Abstract
Aims
To investigate the hypothesis that alteration in histone acetylation/deacetylation triggers aberrant STAT1/MyD88 expression in macrophages from diabetics. Increased STAT1/MyD88 expression is correlated with sterile inflammation in type 1 diabetic (T1D) mice.
Methods
To induce diabetes, we injected low-doses of streptozotocin in C57BL/6 mice. Peritoneal or bone marrow-differentiated macrophages were cultured in 5 mM (low) or 25 mM (high glucose). ChIP analysis of macrophages from nondiabetic or diabetic mice was performed to determine acetylation of lysine 9 in histone H3 in MyD88 and STAT1 promoter regions. Macrophages from diabetic mice were treated with the histone acetyltransferase inhibitor anacardic acid (AA), followed by determination of mRNA expression by qPCR.
Results
Increased STAT1 and MyD88 expression in macrophages from diabetic but not naive mice cultured in low glucose persisted for up to 6 days. Macrophages from diabetic mice exhibited increased activity of histone acetyltransferases (HAT) and decreased histone deacetylases (HDAC) activity. We detected increased H3K9Ac binding to Stat1/Myd88 promoters in macrophages from T1D mice and AA in vitro treatment reduced STAT1 and MyD88 mRNA expression.
Conclusions/interpretation
These results indicate that histone acetylation drives elevated Stat1/Myd88 expression in macrophages from diabetic mice, and this mechanism may be involved in sterile inflammation and diabetes comorbidities.
Keywords: histone acetylation, metabolic memory, diabetes, inflammation, macrophage, epigenetic
Introduction
Sterile inflammation, a chronic low-grade systemic inflammation, is the main cause of comorbidities such as retinopathy and nephropathy that are associated with diabetes. Sterile inflammation is associated with increased systemic levels of pro-inflammatory cytokines, including elevated interleukin (IL)-1β, which drives several comorbidities (Chen and Nunez 2010, Filgueiras, Serezani et al. 2015). The IL-1 receptor is a member of the Toll-Like Receptor (TLR) family, and like most TLR receptors, its signaling pathway is dependent on the adaptor molecule Myeloid Differentiation factor 88 (MyD88) (Takeuchi and Akira 2002).
We have previously shown that macrophages from type 1 diabetic (T1D) mice express increased levels of MyD88 and its transcription factor Signal Transducer and Activator of Transcription (STAT) 1, which is dependent on the production of the inflammatory lipid mediator leukotriene B4 (LTB4). Our results showed that the elevated systemic levels of LTB4 found in T1D mice promote the binding of STAT1 to MyD88 promoter, leading to enhanced MyD88 expression in macrophages. Increased STAT/MyD88 expression potentiates macrophages TLR/IL1β receptors response. Moreover, the sterile inflammation in T1D mice characterized by increased systemic levels of TNF-α and IL-1β, is dependent on the LTB4-induced STAT1/MyD88 axis (Filgueiras, Brandt et al. 2015). Because some long-term diabetes complications develop even in insulin-treated patients who exhibit improved glucose control, it is thought that epigenetic mechanisms associated with diabetes are responsible, a process characterized as “metabolic memory” (Ceriello 2009, Cooper and El-Osta 2010).
Epigenetic mechanisms are typically mediated through chromatin dynamics. Chromatin is a highly organized structure in which DNA is wrapped around a histone octamer unit. The structure of chromatin plays a central role in controlling transcription of genes. Chromatin structure is dynamic, and is tightly regulated by several epigenetic mechanisms including histone modifications. Histones can be acetylated at multiple lysine residues, and the specific combination of acetylated lysines determines the accessibility of the DNA template to the transcriptional machinery enabling gene expression. Thus, acetylation is generally associated with increased transcriptional activity. Histone acetyltransferases (HATs) mediate histone acetylation, whereas, histone deacetylases (HDACs) reduce transcription by removing acetyl groups from lysine residues (Kouzarides 2007, Li, Carey et al. 2007).
Accumulating evidence suggests that in both diabetic animals and human patients, key aspects of regulating the transcription of genes involved in the development of diabetes comorbidities are dependent on epigenetic mechanisms (Reddy, Zhang et al. 2015). Thus, we sought to determine the contribution of epigenetic histone acetylation in increased STAT-1/MyD88 expression associated with development of diabetes in T1D mice.
Material and Methods
Animals
Eight-week-old male C57BL/6 mice were maintained according to National Institutes of Health guidelines for the use of experimental animals with the approval of the Indiana University Committees for the Use and Care of Animals and Ethical Committee for Animal Research of the Institute of Biomedical Sciences of the University of São Paulo.
