and negatively regulates expression of the collagen-binding pilus and the fibronectin-binding adhesins PrtF1 and PrtF2 (Ramirez-Pena et al., 2010, Liu et al., 2012, Danger et al., 2015a, Danger et al., 2015b). FasX activity occurs at the post-transcriptional level (Miller et al., 2014).
Comparisons of FasX abundance between isolates of eight different GAS serotypes identified that M3 isolates have lower levels relative to other serotypes (Perez et al., 2009). Examination of the M3 fas locus uncovered a frameshift mutation in the fasC gene, leading to the premature termination of the gene (Figure 4C) and consequently truncation of the FasC protein (Figure 4D). The mutation, a deletion of one of three adjacent 4 bp (TTTA) repeats, was subsequently identified in all of 125 serotype M3 isolates that were recovered in a temporally (1920s to 2010) and spatially (Europe, North America, Japan, and Russia) diverse manner. Through use of complementation assays it was determined that the fasC mutation was responsible for the low abundance of FasX observed for serotype M3 isolates (Cao et al., 2014).
It was hypothesized that the reduction in FasX abundance in serotype M3 isolates alters the expression of streptokinase, pilus, PrtF1, and PrtF2, and therefore M3 GAS virulence (see green-shaded region of Figure 5). While it has yet to be determined whether the adhesins are differentially regulated due to the fasC mutation, it has been confirmed that complementing this mutation enhances streptokinase expression (Cao et al., 2014). Given that M3 isolates are hyper-virulent during invasive infections it seems counterintuitive that these isolates would harbor a mutation that reduces the expression of this crucial virulence factor. Importantly, while complementing the M3 fasC mutation increases streptokinase levels relative to the parental isolate, the parental isolate actually has higher levels relative to isolates of other serotypes. This is due to the fact that streptokinase expression is increased due to the M3 rocA mutation (Miller et al., 2015a), and that this more than makes up for the “decrease” in expression due to the fasC mutation. Thus, if the fasC mutation contributes to the invasive disease hyper-virulence of M3 isolates then it likely does so through the altered expression of virulence factors other than streptokinase.
The RofA-like protein IV regulator (RivR)
RivR negatively regulates the expression of two virulence factors in serotype M1 GAS strains, the capsule and the protein G-related alpha-2-macroglobulin-binding protein (GRAB) (Trevino et al., 2013). The anti-phagocytic properties of the capsule have already been discussed (Wessels et al., 1991), while GRAB binds the human protease inhibitor alpha-2-macroglobulin and regulates proteolysis at the GAS cell surface (Rasmussen et al., 1999). Analysis of the rivR gene in 125 serotype M3 isolates identified that the oldest isolate, from the 1920s, harbored three non-synonymous SNPs within RivR. The remaining 124 isolates, which were isolated between 1937 and 2010, contained an additional alteration in rivR, a 1 bp deletion within a homopolymeric tract (going from seven T’s to six; Figures 4E and 4F). Through use of complementation assays it was determined that one of the three non-synonymous SNPs, leading to a L242P change, completely disrupts RivR activity in M3 isolates (Cao et al., 2014). Thus, M3 isolates since at least the 1920s harbor null mutations in rivR.
The phenotypic consequences of rivR mutation in serotype M3 isolates are unclear. On the one hand, rivR disruption leads to the enhanced expression of GRAB, as expected (see beige-shaded region of Figure 5). On the other hand, while RivR negatively regulates capsule expression in M1 isolates, no such activity was observed following rivR complementation in an M3 isolate (Cao et al., 2014). Unfortunately, as the mechanism by which RivR regulates capsule expression in M1 isolates has not been delineated this prevents assessment of why capsule is not RivR-regulated in a complemented M3 isolate. What is known however, is that RivR has no regulatory activity in a covR mutant strain, suggesting that RivR functions through CovR. A current hypothesis is that RivR binds to the has promoter and that this enhances CovR binding, leading to repression of capsule expression. Consistent with this is the finding that the has promoter of M3 isolates has multiple genetic alterations relative to that of M1 isolates, possibility providing an explanation as to why RivR does not regulate capsule expression in M3 GAS.
