Abstract
Production of 1,3-propanediol (1,3-PDO) from glycerol is a promising route toward glycerol biorefinery. However, the yield of 1,3-PDO is limited due to the requirement of NADH regeneration via glycerol oxidation process, which generates large amounts of undesired byproducts. Glutamate fermentation by Corynebacterium glutamicum is an important oxidation process generating excess NADH. In this study, we proposed a novel strategy to couple the process of 1,3-PDO synthesis with glutamate production for cofactor regeneration. With the optimization of 1,3-PDO synthesis route, C. glutamicum can efficiently convert glycerol into 1,3-PDO with the yield of ~ 1.0 mol/mol glycerol. Co-production of 1,3-PDO and glutamate was also achieved which increased the yield of glutamate by 18% as compared to the control. Since 1,3-PDO and glutamate can be easily separated in downstream process, this study provides a potential green route for coupled production of 1,3-PDO and glutamate to enhance the economic viability of biorefinery process.
1,3-Propanediol (1,3-PDO) is an important platform chemical which is used in a wide range of areas, including the textile industry, solvent, food, lubricants, and medicine1,2,3,4. Of particular interest is its use as a monomer for the synthesis of polyethers, polyurethanes and polyesters such as polytrimethylene terephthalate (PTT)5. Biological production of 1,3-PDO has received broad interest in recent years. Dupont has paid large efforts to develop a recombinant Escherichia coli which can directly utilize glucose for 1,3-PDO production with high titer and yield1. This process has been commercialized and considered as a milestone of metabolic engineering. In recent years, the fast development of biofuel industry has generated a large amount of crude glycerol as byproduct6,7,8. The excess of crude glycerol produced in the biofuel industry is leading to a dramatic decrease in glycerol price, making it a waste with a disposal cost for many biodiesel plants9,10. Production of 1,3-PDO from cheap and abundant crude glycerol is becoming economically competitive to glucose-based process, representing a promising route toward glycerol biorefinery11.
Several species of microorganisms, including Klebsiella12, Clostridia13, Enterobacter14, Citrobacter15, and Lactobacilli16, can directly convert glycerol into 1,3-PDO via two metabolic steps: the dehydration of glycerol to 3-hydroxypropionaldehyde (3-HPA) by glycerol dehydratase and the reduction of 3-HPA to 1,3-PDO by 1,3-propanediol dehydrogenase (Fig. 1). Since the production of 1,3-PDO is a reduction process, regeneration of NADH via glycerol oxidation is required. Currently established fermentation process can only reach a maximum yield of 0.5–0.6 mol 1,3-PDO/mol glycerol, with about 40–50% of glycerol converted to undesired by-products such as formate, acetate, lactate, and 2,3-butanediol4,5,11. The generation of large amounts of byproducts not only reduce the atom economy but also significantly increase the complexity of downstream process. It is estimated that the downstream process makes more than 50% of the total costs in 1,3-PDO production17. To reduce byproduct formation and increase process economic viability, lignocellulosic hydrolysates were recently used as co-substrates for glycerol fermentation, resulting in an 18–28% increase of 1,3-PDO yield18. Co-production of 1,3-PDO with 3-hydroxypropionic acid (3-HP), an oxidation product of glycerol, for NADH recycling has also been achieved in K. pneumoniae with an overall yield of 0.66 mol 1,3-PDO plus 3-HP per mol glycerol19.
Figure 1. Co-production of 1,3-PDO with glutamate by Corynebacterium glutamicum.
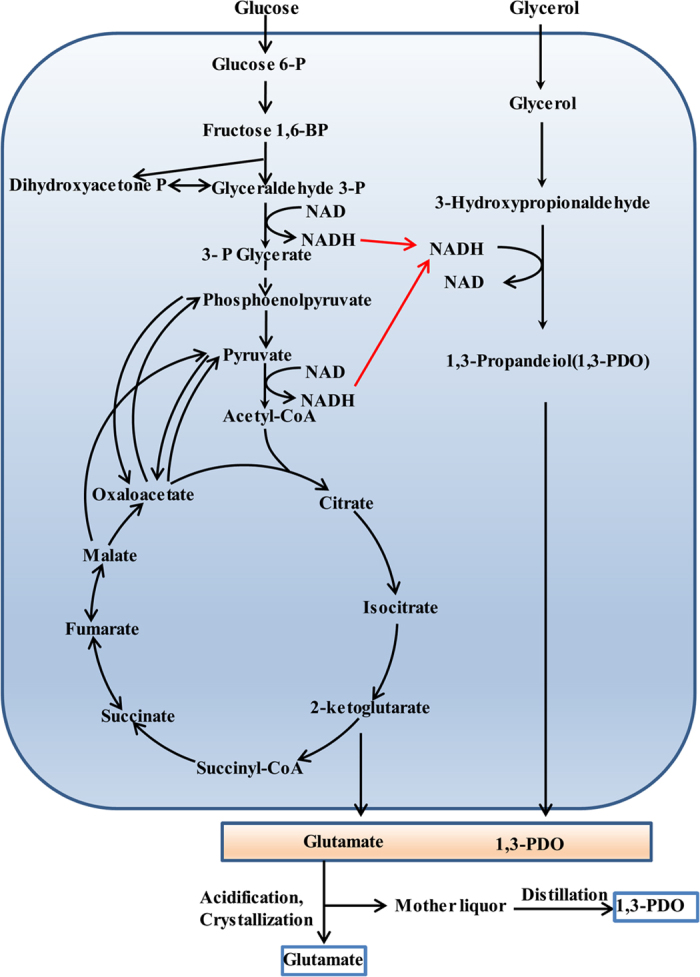
The reducing equivalents generated during glutamate fermentation can be recycled for 1,3-PDO production. 1,3-PDO and glutamate can be easily separated via classical separation processes such as crystallization and distillation.
Corynebacterium glutamicum is a gram-positive bacterium which has been widely used for amino acids production in industry20,21,22,23,24. Recently, applications of this microorganism have been extended for the production of other chemicals, such as isobutanol25, cadaverine26, and ethylene glycol27. Glutamate fermentation by C. glutamicum is a well-established industrial process which produces about 2.5 million tons of glutamate per year28. During glutamate fermentation, about 3 mol NADH/mol glutamate is produced, which need to be oxidized via oxidative phosphorylation28,29. The oxidation of excess NADH via oxidative phosphorylation may reduce the yield of glutamate. It was previously reported that disruption of oxidative phosphorylation via the deletion of H+-ATPase significantly enhanced glutamate production30.
