Abstract
This short “how to” article describes a plot I find useful for assessing fMRI data quality. I discuss the reasoning behind the plot and how it is constructed. I create the plot in scans from several publicly available datasets to illustrate different kinds of fMRI signal variance, ranging from thermal noise to motion artifacts to respiratory-related signals. I also show how the plot can be used to understand the variance removed during denoising. Code to make the plot is provided with the article, and supplemental movies show plots for hundreds of additional subjects.
The plot
For this special issue I was asked to write an unusual kind of paper: a practical “how to” guide that described how I prefer to assess functional magnetic resonance imaging (fMRI) data quality. This paper was not to be a review of denoising techniques but instead a paper more about the process of assessing data: how one decides if a technique has improved the data, and how one decides whether the data are “too noisy” or “good enough”.
I am going to use this opportunity to write a short paper about a single plot. The central element of the plot is a 2-dimensional heatmap of relevant timeseries within a scan, a “flattened out” scan, with time on the x-axis and voxels or regions of interest defining the y-axis. Above the heatmap are some traces conveying information about likely sources and timings of artifacts and unwanted signals. This plot is the single most informative summary of the quality of a scan, and the kinds of variance the scan contains, that I know how to make. I have at different times called these pictures “grayplots”, “staticplots”, and “carpetplots” for reasons that will shortly be clear. Perhaps “voxplot” would be better. Here I will just refer to it as The Plot. I have put a small “demo” on my website1 that includes sample files and code to aid readers who wish to make their own versions of the plot.
The plot works by putting indications of artifact in close spatial proximity to signals of interest so that the eye notes temporal coincidence. I will first discuss some indications of artifacts in scans, and then construction of the plot. Then I will show several versions of the plot in different fMRI datasets that illustrate different kinds of unwanted signals in fMRI data, and I will also show an instance of plots before and after denoising. To conclude I will discuss how examining plots guides my understanding of fMRI variance and signal denoising. In this article I develop the plot and the kinds of variance it illustrates gradually, but readers wishing to gain a quick appreciation for the range of variance that occurs in real data may immediately view plots in several hundred subjects at the website mentioned above.
Indicators of unwanted fMRI signals
Signals in T2*-weighted images have a variety of sources beyond “neural” signals, including thermal noise, head motion, hardware artifacts, respiratory and cardiac cycles, and modulation of arterial pCO2 due to ventilation changes (this list includes major known signal sources but is not exhaustive, see (Murphy et al., 2013)). In task fMRI the timing structure of the task allows the “neural” signal to be extracted using general linear models (with caveats for task-associated artifacts, (Barch et al., 1999; Bullmore et al., 1999)). But in resting state fMRI there is no timing structure and the procedure is reversed: rather than extract the signal, an investigator must try to extract the noise and hope that what remains is “neural” signal.
For a given fMRI scan, several properties can give spatial or temporal indications that unwanted signal may be present in gray matter voxels. First, signals in the white matter and ventricles are typically interpreted as noise. Second, head motion can be measured in any fMRI scan from the scan itself, and head motion is a major cause of artifacts. Traces reflecting both head position (x, y, z, pitch, roll, yaw) and head motion (the first derivative of the position estimates) are useful to index the potential for such artifacts. Third, though these traces are often not available, both respiratory belt traces and pulse oximeter traces can provide useful information about cardiac and respiratory rhythms as well as expected longer-term modulation of arterial pCO2 (and thus blood oxygen level dependent (BOLD) signal) due to changes in ventilation (Birn et al., 2006; Birn et al., 2008; Glover et al., 2000; Wise et al., 2004). Most model-based fMRI denoising techniques utilize some combination of cardiac, respiratory, motion, and nuisance compartment signals (for review, see (Murphy et al., 2013; Power et al., 2015)).
All of the above measures give indications of artifact by temporal coincidence, either immediately (e.g., in the case of head motion) or with some delay (e.g., in the case of ventilatory changes altering pCO2 in the brain). Other criteria can help identify noise, such as the spatial or temporal characteristics of certain signals (e.g., used to bin signals in independent components analysis (ICA)), or the decay properties of certain signals (e.g., in multi-echo imaging). These criteria can be incorporated into the plot in various ways but here I want to focus on simple versions of the plot and will not bring in indicators beyond those listed in the previous paragraph.
Constructing the plot
A basic version of the plot, featured in the next six figures, is constructed as follows. FreeSurfer is used to segment a high-resolution T1-weighted image that is in register with the fMRI data. The FreeSurfer segmentation is resampled to the fMRI resolution, and binary masks are created of major gray matter compartments (cortex, cerebellum, subcortical nuclei), of the white matter, and of the ventricles. I also erode the nuisance compartments several times to derive masks of superficial, deeper, and deepest layers of these compartments (with depth referring to distance from any gray matter, not from the cranium). A set of these masks are shown at the bottom of Figure 1. When I construct the heatmap, I use these masks to sample signals from a scan in the order shown by the color bars at the sides of the heatmaps. Signals of interest (in gray matter) are divided from nuisance signals (in white matter and ventricles) by a thick line, and nuisance signals are ordered by their proximity to gray matter (superficial, deeper, deepest). In all figures the heatmaps show all voxels in the brain, giving a comprehensive picture of the scan in time (and to some extent space due to compartment ordering). By using other partitions and orderings of signals an investigator can represent directionality, functional organization, or other spatial features in the heatmap. I will use only the single just-described ordering in this article.
