Abstract
Saccharomyces cerevisiae encodes two distinct Pif1-family helicases – Pif1 and Rrm3 – which have been reported to play distinct roles in numerous nuclear processes. Here, we systematically characterize the roles of Pif1 helicases in replisome progression and lagging-strand synthesis in S. cerevisiae. We demonstrate that either Pif1 or Rrm3 redundantly stimulate strand-displacement by DNA polymerase δ during lagging-strand synthesis. By analyzing replisome mobility in pif1 and rrm3 mutants, we show that Rrm3, with a partially redundant contribution from Pif1, suppresses widespread terminal arrest of the replisome at tRNA genes. Although both head-on and codirectional collisions induce replication fork arrest at tRNA genes, head-on collisions arrest a higher proportion of replisomes. Consistent with this observation, we find that head-on collisions between tRNA transcription and replication are under-represented in the S. cerevisiae genome. We demonstrate that tRNA-mediated arrest is R-loop independent, and propose that replisome arrest and DNA damage are mechanistically separable.
INTRODUCTION
The eukaryotic replisome faces the formidable task of unwinding and replicating tens- to hundreds of kilobases of DNA, which may contain impediments such as ongoing transcription, stable protein-DNA complexes, DNA lesions, and intramolecular DNA secondary structures1. Replication fork blockage at such impediments can lead to fork collapse and DNA breaks2; however, such arrest can be detrimental even if fork integrity is maintained. Most intuitively, the arrest of two convergent replication forks bordering a region without a licensed replication origin will preclude timely replication of this region, potentially leading to under-replication and attendant downstream problems such as chromosome mis-segregation3. Therefore, continued replication fork progression on challenging templates is fundamentally important for organismal viability, even if the replication fork is able to progress past lesions, which can be repaired after S-phase is complete4.
Pif1 helicases are a conserved family of 5’-3’ helicases capable of removing proteins from DNA, and of unwinding duplex DNA, RNA:DNA hybrids, and G-quadruplexes – stable intramolecular DNA secondary structures resulting from Hoogsteen base pairing between four planar guanine bases5–8. While most metazoans encode only one Pif1, S. cerevisiae encodes two separate helicases: PIF1 and RRM3, which are proposed to have largely distinct functions. Pif1 inhibits telomerase9 and helps prevent genome rearrangements at G-quadruplexes8. Additionally, Pif1 has been identified as an accessory helicase for the lagging-strand DNA polymerase δ (Pol δ)10–12. Rrm3 has predominantly been implicated as a replisome component13,14 that facilitates passage past protein-DNA complexes including tRNA genes and centromeres15, and more recently has been found to contribute to the maintenance of G-quadruplex-forming structures in combination with Pif18.
We have previously described Okazaki fragment sequencing to analyze lagging-strand maturation and replication fork movement genome-wide in S. cerevisiae16,17. Here, we report the systematic analysis of the effects of Pif1 and Rrm3 activity on replisome progression and lagging-strand synthesis. We determine that Pif1 and Rrm3 act redundantly to stimulate strand-displacement by Pol δ during Okazaki fragment maturation. While previous work found a role for Rrm3 but not Pif1 in aiding in fork progression at tRNA genes16, we demonstrate that Pif1 does help to facilitate replisome mobility in cells lacking Rrm3. The absence of both helicases leads to a high rate of terminal replication fork arrest (hereafter referred to as arrest and distinguished from transient replisome pausing, which we refer to as stalling) at tRNA genes (hereafter referred to as tDNAs). Furthermore, we show increased arrest in response to head-on collisions, and provide evidence that tDNAs in S. cerevisiae show a directional bias that reduces the likelihood of such interactions. In contrast to previous reports, we do not detect significant arrest at other likely substrates of Pif1 helicases, e.g. G-quadruplexes and highly transcribed RNA polymerase II genes. Additionally, we find that conditions that increase or decrease the levels of R-loops at tDNAs18 do not impact replisome arrest at these loci.
RESULTS
Assaying lagging-strand synthesis and replisome progression using Okazaki fragment sequencing
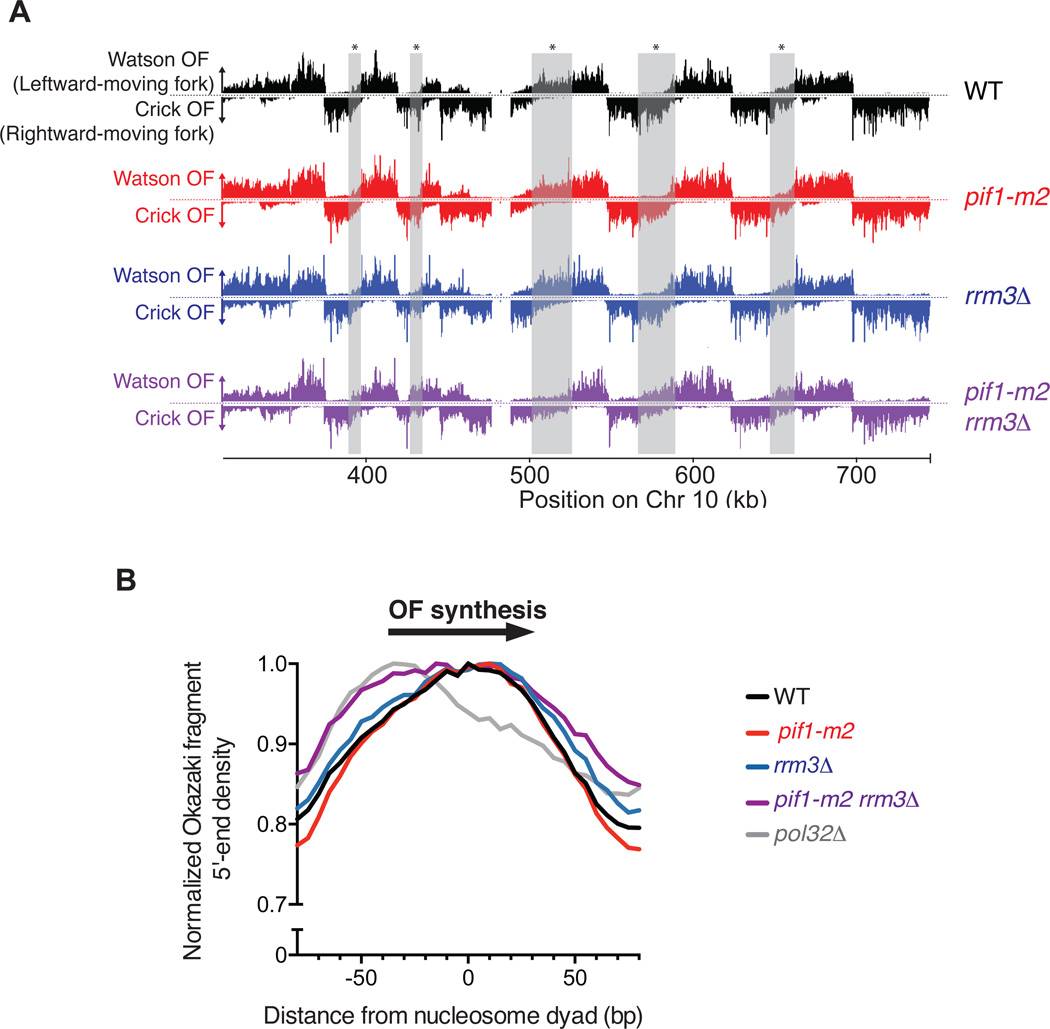
We combined mutants in the Pif1-family helicases (rrm3Δ, pif1-m2, rrm3Δ pif1-m2) with a previously described doxycycline-repressible CDC9 (DNA ligase I) allele to sequence Okazaki fragments genome-wide17 (Figure 1A) and thereby monitor fork progression and lagging strand maturation in S. cerevisiae strains lacking either or both Pif1-family helicase(s). Neither PIF1 nor RRM3 is essential for viability, but Pif1 is needed for stable maintenance of the mitochondrial DNA. To avoid potential artifacts due to mitochondrial defects, we used the well-characterized pif1-m2 allele, which maintains mitochondrial function without detectable Pif1 in the nucleus19–21. Lagging-strand sequencing can be used to infer replication direction because Okazaki fragments are synthesized on the Watson strand by a leftward-moving fork and the Crick stand by a rightward-moving fork16 (Fig. 1A). Origin usage, detectable as an abrupt transition from leftward-to rightward-moving forks, appears similar between all four strains (Figs. 1A and S1), and fork progression appears largely unaltered in the rrm3Δ and pif1-m2 strains. However, the double mutant shows differences in Okazaki fragment distributions between replication origins, suggestive of significantly altered replication fork progression (Fig. 1A – gray boxes).