Based on these observation, herein, we propose a novel process to couple the production of 1,3-PDO with glutamate fermentation in C. glutamicum for efficient cofactor regeneration (Fig. 1). With the introduction of 1,3-PDO synthesis pathway in C. glutamicum, the NADH generated during glutamate fermentation could be recycled for 1,3-PDO production. Theoretically, 1,3-PDO and glutamate can be coproduced with the maximum yield of 1.0 mol PDO/mol glycerol and 1.0 mol glutamate/mol glucose. The produced glutamate and 1,3-PDO can be easily purified via classical separation processes such as crystallization and distillation (Fig. 1). Thus, this process may be integrated into current glutamate production line, increasing the economic viability of glutamate industry. In this study, we first introduced the heterologous 1,3-PDO synthesis pathway in C. glutamicum. The production efficiency of 1,3-PDO was further improved via pathway and culture optimization. We showed that co-production of glutamate and 1,3-PDO in one fermentation was possible and the yield of glutamate could be increased in the coupled process. This is also the first report of 1,3-PDO production by industrial important strain C. glutamicum.
Results
Construction of 1,3-propanediol synthesis pathway in C. glutamicum
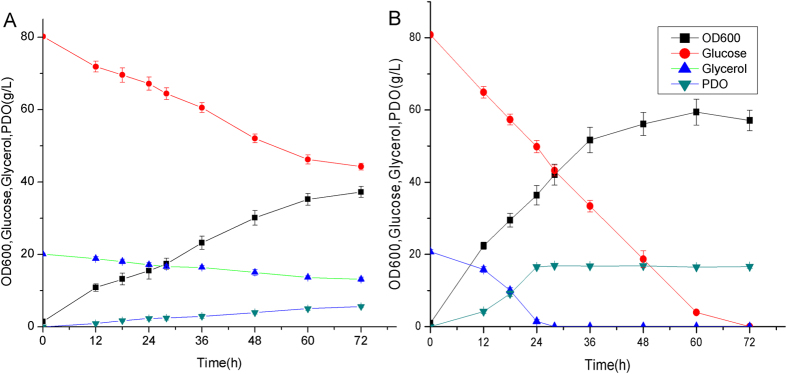
There is no glycerol assimilation pathway in C. glutamicum. Although C. glutamicum possesses a potential glycerol kinase (encoded by glpK, cgp_3198)10, it cannot utilize glycerol as sole carbon source. To efficiently produce 1,3-PDO, we first started by heterologous expression of 1,3-PDO synthesis pathway in C. glutamicum MB001. The pduCDEGH gene from K. penumoniae, encoding diol dehydratase and its activator, was co-expressed with dhaT gene encoding 1,3-PDO dehydrogenase in plasmid pEC-K18mob2 under the control of constitutive lac promoter. The recombinant strain PT01 still cannot utilize glycerol as carbon source, indicating that there is no active glycerol oxidation pathway in C. glutamicum. When cultured in LPG2 medium using glucose and glycerol as co-substrates, strain PT01 can grow and simultaneously utilize glucose and glycerol (Fig. 2). 16.2 g/L of 1,3-PDO was produced from 20.3 g/L of glycerol, with the yield of ~1.0 mol PDO/mol glycerol, suggesting that almost all of glycerol is converted into 1,3-PDO. This was consistent with the reported that C. glutamicum cannot oxidize glycerol due to the extremely low activity of glycerol kinase and glycerol 3-phosphate dehydrogenase10. Therefore, glucose catabolism is utilized for cell growth and regeneration of NADH for 1,3-PDO production. Glucose is mainly converted into CO2 via TCA cycle under aerobic condition in LPG2 medium. Besides CO2, accumulation of acetate (~5.7 g/L) were also observed during the fermentation (Fig. 2).
Figure 2. Cell growth, substrates consumption and products formation of Corynebacterium glutamicum PT01 under aerobic condition.
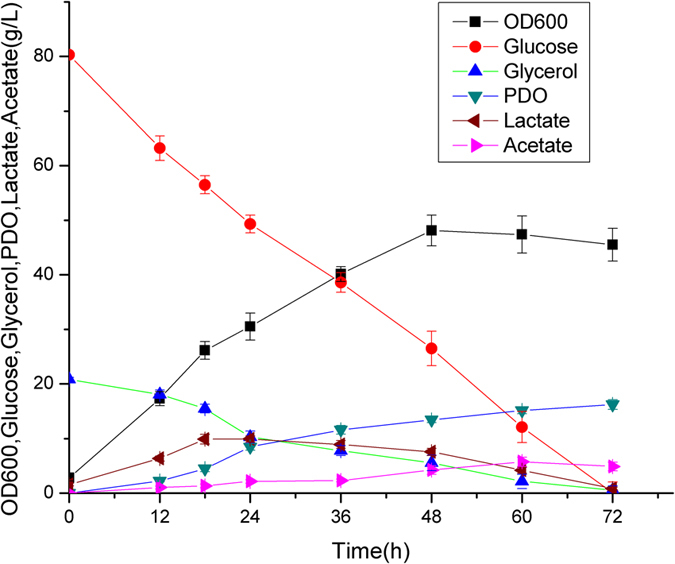
The fermentation was carried out in aerobic condition (500 ml baffled flasks containing 50 ml modified LPG2 medium at 30 °C and 200 rpm).