Diabetes induction
T1D was chemically induced by five sequential daily intraperitoneal injections of a freshly prepared solution of streptozotocin (40 mg/kg) (Filgueiras, Brandt et al. 2015). Blood glucose concentrations were measured 10 days after the last injection of STZ using OneTouch Select glucometer and test strips (Life Scan). Mice were considered diabetic when blood glucose concentrations reached >300 mg/dl on two consecutive days. The control group received five intraperitoneal injections of the vehicle.
Macrophage isolation and differentiation
Resident peritoneal macrophages were isolated by lavage of the peritoneal cavity with cold PBS and cultured as described (Serezani, Lewis et al. 2011). Peritoneal macrophages were treated with the noncompetitive and specific inhibitor of class I and II histone deacetylases trichostatin A (TSA -10 µM) (Vigushin, Ali et al. 2001) or the p300 and PCAF histone acetyltransferase inhibitor anacardic acid (AA- 10 µM or 30 µM) (Mai, Rotili et al. 2006) for 24 hours followed by RNA isolation.
Bone marrow-derived macrophages were obtained as previously described (Davies and Gordon 2005), with minor modifications. In brief, femurs were flushed with DMEM (Dulbecco’s modified eagle medium containing 2mM l-glutamine, 100U/mL penicillinG, and 100mg/mL streptomycin, all from Gibco, Long Island, NY, USA), using a 26-G ×½ G needle. Cells were grown in high (25 mM) or low (5 mM) glucose DMEM containing 20 ng/mL M-CSF (Pepro Tech, Brazil) and 5% of heat-inactivated fetal calf serum (FCS), incubated at 37°C in 5% CO2. On day 3, new fresh DMEM with M-CSF (20 ng/mL) was added. A monolayer of macrophages was scraped on day 6 (96% of the cells were positive for CD11b and F4/80 (Ferracini, Rios et al. 2013).
RNA isolation and qPCR
Total RNA from cultured cells was isolated using Direct-zol RNA MiniPrep (Zymo Research) according to the manufacturer’s instruction. Complementary DNA (cDNA) was synthesized using a reverse transcription system (RevertAid First Strand cDNA Synthesis Kit, Thermo Scientific) and qPCR was performed with Fast SYBR® Green Master Mix (Applied Biosystems) containing primers for Myd88, Stat1, Stat3 and Hprt (all from Integrated DNATechnologies) on the Step One Plus Real-Time PCR Detection System (Applied Biosystems). Relative expression was calculated using the comparative threshold cycle (Ct) and calculated relative to control or wild type (DDCt method).
ChIP assay
ChIP assays were performed with a SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology) according to the manufacturer’s protocol. Cells were fixed and cross-linked with 1% formaldehyde, and chromatin was digested with micrococcal nuclease and sonicated (UP100H; Hielscher) to obtain DNA fragments of about 150 to 900 base pairs. The resulting cross-linked chromatin preparation was subsequently enriched by immunoprecipitation with anti-H3K9Ac (1:100; Cell Signaling Technology) or anti- di-methyl-H3K79 (1:100; Cell Signaling Technology) antibodies. Normal rabbit immunoglobulin G (IgG) (1:100) and anti–histone H3 (1:50) antibodies (Cell Signaling Technology) were used as negative and positive controls, respectively. For each immunoprecipitation, 20 mg of cross-linked chromatin was diluted in ChIP buffer to a final volume of 0.5 ml, mixed with indicated antibodies, and incubated for 4 hours at room temperature with rotation. Immune complexes were captured using 30 ml of ChIP-Grade Protein G Magnetic Beads (Cell Signaling Technology) according to the manufacturer’s protocol. The chromatin was eluted from the beads by adding elution buffer and incubating at 65°C for 30 min followed by digestion with proteinase K for 2 hours at 65°C. Subsequently, DNA was purified using spin columns (Qiagen), and samples were subjected to real-time PCR using the primers for different promoter regions of Stat1, Stat3 or Myd88.