Summary of regulator-encoding gene mutations in serotype M3 GAS
In summary, data are consistent with serotype M3 GAS isolates causing disease over the course of the last 90-plus years all harboring mutations in rocA, rivR, and fasC regulator-encoding genes. Given that only serotype M3 GAS isolates have mutations within all three of these genes, indeed M3 isolates are the only serotype that harbor null mutations in fasC or rivR, we propose that in combination the three regulatory gene mutations are the driving force behind the serotype-specific virulence factor expression profile observed for M3 isolates. Interestingly, serotype M18 GAS isolates are also uniformly rocA mutants (but do not harbor mutations in rivR or fasC at any detectable frequency) but are not associated with causing severe invasive infections (Lynskey et al., 2013). Thus, while rocA mutation is thought to be necessary for invasive disease hyper-virulence, and therefore the association of serotype M3 isolates with particularly severe invasive infections, it is not thought to be sufficient.
Conclusions
For an increasing number of bacterial pathogens, it is becoming apparent that regulatory gene mutations play an integral role in the frequency and severity of infections caused by these pathogens. From disruption of the repressor of toxins (rot) gene in S. aureus USA500 strains (Benson et al., 2014), to disruption of the positive regulatory factor of listeriolysin (prfA) gene in select isolates of Listeria monocytogenes (Becavin et al., 2014, Rupp et al., 2015), to disruption of the regulator of lasB (lasR) gene in Pseudomonas aeruginosa isolates during infection of the cystic fibrosis lung (Smith et al., 2006, Diaz Caballero et al., 2015), these mutations show clinical relevance. Perhaps in no pathogen has the incidence and consequences of natural regulatory gene mutations been more extensively studied than in GAS. Full genome sequences of thousands of GAS isolates that span multiple serotypes, diseases, sites of isolation, geographic locations, and years of isolation have enabled in-depth analyses of strain and serotype-specific variation (Maruyama et al., 2016). As described in this review, mutations in both positive (e.g. Mga) and negative (e.g. CovR) regulators of virulence factor expression are associated with phenotypic variation in GAS. These mutations can dramatically alter disease potential, from promoting invasive infections to promoting asymptomatic carriage. Further, it should also be noted that the mutation-induced differences in the assortment of cell-surface and secreted proteins has the potential to significantly impact the efficacy of the GAS vaccines that are currently being tested.
As we move toward a future where infecting pathogens are whole-genome sequenced within a few hours of the patient arriving at the hospital, it may be prudent to determine the allelic status of the genes described in this review given their impact on disease potential. Such information may inform the clinician with respect to the potential of the strain to disseminate and cause more severe infections, and ultimately may influence treatment options. Even if this information is never used in the clinic, it would be valuable to the study of pathogen evolution and emergence, and we look forward to the challenges of drawing a more direct line between regulatory gene mutations and GAS disease manifestations.
Acknowledgments
Some of the research described in this review was made possible by grant R01AI087747 from the National Institute of Allergy and Infectious Diseases (to P.S.). We apologize to our many colleagues whose research were not referenced due to space limitations.