Pathway optimization to improve strain performance
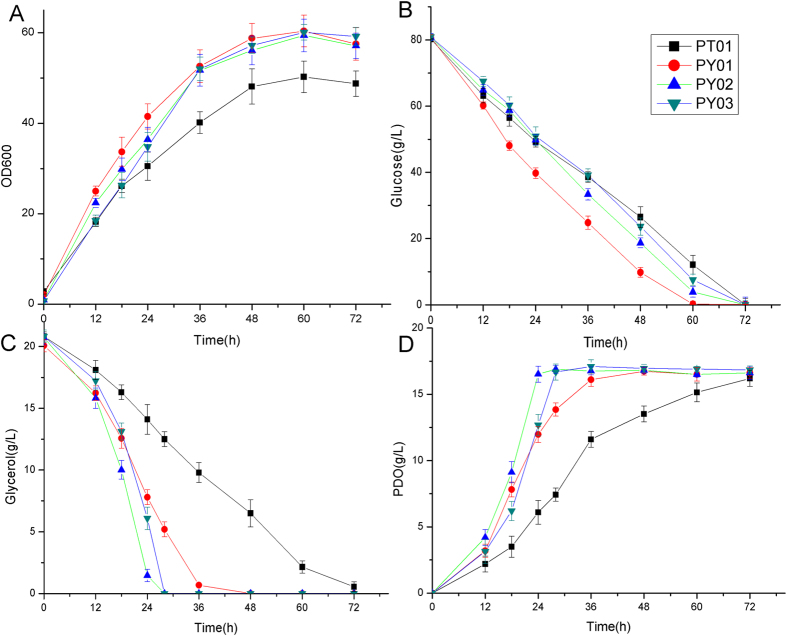
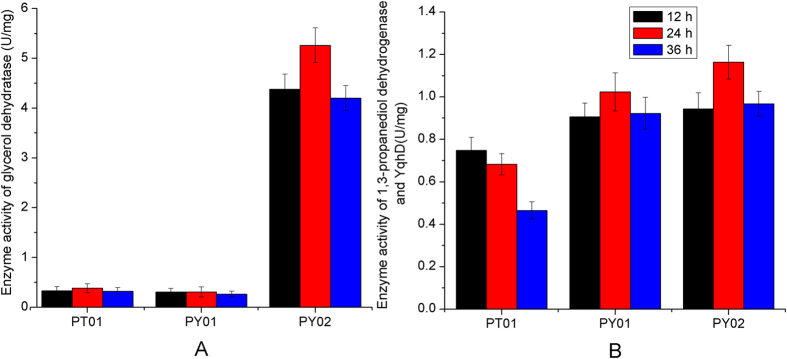
Although strain PT01 can produce 1,3-PDO with high yield, the glycerol consumption rate (0.28 g/L/h) and 1,3-PDO production rate (0.23 g/L/h) were relatively low. To improve the production rate of 1,3-PDO, we attempted to further optimize 1,3-PDO synthesis pathway. 1,3-PDO dehydrogenase is the terminal enzyme of 1,3-PDO synthesis pathway which catalyzes the reduction of 3-HPA. There are two enzymes which have been reported to be able to catalyze this reaction: the NADH-dependent alcohol dehydrogenase encoded by dhaT gene and NADPH-dependent alcohol dehydrogenase encoded by yqhD gene1. We substituted the dhaT gene in the plasmid of pEC-dhaT-pdu by the yqhD gene, constructing the recombinant plasmid of pEC-yqhD-pdu (Supplementary Table 2). The resulting strain PY01 showed significantly improved glycerol consumption rate (0.54 g/L/h vs 0.28 g/L/h) and 1,3-PDO production rate (0.45 g/L/h vs 0.23 g/L/h) (Fig. 3). The yield of 1,3-PDO was not affected (~1.0 mol PDO/mol glycerol). Interestingly, the cell growth and glucose consumption rate were also improved. For the aerobic production of 1,3-PDO in E. coli, YqhD was shown to be more effective than DhaT1. Under aerobic condition, YqhD can efficiently utilize NADPH as cofactor to catalyze the reduction of 3-HPA. The activity of YqhD for reductive reaction is about 50 times higher than oxidative reaction31,32,33. Contrarily, DhaT utilizes NADH as cofactor and has a high activity for oxidative reaction (~25% of activity for 3-HPA reduction)34, which may result in lower efficiency for 1,3-PDO production. Enzyme assay showed that the activity of YqhD in PY01 was significantly higher than DhaT in PT01 especially in the later phase of fermentation (Fig. 4), suggesting that YqhD is also more effective for 1,3-PDO production in C. glutamicum in aerobic condition.
Figure 3.
Pathway optimization for improved 1,3-PDO production: (A) cell growth; (B) glucose consumption; (C) glycerol consumption; (D) 1,3-PDO production. The fermentations were carried out in aerobic condition (500 ml baffled flasks containing 50 ml modified LPG2 medium at 30 °C and 200 rpm).
Figure 4.
Enzyme activities of glycerol dehydratase (A) and 1,3-propanediol dehydrogenases (B) of strain PT01, PY01, and pY02. The culture was carried out in aerobic condition (500 ml baffled flasks containing 50 ml modified LPG2 medium at 30 °C and 200 rpm).
Since glycerol dehydratase controls the entry of glycerol into 1,3-PDO synthesis pathway, we attempted to enhance the activity of glycerol dehydratase by inserting a strong promoter H3635 in front of pduCDEGH (plasmid pEC-yqhD-H36-pdu, Supplementary Table 2). The activity of glycerol dehydratase of the resulting strain PY02 was increased by ~13 fold as compared to strain PY01 (Fig. 4). The activities of 1,3-PDO dehydrogenase of the two strains were comparable (Fig. 4). The glycerol consumption rate and 1,3-PDO production rate were also improved by 50.3% and 48.2%, indicating that glycerol dehydratase is one of the limiting factors for 1,3-PDO production (Fig. 3). Interestingly, the consumption of glucose during 1,3-PDO synthesis (0–36 h) was reduced by 25.4%, suggesting that the oxidation of glucose is more effectively coupled with 1,3-PDO production than strain PY01 since more NADH generated from glucose oxidation was utilized for 1,3-PDO synthesis.
The glycerol facilitator encoded by glpF gene of E. coli is a transporter of glycerol which has been previously implemented in C. glutamicum to improve the assimilation of glycerol10. To examine its effect for 1,3-PDO production, the glpF gene was inserted into plasmid pEC-yqhD-H36-pdu, giving plasmid pEC-yqhD-glpF-H36-pdu (Supplementary Table 2). The resulting strain PY03 showed similar fermentation profiles as strain PY02 (Fig. 3), suggesting that glycerol transport is not a limiting factor under the tested condition. Glycerol transported by unspecific transporters or passive diffusion was enough under the tested condition.
