Abstract
Introduction
Urinary Tract Infections (UTIs) caused by Uropathogenic Escherichia coli (UPEC) are among the most common infections worldwide. It is well-documented that the pathogenesis of UPEC is mediated by the production of a wide variety of Virulence Factors (VFs). Thus, detection of these VFs and evaluation of their association with different clinical types of UTIs could help to understand the role of these factors in pathogenesis of UPEC isolates.
Aim
To investigate the genotypic characteristics of UPEC isolates and to examine the relationship between VFs and different clinical symptoms of UTI.
Materials and Methods
In this cross-sectional study conducted at Pasteur Institute of Iran, a total of 156 UPEC isolated from outpatients and inpatients (symptomatic and asymptomatic UTI patients) visiting general and private hospitals in Tehran, Iran between March 2014 and February 2015 were included. Among them, 49 patients experienced at least one episode of recurrent UTI. A Polymerase Chain Reaction (PCR) assay was developed to detect the presence of different VFs in the isolates. Moreover, Pulsed-Field Gel Electrophoresis (PFGE) was used to characterize clonal relationships among UPEC isolates.
Results
The prevalence of virulence genes ranged from 0% for cdtB to 100% for fimH. The papEF, hlyA and aer genes were found to be significantly more frequent in UPEC isolated from patients with pyelonephritis, while the afa gene, the only indicator of recurrent UTIs, was more prevalent in UPEC isolated from patients with cystitis. In the present study, 34 PFGE clonal groups were found in the UPEC genome.
Conclusion
Our findings showed that from outpatients and patients with pyelonephritis, isolates were more virulent than those isolated from inpatients and cystitis patients, respectively. PFGE displayed a large diversity in the UPEC isolates that could be considered as an evolutionary strategy in the survival of the bacteria.
Keywords: Pulsed-field gel electrophoresis, Urinary tract infection, Uropathogenic Escherichia coli
Introduction
Urinary Tract Infections (UTIs) are known to be one of the most frequently encountered infections both in community and hospital settings [1]. Although UTIs are often considered to be easily treatable infections, an estimated 150-250 million cases of UTI are diagnosed annually worldwide, accounting for more than 7 million physician office visits and over 100,000 hospital admissions in the United States [2,3]. Approximately 40% of women and 12% of men will experience at least one symptomatic UTI within their lifetime, about 25% of whom will suffer from another UTI (recurrent UTI) [3]. Recurrent UTI (RUTI) is a common problem among young and healthy women of all ages, being associated with increased healthcare costs [4]. UTIs are classified as either symptomatic or asymptomatic cases; symptomatic UTI is defined as the presence of local or systemic symptoms, including cystitis and pyelonephritis infections, while Asymptomatic Bacteriuria (ABU) is characterized by the presence of significant bacteria in the urinary tract without clinical symptoms [5,6]. Studies have shown that clinical manifestations of UTI patients are highly variable in severity, depending on both the pathogenic properties of the causative agent and host susceptibility to infection [7,8].
Among the causative agents of UTIs, Uropathogenic Escherichia coli (UPEC) are the most common bacterial pathogens causing community-acquired (70-90%) and nosocomial (40-50%) UTI in all age ranges [9]. The UPEC strains, which are epidemiologically and phylogenetically distinct from the commensal E. coli residing in the gastrointestinal tract, encode a variety of Virulence Factors (VFs), playing an important role in bacterial colonization, pathogenesis and persistence in the urinary tract [8]. The most important VFs associated with UPEC strains include adherence factors (P fimbriae, S and F1C fimbriae, type 1 fimbriae and afimbrial adhesins), toxins (α-haemolysin and cytotoxic necrotizing factor type 1), flagella, iron acquisition systems and polysaccharide coatings (group II and III capsules) [8,10].
Molecular characterization of the VFs and evaluation of genotypic diversity among the UPEC strains support the need of better understanding of the UPEC pathogenesis as well as the development of effective therapies against UTIs.
Materials and Methods
Epidemiological and demographical characteristics and all other clinically relevant information were obtained from patients with UTI by examining the hospital administrative database and/or by interviewing the patients. A random collection of 156 E.coli isolated from urine samples of patients with UTI (females=79.5% and male=20.5%) containing significant counts (≥105CFU/ml) were included in this study. Moreover, only patients aged ≥20 years, who received no antibiotic treatment during the sample collection period or had no antibiotic use within a month prior to hospital admission, were registered in this study. All the isolates were collected from 54 outpatients and 102 inpatients, including 103, 22 and 31 patients with cystitis, pyelonephritis and ABU, respectively. Cystitis was defined by the presence of dysuria and urinary frequency and urgency, while acute pyelonephritis was characterized by the presence of fever (38°C) plus flank pain and/or lumbar tenderness, with or without symptoms of cystitis [3]. On the other hand, ABU was manifested by the presence of bacteriuria without clinical symptoms [11]. During a 6-month follow-up, 49 (31.4%) of UTI cases had at least two episodes of recurrent UTI. Recurrent UTI was defined as ≥2 episodes of UTI in six months or ≥3 UTI during a 12-month period [4]. Ethical approval for this study was granted by the scientific and Ethics Committee of Pasteur Institute of Iran.
Bacterial Identification and DNA Extraction
The isolated bacteria was identified as UPEC on the basis of cultural and biochemical analyses [12]. E.coli isolates were grown on Luria Bertani (LB) broth at 37°C overnight and genomic DNA was extracted using the DNA purification Kit (Roche Diagnostics GmbH, Mannheim, Germany). Purity of the extracted DNA was measured using a NanoDrop™ spectrophotometer. DNA samples with an OD260/OD280 ratio of ≥1.8 were used for further analysis. Moreover, the quality of extracted DNA was examined by 1% agarose gel electrophoresis (Sigma Chemical Co.) [13].