Figure 1. Okazaki fragment Sequencing is a quantitative and genome-wide assay for replisome mobility and lagging strand biogenesis in WT and mutant cells.
(A) Distribution of Watson- and Crick-strand Okazaki fragments across the right arm of chromosome 10 in wild-type, rrm3Δ, pif1-m2, and pif1-m2 rrm3Δ S. cerevisiae strains. Watson strand fragments result from leftward-moving replication forks and are shown above each axis; Crick strand fragments result from rightward-moving forks and are shown below the axis. Grey boxes indicate regions where differences between distributions in wild-type and pif1-m2 rrm3Δ can be readily observed. Data were visualized using Mochiview49. (B) Either Pif1 or Rrm3 is required for normal DNA Pol δ displacement synthesis through nucleosomes. Distribution of Okazaki fragment 5’ termini from the indicated strains around consensus S. cerevisiae nucleosome dyads50. Data are normalized to the maximum value in range and binned to 5bp. Data from the pol32Δ strain are from Smith and Whitehouse17.
Either Pif1 or Rrm3 can act as a lagging strand processivity factor
Lagging-strand synthesis in eukaryotes involves the generation of 5’ flap structures via strand-displacement synthesis by Pol δ, and their cleavage by structure-specific nucleases22. We have previously shown that Okazaki fragment maturation occurs in the context of nascent nucleosomes, and that Okazaki fragment ends are enriched around nucleosome midpoints due to the limited ability of Pol δ to penetrate the protein/DNA complex.17 Nucleosome penetrance during Okazaki fragment processing, which is reduced in strains lacking POL32,17 is a proxy for the level of Pol δ processivity during strand-displacement synthesis. In pif1-m2 and rrm3Δ single mutants, the distribution of Okazaki fragment 5’ and 3’ termini around nucleosomes23 was similar to wild-type (Fig. 1B & S2A). However, in the double mutant strain, both 5’ and 3’ termini showed a pronounced shift toward the replication fork-proximal edge of the nucleosome (Fig. 1B & S2A, purple line). These data are consistent with a reduction in the ability of Pol δ to carry out strand displacement on nucleosomal templates throughout the majority of the genome. Okazaki fragments across all strains were similarly sized and poised for ligation (Fig. S2B), suggesting that the reduction in strand displacement seen in pif1-m2 rrm3Δ cells does not result in a substantial inability to complete lagging-strand processing. We therefore conclude that either Rrm3 or Pif1 is sufficient to act as a processivity factor for DNA Pol δ during lagging-strand biogenesis (Fig. S2C)
tDNAs arrest replisomes in the absence of Rrm3; Pif1 can serve as a backup in the absence of Rrm3 activity
A point replication origin can be identified from Okazaki fragment sequencing data as an abrupt reduction in Watson-strand coverage (moving from left to right) with a corresponding increase in Crick-strand coverage; the magnitude of this change is proportional to origin firing efficiency16,24. A point replication terminator will manifest as the converse change, with Watson signal increasing at the expense of Crick signal (Figure 2A). Okazaki fragments from the pif1-m2 rrm3Δ double mutant showed numerous strand transitions suggestive of termination (Fig. 2B). Because these transitions often occurred close to tDNAs, we undertook a quantitative analysis of fork progression around tDNAs.
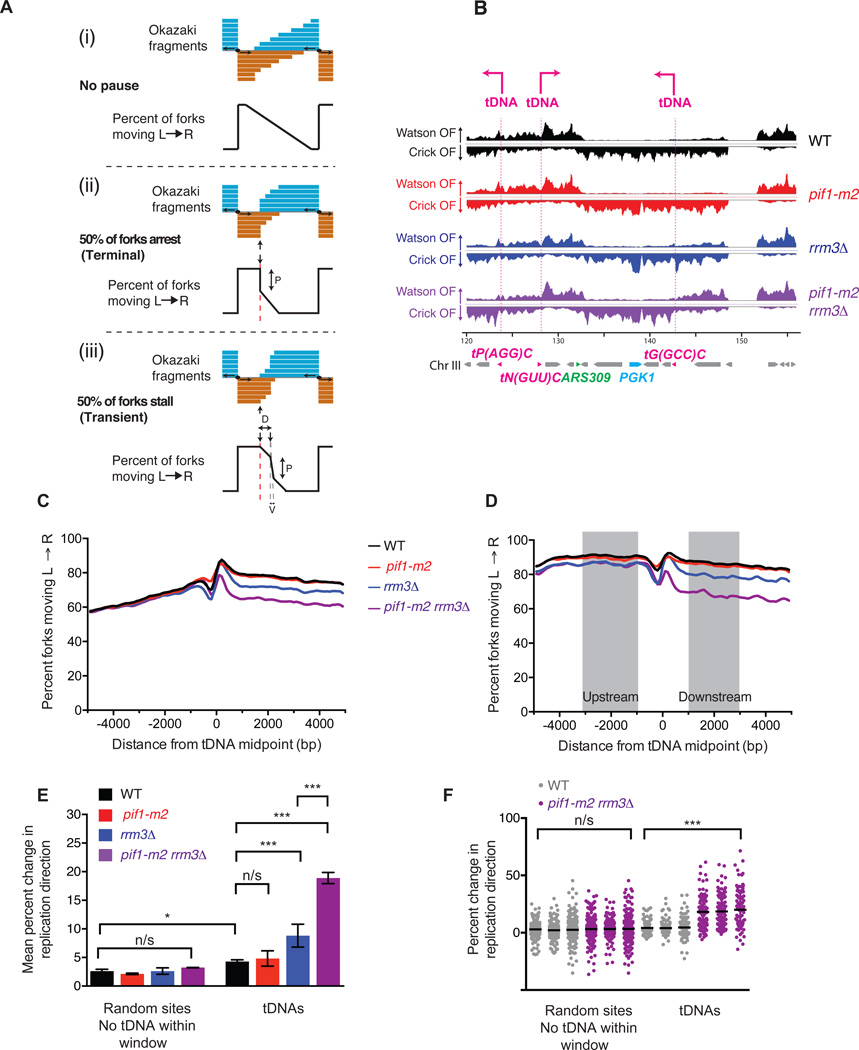
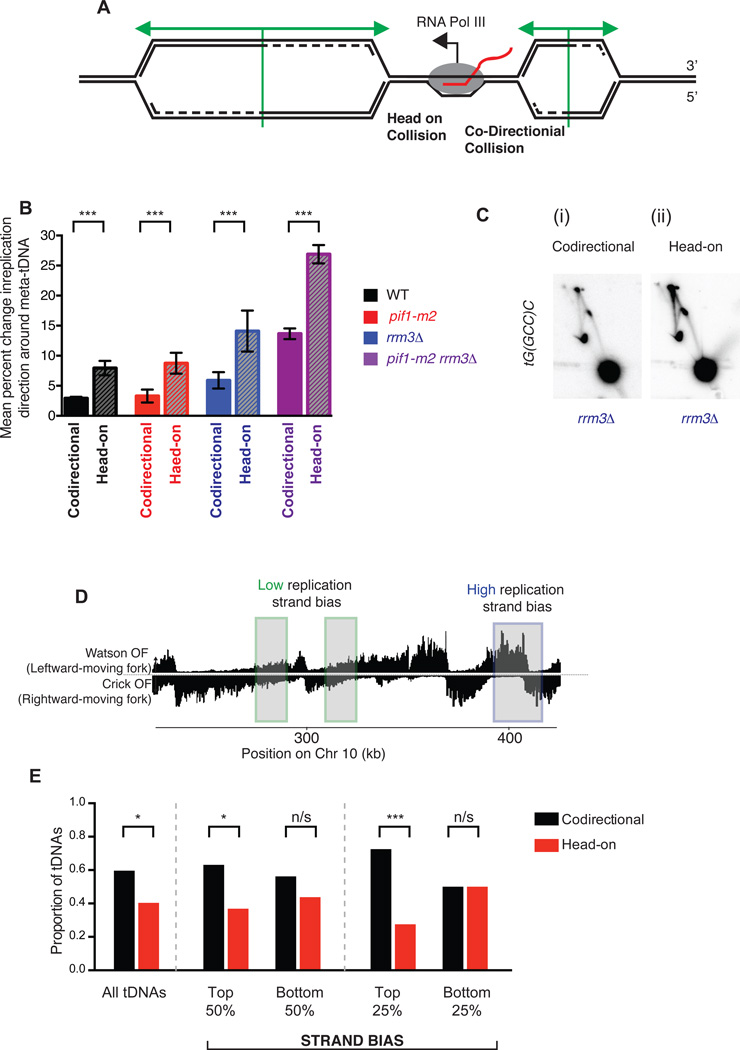
Figure 2. tDNAs are sites of replication fork arrest in rrm3Δ and rrm3Δ pif1-m2 strains.
