Abstract
Steroid receptor coactivator 3 (SRC-3) is a transcriptional coactivator that interacts with nuclear receptors and some other transcription factors to enhance their effects on target gene transcription. We previously reported that SRC-3-deficient (SRC-3−/−) mice are super susceptible to E. coil-induced septic peritonitis due to uncontrolled inflammation and a defect in bacterial clearance. In this study, we observed significant upregulation of SRC-3 in the colonic epithelial cells in response to Citrobacter rodentium infection. Based on these findings, we hypothesized that SRC-3 is involved in host defense against attaching and effacing (A/E) bacterial infection. We compared the responses of SRC-3−/− and wild-type mice to intestinal C. rodentium infection. We found that SRC-3−/− mice exhibited delayed clearance of C. rodentium and more severe tissue pathology after oral infection with C. rodentium compared with wild-type mice. SRC-3−/− mice expressed normal antimicrobial peptides in the colons, but exhibited delayed recruitment of neutrophils into the colonic mucosa. Accordingly, SRC-3−/− mice showed a delayed induction of CXCL2 and CXCL5 in the colonic epithelial cells, which are responsible for neutrophil recruitment. At the molecular level, we found that SRC-3 can activate the NF-κB signaling pathway to promote CXCL2 expression at the transcriptional level. Collectively, we show that SRC-3 contributes to host defense against enteric bacteria at least in part via upregulating CXCL2 expression to recruit neutrophils.
Introduction
The attaching and effacing (A/E) enteric bacteria Enteropathogenic Escherichia coli (EPEC) and Enterohemorrhagic E.coli (EHEC) are human-specific pathogens and they do not cause relevant disease in animal models (1). These bacterial pathogens cause global diarrheal disease and other severe complications in human, especially among infants and children (2–4). Citrobacter rodentium, a natural A/E pathogen of mice, utilizes similar virulence strategies to those employed by EPEC and EHEC to infect cells, so it is widely used to model human infections with EPEC and EHEC (5). Upon infection, C. rodentium can colonize the murine cecum and colon and displace the commensal flora and lead to intestinal inflammation characterized by mucosal hyperplasia, goblet cell depletion and epithelial layer disruption, but is eventually cleared from cecum and colon of C57BL/6 mice in 3–4 weeks (5). C. rodentium infection induces infiltration of immune cells within lamina propria. Innate and adaptive immune systems are involved in eradicating C. rodentium infection. Studies of C. rodentium infection in immunodeficient mice have established that neutrophils are required for early innate immune protection against C. rodentium infection (6–7), and inflammatory monocytes are important in promoting bacterial eradication in the intestine (8). Intestinal monocytes and macrophages are required for initiating adaptive immune responses to C. rodentium (9–10). The adaptive immune component, CD4+ T cell-dependent IgG response, plays an essential role in clearing C.rodentium infection (11). Additionally, nonhematopoietic cells, for example intestinal epithelial cells, can also orchestrate the immune responses against mucosal bacterial infection (7, 12–14). Cathelicidin from intestinal epithelial cells, as an important component of innate antibacterial defense, can protect mice from C. rodentium infection (15). Another nonhematopoietic cells, goblet cells, can produce Muc2 to protect mice from C. rodentium infection by limiting tissue damage and translocation of pathogenic and commensal bacteria across the epithelium (16), suggesting that goblet cells form a critical innate immune player in the control of C. rodentium infection. Therefore, both hematopoietic cells and nonhematopoietic cells are critical for the control of C. rodentium infection.
Steroid receptor coactivator 3 (SRC-3) is a transcriptional coactivator that interacts with nuclear receptors and some other transcription factors to enhance their effects on target gene transcription (17). It has been reported macrophage SRC-3 could enhance LPS-induced expression of proinflammatory cytokines (18). In contrast to this study, our previous study demonstrated that SRC-3−/− mice produced significantly more proinflammatory cytokines and macrophage SRC-3 could inhibit cytokine mRNA translation in response to LPS challenge (19), indicating that macrophage SRC-3 can protect mice by regulating proinflammatory cytokine balance in LPS-induced endotoxic shock. We also reported that SRC-3−/− mice were more susceptible to Escherichia coli-induced septic peritonitis due to an uncontrolled overwhelming inflammation and a defect in bacterial clearance (20), suggesting a critical protective role for SRC-3 in the host defense against bacterial infection. However, its involvement in host defense against A/E enteric bacterial pathogen remains poorly defined.
In this study, we found that SRC-3 was expressed in the colonic epithelial cells and further up-regulated in response to C. rodentium infection. In the light of these results, we hypothesized that SRC-3 plays a protective role in C. rodentium-induced colitis. We tested this hypothesis by comparing the response of SRC-3−/− mice and wild-type mice to C. rodentium challenge. We found that SRC-3−/− mice displayed delayed clearance of bacteria and more severe tissue pathology after oral infection of C. rodentium compared to wild-type mice, and SRC-3 may contribute to host defense against C. rodentium at least in part via upregulating chemokine CXCL2 expression to recruit neutrophils.
Material and Methods
Mice
SRC-3−/− mice (on C57BL/6×129Sv background) were generated as described previously (21). Wild-type mice serve as control mice. 6–8 week old female mice were used in all experiments. Animal experiments were approved by the Laboratory Animal Center of Xiamen University, Xiamen, Fujian, China.
Bacterial strain and infection of mice
C. rodentium strain ATCC51459 was inoculated into Luria-Bertani (LB) and cultured in a shaker for 12 h at 37 °C. Mice were orally inoculated with 0.2 ml of PBS containing 9×108 colony formation unit (CFU) C. rodentium per mouse after fasting for 8 h. The number of alive bacteria was determined by serially diluting and seeding on MacConkey agar plate for 18–24 h.
Bacterial counts
The distal 0.5 cm piece of colon tissue and feces were weighted, and then homogenized in a Tissuelyser-24 for a total 6 min at 30 Hz at room temperature. Tissue homogenates were serially diluted in PBS and plated in triplicate onto MacConkey agar plates. C. rodentium colonies were identified as pink colonies after 18–24 hours of incubation at 37 °C. Spleen and liver colonization were accessed as described for colon specimens. The detection limit of the CFU assay was 103 CFU/g feces or colons and 102 CFU/g spleens or livers.
Tissue collection
Mice were sacrificed at various indicated times. Entire colons, spleens and livers were removed under sterile conditions. The distal 0.5 cm piece of colon tissue was collected in a 1.5 ml microtube containing 1 ml PBS. The adjacent 0.5 cm was fixed in cassettes in 10% neutral-buffer formalin. The next 1 cm was stored in Trizol (Life technologies) at −80 °C for RNA extraction.
Colon culture
The protocols of Siegmund et al. (22) and Gibson et al (23) were modified. Briefly, segments of mid parts of colon were cut longitudinally and cleaned with cold PBS containing 100 U/ml Penicillin and 100 μg/ml Streptomycin. Then these pieces were cultured in 6-well plate in 3 ml serum-free RPMI-1640 (Gibco) supplemented with 100 U/ml Penicillin and 100 μg/ml Streptomycin for 24 h at 37 °C and 5% CO2. Supernatants were centrifuged for 5 min at 16060 × g at 4 °C and collected and stored at −80 °C until analysis.
Histological analysis
To assess tissue pathology, a scoring system was performed using previously described scoring systems (12, 24). In brief, colons were fixed in 10% neutral formalin and embedded in paraffin. Then 5 μm sections were prepared and stained with H&E. Two tissue sections from four to eight mice per group were assessed for submucosal oedema (0 = no change; 1 = mild; 2 = moderate; 3 = profound), goblet cell depletion (scored based on numbers of goblet cells per field averaged from five fields at 400× magnification, where 0 = > 50; 1 = 25–50; 2 = 10–25; 3 = < 10), epithelial hyperplasia (scored based on percentage above the height of the control, where 0 = no change; 1 = 1–50%; 2 = 51–100%; 3 = > 100%), epithelial integrity (0 = no change; 1 = mild epithelial ulceration and crypt destruction; 2 = moderate epithelial ulceration and crypt destruction; 3= severe epithelial ulceration with crypt destruction) and inflammatory cell infiltration (0 = occasional inflammatory cells in the lamina propria; 1 =increased number of inflammatory cells in the lamina propria; 2 = confluent inflammatory cells, extending into the submucosa; 3 = transmural extension of the infiltrate).
