ABSTRACT
Heterodisulfide reductases (Hdr) of the HdrABC class are ancient enzymes and a component of the anaerobic core belonging to the prokaryotic common ancestor. The ancient origin is consistent with the widespread occurrence of genes encoding putative HdrABC homologs in metabolically diverse prokaryotes predicting diverse physiological functions; however, only one HdrABC has been characterized and that was from a narrow metabolic group of obligate CO2-reducing methanogenic anaerobes (methanogens) from the domain Archaea. Here we report the biochemical characterization of an HdrABC homolog (HdrA2B2C2) from the acetate-utilizing methanogen Methanosarcina acetivorans with unusual properties structurally and functionally distinct from the only other HdrABC characterized. Homologs of the HdrA2B2C2 archetype are present in phylogenetically and metabolically diverse species from the domains Bacteria and Archaea. The expression of the individual HdrA2, HdrB2, and HdrB2C2 enzymes in Escherichia coli, and reconstitution of an active HdrA2B2C2 complex, revealed an intersubunit electron transport pathway dependent on ferredoxin or coenzyme F420 (F420H2) as an electron donor. Remarkably, HdrA2B2C2 couples the previously unknown endergonic oxidation of F420H2 and reduction of ferredoxin with the exergonic oxidation of F420H2 and reduction of the heterodisulfide of coenzyme M and coenzyme B (CoMS-SCoB). The unique electron bifurcation predicts a role for HdrA2B2C2 in Fe(III)-dependent anaerobic methane oxidation (ANME) by M. acetivorans and uncultured species from ANME environments. HdrA2B2C2, ubiquitous in acetotrophic methanogens, was shown to participate in electron transfer during acetotrophic growth of M. acetivorans and proposed to be essential for growth in the environment when acetate is limiting.
IMPORTANCE
Discovery of the archetype HdrA2B2C2 heterodisulfide reductase with categorically unique properties extends the understanding of this ancient family beyond CO2-reducing methanogens to include diverse prokaryotes from the domains Bacteria and Archaea. The unprecedented coenzyme F420-dependent electron bifurcation, an emerging fundamental principle of energy conservation, predicts a role for HdrA2B2C2 in diverse metabolisms, including anaerobic CH4-oxidizing pathways. The results document an electron transport role for HdrA2B2C2 in acetate-utilizing methanogens responsible for at least two-thirds of the methane produced in Earth’s biosphere. The previously unavailable heterologous production of individual subunits and the reconstitution of HdrA2B2C2 with activity have provided an understanding of intersubunit electron transfer in the HdrABC class and a platform for investigating the principles of electron bifurcation.
INTRODUCTION
Heterodisulfide reductase (Hdr) was first discovered in CH4-producing species (methanogens) from the domain Archaea where one or the other of two classes (HdrABC or HdrDE) is essential for all methanogenic pathways (1). However, the genomes of diverse species in the domains Bacteria and Archaea are annotated with genes encoding HdrABC homologs, suggesting that these genes play roles in a greater diversity of energy-conserving metabolisms, which include the oxidation of methanol and inorganic sulfur compounds (2–4), the reduction of sulfate and ferric iron (4–6), syntrophic utilization of fatty acids (7), and the anaerobic oxidation of CH4 (8–10). Indeed, the HdrABC class belongs to the core repertoire of the ancient prokaryotic common ancestor consistent with diverse physiological functions of extant species (11).
Although of ancient origin and widespread, the biochemical and physiological understanding of the HdrABC class is restricted to one homolog essential for the pathway of CO2 reduction to CH4. The final step in all methanogenic pathways (equation 1) is the reductive demethylation of methyl coenzyme M (CH3-SCoM) for which coenzyme B (HSCoB) supplies reductant. The heterodisulfide product is reduced by Hdr (equation 2), releasing HSCoB and HSCoM for methylation.
| (1) |
| (2) |
The cytoplasmic HdrABC in H2-oxidizing CO2-reducing species is complexed with the MvhAGD hydrogenase. Electron pairs donated from the hydrogenase are bifurcated by HdrABC, reducing CoMS-SCoB and ferredoxin (Fdx), and Fdx donates electrons for the first step in the reduction of CO2 to produce CH3-SCoM. The bifurcation is a flavin-based coupling of the endergonic reduction of Fdx with the exergonic reduction of CoMS-SCoB. Flavin-based electron bifurcation is considered to be an ancient energy-conserving mechanism (12–14). Importantly, a comprehensive mechanistic understanding of the HdrABC class has been impeded by the unavailability of a recombinantly produced enzyme or an activity assay utilizing a physiological electron donor. Although the obligate two-electron carrier coenzyme F420 (F420) has diverse roles in species from the domains Bacteria and Archaea, roles in electron bifurcation and the donor to HdrABC homologs are still unknown (15).
More-diverse roles are postulated for electron bifurcation catalyzed by HdrABC homologs in phylogenetically and physiologically diverse species in the domains Bacteria and Archaea (12, 13, 16). Bioinformatic analyses of methanogens and nonmethanogens from the domains Bacteria and Archaea predict an HdrABC homolog wherein the HdrA subunit is fused with MvhD (see Fig. S1 in the supplemental material). Genes encoding the homolog, designated HdrA2B2C2, are present in the methanogenic archaeon Methanosarcina acetivorans with metabolic capabilities distinct from those of obligate CO2-reducing methanogens (17, 18). The fused HdrA2 subunit and low sequence identity of all subunits with subunits of the HdrABC homolog from obligate CO2-reducing methanogens (29 to 37%) predict differences in structure and function (19).
Sequence alignments of HdrA2 from M. acetivorans with homologs. The MvhD domain sequences are shown in boldface type. Cysteine residues conserved in HdrA (CAA57039) and MvhD (AAB02349.1) from Methanothermobacter thermautotrophicus are marked with an asterisk, whereas others are marked with a pound symbol. Alignments were made with the COBALT multiple alignment tool. The sequences belong to the following species (the corresponding percent coverage and identity to M. acetivorans HdrA2 are shown before and after the slash, respectively, in parentheses): WP_011022820.1, Methanosarcina acetivorans (100/100); WP_048037482.1, Methanosarcina mazei (100/95); WP_011307542.1, Methanosarcina barkeri (98/94); WP_023843920.1, Methanolobus tindarius (98/67); WP_015324480.1, Methanomethylovorans hollandica (98/67); WP_048194718.1, Methanococcoides methylutens (98/66); WP_013897960.1, Methanosalsum zhilinae (98/65); WP_013194860.1, Methanohalobium evestigatum (98/65); WP_013037418.1, Methanohalophilus mahii (98/65); WP_048089472.1, “Candidatus Methanoperedens nitroreducens” (98/61); WP_014586170.1, Methanosaeta harundinacea (98/61); WP_042684393.1, Methermicoccus shengliensis (98/55); WP_012964674.1, Ferroglobus placidus (98/53); WP_010878733.1, Archaeoglobus fulgidus (97/55); KPL16356.1, Bacteroides sp. strain SM23_62 (97/48); KPV63986.1, “Candidatus Bathyarchaeota archaeon” BA1 (96/49); WP_051309217.1, Desulfobulbus japonicas (96/47); WP_041286569.1, Desulfomonile tiedjei (97/45); KPJ88130.1, Spirochaetes bacterium DG_61 (97/46). Download FIG S1, DOCX file, 0.02 MB (22.4KB, docx) .
Copyright © 2017 Yan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Here we report the individual heterologous production and characterization of HdrA2, HdrB2, and HdrB2C2 from M. acetivorans that form an HdrA2B2C2 complex with Fdx2−- and F420H2-dependent heterodisulfide reductase activity. The experimental approach has advanced an understanding of intersubunit electron transfer for the HdrABC class and uncovered a previously unknown coenzyme F420H2-dependent electron bifurcation that predicts roles for HdrA2B2C2 homologs in pathways of anaerobic CH4 oxidation in M. acetivorans and other species (20–22). The uncommon properties of HdrA2B2C2 identify it as representative of an HdrABC subclass with homologs in diverse species from the domains Bacteria and Archaea.
RESULTS
Bioinformatic analyses.
A search of the nonredundant databases (https://www.ncbi.nlm.nih.gov/pubmed) with HdrA2 as the query retrieved 150 sequences, of which 77 were HdrA2 homologs containing HdrA and MvhD domains with greater than 46% identity and 96% coverage. Figure S1 in the supplemental material shows an alignment of representative sequences. A total of 48 HdrA2 homologs were from acetotrophic and methylotrophic methanogens. None of the homologs were from obligate CO2-reducing methanogens, indicating a role for HdrA2 specific to acetotrophic and methylotrophic methanogens. Of the 77 HdrA2 homologs, 29 were from physiologically and phylogenetically diverse nonmethanogenic species in the domains Bacteria and Archaea. Of the 150 sequences retrieved, 73 were homologs of the canonical HdrA from both methanogenic and nonmethanogenic species. These results indicate that HdrA2 homologs play roles in metabolically diverse species, warranting investigation of an HdrA2 representative chosen from M. acetivorans that grows by converting acetate to CH4 (17).
Recombinant Fdx.
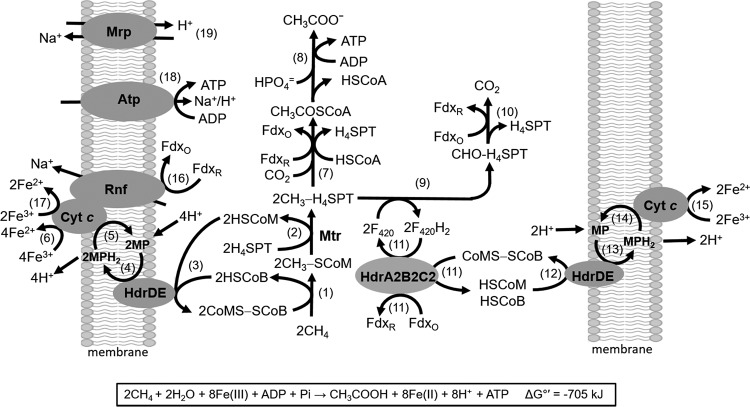
The conversion of acetate to CH4 begins with activation to acetyl coenzyme A (acetyl-CoA) that is cleaved at the C-C and C-S bonds producing methyl and carbonyl groups catalyzed by CO dehydrogenase/acetyl-CoA synthase (CODH/ACS) (23, 24). The methyl group is transferred to HSCoM, and the carbonyl group is oxidized to CO2 with reduction of Fdx. Thus, Fdx is a candidate electron donor/acceptor of HdrA2B2C2. Of multiple Fdx-encoding genes, the Fdx encoded by MA0431 is produced in acetate-grown M. acetivorans and is the electron acceptor for CODH/ACS (25). The Fdx produced in Escherichia coli strain BL21(DE3) ΔiscR (Fig. 1A) showed a prominent absorption band centered at ~390 nm with an A390/A280 ratio of 0.69, indicating a nearly full complement of the predicted two [Fe4S4] clusters (25). The recombinant Fdx was competent in accepting electrons from CODH/ACS at a rate of 55.3 nmol/min/mg.
FIG 1 .