Effect of culture conditions for 1,3-propanediol production
After pathway optimization, we attempted to examine the effect of culture conditions for 1,3-PDO production. Since strain PY02 showed the highest glycerol consumption rate and 1,3-PDO production rate, it was selected for further optimization. Since the activity of glycerol dehydratase is affected by oxygen36, we first examined the effect of aeration for 1,3-PDO production. Since C. glutamicum cannot grow in anaerobic condition, two conditions were examined: aerobic condition and micro-aerobic condition. For aerobic condition, the cultivation was performed in 500 ml baffled flasks containing 50 ml modified LPG2 medium (20 g/L of glycerol) at 200 rpm. For micro-aerobic condition, the cultivation was performed in 500 ml baffled flasks containing 100 ml modified LPG2 medium (20 g/L of glycerol) at 100 rpm. As shown in Fig. 5, the glycerol consumption and 1,3-PDO production were significantly reduced under micro-aerobic condition, indicating that aerobic condition is beneficial for 1,3-PDO production in C. glutamicum. The specific activities of glycerol dehydratase under micro-aerobic condition and aerobic condition were comparable (5.34 ± 0.17 U/mg vs 4.23 ± 0.15 U/mg). However, since the cell grew much faster under aerobic condition, the higher biomass may contribute to the higher glycerol consumption rate.
Figure 5. Effect of aeration for 1,3-PDO production by Corynebacterium glutamicum PY02.
(A) Fermentation profile under micro-aerobic condition. The micro-aerobic culture was carried out in 500 ml baffled flasks containing 100 ml modified LPG2 medium at 30 °C and 100 rpm. (B) Fermentation profile under aerobic condition. The aerobic culture was carried out in 500 ml baffled flasks containing 50 ml modified LPG2 medium at 30 °C and 200 rpm.
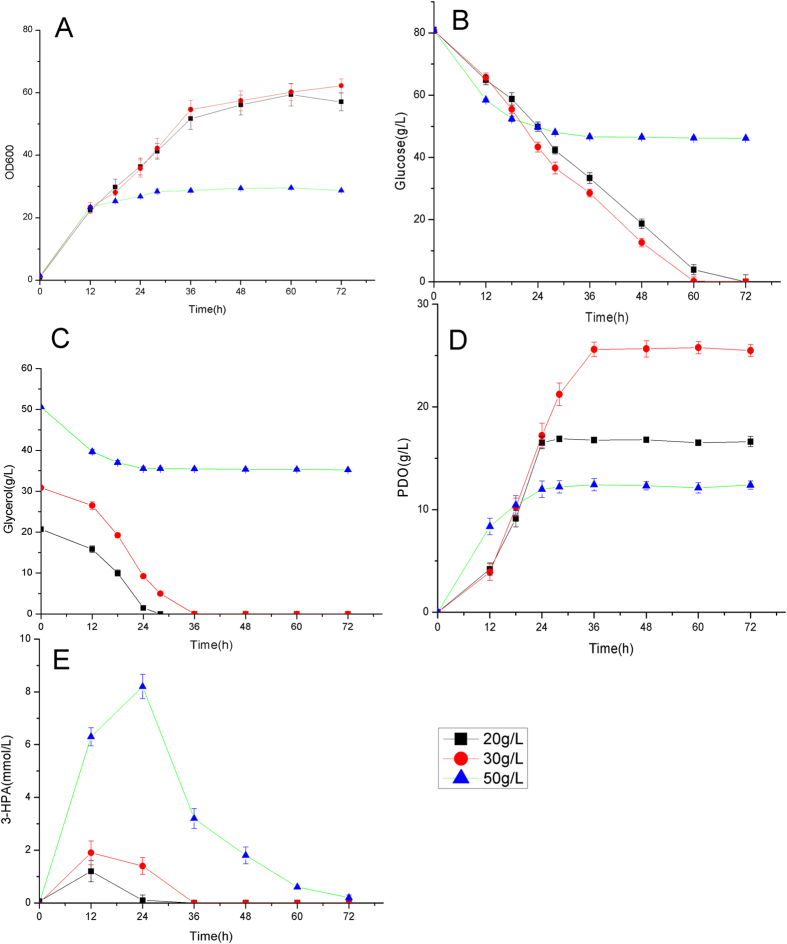
Initial concentration of glycerol was reported to be an important factor affecting the production of 1,3-PDO by K. pneumoniae3,37,38,39,40,41. We examined three different initial concentrations of glycerol (20 g/L, 30 g/L and 50 g/L) with the same glucose concentration (80 g/L) for the culture of C. glutamicum PY02. With the initial concentration of 20 g/L or 30 g/L, glycerol can be completely consumed and the yield of 1,3-PDO is close to 1.0 mol/mol glycerol (Fig. 6). With the initial concentration of 50 g/L, glycerol cannot be completely consumed and cell growth stopped after 24 h. A high accumulation of 3-HPA (8.2 mmol/L) was observed at 24 h (Fig. 6). High concentration of 3-HPA was reported to be very toxic to cell, resulting in the abnormal cessation of fermentation3,37,38,39. Thus, it is important to keep the low initial concentration of glycerol.
Figure 6. Effect of initial glycerol concentration for 1,3-PDO production by Corynebaterium glutamicum PY03.
(A) cell growth; (B) glucose consumption; (C) glycerol consumption; (D) 1,3-PDO formation; (E) 3-hydroxypropionaldehyde (3-HPA) accumulation. The aerobic culture was carried out in 500 ml baffled flasks containing 50 ml modified LPG2 medium at 30 °C and 200 rpm.