Immunohistochemistry
Five μm paraffin sections were deparaffinized and rehydrated. Antigens were retrieved by soaking in preheated citrate buffer (pH 6.0) under microwave heating for 20 min. To quench endogenous peroxidase activity, sections were incubated with 3% hydrogen peroxide for 15 min. Sections were incubated with 5% BSA for 1 h at room temperature to block nonspecific binding sites, and then were incubated with rabbit anti-Citrobacter rodentium antibodies (7, 12–14). Subsequently, sections were incubated with Elivision plus kits (Maixin) at room temperature. After washing, DAB reagent was added to visualize stained proteins.
Cytokine analysis by ELISA
The concentrations of IL-6 (eBioscience), IL-1β (eBioscience), TNF-α (eBioscience), CXCL2 (R&D Systems) and CXCL5 (Abcam) were assayed using ELISA kits according to the manufacturer’s instructions. Supernatants from cultured colons were standardized to the amount of total protein in the supernatant by quantification using the DC Protein Assay (Bio-Rad).
Western blot
Colonic epithelial cells and CMT93 cells were lysed in RIPA buffer (150 mM NaCl, 50 mM Tris, 0.1% SDS, 1 mM EDTA, 1% Triton X-100, 1 mM PMSF and Cocktail). Proteins were quantified with BCA assay. Equal amounts of proteins were loaded onto 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels and transferred onto polyvinylidene difluoride membranes (Millipore), followed by immunoblotting with anti-SRC-3, anti-p-p65, anti-p65, anti-IκBα (cell signaling) and anti-β-actin (Sigma). Western blots were analyzed using a Tonen Image.
Myeloperoxidase Staining
Colons were processed for myeloperoxidase staining by using Hanker-Yates reagent (Polysciences) according to the manufacturer’s instruction. MPO+ cells were counted under a microscope in 5 randomly chosen areas in each specimen at a magnification of 400×.
Quantitative real-time PCR
Total RNA was extracted with Trizol (Life technologies) from cells or tissues in accordance with the manufacturer’s instructions and cDNA was synthesized with ReverTra Ace®qPCR RT Kit (TOYOBO). Real-time PCR was performed with FastStart Universal Probe Master (Rox) (Roche). Expression of each target gene was normalized to that of GAPDH. The sequences of primers used for quantitative real-time PCR can be provided on request.
Isolation of epithelial and lamina propria cells
Epithelial and lamina propria cells were isolated from the lamina propria with a modification of a protocol described previously (25). In brief, mice were sacrificed, and colonic tissues were removed. The colons were washed in PBS containing penicillin and streptomycin (10,000 unit/ml penicillin, 10 mg/ml streptomycin). Colons were cut longitudinally and washed three times in PBS containing penicillin and streptomycin. Tissue was cut into 0.5 cm pieces and incubated in PBS containing 2.5% FCS and 1 mM DTT (Sigma-Aldrich, St. Louis, MO) for 30 min at 37 °C with gentle shaking to remove any mucus. The pieces were incubated twice in PBS containing 5 mM EDTA (Sigma-Aldrich) for 15 min each at 37 °C with gentle shaking, washed three times with PBS. Epithelial cells were pooled from the supernatant after each round of ringing and collected by centrifugation after having passed through a nylon mesh strainer (100 μm pore size, BD). Collected epithelial cells were resuspended in 1 ml of Trizol (Life technologies) for further RNA extraction. Pieces of colons were then washed for 10 min in RPMI-1640 to inactivate EDTA. Thereafter pieces of colons were incubated in RPMI-1640 containing 5% FCS, 0.5 mg/ml collagenase (Sigma-Aldrich) for 2 h at 37 °C with gentle shaking. Supernatants were filtered through a 40 μm nylon sieve and washed in RPMI-1640 containing 5% FCS and centrifuged to pellet the cells. Cells were resuspended in RPMI-1640 containing 10% FCS, washed twice, and used immediately for flow cytometric analysis. Colonic macrophages were purified by positive selection with a magnetic cell separation system (MACS; Miltenyi Biotec, Auburn, CA) with anti-mouse CD11b microbeads (8).
Flow Cytometry
Cells isolated from the lamina propria or peritoneal cavity were pretreated on ice for 30 min with rat IgG (0.2 mg/ml) to block nonspecific binding to Fc receptors at 4 °C. Thereafter, cells were incubated on ice for 30 min with fluorescently conjugated antibodies for cell-surface markers: PECy5-conjugated anti-Gr1 mAb RB6-8C5 (eBioscience, San Diego, CA, USA), PE-conjugated anti-F4/80 mAb BM8 (eBioscience, San Diego, CA, USA) and FITC-conjugated anti-CD11b mAb M1/70 (eBioscience). Isotype controls consisted PE-conjugated IgG2a and PECy5-conjugated IgG2b (eBioscience) and FITC-conjugated IgG2b (eBioscience). Cells were washed, and fixed in 1% paraformaldehyde over night at 4 °C. Analysis of stained cells was performed with a FACScanto flow cytometer (BD Biosciences). Isotype controls were used to set appropriate gates. Data were analyzed with FACS diva (BD Bioscience) and FloJo 6.4.7 (Tree Star, Ashland, OR, USA). For all samples, approximately 10,000 cells were analyzed for plot generation.
In vitro bacterial infection
C. rodentium was inoculated into Luria-Bertani (LB) and cultured in a shaker for 12 h at 37 °C. The bacteria were washed twice with PBS and resuspended with PBS. CMT93 cells were infected with C. rodentium at an MOI of 200 by directly adding C. rodentium to DMEM medium without spinning for indicated times at 37 °C and then 50 μg/ml of gentamycin was added to limit the growth of extracellular bacteria.
Luciferase reporter assay
The CXCL2 promoter fragment −1227 to +14 was ligated into KpnI and XhoI sites of PGL3-basic vector to yield the CXCL2 reporter plasmid according to the method described previously (26). To produce site-specific mutation at NF-κB sites, the transcription factor recognition sites were abolished by a one-step site-directed and site-saturation mutagenesis method (27). The primers designed to introduce site-directed mutation were synthesized as follows: mNF-κB (−144), forward, 5′CCCTGAGCTCAGtcgATTTCCCTGGTCCCCGGGCTTTTC3′, reverse, 5′-GAC-CAGGGAAATcgaCTGAGCTCAGGGTCCCCTCATCAGG-3′; mNF-κB (−742), forward, 5′TGGAAGAGCCTCatAAGTTCCAGAATTTCACAGAGG3′, reverse, 5′ATTCTGGAACTTatGAGGCTCTTCCAGTGTGCTGGTG3′. Lowercase letters indicates mutated sites. In the light of these primers, we yielded the reporter constructs pCXCL2-M1-Luc, pCXCL2-M2-Luc and pCXCL2-M1+M2-Luc. We used DNA sequencing to verify the nucleotide sequences of these constructs. Individual assays were normalized for Renilla luciferase activity and the data are presented as a fold increase in activity relative to the empty-vector control. Luciferase activity was assayed using a dual luciferase reporter assay system (Promega, Madison, WI).
Chromatin immunoprecipitation assay
CMT93 cells were used for chromatin immunoprecipitation (ChIP) assay and processed according to the method described by Abcam (Cambridge, MA). The following primers were used: CXCL2 promoter −742 NF-κB binding site, forward, 5′-CTCTTACAGCTGATGTGGTGCTC-3′, and reverse, 5′-GAGATGTGTCAAAAGCTTAATTC-3′; CXCL2 promoter −144 NF-κB binding site, forward, 5′-AACCCACTCAGCTTAGGGGC-3′, and reverse, 5′-TTGTTGGAGGCACTGAGGC-3′. Anti-SRC-3 antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Statistical analysis
Mann-Whitney test was used to analyze chemokine production and bacterial counts of infected mice in vivo. Other statistical significance was determined by two-tailed t-test in GraphPad Prism 5. Differences were considered significant when P value was <0.05. All values were expressed as mean + SEM or + SD.