UV-Vis spectra of recombinantly produced ferredoxin (Fdx), HdrA2, HdrB2, and HdrB2C2 from M. acetivorans. All proteins were contained in 50 mM Tris buffer (pH 8.0). Spectra were recorded with a Cary 50 Bio UV-Vis spectrophotometer. (A) Spectra of 15.8 μM Fdx. Spectra of Fdx as purified (solid line) and as purified and reduced with sodium dithionite (dotted line) are shown. The CODH/ACS purified from acetate-grown M. acetivorans reduced 33.3 μM Fdx at a rate of 55.3 nmol/min/mg CODH/ACS. (B) Spectra of 12.2 µM HdrA2. Spectra of HdrA2 as purified (solid line), reconstituted with FAD (dotted line), reconstituted with FAD and reduced with sodium dithionite (dashed line). (C) Spectra of 18.3 µM HdrB2. Spectra of HdrB2 as purified (solid line), reduced with 0.1 mM sodium dithionite (dotted line), and the reduced protein after the addition of 0.1 mM CoMS-SCoB (dashed line). (D) Spectra of 11.2 µM HdrB2C2. Spectra of HdrB2C2 as purified (solid line), reduced with 0.1 mM sodium dithionite (dotted line), and the reduced protein after the addition of 0.1 mM CoMS-SCoB (dashed line).
Recombinant HdrA2, HdrB2, HdrC2, and HdrB2C2.
HdrA2, HdrB2, HdrC2, and HdrB2C2 were produced in E. coli strain BL21(DE3) ΔiscR. All except HdrC2 were present in the cytoplasm and purified to homogeneity (Fig. S2). HdrC2 was present in inclusion bodies and not purified. When coproduced with HdrB2, HdrC2 was present in the cytoplasm and copurified with HdrB2 to homogeneity (Fig. S2). The results indicate that either HdrC2 was misfolded or the presence of HdrB2 is required for HdrC2 to fold properly. Regardless, the results indicate that HdrB2 and HdrC2 form an HdrB2C2 complex.
SDS-PAGE of purified recombinant proteins produced in Escherichia coli and CODH/ACS purified from Methanosarcina acetivorans. The Bolt 4 to 12% gels (Invitrogen) were stained with Coomassie brilliant blue. The numbers at the sides of the gels refer to the molecular masses (in kilodaltons) of adjacent standards. Amounts loaded: HdrA2 and HdrB2 (10 μg); HdrB2C2 and CODH/ACS (25 μg). Download FIG S2, DOCX file, 0.2 MB (158.9KB, docx) .
Copyright © 2017 Yan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
HdrA2 migrated in SDS-polyacrylamide gels with an apparent molecular mass of 87 kDa (Fig. S2) consistent with the molecular mass of 86.9 kDa calculated for the HdrA-MvhD fusion. The UV-visible (UV-Vis) spectrum of the protein as purified (as-purified protein) shows prominent absorption bands centered at ~320 and ~420 nm attributed to Fe-S clusters (Fig. 1B). Reconstitution of the as-purified protein with flavin adenine dinucleotide (FAD) resulted in the enhancement of an absorption band centered at ~560 nm attributed to flavin. Indeed, upon reconstitution, the flavin content increased from 0.32 ± 0.06 (n = 3) to 1.2 ± 0.1 (n = 3) mol per mol of HdrA2. The FAD-reconstituted preparation had 20.6 ± 2.4 atoms of nonheme iron and 18.6 ± 1.3 atoms of acid-labile sulfur per molecule of HdrA2. The ε420 was found to be 82.3 mM−1 cm−1. These results are consistent with each molecule of HdrA2 containing one FAD molecule and four Fe4S4 clusters in the HdrA domain, and one Fe2S2 cluster in the MvhD domain, predicted by motifs in the deduced sequence (Fig. S3).
Sequence alignments of HdrA2B2C2 from Methanosarcina acetivorans with HdrABC and MvhD of Methanothermobacter thermautotrophicus. (A) M.t. HdrA, HdrA from M. thermautotrophicus (CAA57039); M.a. HdrA2, HdrA2 from M. acetivorans (AAM06247.1); M.t. MvhD, MvhD from M. thermautotrophicus (AAB02349.1). The overall sequences of HdrA and MvhD were found to be 50 and 49% identical with the corresponding domains in HdrA2. The sequence comparisons revealed four Fe4S4-binding motifs (CX2CX2CX3CP) common to HdrA of M. thermautotrophicus and the HdrA domain (residues 1 to 649) of HdrA2. A motif (CX2CX25CX24CX4C) predicted to ligate the Fe2S2 cluster in MvhD from M. thermautotrophicus (62) was found to be conserved in the MvhD domain of M. acetivorans HdrA2 (residues 650 to 793). (B) M.t. HdrB, HdrB M. thermautotrophicus (CAA57038.1); M.a. HdrB2, HdrB2 from M. acetivorans (AAM07582.1). The HdrB2 sequence has 55% identity with HdrB and contains two conserved motifs (CX31–39CCX35–36CX2C) of which one is predicted to ligate the active site [Fe4S4]3+ cluster in obligate CO2-reducing methanogens. (C) M.a. HdrC2, HdrC2 from M. acetivorans (AAM07581.1); M.t. HdrC, HdrC from M. thermautotrophicus (AAB86344.1). The sequence of HdrC2 has 43% identity with HdrC. The comparison shows two Fe4S4-binding motifs of HdrC2 conserved in HdrC from M. thermautotrophicus. Alignments were made with COBALT. Download FIG S3, DOCX file, 0.01 MB (15.4KB, docx) .
Copyright © 2017 Yan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
HdrB2 migrated in SDS-polyacrylamide gels with an apparent molecular mass of 33 kDa in agreement with a calculated molecular mass of 32.8 kDa (Fig. S2). The UV-Vis spectrum of as-purified preparations showed absorption bands centered at ~320, ~420, and ~580 nm (Fig. 1C). The protein contained 5.0 ± 0.1 (n = 3) nonheme iron atoms and 5.1 ± 0.2 (n = 3) acid-labile sulfur atoms per molecule of HdrB2. The ε420 was found to be 15.9 mM−1 cm−1. These results are consistent with the presence of an Fe4S4 cluster ligated by one of two cysteine-rich motifs (CX31–39CCX35–36CX2C) (Fig. S3) that are conserved in HdrB of obligate CO2-reducing methanogens and proposed to ligate a novel active site [Fe4S4]3+ cluster (26). The addition of CoMS-SCoB to dithionite-reduced HdrB2 increased the absorbance at ~320 and ~420 nm, indicating that the protein was competent to reduce the heterodisulfide to HSCoM and HSCoB (Fig. 1C). In an apparent anomaly, no significant forward activity (reduction of CoMS-SCoB with reduced methyl viologen [MV]) was detected, either with HdrB2 alone or in combination with HdrA2 and Fdx. However, reverse activity with HdrB2 alone was measurable, and there was no significant change in the Vmax when HdrA2 was present (Table 1). To our knowledge, this is the first heterologously produced and catalytically active HdrB from any source.
TABLE 1 .
Kinetic constants of forward and reverse heterodisulfide reductase activities for combinations of HdrA2, HdrB2, and HdrB2C2
| Subunit(s) | Heterodisulfide reductase activity | Vmax (μmol/min/mg) | Km (mM) |
|---|---|---|---|
| HdrB2C2 | Reverse | 0.6 ± 0.1 | 1.4 ± 0.2 HSCoM |
| 1.2 ± 0.2 HSCoB | |||
| HdrB2C2 HdrA2 | Forward | 2.2 ± 0.3 | 0.05 ± 0.01 CoMS-SCoB |
| 4.2 ± 0.2a | |||
| Reverse | 0.63 ± 0.1b | NDc | |
| HdrB2 | Reverse | 0.5 ± 0.1 | 1.2 ± 0.3 HSCoM |
| 1.1 ± 0.2 HSCoB | |||
| HdrB2 HdrA2 | Reverse | 0.6 ± 0.1b | ND |
Ferredoxin (50 μg) was added to the reaction mixture for forward activity.
Determined with saturating amounts (5 × Km) of HSCoM and HSCoB.
ND, not determined.
The HdrC2 of purified HdrB2C2 migrated in SDS-polyacrylamide gels with an apparent molecular mass of 18 kDa in agreement with a calculated molecular mass of 18.1 kDa (Fig. S2). The UV-Vis spectrum of as-purified HdrB2C2 showed absorption bands centered at ~320, ~420, and ~580 nm (Fig. 1D). Preparations contained 14.6 ± 1.0 (n = 3) nonheme iron atoms and 11.9 ± 0.8 (n = 3) acid-labile sulfur atoms per molecule of HdrB2C2. The ε420 was found to be 45.7 mM−1 cm−1. These results are consistent with the presence of three Fe4S4 clusters in the HdrB2C2 complex which is predicted from results obtained for HdrB2 in addition to the sequence of HdrC2 that contains two canonical Fe4S4-binding motifs (Fig. S3). Thus, the results indicate that as-purified HdrB2C2 had a full complement of iron-sulfur centers.
The addition of CoMS-SCoB to dithionite-reduced HdrB2C2 increased the absorbance at ~420 nm, indicating that the complex was active in reducing the heterodisulfide to HSCoM and HSCoB (Fig. 1D). No significant forward activity was detected unless HdrA2 was present, a result indicating that HdrB2C2 is incapable of accepting electrons from MV and that HdrA2 mediates electron transfer between MV and HdrB2C2. Kinetic analyses (Table 1) showed that Vmax nearly doubled in the presence of both HdrA2 and Fdx. These results, and the finding that HdrB2 in combination with HdrA2 was incapable of catalyzing MV-dependent activity, is consistent with a role for HdrC2 in mediating electron transfer from HdrA2 to HdrB2. Kinetic constants for the reverse activity of HdrB2C2 (Table 1) were not significantly different from those determined for HdrB2, which indicates that HdrC2 plays no role in catalysis by HdrB2.
Reconstitution of an active HdrA2B2C2 complex.