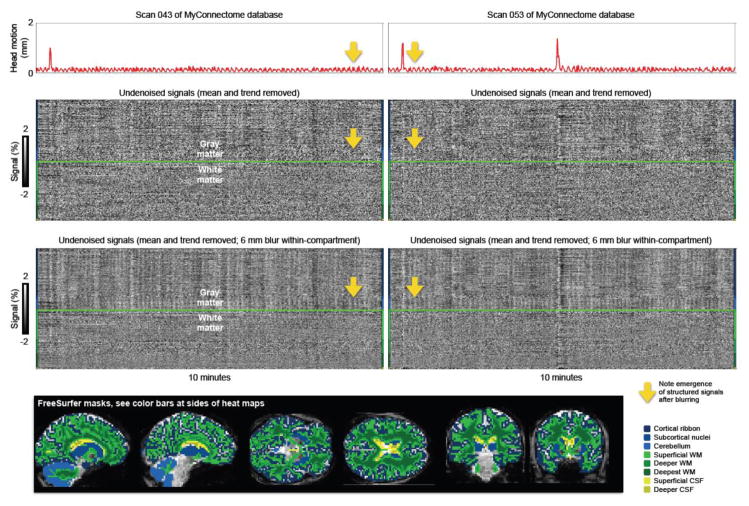
Figure 1. Thermal noise can obscure structured signals.
Two scans of the MyConnectome database are shown with motion traces at top, undenoised signals at middle, and blurred but otherwise undenoised signals at the bottom. These data are 2.4 × 2.4 × 2.4 mm voxels acquired every 1.16 seconds. The blurring occurs within brain compartment masks defined by FreeSurfer so that signals are not combined across brain compartments. Blurring averages away thermal noise, revealing structured signals. Note that periodic head motion is present and that similarly periodic signals become visible in the gray matter upon reduction of thermal noise (yellow arrows). See Online Movie 1 for such plots of all MyConnectome scans.
Above the heatmap I place one or more traces. Here I will use only one or two traces at a time, at most three: I will always show head motion in red, respiration in blue, and heart rate in green. Head motion is calculated as Framewise Displacement, following (Power et al., 2012). The respiratory traces are just respiratory belt records. Heart rate is derived from the time between peaks in pulse oximeter traces. The utility of the plot is constrained by the goodness of the information carried by the traces, and each of the traces mentioned above can vary widely in quality and utility depending on how the trace is obtained and handled by the investigator. I will talk below about obtaining high-quality and informative traces.
The scans shown in the plots
The scans shown are publicly available or previously published resting state fMRI scans from the Autism Brain Imaging Data Exchange consortium2 (ABIDE), from Alex Martin’s group at the National Institutes for Health (NIH) (Gotts et al., 2012), from the Human Connectome Project (HCP) (Glasser et al., 2013), and from the MyConnectome Project (Poldrack et al., 2015). These scans are drawn from about 1500 scans that I routinely use for various purposes, and I am using multiple sites simply so that multiple kinds of data are shown with different properties. About 30 scans are shown in figures. These scans were selected by me because they nicely illustrate something I would like to show. But the effects I will illustrate are neither subtle nor rare. Many other scans could have been chosen instead and I put a few hundred additional plots in some online movies (at the website mentioned above) just to show that the phenomena of interest can be seen in many scans.
I perform no denoising and only minimal image processing. My aim is primarily to illustrate signal variance “as it is” when it comes off the scanner. The HCP and MyConnectome data were provided already preprocessed and in register with T1-weighted images (the HCP data came in undenoised and denoised formats via pre-FIX-ICA and post-FIX-ICA scans, the MyConnectome data were preprocessed exactly as described in (Laumann et al., 2015)). The other fMRI datasets underwent standard preprocessing including slice time correction, motion correction, and registration to a target atlas, as described in (Power, submittedA). For the heatmaps, mean and linear trend terms are removed from each voxel’s timeseries in each run. As I discuss next, I also perform within-compartment-mask blurring to reduce thermal noise, using AFNI’s 3dBlurInMask command with a 6 mm FWHM Gaussian kernel and the FreeSurfer-derived masks mentioned above. Plots without and with such blurring are provided in the online movies mentioned below, illustrating the minor or major role thermal noise has in plots generated from various kinds of scans.
Recognizing noise in fMRI scans
I would like to use the plot to build a visual vocabulary of the variance present in fMRI scans, focusing on thermal noise, motion artifacts, hardware artifacts, and certain physiological signals. These are all prominent sources of variance that can differ greatly across subjects and sites.
Thermal noise, unlike all other signal sources listed above, has no spatial or temporal structure. In the plot, thermal noise in fMRI signals looks like “static” in the heatmap. If structured signals are small relative to thermal noise, “static” can be a major impression made by the plot. I often get this impression from high-resolution fMRI data such as scans from the MyConnectome or HCP databases, which use 2.4 mm and 2.0 mm isotropic voxel acquisitions, respectively. If spatial blurring is performed the thermal noise averages away to reveal structured signals, as shown in Figure 1. Note that the “static” in heat maps will be an error term in statistical models of the signals – it is random and cannot be modeled. To facilitate an appreciation of the amount of thermal noise in fMRI scans, in the online movies mentioned below, plots are always presented for scans both before and after spatial blurring. The movies illustrate hundreds of scans from several sites ranging from relatively high resolution HCP data (much thermal noise) to relatively low-resolution scans in the ABIDE database (3 mm isotropic (or larger) voxels at acquisition, with relatively little thermal noise).
Motion artifacts are quite visible in the plot. In fact, I first began making the plot to try to better see what motion was doing to my fMRI data (I put one plot in (Power et al., 2012), as Supplemental Figure 4; within two years the plot had become so integral to my approach to assessing data that I placed 120 plots in the Supplemental Cohort Illustration of (Power et al., 2014)). I would like to make 3 points about motion in Figure 1.