Results
Persistent increase in MyD88 and STAT1 gene expression in macrophages from diabetic mice
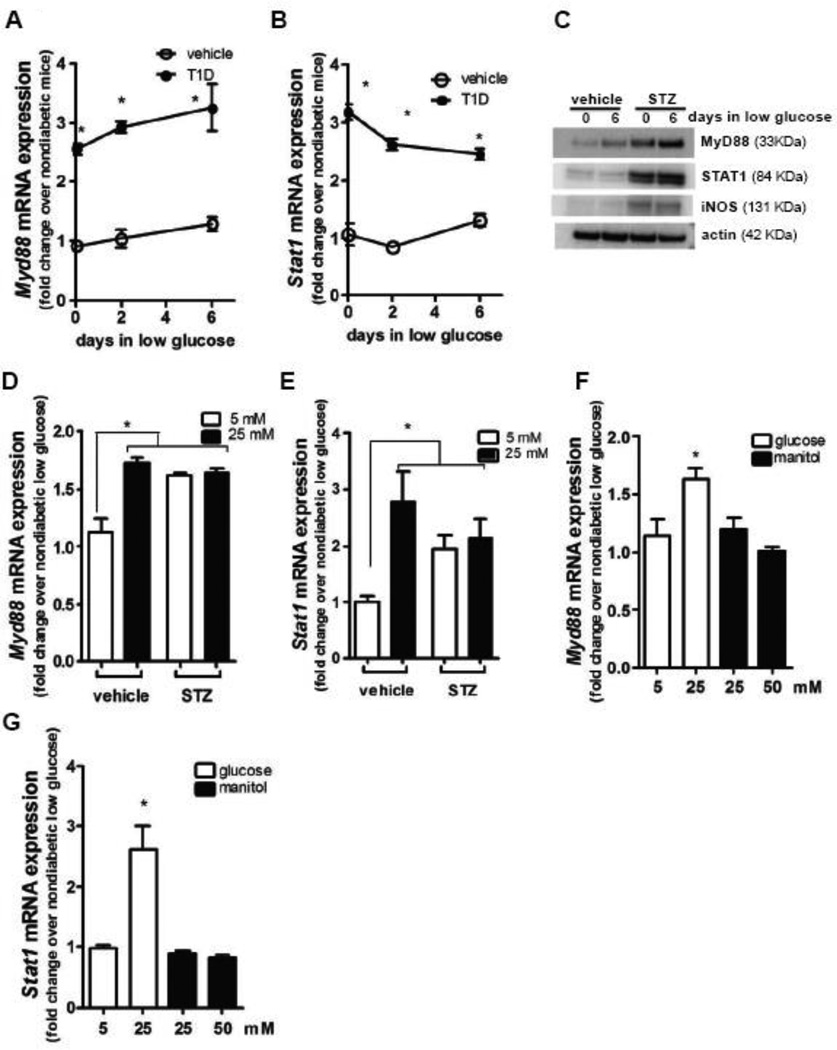
We have previously shown that peritoneal macrophages from T1D mice exhibited higher expression of MyD88 and STAT1 than macrophages from nondiabetic animals (Filgueiras, Brandt et al. 2015), although the mechanisms remain unknown. Initially, we sought to determine whether the observed increase in STAT1/MyD88 expression in macrophages from T1D mice could be restored to normal levels by culturing cells in low glucose medium. The results showed that expression was not normalized, as increased STAT1/MyD88 mRNA and protein expression persisted for at least 6 days in T1D macrophages incubated in low glucose medium (Fig 1A to C). Because diabetes skews macrophages toward pro-inflammatory phenotype (Kanter, Kramer et al. 2012, Filgueiras, Brandt et al. 2015), we also analyzed iNOS expression and found that after 6 days in low glucose medium, macrophages from T1D mice still presented increased iNOS expression (Fig 1C).
Figure 1.
Resident peritoneal macrophages from T1D mice and nondiabetic mice were cultured in low glucose (5 mM) for 6 days. At 2 and 6 day time points, Myd88 (A) and Stat1 (B) mRNA expression were determined by qPCR, and MyD88, STAT1, iNOS and actin protein expression were determined after 6 days of culture by immunoblotting (C). Blots shown are from one representative experiment (n=3). Bone marrow-derived macrophages from nondiabetic and T1D mice were differentiated in 5 mM or 25 mM of glucose, and Myd88 (D) and Stat1 (E) expression was determined by qPCR. Bone marrow-derived macrophages from nondiabetic were differentiated in low 5 mM or 25 mM of glucose or Data are expressed as mean ± SEM from at least 3 independent experiments; *p<0.05 compared to macrophages from nondiabetic mice or compared to macrophages differentiated from bone marrow of nondiabetic mice in 5 mM glucose.
We then tested whether increased Stat1/Myd88 expression by macrophages in T1D mice was initiated in precursor cells in bone marrow or was restricted to mature macrophages. When differentiated in low glucose medium, macrophages derived from bone marrow of T1D mice exhibited increased Stat1/Myd88 levels, whereas, macrophages differentiated from bone marrow of nondiabetic mice did not. When differentiated in high glucose medium Stat1/Myd88 expression by macrophages differentiated from bone marrow of T1D mice was not higher than expression by T1D macrophages differentiated in low glucose medium. However, Stat1/Myd88 expression by macrophages differentiated from bone marrow of nondiabetic mice in high glucose medium was higher than expression by macrophages differentiated in low glucose (Fig 1D and E). This was not observed when manitol was added for osmotic control (Fig 1F and G), indicating that indeed hyperglycemia promotes expression of these genes.