Cofactor coupling for simultaneous production of 1,3-propanediol and glutamate
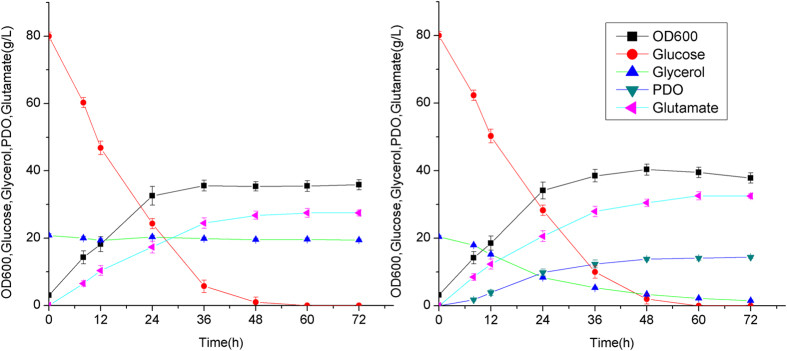
In the previous sessions, 1,3-PDO production was not coupled with glutamate production. Glucose was only used for cell growth and generation of reducing equivalent. Glucose was mainly catabolized to CO2 and other byproducts such as acetate. To couple the production of 1,3-PDO with glutamate, the pEC-yqhD-H36-pdu plasmid was transformed into C. glutamicum OD01, generating strain PY04. C. glutamicum OD01 is a modified MB001 strain with the deletion of odhA gene, which can accumulate high amount of glutamate without induction29. Strain PY04 and OD01 were both cultured in LPG2 medium with 80 g/L of glucose and 20 g/L of glycerol under aerobic condition. As shown in Fig. 7, strain PY04 can consume both glucose and glycerol for glutamate and 1,3-PDO production. 14.4 g/L of 1,3-PDO was produced with the yield of 0.89 mol 1,3-PDO/mol glycerol. Strain PY04 also produced 32.5 g/L of glutamate with the yield of 0.50 mol/mol glucose. The yield of glutamate by strain PY04 was increased by 18.2% as compared to the control strain OD01, suggesting that 1,3-PDO synthesis is beneficial for glutamate production. The intracellular redox state of engineered strains was investigated during the cultivation. As shown in Fig. 8, the ratio of NADH/NAD in strain PY04 was significantly lower than that of strain OD01, indicating that NADH generated from glutamate synthesis was utilized for 1,3-PDO production. The amount of NADH used for 1,3-PDO formation accounted for ~28.4% of NADH generated during glutamate synthesis.
Figure 7. Co-production of 1,3-PDO and glutamate by C. glutamicum OD01 and PY04.
(A) Fermentation profile of strain OD01; (B) Fermentation profile of strain PY04. The aerobic culture was carried out in 500 ml baffled flasks containing 50 ml modified LPG2 medium at 30 °C and 200 rpm. The pH was adjusted to 7.0 every 4 h by adding 5 M NaOH.
Figure 8. Intracellular concentration of NADH/NAD of strain OD01 and PY04.
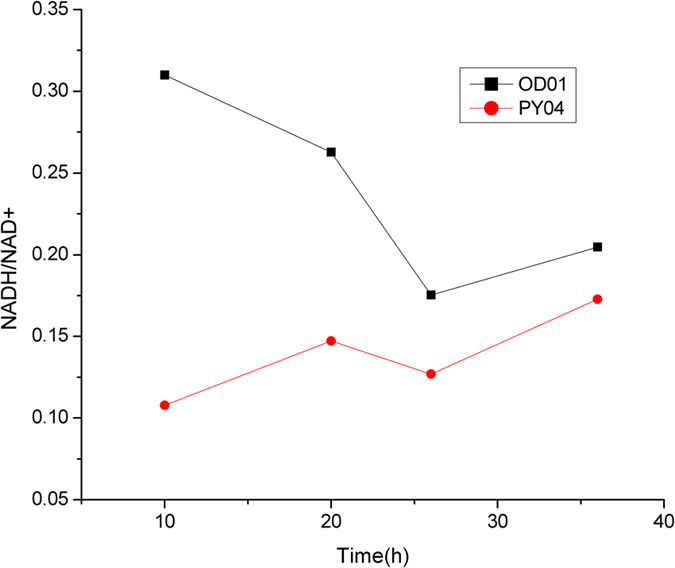
The aerobic culture was carried out in 500 ml baffled flasks containing 50 ml modified LPG2 medium at 30 °C and 200 rpm. The pH was adjusted to 7.0 every 4 h by adding 5 M NaOH.
Discussion
Microbial production of 1,3-PDO from renewable resources has been widely investigated in recent years4,5,11. Due to the reduction of glycerol price9,10, direct conversion of glycerol into 1,3-PDO is becoming an appealing approach for 1,3-PDO production. Among the natural 1,3-PDO producers, K. pneumoniae was mostly studied due to its high productivity12,42. However, the industrial application of K. pneumoniae is limited since it is an opportunistic pathogen. Thus, development of a safe 1,3-PDO producer is highly desirable. E. coli as a model organism has been engineered for 1,3-PDO production from glycerol43. In this study, C. glutamicum, a GRAS (generally regarded as safe) strain, was engineered for the first time to produce 1,3-PDO from glycerol. C. glutamicum is a widely used industrial workhorse which is currently utilized for production of million tons of amino acids including glutamate and lysine20,21,22,23,24. C. glutamicum cannot oxidize glycerol10, thus providing a good opportunity to build a strain with high yield of 1,3-PDO. With the introduction of 1,3-PDO synthesis pathway, the engineered C. glutamicum strains can convert almost all of glycerol into 1,3-PDO (~1.0 mol PDO/mol glycerol). The conversion of glycerol to 1,3-PDO is only catalyzed by two enzymes. However, the balance of enzyme activities between the two enzymes is very important. Increasing the expression of diol dehydratase accelerated glycerol consumption rate and 1,3-PDO formation rate. However, the toxic intermediate 3-HPA was accumulated at high initial glycerol concentration which caused the abnormal cessation of fermentation. 3-HPA accumulation was also observed in Enterobacterial agglomerans and K. pneumoniae3,37,38,39. Imbalance of activities of diol dehydratase and 1,3-PDO dehydrogenase were also observed during 3-HPA accumulation3,37,38,39. Two potential reasons may cause this phenomenon. First, high glycerol concentration could enhance the rate of glycerol dehydration since the Km value of glycerol for diol dehydratase was reported to be relatively high (about 16 mM)44. Second, high ratio of NAD/NADH was observed at high glycerol concentration which may also result in the lower rate of 3-HPA reduction39. Lim et al. reported that application of UTR (untranslated region) engineering to precisely control the expression of glycerol dehydrogenase for optimal metabolic balance could significantly enhance the production of 3-HP45. Similar strategy could also be applied for improving the production of 1,3-PDO.