Detection of Virulence Genes
Polymerase Chain Reaction (PCR) (eppendorf thermocycler) was used to detect the presence of virulence genes fimH, papEF, sfa/foc, afa, hlyA, cnf1, iron, aer, iuc, fliC, usp and cdtB. Specific primers (Genfanavaran, Tehran, Iran) [Table/Fig-1] [11,14,15] were used to amplify the genes encoding urovirulence factors. Amplification reactions were carried out in a total volume of 25μl, containing 1μl of DNA template, 2.5μl of 10x reaction buffer, 1μl of dNTPs (10mM), 1μl of Mgcl2 (50mM), 1μl of each primer (10pmol), and 1U Taq DNA polymerase (Fermentas, Lithuania). The PCR condition was as follows: an initial denaturation at 94°C for 5 min, followed by 30 cycles, each consisting of denaturation at 94°C for 1min; annealing at specific temperature [Table/Fig-1] for 1min; and elongation at 72°C for 1min and a final extension step at 72°C for 5min. After amplification, 5μl sample of each reaction was electrophoresed on a 1% agarose gel to confirm the presence of expected PCR products. After staining with ethidium bromide, banding patterns were visualized and photographed using a GelDoc system (UVP, Upland, Calif.) under UV fluorescence. A 100-bp and 1kbp DNA ladders (Fermentas, Lithuania), as a size standard, were used for band-size comparison of the PCR products. All amplification procedures were performed in duplicate.
[Table/Fig-1]:
Primers used to amplify virulence associated genes.
| Target gene | Primer sequence (5′→3′) | Size of PCR product (bp) | Annealing temp (°C) | Reference |
|---|---|---|---|---|
| papEF | F: GCAACAGCAACGCTGGTTGCATCAT R: AGAGAGAGCCACTCTTATACGGACA |
336 | 62 | [14] |
| sfa/foc | F: CTCCGGAGAACTGGGTGCATCTTAC R: CGGAGGAGTAATTACAAACCTGGCA |
410 | 60 | [11] |
| afa | F: GCTGGGCAGCAAACTGATAACTCTC R: CATCAAGCTGTTTGTTCGTCCGCCG |
750 | 62 | [15] |
| fimH | F: ATGAAACGAGTTATTACC R: TTGATAAACAAAAGTCAC |
900 | 58 | Present study |
| hlyA | F: AACAAGGATAAGCACTGTTCTGGCT R: ACCATATAAGCGGTCATTCCCGTCA |
1177 | 58 | [14] |
| cnf1 | F: AAGATGGAGTTTCCTATGCAGGAG R: CATTCAGAGTCCTGCCCTCATTATT |
498 | 59 | [14] |
| cdtB | Fa1: AAATCACCAAGAATCATCCAGTTA Ra2: AAATCTCCTGCAATCATCCAGTTTA |
430 | 58 | [11] |
| Fs1: GAAAGTAAATGGAATATAAATGTCCG Rs2:GAAAATAAATGGAACACACATGTCCG |
63 | |||
| iron | F: AAGTCAAAGCAGGGGTTGCCCG R: GACGCCGACATTAAGACGCAG |
665 | 59 | [15] |
| aer | F: TACCGGATTGTCATATGCAGACCGT R: AATATCTTCCTCCAGTCCGGAGAAG |
602 | 60 | [14] |
| iuc | F: ATGAGAATCATTATTGACATAATTG R: CTCACGGGTGAAAATATTTT |
1482 | 48 | [15] |
| fliC | F: ATGCCATGGCGATGGCACAAGTCATTAAT R: CCCAAGCTTACCCTGCAGCAGAGACAG |
1000-2000 | 58 | Present study |
| usp | F: ACATTCACGGCAAGCCTCAG R: AGCGAGTTCCTGGTGAAAGC |
448 | 58 | [15] |
Genomic Fingerprinting
Based on the UPEC patterns and patient profiles, Pulsed-Field Gel Electrophoresis (PFGE) was performed on 40 representative UPEC isolates according to the Centers for Disease Control and Prevention Pulse Net protocol [16]. Briefly, the isolates were cultured on Trypticase Soy Agar (TSA) (Merck, Darmstadt, Germany) at 37°C for 18h. Bacterial cultures were embedded in 1% agarose plugs (SeaKem Gold agarose; Cambrex, Rockland, ME), lysed, washed and digested separately with the restriction enzyme XbaI (Fermentas, Lithuania) for at least 2 hour at 37°C. Restricted DNA fragments were separated by electrophoresis for 19 hour in 1% agarose gels by electrophoresis using a CHEF-DR III system (Bio-Rad, Richmond, CA, USA). Voltage was set at 6V/cm with switch times of 2.2s to 54.2s. XbaI-digested Salmonella braenderup H9812 DNA was used as a reference size standard. After electrophoresis, the gels were stained with ethidium bromide, and fragments were photographed under U.V. illumination using a Gel Doc 2000 system (UVP, Upland, Calif.). PFGE patterns were then analysed and interpreted using the Gel compare II, version 6.5 software (Applied Maths, St Martens-Latem, Belgium). Clonal relationships between isolates, based on band position, were assessed by using the Un-weighted Pair Group Method with Arithmetic mean (UPGMA) [16]. In this light, isolates were considered to be derived from the same cluster when their dice similarity index was ≥80%. Moreover, the similarity of 40 representative UPEC isolates was assessed according to the presence or absence of genotypic factors, by which a dendrogram was generated using binary patterns of VF genes for each isolate.
Statistical Analysis
Statistical analysis was performed using SPSS software version 19.0 for windows (IBM, Chicago, USA). Chi-square and Fisher’s-exact tests (2-tailed) were used to compare proportions. Furthermore, pair wise correlations were assessed using Phi coefficients. The p-value less than 0.05 was considered to be statistically significant.
Results
Distribution of Virulence Genes
Overall, the fimH gene was the most frequent virulence gene found in the UPEC isolates studied (100%), while the cdtB gene was found to be negative in all of the isolates (0%) [Table/Fig-2]. Among toxin and iron-related genes studied, the cnf1 (36.5%) and aer (73.1%) genes were the most prevalent ones [Table/Fig-2]. Relative prevalence ratios of papEF, sfa/foc, hlyA and iron genes were 2.2 to 3.5 times more prevalent among outpatient compared with inpatient isolates [Table/Fig-2].