Together these results suggest that diabetic condition promote epigenetic mechanisms that induce a persistent increase in macrophage Stat1 and Myd88 gene expression.
Imbalance in HAT/HDAC activities in macrophages from T1D mice correlates with increased MyD88 expression
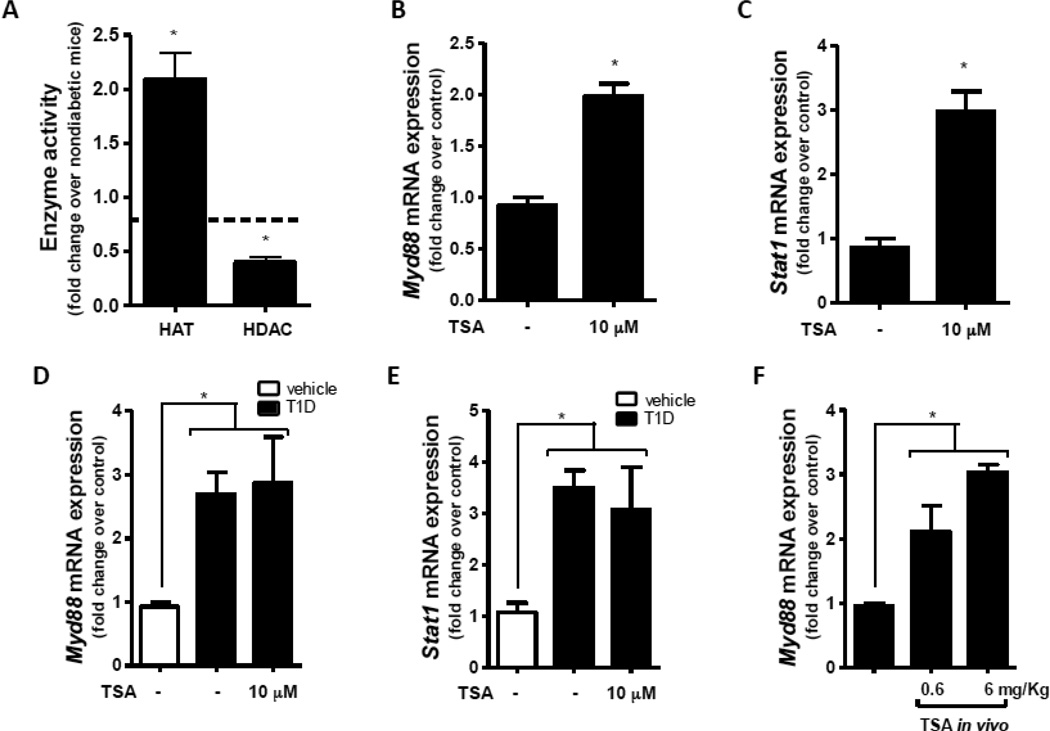
To address the possibility that these effects of glucose levels during macrophage differentiation on STAT1/MyD88 expression involved epigenetic regulation, we investigated the activity of enzymes involved in histone acetylation. We found that total HAT activity was increased and total HDAC activity was decreased in macrophages from T1D mice (Fig 2A) relative to normal macrophages. When we treated macrophages from nondiabetic mice with the potent HDAC activity inhibitor Trichostatin A (TSA), which favors histone acetylation (Vigushin, Ali et al. 2001), we found that expression of Myd88 (Fig 2B) and Stat1 (Fig 2C) were enhanced. This was not observed when T1D macrophages were treated with TSA (Fig. 1D and E). We observed a similar effect when nondiabetic mice were treated in vivo with TSA (Fig. 2F). These results are consistent with the possibility that epigenetic histone acetylation drives increased Stat1/Myd88 expression in T1D.
Figure 2.
Activity of HAT and HDAC was analyzed in the nuclear fraction of peritoneal macrophages from T1D and nondiabetic mice as described in Materials and Methods (A). Macrophages from nondiabetic mice were cultured with or without TSA (10 µM) for 24 h, and Myd88 (B) and Stat1 (C) expression were determined by qPCR. Nondiabetic mice (n=4 mice per group) were treated in vivo with TSA (0.6 or 6 mg/Kg) for 24 h; resident peritoneal macrophages were harvested, and Myd88 (D) expression was determined by qPCR. Data are expressed as mean ± SEM from at least 3 independent experiments; *p<0.05 compared to macrophages from nondiabetic mice or compared to peritoneal macrophages isolated from vehicle treated cells.