(A) Expected Okazaki fragment distributions through a region that (i) allows unimpaired replisome movement, (ii) arrests the replisome, or (iii) transiently stalls the replisome. The magnitude of the strand transition, P, is proportional to the number of forks stalling or arresting: in the event of a transient pause, this transition will be offset from the element by a distance, D, and broadened over a window, V, dependent on pause duration and fork speed. (B) Distribution of Okazaki fragments across a 40 kb region of S. cerevisiae chromosome 3. Locations of tDNAs are indicated. (C) Analysis of replisome direction around all 275 tDNAs in the nuclear genome. tDNAs replicated predominantly by rightward- or leftward-moving forks were analyzed separately, and the data superimposed such that the direction of fork motion is always from left to right (see methods). (D) Analysis of replisome direction, as in C, at the 93 tDNAs without a replication origin or sequence gap within ± 5 kb. The change in replication fork direction between 1–3 kb up- and downstream of a meta-element is used to calculate termination signal (see methods). (E) Grand mean ± SD from three independent experiments of the change in replication direction at tDNAs and filtered random sites in three independently isolated strains of each genotype. Significance was determined by Monte Carlo resampling;; *** indicates a p<0.0001; * indicates 0.0001 < p < 0.05; n/s indicates p>0.05. (F) Change in replication direction individual tDNAs (n=93) and random sites (n=292) in three independent experiments. Black bars indicate the mean. Significance was determined by Monte Carlo resampling; *** indicates p<0.0001
The expected Okazaki fragment distribution around a site of replication fork arrest (terminal) or stalling (transient) affecting 50% of replication forks is schematized in Fig. 2A. Fork arrest or very long-lived replisome stalling events – those of sufficient duration to allow complete replication of the downstream region by a convergent replisome – should produce distributions with abrupt strand transitions directly at the termination element but otherwise unaffected up-or downstream (compare Figure 2A(ii) and (iii)). We separately analyzed Okazaki fragments around tDNAs predominantly replicated by leftward- or rightward-moving forks (see methods and Fig. S3A), and reversed the data from the leftward-moving set such that all forks were effectively moving from left to right. Meta-analysis of effective replication direction around all tDNAs reveals a dramatic change in fork mobility in rrm3Δ and pif1-m2 rrm3Δ strains (Fig. 2C), consistent with a high degree of replisome arrest at tDNAs. The transition was sharp and centered on the meta-tDNA midpoint – the predicted behavior for a site of fork arrest as opposed to a termination zone or stall (Figure 2C). However, the change in replisome direction observed upstream of the meta-tDNA, which is due to proximal replication origins (tDNAs are located, on average, near replication origins, Figs. S3 B–D and Table S1 and S2), precluded the quantitation of these changes in replisome mobility. To avoid this complication, we selected a set of 93 tDNAs for quantitative analysis, excluding those with an active replication origin16 or sequence gap within the region ±5 kb from the tDNA midpoint (see Methods and Table S1). While we use this stringent set of tDNAs for quantification, we note that there is clear evidence for fork arrest at essentially all 275 tDNAs in the pif1-m2 rrm3Δ strain (Figs 2C and S4A&B).
Because of fluctuations in sequence coverage in the immediate vicinity of tDNAs, we used the mean replication direction in the regions 1–3 kb up- and downstream of the tDNA midpoint (Fig. 2D) to calculate the proportion of replisomes terminating within ±1 kb of the each tDNA in each of three independent isolates of each strain (Figure 2E). rrm3Δ, and to an even greater extent pif1-m2 rrm3Δ strains, show robust and significant arrest, with nearly 20% of replication forks arresting and being rescued by a convergent fork in the double mutant (Figure 2E). Mean values across the three datasets were highly reproducible (Fig. 2F). Therefore, we conclude that Rrm3 normally suppresses replisome arrest at tDNAs. Surprisingly, Pif1 can partially compensate for the absence of Rrm3 at tDNAs, and in the absence of both helicases, arrest is pronounced.
To determine whether the observed replisome arrest was specific to tDNAs, we analyzed replication direction around a set of random sites filtered equivalently to our tDNA set to remove those with proximal replication origins and/or sequence gaps (Table S3). In addition, due to the strong stalling signal at tDNAs, we excluded sites within 5 kb of a tDNA midpoint. Quantitation of replisome stalling revealed no replication termination above WT level in any mutant strain at random sites (Fig. 2E–F & S3E–H). There is a small (≈2%) but significant increase in fork arrest at tDNAs compared to random sites in WT cells (Fig. 2E), in agreement with previous estimates that ~1% of replisomes arrest at tDNAs in wild-type yeast25.
Validation of replication fork arrest at tDNAs by 2-D gel
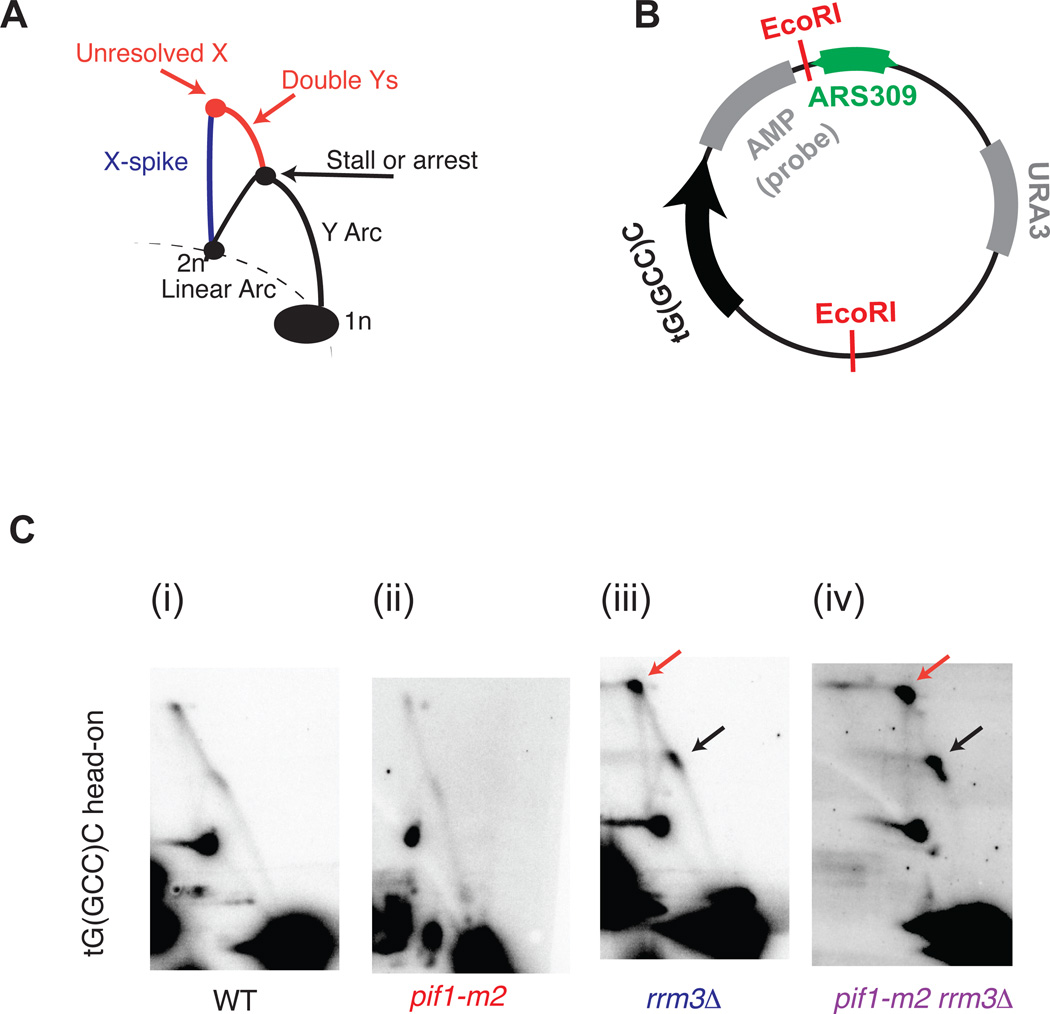
To validate our Okazaki fragment data, we assayed replisome mobility around a single tDNA by neutral-neutral 2-D gel agarose electrophoresis. We identified a site on chromosome III where a rightward moving replication fork stalls at a tDNA oriented head-on with respect to replication (Figure 2A). Previous work has implicated the adjacent highly transcribed PGK1 gene as a replisome stalling element in rrm3Δ cells through increased occupancy of DNA Polymerase ε26. We cloned the origin that drives replication toward this site (ARS309) and the tDNA itself (tG(GCC)C) into a plasmid such that replication and transcription occur in the head-on orientation as at the native locus (Fig. 3B). Stalling or arrest of the leftward-moving replication fork should give rise to a dark spot on the Y-arc; arrest of the leftward-moving fork should allow the rightward-moving fork to progress past the second EcoRI site, generating double-Y structures and a specific X spot (Fig. 3A). In agreement with our genome-wide data, we see a slight or no enrichment of replication intermediates on the Y-arc in the WT and pif1-m2 cells, a moderate enrichment in the rrm3Δ cells, and a strong enrichment in the pif1m2-rrm3Δ cells (Figure 3C, black arrows) suggesting that Pif1 can indeed act as a backup for Rrm3 activity. In all cases, and consistent with arrest, an X-spot signal is observed (Figure 3C, red arrows).