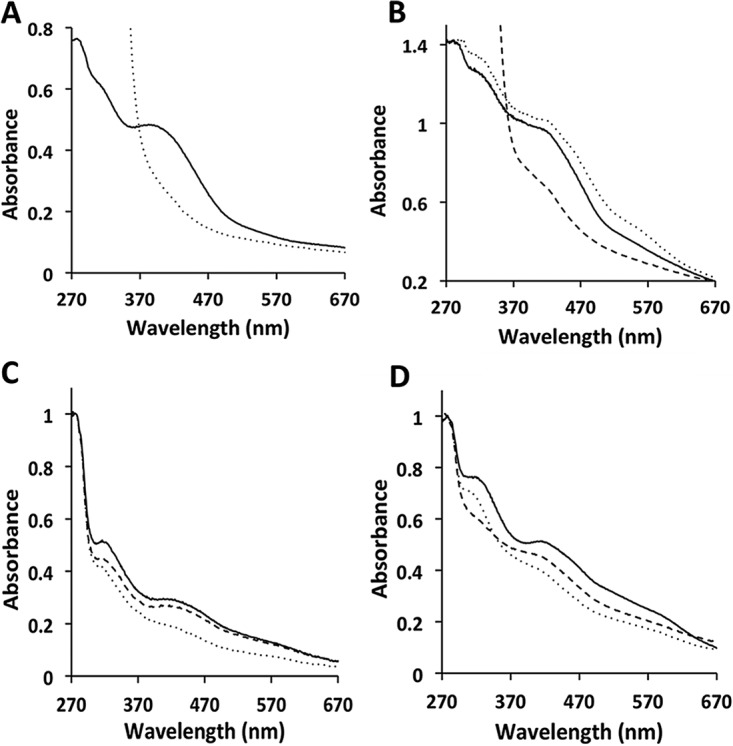
Figure 2A shows representative time courses for reduction of CoMS-SCoB catalyzed by various combinations of HdrA2, HdrB2, and HdrB2C2 in the presence of CO, CODH/ACS, and Fdx as electron donors. Substantial initial rates were observed only with HdrB2C2, which indicates that Fdx mediates direct electron transfer from CODH/ACS to HdrB2C2. Rates with only HdrB2 were inconsequential, indicating a role for HdrC2 in the transfer of electrons from Fdx to HdrB2. Rates of Fdx-dependent reduction of HdrB2 or HdrB2C2 (Fig. S4) showed that HdrB2C2 was reduced at a rate fivefold greater than for HdrB2, providing further support for the role of HdrC2 in mediating electron transfer from Fdx to HdrB2. In contrast, the combination of HdrA2 and HdrB2 enhanced the initial rate of CoMS-SCoB to levels comparable with only HdrB2C2 (Fig. 2A). This result indicates that HdrA2 mediates electron transfer from Fdx to catalytic HdrB2 without HdrC2 participation. Reaction mixtures containing a combination of HdrA2 and HdrB2C2 catalyzed the reduction of CoMS-SCoB at initial rates approximately sixfold greater than the rate observed for the combination of HdrA2 and HdrB2 (Fig. 2A). These results indicate that HdrC2 mediates electron transfer from HdrA2 to HdrB2 at a rate substantially greater than the transfer of electrons from HdrA2 directly to HdrB2. Although hdrA2 (MA2868) is located distant from hdrB2 and hdrC2 (MA4237-MA4236), the results establish formation of an HdrA2B2C2 complex catalyzing the most efficient oxidation of Fdx and reduction of CoMS-SCoB. Nevertheless, gel filtration column chromatography of the reconstituted HdrA2B2C2 resulted in separation of HdrA2 from HdrB2C2, indicating a weakly bound catalytic complex.
FIG 2 .
Reconstitution of a system catalyzing the CO-dependent reduction of CoMS-SCoB. (A) Time course of free thiol production from CoMS-SCoB dependent on combinations of HdrA2, HdrB2, and HdrB2C2. The complete reaction mixture (CRM) (0.5 ml) contained 0.1 mg CODH/ACS, 1.5 μM Fdx, 0.8 μM HdrA2, 1.1 μM HdrB2C2 or HdrB2, and 0.1 mM CoMS-SCoB in 50 mM MOPS buffer (pH 7.0). The atmosphere was 100% CO, and the temperature was 20°C. Reactions were initiated by the addition of 0.1 mg CODH/ACS. Initial rates are shown in parentheses where applicable. Time course of free thiol production from CoMS-SCoB in CRM with HdrB2 (0.73 mol/min/mol HdrB2) (▲), CRM with HdrB2 minus HdrA2 (■), CRM with HdrB2C2 (3.6 mol/min/mol HdrB2C2) (□), CRM with HdrB2C2 minus HdrA2 (0.62 mol/min/mol HdrB2C2) (•), CRM minus HdrB2 or HdrB2C2 (⧫), or CRM with HdrB2C2 or HdrB2 minus Fdx (Δ). All activities were dependent on the presence of CODH/ACS and CoMS-SCoB (not shown). (B) Oxidation of ferredoxin catalyzed by HdrA2B2C2 in the presence of CoMS-SCoB. The basal reaction mixture (0.5 ml) contained 9.4 nmol recombinant Fdx and 0.1 mg CODH/ACS in 50 mM MOPS buffer (pH 7.0) with an Ar atmosphere. Alterations were made to the basal mixture in the following order: none (solid line), CO was added to reduce Fdx (dashed line), CO was replaced with Ar and 0.5 μM HdrA2 plus 0.8 μM HdrB2C2 were added (dotted line), 0.1 mM CoMS-SCoB was added (dashed and dotted line), the reaction mixture was exposed to air (dashed and double-dotted line).
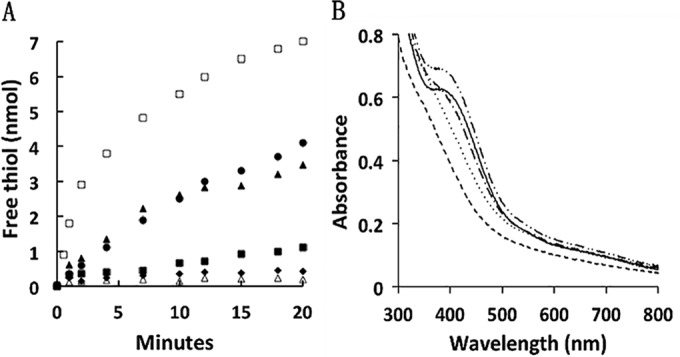
Fdx-dependent reduction of HdrA2, HdrB2C2, and HdrB2. It was previously shown that Fdx is an electron acceptor of CO-reduced CODH/ACS (25) that was used to evaluate the ability of recombinant Fdx to reduce HdrA2, HdrB2, and HdrB2C2. The atmosphere was 100% CO, and the temperature was 20°C. Reduction of HdrA2 and HdrB2C2 was monitored at 410 nm with a Cary 50 Bio UV-Vis spectrophotometer. (A) HdrA2 reduction. The reaction mixtures (0.5 ml) contained 0.1 mg CODH/ACS, 15 μM HdrA2, and the indicated concentrations of Fdx in 50 mM MOPS buffer (pH 7.0). Reactions were initiated by the addition of HdrA2 after preincubation of the reaction mixture with CO to fully reduce Fdx. Key to curves or traces: a, minus Fdx; b, 2.2 μM Fdx; c, 4.4 μM Fdx (2.3 nmol HdrA2 reduced/min). (B) HdrB2C2 and HdrB2 reduction. The reaction mixtures (0.5 ml) contained 0.1 mg CODH/ACS, 22.8 μM HdrB2C2 or HdrB2, and the indicated concentrations of Fdx in 50 mM MOPS buffer (pH 7.0). Reactions were initiated by the addition of HdrB2C2 or HdrB2 after preincubation of the reaction mixtures with CO to fully reduce Fdx. Key to curves or traces: a, reaction mixture containing either HdrB2C2 or HdrB2 minus Fdx; b, reaction mixture containing HdrB2 and 4.4 μM Fdx (0.3 nmol HdrB2 reduced/min); c, reaction mixture containing HdrB2C2 and 2.2 μM Fdx; d, reaction mixture containing HdrB2C2 and 4.4 μM Fdx (1.5 nmol HdrB2C2 reduced/min). Comparison of traces b and d indicates that HdrC2 is important for maximizing electron transfer from Fdx to the catalytic HdrB2. Download FIG S4, DOCX file, 0.1 MB (55.7KB, docx) .
Copyright © 2017 Yan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The redox potential for the CO/CO2 couple (−558 mV) predicts two-electron reduction of Fdx (−520 mV) by CODH/ACS (27). Indeed, results shown in Fig. 2B confirm reduction to Fdx2− in the presence of CODH/ACS and 1.0 atm of CO. However, Fdx2− was only partially oxidized by HdrA2B2C2 in the presence of CoMS-SCoB. A total of 10.1 ± 1.1 (n = 3) nmol of free thiols were produced from CoMS-SCoB in the reaction mixture, which contained 9.4 nmol Fdx2−. The results indicate that the physiologically relevant reduction of CoMS-SCoB catalyzed by CODH/ACS and HdrA2B2C2 proceeds according to equations 3 and 4 that are summed in equation 5.
| (3) |
| (4) |
| (5) |
Interaction of F420 with HdrA2 and HdrA2B2C2.
F420 is an electron carrier with a multitude of functions in methanogenic and nonmethanogenic species in the domains Bacteria and Archaea that also contain genes encoding HdrA2B2C2 (15). F420 is an obligatory two-electron carrier that requires Fdx:F420 oxidoreductases to contain a flavin that accepts an electron from Fdx and generates the hydride for transfer to F420. Thus, the flavin-containing HdrA2 is a candidate for interacting with F420. Indeed, HdrA2 catalyzed the Fdx-dependent reduction of F420 with a Km for F420 of 6.4 µM (Fig. S5), identifying a novel function for heterodisulfide reductases. NAD did not substitute for F420, although an HdrA homolog from the acetogen Moorella thermoacetica reduces the artificial electron acceptor benzyl viologen with NADH (28).
Fdx:F420 oxidoreductase activity of HdrA2. The atmosphere was 100% CO, and the temperature was 20°C. Reduction of F420 was monitored at 420 nm with a Cary 50 Bio UV-Vis spectrophotometer. The reactions were initiated by the addition of F420 after preincubation with CO to fully reduce Fdx and HdrA2. (A) Fdx- and HdrA2-dependent reduction of coenzyme F420. The reaction mixtures (0.5 ml) contained 0.1 mg CODH/ACS, 2.2 μM Fdx, 15 μM F420, and the indicated concentrations of HdrA2 in 50 mM MOPS buffer (pH 7.0). Key to curves or traces: (a) minus HdrA2; (b) minus Fdx; (c) 0.8 μM HdrA2 (5.1 nmol of F420 reduced/min/mg of HdrA2); (d) 3.2 μM HdrA2 (6.2 nmol of F420 reduced/min/mg of HdrA2). (B) Kinetic parameters for Fd:F420 oxidoreductase activity of HdrA2. The reaction mixtures (0.5 ml) contained 0.1 mg CODH/ACS, 2.2 μM Fdx, 2.1 μM HdrA2, and different amounts of F420 in 50 mM MOPS buffer (pH 7.0). 1 mU indicates that 1 nmol of F420 is reduced per minute. (Inset) Double-reciprocal plot. Download FIG S5, DOCX file, 0.1 MB (62.4KB, docx) .
Copyright © 2017 Yan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The ability of HdrA2 to interact with F420 prompted asking whether HdrA2 together with HdrB2C2 is able to bifurcate electron pairs from F420H2 for reduction of Fdx and CoMS-SCoB. Figure 3A and B are representative of results showing a dependence on Fdx for free thiol (HSCoM and HSCoB) formation and F420H2 oxidation in the presence of CoMS-SCoB. A replot (Fig. 3C) of the replicated data shows one free thiol produced from CoMS-SCoB for each F420H2 oxidized, a result consistent with simultaneous reduction of Fdx. These results show that HdrA2B2C2 catalyzes a thermodynamically favorable (ΔG°′ =−38.6 kJ), flavin-based, coupling of the endergonic one-electron reduction of Fdx (E0′ of ca. −520 mV) with F420H2 (E0′ of −380 mV) to the exergonic reduction of CoMS-SCoB (E0′ of −140 mV) (Table S2) as previously predicted (29). Considering the inability of HdrA2B2C2 to utilize Fdx1− as an electron donor (Fig. 2B), the results indicate the bifurcation proceeds according to equation 6. Figure 3D shows the dependence of F420H2 oxidation on two limiting concentrations of CoMS-SCoB and a fixed nonlimiting concentration of Fdx that is stoichiometrically consistent with equation 6.