References
- Alam FM, Turner CE, Smith K, Wiles S, Sriskandan S. Inactivation of the CovR/S virulence regulator impairs infection in an improved murine model of Streptococcus pyogenes naso-pharyngeal infection. PLoS One. 2013;8:e61655. doi: 10.1371/journal.pone.0061655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becavin C, Bouchier C, Lechat P, Archambaud C, Creno S, Gouin E, Wu Z, Kuhbacher A, Brisse S, Pucciarelli MG, Garcia-del Portillo F, Hain T, Portnoy DA, Chakraborty T, Lecuit M, Pizarro-Cerda J, Moszer I, Bierne H, Cossart P. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. mBio. 2014;5 doi: 10.1128/mBio.00969-14. e00969-00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson MA, Ohneck EA, Ryan C, Alonzo F, 3rd, Smith H, Narechania A, Kolokotronis SO, Satola SW, Uhlemann AC, Sebra R, Deikus G, Shopsin B, Planet PJ, Torres VJ. Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element. Molecular microbiology. 2014;93:664–681. doi: 10.1111/mmi.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Carroll RK, Shea PR, Sitkiewicz I, Martinez-Gutierrez JC, Low DE, McGeer A, Willey BM, Green K, Tyrrell GJ, Goldman TD, Feldgarden M, Birren BW, Fofanov Y, Boos J, Wheaton WD, Honisch C, Musser JM. Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics. Proc Natl Acad Sci U S A. 2010;107:4371–4376. doi: 10.1073/pnas.0911295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Richter EW, Nagiec MJ, Sumby P, Porcella SF, DeLeo FR, Musser JM. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc Natl Acad Sci U S A. 2006;103:7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas I, Scott JR. Identification of rocA, a positive regulator of covR expression in the group A streptococcus. Journal of bacteriology. 2003;185:3081–3090. doi: 10.1128/JB.185.10.3081-3090.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao TN, Liu Z, Cao TH, Pflughoeft KJ, Trevino J, Danger JL, Beres SB, Musser JM, Sumby P. Natural disruption of two regulatory networks in serotype M3 group A Streptococcus isolates contributes to the virulence factor profile of this hypervirulent serotype. Infect Immun. 2014;82:1744–1754. doi: 10.1128/IAI.01639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll RK, Musser JM. From transcription to activation: how group A streptococcus, the flesh-eating pathogen, regulates SpeB cysteine protease production. Mol Microbiol. 2011;81:588–601. doi: 10.1111/j.1365-2958.2011.07709.x. [DOI] [PubMed] [Google Scholar]
- Cheung GY, Wang R, Khan BA, Sturdevant DE, Otto M. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infection and immunity. 2011;79:1927–1935. doi: 10.1128/IAI.00046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman G, Tanna A, Efstratiou A, Gaworzewska ET. The serotypes of Streptococcus pyogenes present in Britain during 1980–1990 and their association with disease. Journal of medical microbiology. 1993;39:165–178. doi: 10.1099/00222615-39-3-165. [DOI] [PubMed] [Google Scholar]
- Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham MW. Rheumatic fever, autoimmunity, and molecular mimicry: the streptococcal connection. International reviews of immunology. 2014;33:314–329. doi: 10.3109/08830185.2014.917411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danger JL, Cao TN, Cao TH, Sarkar P, Trevino J, Pflughoeft KJ, Sumby P. The small regulatory RNA FasX enhances group A Streptococcus virulence and inhibits pilus expression via serotype-specific targets. Mol Microbiol. 2015a;96:249–262. doi: 10.1111/mmi.12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danger JL, Makthal N, Kumaraswami M, Sumby P. The FasX Small Regulatory RNA Negatively Regulates the Expression of Two Fibronectin-Binding Proteins in Group A Streptococcus. J Bacteriol. 