Figure 3. 2-dimensional gel validation of replication fork arrest in rrm3Δ and rrm3Δ pif1-m2 strains.
(A) Schematic of intermediates, resolved in a neutral-neutral 2-dimensional agarose gel, resulting from the replication of plasmid (B) in asynchronous cultures. (C) Neutral-neutral 2-Dimensional gel analysis of the indicated plasmid in WT, pif1-m2, rrm3Δ and pif1-m2 rrm3Δ cells, as indicated. 2-D gel signals consistent with replisome stalling and/or arrest are indicated in colors corresponding to the schematic in Fig. 3A. Plasmids were cut with the indicated restriction enzymes and crosslinked with psoralen prior to electrophoresis.
Highly transcribed RNA Pol II genes and G-quadruplexes do not induce significant replisome arrest in pif1 and/or rrm3 mutant strains
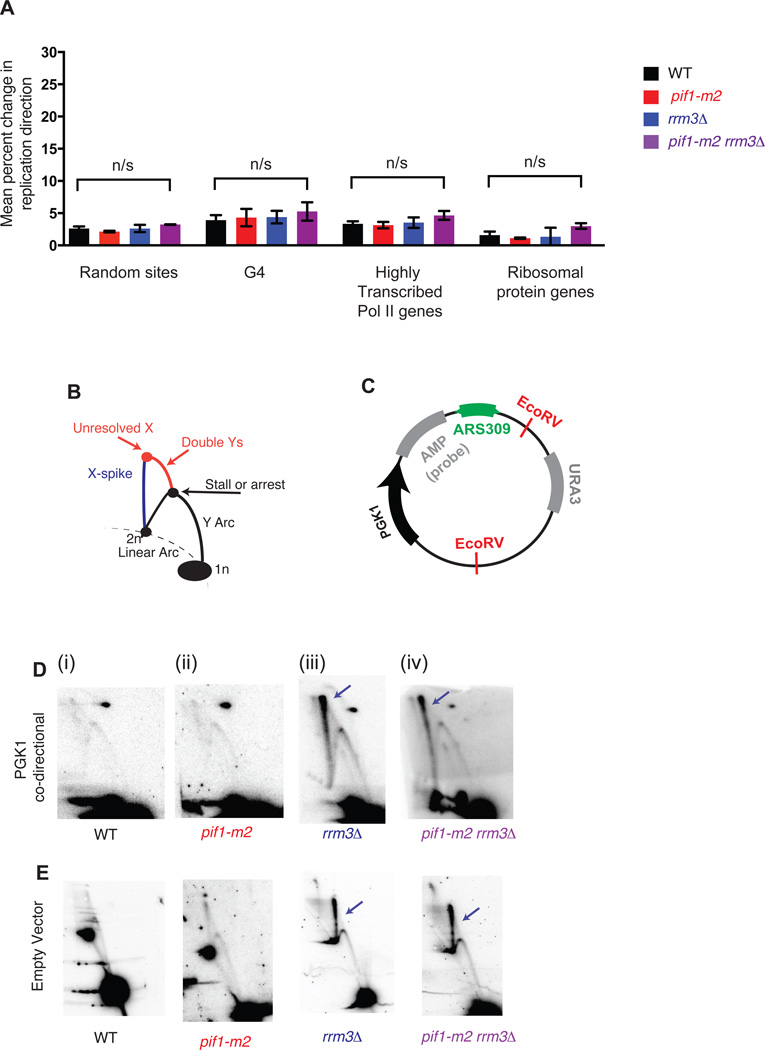
Previous work has implicated Pif1, Rrm3, or both proteins in aiding replication fork progression through highly transcribed RNA Polymerase II (RNAP2) genes13,15,26 and G-quadruplex structures in vivo8,27. Therefore, we analyzed replisome mobility around highly transcribed RNAP2 genes (from28, Table S4), and the ribosomal protein genes as a representative group of coordinately regulated, highly transcribed RNAP2 genes (from29 Table S4), and G-quadruplexes (from30, Table S5), again excluding those within ± 5kb of an origin, sequence gap, or tDNA. Strikingly, neither RNAP2 genes nor G-quadruplexes were associated with changes in replisome mobility as monitored by Okazaki fragment distributions (Fig. 4A). We considered many subsets of both RNAP2 genes and G-quadruplex structures, including G-quadruplexes that were previously shown to be enriched in Pif1 binding27, and did not identify a biologically relevant subset that showed robust and significant stalling or arrest when sites that overlap with tDNAs were excluded (Figure S5C). Importantly, we find that tDNAs are not more likely than random sites to contain G-quadruplex forming sequences (Table S2). In contrast to previous reports27, we see no evidence for changes in replisome progression at G-quadruplex sites even in cells grown in the presence of 25 mM hydroxyurea to deplete dNTP pools (Figure S5D). Moreover, we see no significant orientation effect for either RNAP2 genes or G-quadruplex structures (Figure S6) even though leading-strand G-quadruplexes have been shown to induce more genome instability than lagging strand sites31. We can identify a small proportion of G-quadruplexes that show a significant arrest signal, but random sites also show this effect, arguing that G-quadruplexes act similarly to randomly selected genomic sites and that any apparent arrest signal at a minority of sites is due to noise in the data (see Methods and Fig. S5E).
Figure 4. G-quadruplexes and highly transcribed RNA Polymerase II genes do not contribute to significant replication fork stalling or arrest genome wide.