When glucose was utilized as carbon source for cell growth and NADH generation, about ~50 g/L of glucose was consumed for the conversion of 30 g/L of glycerol (Fig. 5). To increase the process economy, we propose that glucose can be oxidized into glutamate, another important industrial product. The cofactor can be recycled for the co-production of 1,3-PDO and glutamate. It was shown that this coupled process increased the yield of glutamate (Fig. 7). Although the titers of 1,3-PDO and glutamate production were not very high, they may be further improved by combining system metabolic engineering strategies and process optimization. Since glutamate and 1,3-PDO can be easily separated in downstream processes, this study provides a promising alternative for 1,3-PDO and glutamate production with high atom economy. The same strategy may be applied for the production of other oxidized and reduced products.
Methods
Bacterial strains and plasmids
Strains and plasmids used in this study are listed in Table 1. E. coli DH5α was used for routine cloning procedures. C. glutamicum MB001, a prophage-free strain derived from C. glutamicum ATCC 13032, was used as a background strain46. C. glutamicum OD01, which can produce glutamate without induction, is derived from strain MB001 with the deletion of odhA gene encoding a subunit of α-ketoglutarate dehydrogenase29. pEC-K18mob2 is a E. coli/C. glutamicum shuttle vector used for gene overexpression47.
Table 1. Strains and plasmids used for this study.
| Strain or plasmid | Description | Reference |
|---|---|---|
| MB001 | Prohage-free variant of Corynebacterium glutamicum ATCC 13032 | 13 |
| OD01 | MB001 with the deletion of odhA gene, which can produce glutamate without induction | Lab collection |
| PT01 | MB001/pEC-dhaT-pduCDEGH | This study |
| PY01 | MB001/pEC-yqhD-pduCDEGH | This study |
| PY02 | MB001/pEC-yqhD-H36-pduCDEGH | This study |
| PY03 | MB001/pEC-yqhD-glpF-H36-pduCDEGH | This study |
| PY04 | OD01/pEC-yqhD-H36-pduCDEGH | This study |
| Plasmids | ||
| pEC-K18mob2 | E. coli/C. glutamicum shuttle vector, Kmr, ori pGA1 | 15 |
| pEC-dhaT-pduCDEGH | pEC-K18mob2 containing dhaT and pduCDEGH gene from K. pneumoniae | This study |
| pEC-yqhD-pduCDEGH | pEC-K18mob2 containing yqhD gene from E. coli and pduCDEGH gene from K. pneumoniae | This study |
| pEC-yqhD-H36-pduCDEGH | pEC-yqhD-pduCDEGH with the insertion of H36 promoter in front of pduCDEGH | This study |
| pEC-yqhD-glpF-H36-pduCDEGH | pEC-yqhD-H36-pduCDEGH with the insertion of glpF gene between yqhD and H36 | This study |
Plasmids and strains construction
The yqhD gene encoding alcohol dehydrogenase and pduCDEGH gene encoding diol dehydratase and its activator were amplified from the genome of E. coli K12 and K. pneumoniae DSM 2026 using primers 11-F/11-R and 12-F/12-R. The two fragments were inserted into the restriction site of EcoRI/XbaI of pEC-K18mob2 by Gibson Assembly Master Kit (NEB)48, giving recombinant plasmid pEC-yqhD-pdu. To construct plasmid pEC-dhaT-pdu, the dhaT gene encoding 1,3-PDO dehydrogenase was PCR amplified from the genome of K. pneumoniae DSM 2026 using primers 13-F/13-R. The backbone of pEC-yqhD-pdu was also PCR amplified using primers 14-F/14-R. The dhaT fragment and pEC-yqhD-pdu backbone were assembled into plasmid pEC-dhaT-pdu by Gibson Assembly Master Kit (NEB). To insert an artificial constitutive promoter H3635 in front of pduCDEGH, the H36 promoter was amplified from plasmid pEC-H3627 using primers 15-F/15-R. The backbone of pEC-yqhD-pdu was also PCR amplified using primers 16-F/16-R. The two fragments were assembled by Gibson Assembly Master Kit (NEB), giving plasmid pEC-yqhD-H36-pdu. To construct plasmid pEC-glpF-yqhD-H36-pdu, the glpF gene encoding glycerol facilitator was PCR amplified from the genome of K. pneumoniae DSM 2026 using primers 17-F/17-R. The fragment was inserted into the restriction site of EcoRI of pEC-yqhD-H36-pdu by Gibson Assembly Master Kit (NEB).
The recombinant plasmids were transformed into C. glutamicum strains MB001 and OD01 using electroporation as described by van der Rest et al.49. The correct recombinants were verified by colony PCR and sequence analysis. All of the primers used in this study were listed in Supplementary Table 1.
Culture condition
Lysogenic broth (LB) medium was used for the routine culture of C. glutamicum strains. Production of 1,3-PDO by C. glutamicum was performed in 500 ml baffled flasks. For aerobic cultivation, the culture was carried out in 500 ml baffled flasks containing 50 ml modified LPG2 medium at 30 °C and 200 rpm. For micro-aerobic cultivation, the culture was carried out in 500 ml baffled flasks containing 100 ml modified LPG2 medium at 30 °C and 100 rpm. The modified LPG2 medium consists (per liter): 80 g glucose, 10–50 g glycerol, 10 g corn steep liquor, 20 g (NH4)2SO4, 4.5 g urea, 0.5 g MgSO4·7H2O, 0.5 g KH2PO4, 10 mg FeSO4·7H2O, 10 mg MnSO4·H2O, 0.2 mg biotin, 5 mg thiamine-HCl, 50 μM of vitamin B12, and 30 g CaCO3. When appropriate, the medium was supplemented with 25 μg/ml kanamycin. For the coproduction of 1,3-PDO and glutamate, the pH decreased dramatically during the cultivation. Thus, we manually adjusted the pH to 7.0 every 4 h by adding 5 M NaOH. All experiments were repeated for three times.
Enzyme assays
The activity of glycerol dehydratase was assayed by the 3-methyl-2-benzothiazolinone hydrazone (MBTH) method as described by Toraya et al.50. The reaction (in a total volume of 1 mL) contains 0.05 M KCl, 0.2 M 1,2-propanediol, 15 μM coenzyme B12, and 0.035 M potassium phosphate buffer solution (pH 7.0). The assay was started by the addition of cell extract and incubated at 37 °C. After 10 min incubation, the reaction was stopped by adding 1 mL of 0.1 M potassium citrate buffer (pH 3.6). For developing the color, 0.5 mL of 0.1% MBTH solution was added and the mixture was incubated again for 15 min at 37 °C. The color was detected at 305 nm.