First, 3 large motions are seen (1 in the first scan, 2 in the second), each with concomitant disruptions of signals across gray and white matter voxels. Such motion artifacts have been comprehensively described in earlier work (e.g., (Power et al., 2014; Satterthwaite et al., 2013; Yan et al., 2013)) and are present in many figures.
Second, a feature of considerable interest is that small periodic motions are seen throughout the MyConnectome scans, with approximately 8 peaks per minute (~0.13 Hz), a frequency consistent with respiration (no respiratory traces exist for these scans). Such small periodic movements are seen in all MyConnectome fMRI scans examined by the author (N = 84 sessions), which were acquired in a single subject over a year-long period (to see such movements in all scans, watch Online Movie 1, which shows a version of Figure 1 for each MyConnectome scan). These data suggest that head motion related to breathing may play a major role in establishing typical amounts of motion in a subject. These periodic motions are not apparent in datasets with multi-second intervals between signal acquisitions (see plots that follow in other datasets), possibly because such motion is aliased.
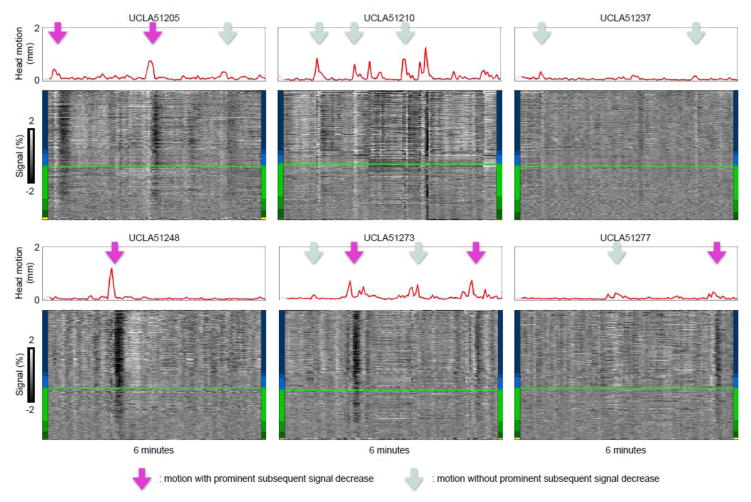
Third, note that the first two large motions in Figure 1 have no visible long-term sequellae, but the third large motion is followed by a signal decrease throughout the brain (the vertical black band) that lasts over 20 seconds. Motion often occurs without the subsequent decreases, and decreases are also seen without preceding motion. Intermittent pairing of motion and subsequent signal decreases is a feature of all datasets I have ever examined. Figures 2, S1, and S2 show such pairings in 3 sites of the ABIDE database (or see Online Movie 2, which shows plots for all subjects from these ABIDE sites). The different timescales of the sometimes-paired phenomena suggest that two different mechanisms may be at work, perhaps caused by the same original event when they are paired.
Figure 2. Motion is sometimes paired with prolonged signal decreases.
Scans from six subjects of the University of California at Los Angeles (UCLA) site of the ABIDE database are shown following the conventions of Figure 1. These data are 3 × 3 × 4 mm voxels acquired every 3 seconds. See Figure S1 and S2 for similar examples in the Utah School of Medicine (USM) and New York University (NYU) ABIDE scans, or Online Movie 2 for plots of all subjects from these ABIDE sites. Purple arrows denote motion with prominent signal decreases afterwards, light blue arrows denote motion without such prominent signal decreases.
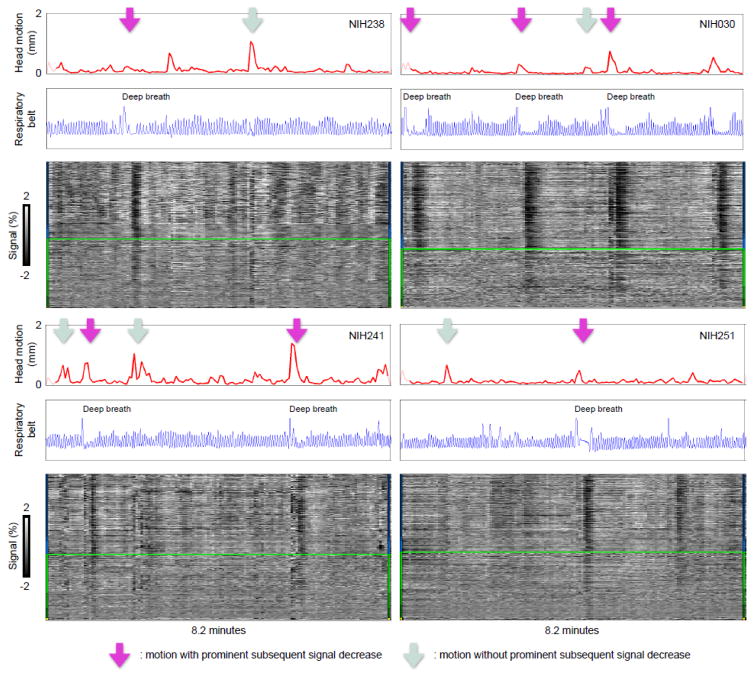
By adding respiratory traces to the plot we gain new insight and identify a new kind of variance: Figure 3 shows in NIH scans that deep inspirations typically mark motions paired with subsequent decreases, but seem not to mark motions unpaired with subsequent decreases. In other words, instances of motion paired with subsequent signal decreases likely represent deep breaths – sighs and yawns – which cause both immediate motion artifacts as the head moves and then prolonged pCO2-related signal changes due to a transient change in ventilation. fMRI studies that involve instructed deep breaths report similar waveforms: modest initial elevations peaking ~5 seconds after inspiration followed by larger signal decreases with troughs ~15–20 seconds after inspiration (Birn et al., 2008; Chang and Glover, 2009).