[Table/Fig-2]:
Prevalence of urovirulence genes among UPEC isolates, grouped according to inpatient- vs. outpatient origin.
| Virulence genes | No. (%) of positive isolates in patients with UTI | Total No. (%) of positive isolates (n=156) |
||
|---|---|---|---|---|
| Inpatients (n=102) |
Outpatients (n=54) |
|||
| Adhesin genes | papEF | 25 (24.5) | 31 (57.4) | 56 (35.9) |
| sfa/foc | 7 (6.9) | 13 (24.1) | 20 (12.8) | |
| afa | 24 (23.5) | 22 (40.7) | 46 (29.5) | |
| fimH | 102 (100) | 54 (100) | 156 (100) | |
| Toxin genes | hlyA | 22 (21.6) | 26 (48.1) | 48 (30.8) |
| cnf1 | 29 (28.4) | 28 (51.8) | 57 (36.5) | |
| cdtB | 0 (0) | 0 (0) | 0 (0) | |
| Iron-related genes | iron | 26 (25.5) | 32 (59.2) | 58 (37.2) |
| aer | 71 (69.6) | 43 (79.6) | 114 (73.1) | |
| iuc | 41 (40.2) | 34 (63) | 75 (48.1) | |
| Flagella gene | fliC | 64 (62.7) | 41 (75.9) | 105 (67.3) |
| Other genes | usp | 49 (48.0) | 28 (51.8) | 77 (49.4) |
Pap, Pyelonephritis-associated pili; sfa, S fimbrial adhesin; afa, Afimbrial adhesin; fimH, Type 1 fimbriae; hlyA, α-haemolysin; cnf1, cytotoxic necrotizing factor type 1; cdtB, cytolethal distending toxin; aer, aerobactin; iuc, Iron uptake chelate; fliC, flagellin gene; usp, uropathogenic specific protein.
Comparison of the frequency of urovirulence genes with clinical diagnosis of UTI indicated that papEF, hlyA and aer genes were significantly more frequent among the pyelonephritis isolates, when compared to cystitis and asymptomatic bacteriuria isolates [Table/Fig-3]. Furthermore, iuc was found to be more frequently associated with pyelonephritis than cystitis isolates (p=0.028). However, fliC and afa genes were more detected in cystitis isolates than pyelonephritis ones (p=0.015 and p=0.05, respectively). In addition, the results from this study demonstrated that there is a statistical significant difference between the presence of the afa gene and a history of recurrent UTI in patients (p=0.004). In contrast, two VFs, including cnf1 (p=0.02) and hlyA (p=0.001), were associated with the absence of RUTI [Table/Fig-4].
[Table/Fig-3]:
Distribution of virulence factors (VFs) among UPEC isolates from patients with cystitis, pyelonephritis, and asymptomatic bacteriuria.
| VF | Prevalence of VF, No. (%) of isolates | p-valuesa for comparison of | |||||
|---|---|---|---|---|---|---|---|
| Cystitis (n=103) |
Pyelone-phritis (n=22) |
ABU (n=31) |
All 3 clinical groups | Cystitis vs. pyelone-phritis | Cystitis vs. ABU | Pyelone-phritis vs. ABU | |
| papEF | 34 (33.0) | 18 (81.8) | 4 (12.9) | 0.008* | 0.012* | NS | 0.015* |
| sfa/foc | 12 (11.6) | 4 (18.2) | 4 (12.9) | NS | NS | NS | NS |
| afa | 36 (34.9) | 3 (13.6) | 7 (22.6) | NS | 0.05 | 0.041* | NS |
| hlyA | 27 (26.2) | 15 (68.2) | 6 (19.4) | 0.031* | 0.019* | NS | 0.025* |
| cnf1 | 35 (34) | 14 (63.6) | 8 (25.8) | NS | NS | NS | NS |
| iron | 39 (37.9) | 9 (40.9) | 10 (32.3) | NS | NS | NS | NS |
| aer | 81 (78.6) | 20 (90.9) | 13 (41.9) | 0.001* | NS | 0.01* | 0.023* |
| iuc | 45 (43.7) | 17 (77.3) | 13 (41.9) | NS | 0.028* | NS | NS |
| fliC | 87 (84.5) | 13 (59.1) | 5 (16.1) | NS | 0.015* | NS | 0.04* |
| usp | 54 (52.4) | 16 (72.7) | 7 (22.6) | NS | NS | NS | NS |
a, p-values (by 2-tailed Fisher’s exact test) are shown where p is <0.05.
*, Significant
NS, Not significant
ABU, Asymptomatic bacteriuria
[Table/Fig-4]:
![[Table/Fig-4]:](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/1cc2/5296426/f7307f194669/jcdr-10-DC01-g001.jpg)
Distribution (percentage) of gene encoding virulence factors among UPEC isolates from patients with or without recurrent UTI.
RUTI, Recurrent urinary tract infection
*, Significant at p<0.05
Association between Virulence Genes
Pairwise comparisons of virulence genes were plotted against each other and a wide variety of distinctive associations was found between the VFs. Interestingly, there was a significant correlation between hlyA and all VFs surveyed in this study [Table/Fig-5]. The co-occurrence analysis revealed that hlyA was positively associated with the occurrence of papEF, sfa/foc, cnf1, iron and usp, but negatively did with afa, aer, iuc and fliC genes. Interestingly, a negative association was detected between the fliC gene and papEF, hlyA, cnf1 and usp genes [Table/Fig-5].
[Table/Fig-5]:
Pairwise statistical associations between virulence genes of UPEC isolates.
| sfa/foc | afa | hlyA | cnf1 | iron | aer | iuc | fliC | usp | |
|---|---|---|---|---|---|---|---|---|---|
| papEF | +** | NS | +** | +** | +* | NS | -* | -** | +* |
| sfa/foc | NS | +** | +** | +** | -** | NS | NS | +** | |
| afa | -** | -** | NS | +* | NS | NS | NS | ||
| hlyA | +** | +** | -** | -* | -** | +** | |||
| cnf1 | NS | NS | NS | -** | +** | ||||
| iron | NS | NS | NS | +* | |||||
| aer | +** | NS | NS | ||||||
| iuc | NS | NS | |||||||
| fliC | -** |
Significant levels: **p≤0.01, *0.01<p<0.05
+, Positive correlation
-, Negative correlation
NS, Not significant
Genotypic Virulence Patterns
The UPEC isolates exhibited 74 different virulence patterns, 44 and 13 of which displayed single and duplicate patterns, respectively (data not shown). The UPEC isolates showed 17 most common virulence patterns, referred to as UP (UPEC pattern) [Table/Fig-6]. UP 1, which is characterized by the presence of only the fimH and fliC genotypic markers, was the most predominant pattern found in 12 isolates. In addition, the occurrence of multiple virulence markers (isolates with three or more virulence markers) was observed in 141 (90.4%) of the UPEC isolates tested (results not shown).