(A) Grand mean ± SD from three independent experiments of the change in replication direction, interpreted as replisome stalling or arrest, around random sites (n=292), G-quadruplexes (n=154), highly transcribed RNA Pol II genes (n=172) and ribosomal protein genes (n=56) as in Fig. 2E. Loci were filtered as for the tDNA analysis in Fig. 2D–F, excluding sites within ±5 kb of a sequence gap, replication origin, or tDNA. Significance was determined by Monte Carlo resampling; *** indicates a p<0.0001; n/s indicates p>0.05. (B) Schematic of intermediates, resolved in a neutral-neutral 2-dimensional agarose gel, resulting from the replication of plasmid (C) in asynchronous cultures. (D–E) Neutral-neutral 2-Dimensional gel analysis of a plasmid with or without a PGK1 insert in asynchronous WT, pif1-m2, rrm3Δ or pif1-m2 rrm3Δ, as indicated. 2D gel signals consistent with changes in replication fork mobility or resolution are indicated in colors corresponding to the schematic in Fig. 3A. Plasmids were cut with the indicated restriction enzymes and crosslinked with psoralen prior to electrophoresis. The spot on the upper right in (D) is due to background hybridization of the probe.
To confirm unperturbed repisome progression through highly transcribed RNAP2 genes, we cloned PGK1 and its promoter into the previously described ARS309-containing plasmid for 2-D gel analysis. PGK1, unlike the adjacent tG(GCC)C, did not affect Okazaki fragment distributions (Fig. 2B). We did not observe effects on replisome mobility at PGK1 by 2-D gel in any of the four strains analyzed (Figure 4D and cf. Fig. 3C). However, a strong X-spike signal was observed in both rrm3Δ and pif1-m2 rrm3Δ (Figure 4D, blue arrows). To test the hypothesis that this X-spike represented a defect in the resolution of convergent replication forks in these strains, and that the defect in replication termination was not due to conflicts with transcription, we analyzed a plasmid lacking PGK1, whose left half is therefore transcriptionally silent; the X-spike is still apparent in both rrm3Δ and pif1-m2 rrm3Δ strains even when the replication fork encounters no transcribing RNA polymerase (Fig. 4E, iii & iv).
Replication fork arrest at tDNAs is partially orientation dependent
There have been conflicting reports about whether tDNAs impede the replication machinery in WT cells. tRNA genes have been found by 2-D gel to act as replication fork blocks regardless of orientation the absence of Rrm313,15, whereas the original report of tDNA-mediated stalling found that only head-on tDNAs stalled or arrested the wild-type replisome32. We separated the 93 tDNAs in our analysis into those replicated head-on or co-directionally with respect to RNA Pol III transcription (Figure 5A and see methods). In all strains, head-on collisions were significantly more likely to arrest replisomes than co-oriented collisions (Fig. 5B). Interestingly, while we find that while the proportion of replisomes that arrest at tDNAs increases upon deletion of RRM3, and further on additional mutation of PIF1, the relative ratio of head-on vs co-directional arrest remains constant. For validation, we switched the orientation of the tDNA in the plasmid-based 2-D gel assay in the rrm3Δ strain; more arrest, reflected by more intense localized signal on the Y-arc and X-spot, is observed when the tDNA is in the head-on orientation (Figure 4C). We conclude that tDNAs in either orientation can induce replisome arrest, but that this arrest is approximately twice as frequent at tDNAs oriented head-on relative to replication. While Pif1 helicases reduce the overall frequency of fork arrest, they do not preferentially impact one orientation relative to the other.
Figure 5. All tDNAs act as replication terminators, but head-on orientation between replication and transcription machinery increases fork arrest.
(A) Schematic of two replication forks approaching a highly transcribed tDNA. The fork moving from left to right will meet the transcribing RNA Polymerase III molecule in a head-on fashion. (B) Grand mean ± SD from three independent experiments of the change in replication direction, interpreted as indicative of replisome arrest, at tDNAs transcribed co-directionally or head-on relative to the predominant direction in which they are replicated (see methods). Termination signal was calculated as in Figure 2E. Significance was determined by Monte Carlo resampling; *** indicates a p<0.0001 (C) Neutral-neutral 2-D agarose gel electrophoresis of the plasmid shown in Fig. 3B, but with tG(GCC)C in either the codirectional or head-on orientation as indicated, purified from an asynchronous S. cerevisiae culture. (D) Schematic of chromosomal regions of high (blue) and low (green) replication strand bias. (E) Bar plot indicating the proportion of tDNAs oriented head-on or codirectionally relative to replication, separated into bins (above or below the median strand bias for the 275 nuclear tDNAs) as indicated by strand bias as shown in D. All tDNAs (n=275), high and low strand bias (n=137) and top and bottom quartiles (n=68) were calculated and p-values for cumulative binomial; *** indicates a p<0.0001; * indicates 0.0001 < p < 0.05; n/s indicates p>0.05.
We interrogated tDNA orientation with respect to replication to search for an evolutionary signature of increased replisome arrest due to head-on tDNA transcription. Examination of all 275 tDNAs revealed significant overrepresentation of co-oriented transcription/replication conflicts (Fig. 5E). We then sought to analyze tDNAs with high and low replication strand bias (Fig. 5D) to determine whether the likelihood of the conflict being always co-directional or head-on could predict the orientation of the tDNA. Indeed, in regions of high replication strand bias, where a tDNA is likely to be replicated in only one direction in almost all cells, there is an increased bias for co-oriented transcription/replication conflicts (Fig. 5E). Logistic regression confirmed that increased strand bias leads to a significant decrease in the likelihood of a head-on orientation between transcription and replication (p=0.01, O.R. = 0.977). This observation suggests an evolutionary pressure to co-orient tDNAs with replisome movement in S. cerevisiae and is consistent with head-on tDNAs behaving as sites of detectable replisome arrest in wild-type cells (Fig. 5B). Equivalent analysis of protein-coding genes did not indicate a directional bias, consistent with previous reports33 and our inability to detect orientation-specific replisome mobility changes at these sites (Figure S6).
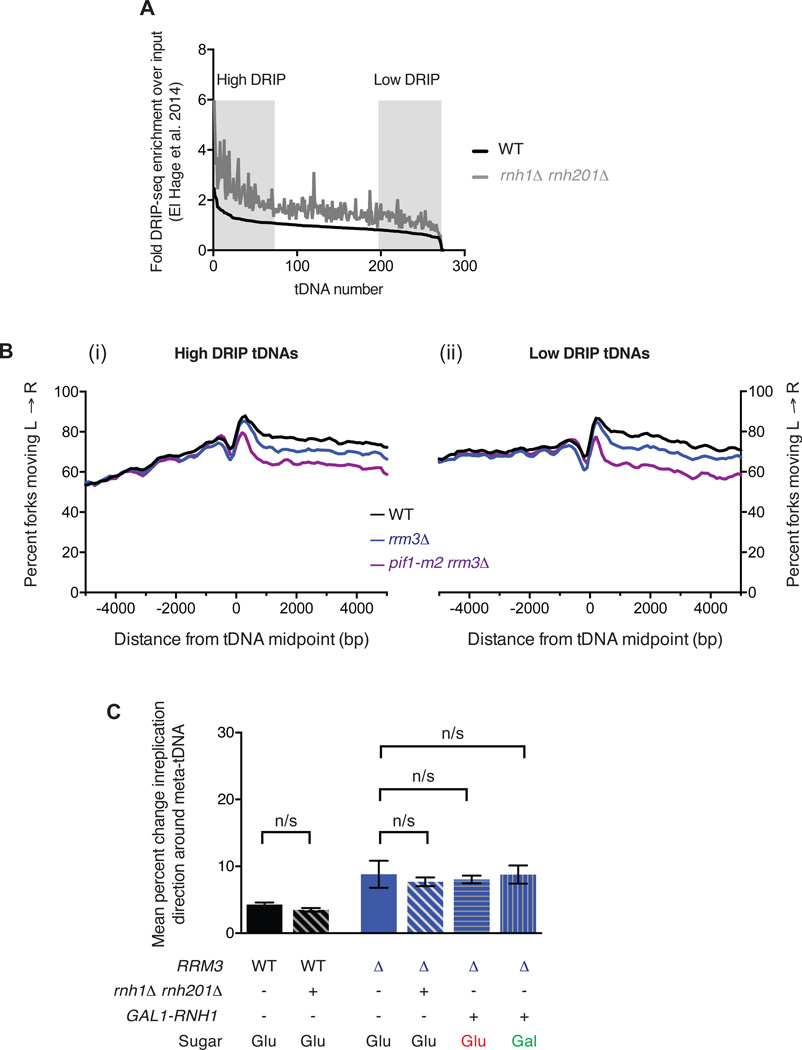
R-loops do not significantly contribute to replication fork arrest at tDNAs
R-loops are RNA:DNA hybrid structures that form in vivo when transcribed RNA reanneals with the template strand18,34. Importantly, R-loops are a directional feature: a Watson strand gene generates an R-loop on the lagging strand for a leftward moving fork, and a Crick strand gene generates a leading-strand R-loop (see Figure 5A). The RNA strand of R-loops is degraded by RNase H1; modulating the levels of RNase H1 and H2 in the cell can increase or decrease the level of R-loops35,36. In particular, a rnh1Δ rnh201Δ strain was recently shown by DRIP-seq to have increased R-loop occupancy at tDNAs18. We sorted tDNAs by their previously reported levels of R-loops in wild-type cells (Figure 6A), and analyzed replisome arrest at high-DRIP (top quartile) vs low-DRIP (bottom quartile) tDNAs. Both tDNA subsets show strong replisome arrest in both rrm3Δ and pif1-m2 rrm3Δ strains (Fig. 6C). Direct quantitation of changes in replisome mobility at high- vs low-DRIP tDNAs is precluded by the distinct fork direction gradients observed upstream of the two subsets. Therefore, we sought to modulate R-loop levels in the context of wild-type and rrm3Δ strains and assay the impact of this perturbation on replisome mobility. We constructed rnh1Δ rnh201Δ and rnh1Δ rnh201Δ rrm3Δ strains with repressible CDC9, and carried out triplicate replicates as in previous analyses. Separately, we placed an inducible GAL1,10 promoter upstream of RNH1 in rrm3Δ cells. Analysis of replication fork arrest in strains with increased or decreased RNaseH activity indicated no significant difference in either a wild-type or rrm3Δ background (Fig. 6D). We conclude that R-loops are extremely unlikely to underlie the orientation-based asymmetry of replisome arrest at tDNAs.