Results
SRC-3 is mainly expressed in mouse colonic epithelial cells and further upregulated in response to C. rodentium infection
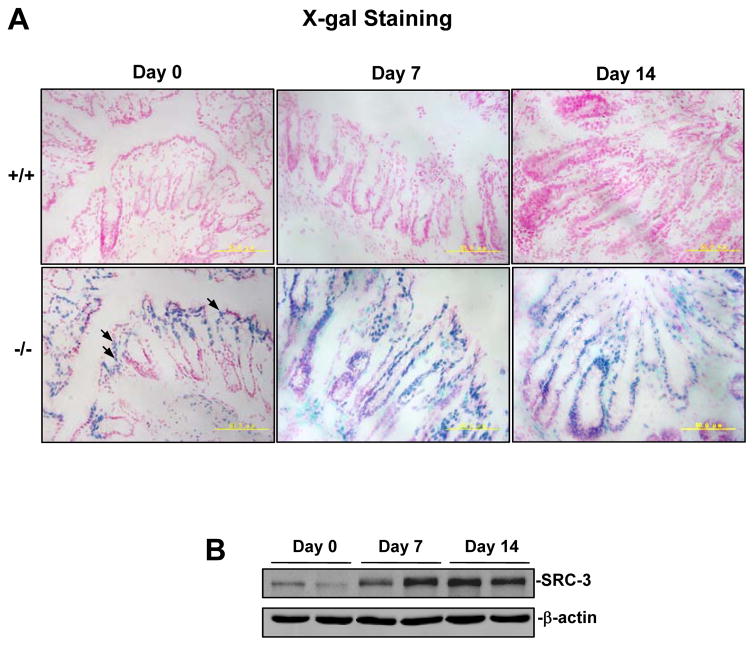
In SRC-3−/− mice, the lacZ coding sequence was fused to the 10th amino acid of SRC-3 to allow the endogenous SRC-3 promoter to control the lacZ expression (21). We therefore could perform X-gal staining to investigate SRC-3 expression patterns in the colons of SRC-3−/− mice. As shown in Fig. 1A, without C. rodentium infection, SRC-3 expression was mainly detected in the colon enterocytes of SRC-3−/− mice as indicated by X-gal staining (blue); after intestinal C. rodentium infection for 7 or 14 days, SRC-3 expression was detected in most of colonic epithelial cells, suggesting that the SRC-3 in the colonic epithelial cell can be induced by C. rodentium infection. Consistently, Western blot analysis showed that SRC-3 expression was significantly upregulated in the colonic epithelial cells isolated from SRC-3+/+ mice at 7 and 14 days after intestinal C. rodentium infection (Fig. 1B). These results demonstrate that the increase of SRC-3 in the colonic epithelial cells after C. rodentium infection was due to both an increase in SRC-3-positive epithelial cell number and an increase on a “per cell” basis. Since colonic epithelial cells are target cells of C. rodentium, upregulation of SRC-3 expression in the colonic epithelial cells after C. rodentium infection indicates that the SRC-3 in the colonic epithelial cells may be involved in the host defense against C. rodentium infection.
Figure 1. SRC-3 is highly expressed in the colonic epithelial cells and upregulated in response to C. rodentium infection.
A, SRC-3 was highly expressed in the colonic epithelial cells and further upregulated in response to C. rodentium infection. Sections of frozen colon tissues from SRC-3+/+ (n=4) and SRC-3−/− mice (n=4) were stained with X-gal, arrows indicated the representative positive staining cells (blue). B, SRC-3 was upregulated in the colonic epithelial cells in response to C. rodentium infection. The protein levels of SRC-3 in the colonic epithelial cells of uninfected wild-type mice or wild-type mice infected with C. rodentium for 7 and 14 days were detected by Western blot analysis. Results are representative of three independent experiments.
Delayed clearance of C. rodentium by SRC-3−/− mice
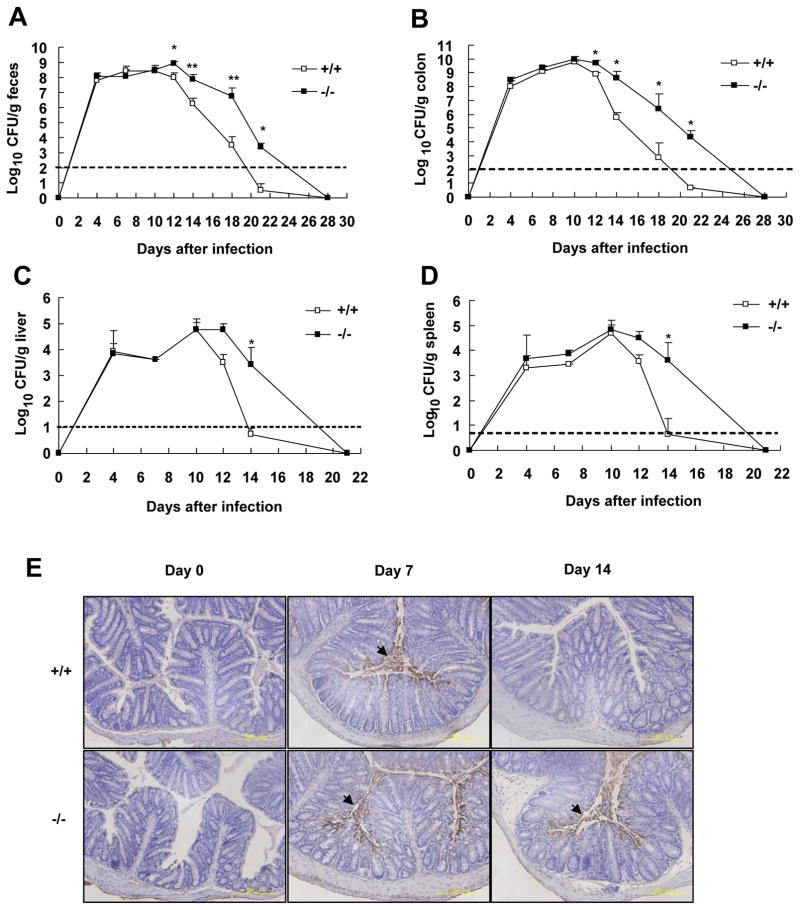
To assess the role of SRC-3 in the host defense against C. rodentium, we infected wild-type and SRC-3−/− mice with C. rodentium orally and monitored the numbers of pathogenic bacteria in the feces, colons, spleens and livers for up to 28 days. As shown in Fig. 2A, C. rodentium in the feces of wild-type mice reached the maximal plateau level (108 colony-forming units (CFU)/g) on days 7–12 and then declined over time to become almost undetectable by day 21 after infection. In contrast, although the number of C. rodentium in the feces of SRC-3−/− mice reached the maximal plateau level similar to wild-type mice on days 7–12, the bacterial number in the feces of SRC-3−/− mice declined much slower than that of wild-type mice after then (Fig. 2A). Similar kinetic changes of bacterial number were observed in the colons, livers and spleens (Fig. 2B–D). Immunohistochemistry staining of C. rodentium in the colons of wild-type and SRC-3−/− mice confirmed the kinetic change of bacterial number in the colons (Fig. 2E). These results indicate that SRC-3 deficiency does not affect the expansion and systemic spreading of C. rodentium at the early phase of infection, but it severely impairs the clearance of C. rodentium at the late phase of infection.
Figure 2. Delayed clearance of C. rodentium by SRC-3−/− mice.
Bacterial numbers in feces (A), colon (B), liver (C) and spleen (D) from wild-type or SRC-3−/− mice infected orally with C. rodentium were determined by CFU assays at the indicated time after infection. The transverse bar is the detection limit. Data are the means + SEM (n=5–10). *p<0.05; **p<0.01. Results are representative of two independent experiments. E, Localization of C. rodentium in the colons of wild-type and SRC-3−/− mice by immunohistochemistry staining. Images are representative of three separate experiments, n=4–5 per group. Arrows indicate immunopositive signals.