FIG 3 .
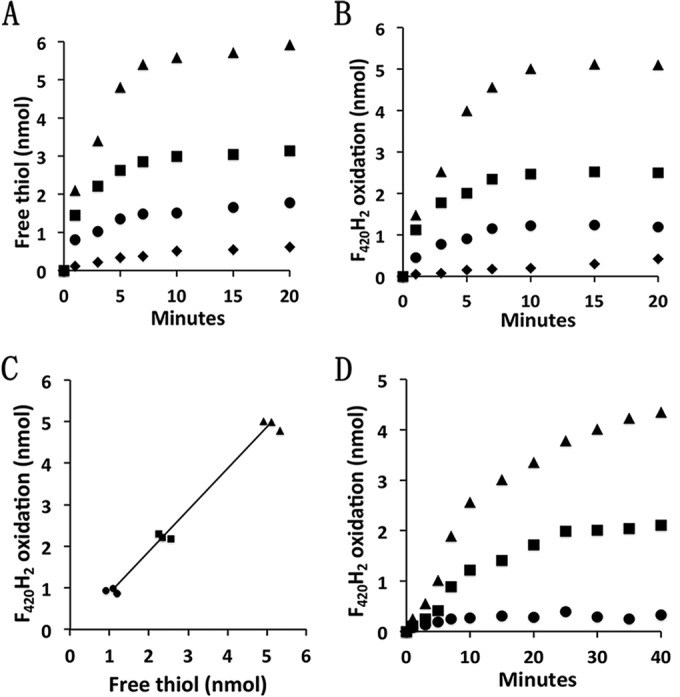
Electron bifurcation of electron pairs from F420H2 to Fdx and CoMS-SCoB. The reaction mixtures (0.5 ml) contained 1.6 μM HdrA2, 1.8 μM HdrB2C2, 12.5 μM F420H2, and the indicated amounts of CoMS-SCoB or Fdx in 50 mM MOPS buffer (pH 7.0). The atmosphere was 100% N2, and the temperature was 21°C. Reactions were initiated by the addition of HdrA2 or HdrB2C2. (A) Time course for Fdx-dependent production of free thiols in the presence of 50 μM CoMS-SCoB. Symbols: ▲, 5.2 nmol Fdx; ■, 2.6 nmol Fdx; •, 1.3 nmol Fdx; ⧫, no Fdx. (B) Time course for Fdx-dependent oxidation of F420H2 in the presence of 50 μM CoMS-SCoB. The symbols are the same as those used for panel A. (C) Amounts of free thiol produced versus the amounts of F420H2 oxidized in the presence of 1.3, 2.6, and 5.2 nmol Fdx. The amounts in the absence of Fdx were subtracted as a blank control. The results from three replicate experiments are shown. Symbols: ▲, 5.2 nmol Fdx; ■, 2.6 nmol Fdx; •, 1.3 nmol Fdx. (D) CoMS-SCoB-dependent F420H2 oxidation in the presence of 5.2 nmol Fdx. Symbols: ▲, 2.4 nmol CoMS-SCoB; ■, 1.2 nmol CoMS-SCoB; •, no CoMS-SCoB added.
| (6) |
Fdx:CoMS-SCoB oxidoreductase activity in cytoplasmic and membrane fractions of acetate-grown M. acetivorans.
During growth of M. acetivorans with acetate, the reduced Fdx generated by oxidation of the carbonyl group donates its electrons to a membrane-bound electron transfer chain culminating with membrane-bound HdrDE that reduces CoMS-SCoB to the corresponding sulfhydryl forms of the cofactors (equation 2). The membrane-bound electron transport is coupled to H+ and Na+ translocation, forming gradients that drive ATP synthesis (30). Acetate-grown cells also upregulate genes encoding a homolog of HdrABC from CO2-reducing methanogens (HdrA1B1C1), HdrA2 and HdrB2C2 (18, 19, 31). Thus, it is hypothesized that either HdrA1B1C1 or HdrA2B2C2 catalyze cytoplasmic Fdx:CoMS-SCoB oxidoreductase activity in acetate-grown cells (19, 25, 31) for which biochemical evidence of cytoplasmic oxidoreductase activity is shown in Table 2. Of the total heterodisulfide reductase activity present in the extracts, a corresponding amount, namely, 56 and 29%, was recovered in the cytoplasmic and membrane fractions, respectively. The cytoplasmic fraction contained approximately 50% of the total oxidoreductase activity in extracts. Less than 10% of the total activity was recovered in the membrane fraction with the location of the remaining 40% unexplained. Nonetheless, the results support a cytoplasmic Fdx:CoMS-SCoB oxidoreductase system that accounts for approximately half of the total activity in acetate-grown cells.
TABLE 2 .
Fdx:heterodisulfide oxidoreductase and heterodisulfide reductase activities in acetate-grown Methanosarcina acetivorans
| Fraction | Fdx:heterodisulfide oxidoreductase activitya |
Heterodisulfide reductase activityb |
||||
|---|---|---|---|---|---|---|
| CO/CODH-dependent |
NADPH/FNR-dependent |
Total activity (U) | Sp act (U/mg) | |||
| Total activity (mU) | Sp act (mU/mg) | Total activity (mU) | Sp act (mU/mg) | |||
| Extractc | 2,839 ± 300 | 17.0 ± 1.8 | 2,956 ± 301 | 17.7 ± 1.8 | 119 ± 12.8 | 0.73 ± 0.08 |
| Cytoplasmic | 1,579 ± 75 | 10.6 ± 0.5 | 1,401 ± 164 | 9.4 ± 1.1 | 66.5 ± 5.9 | 0.44 ± 0.04 |
| Membrane | 193 ± 2 | 8.4 ± 0.1 | 223 ± 16 | 9.7 ± 0.7 | 34.4 ± 0.6 | 1.48 ± 0.03 |
One unit is defined as micromoles of sulfhydryl produced per minute.
One unit is defined as micromoles of reduced methyl viologen oxidized per minute.
A sample from the same extract was loaded onto the sucrose gradient for preparation of cytoplasmic and membrane fractions.
DISCUSSION
The results have produced several milestones that provide a greater comprehensive understanding of the HdrABC class. Fusion of the MvhD homolog to the HdrA domain of HdrA2 is an unusual feature that, together with unusual catalytic capabilities reported here, identifies HdrA2B2C2 as representative of a previously unrecognized HdrABC subclass distributed in diverse species from the domains Bacteria and Archaea.
Intersubunit electron transfer.
The individual expression and characterization of recombinant HdrA2, HdrB2, and HdrB2C2 provided an experimental approach to identify roles for each subunit previously unknown for any HdrABC homolog (13). The results show that the flavin-containing HdrA2 interacts with F420 and Fdx and that HdrA2B2C2 bifurcates electron pairs from F420H2 directed to the reduction of Fdx and CoMS-SCoB. It was also shown that, although HdrC2 has no role in catalysis, this subunit mediates electron transfer from HdrA2 to the catalytic HdrB2. These results establish the path of electron transfer from Fdx to CoMS-SCoB (Fig. 4) that in all probability generally applies to the HdrABC class.
FIG 4 .
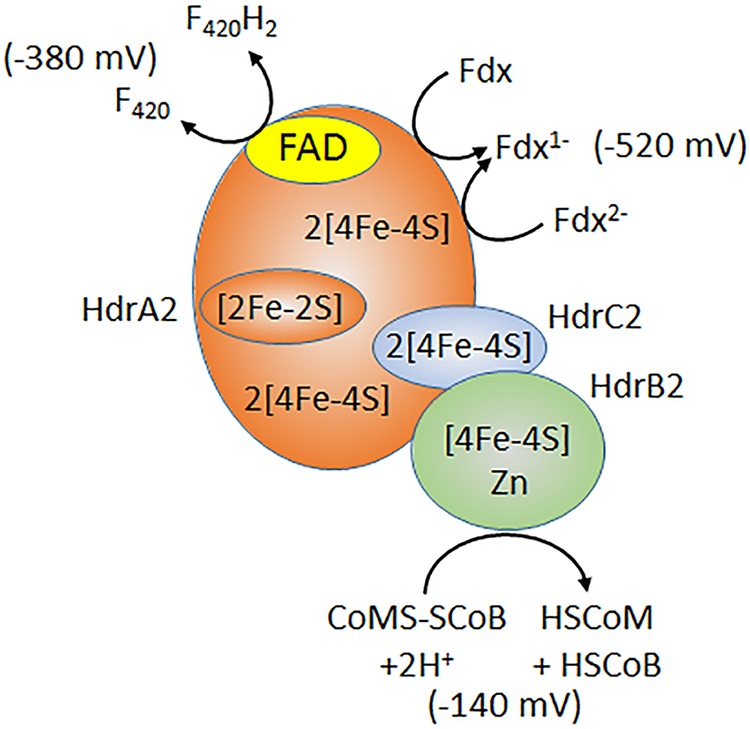
Electron transfer reactions catalyzed by HdrA2B2C2. The redox potentials are standard potentials at pH 7.0 described elsewhere (29). The value for Fdx from M. acetivorans is assumed to be similar to estimates (29, 61).
Role for the Fdx:CoMS-SCoB oxidoreductase activity of HdrAB2C2 in the pathway for conversion of acetate to CH4.
Coenzyme F420 does not participate in the pathway of acetate conversion to methane, ruling out a role for the F420H2 bifurcating activity of HdrA2B2C2. However, a role is envisioned for the Fdx:CoMS-SCoB oxidoreductase activity of HdrAB2C2 as shown in Fig. 5. The canonical HdrABC class is a cytoplasmic enzyme that contains flavin, whereas the HdrDE class is membrane bound, flavin free, and contains heme (32, 33). The cytoplasmic HdrABC class is considered specific to obligatory CO2-reducing methanogens, and the membrane-bound HdrDE class is considered specific to pathways converting acetate and methylotrophic substrates (methanol, methylamines, and methylsulfide) to CH4. However, homologs of the cytoplasmic HdrABC class are also encoded in the genomes of acetotrophic and methylotrophic species from the order Methanosarcinales consistent with an auxiliary function. Our results indicate HdrA2B2C2 is responsible for cytoplasmic Fdx:CoMS-SCoB oxidoreductase activity of M. acetivorans when metabolizing acetate to CH4. This conclusion is further supported by the reported upregulation of hdrA2, hdrB2, and hdrC2 in response to growth with acetate and genetic analyses indicating a role for HdrA2 while excluding a role for the canonical HdrA1B1C1 (19, 31, 34). Although HdrB2C2 was shown to catalyze the transfer of electrons from Fdx to CoMS-SCoB, the HdrA2B2C2 complex was shown to be more efficient, which suggests a greater role in cytoplasmic electron transport during conversion of acetate to CH4. Genes encoding HdrA2B2C2 of M. acetivorans constitute a clade with homologs of other acetotrophic methanogens, further supporting a role in the pathway of acetate conversion to methane (19).