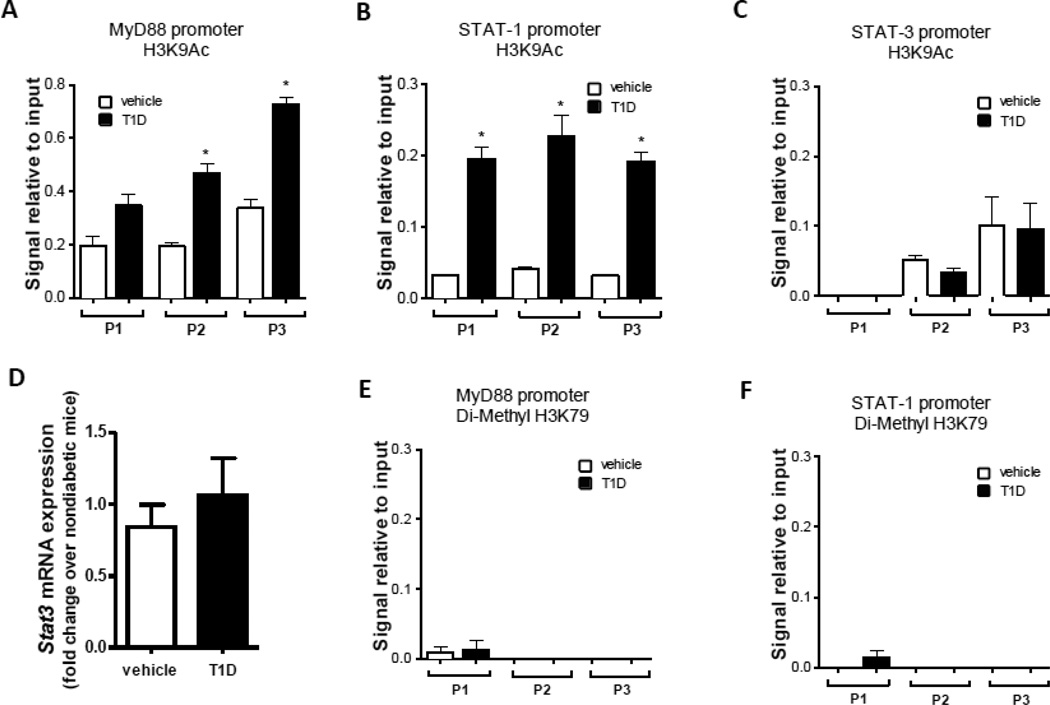
Acetylation in lysine 9 residues of histone H3 on Myd88 and Stat1 promoter region in macrophages from diabetic mice
We next determined whether the lysine 9 residues of histone H3 (H3K9) associated with promoter regions of Myd88 and Stat1 were acetylated in macrophages during T1D; acetylation of H3K9 (H3K9Ac) is a key histone modification involved in transcription (Reddy, Zhang et al. 2015). In macrophages from T1D but not nondiabetic mice, we found H3K9Ac in 2 different regions of the MyD88 promoter (Fig. 3A) and in 3 different regions of the Stat1 promoter (Fig. 3B). In this same analysis, we did not detect any difference in H3K9Ac between macrophages from T1D and nondiabetic mice in three tested regions of the STAT3 promoter (Fig. 3C), a gene that is not modulated in STZ-induced T1D (Fig 3D). Similarly, when we assessed another histone modification, H3K79 dimethylation, we found no differences between macrophages from T1D and nondiabetic mice (Fig. 3E and F). Thus, these results suggest that H3K9Ac on the MyD88 promoter is specifically associated with T1D.
Figure 3.
Formaldehyde-fixed chromatin from macrophages from T1D and nondiabetic mice was immunoprecipitated (IP) to enrich (A–C) H3K9Ac and (D, E) Di-Methyl H3K79 complex using specific antibodies followed by qPCR as described in Materials and Methods. Stat3 expression was determined in macrophages from T1D mice and nondiabetic mice by qPCR (D). Data are expressed as means ± SEM from 3 independent experiments; *p<0.05 compared to macrophages from nondiabetic mice.