SRC-3−/− mice display more severe colonic pathology than wild-type mice after C. rodentium infection
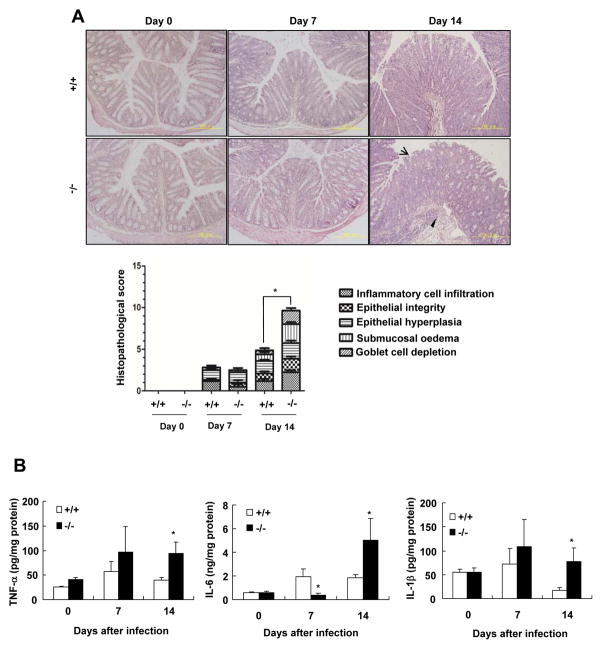
Oral infection with C. rodentium induces inflammation and colonic pathology characterized by inflammatory cell infiltration, hyperplasia, disruption of epithelial cells, submucosa oedema, and goblet cell depletion (23). We performed H&E staining to assess C. rodentium-induce colonic pathology in SRC-3−/− mice and wild-type mice. In the absence of C. rodentium infection, neither SRC-3−/− mice nor wild-type mice exhibited any colonic pathology (Fig. 3A and 3B); On day 7 after infection, both SRC-3−/− and wild-type mice showed comparable mild colonic pathology characterized by increased inflammatory cell infiltration, disruption of epithelial integrity, epithelial hyperplasia, submucosal oedema, and goblet cell depletion (Fig. 3A); However, on day 14 after infection, SRC-3−/− mice exhibited more severe colonic pathology than wild-type mice (Fig. 3A). These results indicate that SRC-3 can protect mice from C. rodentium-induced colonic pathology at the late phase of infection.
Figure 3. SRC-3−/− mice display more severe colonic pathology compared with wild-type mice after C. rodentium infection.
A, H&E staining of colon slides from uninfected or infected wild-type and SRC-3−/− mice. Arrow heads and arrows denote marked submucosal inflammatory cellular infiltrates/edema and epithelial damage/inflammation in the mucosa, respectively. A 200 × magnification is shown. Histopathalogical scores of wild-type and SRC-3−/− mice at day 0, 7 and 14 after infection. Data represent means + SEM (n=4–10). B, The expression of inflammatory cytokines was higher in the colon of SRC-3−/− mice on day 14 after C. rodentium infection. Data are the means + SEM (n=4–10). *p<0.05. Results are representative of two independent experiments.
Furthermore, we measured the levels of proinflammatory cytokines after oral infection with C. rodentium. The mRNA levels of IL-22, IL-17A and IFNγ were comparable in the colons of wild-type and SRC-3−/− mice on days 0, 7 and 14 after C. rodentium infection (Data not shown). The concentrations of TNF-α and IL-1β were comparable in the colons of wild-type and SRC-3−/− mice on day 0 and day 7 after infection (Fig. 3B), while the concentrations of IL-6 in the colons of wild-type mice were higher than SRC-3−/− mice on day 7 after infection (Fig. 3B). However, on day 14 after infection, the concentrations of TNF-α, Il-6 and IL-1β in the colons of SRC-3−/− mice were significantly higher than that of wild-type mice (Fig. 3B), indicating that SRC-3−/− mice suffer more severe colonic inflammation at the late phase of C. rodentium infection.
SRC-3 deficiency does not impair the expression of antimicrobial genes in mice infected with C. rodentium
Antimicrobial genes such as Reg family of antimicrobial proteins, cathelicidin-related antimicrobial peptide (mCRAMP), and β-defensins, play important roles in host defense against C. rodentium (28) (15, 29). To determine whether SRC-3 protects mice from C. rodentium infection by regulating the expression of antimicrobial genes, we analyzed the expression of some antimicrobial genes involved in host defense against C. rodentium in the colons of SRC-3−/− mice and wild-type mice. As showed in Fig. S1, SRC-3 deficiency did not impair the expression Reg3β, Reg3γ, mCRAMP, defensinβ1, defensinβ3 and defensinβ4 in the colons of mice infected by C. rodentium, indicating that impaired clearance of C. rodentium in SRC-3−/− mice is not due to the defects in the expression of antimicrobial genes.
Recruitment of CD11b+F4/80−Gr1hi neutrophils to the colon is delayed in SRC-3−/− mice after C. rodentium infection
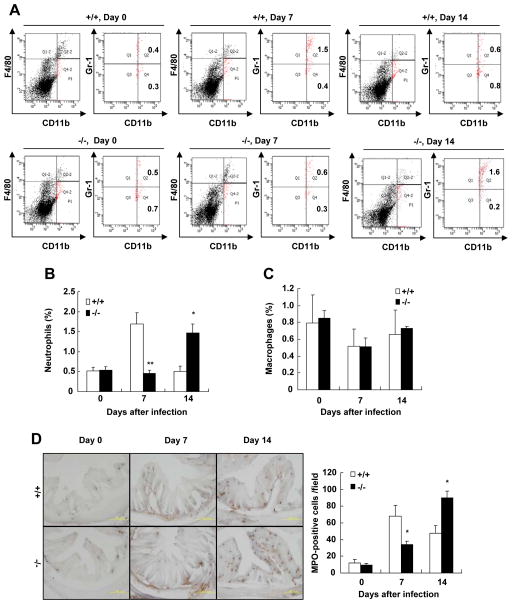
Because neutrophils and monocytes/macrophages are important for promoting bacterial eradication in the colon, we assessed the influx of neutrophils and monocytes in the colon lamina propria of SRC-3−/− mice and wild-type mice after oral infection with C. rodentium by flow cytometry. Lamina propria cells were stained for the macrophage maker F4/80, the leukocyte marker CD11b which is shared among macrophages, neutrophils and some DCs, and Gr1 which is expressed in neutrophils, inflammatory macrophages and some populations of DCs (30). In the absence of C. rodentium infection, the number of neutrophils (CD11b+F4/80−Gr1hi) was comparable in the colons of SRC-3−/− mice and wild-type mice (Fig. 4A and 4B). On day 7 after C. rodentium infection, there were significantly more neutrophils recruited to the colons of wild-type mice (1.7%) versus SRC-3−/− mice (0.5%) (Fig. 4A and 4B). However, on day 14 after C. rodentium infection, the influx of neutrophils to the colons of wild-type mice was decreased to 0.5%, whereas the influx of neutrophils to the colons of SRC-3−/− mice was increased to 1.4% (Fig. 4A and 4B). The number of CD11b+F4/80+Gr-1hi inflammatory macrophages was comparable in the colons of SRC-3−/− mice and wild-type mice during C. rodentium infection (Fig. 4C). These results indicate that SRC-3 deficiency causes a delay of neutrophil recruitment to the colon, but has no effect on the recruitment of monocytes to the colon in response to C. rodentium infection.
Figure 4. Recruitment of CD11b+F4/80−Gr-1hi neutrophils to the colons is delayed in SRC-3−/− mice after C. rodentium infection.
A, Cells were isolated from the colon of wild-type and SRC-3−/− mice on days 0, 7 and 14 after infection and stained for CD11b, F4/80 and Gr-1 mAb. CD11b+F4/80− cells (total fraction) were gated and percentage of CD11b+F4/80−Gr-1hi cells in the gated population was determined. B, Quantitation of CD11b+F4/80−Gr-1hi neutrophils in the colons of wild-type and SRC-3−/− mice on days 0, 7 and 14 after infection, n=4–5. C, Quantitation of CD11b+F4/80+Gr-1hi inflammatory macrophages in the colons of wild-type and SRC-3−/− mice on days 0, 7 and 14 after infection, n=4–5. D, Examination and quantitation of MPO+ neutrophils. Left panel: Frozen sections of the colon were prepared and stained with Hanker-Yates regent. Brown signal denote MPO+ cells. A 200 × magnification is shown. Right Panel: Quantitation of MPO+ cells. Data are the means + SEM (n=5). *p<0.05; **p<0.01. Results are representative of two independent experiments.