Figure 6. R-loops do not mediate replication fork arrest at tDNAs.
A. R-loop load at all 275 tDNAs determined by DRIP-seq18 in WT and rnh1Δ rnh201Δ cells. The level of R-loops over background was determined for a 400bp window around the tDNA midpoint. tDNAs were sorted by the level of R-loops in the WT strain and the levels in both the WT (black) and the rnh1Δ rnh201Δ double mutant (gray) were plotted. Regions corresponding to high- and low-DRIP in Fig. 6B are indicated. (B) Analysis of replisome direction around (i) high- and (ii) low-DRIP-seq tDNAs (n=68) as identified in A. (C). Grand mean ± SD from three independent experiments of the change in replication direction, interpreted as indicative of replisome arrest, at the 93 tDNAs from Fig. 2E in the indicated strains. Cells were grown in either YEP + dextrose (Glu) or galactose (Gal) as specified. Significance was determined by Monte Carlo resampling; n/s indicates p>0.05.
DISCUSSION
Pif1 helicases and Okazaki fragment maturation
We show that Rrm3 can stimulate Pol δ strand displacement on the lagging strand (Fig. 1). Previous genetic experiments implicated only Pif1 but not Rrm3 in lagging-strand biogenesis: pif1Δ or pif1-m2 act as suppressors of dna2 mutations, and can restore viability to dna2Δ strains11, and Pif1 stimulates strand displacement by Pol δ in vitro12. These results have been interpreted to support a two-nuclease model of Okazaki fragment processing in which Dna2 is responsible for the cleavage of Fen1-resistant long 5’-flaps generated by excessive strand displacement22. Conversely, rrm3Δ is synthetic lethal with dna2 mutants10. Because Rrm3 is able to promote Pol δ processivity, we think it unlikely that genome-wide Okazaki fragment processing underlies the genetic interactions between DNA2 and PIF1/RRM3. While Rrm3 can apparently substitute for Pif1 during bulk Okazaki fragment processing, it cannot analogously replace Pif1 during Break-induced replication – another Pol δ-dependent synthesis reaction37; determining the molecular basis for this differential helicase requirement will provide insights into both canonical and break-induced replication.
tDNAs are the predominant site of replication fork arrest in cells lacking Rrm3 or Rrm3 and Pif1
As previously reported, we find that Rrm3 contributes significantly to replisome passage through tDNAs in S. cerevisiae, but we report a novel role for Pif1, which can partially compensate for the absence of Rrm3 at these sites. In the absence of both helicases, all tDNAs have the potential to stably arrest the replication fork. Indeed, the arrest signal observed at tDNAs is sufficiently pronounced that analysis of replisome mobility at non-tDNA elements requires the removal of tDNA-proximal sites (Fig. S5H). While it is difficult to conclusively show that there is no replisome arrest at non-tDNA loci in cells lacking Pif1 and/or Rrm3, the signal at tDNAs clearly reflects the vast majority of fork stalling or arrest genome-wide and is evident even in WT cells when the tDNA is oriented head-on with respect to fork progression (Fig. 5B). Therefore, while we cannot formally conclude that G-quadruplexes and RNAP2 genes do not induce any replisome stalling or arrest in pif1 and/or rrm3 mutant cells, we establish that any such changes in fork mobility are extremely rare compared to those observed at tDNAs.
Some previous reports of tDNA-mediated replisome arrest suggested positive supercoiling ahead of the tDNA as a contributing mechanism due to relatively de-localised stalling by 2-D gel and a strong orientation bias32. Our data indicate that both co-oriented and head-on collisions lead to significant fork arrest centered precisely on the tDNA (Fig. 4B). R-loops do not contribute significantly to the observed arrest (Fig. 6), and a parsimonious interpretation of our data is that the RNAP3 transcription complex itself represents an asymmetric barrier to the passage of the replication fork. The leading edge of RNA polymerase has previously been identified as a replication barrier in prokaryotes38. Moreover, the observation that both Pif1 and Rrm3 suppress replisome arrest equivalently for both tDNA orientations lends further support to the characterization of these proteins as replisome-accessory ‘sweepases’.
Redundant Pif1 helicase activities and S-phase transcription
Fast-growing eukaryotes inevitably face conflicts between replication and transcription because the complete cessation of RNAP1 and RNAP3 transcription during S-phase – one third of the 90-minute cell cycle in S. cerevisiae39 – would limit growth rate by reducing protein-synthesis capacity. Reduced need for RNAP3 transcription during S-phase in slower-growing cells of higher eukaryotes may underlie the mild phenotypes observed in PIF1 knockout mice40. However, metazoan Pif1 may have a more significant impact on replication dynamics and genome integrity in transformed cells41.
The S. cerevisiae rDNA repeats, transcribed by RNAP1, are unidirectionally replicated to avoid head-on collisions between replication and transcription42,43. Unlike the rDNA, tDNAs are dispersed throughout the S. cerevisiae genome, but head-on collisions have been disfavored by evolution – presumably because they have a greater capacity to arrest replisomes than do co-directional collisions (Fig. 5B). However, we note that S. cerevisiae has maintained a fraction of tDNAs in the head-on orientation (Fig. 5E). It was recently proposed that Bacillus subtilis maintains some essential genes in the normally disfavored head-on orientation to speed their evolution by stimulating replication-transcription conflicts44, which lead to TLS-dependent mutagenesis45. A possible extension of this hypothesis to budding yeast would be that fork arrest at tDNAs could stimulate recombination between transposons adjacent to tDNAs, thereby stimulating genomic rearrangements.