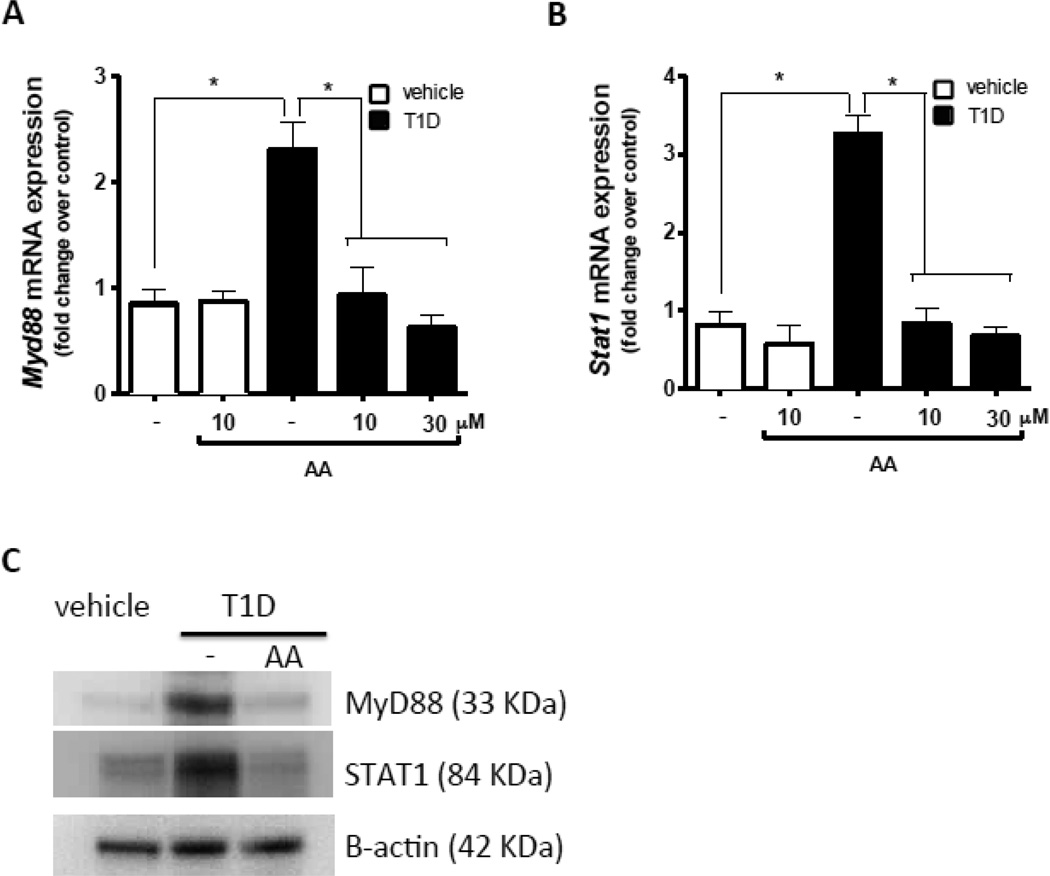
Increased HAT activity in T1D mice leads to enhanced MyD88 and STAT1 expression in macrophages
We next sought to determine whether increased expression of MyD88 and STAT1 in macrophages from diabetic mice was a consequence of imbalanced HAT and HDAC activity that resulted in increased histone acetylation. Macrophages from nondiabetic and T1D mice were treated in vitro with the HAT inhibitor anacardic acid (Mai, Rotili et al. 2006). We found that anacardic acid treatment reduced mRNA (Fig. 4A and B) and protein (Fig. C) expression of Myd88 and Stat1 mRNA by T1D macrophages to normal levels. These results provide direct evidence that the increased expression of STAT1 and MyD88 in macrophages from T1D mice can be blocked by inhibiting H3K9Ac.
Figure 4.
Macrophages from T1D mice (n=4) and nondiabetic mice (n=5) were cultured with or without anacardic acid (AA) for 24 h. Myd88 (A) and Stat1 (B) expression were determined by qPCR, and MyD88, STAT1 and actin protein expression were determined by immunoblotting (C). Immunoblots are shown from one representative experiment (n=3). Data are expressed as means ± SEM from 3 independent experiments; *p<0.05 compared to macrophages from nondiabetic mice.
Discussion
Epigenetic mechanisms are involved in inflammatory diseases such as arthritis and asthma (Ho 2010, Viatte, Plant et al. 2013). Here, we demonstrated an essential role of histone acetylation in the increased expression of MyD88 and STAT1 in macrophages from diabetic mice. Macrophages from diabetic mice presented increased HAT and decreased HDAC activity. Modulation of HAT and HDAC activity was associated with enhanced Myd88 and Stat1 gene expression. ChIP analysis showed that macrophages from diabetic mice presented acetylation in the promoter region of theses genes in H3K9. Moreover, when macrophages were treated with HAT inhibitor the gene expression of Myd88 and Stat1 was inhibited to the same level found in macrophages from non-diabetic animals.
An increasing body of evidence suggests that epigenetic modifications, subsequent to a priming event, are involved in reprogramming innate immune cells in normal and pathologic immune responses. Upon restimulation, macrophages exhibit enhanced responses, which boost host defense and inflammation, a process termed innate immune memory (Netea, Latz et al. 2015). In Alzheimer’s disease, there is evidence that chronic inflammation promotes innate reprogramming of microglia and astrocytes leading to uncontrolled NLRP3 activation and IL-1β production (Miao, Chen et al. 2014). Because diabetes is associated with chronic inflammation, it is to be expected that macrophages would be reprogramed, resulting in increased sterile inflammation.