Furthermore, histochemistry staining of neutrophils (MPO-positive cells) confirmed the results of flow cytometry (Fig. 4D). These results suggest that SRC-3 plays an important role in the recruitment of neutrophils to the colon in response to C. rodentium infection.
Induction of colonic expression of CXCL2 and CXCL5 is delayed in SRC-3−/− mice after C. rodentium infection
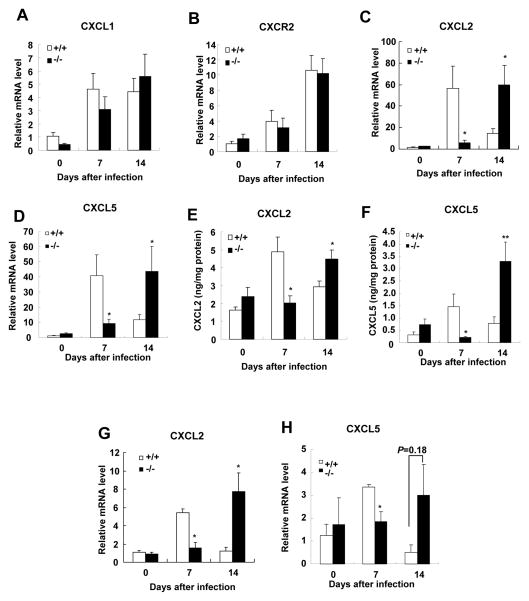
Chemokines drive the recruitment of immune cells such as neutrophils, monocytes, and lymphocytes, which are required for clearing C. rodentium in response to bacterial infection (31–32). CXCL1, CXCL2, and CXCL5 are chemokines involved in neutrophil recruitment and highly induced upon C. rodentium infection (33). We therefore investigated the expression of these chemokines and their receptor CXCR2 in the colons of SRC-3−/− and wild-type mice upon C. rodentium infection (Fig. 5). The expression levels of CXCL1 and CXCR2 mRNAs were comparable in the colons of SRC-3−/− and wild-type mice on days 0, 7 and 14 after C. rodentium infection (Fig. 5A and 5B). On day 7 after infection, the mRNA levels of CXCL2 and CXCL5 were significantly increased in the colons of wild-type mice compared to SRC-3−/− mice (Fig. 5C and 5D). Interestingly, on day 14 after infection, when the mRNA expression of CXCL2 and CXCL5 was declined in the colons of wild-type mice, the mRNA expression of CXCL2 and CXCL5 was significantly increased in the colons of SRC-3−/− mice (Fig. 5C and 5D). In addition, the expression pattern of colonic proteins of CXCL2 and CXCL5 was consistent with the expression pattern of their mRNAs (Fig. 5E and 5F). These results indicate that SRC-3 deficiency causes a delayed induction of the expression of CXCL2 and CXCL5 in the colon in response to C. rodentium infection.
Figure 5. Induction of colonic CXCL2 and CXCL5 is delayed in SRC-3−/− mice after C. rodentium infection.
A–D, Quantitative RT-PCR of CXCL1 (A), CXCR2 (B), CXCL2 (C) and CXCL5 (D) in the distal colons of wild-type and SRC-3−/− mice after C. rodentium infection. Data are the means + SEM (n=8–13). E and F, Quantitation of the protein levels of CXCL2 and CXCL5 in the colons. Colons from wild-type and SRC-3−/− mice were cultured for 24 hours and colon supernatants were analyzed for protein levels of CXCL2 and CXCL5 by ELISA and standardized to levels of total protein. Data are the means + SEM (n=8–13). G and H, Quantitative RT-PCR of CXCL2 and CXCL5 in isolated colonic epithelial cells of wild-type and SRC-3−/− mice after C. rodentium infection. Data are the means + SEM (n=4–5). *p<0.05. Results are representative of two independent experiments.
Since SRC-3 is mainly expressed in the colonic epithelial cells (Fig. 1A), and CXCL2 and CXCL5 from colonic epithelial cells plays an essential role in neutrophil recruitment after C. rodentium infection, we wonder whether the expression of CXCL2 and CXCL5 in the colonic epithelial cells is affected by SRC-3 deficiency. We isolated epithelial cells from colons of SRC-3−/− and wild-type mice on days 0, 7 and 14 after C. rodentium infection and detected the expression of CXCL2 and CXCL5 by real-time PCR. As shown in Fig. 5G, when CXCL2 expression in the colonic epithelial cells from wild-type mice was induced on day 7 after C. rodentium infection, it remained unchanged in SRC-3−/− mice; however, when CXCL2 expression in the colonic epithelial cells from wild-type mice was reduced to near basal level, it was significantly induced in the colonic epithelial cells of SRC-3−/− mice on day 14 after C. rodentium infection. Compared to CXCL2 mRNA expression, similar kinetic changes of CXCL5 mRNA expression were also observed in the colonic epithelial cells during C. rodentium infection (Fig. 5H). We also isolated macrophages from colons on day 7 after C. rodentium infection using a magnetic cell separation system with anti-mouse CD11b microbeads, and detected the mRNA expression of CXCL1, CXCL2 and CXCL5. The mRNA levels of CXCL1, CXCL2 and CXCL5 of macrophages were comparable in the colons of wild-type mice and SRC-3−/− mice on day 7 after infection (Data not shown). These results indicate that SRC-3 deficiency in the colonic epithelial cells causes a delayed induction of CXCL2 and CXCL5 during C. rodentium infection.
SRC-3 regulates the expression of CXCL2 by enhancing NF-κB signaling
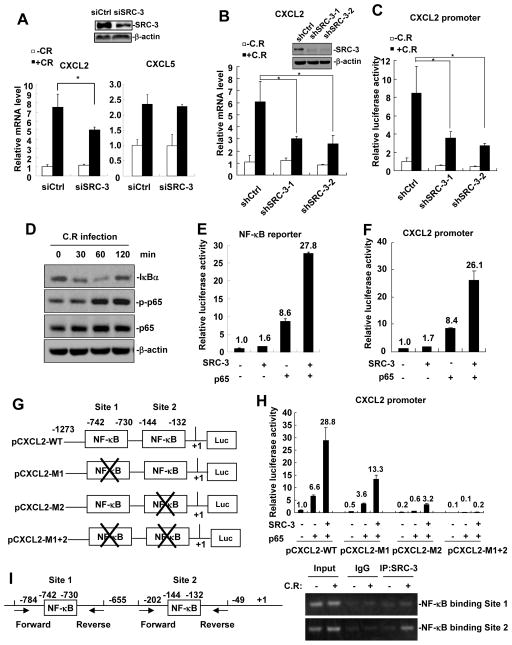
Since the expression of CXCL2 and CXCL5 was reduced in SRC-3−/− colonic epithelial cells, the expression of CXCL2 and CXCL5 may be regulated by SRC-3 in the colonic epithelial cells. To determine whether SRC-3 can regulate the expression of CXCL2 and CXCL5 in mouse colon cancer cell line CMT93, we transfected SRC-3 specific siRNA to knock down SRC-3 in CMT93 cells and examined the effects of SRC-3 knockdown on the expression of CXCL2 and CXCL5 in response to C. rodentium infection. As shown in Fig. 6A, C. rodentium infection could induce a 7-fold increase in CXCL2 expression in control CMT93 cells, whereas the induction of CXCL2 expression was significantly reduced in SRC-3-knockdown CMT93 cells. However, the induction of CXCL5 was not affected by SRC-3 knockdown (Fig. 6A). The impairment of CXCL2 induction by C. rodentium infection was also observed in two SRC-3-knockdown stable CMT93 cell lines (shSRC-3-1 and shSRC-3-2) (Fig. 6B). The number of C. rodentium attached to shCtrl and shSRC-3 CMT93 cells at 2 h post-infection was comparable (Fig. S2A), ruling out the possibility that difference in CXCL2 expression is due to reduced bacterial attachment to the SRC-3-knockdown cells.