The activity of 1,3-PDO dehydrogenase (DhaT and YqhD) was assayed using the reverse reaction34,51. The reaction mixture contained 30 mM ammonium sulfate, 0.1 M 1,3-PDO, 2 mM NAD (or NADP for YqhD), and 0.1 M potassium carbonate buffer solution (pH 9.0). The reaction was started by adding cell extract and the increase of NADH at 340 nm was monitored with a spectrophotometer.
All of the enzyme assays were repeated for three times.
Analytical method
The cell density was determined by monitoring the absorbance at 600 nm using a spectrophotometer. Glucose, glycerol, 1,3-PDO and other organic acids were quantified by using High performance liquid chromatography (HPLC) equipped with an Aminex HPX-87H Column (300 × 7.8 mm) using 0.005 M H2SO4 as the mobile phase with a flow rate of 0.6 mL/min, and detection via refractive index or UV absorption at 210 nm52. Glutamate concentration was determined by a glutamate biosensor (SBA-40C, Shandong Science Academy, Jinan, China)53. The concentration of 3-HPA was measured according to the method described by Krauter et al.54. A mixture of 200 μL sample, 150 μL tryptophan-reagent (10 mM DL-tryptophan, 50 mM HCl) and 600 μL HCl (37%) was mixed, incubated at 37 °C for 20 min and measured at 650 nm. 3-HPA was chemically synthesized from acrolein by the method reported by Hall and Stern55. NADH and NAD concentrations were determined using NAD/NADH assay kit (Sigma, USA) following the manufacturer’s protocol.
Additional Information
How to cite this article: Huang, J. et al. Cofactor recycling for coproduction of 1,3-propanediol and glutamate by metabolically engineered Corynebacterium glutamicum. Sci. Rep. 7, 42246; doi: 10.1038/srep42246 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant no. 21406131), the Science and Technology Planning Project of Guangdong Province (Grant no. 2015A020215001), the Tsinghua University Initiative Scientific Research Program (Grant no. 2015THZ02-1), and the Suzhou-Tsinghua Innovation leading Project.
Footnotes
The authors declare no competing financial interests.
Author Contributions Z.C. conceived and designed the experiments. J.H.H. and Y.W. performed the experiments. J.H.H., W.J.W. and Y.Z. analyzed the data. Z.C. and J.H.H. wrote the paper. All authors reviewed the manuscript.
References
- Nakamura C. E. & Whited G. M. Metabolic engineering for the microbial production of 1,3-propanediol. Curr. Opin. Biotechnol. 14, 454–459 (2003). [DOI] [PubMed] [Google Scholar]
- Chen Z., Geng F. & Zeng A.-P. Protein design and engineering of a de novo pathway for microbial production of 1,3-propanediol from glucose. Biotechnol. J. 10, 284–289 (2015). [DOI] [PubMed] [Google Scholar]
- Chen Z., Liu H. & Liu D. Metabolic pathway analysis of 1,3-propanediol production with a genetically modified Klebsiella pneumoniae by overexpressing an endogenous NADPH-dependent alcohol dehydrogenase. Biochem. Eng. J. 54, 151–157 (2011). [Google Scholar]
- Saxena R. K., Anand P., Saran S. & Isar J. Microbial production of 1,3-propanediol: Recent developments and emerging opportunities. Biotechnol. Adv. 27, 895–913 (2009). [DOI] [PubMed] [Google Scholar]
- Celińska E. Debottlenecking the 1,3-propanediol pathway by metabolic engineering. Biotechnol. Adv. 28, 519–30 (2010). [DOI] [PubMed] [Google Scholar]
- Chen Z., Liu H., Zhang J. & Liu D. Elementary mode analysis for the rational design of efficient succinate conversion from glycerol by Escherichia coli. J. Biomed. Biotechnol. 2010, 518743 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdani S. S. & Gonzalez R. Anaerobic fermentation of glycerol: a path to economic viability for the biofuels industry. Curr. Opin. Biotechnol. 18, 213–9 (2007). [DOI] [PubMed] [Google Scholar]
- Chen Z. & Liu D. Toward glycerol biorefinery: metabolic engineering for the production of biofuels and chemicals from glycerol. Biotechnol. Biofuels 9, 205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Ge X., Cui S. & Li Y. Value-added processing of crude glycerol into chemicals and polymers. Bioresour. Technol. 215, 144–154 (2016). [DOI] [PubMed] [Google Scholar]
- Meiswinkel T. M., Rittmann D., Lindner S. N. & Wendisch V. F. Crude glycerol-based production of amino acids and putrescine by Corynebacterium glutamicum. Bioresour. Technol. 145, 254–258 (2013). [DOI] [PubMed] [Google Scholar]
- Kaur G., Srivastava A. K. & Chand S. Advances in biotechnological production of 1,3-propanediol. Biochem. Eng. J. 64, 106–118 (2012). [Google Scholar]
- Zhang Q. & Xiu Z. Metabolic Pathway analysis of glycerol metabolism in Klebsiella pneumoniae incorporating oxygen regulatory system. Biotechnol. Prog. 25, 103–115 (2009). [DOI] [PubMed] [Google Scholar]
- Ren C., Wen Z., Xu Y., Jiang W. & Gu Y. Clostridia: a flexible microbial platform for the production of alcohols. Curr. Opin. Chem. Biol. 35, 65–72 (2016). [DOI] [PubMed] [Google Scholar]
- Gonzalez R., Murarka A., Dharmadi Y. & Yazdani S. S. A new model for the anaerobic fermentation of glycerol in enteric bacteria: trunk and auxiliary pathways in Escherichia coli. Metab. Eng. 10, 234–45 (2008). [DOI] [PubMed] [Google Scholar]
- Maervoet V. E. T., De Maeseneire S. L., Avci F. G., Beauprez J. & Soetaert W. K. High yield 1,3-propanediol production by rational engineering of the 3 -hydroxypropionaldehyde bottleneck in Citrobacter werkmanii. Microb. Cell Fact. 15, 23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan H., Kandasamy V., Ramakrishnan G. G. & Ramachandran K. B. Glycerol conversion to 1, 3-Propanediol is enhanced by the expression of a heterologous alcohol dehydrogenase gene in Lactobacillus reuteri. AMB Express 1, 37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiu Z. L. & Zeng A. P. Present state and perspective of downstream processing of biologically produced 1,3-propanediol and 2,3-butanediol. Appl. Microbiol. Biotechnol. 78, 917–926 (2008). [DOI] [PubMed] [Google Scholar]