Figure 3. Deep breaths iscriminate motions with subsequent signal decreases from motions without subsequent signal decreases.
Scans from 4 subjects of the NIH cohort are shown following the conventions of Figure 2. Respiratory belt traces are in arbitrary units. These data are 1.7 × 1.7 × 3 mm voxels acquired every 3.5 seconds. As in Figure 2, purple arrows denote motion with prominent signal decreases afterwards, light blue arrows denote motion without such prominent signal decreases.
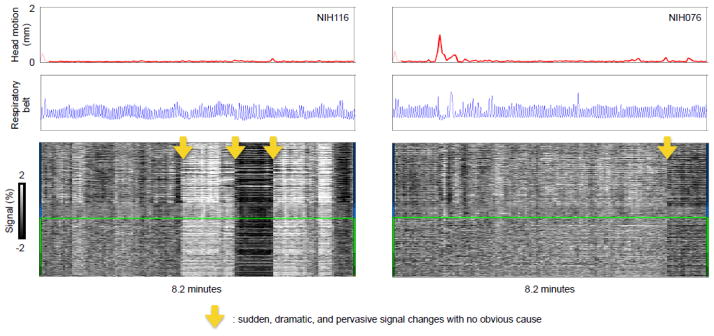
The final element of variance that will be illustrated here is shown in Figure 4: highly unusual signal changes that are sudden, that pervade the brain, and that are large in magnitude, with no obvious cause. These are likely hardware-related artifacts (e.g., these NIH data were collected using a head coil that occasionally malfunctioned, described in (Jo et al., 2010)). The brains in these scans look unremarkable and the scans are of high quality with respect to motion and physiological records. Only the temporal aspects of the signals betray an abnormality.
Figure 4. Abnormalities without obvious causes may be hardware-related artifacts.
Scans from 2 subjects of the NIH cohort are shown following the conventions of Figure 3, illustrating sudden, dramatic, and pervasive signal changes with no obvious cause (yellow arrows).
Considerations about which traces to put in the plot
Many kinds of traces can be incorporated into the plot, but the utility of these traces lies 1) in their ability to provide new information to contextualize the signals present in the heat map, and 2) the goodness of the information in the trace. Beyond the traces already illustrated, a large variety of other traces could be used, for example skin conductance measures, task block timings, or data quality measures. I offer comments on three kinds of traces: quality traces, motion traces, and physiological traces.
A variety of “quality” traces can be placed in the plot, such as DVARS (the RMS value across the brain of differentiated timeseries (Smyser et al., 2010)), the standard deviation of the fMRI image, or mean compartment signals such as white matter signals or the global signal. These traces will tend to reflect what is already apparent in the heatmap since one can visually see the magnitude, extent, and uniformity of signal changes across the brain.
Motion traces give new information not implicit in the heatmap. But motion traces can vary in utility. For example, if motion is calculated in a scan before and then after temporal interpolation (e.g., despiking or slice time correction), large discrepancies in the motion traces are produced at times of motion simply because temporal interpolation logically tends to reduce brain motion. If the truth of this statement is not apparent, consider the following thought experiment. A small Lego, occupying a single voxel, is at location A for an entire phantom scan except for a single volume X in the middle of the scan during which the Lego is temporarily moved to location B. At voxel A, signal intensity will be high at all times except during volume X, and conversely the signal at voxel B will be low at all times other than volume X. During temporal interpolation, at voxel A both despiking and slice time correction routines will draw high signal values from times before and after volume X and the post-interpolation value will therefore be raised from the original low value to some higher value. Complementary signal decreases will occur at voxel B. The net effect is to move the apparent position of the Lego in the image at volume X. The point is that if a voxel represents a point in space, and not necessarily a fixed location in the brain, then temporal interpolation will tend to dampen movement in and out of the voxel’s “field of view” because such movement involves changes in image intensity as different tissues enter and exit the voxel. With regard to the plot the point is that it becomes more difficult to identify motions and their accompanying signal disruptions when motion traces derived after temporal interpolation are used. These points, and others pertaining to motion estimation, are discussed in (Power, submittedA).
Physiological graces give new information not implicit in the heatmap, but the quality of these traces varies widely. In the spirit of a “how to” article I would like to discuss the acquisition of these traces in some detail because there are important considerations regarding their quality that may not be evident to readers without personal experience in the measures. I have dealt with physiological records from several sites, probably around 200 sets of records in total. A large fraction of physiological records are unusable due to artifacts or poor signals, perhaps 10–25% of records in my experience. This loss is unfortunate because 1) it reduces data that qualifies for analysis and 2) a few double-checks prior to starting the scan might have improved or salvaged some of those records.
Physiological signals in a compliant subject should be smooth and large-amplitude. When placing a respiratory belt, watch the subject’s abdomen and put the strap snugly but not tightly around the part that moves up and down most as they lie on the scanner bed. This location in thinner subjects is often near the belly button, and in heavier subjects it may be higher towards the chest. The trace should look something like a sine wave if the subject is breathing regularly, similar to what is shown in Figure S3. The trace should not be spiky, flat, or jerky – it should smoothly reflect respiratory patterns. The pulse oximeter should be fastened on a digit that can remain still during the scan and should be wrapped to shield the detector from ambient light (a bedsheet or gown will suffice). Within seconds after the hand is at rest a regular, smooth waveform should be present, similar to what is shown in Figure S3. If the trace is spiky, flat, or jerky, try repositioning the device – the beam should be centered on a fingernail. Some scanners such as the Siemens Prisma display respiratory and pulse oximeter waveforms on a panel above the bore, making it easy to know the quality of the data before the subject is placed in the bore.