Figure 6. SRC-3 regulates the expression of CXCL2 in CMT93 cells via enhancing NF-κB signaling.
A, siSRC-3-mediated knockdown of SRC-3 in CMT93 cells reduced C. rodentium-induced CXCL2 expression, but not CXCL5 expression. Data are presented as the means + SD (n = 3). *p<0.05. B, C. rodentium-induced CXCL2 expression was reduced in two SRC-3-knockdown stable CMT93 cell lines (shSRC-3-1 and shSRC-3-2). Data are presented as the means + SD (n = 3). *p<0.05. C, CXCL2 promoter activity was decreased in shSRC-3-1 and shSRC-3-2 CMT 93 cells infected with C. rodentium for 6 hours at MOI 200. Data are presented as the means + SD (n = 3). D, C. rodentium infection induced IκBα degradation and p65 phosphorylation. E, SRC-3 cooperated with p65 to enhance the activity of NF-κB reporter. F, SRC-3 cooperated with p65 to enhance the activity of CXCL2 reporter. G, Schematic representation of the CXCL2 promoter. Two NF-κB binding sites were shown. × denotes the mutation of NF-κB binding site. H, Mutation analysis of the role of two NFκ-B binding sites in NF-κB-mediated activation of CXCL2 promoter. CMT 93 cells were transfected with control or mutated CXCL2 promoter, SRC-3 expression plasmids and p65 expression plasmids. CXCL2 promoter activity was assayed 24 hours post transfection. Data are presented as the means + SD (n = 3). I, SRC-3 was recruited to the CXCL2 promoter upon C. rodentium infection. Left panel: Positions of the sub-fragments detected in ChIP assays. Right panel: ChIP assay showed that SRC-3 was recruited to the CXCL2 promoter upon C. rodentium infection.
Furthermore, we examined whether SRC-3 could promote CXCL2 transcription by transfecting CXCL2 promoter/reporter into control and SRC-3-knockdown CMT93 cells. As shown in Fig. 6C, C. rodentium infection induced CXCL2 promoter activity to 8-fold in control CMT93 cells, whereas the induction of CXCL2 promoter activity was significantly reduced in SRC-3-knockdown CMT93 cells. These results suggest that SRC-3 can promote CXCL2 expression at the transcriptional levels.
It has been reported that CXCL2 expression can be induced by NF-κB (34). Given that C. rodentium could attach to CMT93 cells (Fig S2B) to induce NF-κB activation as demonstrated by induction of IκBα degradation and phosphorylation of p65 subunit of NF-κB (Fig. 6D), and SRC-3 could interact with p65 (Fig. S3) to enhance p65-mediated activation of NF-κB reporter (Fig. 6E), we hypothesized that SRC-3 could enhance NF-κB-induced CXCL2 expression. To test this hypothesis, we transfected CMT93 cells with CXCL2 promoter/reporter together with the expression vectors of SRC-3, p65, and both SRC-3 and p65, respectively. Transfection of p65 could induce CXCL2 promoter activity to 8.4 fold in CMT93 cells (Fig. 6F), indicating that CXCL2 transcription can be activated by NF-κB as expected; although transfection of SRC-3 could only induce CXCL2 promoter activity to 1.7 fold, co-transfection of SRC-3 and p65 could dramatically induce CXCL2 promoter activity to 26 fold, suggesting that SRC-3 indeed can enhance NF-κB-induced CXCL2 expression.
There are two NF-κB binding sites on CXCL2 promoter (Fig. 6G). We performed mutation analysis to determine the role of these NF-κB binding sites in NF-κB-mediated activation of CXCL2 promoter. As shown in Fig. 6H, co-transfection of SRC-3 and p65 induced the activity of wild-type CXCL2 promoter (CXCL2-WT) to 28.8 fold, but only induced the promoter activity of Site 1-mutated (CXCL2-M1), Site 2-mutated (CXCL2-M1), and Site 1/2-double-mutated (CXCL2-M1+2) to 13.3, 3.2, and 0.2 fold, respectively, suggesting that both NF-κB binding sites contributes to the NF-κB-mediated activation of CXCL2 promoter, but NF-κB binding Site 2 plays a major role. We next determined whether SRC-3 can be recruited to the NF-κB binding sites on CXCL2 promoter after NF-κB activation by C. rodentium infection. As shown in Fig. 6I, SRC-3 mainly bound to NF-κB binding Site 2 after C. rodentium infection, suggesting that SRC-3 is mainly recruited to NF-κB binding site 2 to enhance NF-κB-mediated activation of CXCL2 promoter upon C. rodentium infection.
Discussion
In this study, we showed that SRC-3−/− mice exhibited increased intestinal C. rodentium load and more severe intestinal inflammation and pathology after oral infection with C. rodentium compared to wild-type mice. A/E pathogens including EPEC and C. rodentium use A/E lesions to colonize the intestine of host and use virulence factors such as EspF to induce epithelial cell death and then cause the epithelial barrier breakdown (35). In addition, C. rodentium infection can trigger host Th1 immune response to induce immune-driven injury of intestinal tissue (36–37). Therefore, more enteric bacteria in SRC-3−/− mice on day 14 after C. rodentium infection could cause more severe intestinal inflammation and more serious intestinal tissue damage.
C. rodentium infection can be divided into two phases: the early phase (before 8 days after C. rodentium infection) and the late phase (between 8 and 16 days after C. rodentium infection) (38). Neutrophils play an important role in immunity to infection not only by ingesting and killing invading pathogens, but also by formation of neutrophils extracellular traps (NETs) which is an alternative to death by necrosis or apoptosis (39–40). Depletion of neutrophils with specific Ly6G antibody results in increased bacteria burden, increased colon pathology and accelerated mortality at the early phase of C. rodentium infection, indicating that neutrophils are necessary for the early innate phase of the mucosal immune response (6–7). In this study, we found when intestinal bacterial burden was rapidly increased on day 7 after C. rodentium infection in both wild-type and SRC-3−/− mice, the recruitment of neutrophils into colonic lamina propria of SRC-3−/− mice was impaired compared to wild-type mice. Therefore, impaired intestinal neutrophil recruitment in SRC-3−/− mice at the early phase of C. rodentium infection may be responsible for the increased bacteria burden and severe tissue pathology on day 14 after C. rodentium infection. Interestingly, the recruitment of intestinal neutrophils in SRC-3−/− mice could be resumed on day 14 after C. rodentium infection, resulting in the infection control after that. In general, SRC-3 deficiency causes delayed intestinal neutrophil recruitment and corresponding delayed control of C. rodentium infection.
Neutrophil recruitment is driven by chemokines such as CXCL1, CXCL2, and CXCL5 (41). The lack of a proper expression of chemokines is often associated with impaired neutrophil recruitment. In this study, we found that the induction of CXCL2 and CXCL5 in the colons and colonic epithelial cells of SRC-3−/− mice was impaired at the early phase of C. rodentium infection, but resumed at the late phase of C. rodentium infection compared with wild-type mice. These results suggest that the expression of CXCL2 and CXCL5 is dependent on SRC-3 at the early phase of C. rodentium infection, but independent on SRC-3 at the late phase of C. rodentium infection. The expression pattern of CXCL2 and CXCL5 is correlated with the pattern of neutrophil recruitment in both wild-type and SRC-3−/− mice, indicating that CXCL2 and CXCL5 play essential roles in neutrophil recruitment at the early phase of C. rodentium infection.