2015b;197:3720–3730. doi: 10.1128/JB.00530-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Ake FM, Derkaoui M, Zebre AC, Cao TN, Bouraoui H, Kentache T, Mokhtari A, Milohanic E, Joyet P. The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiology and molecular biology reviews : MMBR. 2014;78:231–256. doi: 10.1128/MMBR.00001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Caballero J, Clark ST, Coburn B, Zhang Y, Wang PW, Donaldson SL, Tullis DE, Yau YC, Waters VJ, Hwang DM, Guttman DS. Selective Sweeps and Parallel Pathoadaptation Drive Pseudomonas aeruginosa Evolution in the Cystic Fibrosis Lung. mBio. 2015;6 doi: 10.1128/mBio.00981-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engleberg NC, Heath A, Miller A, Rivera C, DiRita VJ. Spontaneous mutations in the CsrRS two-component regulatory system of Streptococcus pyogenes result in enhanced virulence in a murine model of skin and soft tissue infection. The Journal of infectious diseases. 2001;183:1043–1054. doi: 10.1086/319291. [DOI] [PubMed] [Google Scholar]
- Flores AR, Jewell BE, Yelamanchili D, Olsen RJ, Musser JM. A Single Amino Acid Replacement in the Sensor Kinase LiaS Contributes to a Carrier Phenotype in Group A Streptococcus. Infect Immun. 2015;83:4237–4246. doi: 10.1128/IAI.00656-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores AR, Olsen RJ, Wunsche A, Kumaraswami M, Shelburne SA, 3rd, Carroll RK, Musser JM. Natural variation in the promoter of the gene encoding the Mga regulator alters host-pathogen interactions in group a Streptococcus carrier strains. Infect Immun. 2013;81:4128–4138. doi: 10.1128/IAI.00405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friaes A, Pato C, Melo-Cristino J, Ramirez M. Consequences of the variability of the CovRS and RopB regulators among Streptococcus pyogenes causing human infections. Scientific reports. 2015;5:12057. doi: 10.1038/srep12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MR, Smoot LM, Migliaccio CA, Virtaneva K, Sturdevant DE, Porcella SF, Federle MJ, Adams GJ, Scott JR, Musser JM. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc Natl Acad Sci U S A. 2002;99:13855–13860. doi: 10.1073/pnas.202353699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green NM, Zhang S, Porcella SF, Nagiec MJ, Barbian KD, Beres SB, LeFebvre RB, Musser JM. Genome sequence of a serotype M28 strain of group a streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J Infect Dis. 2005;192:760–770. doi: 10.1086/430618. [DOI] [PubMed] [Google Scholar]
- Gryllos I, Levin JC, Wessels MR. The CsrR/CsrS two-component system of group A Streptococcus responds to environmental Mg2+ Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4227–4232. doi: 10.1073/pnas.0636231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryllos I, Tran-Winkler HJ, Cheng MF, Chung H, Bolcome R, 3rd, Lu W, Lehrer RI, Wessels MR. Induction of group A Streptococcus virulence by a human antimicrobial peptide. Proc Natl Acad Sci U S A. 2008;105:16755–16760. doi: 10.1073/pnas.0803815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusa AA, Gao J, Stringer V, Churchward G, Scott JR. Phosphorylation of the group A Streptococcal CovR response regulator causes dimerization and promoter-specific recruitment by RNA polymerase. Journal of bacteriology. 2006;188:4620–4626. doi: 10.1128/JB.00198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerstrom TG, Horton LB, Swick MC, Joachimiak A, Osipiuk J, Koehler TM. Crystal structure of Bacillus anthracis virulence regulator AtxA and effects of phosphorylated histidines on multimerization and activity. Mol Microbiol. 2015;95:426–441. doi: 10.1111/mmi.12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heithoff DM, Shimp WR, House JK, Xie Y, Weimer BC, Sinsheimer RL, Mahan MJ. Intraspecies variation in the emergence of hyperinfectious bacterial strains in nature. PLoS pathogens. 2012;8:e1002647. doi: 10.1371/journal.ppat.1002647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollands A, Pence MA, Timmer AM, Osvath SR, Turnbull L, Whitchurch CB, Walker MJ, Nizet V. Genetic switch to hypervirulence reduces colonization phenotypes of the globally disseminated group A streptococcus M1T1 clone. J Infect Dis. 2010;202:11–19. doi: 10.1086/653124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondorp ER, Hou SC, Hause LL, Gera K, Lee CE, McIver KS. PTS phosphorylation of Mga modulates regulon expression and virulence in the group A streptococcus. Mol Microbiol. 2013;88:1176–1193. doi: 10.1111/mmi.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondorp ER, McIver KS. The Mga virulence regulon: infection where the grass is greener. Mol Microbiol. 2007;66:1056–1065. doi: 10.1111/j.1365-2958.2007.06006.x. [DOI] [PubMed] [Google Scholar]