Reconciling the absence of replication fork stalling with increased rates of genomic instability at G-quadruplexes and R-loops
We provide evidence against substantial, prolonged replisome stalling or arrest at G-quadruplexes, even in a pif1-m2 rrm3Δ strain grown in the presence of low concentrations of hydroxyurea (Fig 4 and S5D). While predicted G-quadruplex sequences exist in the S. cerevisiae genome, it is unclear how many actually form stable intramolecular structures in vivo in the absence of G4-stabilizing drugs46. However, Pif1 helicases from both eukaryotes and prokaryotes preferentially unwind G-quadruplexes in vitro8. Moreover, mutation rate, microsatellite instability, and gross chromosomal rearrangements around model G-quadruplexes increase in the absence of Pif1 activity8,27,31. Therefore, G-quadruplexes are unstable in the absence of Pif1 even under conditions where they do not broadly impact replisome mobility. It is possible that fork blockage at G-quadruxplexes remains below the level of detection of our assay, but that techniques that select for rare events may identify biologically relevant but extremely rare fork impediments, or damage that is independent of replisome stalling. Even so, since Pif1 recruitment to G-quadruplexes has been reported to occur long after replisome passage27, we propose that genome instability due to these structures is separable from replication fork stalling, and rather reflects a post-replicative repair phenomenon. Similarly, R-loops are associated with DNA damage, but our data suggest that they do not significantly stall the replication fork even in the absence of Rrm3 (Fig. 6). Recent experiments in human cells have demonstrated that R-loop structures are repaired via a NER-like pathway prone to double-stranded breaks that do not require the collision of a replication fork and instead are the direct result of R-loop processing events47. Our data support a model in which replisome stalling and DNA damage due to both R-loop and G-quadruplex formation are mechanistically separable, and underscore the importance of using a damage-independent assay to directly evaluate replisome mobility.
ONLINE METHODS
In vivo methods
All S. cerevisiae strains are derived from W303 RAD5+, and were grown in YEPD at 30°C unless otherwise noted. To minimize the accumulation of suppressors in pif1-m2 rrm3Δ, which has been reported to show elevated genomic instability8, we froze stocks of all strains after the minimum feasible number of divisions and conducted all experiments using freshly streaked cells from these stocks. The three wild-type, rrm3Δ, pif1-m2 and pif1-m2 rrm3Δ strains used were independently derived strains taken from segregants from three single tetrads following sporulation of a pif1-m2/PIF1 rrm3/RRM3 CDC9REP/CDC9REP diploid. Strains that used a Gal1,10 promoter to drive the expression of RNH1 were grown overnight in the presence of galactose (2%) prior to ligase repression.
Following growth to mid-log phase, DNA ligase expression was repressed in asynchronous cultures by treatment with 40 mg/L doxycycline for 2.5h. For replication stress experiments, hydroxyurea was added to a final concentration of 25 mM 1h prior to addition of doxycycline, which was added to cultures at the same concentration for 2h. Okazaki fragments were labeled, purified, and deep-sequenced as previously described17. Paired end sequencing (2 × 50bp) was carried out on the Illumina Hi-seq 2500 platform.
Computational methods
Reads were mapped to the S288c reference genome48 using Bowtie (v2.2.3) and converted into bamfiles using samtools, removing PCR duplicates with the MarkDuplicates function in picard tools. In-house python and command line scripts were used to convert to 1-based bedpe and sorted bedfiles, with the strand and 5’- and 3’-end of each read indicated. We used bedtools to convert bedfiles into stranded sgr files giving per base read coverage over the genome; Python was used to calculate the percent of total reads mapping to either the Watson or Crick strand. All calculations were carried out using un-smoothed data. 1 was added to all values to avoid dividing by 0. Datasets were internally normalized by calculating the percent reads mapping to the Watson or Crick strand (i.e. a value independent of read depth) and only combined after analysis when we calculated the grand mean and p-value for fork arrest (see below).
For the analysis of fork progression at sites of interest (tDNAs, RNA Pol II genes, G-quadruplex forming sequences, random genomic positions), we determined the average fork movement at that site (leftward or rightward) by the percent Watson strand hits within 1kb of the site (>50% indicates leftward-moving; <50% indicates rightward-moving). Random sites were generated by sampling lines from an sgr file to ensure equal coverage per base per chromosome. We calculated the percent of rightward-moving forks by the percent of reads mapping to the Crick strand. We then analyzed leftward- and rightward-moving forks separately so that we could reverse the direction of leftward-moving forks and sum the average percent of forks moving unidirectionally over a 10kB window of analysis (i.e. value = average(rev(percentWatson(left-to-right)) + (100−(percentWatson(right-to-left))), see Figure 2C and D and Figure S3A).
To calculate the percent change in fork direction at each site, we used a window 1–3kB upstream and downstream of the site (see Figure 2D). The change in mean reads mapping to the Watson strand over this window indicates the percent of forks that have stalled or arrested at this site (see Figure 2B). Bar graphs indicate the grand mean of three independent experiments and the standard deviation of the three independent means. For all classes of sites (i.e. random sites versus G-quadruplex sites), we excluded sites that have an origin or tDNA within the 10kB window of analysis or that have a 100bp bin with no data unless otherwise noted. Sites were binned into co-directional and head-on by the average fork direction within 1kb of the site, as described above, and the direction of transcription (or the strand of the G-quadruplex forming structure). For RNA Pol II genes, we took the top 10% of genes expressed in YEP28 as ‘highly transcribed,’ but had similar results with more or less stringent subsets. We calculated the termination signal at the transcription start site, midpoint, and transcription termination site of the gene, but only report the transcription start site as the data were similar (i.e. no significant differences between strains).
Drip-seq data18 were accessed via Geo and analyzed using an in-house pipeline. DRiP-seq signal around tDNAs was calculated using a 400bp window and tDNAs were sorted by their integrated DRiP-seq signal in WT (see Figure 6A).
Statistical methods
We evaluated the significance of the change in replication direction between datasets and sites by Monte Carlo resampling. Briefly, we compared the grand mean of two treatments (e.g. WT versus pif1-m2 rrm3Δ at 93 tDNAs) before creating a dataset that conformed to the null hypothesis by randomly assigning data into the two treatments. Data were randomly assigned 10,000 times, and the difference in grand mean calculated for each sampling; the number of times that the resampled data (i.e. the null hypothesis) recreated the difference in means calculated for the data denoted the p-value. Monte Carlo resampling was done many times (at 10,000 trials per run) until a p-value was converged upon. For datasets with more than 93 sites, we randomly sampled 93 of the total sites before randomly assigning data into the two treatments; when the p-value varied widely between runs (due to different lines in the file being tested), data were considered insignificant (p>0.05) if more than half of the trials returned insignificant p-values; in these cases, we looked for smaller numbers of sites that were driving the potentially significant differences (see Figure S5E). In Figures, (*) denotes a significant p-value between 0.05 and 0.0002; (***) denotes a p-value less than 0.0001, wherein our many trials of 10,000 resamplings never produced a single run where the observed difference in means could be explained by random sampling.
2-Dimensional gel analysis
The plasmid for our 2-Dimensional gel analyses (Figure 3B) was cloned by standard techniques from pRS426, which contained a Ura marker and 2µ yeast origin of replication. We replaced the entire 2µ origin sequence with ARS309 and inserted either Pgk1 or tG(TCC)C with 250bp of their native upstream promoter sequences to drive expression. 100ml cells were grown in -URA media to mid-log phase, collected by centrifugation, and resuspended in two 0.75ml aliquots in 50mM Tris HCl pH 8.0, 50mM EDTA, 100µg/ml psoralen before 12min crosslinking at 365nm. Cells were then pelleted, washed in 0.5ml TE, pelleted again and stored at −20°C until analysis.
DNA was purified by standard techniques. Briefly, cell pellets were resuspended in 0.5ml TE buffer with 1:100 β-ME and 250µg/ml zymolyase (T100) and incubated for 30 minutes before the addition of 100µl lysis buffer (500mM Tris HCl pH 8.0, 0.25M EDTA, 3% SDS) and 20µl Proteinase K (Roche). After 2h at 50°C, 150uL 5M potassium acetate was added and cell debris was pelleted for 30min at top speed at 4°C. The supernatant was precipitated with ethanol, washed, dried and resuspended in 750µl TE with RNase A (50µg/ml) for at least 1h at 37°C before phenol:cholorform extraction and isopropanol precipitation. Pellets were resuspended in 150µl TE o/n and digested o/n with EcoRI-HF (tG(TCC)C or empty vector) or EcoRV (Pgk1) in 600µl total volume with 400 units of enzyme. DNA was precipitated prior to loading on a 0.4% agarose gel run at room temperature (22V, 18–24h). Slices containing the size of interest were cut, turned counter-clockwise 90°, and run on a 0.95% agarose gel containing 0.3µg/ml Ethidium Bromide (130V, 18h, 4°C). Arcs were visualized, cut, nicked by autocrosslinking (Strata-linker), depurinated by treatment with acid (0.25N HCl), treated with denaturing solution (0.5N NaOH) and equilibrated in blotting solution (1.5MNaCl, 0.25N NaOH) before transfer onto Hybond N+ nylon membrane o/n. Blots were neutralized in 50mM sodium phosphate (pH 7.2), equilibrated at 65°C in hybridization buffer (0.25M Na phosphate pH 7.2, 0.25 M NaCl, 1mM EDTA, 7% SDS, 5% Dextran Suflate) before the addition of radiolabeled (Invitrogen Random Primers DNA Labeling Kit) probe (against the Ampr gene of the plasmid, see Figure 3B). Blots were washed 5× 100ml in low (2× SSC, 0.1%SDS) and high (0.1× SSC, 0.1% SDS) stringency washes, patted dry, and exposed to phosphoimager screens. 2-demensional gel analyses are representative of at least two independent experiments.