FIG 5 .
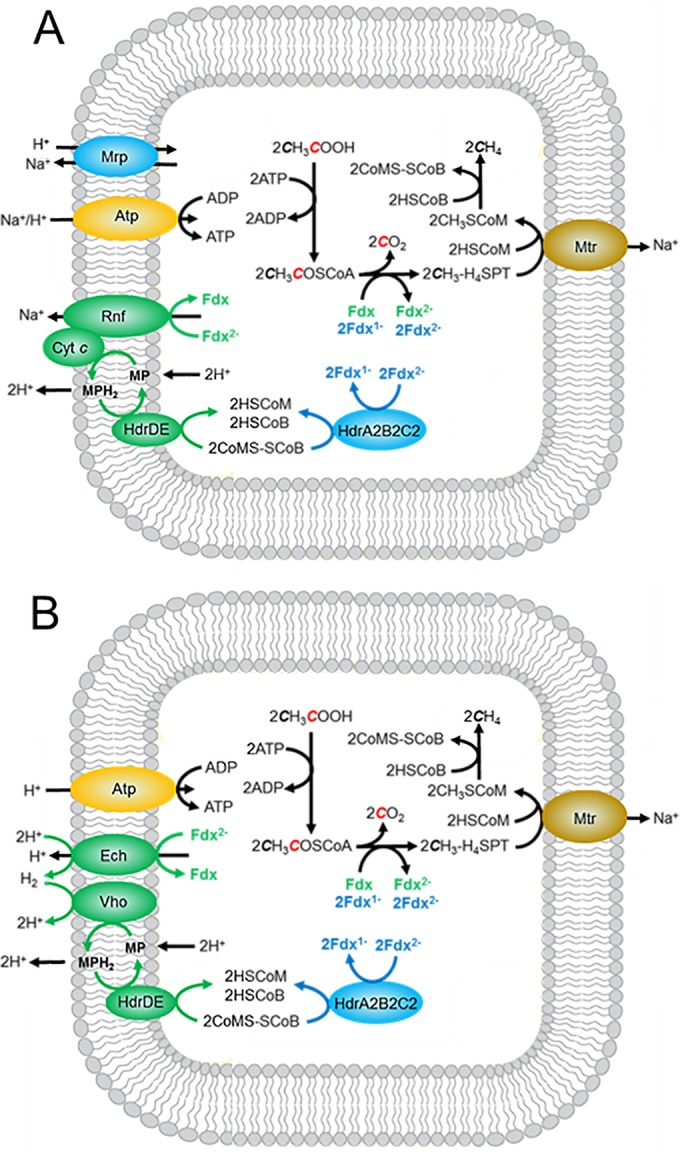
Pathways of acetate conversion to methane by Methanosarcina species showing electron transport chains diverging from reduced ferredoxin. (A) Pathway not requiring H2. (B) Pathway requiring H2. Ech, proton-pumping H2-evolving hydrogenase; Vho, H2 uptake hydrogenase; Mrp, multisubunit Na+/H+ antiporter; Atp, ATP synthase; Rnf, Rnf complex; Cyt c, multiheme cytochrome c; Fdx, ferredoxin; H4SPT, tetrahydrosarcinapterin; MP, methanophenazine; HdrDE, membrane-bound heterodisulfide reductase; HdrA2B2C2, cytoplasmic heterodisulfide reductase; HSCoM, coenzyme M; HSCoB, coenzyme B; Mtr, methyltransferase.
The hdrA2 gene in M. acetivorans is in an operon with a gene encoding a putative polyferredoxin that is upregulated during acetotrophic growth, which suggests a potential role in mediating electron transfer to HdrA2B2C2 (18, 19, 31). Attempts to produce the recombinant protein in E. coli strain BL21(DE3) ΔiscR resulted in preparations that contained only a partial complement of Fe-S clusters that resisted reconstitution. The preparations were unable to replace Fdx or stimulate reduction of CoMS-SCoB in complete reaction mixtures. Therefore, no definitive conclusions can be drawn regarding the role of the polyferredoxin except that it is not necessary for the transfer of electrons from CODH/ACS to HdrA2B2C2.
A few Methanosarcina species contain a hydrogenase (Ech) that oxidizes the Fdx with production of H2 and translocation of H+ (24). A second hydrogenase (Vho) oxidizes H2 and reduces methanophenazine (MP), a quinone-like electron carrier that donates electrons to HdrDE and translocates an additional 4H+. Thus, together with the membrane-bound methyltransferase (Mtr), four coupling sites produce Na+ and H+ gradients that drive ATP synthesis. Most Methanosarcina species, represented by M. acetivorans, do not contain functional hydrogenases. Instead, M. acetivorans contains the membrane-bound Rnf complex that accepts electrons from Fdx and reduces MP mediated by cytochrome c (25). The membrane-bound transfer of electrons from Fdx to CoMS-SCoB supports Na+ translocation by the Rnf complex and H+ translocation by HdrDE (35). Thus, a possible three coupling sites generate Na+ and H+ gradients that together drive ATP synthesis in M. acetivorans (30). The free energy available from conversion of acetate to CH4 and CO2 under standard conditions of equimolar reactants and products (ΔG°′ = −36 kJ) provides only a marginal amount of energy for growth considering the ATP requirement for activating acetate to acetyl-CoA in the first step in the pathway (ΔG°′ = +31.8 kJ) (36). Thus, growth with acetate is at the extreme thermodynamic limit, requiring extraordinary mechanisms for maximizing the thermodynamic efficiency. Our finding of a cytoplasmic Fdx:CoMS-SCoB oxidoreductase system in acetate-grown M. acetivorans, and reconstitution of an active HdrA2B2C2, supports the divergent electron transport pathways from Fdx to CoMS-SCoB shown in Fig. 5A. Methanosarcina mazei and Methanosarcina barkeri, species that produce and consume H2 during growth on acetate, also encode HdrA2, HdrB2, and HdrC2 homologs with greater than 80% amino acid sequence identity, predicting these species also synthesize HdrA2B2C2 participating in a soluble electron transport pathway (Fig. 5B). Acetyl-CoA-dependent methanogenesis catalyzed by M. barkeri cell lysate is not dependent on the membrane fraction consistent with a cytoplasmic electron transport system (37). For Methanosarcina species with documented respiratory control (38), HdrA2B2C2 provides a mechanism for modulating cytoplasmic versus membrane-bound electron transport proportional to ATP demand. We posit that the HdrA2B2C2-dependent electron transport allows cells to maximize thermodynamic efficiency by circumventing membrane-bound electron transport when nonstandard concentrations of acetate encountered in the environment constrain the free energy available for multiple ion translocation sites. With this mechanism, Methanosarcina species could maximize the thermodynamic efficiency and metabolize acetate more rapidly, thereby outcompeting others for acetate. Indeed, M. acetivorans must compete with the more thermodynamically favorable acetate utilization by sulfate-reducing species (CH3COO− + SO42− → 2HCO3− + HS−; ΔG°′ = −71.7 kJ) in the marine environment where M. acetivorans was isolated (17, 39).
A proposed role for the F420H2 bifurcation activity of HdrA2B2C2 homologs in Fe(III)-dependent anaerobic CH4 oxidation (ANME).
Although the F420H2 bifurcation activity of HdrA2B2C2 is unanticipated in the pathway of acetate conversion to methane, a role for this activity can be envisioned for homologs in pathways of Fe(III)-dependent anaerobic methane oxidation as shown in Fig. 6. For more than a decade, it was assumed that ANME required a consortium of at least two metabolic groups. However, recent reports indicate uncultured Archaea species in ANME group 2 (ANME-2) environments oxidize CH4 alone, albeit dependent on Fe(III) as a direct electron acceptor (40, 41). Furthermore, genomic analyses of an ANME-2a environment implicate a role for HdrABC homologs and acetate as a product (9). Homologs of HdrA2 and HdrB2 are encoded in the genome of “Candidatus Methanoperedens nitroreducens” (see Fig. S1 in the supplemental material) consistent with roles in the NO3−-dependent ANME pathway of this and possibly other ANME species dependent on external electron acceptors (42). Notably, Fe(III)-dependent oxidation of CH4 was recently reported for an enrichment culture containing archaea of the order Methanosarcinales related to “Candidatus Methanoperedens nitroreducens” (43). Isolated ANME species have not been reported; however, M. acetivorans is capable of Fe(III)-dependent conversion of CH4 to acetate and is phylogenetically related to uncultured species identified in ANME-2 consortia (44). Our current understanding leads to a proposed ANME pathway for M. acetivorans involving HdrA2B2C2 (Fig. 6; Table S2) that may also be operable in ANME-2 environments.
FIG 6 .
Pathway proposed for anaerobic CH4 oxidation (ANME) by M. acetivorans. The reaction numbers are shown in parentheses in the figure. CoA-SH, coenzyme A; H4SPT, tetrahydrosarcinapterin; FdxR, reduced ferredoxin; FdxO, oxidized ferredoxin; HSCoM, coenzyme M; HSCoB, coenzyme B; MP, methanophenazine; HdrDE, membrane-bound heterodisulfide reductase; HdrA2B2C2, cytoplasmic heterodisulfide reductase; Rnf, Rnf complex; Cyt c, cytochrome c; Atp, ATP synthase; Mrp, sodium/proton antiporter; Mtr, membrane-bound methyltransferase. Enzymes and thermodynamic calculations for each reaction are shown in Table S2.
The core of the proposed ANME pathway (reactions 1 to 5, 7, 8, 16, 18, and 19) is a reversal of the established acetate-utilizing CH4-producing pathway in M. acetivorans (45). Table S2 lists thermodynamic calculations for reactions shown in Fig. 6. Reactions 1 and 2 in the ANME pathway are thermodynamically unfavorable (ΔG°′ = +121 kJ), requiring oxidation of HSCoM and HSCoB coupled to reduction of Fe(III) (reactions 1 to 6), yielding an overall ΔG°′ of −176 kJ (Table S2). The oxidation is catalyzed by HdrDE with transfer of electrons to methanophenazine (MP) and cytochrome c where Fe(III) is the terminal electron acceptor, thereby pulling reactions 1 and 2. Scalar proton translocation is accomplished by a “Q loop” mechanism involving MP and driven by reactions 3 to 6 (ΔG°′ = −297 kJ). Acetate-grown M. acetivorans is rich in multiheme cytochrome c that shuttles electrons between the Rnf complex and MP (25, 46). Reduction of Fe(III) at cytochrome c of M. acetivorans is consistent with the documented role of multiheme c-type cytochromes as electron shuttles to Fe(III) minerals outside microbial cells (47, 48). Moreover, metagenomic analyses of ANME environments reveal homologs of genes encoding c-type cytochromes and Rnf complexes hypothesized to function in ANME pathways (9, 41, 49).