- Ikebe T, Ato M, Matsumura T, Hasegawa H, Sata T, Kobayashi K, Watanabe H. Highly frequent mutations in negative regulators of multiple virulence genes in group A streptococcal toxic shock syndrome isolates. PLoS pathogens. 2010;6:e1000832. doi: 10.1371/journal.ppat.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansal RG, McGeer A, Low DE, Norrby-Teglund A, Kotb M. Inverse relation between disease severity and expression of the streptococcal cysteine protease, SpeB, among clonal M1T1 isolates recovered from invasive group A streptococcal infection cases. Infect Immun. 2000;68:6362–6369. doi: 10.1128/iai.68.11.6362-6369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinzing DC, Ishmael N, Dunning Hotopp JC, Tettelin H, Shields KR, Madoff LC, Puopolo KM. The two-component response regulator LiaR regulates cell wall stress responses, pili expression and virulence in group B Streptococcus. Microbiology. 2013;159:1521–1534. doi: 10.1099/mic.0.064444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause RM, Rammelkamp CH, Jr, Denny FW, Jr, Wannamaker LW. Studies of the carrier state following infection with group A streptococci. 1. Effect of climate. The Journal of clinical investigation. 1962;41:568–574. doi: 10.1172/JCI104510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreikemeyer B, Boyle MD, Buttaro BA, Heinemann M, Podbielski A. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol Microbiol. 2001;39:392–406. doi: 10.1046/j.1365-2958.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- Kwinn LA, Nizet V. How group A Streptococcus circumvents host phagocyte defenses. Future microbiology. 2007;2:75–84. doi: 10.2217/17460913.2.1.75. [DOI] [PubMed] [Google Scholar]
- Li J, Liu G, Feng W, Zhou Y, Liu M, Wiley JA, Lei B. Neutrophils select hypervirulent CovRS mutants of M1T1 group A Streptococcus during subcutaneous infection of mice. Infect Immun. 2014;82:1579–1590. doi: 10.1128/IAI.01458-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman TD, Michel JB, Aingaran M, Potter-Bynoe G, Roux D, Davis MR, Jr, Skurnik D, Leiby N, LiPuma JJ, Goldberg JB, McAdam AJ, Priebe GP, Kishony R. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nature genetics. 2011;43:1275–1280. doi: 10.1038/ng.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Trevino J, Ramirez-Pena E, Sumby P. The small regulatory RNA FasX controls pilus expression and adherence in the human bacterial pathogen group A Streptococcus. Mol Microbiol. 2012;86:140–154. doi: 10.1111/j.1365-2958.2012.08178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynskey NN, Goulding D, Gierula M, Turner CE, Dougan G, Edwards RJ, Sriskandan S. RocA truncation underpins hyper-encapsulation, carriage longevity and transmissibility of serotype M18 group A streptococci. PLoS pathogens. 2013;9:e1003842. doi: 10.1371/journal.ppat.1003842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynskey NN, Turner CE, Heng LS, Sriskandan S. A truncation in the regulator RocA underlies heightened capsule expression in serotype M3 group A streptococci. Infect Immun. 2015;83:1732–1733. doi: 10.1128/IAI.02892-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon WR, Gibson CM, Caparon MG. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. The EMBO journal. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama F, Watanabe T, Nakagawa I. Streptococcus pyogenes Genomics. In: Ferretti JJ, Stevens DL, Fischetti VA, editors. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. Oklahoma City (OK): University of Oklahoma Health Sciences Center (c) The University of Oklahoma Health Sciences Center; 2016. [Google Scholar]