[Table/Fig-6]:
Common virulence patterns identified among the UPEC isolates.
| Pattern | Virulence factor | NO. (%) of isolates | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| papEF | sfa/foc | afa | fimH | hlyA | cnf1 | cdtB | iron | aer | iuc | fliC | usp | ||
| UP 1 | - | - | - | + | - | - | - | - | - | - | + | - | 12 (7.7) |
| UP 2 | + | - | - | + | + | + | - | - | + | - | - | + | 8 (5.1) |
| UP 3 | - | - | - | + | - | - | - | - | + | + | + | + | 6 (3.8) |
| UP 4 | - | - | + | + | - | - | - | - | + | - | + | - | 6 (3.8) |
| UP 5 | - | - | - | + | - | - | - | - | + | + | + | - | 6 (3.8) |
| UP 6 | - | - | + | + | - | - | - | - | + | + | + | - | 6 (3.8) |
| UP 7 | - | - | + | + | - | - | - | + | + | + | - | + | 5 (3.2) |
| UP 8 | - | - | - | + | - | - | - | - | + | - | + | + | 4 (2.6) |
| UP 9 | - | - | - | + | - | + | - | - | + | + | + | - | 4 (2.6) |
| UP 10 | - | - | - | + | - | - | - | + | + | + | + | + | 4 (2.6) |
| UP 11 | + | + | - | + | + | + | - | + | - | - | + | + | 4 (2.6) |
| UP 12 | - | - | - | + | - | + | - | - | + | + | + | + | 4 (2.6) |
| UP 13 | - | - | - | + | - | - | - | - | + | - | + | - | 4 (2.6) |
| UP 14 | + | - | + | + | - | - | - | + | + | + | + | - | 4 (2.6) |
| UP 15 | + | + | - | + | + | + | - | + | - | - | - | + | 3 (1.9) |
| UP 16 | - | - | - | + | - | - | - | + | + | - | - | - | 3 (1.9) |
| UP 17 | - | + | - | + | + | + | - | + | - | - | - | + | 3 (1.9) |
UP, UPEC pattern
Analysis of Genomic Fingerprint Profiles of UPEC Isolates by PFGE
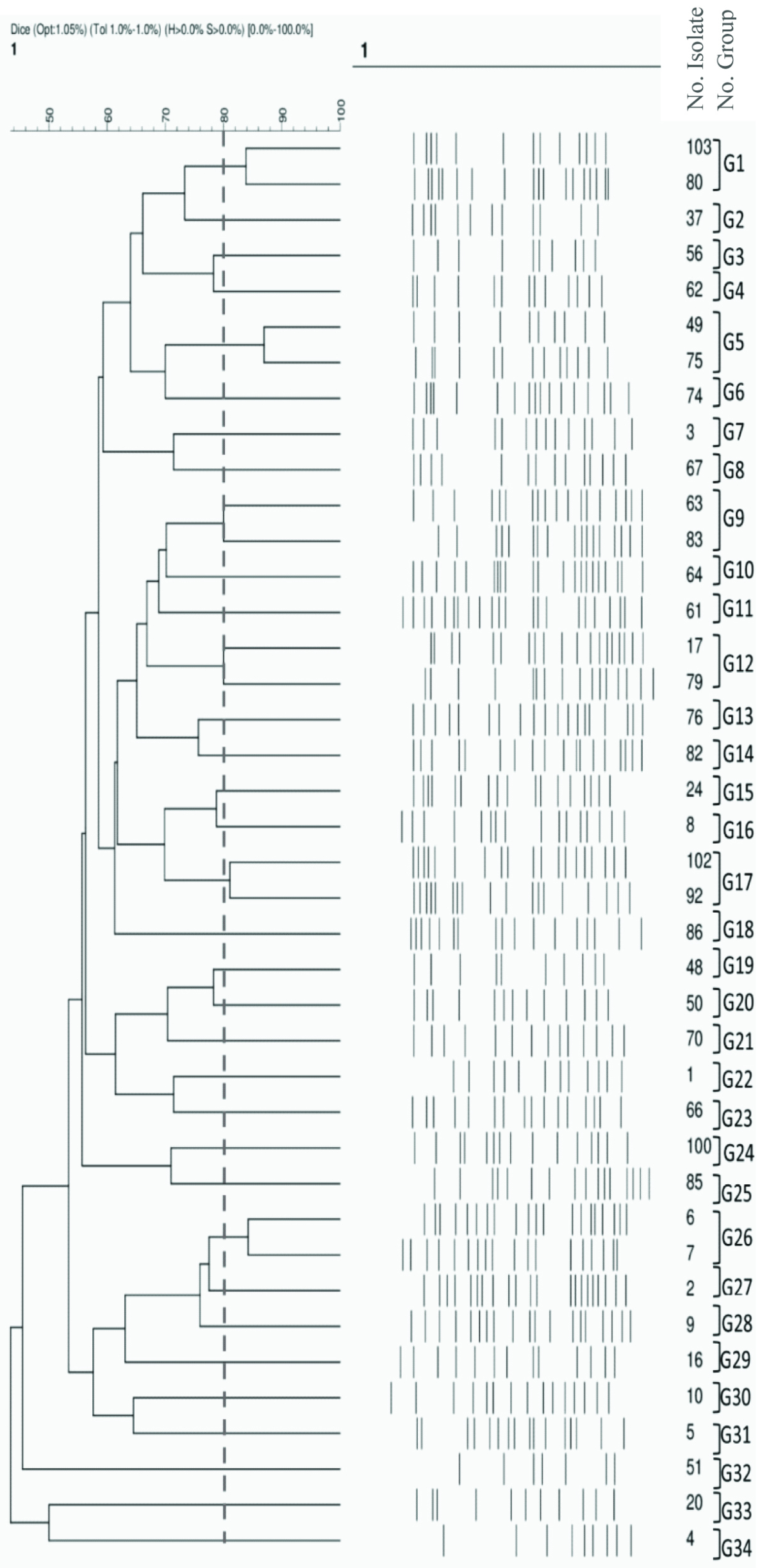
In our study, 40 representative UPEC isolates were clustered into 34 PFGE clonal groups based on the drawn dendrogram (G1 to G34, [Table/Fig-7]). A majority of the isolates showed 15 to 20 bands, although the isolates with less than 10 bands had the least percentage. The similarity range had been between 46-87%. Furthermore, the maximum and minimum numbers of isolates were found in six PFGE groups, each consisting of two isolates, and 28 PFGE groups, each consisting of one isolate, respectively [Table/Fig-7].