Some signal irregularities during a scan are unavoidable. It is normal for respiratory traces to hit ceiling during very large breaths (look closely at the deep breaths in Figure 3). Likewise it is normal for there to be temporary disruptions in a pulse oximeter trace when the subject moves their finger. But if pulse oximeter records from much of a scan are noisy the device probably needs to be better-secured on the finger (tape can be useful).
The need for smooth, large-amplitude signals stems from the fact that peak-finding underlies many uses of physiological traces. From pulse oximeter traces, heart rate is derived from the times between peaks, as is phase of the cardiac cycle. And accessory measures such as peak amplitude depend upon proper recognition of the main peaks. Incorrect peak detection can create large, spurious changes in heart rate and other measures (see Figure S4). Incorrect peak detection often occurs during periods of signal artifacts, and it can even happen during “good” signal periods. In my experience, large incorrect outliers occur at least once and often multiple times in approximately 25–50% of traces I examine. If not recognized and corrected, these large deviations can strongly influence regressions or other procedures that depend on these measures. In my view it is inadvisable to use pulse oximeter records without personally assessing the peaks and the resulting derived measures (like heart rate). I always visually assess a trace and its derived measures trace before using them; code to facilitate this process is in one of the “demos” on my website. Because the pulse oximeter waveform is so stereotyped and regular (I can recall only two “true” irregularities in cardiac rhythms in my traces) it is relatively easy to assess the validity of peaks and the pulse oximeter trace. Respiratory waveforms are sometimes regular but also sometimes very irregular and I generally do not personally intervene in these traces other than to discard entire traces if it is obvious that the signal is noisy or lost. As will become apparent below, it is sensible to use vetted heart rate traces to help assess the goodness of respiratory traces since there are often obvious similarities between the traces.
Respiratory and cardiac traces are used in several ways for denoising. Uses range from removing immediate effects of respiratory and cardiac cycles via RETROICOR (Glover et al., 2000) to removing immediate effects of the amount of respired air (via measures of respiration volume per time (RVT), (Birn et al., 2006) or respiratory variance (RV) (Chang and Glover, 2009)), to removing delayed effects of heart rate and respiration (Birn et al., 2008; Chang et al., 2009)). Sometimes in publications describing these techniques it is mentioned that cardiac and respiratory records are somewhat collinear, but I have been surprised at the extent of correspondence between these traces in my plots, and I wish to highlight the degree to which these measures can be intertwined.
In many subjects there are strong interrelationships between heart rate and respiration, between respiration and motion, and between motion and heart rate. Several instances of strong relationships between cardiac rhythms and respiratory patterns are shown in Figure 5. This effect is seen in many subjects (see Figure S4 for 3 more examples, or Online Movie 3 for plots in all NIH subjects). A different manifestation of respiratory-cardiac relationships is that deep inspirations (often accompanied by motion) can produce transient elevations in heart rate such as the instance in the top subject of Figure 5. With regard to motion and breathing, in Figure 1 we saw small periodic motions that seem likely to reflect breathing. In other figures we saw many linked instances of large head motion and deep inspirations. Thus, at many times, in many subjects, modulation of heart rate accompanies modulation of respiratory rate, which is also sometimes accompanied by head motion. Disentangling the unique contributions of each of these sources of variance is a non-trivial task.
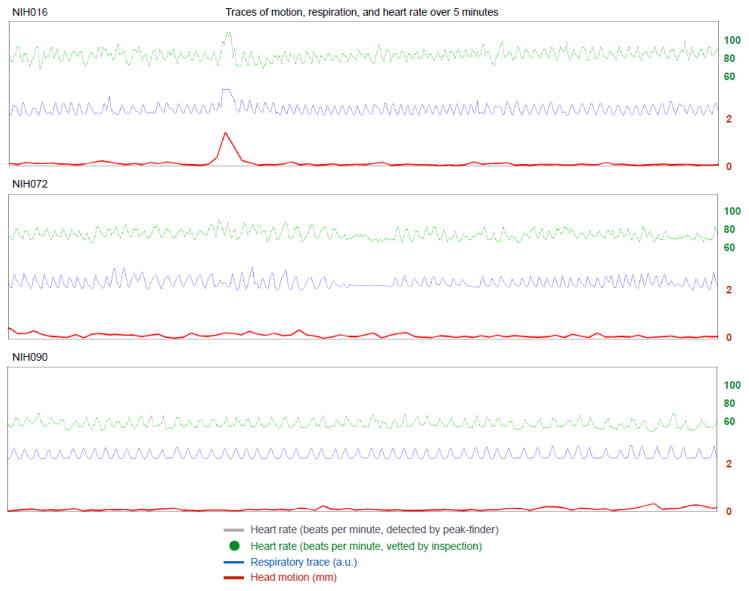
Figure 5. Interrelationships among motion, respiration, and heart rate.
For 3 NIH subjects, 5 minutes of heart rate, respiratory, and head motion traces are shown. Note the slightly lagged modulation of heart rate by respiratory cycle; this finding is called “sinus arrhythmia”, a normal finding in most children and adults. Also note that transient elevations in heart rate accompany deep inspiration, which in turn is often marked by head motion. See additional examples of these plots in Figure S4 and Online Movie 3 for plots of all NIH subjects.