The product of reaction 2 (CH3-H4SPT) is metabolized by divergent pathways leading to either acetate or CO2. The acetogenic pathway begins with reaction 7 (ΔG°′ = −40.5 kJ) requiring reduced Fdx for reduction of CO2 that provides the carbonyl group of acetyl-CoA. The requisite reduced Fdx is generated in the pathway oxidizing the methyl group of CH3-H4SPT to CO2 (reactions 9 to 15). The oxidation pathway is identical to that which functions in the dismutation of methylotrophic substrates to CO2 and CH4 wherein reaction 9 involves multiple steps, including two oxidations dependent on F420 as the electron acceptor (50). The F420H2 is oxidized by HdrA2B2C2 with bifurcation of electrons directed to Fdx and CoMS-SCoB (reaction 11). The bifurcation (ΔG°′ = −38.6 kJ) is coupled to reactions 12 to 15 (ΔG°′ = −148.5 kJ), reducing Fe(III) with an overall ΔG°′ of −187.1 kJ. Reactions 12 to 15 are catalyzed by the same proteins and electron carriers as those used in reactions 3 to 6 albeit with different stoichiometry. Scalar proton translocation generates a proton gradient (high outside) accomplished by a “Q loop” mechanism involving MP. The exergonic oxidation of Fdx and reduction of Fe(III) in reactions 16 and 17 (ΔG°′ = −249.3 kJ) drives a vectorial translocation of Na+ consistent with the previously reported reduction of cytochrome c and pumping of Na+ by Rnf (25, 35). The thermodynamically unfavorable reaction 2 (ΔG°′ = +29.2 kJ) is catalyzed by the membrane-bound methyltransferase (Mtr) and driven by the Na+ gradient. ATP synthesis is catalyzed by the ATP synthase (reaction 18) dependent on the Na+ and H+ gradients (30). ATP is also synthesized by substrate level phosphorylation (reaction 8) catalyzed by phosphotransacetylase and acetate kinase. As previously proposed, multisubunit Na+/H+ antiporter (Mrp) functions to adjust the ratio of Na+/H+ (reaction 19) optimal for the ATP synthase and methyltransferase (51, 52). Finally, the stoichiometry shown in Fig. 6 assumes a low availability of Fe3+ that limits the Fe(III)-dependent oxidation of Fdx (reactions 16 and 17), allowing for the Fdx-dependent synthesis of acetate (reactions 7 and 8). Reactions 16 and 17 are more thermodynamically favorable than reactions 7 and 8 (Table S2); therefore, complete oxidation of CH4 to CO2 would be expected when Fe3+ is nonlimiting. Thus, the availability of Fe3+ in the native environment is expected to modulate the stoichiometry of CH4 oxidation to acetate and CO2. It is also possible that reactions 7 and 10, or reactions 7 and 11, are spatially coupled, circumventing the Fe(III)-dependent oxidation of reduced Fdx (FdxR) by reactions 16 and 17, resulting in the stoichiometry shown in Fig. 6 when Fe(III) is nonlimiting.
Additional electron bifurcating roles are possible for HdrA2B2C2 homologs in nonmethanogenic species with diverse metabolisms (Fig. S1) that may replace F420H2 (E0′ = −380 mV) with the analogous two-electron carrier NADH (E0′ = −320 mV) and replace CoMS-SCoB with disulfides such as the DsrC protein postulated for the FlxlABCD-HdrABC bifurcating complex of Desulfovibrio vulgaris (12).
Conclusions.
HdrA2B2C2 represents a subclass of the HdrABC class with homologs in diverse species of the domains Bacteria and Archaea. Results revealed a previously unknown electron bifurcation system and intersubunit electron transport pathway generally applicable to the HdrABC class. Properties of HdrA2B2C2 predict that homologs participate in anaerobic CH4 oxidation pathways and that HdrA2B2C2 is essential for optimal growth of acetotrophic methanogens in native environments.
MATERIALS AND METHODS
Cell growth and materials.
Acetate-grown Methanosarcina acetivorans was mass cultured and harvested as previously described (53). E. coli strain BL21(DE3) ΔiscR was a gift from J. Golbeck. HSCoB and CoMS-SCoB was a gift from T. Wood. All chromatography columns, resins, and prepacked columns were purchased from GE Healthcare. Purification of F420 from methanol-grown M. acetivorans cells and preparation of F420H2 was performed as described elsewhere (54). All other chemicals were purchased from Sigma-Aldrich or VWR International.
Cloning of M. acetivorans genes.
Genes encoding Fdx (MA0431), HdrA2 (MA2868), and HdrB2 (MA4237) were amplified by PCR from genomic DNA using primers shown in Table S1 in the supplemental material and cloned into pET22b (Novagen) using In-Fusion (Clontech) for expression of the proteins with a C-terminal His6 tag. Genes encoding HdrC2 (MA4236) and HdrB2 (MA4237) were amplified by PCR and cloned into the first and second site, respectively, of pETDuet (Novagen) for coexpression with a C-terminal His6 tag on HdrB2. All constructs were validated by sequencing.
Primers used for amplification of genes and construction of plasmids. Download TABLE S1, DOCX file, 0.01 MB (12.3KB, docx) .
Copyright © 2017 Yan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Standard Gibbs free energy values for reactions in the Fe(III)-dependent ANME pathway proposed for Methanosarcina acetivorans. Download TABLE S2, DOCX file, 0.02 MB (16.5KB, docx) .
Copyright © 2017 Yan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gibbs free energy of metabolite formation (ΔfGt) calculated at 25°C, pH 7, and ionic concentration of 0.25 M. Download TABLE S3, DOCX file, 0.03 MB (33.3KB, docx) .
Copyright © 2017 Yan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression and purification of recombinant proteins.
The expression plasmids (Table S1) were transformed into E. coli strain BL21(DE3) ΔiscR and grown with 100 µg/ml ampicillin. A single colony was used to inoculate 100 ml of LB medium buffered with 50 mM morpholinepropanesulfonic acid (MOPS) (pH 7.4) and supplemented with 100 μg/ml of ampicillin that was incubated at 37°C for 12 h. The starter culture was subcultured (1:50 dilution) in 3 liters of the above medium supplemented with 1 mM ferric ammonium citrate that was contained in a spinner flask (Chemglass) modified to allow sparging with 100% Ar. Cultures were grown aerobically at 37°C to an optical density at 600 nm (OD600) of 0.6 that was then supplemented with 1 mM cysteine and induced with 200 µM isopropyl-β-d-thiogalactopyranoside (IPTG). Anaerobic metabolism was facilitated by the addition of glucose (0.5%, wt/vol) and sodium fumarate (25 mM) followed by incubation at 21°C for 20 h with sparging. Cells were harvested by centrifugation in sealed tubes containing 95% N2–5% H2, resuspended (1:5, wt/vol) in 50 mM Tris (pH 8.0) containing 10% glycerol (buffer C), and stored at −80°C until use. The anaerobic purification of recombinant proteins is described in Text S1. Purified HdrA2 was reconstituted with FAD by anaerobic incubation at 4°C for 10 h in buffer containing 0.1 mM FAD and passed through a PD-10 column to remove excess flavin.
Supplemental Materials and Methods. Download TEXT S1, DOCX file, 0.02 MB (20.6KB, docx) .
Copyright © 2017 Yan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Purification of CODH/ACS.
The five-subunit enzyme was purified from acetate-grown M. acetivorans as described previously with modifications noted in Text S1 (55). A typical preparation reduced the recombinant Fdx (33.3 μM) at a rate of 55.3 nmol/min/mg CODH/ACS.
Enzyme assays.
All enzyme assays were performed at 21°C in stoppered cuvettes containing the indicated atmosphere. Heterodisulfide reductase forward and reverse activities were performed as described elsewhere with modifications noted in Text S1 (56). Fdx:CoMS-SCoB oxidoreductase activity was performed as described in Text S1.
Other analytical procedures.
All protein concentrations, except for Fdx, were determined with either the Bradford assay kit (Bio-Rad Laboratories) or Pierce assay kit (Thermo Scientific) that gave similar results. The protein concentration for Fdx was determined using the ε390 of 30 mM−1 cm−1 (57). Iron and acid-labile sulfur contents were determined as described elsewhere (58, 59). The flavin content of HdrA2 was determined by UV-Vis and fluorescence spectrometry as described elsewhere (60). For the electron bifurcation experiments, F420H2 oxidation was determined by monitoring the fluorescence intensity with excitation at 420 and emission at 480 nm.
ACKNOWLEDGMENTS
We thank Hadi Nazem-Bokaee for thermodynamic calculations, Karim Walters for assistance in anaerobic expression of proteins, and Prashanti Iyer for coenzyme F420.
Approximately 65% of this work was supported by the U.S. Department of Energy ARPA-e 0881-1525, 25% by the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy through grant DE-FG02-95ER20198 MOD16, and 10% by the Person Endowment to J.G.F.
Footnotes
Citation Yan Z, Wang M, Ferry JG. 2017. A ferredoxin- and F420H2-dependent, electron-bifurcating, heterodisulfide reductase with homologs in the domains Bacteria and Archaea. mBio 8:e02285-16. https://doi.org/10.1128/mBio.02285-16.