[Table/Fig-7]:
Phylogenetic tree diagram. XbaI-digested DNA from the 40 representative UPEC isolates included in this study. Dendrogram was constructed based on UPGMA by using Dice coefficient with a 1.0% band position tolerance. The scale above the dendrogram indicates percentage of similarity. The dotted line indicates 80% similarity. G, Group; UP, UPEC Pattern; RUTI, Recurrent Urinary Tract Infection.
 |
Patient status | Type of UTI | RUTI | UP. No |
| Inpatient | Cystitis | Absence | UP13 | |
| Inpatient | Pyelonephritis | Absence | Other UP | |
| Inpatient | Cystitis | Present | UP7 | |
| Outpatient | Pyelonephritis | Present | Other UP | |
| Outpatient | Pyelonephritis | Absence | Other UP | |
| Inpatient | Cystitis | Absence | Other UP | |
| Inpatient | Cystitis | Present | UP8 | |
| Outpatient | Pyelonephritis | Absence | Other UP | |
| Outpatient | Pyelonephritis | Absence | UP15 | |
| Inpatient | ABU | Present | Other UP | |
| Inpatient | Cystitis | Absence | Other UP | |
| Inpatient | Cystitis | Absence | UP3 | |
| Outpatient | Cystitis | Absence | Other UP | |
| Inpatient | ABU | Present | Other UP | |
| Inpatient | Cystitis | Absence | Other UP | |
| Outpatient | Pyelonephritis | Absence | UP15 | |
| Inpatient | Cystitis | Absence | UP8 | |
| Inpatient | ABU | Absence | UP3 | |
| Inpatient | Cystitis | Absence | Other UP | |
| Inpatient | Cystitis | Absence | Other UP | |
| Inpatient | Pyelonephritis | Present | UP4 | |
| Inpatient | Cystitis | Absence | UP10 | |
| Outpatient | Pyelonephritis | Absence | UP9 | |
| Inpatient | Cystitis | Absence | UP17 | |
| Outpatient | Pyelonephritis | Present | Other UP | |
| Inpatient | ABU | Absence | Other UP | |
| Outpatient | Pyelonephritis | Present | Other UP | |
| Inpatient | Cystitis | Absence | Other UP | |
| Outpatient | Cystitis | Present | UP4 | |
| Inpatient | ABU | Absence | UP3 | |
| Inpatient | Pyelonephritis | Absence | Other UP | |
| Inpatient | Cystitis | Absence | Other UP | |
| Inpatient | ABU | Absence | UP8 | |
| Outpatient | Pyelonephritis | Absence | Other UP | |
| Inpatient | Cystitis | Absence | UP12 | |
| Inpatient | Cystitis | Absence | UP3 | |
| Outpatient | Pyelonephritis | Present | Other UP | |
| Inpatient | Cystitis | Present | Other UP | |
| Outpatient | Pyelonephritis | Absence | UP4 | |
| Inpatient | Pyelonephritis | Absence | Other UP |
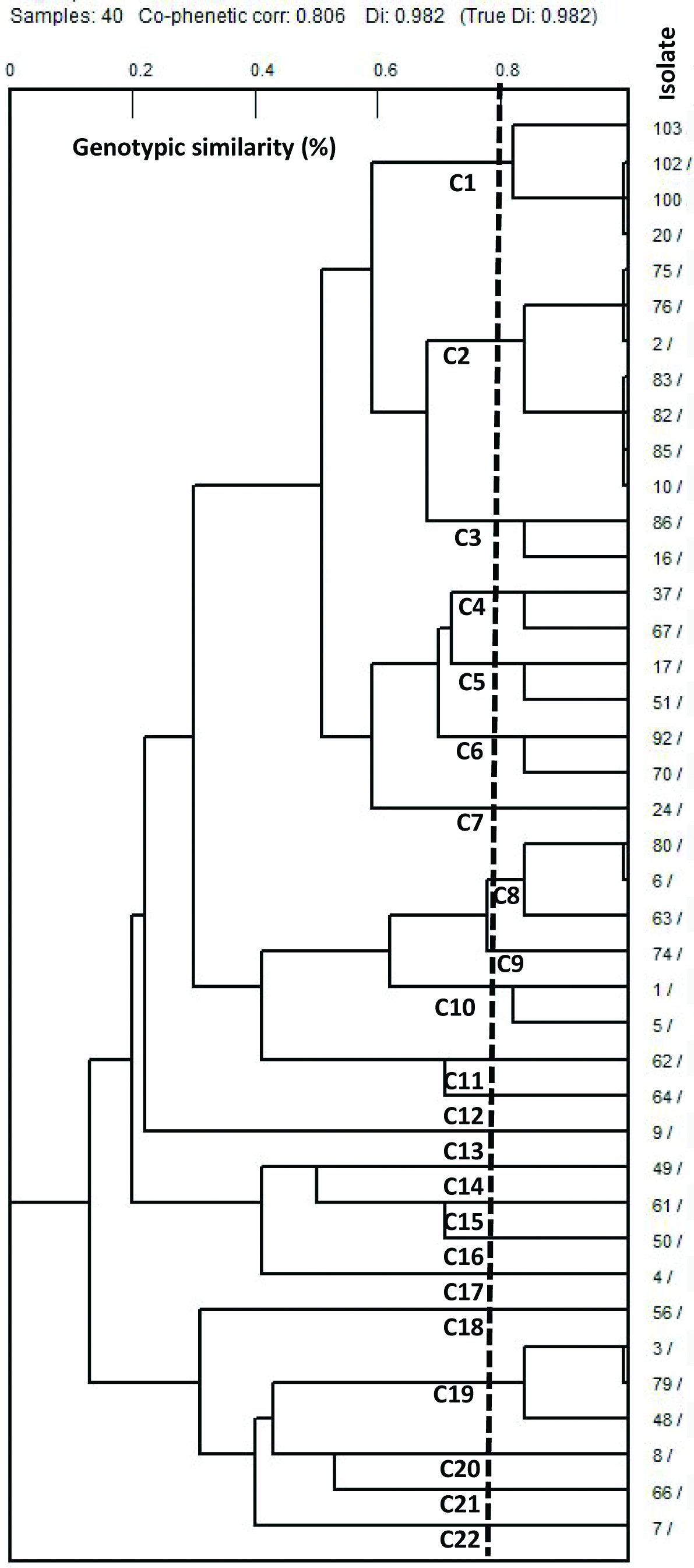
Cluster Analysis of VF Profiles
According to the virulence gene patterns and patient profiles, 40 UPEC isolates analysed with PFGE were sorted into 22 different clusters, from which 9 common clusters, containing 2 to 7 isolates, and 13 unique single-isolate clusters were found using a similarity threshold of 80% (C1 to C22) [Table/Fig-8]. The predominant cluster (C2) was found in 7 isolates (17.5%) with an average of 4.6 virulence determinant genes. The C2 cluster was divided into two subclusters based on the presence of the iuc gene. Importantly, a high similarity was observed between 14 isolates.