- Mayfield JA, Liang Z, Agrahari G, Lee SW, Donahue DL, Ploplis VA, Castellino FJ. Mutations in the control of virulence sensor gene from Streptococcus pyogenes after infection in mice lead to clonal bacterial variants with altered gene regulatory activity and virulence. PLoS One. 2014;9:e100698. doi: 10.1371/journal.pone.0100698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIver KS. Stand-alone response regulators controlling global virulence networks in streptococcus pyogenes. Contrib Microbiol. 2009;16:103–119. doi: 10.1159/000219375. [DOI] [PubMed] [Google Scholar]
- Miller EW, Cao TN, Pflughoeft KJ, Sumby P. RNA-mediated regulation in Gram-positive pathogens: an overview punctuated with examples from the group a Streptococcus. Mol Microbiol. 2014;94:9–20. doi: 10.1111/mmi.12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EW, Danger JL, Ramalinga AB, Horstmann N, Shelburne SA, Sumby P. Regulatory rewiring confers serotype-specific hyper-virulence in the human pathogen group A Streptococcus. Mol Microbiol. 2015a;98:473–489. doi: 10.1111/mmi.13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EW, Pflughoeft KJ, Sumby P. Reply to "A truncation in the regulator RocA underlies heightened capsule expression in serotype M3 group A streptococci". Infect Immun. 2015b;83:1734. doi: 10.1128/IAI.03162-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell ES. Frequency of serotypes of Streptococcus pyogenes in different diseases. Journal of clinical pathology. 1962;15:231–234. doi: 10.1136/jcp.15.3.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely MN, Lyon WR, Runft DL, Caparon M. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. Journal of bacteriology. 2003;185:5166–5174. doi: 10.1128/JB.185.17.5166-5174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Garbe J, Collin M. Cysteine proteinase SpeB from Streptococcus pyogenes - a potent modifier of immunologically important host and bacterial proteins. Biological chemistry. 2011;392:1077–1088. doi: 10.1515/BC.2011.208. [DOI] [PubMed] [Google Scholar]
- Nicol MP, Wilkinson RJ. The clinical consequences of strain diversity in Mycobacterium tuberculosis. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2008;102:955–965. doi: 10.1016/j.trstmh.2008.03.025. [DOI] [PubMed] [Google Scholar]
- Olsen RJ, Musser JM. Molecular pathogenesis of necrotizing fasciitis. Annual review of pathology. 2010;5:1–31. doi: 10.1146/annurev-pathol-121808-102135. [DOI] [PubMed] [Google Scholar]
- Olsen RJ, Raghuram A, Cantu C, Hartman MH, Jimenez FE, Lee S, Ngo A, Rice KA, Saddington D, Spillman H, Valson C, Flores AR, Beres SB, Long SW, Nasser W, Musser JM. The majority of 9,729 group A streptococcus strains causing disease secrete SpeB cysteine protease: pathogenesis implications. Infect Immun. 2015;83:4750–4758. doi: 10.1128/IAI.00989-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez N, Trevino J, Liu Z, Ho SCM, Babitzke P, Sumby P. A Genome-Wide Analysis of Small Regulatory RNAs in the Human Pathogen Group A Streptococcus. PLoS ONE. 2009;4:e7668. doi: 10.1371/journal.pone.0007668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph AP, Carapetis JR. Group a streptococcal diseases and their global burden. Current topics in microbiology and immunology. 2013;368:1–27. doi: 10.1007/82_2012_280. [DOI] [PubMed] [Google Scholar]
- Ramirez-Pena E, Trevino J, Liu Z, Perez N, Sumby P. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol Microbiol. 2010;78:1332–1347. doi: 10.1111/j.1365-2958.2010.07427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M, Muller HP, Bjorck L. Protein GRAB of streptococcus pyogenes regulates proteolysis at the bacterial surface by binding alpha2-macroglobulin. The Journal of biological chemistry. 1999;274:15336–15344. doi: 10.1074/jbc.274.22.15336. [DOI] [PubMed] [Google Scholar]