During C. rodentium infection, colonic epithelial CXCL2 expression can be induced by NF-κB signaling activated by C. rodentium via pattern recognition receptors(24, 42), or by lymphotoxin via lymphotoxin beta receptor(7). It is known that SRC-3 can serve as a coactivator for NF-κB to enhance its transcriptional activity (43–44). Our results showed that SRC-3 could be recruited to the NF-κB binding site on NF-κB promoter and SRC-3 could cooperate with NF-κB to enhance CXCL2 transcription, suggesting that SRC-3 regulates CXCL2 expression at the transcriptional level at least in part through activating NF-κB signaling. Intriguingly, although induction of CXCL2 expression in the colon of SRC-3−/− mice was impaired on day 7 after C. rodentium infection, induction of CXCL2 expression in the colons of SRC-3−/− mice could be resumed on day 14 after C. rodentium infection, indicating that induction of CXCL2 expression in the colons of SRC-3−/− mice on day 14 after C. rodentium infection is SRC-3-independent. The possible explanations are that more severe inflammation and tissue damage in the colons of SRC-3−/− mice on day 14 after C. rodentium infection may induce stronger NF-κB signaling, or induction of other SRC family members such as SRC-1(Fig. S4A), which can also cooperate with NF-κB to enhance CXCL2 expression (Fig. S4B&C), may compensate for SRC-3 function.
Although the induction of CXCL5 in the colonic epithelial cells of SRC-3−/− mice was impaired at the early phase of C. rodentium infection, siSRC-3-mediated knockdown of SRC-3 did not reduce C. rodentium-induced CXCL5 expression in mouse colon cancer cell line CMT93. The inconsistency could be explained by several possibilities. 1) To some extent, the characteristics of colonic epithelial cells are different from CMT93 cells, so the effect of SRC-3 on C. rodentium-induced CXCL5 expression in the colonic epithelial cells can not be mimicked in CMT93 cells. 2) Some factors needed for SRC-3 to enhance C. rodentium-induced CXCL5 expression in the colonic epithelial cells in vivo are absent in vitro. 3) The siSRC-3-mediated knockdown of SRC-3 in CMT93 cells is not sufficient to reduce C. rodentium-induced CXCL5 expression, although it is sufficient to reduce C. rodentium-induced CXCL2 expression. Further studies are needed to reveal the mechanism by which SRC-3 deficiency affects C. rodentium-induced CXCL5 expression in vivo.
Although the significant difference in the expression of colonic CXCL2 and CXCL5 was observed when compared them in wild-type and SRC-3−/− mice at days 7 and 14 after C. rodentium infection, the expression of colonic CXCR2 was comparable in wild-type and SRC-3−/− mice. In addition to neutrophils, it has been shown that CXCR2 is also expressed in T lymphocytes and mast cells (45), which are also required for C. rodentium clearance (46–47). T cells such as CD4+ T cells and Th22 cells play a critical role in C. rodentium clearance at the late-phase infection of C. rodentium (48). In the light of these studies, it is interesting to determine whether differential expression of colonic CXCL2 and CXCL5 in wild-type and SRC-3−/− mice affects the function of T cells at the late-phase infection of C. rodentium.
We previously reported that SRC-3−/− mice exhibited more susceptible to E. coli-induced septic peritonitis due to an uncontrolled overwhelming inflammation and a defect in bacterial clearance (20). The defect in bacterial clearance is at least in part due to the decreased E. coli phagocytosis by SRC-3−/− macrophages and increased SRC-3−/− macrophage apoptosis, indicating that SRC-3 exerts its anti-bacterial function by enhancing bacterial phagocytosis and macrophage survival during peritoneal E. coli infection. In this study, we showed that SRC-3 exerted its anti-bacterial function by recruiting neutrophils via upregulation of CXCL2 expression during intestinal C. rodentium infection. Therefore, our studies demonstrate that SRC-3 can exert its anti-bacterial function via different mechanisms in face of different bacterial infection, highlighting the important role of SRC-3 in host defense again bacterial infection.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 31170819 to C.Y.), National Basic Research Program of China (973 Program, No. 2015CB553800 to C.Y.), and Institute Merieux (No.17102012 to C.Y.). J.X. was partially supported by grants CA112403 and CA193455 from National Institutes of Health, USA.
Abbreviations used in this paper
- CFU
colony formation unit
- ChIP
chromatin immunoprecipitation
- SRC-3
steroid receptor coactivator 3
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Wales AD, Woodward MJ, Pearson GR. Attaching-effacing bacteria in animals. J Comp Pathol. 2005;132:1–26. doi: 10.1016/j.jcpa.2004.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 3.Mead PS, Griffin PM. Escherichia coli O157:H7. Lancet. 1998;352:1207–1212. doi: 10.1016/S0140-6736(98)01267-7. [DOI] [PubMed] [Google Scholar]
- 4.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mundy R, MacDonald TT, Dougan G, Frankel G, Wiles S. Citrobacter rodentium of mice and man. Cell Microbiol. 2005;7:1697–1706. doi: 10.1111/j.1462-5822.2005.00625.x. [DOI] [PubMed] [Google Scholar]
- 6.Lebeis SL, Bommarius B, Parkos CA, Sherman MA, Kalman D. TLR signaling mediated by MyD88 is required for a protective innate immune response by neutrophils to Citrobacter rodentium. J Immunol. 2007;179:566–577. doi: 10.4049/jimmunol.179.1.566. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Koroleva EP, Kruglov AA, Kuprash DV, Nedospasov SA, Fu YX, Tumanov AV. Lymphotoxin beta receptor signaling in intestinal epithelial cells orchestrates innate immune responses against mucosal bacterial infection. Immunity. 2010;32:403–413. doi: 10.1016/j.immuni.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim YG, Kamada N, Shaw MH, Warner N, Chen GY, Franchi L, Nunez G. The Nod2 sensor promotes intestinal pathogen eradication via the chemokine CCL2-dependent recruitment of inflammatory monocytes. Immunity. 2011;34:769–780. doi: 10.1016/j.immuni.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schreiber HA, Loschko J, Karssemeijer RA, Escolano A, Meredith MM, Mucida D, Guermonprez P, Nussenzweig MC. Intestinal monocytes and macrophages are required for T cell polarization in response to Citrobacter rodentium. J Exp Med. 2013;210:2025–2039. doi: 10.1084/jem.20130903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maaser C, Housley MP, Iimura M, Smith JR, Vallance BA, Finlay BB, Schreiber JR, Varki NM, Kagnoff MF, Eckmann L. Clearance of Citrobacter rodentium requires B cells but not secretory immunoglobulin A (IgA) or IgM antibodies. Infect Immun. 2004;72:3315–3324. doi: 10.1128/IAI.72.6.3315-3324.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bry L, Brigl M, Brenner MB. CD4+-T-cell effector functions and costimulatory requirements essential for surviving mucosal infection with Citrobacter rodentium. Infect Immun. 2006;74:673–681. doi: 10.1128/IAI.74.1.673-681.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang YJ, Otsuka M, van den Berg A, Hong L, Huang Z, Wu X, Zhang DW, Vallance BA, Tobias PS, Han J. Epithelial p38alpha controls immune cell recruitment in the colonic mucosa. PLoS Pathog. 2010;6:e1000934. doi: 10.1371/journal.ppat.1000934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 2013;145:396–406. doi: 10.1053/j.gastro.2013.04.056. [DOI] [PubMed] [Google Scholar]
- 14.Xue X, Ramakrishnan S, Anderson E, Taylor M, Zimmermann EM, Spence JR, Huang S, Greenson JK, Shah YM. Endothelial PAS domain protein 1 activates the inflammatory response in the intestinal epithelium to promote colitis in mice. Gastroenterology. 2013;145:831–841. doi: 10.1053/j.gastro.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iimura M, Gallo RL, Hase K, Miyamoto Y, Eckmann L, Kagnoff MF. Cathelicidin mediates innate intestinal defense against colonization with epithelial adherent bacterial pathogens. J Immunol. 2005;174:4901–4907. doi: 10.4049/jimmunol.174.8.4901. [DOI] [PubMed] [Google Scholar]