[Table/Fig-8]:
Clustering of the forty UPEC isolates from patients with UTI based on the UPGMA method derived from analysis of presence or absence of the virulence genes. Each row shows the genotypic results of a single isolate.
 |
Virulence Profile | |||||||||||
| fimH | hlyA | cnf1 | usp | Iron | Iuc | aer | sfa/foc | afa | papEF | fliC | cdtB | |
| 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | |
| 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | |
| 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | |
| 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | |
| 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | |
| 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | |
| 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | |
| 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | |
| 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | |
| 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | |
| 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | |
| 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | |
| 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | |
| 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | |
| 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | |
| 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | |
| 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | |
| 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | |
| 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | |
| 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | |
| 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |
| 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |
| 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |
| 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | |
| 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | |
| 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | |
| 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | |
| 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | |
| 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | |
| 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | |
| 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | |
| 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | |
| 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | |
| 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | |
| 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | |
| 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | |
| 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | |
C: Cluster, 1: Presence of virulence gene, 0: absence of virulence gene.
Discussion
Uropathogenic Escherichia coli (UPEC) strains are associated with increased morbidity and mortality in both community and hospitalized patients [1]. In light of this, there is a pressing need to identify virulence genetic determinants and clonal relationships for a better understanding of UPEC pathogenesis and epidemiology, which may be helpful in the development of effective treatment.
In agreement with other reports, we found that the fimH gene is the most common virulence gene among the UPEC isolated from patients with UTI [6,13,15]. Taken together, the ability of type 1 fimbriae to exert FimH adhesin, makes it one of the most important factors for UPEC colonization and invasiveness [3,8]. More importantly, our findings in line with other studies were unable to detect the cdtB gene among UTI samples and reported that cdtB has primarily been considered as an enteric virulence factor [17,18]. In our study, the prevalence of the pap and sfa adhesin genes was 35.9% and 12.8%, respectively [Table/Fig-2]. Other studies have shown the frequency of pap and sfa varying from 0 to 70% and from 0 to 74%, respectively, in UPEC isolated from different UTI patients [19–21]. Although other studies have reported the low prevalence (frequency of 2 to 27%) of afa gene among UPEC adhesins [19,20], but our results showed the higher prevalence of afa as compared with sfa adhesin gene (29.5 versus 12.8%). It is possible that Afa adhesin has more important role in pathogenesis of our UPEC collection. In accordance with most of the other reports [19,20], cnf1 toxin gene was more prevalent than hlyA gene (36.8 versus 30.8%) in our UPEC isolates. Frequencies of cnf1 ranging from 0 to 57%, and from 0 to 50% for hly gene have been reported from Iran and other countries [15,19,20]. In our study, prevalence of aer as the most prevalent iron gene receptor (73.1%) was similar to those found in other study [22], although the variation in frequencies of iron acquisition genes also has been observed in different studies [15,20,23]. Uropathogenic specific protein (usp) encoded by UPEC may act as a bacteriocin and enhances their infectivity in the urinary tract [23]. Here, we detected the usp gene in 77 (49.4%) of UPEC isolates and the prevalence of usp gene has been documented between 22 and 85% in other studies [23,24]. It is suggestive that the differences found in the prevalence of VFs may be explained by the differences in geographical regions, sampling methods, public health levels and food diets.
Consistent with other studies, our findings showed that the majority of UPEC isolates harbors multiple virulence factors, which are essential for pathogenicity in the urinary tract [20,22]. Furthermore, atleast one of the virulence factors was observed in all 156 isolates, whereas in studies carried out by Olivera et al., and Santo et al., 10% and 51% of UPEC strains were negative for any of the virulence factors tested, respectively [20,22]. This reflects the heterogeneity and complexity in the distribution of VFs among UPEC isolated from different geographical regions.
Based on the number of virulence genes and their distribution, several distinct patterns were observed among the UPEC isolates, consistent with other studies [11,20]. In the present study, a majority of the virulence gene patterns of UPEC revealed unique patterns (59.46%), thus demonstrating high heterogeneity of the virulence genes in the isolates. In comparison, Olivera et al., showed that the virulence genes of UPEC strains were distributed in 50 distinct patterns, in which most of the UPEC strains had a unique pattern [20].
Similar to other studies, we found the higher virulence potential of UPEC isolated from outpatients compared with inpatients [25,26]. In comparison with compromised patients, ambulatory patients usually are in a good general state of health. Therefore, they are prone to undergo a high-risk UTI caused by high virulence isolates, because the virulence factors are less necessary when host defense is compromised [25].
Although, consistent with other studies [11,20] we could not characterize the UPEC strains causing acute cystitis or pyelonephritis with a distinct set of virulence factors, significant differences were found between some VFs and different clinical types of UTI. In general, clinical isolates associated with pyelonephritis were more virulent than those associated with cystitis and especially those causing ABU. This may be associated with the fact that a low level of virulence is required to produce cystitis than pyelonephritis. Similar findings were reported by other researches and suggest that with knowledge of the UPEC virulence factor profiling we can gain some information about the clinical course of UTIs [27,28].
Generally, it was also observed that UPEC isolated from patients with symptomatic bacteriuria expresses higher levels of virulence than those isolated from patients with asymptomatic UTI, confirming previous observation conducted by Salvador et al., [29]. This highlights the important role of UPEC virulence factors in severity of clinical symptoms in patients with UTI.
Among the virulence genes, papEF, hlyA, aer and cnf1 were more prevalent in isolates causing acute pyelonephritis than those causing cystitis and ABU. This suggests that, in accordance with the other studies, these VFs may play important roles in the development of upper UTI, particularly in acute kidney injury and could be the indicators of pyelonephritis isolates [27,28]. However, similar to a study performed by Tarchouna et al., this study found a significant increase in the percentage of afa and fliC genes in isolates from patients with cystitis compared with those from pyelonephritis, reflecting that these virulence genes could be used as the promising indicators for cystitis isolates [30].