The occurrence of chromatin modifications have been described in diabetes; these include H3K4 monomethylation in the promoter of p65 in response to hyperglycemia, which leads to increased expression of p65-responsive genes in endothelial cells (Reddy, Zhang et al. 2015). In the present study, we found that T1D promoted H3K9Ac in the promoters of Myd88 and Stat1 in macrophages, leading to enhanced expression of these genes. This same epigenetic mark is found in COX-2 and TNF-a promoters in monocytes from diabetic patients (Reddy, Zhang et al. 2015). Moreover, monocytes from diabetic patients exhibit a higher than normal average number of regions enriched in H3K9Ac (Miao, Chen et al. 2014). It was uncertain, however, whether this acetylation mark occurred in the promoter region of MyD88 and STAT1 in macrophages from people with diabetes patients.
The observation that culturing macrophages in low glucose medium did not restore Stat1/Myd88 expression to normal levels indicates that their elevated expression is associated with metabolic memory. The concept of “metabolic memory” came from the observation that diabetic complications persist long after attainment of glycemic control. As a consequence of hyperglycemia and oxidative stress, epigenetic mechanisms that alter chromatin structure drive persistent gene expression, which results in long lasting chronic inflammation (Reddy, Zhang et al. 2015). These chromatin alterations associated with chronic inflammation are present in mature macrophages and in macrophage precursors in bone marrow.
Increased Stat1/Myd88 expression in T1D mice is associated with sterile inflammation, and several studies have shown that this inflammation is a primary cause of diabetes comorbidities (Filgueiras, Brandt et al. 2015, Filgueiras, Serezani et al. 2015). The fact that HAT inhibitor restored normal Stat1/Myd88 expression in macrophages from T1D mice suggests that therapeutic strategies aimed at reducing Stat1/Myd88 expression in macrophages could provide a means to address diabetes comorbidities.
Table 1.
Primers used for qPCR.
| Gene | Primer sequence |
|---|---|
| Hprt | F: AGCAGGTCAGCAAAGAACT |
| R: CCTCATGGACTGATTATGGACA | |
| Myd88 | F: TAGTCGCAGACAGTGATGAAC |
| R: CTGCAGAGCAAGGAATGTGA | |
| Stat1 | F: GACTTCAGACACAGAAATCAACTC |
| R: TTGACAAAGACCACGCCTT | |
| Stat3 | F: CATGTCAAACGTGAGCGACT |
| R: GGAAATAACGGTGAAGGTGCT |
Table 2.
Primers used for ChIP on the murine Stat1 promoter.
| Region | Region in murine Stat1 promoter |
Primer sequence |
|---|---|---|
| P1 | +52 to + 391 | F: GCGCAAGCCTATAGTTTCCA |
| R: GTCTGGGCAAATCTCTCTGC | ||
| P2 | +51 to +522 | F: GCGCAAGCCTATAGTTTCCA |
| R: TCTCTCTGCGGGATGACAG | ||
| P3 | −154 to −753 | F: GTCAGTGGAGGTCCGAAGAG |
| R: CGAGTCTGGGCAAATCTCTC |
Table 3.
Primers used for ChIP on the murine Myd88 promoter.
| Region | Region in murine Myd88 promoter |
Primer sequence |
|---|---|---|
| P1 | −93 to + 81 | F: CACAAGTGGGTTGACTTTTAGGCT |
| R: GGCAGGCAACCCTGGGCCCCCGG | ||
| P2 | −569 to −280 | F: AATAGATTAACCAAGTGAATTAA |
| R: TATTCTGGTAGTAGGGAGGGAAGAG | ||
| P3 | −1053 to −877 | F: GAAGGAGGTTTCCCAAACTTCTGGTT |
| R: ACCCGGGGCTGAGCACAGCAAA |
Table 4.
Primers used for ChIP on the murine Stat3 promoter.
| Region | Region in murine Stat3 promoter |
Primer sequence |
|---|---|---|
| P1 | −140 to −470 | F: GGTGACACCTGGGGACCGCCTAAG |
| R: AAAAACGCCTCTAGGAGAGAAGGC | ||
| P2 | cre binding | F: GGGGGAGGGAGGAGACATTA |
| R: GAGCCGTATCAGGGCATTTA | ||
| P3 | −1600 | F: CCATTGGGGGATTATCTTTG |
| R: TGGTGTTTGTATTGCTGGTGA |
Acknowledgments
Funding: This work was supported by NIH grants (HL-103777-01 and HL-12159- 01) and Fundação de Amparo a Pesquisa do Estado de São Paulo
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interest: The authors declare that they have no competing interest.