- 16.Bergstrom KS, Kissoon-Singh V, Gibson DL, Ma C, Montero M, Sham HP, Ryz N, Huang T, Velcich A, Finlay BB, Chadee K, Vallance BA. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 2010;6:e1000902. doi: 10.1371/journal.ppat.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Wu RC, O’Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9:615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Liu YH, Ou S, Dai XM, Wang JP, Su YP. Steroid receptor coactivator-3 differentially regulates the inflammatory response in peritoneal macrophages. Mol Med Rep. 2012;5:1099–1105. doi: 10.3892/mmr.2012.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu C, York B, Wang S, Feng Q, Xu J, O’Malley BW. An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response. Mol Cell. 2007;25:765–778. doi: 10.1016/j.molcel.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Q, Chen T, Xu Y, Zhu J, Jiang Y, Zhao Y, Xu J, Yu C. Steroid receptor coactivator 3 is required for clearing bacteria and repressing inflammatory response in Escherichia coli-induced septic peritonitis. J Immunol. 2010;185:5444–5452. doi: 10.4049/jimmunol.0903802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, Liao L, Ning G, Yoshida-Komiya H, Deng C, O’Malley BW. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc Natl Acad Sci U S A. 2000;97:6379–6384. doi: 10.1073/pnas.120166297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegmund B, Lehr HA, Fantuzzi G, Dinarello CA. IL-1 beta -converting enzyme (caspase-1) in intestinal inflammation. Proc Natl Acad Sci U S A. 2001;98:13249–13254. doi: 10.1073/pnas.231473998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibson DL, Ma C, Rosenberger CM, Bergstrom KS, Valdez Y, Huang JT, Khan MA, Vallance BA. Toll-like receptor 2 plays a critical role in maintaining mucosal integrity during Citrobacter rodentium-induced colitis. Cell Microbiol. 2008;10:388–403. doi: 10.1111/j.1462-5822.2007.01052.x. [DOI] [PubMed] [Google Scholar]
- 24.Khan MA, Ma C, Knodler LA, Valdez Y, Rosenberger CM, Deng W, Finlay BB, Vallance BA. Toll-like receptor 4 contributes to colitis development but not to host defense during Citrobacter rodentium infection in mice. Infect Immun. 2006;74:2522–2536. doi: 10.1128/IAI.74.5.2522-2536.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunay IR, Damatta RA, Fux B, Presti R, Greco S, Colonna M, Sibley LD. Gr1+ inflammatory monocytes are required for mucosal resistance to the pathogen Toxoplasma gondii. Immunity. 2008;29:306–317. doi: 10.1016/j.immuni.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee KW, Lee Y, Kwon HJ, Kim DS. Sp1-associated activation of macrophage inflammatory protein-2 promoter by CpG-oligodeoxynucleotide and lipopolysaccharide. Cell Mol Life Sci. 2005;62:188–198. doi: 10.1007/s00018-004-4399-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Research. 2004;32(14):e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 29.Ishigame H, Kakuta S, Nagai T, Kadoki M, Nambu A, Komiyama Y, Fujikado N, Tanahashi Y, Akitsu A, Kotaki H, Sudo K, Nakae S, Sasakawa C, Iwakura Y. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity. 2009;30:108–119. doi: 10.1016/j.immuni.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 30.Tacke F, Randolph GJ. Migratory fate and differentiation of blood monocyte subsets. Immunobiology. 2006;211:609–618. doi: 10.1016/j.imbio.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 31.Johnston B, Butcher EC. Chemokines in rapid leukocyte adhesion triggering and migration. Semin Immunol. 2002;14:83–92. doi: 10.1006/smim.2001.0345. [DOI] [PubMed] [Google Scholar]
- 32.Kinnebrew MA, Pamer EG. Innate immune signaling in defense against intestinal microbes. Immunol Rev. 2012;245:113–131. doi: 10.1111/j.1600-065X.2011.01081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spehlmann ME, Dann SM, Hruz P, Hanson E, McCole DF, Eckmann L. CXCR2-dependent mucosal neutrophil influx protects against colitis-associated diarrhea caused by an attaching/effacing lesion-forming bacterial pathogen. J Immunol. 2009;183:3332–3343. doi: 10.4049/jimmunol.0900600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim DS, Han JH, Kwon HJ. NF-kappaB and c-Jun-dependent regulation of macrophage inflammatory protein-2 gene expression in response to lipopolysaccharide in RAW 264.7 cells. Mol Immunol. 2003;40:633–643. doi: 10.1016/j.molimm.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 35.McNamara BP, Koutsouris A, O’Connell CB, Nougayrede JP, Donnenberg MS, Hecht G. Translocated EspF protein from enteropathogenic Escherichia coli disrupts host intestinal barrier function. J Clin Invest. 2001;107:621–629. doi: 10.1172/JCI11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Higgins LM, Frankel G, Douce G, Dougan G, MacDonald TT. Citrobacter rodentium infection in mice elicits a mucosal Th1 cytokine response and lesions similar to those in murine inflammatory bowel disease. Infect Immun. 1999;67:3031–3039. doi: 10.1128/iai.67.6.3031-3039.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruewer M, Luegering A, Kucharzik T, Parkos CA, Madara JL, Hopkins AM, Nusrat A. Proinflammatory cytokines disrupt epithelial barrier function by apoptosis-independent mechanisms. J Immunol. 2003;171:6164–6172. doi: 10.4049/jimmunol.171.11.6164. [DOI] [PubMed] [Google Scholar]
- 38.Honda K. IL-22 from T cells: better late than never. Immunity. 2012;37:952–954. doi: 10.1016/j.immuni.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 39.Appelberg R. Neutrophils and intracellular pathogens: beyond phagocytosis and killing. Trends Microbiol. 2007;15:87–92. doi: 10.1016/j.tim.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 40.Borregaard N. Neutrophils, from marrow to microbes. Immunity. 2010;33:657–670. doi: 10.1016/j.immuni.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 41.De Filippo K, Dudeck A, Hasenberg M, Nye E, van Rooijen N, Hartmann K, Gunzer M, Roers A, Hogg N. Mast cell and macrophage chemokines CXCL1/CXCL2 control the early stage of neutrophil recruitment during tissue inflammation. Blood. 2013;121:4930–4937. doi: 10.1182/blood-2013-02-486217. [DOI] [PubMed] [Google Scholar]
- 42.De Filippo K, Henderson RB, Laschinger M, Hogg N. Neutrophil chemokines KC and macrophage-inflammatory protein-2 are newly synthesized by tissue macrophages using distinct TLR signaling pathways. J Immunol. 2008;180:4308–4315. doi: 10.4049/jimmunol.180.6.4308. [DOI] [PubMed] [Google Scholar]
- 43.Werbajh S, Nojek I, Lanz R, Costas MA. RAC-3 is a NF-kappa B coactivator. FEBS Lett. 2000;485:195–199. doi: 10.1016/s0014-5793(00)02223-7. [DOI] [PubMed] [Google Scholar]
- 44.Wu RC, Qin J, Hashimoto Y, Wong J, Xu J, Tsai SY, Tsai MJ, O’Malley BW. Regulation of SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1) Coactivator activity by I kappa B kinase. Mol Cell Biol. 2002;22:3549–3561. doi: 10.1128/MCB.22.10.3549-3561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lippert U, Zachmann K, Henz BM, Neumann C. Human T lymphocytes and mast cells differentially express and regulate extra- and intracellular CXCR1 and CXCR2. Exp Dermatol. 2004;13:520–525. doi: 10.1111/j.0906-6705.2004.00182.x. [DOI] [PubMed] [Google Scholar]
- 46.Simmons CP, Clare S, Ghaem-Maghami M, Uren TK, Rankin J, Huett A, Goldin R, Lewis DJ, MacDonald TT, Strugnell RA, Frankel G, Dougan G. Central role for B lymphocytes and CD4+ T cells in immunity to infection by the attaching and effacing pathogen Citrobacter rodentium. Infect Immun. 2003;71:5077–5086. doi: 10.1128/IAI.71.9.5077-5086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei OL, Hilliard A, Kalman D, Sherman M. Mast cells limit systemic bacterial dissemination but not colitis in response to Citrobacter rodentium. Infect Immun. 2005;73:1978–1985. doi: 10.1128/IAI.73.4.1978-1985.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basu R, O’Quinn DB, Silberger DJ, Schoeb TR, Fouser L, Ouyang W, Hatton RD, Weaver CT. Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity. 2012;37:1061–1075. doi: 10.1016/j.immuni.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.