Association analysis between VFs demonstrated that there is a positive correlation between hlyA and iron genes. It has previously been reported that the production of haemolysin is known to be one alternative mechanism for bacterial iron acquisition, allowing for iron release from haemoglobin [31]. The relationships between two or more UPEC virulence genes, especially papEF, hlyA, cnf1 and sfa/foc, were observed both in this study and in other studies [20,23]. In addition, a strong positive correlation found between cnf1 and sfa/foc could indicate the genetic linkages between them, suggesting the presence of a pathogenicity islands.
In agreement with our results, Foxman et al., also reported that the only significant predictor of recurrent UTI was afa, whereas cnf1, hlyA, and sfa/foc genes were associated with the absence of recurrence in UTI patients [32]. On one hand, most recurrent UTIs presented as acute cystitis and, on the other hand, afa adhesin production was more frequent in UPEC isolated from patients with cystitis than those isolated from patients with pyelonephritis and ABU, demonstrating that afa may play an important role in the development of recurrent UTIs.
PFGE as a powerful typing method, was used to study genomic fingerprint profiles. As molecular typing assessed by PFGE and VF profiles demonstrated the high genetic diversity among the UPEC isolates studied, the present study was unable to find any relationship between UPEC causing cystitis and pyelonephritis. Anvarinejad et al., consistent with our findings, also reported the lowest similarity of molecular typing among UPEC isolates in Iran [12]. Unique PFGE patterns and high degree of diversity among UPEC isolates were also reported by Xie et al., [33]. This may be related to the different sources of UTI in community- and hospital-acquired isolates.
Conclusion
In conclusion, fimH, aer and fliC were found to be the most prevalent virulence genes among the UPEC isolates. Moreover, a high level of genetic diversity was observed in UPEC isolates, providing further evidences for the genomic diversity of the isolates. Despite this, further epidemiological studies are needed to determine the prevalence of these and other UPEC VFs and to verify the associations between VFs and different clinical types of UTI. Finally, this study could pave the new way to understand the role of these VFs in causing UTIs, which in turn may lead to the development of vaccines to prevent the infections.
Acknowledgments
The authors would like to thank the staff of Molecular Biology Dept., Pasteur Institute of Iran, for their important technical assistance. Additionally, we are grateful to Dr. Mohammad Katouli and Elham Darzi for clustering data analysis. This work was financially supported by Pasteur Institute of Iran.
Financial or Other Competing Interests
As declared above.
References
- [1].Alqasim A, Emes R, Clark G, Newcombe J, La Ragione R, McNally A. Phenotypic microarrays suggest Escherichia coli ST131 is not a metabolically distinct lineage of extra-intestinal pathogenic E. coli. PLoS One. 2014;9(2):e88374. doi: 10.1371/journal.pone.0088374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ivancic V, Mastali M, Percy N, Gornbein J, Babbitt JT, Li Y, et al. Rapid antimicrobial susceptibility determination of uropathogens in clinical urine specimens by use of ATP bioluminescence. J Clin Microbiol. 2008;46(4):1213–19. doi: 10.1128/JCM.02036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dhakal BK, Kulesus RR, Mulvey MA. Mechanisms and consequences of bladder cell invasion by uropathogenic Escherichia coli. Eur J Clin Invest. 2008;38(Suppl 2):2–11. doi: 10.1111/j.1365-2362.2008.01986.x. [DOI] [PubMed] [Google Scholar]
- [4].Scholes D, Hooton TM, Roberts PL, Stapleton AE, Gupta K, Stamm WE. Risk factors for recurrent urinary tract infection in young women. J Infect Dis. 2000;182(4):1177–82. doi: 10.1086/315827. [DOI] [PubMed] [Google Scholar]
- [5].Saint S. Clinical and economic consequences of nosocomial catheter-related bacteriuria. Am J Infect Control. 2000;28(1):68–75. doi: 10.1016/s0196-6553(00)90015-4. [DOI] [PubMed] [Google Scholar]
- [6].Mabbett AN, Ulett GC, Watts RE, Tree JJ, Totsika M, Ong CL, et al. Virulence properties of asymptomatic bacteriuria Escherichia coli. Int J Med Microbiol. 2009;299(1):53e63. doi: 10.1016/j.ijmm.2008.06.003. [DOI] [PubMed] [Google Scholar]
- [7].Svanborg C, Bergsten G, Fischer H, Godaly G, Gustafsson M, Karpman D, et al. Uropathogenic Escherichia coli as a model of host-parasite interaction. Curr Opin Microbiol. 2006;9(1):33–39. doi: 10.1016/j.mib.2005.12.012. [DOI] [PubMed] [Google Scholar]
- [8].Bien J, Sokolova O, Bozko P. Role of uropathogenic Escherichia coli virulence factors in development of urinary tract infection and kidney damage. Int J Nephrol. 2012;2012:681473. doi: 10.1155/2012/681473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wiles TJ, Kulesus RR, Mulvey MA. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol. 2008;85(1):11–19. doi: 10.1016/j.yexmp.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yamamoto S. Molecular epidemiology of uropathogenic Escherichia coli. J Infect Chaemother. 2007;13(2):68–73. doi: 10.1007/s10156-007-0506-y. [DOI] [PubMed] [Google Scholar]
- [11].Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181(1):261–72. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- [12].Anvarinejad M, Sh F, Ranjbar R, Giammanco G, Alborzi A, Japoni A. Genotypic analysis of E. coli strains isolated from patients with cystitis and pyelonephritis. Iran Red Crescent Med J. 2012;14(7):408. [PMC free article] [PubMed] [Google Scholar]
- [13].Asadi KM, Oloomi M, Habibi M, Bouzari S. Cloning of fimH and fliC and expression of the fusion protein FimH/FliC from Uropathogenic Escherichia coli (UPEC) isolated in Iran. Iran J Microbiol. 2012;4(2):55–62. [PMC free article] [PubMed] [Google Scholar]