It seems prudent to add three pieces of related information. First, modulation of heart rate by respiratory phase is a well-established clinical phenomenon called “sinus arrhythmia” that is thought to occur primarily due to changes in intrathoracic pressure. Despite the name, sinus arrhythmia is a benign and common finding. Second, in the critical care literature, modulation of some cardiovascular parameters by respiratory phase is dependent on volume status (Cannesson et al., 2007), and this may help explain why some subjects exhibit stronger modulation of heart rate by respiration than other subjects (i.e., some subjects are dehydrated, others are not). Third, anecdotally, I have inspected over 100 scans in the manner shown in Figures 1–3 for evidence of major heart rate effects in isolation from respiratory or motion effects, and I am not (yet) able to recognize an effect (readers may similarly examine plots in Online Movie 3).
Assessing the efficacy of denoising techniques via the plot
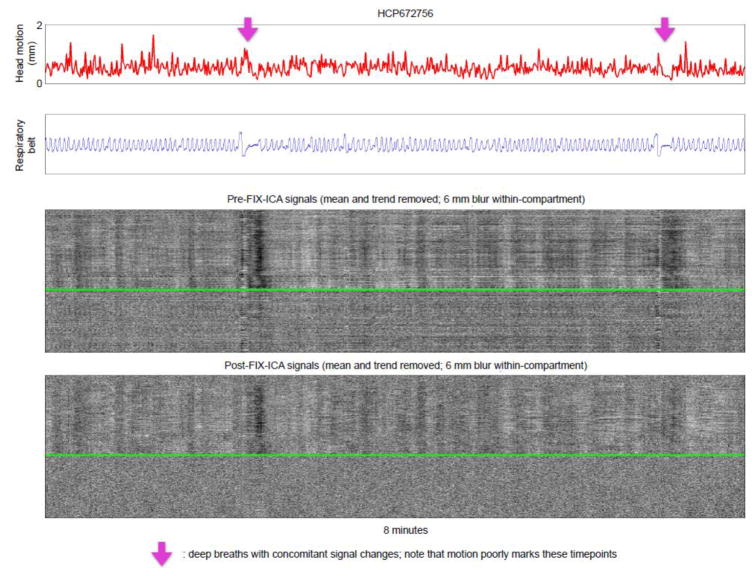
In my view, the central questions of denoising are 1) what artifact was in the data to begin with, 2) what artifact remains in the data after denoising, and 3) what “neural” signal was removed during denoising? The plot can help address the first two points. If an investigator believes that he or she has developed a visual vocabulary for motion artifact, for hardware artifacts, and for respiratory-related variance, then pictures of signals before and after a denoising procedure become a valuable part of data assessment. In Figure 6, a single scan of the HCP database is shown, illustrating several head movements with paired inspirations. Motion- and respiratory-related variance is attenuated but not fully removed by FIX-ICA. Figures S5 and S6 show two further examples of such plots, and Online Movie 4 shows similar plots in all HCP subjects of the “40 Unrelated Subjects” HCP download. I have used plots to assess a variety of denoising techniques, and there is much to say in this regard, but I do not want this paper to become about denoising techniques and will therefore not develop this topic further. In other work I address denoising-related issues more substantively (Power, submittedB).
Figure 6. Examining single scans before and after a denoising step.
A scan from the HCP dataset is shown before and after FIX-ICA. Purple arrows denote breaths (see blue respiratory traces). These data are 2 × 2 × 2 mm voxels acquired every 0.72 seconds. Only 8 minutes of the 15-minute scan are shown because the data are highly sampled in time and the image becomes “cramped” if more time is included. See Figure S5 and S6 for additional examples, or Online Movie 4 for plots of all HCP subjects.
Readers will note that there is much motion in Figure 6. In my experience, HCP data often yields surprisingly high motion estimates, seemingly throughout a scan (see the 38 plots in Online Movie 4). This constant motion sometimes makes it hard to discern instances of large head movement from the motion trace, but large motions can be rather clearly identified in plots via adjunct information in the heatmaps: note the disruptions of signals across the brain marked by purple arrows in Figure 6, disruptions that are characteristic of motion artifact. Although the constant motion (according to FD traces) in HCP data sometimes includes periodic respiration-suggesting humps like those shown for the MyConnectome data in Figure 1, in most instances there is no apparent structure to the motion that I can discern. I am unable to account for the unusual properties of motion estimates in HCP data. It is possible that by shifting sampling from 1160 ms intervals in MyConnectome data to 720 ms intervals in HCP data that periodic motion becomes so finely sampled that motion between samples no longer stands out from “noise” in motion estimation, but such an explanation would suggest that the entire scan would then exhibit very low motion estimates, not high estimates. Another possibility is that some sequence- or hardware-specific property of the HCP scans is responsible for the atypical motion estimates.
Strengths and limitations of the plot
I hope that I have shown that the plot can make plain multiple interesting features of a scan. The reader by now has seen dozens of scans in this format, and has begun to recognize common patterns across scans. I would like to close by commenting on some strengths and limitations of the plot.
The plot’s strength is that it is a compact but fairly comprehensive representation of a scan, capable of producing an immediate visual impression. It is possible to examine many plots in a short amount of time, which is conducive to pattern recognition. And pattern recognition is helpful both for recognizing commonalities across scans (which I have stressed here) and differences between scans (which I have not stressed as much). As an example of the latter approach, multi-echo denoising separates fMRI variance into S0-dependent and R2*-dependent elements, and plots constructed from these two elements have striking and informative differences (not detailed here).