References
- Ceriello A. "Hypothesis: the "metabolic memory", the new challenge of diabetes". Diabetes Res Clin Pract. 2009;86(Suppl 1):S2–S6. doi: 10.1016/S0168-8227(09)70002-6. [DOI] [PubMed] [Google Scholar]
- Chen GY, Nunez G. "Sterile inflammation: sensing and reacting to damage". Nat Rev Immunol. 2010;10(12):826–837. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper ME, El-Osta A. "Epigenetics: mechanisms and implications for diabetic complications". Circ Res. 2010;107(12):1403–1413. doi: 10.1161/CIRCRESAHA.110.223552. [DOI] [PubMed] [Google Scholar]
- Davies JQ, Gordon S. "Isolation and culture of murine macrophages". Methods Mol Biol. 2005;290:91–103. doi: 10.1385/1-59259-838-2:091. [DOI] [PubMed] [Google Scholar]
- Ferracini M, Rios FJ, Pecenin M, Jancar S. "Clearance of apoptotic cells by macrophages induces regulatory phenotype and involves stimulation of CD36 and platelet-activating factor receptor". Mediators Inflamm. 2013;2013:950273. doi: 10.1155/2013/950273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filgueiras LR, Brandt SL, Wang S, Wang Z, Morris DL, Evans-Molina C, Mirmira RG, Jancar S, Serezani CH. "Leukotriene B4-mediated sterile inflammation promotes susceptibility to sepsis in a mouse model of type 1 diabetes". Sci Signal. 2015;8(361):ra10. doi: 10.1126/scisignal.2005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filgueiras LR, Serezani CH, Jancar S. "Leukotriene B4 as a Potential Therapeutic Target for the Treatment of Metabolic Disorders". Front Immunol. 2015;6:515. doi: 10.3389/fimmu.2015.00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SM. "Environmental epigenetics of asthma: an update". J Allergy Clin Immunol. 2010;126(3):453–465. doi: 10.1016/j.jaci.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanter JE, Kramer F, Barnhart S, Averill MM, Vivekanandan-Giri A, Vickery T, Li LO, Becker L, Yuan W, Chait A, Braun KR, Potter-Perigo S, Sanda S, Wight TN, Pennathur S, Serhan CN, Heinecke JW, Coleman RA, Bornfeldt KE. "Diabetes promotes an inflammatory macrophage phenotype and atherosclerosis through acyl-CoA synthetase 1". Proc Natl Acad Sci U S A. 2012;109(12):E715–E724. doi: 10.1073/pnas.1111600109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. "Chromatin modifications and their function". Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. "The role of chromatin during transcription". Cell. 2007;128(4):707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Mai A, Rotili D, Tarantino D, Ornaghi P, Tosi F, Vicidomini C, Sbardella G, Nebbioso A, Miceli M, Altucci L, Filetici P. "Small-molecule inhibitors of histone acetyltransferase activity: identification and biological properties". J Med Chem. 2006;49(23):6897–6907. doi: 10.1021/jm060601m. [DOI] [PubMed] [Google Scholar]
- Miao F, Chen Z, Genuth S, Paterson A, Zhang L, Wu X, Li SM, Cleary P, Riggs A, Harlan DM, Lorenzi G, Kolterman O, Sun W, Lachin JM, Natarajan R, Group DER. "Evaluating the role of epigenetic histone modifications in the metabolic memory of type 1 diabetes". Diabetes. 2014;63(5):1748–1762. doi: 10.2337/db13-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Latz E, Mills KH, O'Neill LA. "Innate immune memory: a paradigm shift in understanding host defense". Nat Immunol. 2015;16(7):675–679. doi: 10.1038/ni.3178. [DOI] [PubMed] [Google Scholar]
- Reddy MA, Zhang E, Natarajan R. "Epigenetic mechanisms in diabetic complications and metabolic memory". Diabetologia. 2015;58(3):443–455. doi: 10.1007/s00125-014-3462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serezani CH, Lewis C, Jancar S, Peters-Golden M. "Leukotriene B4 amplifies NF-kappaB activation in mouse macrophages by reducing SOCS1 inhibition of MyD88 expression". J Clin Invest. 2011;121(2):671–682. doi: 10.1172/JCI43302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. "MyD88 as a bottle neck in Toll/IL-1 signaling". Curr Top Microbiol Immunol. 2002;270:155–167. doi: 10.1007/978-3-642-59430-4_10. [DOI] [PubMed] [Google Scholar]
- Viatte S, Plant D, Raychaudhuri S. "Genetics and epigenetics of rheumatoid arthritis". Nat Rev Rheumatol. 2013;9(3):141–153. doi: 10.1038/nrrheum.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigushin DM, Ali S, Pace PE, Mirsaidi N, Ito K, Adcock I, Coombes RC. "Trichostatin A is a histone deacetylase inhibitor with potent antitumor activity against breast cancer in vivo". Clin Cancer Res. 2001;7(4):971–976. [PubMed] [Google Scholar]