REFERENCES
- 1.Hedderich R, Thauer RK. 1988. Methanobacterium thermoautotrophicum contains a soluble enzyme system that specifically catalyzes the reduction of the heterodisulfide of coenzyme M and 7-mercaptoheptanoylthreonine phosphate with H2. FEBS Lett 234:223–227. doi: 10.1016/0014-5793(88)81339-5. [DOI] [Google Scholar]
- 2.Christel S, Fridlund J, Buetti-Dinh A, Buck M, Watkin EL, Dopson M. 2016. RNA transcript sequencing reveals inorganic sulfur compound oxidation pathways in the acidophile Acidithiobacillus ferrivorans. FEMS Microbiol Lett 363:pii=fnw057. doi: 10.1093/femsle/fnw057. [DOI] [PubMed] [Google Scholar]
- 3.Justice NB, Norman A, Brown CT, Singh A, Thomas BC, Banfield JF. 2014. Comparison of environmental and isolate Sulfobacillus genomes reveals diverse carbon, sulfur, nitrogen, and hydrogen metabolisms. BMC Genomics 15:1107. doi: 10.1186/1471-2164-15-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osorio H, Mangold S, Denis Y, Ñancucheo I, Esparza M, Johnson DB, Bonnefoy V, Dopson M, Holmes DS. 2013. Anaerobic sulfur metabolism coupled to dissimilatory iron reduction in the extremophile Acidithiobacillus ferrooxidans. Appl Environ Microbiol 79:2172–2181. doi: 10.1128/AEM.03057-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otwell AE, Callister SJ, Zink EM, Smith RD, Richardson RE. 2016. Comparative proteomic analysis of Desulfotomaculum reducens MI-1: insights into the metabolic versatility of a Gram-positive sulfate- and metal-reducing bacterium. Front Microbiol 7:191. doi: 10.3389/fmicb.2016.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Möller-Zinkhan D, Börner G, Thauer RK. 1989. Function of methanofuran, tetrahydromethanopterin, and coenzyme F420 in Archaeoglobus fulgidus. Arch Microbiol 152:362–368. doi: 10.1007/BF00425174. [DOI] [Google Scholar]
- 7.McInerney MJ, Rohlin L, Mouttaki H, Kim U, Krupp RS, Rios-Hernandez L, Sieber J, Struchtemeyer CG, Bhattacharyya A, Campbell JW, Gunsalus RP. 2007. The genome of Syntrophus aciditrophicus: life at the thermodynamic limit of microbial growth. Proc Natl Acad Sci U S A 104:7600–7605. doi: 10.1073/pnas.0610456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arshad A, Speth DR, de Graaf RM, Op den Camp HJ, Jetten MS, Welte CU. 2015. A metagenomics-based metabolic model of nitrate-dependent anaerobic oxidation of methane by Methanoperedens-like Archaea. Front Microbiol 6:1423. doi: 10.3389/fmicb.2015.01423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang FP, Zhang Y, Chen Y, He Y, Qi J, Hinrichs KU, Zhang XX, Xiao X, Boon N. 2014. Methanotrophic archaea possessing diverging methane-oxidizing and electron-transporting pathways. ISME J 8:1069–1078. doi: 10.1038/ismej.2013.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thauer RK. 2011. Anaerobic oxidation of methane with sulfate: on the reversibility of the reactions that are catalyzed by enzymes also involved in methanogenesis from CO2. Curr Opin Microbiol 14:292–299. doi: 10.1016/j.mib.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Sousa FL, Nelson-Sathi S, Martin WF. 2016. One step beyond a ribosome: the ancient anaerobic core. Biochim Biophys Acta 1857:1027–1038. doi: 10.1016/j.bbabio.2016.04.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramos AR, Grein F, Oliveira GP, Venceslau SS, Keller KL, Wall JD, Pereira IA. 2015. The FlxABCD-HdrABC proteins correspond to a novel NADH dehydrogenase/heterodisulfide reductase widespread in anaerobic bacteria and involved in ethanol metabolism in Desulfovibrio vulgaris Hildenborough. Environ Microbiol 17:2288–2305. doi: 10.1111/1462-2920.12689. [DOI] [PubMed] [Google Scholar]
- 13.Buckel W, Thauer RK. 2013. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim Biophys Acta 1827:94–113. doi: 10.1016/j.bbabio.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Costa KC, Wong PM, Wang T, Lie TJ, Dodsworth JA, Swanson I, Burn JA, Hackett M, Leigh JA. 2010. Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc Natl Acad Sci U S A 107:11050–11055. doi: 10.1073/pnas.1003653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greening C, Ahmed FH, Mohamed AE, Lee BM, Pandey G, Warden AC, Scott C, Oakeshott JG, Taylor MC, Jackson CJ. 2016. Physiology, biochemistry, and applications of F420- and Fo-dependent redox reactions. Microbiol Mol Biol Rev 80:451–493. doi: 10.1128/MMBR.00070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grein F, Ramos AR, Venceslau SS, Pereira IA. 2013. Unifying concepts in anaerobic respiration: insights from dissimilatory sulfur metabolism. Biochim Biophys Acta 1827:145–160. doi: 10.1016/j.bbabio.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Sowers KR, Baron SF, Ferry JG. 1984. Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl Environ Microbiol 47:971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Li Q, Rohlin L, Kim U, Salmon K, Rejtar T, Gunsalus RP, Karger BL, Ferry JG. 2007. Quantitative proteomic and microarray analysis of the archaeon Methanosarcina acetivorans grown with acetate versus methanol. J Proteome Res 6:759–771. doi: 10.1021/pr060383l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buan NR, Metcalf WW. 2010. Methanogenesis by Methanosarcina acetivorans involves two structurally and functionally distinct classes of heterodisulfide reductase. Mol Microbiol 75:843–853. doi: 10.1111/j.1365-2958.2009.06990.x. [DOI] [PubMed] [Google Scholar]
- 20.Soo VW, McAnulty MJ, Tripathi A, Zhu F, Zhang L, Hatzakis E, Smith PB, Agrawal S, Nazem-Bokaee H, Gopalakrishnan S, Salis HM, Ferry JG, Maranas CD, Patterson AD, Wood TK. 2016. Reversing methanogenesis to capture methane for liquid biofuel precursors. Microb Cell Fact 15:11. doi: 10.1186/s12934-015-0397-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moran JJ, House CJ, Thomas B, Freeman KH. 2007. Products of trace methane oxidation during nonmethyltrophic growth by Methanosarcina. J Geophys Res 112:1–7. [Google Scholar]
- 22.Moran JJ, House CH, Freeman KH, Ferry JG. 2005. Trace methane oxidation studied in several Euryarchaeota under diverse conditions. Archaea 1:303–309. doi: 10.1155/2005/650670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferry JG. 2010. How to make a living by exhaling methane. Annu Rev Microbiol 64:453–473. doi: 10.1146/annurev.micro.112408.134051. [DOI] [PubMed] [Google Scholar]
- 24.Welte C, Deppenmeier U. 2014. Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim Biophys Acta 1837:1130–1147. doi: 10.1016/j.bbabio.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Wang M, Tomb JF, Ferry JG. 2011. Electron transport in acetate-grown Methanosarcina acetivorans. BMC Microbiol 11:165. doi: 10.1186/1471-2180-11-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamann N, Mander GJ, Shokes JE, Scott RA, Bennati M, Hedderich R. 2007. A cysteine-rich CCG domain contains a novel [4Fe-4S] cluster binding motif as deduced from studies with subunit B of heterodisulfide reductase from Methanothermobacter marburgensis. Biochemistry 46:12875–12885. doi: 10.1021/bi700679u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grahame DA, Demoll E. 1995. Substrate and accessory protein requirements and thermodynamics of acetyl-CoA synthesis and cleavage in Methanosarcina barkeri. Biochemistry 34:4617–4624. doi: 10.1021/bi00014a015. [DOI] [PubMed] [Google Scholar]
- 28.Mock J, Wang S, Huang H, Kahnt J, Thauer RK. 2014. Evidence for a hexaheteromeric methylenetetrahydrofolate reductase in Moorella thermoacetica. J Bacteriol 196:3303–3314. doi: 10.1128/JB.01839-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Catlett JL, Ortiz AM, Buan NR. 2015. Rerouting cellular electron flux to increase the rate of biological methane production. Appl Environ Microbiol 81:6528–6537. doi: 10.1128/AEM.01162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlegel K, Leone V, Faraldo-Gómez JD, Müller V. 2012. Promiscuous archaeal ATP synthase concurrently coupled to Na+ and H+ translocation. Proc Natl Acad Sci U S A 109:947–952. doi: 10.1073/pnas.1115796109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohlin L, Gunsalus RP. 2010. Carbon-dependent control of electron transfer and central carbon pathway genes for methane biosynthesis in the Archaean, Methanosarcina acetivorans strain C2A. BMC Microbiol 10:62. doi: 10.1186/1471-2180-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heiden S, Hedderich R, Setzke E, Thauer RK. 1994. Purification of a two-subunit cytochrome-b-containing heterodisulfide reductase from methanol-grown Methanosarcina barkeri. Eur J Biochem 221:855–861. doi: 10.1111/j.1432-1033.1994.tb18800.x. [DOI] [PubMed] [Google Scholar]
- 33.Kaster AK, Moll J, Parey K, Thauer RK. 2011. Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc Natl Acad Sci U S A 108:2981–2986. doi: 10.1073/pnas.1016761108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Q, Li L, Rejtar T, Karger BL, Ferry JG. 2005. Proteome of Methanosarcina acetivorans. Part I, an expanded view of the biology of the cell. J Proteome Res 4:112–128. doi: 10.1021/pr049832c. [DOI] [PubMed] [Google Scholar]
- 35.Schlegel K, Welte C, Deppenmeier U, Müller V. 2012. Electron transport during aceticlastic methanogenesis by Methanosarcina acetivorans involves a sodium-translocating Rnf complex. FEBS J 279:4444–4452. doi: 10.1111/febs.12031. [DOI] [PubMed] [Google Scholar]
- 36.Thauer RK, Jungermann K, Decker K. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev 41:100–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischer R, Thauer RK. 1990. Ferredoxin-dependent methane formation from acetate in cell extracts of Methanosarcina barkeri (strain MS). FEBS Lett 269:368–372. doi: 10.1016/0014-5793(90)81195-T. [DOI] [PubMed] [Google Scholar]
- 38.Deppenmeier U, Blaut M, Mahlmann A, Gottschalk G. 1990. Reduced coenzyme F420:heterodisulfide oxidoreductase, a proton-translocating redox system in methanogenic bacteria. Proc Natl Acad Sci U S A 87:9449–9453. doi: 10.1073/pnas.87.23.9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Widdel F, Pfennig N. 1981. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. I. Isolation of new sulfate-reducing bacteria enriched with acetate from saline environments. Description of Desulfobacter postgatei gen. nov., sp. nov. Arch Microbiol 129:395–400. doi: 10.1007/BF00406470. [DOI] [PubMed] [Google Scholar]
- 40.Scheller S, Yu H, Chadwick GL, McGlynn SE, Orphan VJ. 2016. Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science 351:703–707. doi: 10.1126/science.aad7154. [DOI] [PubMed] [Google Scholar]
- 41.McGlynn SE, Chadwick GL, Kempes CP, Orphan VJ. 2015. Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526:531–535. doi: 10.1038/nature15512. [DOI] [PubMed] [Google Scholar]