Another strength of the plot is that it shows noise, and noise removal, in a scan-specific and model-free way. Often, denoising “success” is expressed as percent variance explained. But that outcome measure is incomplete, because variance explained, in my view, only really gains meaning in the context of how much variance “ought” to have been explained. Examination of plots before and after denoising can provide critical information regarding which variance “ought” to be explained, and whether it is removed by a denoising technique.
A limitation of the plot is that by flattening the image, it loses spatial specificity. Various signal orderings on the y-axis of the heatmap can deliver certain kinds of specificity, but there is a basic limit to what can be shown. In my own work, I actually animate the plots, with additional panels of slices or surfaces showing signals in time, thus adding spatial specificity to the plot (which is then really a movie).
A related topic, raised by Reviewers of this article, is how the plot can complement aspects of ICA-based fMRI denoising. ICA signal separation is typically accomplished by identifying spatial distinctions among signals. Each component signal thus has a particular spatial localization that is not conveniently represented by the plot. However, the plot can be used in simple ways to inform ICA-based denoising. One way, shown in Figure 6, is to show “undenoised” and “denoised” timeseries. Another approach, mentioned above for multi-echo data, is to create plots of discarded signals and plots of retained signals. A third way is to incorporate into the plot an additional heatmap showing the timeseries of each component so that one sees how signals of varying kinds are distributed across components. I have used multi-echo ICA, FIX-ICA, and other forms of ICA in my own work. I find it helpful not only to examine the spatial maps and other features of the ICA components, but also plots such as those mentioned above, in order to understand what kinds of variance are and are not being removed during denoising. For example, as illustrated in Figure 6, FIX-ICA usually categorizes much respiratory-related variance as “signal”, a fact that is easily appreciated in the plot but which is often less obvious in any individual component map.
I want to close by emphasizing that the plot is just a visualization tool. It is not a statistical description of the qualities of fMRI data. It is not even an analysis - it is just a way to look at data. But in an era of “big data”, where it becomes increasingly common for a single investigator to utilize hundreds or even thousands of scans for a single paper, methods that help an investigator “lay eyes” on individual scans are much needed. The plot serves this need. And in my experience, looking carefully at data is an excellent way to prompt ideas that can evolve into more substantive analyses.
Supplementary Material
Figure S1: Motion is sometimes paired with prolonged signal decreases.
Scans from four subjects of the Utah School of Medicine (USM) site of the ABIDE database are shown following the conventions of Figure 2. These data are 3.4 × 3.4 × 4 mm voxels acquired every 2 seconds.
Figure S2: Motion is sometimes paired with prolonged signal decreases.
Scans from six subjects of the New York University (NYU) site of the ABIDE database are shown following the conventions of Figure 2. These data are 3 × 3 × 3 mm voxels acquired every 2 seconds.
Figure S3: What physiological traces “should” look like.
Top, an unprocessed respiratory trace. Middle, an unprocessed pulse oximeter trace. Bottom, instantaneous heart rate calculated from the time between peaks in the pulse oximeter trace. The modulation of heart rate by respiratory phase is called “sinus arrhythmia”; it is a normal finding seen in most children and young adults.
Figure S4: More examples of interrelationships among motion, respiration, and heart rate.
Plots in 3 additional subjects following conventions of Figure 5.
Figure S5: Examining single scans before and after a denoising step.
Plot follows the conventions of Figure 6, using a different subject.
Figure S6: Examining single scans before and after a denoising step.
Plot follows the conventions of Figure 6, using a different subject.
Highlights.
Presents a plot that summarizes some important features of single fMRI scans
Illustrates multiple kinds of noise and unwanted signals using the plot
Illustrates signal denoising effects with the plot
Includes software tools to make the plot
Illustrates the plot in hundreds of subjects from several different sites
Acknowledgments
I am grateful to Alex Martin for his support and feedback as I developed the infrastructure that led to the plots in this paper. I thank Tim Laumann for help with the MyConnectome scans, and Mark Plitt, Kevin Tran, David Godlove, and the Biowulf staff at the NIH for their computing expertise. I thank Maureen Ritchey and the Reviewers for comments that improved the manuscript. This work was supported by the Intramural Research Program, National Institute of Mental Health/NIH (ZIAMH002920; NCT01031407).