Supplementary Material
Acknowledgments
We thank V. Zakian and members of the Zakian lab for the 2-D gel protocol, for communicating data prior to publication, and for helpful discussions. We additionally thank S. Ercan, A. Hochwagen, H. Klein, and members of the Smith lab for insightful discussions and critical reading of the manuscript, D. Tranchina for help with statistical analyses, and V. Subramanian for assistance with 2-D gel electrophoresis. This work was supported by NIH grant R01 GM114340, a March of Dimes Basil O’Connor Starter Scholar award (FY15-BOC-2141) and the Searle Scholars program (all to D.J.S). J.S.O is supported by an American Cancer Society - New York Cancer Research Fund postdoctoral fellowship (PF-16-096-01-DMB).
Footnotes
AUTHOR CONTRIBUTIONS
J.S.O., J.K. and R.Y. generated data; J.S.O., J.K. and D.J.S. analyzed data and interpreted results; J.S.O. and D.J.S. wrote the manuscript with input from J.K. and R.Y.
COMPETING FINANCIAL INTERESTS
None declared.
DATA AVAILABILITY
Raw and processed sequencing data have been deposited to the Gene Expression Omnibus under accession number GSE71973.
REFERENCES
- 1.Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 3.Dulev S, et al. Essential global role of CDC14 in DNA synthesis revealed by chromosome underreplication unrecognized by checkpoints in cdc14 mutants. Proc Natl Acad Sci U S A. 2009;106:14466–14471. doi: 10.1073/pnas.0900190106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waters LS, Walker GC. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G(2)/M phase rather than S phase. Proc Natl Acad Sci U S A. 2006;103:8971–8976. doi: 10.1073/pnas.0510167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochman ML, Sabouri N, Zakian VA. Unwinding the functions of the Pif1 family helicases. DNA Repair (Amst) 2010;9:237–249. doi: 10.1016/j.dnarep.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boule JB, Zakian VA. Roles of Pif1-like helicases in the maintenance of genomic stability. Nucleic Acids Res. 2006;34:4147–4153. doi: 10.1093/nar/gkl561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boule JB, Zakian VA. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 2007;35:5809–5818. doi: 10.1093/nar/gkm613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paeschke K, et al. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou J, Monson EK, Teng SC, Schulz VP, Zakian VA. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science. 2000;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- 10.Budd ME, et al. A network of multi-tasking proteins at the DNA replication fork preserves genome stability. PLoS Genet. 2005;1:e61. doi: 10.1371/journal.pgen.0010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budd ME, Reis CC, Smith S, Myung K, Campbell JL. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol Cell Biol. 2006;26:2490–2500. doi: 10.1128/MCB.26.7.2490-2500.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pike JE, Burgers PM, Campbell JL, Bambara RA. Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J Biol Chem. 2009;284:25170–25180. doi: 10.1074/jbc.M109.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azvolinsky A, Dunaway S, Torres JZ, Bessler JB, Zakian VA. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 2006;20:3104–3116. doi: 10.1101/gad.1478906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 2005;19:1905–1919. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivessa AS, et al. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 16.McGuffee SR, Smith DJ, Whitehouse I. Quantitative, Genome-Wide Analysis of Eukaryotic Replication Initiation and Termination. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–438. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El Hage A, Webb S, Kerr A, Tollervey D. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet. 2014;10:e1004716. doi: 10.1371/journal.pgen.1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulz VP, Zakian VA. The saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 20.Myung K, Chen C, Kolodner RD. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- 21.Chen X, et al. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat Struct Mol Biol. 2011;18:1015–1019. doi: 10.1038/nsmb.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balakrishnan L, Bambara RA. Okazaki fragment metabolism. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang C, Pugh BF. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 2009;10:R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petryk N, et al. Replication landscape of the human genome. Nat Commun. 2016;7:10208. doi: 10.1038/ncomms10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sekedat MD, et al. GINS motion reveals replication fork progression is remarkably uniform throughout the yeast genome. Mol Syst Biol. 2010;6:353. doi: 10.1038/msb.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell. 2009;34:722–734. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paeschke K, Capra JA, Zakian VA. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelechano V, Wei W, Steinmetz LM. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497:127–131. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu H, Li X. Transcriptional regulation in eukaryotic ribosomal protein genes. Genomics. 2007;90:421–423. doi: 10.1016/j.ygeno.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 30.Capra JA, Paeschke K, Singh M, Zakian VA. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput Biol. 2010;6:e1000861. doi: 10.1371/journal.pcbi.1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopes J, et al. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011;30:4033–4046. doi: 10.1038/emboj.2011.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deshpande AM, Newlon CS. DNA replication fork pause sites dependent on transcription. Science. 1996;272:1030–1033. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- 33.Raghuraman MK, et al. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- 34.Kaback DB, Rosbash M, Davidson N. Determination of cellular RNA concentrations by electron microscopy of R loop-containing DNA. Proc Natl Acad Sci U S A. 1981;78:2820–2824. doi: 10.1073/pnas.78.5.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santos-Pereira JM, et al. The Npl3 hnRNP prevents R-loop-mediated transcription-replication conflicts and genome instability. Genes Dev. 2013;27:2445–2458. doi: 10.1101/gad.229880.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sollier J, et al. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell. 2014;56:777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vasianovich Y, Harrington LA, Makovets S. Break-induced replication requires DNA damage-induced phosphorylation of Pif1 and leads to telomere lengthening. PLoS Genet. 2014;10:e1004679. doi: 10.1371/journal.pgen.1004679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mirkin EV, Castro Roa D, Nudler E, Mirkin SM. Transcription regulatory elements are punctuation marks for DNA replication. Proc Natl Acad Sci U S A. 2006;103:7276–7281. doi: 10.1073/pnas.0601127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brewer BJ, Chlebowicz-Sledziewska E, Fangman WL. Cell cycle phases in the unequal mother/daughter cell cycles of Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2529–2531. doi: 10.1128/mcb.4.11.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snow BE, et al. Murine Pif1 interacts with telomerase and is dispensable for telomere function in vivo. Mol Cell Biol. 2007;27:1017–1026. doi: 10.1128/MCB.01866-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gagou ME, et al. Human PIF1 helicase supports DNA replication and cell growth under oncogenic-stress. Oncotarget. 2014;5:11381–11398. doi: 10.18632/oncotarget.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brewer BJ, Fangman WL. A replication fork barrier at the 3’ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 43.Kobayashi T, Horiuchi T. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells. 1996;1:465–474. doi: 10.1046/j.1365-2443.1996.d01-256.x. [DOI] [PubMed] [Google Scholar]
- 44.Paul S, Million-Weaver S, Chattopadhyay S, Sokurenko E, Merrikh H. Accelerated gene evolution through replication-transcription conflicts. Nature. 2013;495:512–515. doi: 10.1038/nature11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Million-Weaver S, et al. An underlying mechanism for the increased mutagenesis of lagging-strand genes in Bacillus subtilis. Proc Natl Acad Sci U S A. 2015;112:E1096–E1105. doi: 10.1073/pnas.1416651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piazza A, et al. Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J. 2015;34:1718–1734. doi: 10.15252/embj.201490702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sollier J, et al. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell. 2014;56:777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engel SR, et al. The reference genome sequence of Saccharomyces cerevisiae: then and now. G3 (Bethesda) 2014;4:389–398. doi: 10.1534/g3.113.008995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Homann OR, Johnson AD. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol. 2010;8:49. doi: 10.1186/1741-7007-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.