- [14].Yamamoto S, Terai A, Yuri K, Kurazono H, Takeda Y, Yoshida O. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol. 1995;12(2):85–90. doi: 10.1111/j.1574-695X.1995.tb00179.x. [DOI] [PubMed] [Google Scholar]
- [15].Momtaz H, Karimian A, Madani M, Safarpoor Dehkordi F, Ranjbar R, Sarshar M, et al. Uropathogenic Escherichia coli in Iran: serogroup distributions, virulence factors and antimicrobial resistance properties. Ann Clin Microbiol Antimicrob. 2013;12:8. doi: 10.1186/1476-0711-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. Center for Disease Control and Prevention (CDC). Standard operating procedure for pulsenet PFGE Escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri Available: http://www.pulsenetinternational.org/SiteCollectionDocuments/pfge/5%201_5%202_5%204_PNetStand_Ecoli_with_Sflexner.pdf. [Accessed 2013 Mar 24. 2013]
- [17].Yun KW, Kim HY, Park HK, Kim W, Lim IS. Virulence factors of uropathogenic Escherichia coli of urinary tract infections and asymptomatic bacteriuria in children. J Microbiol Immunol Infect. 2014;47(6):455–61. doi: 10.1016/j.jmii.2013.07.010. [DOI] [PubMed] [Google Scholar]
- [18].Mirzarazi M, Rezatofighi SE, Pourmahdi M, Mohajeri MR. Occurrence of genes encoding enterotoxins in uropathogenic Escherichia coli isolates. Braz J Microbiol. 2015;46(1):155–59. doi: 10.1590/S1517-838246120130860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dormanesh B, Safarpoor Dehkordi F, Hosseini S, Momtaz H, Mirnejad R, Hoseini MJ, et al. Virulence factors and o-serogroups profiles of uropathogenic Escherichia coli isolated from Iranian pediatric patients. Iran Red Crescent Med J. 2014;16(2):e14627. doi: 10.5812/ircmj.14627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Oliveira FA, Paludo KS, Arend LN, Farah SM, Pedrosa FO, Souza EM, et al. Virulence characteristics and antimicrobial susceptibility of uropathogenic Escherichia coli strains. Genet Mol Res. 2011;10(4):4114–25. doi: 10.4238/2011.October.31.5. [DOI] [PubMed] [Google Scholar]
- [21].Lopez-Banda DA, Carrillo-Casas EM, Leyva-Leyva M, Orozco-Hoyuela G, Manjarrez-Hernandez AH, Arroyo-Escalante S, et al. Identification of virulence factors genes in Escherichia coli isolates from women with urinary tract infection in Mexico. Biomed Res Int. 2014;2014:959206. doi: 10.1155/2014/959206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Santo E, Macedo C, Marin JM. Virulence factors of uropathogenic Escherichia coli from a university hospital in Ribeirao Preto, Sao Paulo, Brazil. Rev Inst Med Trop Sao Paulo. 2006;48(4):185–88. doi: 10.1590/s0036-46652006000400002. [DOI] [PubMed] [Google Scholar]
- [23].Tiba MR, Yano T, Leite Dda S. Genotypic characterization of virulence factors in Escherichia coli strains from patients with cystitis. Rev Inst Med Trop Sao Paulo. 2008;50(5):255–60. doi: 10.1590/s0036-46652008000500001. [DOI] [PubMed] [Google Scholar]
- [24].Kurazono H, Yamamoto S, Nakano M, Nair GB, Terai A, Chaicumpa W, et al. Characterization of a putative virulence island in the chromosome of uropathogenic Escherichia coli possessing a gene encoding a uropathogenic-specific protein. Microb Pathog. 2000;28(3):183–89. doi: 10.1006/mpat.1999.0331. [DOI] [PubMed] [Google Scholar]
- [25].Skjot-Rasmussen L, Ejrnaes K, Lundgren B, Hammerum AM, Frimodt-Moller N. Virulence factors and phylogenetic grouping of Escherichia coli isolates from patients with bacteraemia of urinary tract origin relate to sex and hospital- vs. community-acquired origin. Int J Med Microbiol. 2012;302(3):129–34. doi: 10.1016/j.ijmm.2012.03.002. [DOI] [PubMed] [Google Scholar]
- [26].Toval F, Kohler CD, Vogel U, Wagenlehner F, Mellmann A, Fruth A, et al. Characterization of Escherichia coli isolates from hospital inpatients or outpatients with urinary tract infection. J Clin Microbiol. 2014;52(2):407–18. doi: 10.1128/JCM.02069-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Idress M, Mussarat U, Badshah Y, Qamar R, Bokhari H. Virulence factors profile of drug-resistant Escherichia coli isolates from urinary tract infections in Punjab, Pakistan. Eur J Clin Microbiol Infect Dis. 2010;29(12):1533–37. doi: 10.1007/s10096-010-1036-6. [DOI] [PubMed] [Google Scholar]
- [28].Ruiz J, Simon K, Horcajada JP, Velasco M, Barranco M, Roig G, et al. Differences in virulence factors among clinical isolates of Escherichia coli causing cystitis and pyelonephritis in women and prostatitis in men. J Clin Microbiol. 2002;40(12):4445–49. doi: 10.1128/JCM.40.12.4445-4449.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Salvador E, Wagenlehner F, Kohler CD, Mellmann A, Hacker J, Svanborg C, et al. Comparison of asymptomatic bacteriuria Escherichia coli isolates from healthy individuals versus those from hospital patients shows that long-term bladder colonization selects for attenuated virulence phenotypes. Infect Immun. 2012;80(2):668–78. doi: 10.1128/IAI.06191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tarchouna M, Ferjani A, Ben-Selma W, Boukadida J. Distribution of uropathogenic virulence genes in Escherichia coli isolated from patients with urinary tract infection. Int J Infect Dis. 2013;17(6):e450–53. doi: 10.1016/j.ijid.2013.01.025. [DOI] [PubMed] [Google Scholar]
- [31].Caza M, Kronstad JW. Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front Cell Infect Microbiol. 2013;3:80. doi: 10.3389/fcimb.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Foxman B, Zhang L, Tallman P, Palin K, Rode C, Bloch C, et al. Virulence characteristics of Escherichia coli causing first urinary tract infection predict risk of second infection. J Infect Dis. 1995;172(6):1536–41. doi: 10.1093/infdis/172.6.1536. [DOI] [PubMed] [Google Scholar]
- [33].Xie J, Foxman B, Zhang L, Marrs CF. Molecular epidemiologic identification of Escherichia coli genes that are potentially involved in movement of the organism from the intestinal tract to the vagina and bladder. J Clin Microbiol. 2006;44(7):2434–41. doi: 10.1128/JCM.00397-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