- Reyes J, Panesso D, Tran TT, Mishra NN, Cruz MR, Munita JM, Singh KV, Yeaman MR, Murray BE, Shamoo Y, Garsin D, Bayer AS, Arias CA. A liaR deletion restores susceptibility to daptomycin and antimicrobial peptides in multidrug-resistant Enterococcus faecalis. J Infect Dis. 2015;211:1317–1325. doi: 10.1093/infdis/jiu602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribardo DA, Lambert TJ, McIver KS. Role of Streptococcus pyogenes two-component response regulators in the temporal control of Mga and the Mga-regulated virulence gene emm. Infect Immun. 2004;72:3668–3673. doi: 10.1128/IAI.72.6.3668-3673.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribardo DA, McIver KS. Defining the Mga regulon: Comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A streptococcus. Molecular microbiology. 2006;62:491–508. doi: 10.1111/j.1365-2958.2006.05381.x. [DOI] [PubMed] [Google Scholar]
- Rupp S, Aguilar-Bultet L, Jagannathan V, Guldimann C, Drogemuller C, Pfarrer C, Vidondo B, Seuberlich T, Frey J, Oevermann A. A naturally occurring prfA truncation in a Listeria monocytogenes field strain contributes to reduced replication and cell-to-cell spread. Veterinary microbiology. 2015;179:91–101. doi: 10.1016/j.vetmic.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Sanson M, Makthal N, Gavagan M, Cantu C, Olsen RJ, Musser JM, Kumaraswami M. Phosphorylation events in the multiple gene regulator of group A Streptococcus significantly influence global gene expression and virulence. Infect Immun. 2015a;83:2382–2395. doi: 10.1128/IAI.03023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanson M, O'Neill BE, Kachroo P, Anderson JR, Flores AR, Valson C, Cantu CC, Makthal N, Karmonik C, Fittipaldi N, Kumaraswami M, Musser JM, Olsen RJ. A naturally occurring single amino acid replacement in multiple gene regulator of group A Streptococcus significantly increases virulence. Am J Pathol. 2015b;185:462–471. doi: 10.1016/j.ajpath.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen C, Blom J, Claus H, Schramm-Gluck A, Brandt P, Muller T, Goesmann A, Joseph B, Konietzny S, Kurzai O, Schmitt C, Friedrich T, Linke B, Vogel U, Frosch M. Whole-genome comparison of disease and carriage strains provides insights into virulence evolution in Neisseria meningitidis. Proc Natl Acad Sci U S A. 2008;105:3473–3478. doi: 10.1073/pnas.0800151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J, Nguyen SV, King CJ, Hendrickson C, McShan WM. Phage-Like Streptococcus pyogenes Chromosomal Islands (SpyCI) and Mutator Phenotypes: Control by Growth State and Rescue by a SpyCI-Encoded Promoter. Frontiers in microbiology. 2012;3:317. doi: 10.3389/fmicb.2012.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh N, Leonard E, Martin JM. Prevalence of streptococcal pharyngitis and streptococcal carriage in children: a meta-analysis. Pediatrics. 2010;126:e557–e564. doi: 10.1542/peds.2009-2648. [DOI] [PubMed] [Google Scholar]
- Shea PR, Beres SB, Flores AR, Ewbank AL, Gonzalez-Lugo JH, Martagon-Rosado AJ, Martinez-Gutierrez JC, Rehman HA, Serrano-Gonzalez M, Fittipaldi N, Ayers SD, Webb P, Willey BM, Low DE, Musser JM. Distinct signatures of diversifying selection revealed by genome analysis of respiratory tract and invasive bacterial populations. Proc Natl Acad Sci U S A. 2011;108:5039–5044. doi: 10.1073/pnas.1016282108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, 3rd, Granville C, Tokuyama M, Sitkiewicz I, Patel P, Musser JM. Growth characteristics of and virulence factor production by group A Streptococcus during cultivation in human saliva. Infect Immun. 2005;73:4723–4731. doi: 10.1128/IAI.73.8.4723-4731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, 3rd, Keith D, Horstmann N, Sumby P, Davenport MT, Graviss EA, Brennan RG, Musser JM. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc Natl Acad Sci U S A. 2008;105:1698–1703. doi: 10.1073/pnas.0711767105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanier JG, Jones SJ, Cleary P. Small DNA deletions creating avirulence in Streptococcus pyogenes. Science. 1984;225:935–938. doi: 10.1126/science.6089334. [DOI] [PubMed] [Google Scholar]
- Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS pathogens. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino J, Liu Z, Cao TN, Ramirez-Pena E, Sumby P. RivR is a negative regulator of virulence factor expression in group A Streptococcus. Infect Immun. 2013;81:364–372. doi: 10.1128/IAI.00703-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino J, Perez N, Ramirez-Pena E, Liu Z, Shelburne SA, 3rd, Musser JM, Sumby P. CovS simultaneously activates and inhibits the CovR-mediated repression of distinct subsets of group A Streptococcus virulence factor-encoding genes. Infect Immun. 2009;77:3141–3149. doi: 10.1128/IAI.01560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlemann AC, Otto M, Lowy FD, DeLeo FR. Evolution of community- and healthcare-associated methicillin-resistant Staphylococcus aureus. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2014;21:563–574. doi: 10.1016/j.meegid.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega LA, Malke H, McIver KS. Virulence-Related Transcriptional Regulators of Streptococcus pyogenes. In: Ferretti JJ, Stevens DL, Fischetti VA, editors. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. Oklahoma City (OK): University of Oklahoma Health Sciences Center (c) The University of Oklahoma Health Sciences Center; 2016. [Google Scholar]
- Velarde JJ, Ashbaugh M, Wessels MR. The human antimicrobial peptide LL-37 binds directly to CsrS, a sensor histidine kinase of group A Streptococcus, to activate expression of virulence factors. The Journal of biological chemistry. 2014;289:36315–36324. doi: 10.1074/jbc.M114.605394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MJ, Hollands A, Sanderson-Smith ML, Cole JN, Kirk JK, Henningham A, McArthur JD, Dinkla K, Aziz RK, Kansal RG, Simpson AJ, Buchanan JT, Chhatwal GS, Kotb M, Nizet V. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat Med. 2007;13:981–985. doi: 10.1038/nm1612. [DOI] [PubMed] [Google Scholar]
- Wessels MR, Bronze MS. Critical role of the group A streptococcal capsule in pharyngeal colonization and infection in mice. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:12238–12242. doi: 10.1073/pnas.91.25.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels MR, Moses AE, Goldberg JB, DiCesare TJ. Hyaluronic acid capsule is a virulence factor for mucoid group A streptococci. Proc Natl Acad Sci U S A. 1991;88:8317–8321. doi: 10.1073/pnas.88.19.8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Jelsbak L, Marvig RL, Damkiaer S, Workman CT, Rau MH, Hansen SK, Folkesson A, Johansen HK, Ciofu O, Hoiby N, Sommer MO, Molin S. Evolutionary dynamics of bacteria in a human host environment. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7481–7486. doi: 10.1073/pnas.1018249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young BC, Golubchik T, Batty EM, Fung R, Larner-Svensson H, Votintseva AA, Miller RR, Godwin H, Knox K, Everitt RG, Iqbal Z, Rimmer AJ, Cule M, Ip CL, Didelot X, Harding RM, Donnelly P, Peto TE, Crook DW, Bowden R, Wilson DJ. Evolutionary dynamics of Staphylococcus aureus during progression from carriage to disease. Proc Natl Acad Sci U S A. 2012;109:4550–4555. doi: 10.1073/pnas.1113219109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Olsen RJ, Nasser W, Beres SB, Vuopio J, Kristinsson KG, Gottfredsson M, Porter AR, DeLeo FR, Musser JM. A molecular trigger for intercontinental epidemics of group A Streptococcus. The Journal of clinical investigation. 2015;125:3545–3559. doi: 10.1172/JCI82478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkernagel AS, Timmer AM, Pence MA, Locke JB, Buchanan JT, Turner CE, Mishalian I, Sriskandan S, Hanski E, Nizet V. The IL-8 protease SpyCEP/ScpC of group A Streptococcus promotes resistance to neutrophil killing. Cell host & microbe. 2008;4:170–178. doi: 10.1016/j.chom.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]