- 42.Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW. 2013. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500:567–570. doi: 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- 43.Ettwig KF, Zhu B, Speth D, Keltjens JT, Jetten MS, Kartal B. 2016. Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc Natl Acad Sci U S A 113:12792–12796. doi: 10.1073/pnas.1609534113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orphan VJ, Hinrichs KU, Ussler W III, Paull CK, Taylor LT, Sylva SP, Hayes JM, Delong EF. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl Environ Microbiol 67:1922–1934. doi: 10.1128/AEM.67.4.1922-1934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferry JG. 2015. Acetate metabolism in anaerobes from the domain Archaea. Life 5:1454–1471. doi: 10.3390/life5021454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Q, Li L, Rejtar T, Lessner DJ, Karger BL, Ferry JG. 2006. Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans. J Bacteriol 188:702–710. doi: 10.1128/JB.188.2.702-710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi L, Squier TC, Zachara JM, Fredrickson JK. 2007. Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Mol Microbiol 65:12–20. doi: 10.1111/j.1365-2958.2007.05783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber KA, Achenbach LA, Coates JD. 2006. Microorganisms pumping iron: anaerobic microbial iron oxidation and reduction. Nat Rev Microbiol 4:752–764. doi: 10.1038/nrmicro1490. [DOI] [PubMed] [Google Scholar]
- 49.Hallam SJ, Putnam N, Preston CM, Detter JC, Rokhsar D, Richardson PM, DeLong EF. 2004. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305:1457–1462. doi: 10.1126/science.1100025. [DOI] [PubMed] [Google Scholar]
- 50.Deppenmeier U. 2004. The membrane-bound electron transport system of Methanosarcina species. J Bioenerg Biomembr 36:55–64. doi: 10.1023/B:JOBB.0000019598.64642.97. [DOI] [PubMed] [Google Scholar]
- 51.Jasso-Chavez R, Diaz-Perez C, Rodriguez-Zavala JS, Ferry JG. 2016. Functional role of MrpA in the MrpABCDEFG Na+/H+ antiporter complex from the archaeon Methanosarcina acetivorans. J Bacteriol 199:e00662-16. doi: 10.1128/jb.00662-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jasso-Chávez R, Apolinario EE, Sowers KR, Ferry JG. 2013. MrpA functions in energy conversion during acetate-dependent growth of Methanosarcina acetivorans. J Bacteriol 195:3987–3994. doi: 10.1128/JB.00581-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sowers KR, Nelson MJK, Ferry JG. 1984. Growth of acetotrophic, methane-producing bacteria in a pH auxostat. Curr Microbiol 11:227–229. doi: 10.1007/BF01567165. [DOI] [Google Scholar]
- 54.Welte C, Deppenmeier U. 2011. Proton translocation in methanogens. Methods Enzymol 494:257–280. doi: 10.1016/B978-0-12-385112-3.00013-5. [DOI] [PubMed] [Google Scholar]
- 55.Terlesky KC, Nelson MJK, Ferry JG. 1986. Isolation of an enzyme complex with carbon monoxide dehydrogenase activity containing corrinoid and nickel from acetate-grown Methanosarcina thermophila. J Bacteriol 168:1053–1058. doi: 10.1128/jb.168.3.1053-1058.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hedderich R, Berkessel A, Thauer RK. 1989. Catalytic properties of the heterodisulfide reductase involved in the final step of methanogenesis. FEBS Lett 255:67–71. doi: 10.1016/0014-5793(89)81062-2. [DOI] [Google Scholar]
- 57.Hong JS, Rabinowitz JC. 1970. Molar extinction coefficient and iron and sulfide content of clostridial ferredoxin. J Biol Chem 245:4982–4987. [PubMed] [Google Scholar]
- 58.Eskelinen S, Haikonen M, Räisänen S. 1983. Ferene-S as the chromogen for serum iron determinations. Scand J Clin Lab Invest 43:453–455. doi: 10.3109/00365518309168286. [DOI] [PubMed] [Google Scholar]
- 59.Siegel LM. 1965. A direct microdetermination for sulfide. Anal Biochem 11:126–132. doi: 10.1016/0003-2697(65)90051-5. [DOI] [PubMed] [Google Scholar]
- 60.Kozioł J. 1971. Fluorometric analyses of riboflavin and its coenzymes. Methods Enzymol 18:253–285. [Google Scholar]
- 61.Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 62.Stojanowic A, Mander GJ, Duin EC, Hedderich R. 2003. Physiological role of the F420-non-reducing hydrogenase (Mvh) from Methanothermobacter marburgensis. Arch Microbiol 180:194–203. doi: 10.1007/s00203-003-0577-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignments of HdrA2 from M. acetivorans with homologs. The MvhD domain sequences are shown in boldface type. Cysteine residues conserved in HdrA (CAA57039) and MvhD (AAB02349.1) from Methanothermobacter thermautotrophicus are marked with an asterisk, whereas others are marked with a pound symbol. Alignments were made with the COBALT multiple alignment tool. The sequences belong to the following species (the corresponding percent coverage and identity to M. acetivorans HdrA2 are shown before and after the slash, respectively, in parentheses): WP_011022820.1, Methanosarcina acetivorans (100/100); WP_048037482.1, Methanosarcina mazei (100/95); WP_011307542.1, Methanosarcina barkeri (98/94); WP_023843920.1, Methanolobus tindarius (98/67); WP_015324480.1, Methanomethylovorans hollandica (98/67); WP_048194718.1, Methanococcoides methylutens (98/66); WP_013897960.1, Methanosalsum zhilinae (98/65); WP_013194860.1, Methanohalobium evestigatum (98/65); WP_013037418.1, Methanohalophilus mahii (98/65); WP_048089472.1, “Candidatus Methanoperedens nitroreducens” (98/61); WP_014586170.1, Methanosaeta harundinacea (98/61); WP_042684393.1, Methermicoccus shengliensis (98/55); WP_012964674.1, Ferroglobus placidus (98/53); WP_010878733.1, Archaeoglobus fulgidus (97/55); KPL16356.1, Bacteroides sp. strain SM23_62 (97/48); KPV63986.1, “Candidatus Bathyarchaeota archaeon” BA1 (96/49); WP_051309217.1, Desulfobulbus japonicas (96/47); WP_041286569.1, Desulfomonile tiedjei (97/45); KPJ88130.1, Spirochaetes bacterium DG_61 (97/46). Download FIG S1, DOCX file, 0.02 MB (22.4KB, docx) .
Copyright © 2017 Yan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
SDS-PAGE of purified recombinant proteins produced in Escherichia coli and CODH/ACS purified from Methanosarcina acetivorans. The Bolt 4 to 12% gels (Invitrogen) were stained with Coomassie brilliant blue. The numbers at the sides of the gels refer to the molecular masses (in kilodaltons) of adjacent standards. Amounts loaded: HdrA2 and HdrB2 (10 μg); HdrB2C2 and CODH/ACS (25 μg). Download FIG S2, DOCX file, 0.2 MB (158.9KB, docx) .
Copyright © 2017 Yan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequence alignments of HdrA2B2C2 from Methanosarcina acetivorans with HdrABC and MvhD of Methanothermobacter thermautotrophicus. (A) M.t. HdrA, HdrA from M. thermautotrophicus (CAA57039); M.a. HdrA2, HdrA2 from M. acetivorans (AAM06247.1); M.t. MvhD, MvhD from M. thermautotrophicus (AAB02349.1). The overall sequences of HdrA and MvhD were found to be 50 and 49% identical with the corresponding domains in HdrA2. The sequence comparisons revealed four Fe4S4-binding motifs (CX2CX2CX3CP) common to HdrA of M. thermautotrophicus and the HdrA domain (residues 1 to 649) of HdrA2. A motif (CX2CX25CX24CX4C) predicted to ligate the Fe2S2 cluster in MvhD from M. thermautotrophicus (62) was found to be conserved in the MvhD domain of M. acetivorans HdrA2 (residues 650 to 793). (B) M.t. HdrB, HdrB M. thermautotrophicus (CAA57038.1); M.a. HdrB2, HdrB2 from M. acetivorans (AAM07582.1). The HdrB2 sequence has 55% identity with HdrB and contains two conserved motifs (CX31–39CCX35–36CX2C) of which one is predicted to ligate the active site [Fe4S4]3+ cluster in obligate CO2-reducing methanogens. (C) M.a. HdrC2, HdrC2 from M. acetivorans (AAM07581.1); M.t. HdrC, HdrC from M. thermautotrophicus (AAB86344.1). The sequence of HdrC2 has 43% identity with HdrC. The comparison shows two Fe4S4-binding motifs of HdrC2 conserved in HdrC from M. thermautotrophicus. Alignments were made with COBALT. Download FIG S3, DOCX file, 0.01 MB (15.4KB, docx) .
Copyright © 2017 Yan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fdx-dependent reduction of HdrA2, HdrB2C2, and HdrB2. It was previously shown that Fdx is an electron acceptor of CO-reduced CODH/ACS (25) that was used to evaluate the ability of recombinant Fdx to reduce HdrA2, HdrB2, and HdrB2C2. The atmosphere was 100% CO, and the temperature was 20°C. Reduction of HdrA2 and HdrB2C2 was monitored at 410 nm with a Cary 50 Bio UV-Vis spectrophotometer. (A) HdrA2 reduction. The reaction mixtures (0.5 ml) contained 0.1 mg CODH/ACS, 15 μM HdrA2, and the indicated concentrations of Fdx in 50 mM MOPS buffer (pH 7.0). Reactions were initiated by the addition of HdrA2 after preincubation of the reaction mixture with CO to fully reduce Fdx. Key to curves or traces: a, minus Fdx; b, 2.2 μM Fdx; c, 4.4 μM Fdx (2.3 nmol HdrA2 reduced/min). (B) HdrB2C2 and HdrB2 reduction. The reaction mixtures (0.5 ml) contained 0.1 mg CODH/ACS, 22.8 μM HdrB2C2 or HdrB2, and the indicated concentrations of Fdx in 50 mM MOPS buffer (pH 7.0). Reactions were initiated by the addition of HdrB2C2 or HdrB2 after preincubation of the reaction mixtures with CO to fully reduce Fdx. Key to curves or traces: a, reaction mixture containing either HdrB2C2 or HdrB2 minus Fdx; b, reaction mixture containing HdrB2 and 4.4 μM Fdx (0.3 nmol HdrB2 reduced/min); c, reaction mixture containing HdrB2C2 and 2.2 μM Fdx; d, reaction mixture containing HdrB2C2 and 4.4 μM Fdx (1.5 nmol HdrB2C2 reduced/min). Comparison of traces b and d indicates that HdrC2 is important for maximizing electron transfer from Fdx to the catalytic HdrB2. Download FIG S4, DOCX file, 0.1 MB (55.7KB, docx) .
Copyright © 2017 Yan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fdx:F420 oxidoreductase activity of HdrA2. The atmosphere was 100% CO, and the temperature was 20°C. Reduction of F420 was monitored at 420 nm with a Cary 50 Bio UV-Vis spectrophotometer. The reactions were initiated by the addition of F420 after preincubation with CO to fully reduce Fdx and HdrA2. (A) Fdx- and HdrA2-dependent reduction of coenzyme F420. The reaction mixtures (0.5 ml) contained 0.1 mg CODH/ACS, 2.2 μM Fdx, 15 μM F420, and the indicated concentrations of HdrA2 in 50 mM MOPS buffer (pH 7.0). Key to curves or traces: (a) minus HdrA2; (b) minus Fdx; (c) 0.8 μM HdrA2 (5.1 nmol of F420 reduced/min/mg of HdrA2); (d) 3.2 μM HdrA2 (6.2 nmol of F420 reduced/min/mg of HdrA2). (B) Kinetic parameters for Fd:F420 oxidoreductase activity of HdrA2. The reaction mixtures (0.5 ml) contained 0.1 mg CODH/ACS, 2.2 μM Fdx, 2.1 μM HdrA2, and different amounts of F420 in 50 mM MOPS buffer (pH 7.0). 1 mU indicates that 1 nmol of F420 is reduced per minute. (Inset) Double-reciprocal plot. Download FIG S5, DOCX file, 0.1 MB (62.4KB, docx) .
Copyright © 2017 Yan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used for amplification of genes and construction of plasmids. Download TABLE S1, DOCX file, 0.01 MB (12.3KB, docx) .
Copyright © 2017 Yan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Standard Gibbs free energy values for reactions in the Fe(III)-dependent ANME pathway proposed for Methanosarcina acetivorans. Download TABLE S2, DOCX file, 0.02 MB (16.5KB, docx) .
Copyright © 2017 Yan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gibbs free energy of metabolite formation (ΔfGt) calculated at 25°C, pH 7, and ionic concentration of 0.25 M. Download TABLE S3, DOCX file, 0.03 MB (33.3KB, docx) .
Copyright © 2017 Yan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental Materials and Methods. Download TEXT S1, DOCX file, 0.02 MB (20.6KB, docx) .
Copyright © 2017 Yan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.