Footnotes
Conflict of interest
The author declares no conflicts of interest with regard to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Barch DM, Sabb FW, Carter CS, Braver TS, Noll DC, Cohen JD. Overt verbal responding during fMRI scanning: empirical investigations of problems and potential solutions. Neuroimage. 1999;10:642–657. doi: 10.1006/nimg.1999.0500. [DOI] [PubMed] [Google Scholar]
- Birn RM, Diamond JB, Smith MA, Bandettini PA. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. Neuroimage. 2006;31:1536–1548. doi: 10.1016/j.neuroimage.2006.02.048. [DOI] [PubMed] [Google Scholar]
- Birn RM, Smith MA, Jones TB, Bandettini PA. The respiration response function: the temporal dynamics of fMRI signal fluctuations related to changes in respiration. Neuroimage. 2008;40:644–654. doi: 10.1016/j.neuroimage.2007.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore ET, Brammer MJ, Rabe-Hesketh S, Curtis VA, Morris RG, Williams SC, Sharma T, McGuire PK. Methods for diagnosis and treatment of stimulus-correlated motion in generic brain activation studies using fMRI. Hum Brain Mapp. 1999;7:38–48. doi: 10.1002/(SICI)1097-0193(1999)7:1<38::AID-HBM4>3.0.CO;2-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannesson M, Attof Y, Rosamel P, Desebbe O, Joseph P, Metton O, Bastien O, Lehot JJ. Respiratory variations in pulse oximetry plethysmographic waveform amplitude to predict fluid responsiveness in the operating room. Anesthesiology. 2007;106:1105–1111. doi: 10.1097/01.anes.0000267593.72744.20. [DOI] [PubMed] [Google Scholar]
- Chang C, Cunningham JP, Glover GH. Influence of heart rate on the BOLD signal: the cardiac response function. Neuroimage. 2009;44:857–869. doi: 10.1016/j.neuroimage.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH. Relationship between respiration, end-tidal CO2, and BOLD signals in resting-state fMRI. Neuroimage. 2009;47:1381–1393. doi: 10.1016/j.neuroimage.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, Van Essen DC, Jenkinson M Consortium WUMH. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn Reson Med. 2000;44:162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Gotts SJ, Simmons WK, Milbury LA, Wallace GL, Cox RW, Martin A. Fractionation of social brain circuits in autism spectrum disorders. Brain. 2012;135:2711–2725. doi: 10.1093/brain/aws160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo HJ, Saad ZS, Simmons WK, Milbury LA, Cox RW. Mapping sources of correlation in resting state FMRI, with artifact detection and removal. Neuroimage. 2010;52:571–582. doi: 10.1016/j.neuroimage.2010.04.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumann TO, Gordon EM, Adeyemo B, Snyder AZ, Joo SJ, Chen MY, Gilmore AW, McDermott KB, Nelson SM, Dosenbach NU, Schlaggar BL, Mumford JA, Poldrack RA, Petersen SE. Functional System and Areal Organization of a Highly Sampled Individual Human Brain. Neuron. 2015;87:657–670. doi: 10.1016/j.neuron.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Bandettini PA. Resting-state fMRI confounds and cleanup. Neuroimage. 2013;80:349–359. doi: 10.1016/j.neuroimage.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, Laumann TO, Koyejo O, Gregory B, Hover A, Chen MY, Gorgolewski KJ, Luci J, Joo SJ, Boyd RL, Hunicke-Smith S, Simpson ZB, Caven T, Sochat V, Shine JM, Gordon E, Snyder AZ, Adeyemo B, Petersen SE, Glahn DC, Reese Mckay D, Curran JE, Goring HH, Carless MA, Blangero J, Dougherty R, Leemans A, Handwerker DA, Frick L, Marcotte EM, Mumford JA. Long-term neural and physiological phenotyping of a single human. Nat Commun. 2015;6:8885. doi: 10.1038/ncomms9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Mitra A, Laumann TO, Snyder AZ, Schlaggar BL, Petersen SE. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage. 2014;84:320–341. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Schlaggar BL, Petersen SE. Recent progress and outstanding issues in motion correction in resting state fMRI. Neuroimage. 2015;105:536–551. doi: 10.1016/j.neuroimage.2014.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JDP, Kundu M, Bandettini P, Martin PA. Head motion is underestimated in fMRI scans following common preprocessing steps submitted A. [Google Scholar]
- Power JDP, Laumann M, Martin TA. Sources and implications of whole-brain fMRI signals in humans. doi: 10.1016/j.neuroimage.2016.09.038. submittedB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Elliott MA, Gerraty RT, Ruparel K, Loughead J, Calkins ME, Eickhoff SB, Hakonarson H, Gur RC, Gur RE, Wolf DH. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage. 2013;64:240–256. doi: 10.1016/j.neuroimage.2012.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyser CD, Inder TE, Shimony JS, Hill JE, Degnan AJ, Snyder AZ, Neil JJ. Longitudinal analysis of neural network development in preterm infants. Cereb Cortex. 2010;20:2852–2862. doi: 10.1093/cercor/bhq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RG, Ide K, Poulin MJ, Tracey I. Resting fluctuations in arterial carbon dioxide induce significant low frequency variations in BOLD signal. Neuroimage. 2004;21:1652–1664. doi: 10.1016/j.neuroimage.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Yan CG, Cheung B, Kelly C, Colcombe S, Craddock RC, Di Martino A, Li Q, Zuo XN, Castellanos FX, Milham MP. A comprehensive assessment of regional variation in the impact of head micromovements on functional connectomics. Neuroimage. 2013;76:183–201. doi: 10.1016/j.neuroimage.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Motion is sometimes paired with prolonged signal decreases.
Scans from four subjects of the Utah School of Medicine (USM) site of the ABIDE database are shown following the conventions of Figure 2. These data are 3.4 × 3.4 × 4 mm voxels acquired every 2 seconds.
Figure S2: Motion is sometimes paired with prolonged signal decreases.
Scans from six subjects of the New York University (NYU) site of the ABIDE database are shown following the conventions of Figure 2. These data are 3 × 3 × 3 mm voxels acquired every 2 seconds.
Figure S3: What physiological traces “should” look like.
Top, an unprocessed respiratory trace. Middle, an unprocessed pulse oximeter trace. Bottom, instantaneous heart rate calculated from the time between peaks in the pulse oximeter trace. The modulation of heart rate by respiratory phase is called “sinus arrhythmia”; it is a normal finding seen in most children and young adults.
Figure S4: More examples of interrelationships among motion, respiration, and heart rate.
Plots in 3 additional subjects following conventions of Figure 5.
Figure S5: Examining single scans before and after a denoising step.
Plot follows the conventions of Figure 6, using a different subject.
Figure S6: Examining single scans before and after a denoising step.
Plot follows the conventions of Figure 6, using a different subject.