- Xin B. et al. Co-utilization of glycerol and lignocellulosic hydrolysates enhances anaerobic 1,3-propanediol production by Clostridium diolis. Sci. reports 6, 19044 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Li Z., Shimizu K. & Ye Q. Co-production of 3-hydroxypropionic acid and 1,3-propanediol by Klebseilla pneumoniae expressing aldH under microaerobic conditions. Bioresour. Technol. 128, 505–12 (2013). [DOI] [PubMed] [Google Scholar]
- Chen L., Chen Z., Zheng P., Sun J. & Zeng A.-P. Study and reengineering of the binding sites and allosteric regulation of biosynthetic threonine deaminase by isoleucine and valine in Escherichia coli. Appl. Microbiol. Biotechnol. 97, 2939–49 (2013). [DOI] [PubMed] [Google Scholar]
- Geng F., Chen Z., Zheng P., Sun J. & Zeng A.-P. Exploring the allosteric mechanism of dihydrodipicolinate synthase by reverse engineering of the allosteric inhibitor binding sites and its application for lysine production. Appl. Microbiol. Biotechnol. 97, 1963–71 (2013). [DOI] [PubMed] [Google Scholar]
- Peters-Wendisch P. et al. Metabolic engineering of Corynebacterium glutamicum for L-serine production. Appl. Environ. Microbiol. 71, 7139–7144 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stäbler N., Oikawa T., Bott M. & Eggeling L. Corynebacterium glutamicum as a host for synthesis and export of D-Amino Acids. J. Bacteriol. 193, 1702–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Bommareddy R. R., Frank D., Rappert S. & Zeng A.-P. Deregulation of feedback inhibition of phosphoenolpyruvate carboxylase for improved lysine production in Corynebacterium glutamicum. Appl. Environ. Microbiol. 80, 1388–93 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blombach B. et al. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl. Environ. Microbiol. 77, 3300–10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschke N., Schröder H. & Wittmann C. Metabolic engineering of Corynebacterium glutamicum for production of 1,5-diaminopentane from hemicellulose. Biotechnol. J. 6, 306–317 (2011). [DOI] [PubMed] [Google Scholar]
- Chen Z., Huang J., Wu Y. & Liu D. Metabolic engineering of Corynebacterium glutamicum for the de novo production of ethylene glycol from glucose. Metab. Eng. 33, 12–18 (2016). [DOI] [PubMed] [Google Scholar]
- Chinen A., Kozlov Y. I., Hara Y., Izui H. & Yasueda H. Innovative metabolic pathway design for efficient l-glutamate production by suppressing CO2 emission. J. Biosci. Bioeng. 103, 262–269 (2007). [DOI] [PubMed] [Google Scholar]
- Asakura Y. et al. Altered Metabolic Flux due to Deletion of odhA causes L-Glutamate Overproduction in Corynebacterium glutamicum. Appl. Environ. Microbiol. 73, 1308–1319 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki R., Wada M., Takesue N., Tanaka K. & Yokota A. Enhanced glutamic acid production by a H+-ATPase-defective mutant of Corynebacterium glutamicum. Biosci. Biotechnol. Biochem. 69, 1466–1472 (2005). [DOI] [PubMed] [Google Scholar]
- Wang W., Sun J., Hartlep M., Deckwer W.-D. & Zeng A.-P. Combined use of proteomic analysis and enzyme activity assays for metabolic pathway analysis of glycerol fermentation by Klebsiella pneumoniae. Biotechnol. Bioeng. 83, 525–36 (2003). [DOI] [PubMed] [Google Scholar]
- Pérez J. M., Arenas F. a., Pradenas G. a., Sandoval J. M. & Vásquez C. C. Escherichia coli YqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J. Biol. Chem. 283, 7346–53 (2008). [DOI] [PubMed] [Google Scholar]
- Sulzenbacher G. et al. Crystal structure of E.coli alcohol dehydrogenase YqhD: evidence of a covalently modified NADP coenzyme. J. Mol. Biol. 342, 489–502 (2004). [DOI] [PubMed] [Google Scholar]
- Ahrens K., Menzel K., Zeng A.-P. & Deckwer W.-D. Kinetic,dynamic and pathway studies of glycerol metabolism by Klebsiella pneumoniae in anaerobic continuous culture: III. enzymes and fluxes of glycerol dissimilation and 1,3-propanediol formation. Biotechnol. Bioeng. 59, 544–552 (1998). [DOI] [PubMed] [Google Scholar]
- Yim S. S., An S. J., Kang M., Lee J. & Jeong K. J. Isolation of fully synthetic promoters for high-level gene expression in Corynebacterium glutamicum. Biotechnol. Bioeng. 110, 2959–2969 (2013). [DOI] [PubMed] [Google Scholar]
- Chen Z., Liu H., Zhang J. & Liu D. Cell physiology and metabolic flux response of Klebsiella pneumoniae to aerobic conditions. Process Biochem. 44, 862–868 (2009). [Google Scholar]
- Chen Z., Liu H. & Liu D. Regulation of 3-hydroxypropionaldehyde accumulation in Klebsiella pneumoniae by overexpression of dhaT and dhaD genes. Enzyme Microb. Technol. 45, 305–309 (2009). [Google Scholar]
- Barbirato F. et al. 3-Hydroxypropionaldehyde, an inhibitory metabolite of glycerol fermentation to 1, 3-propanediol by Enterobacterial species. Appl. Enviromental Microbiol. 62, 3–7 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbirato F., Soucaille P. & Bories A. Physiologic mechanisms involved in accumulation of 3- hydroxypropionaldehyde during fermentation of glycerol by Enterobacter agglomerans. Appl. Environ. Microbiol. 62, 4405–4409 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. & Zeng A.-P. Protein engineering approaches to chemical biotechnology. Curr. Opin. Biotechnol. 42, 198–205 (2016). [DOI] [PubMed] [Google Scholar]
- Chen Z. & Zeng A.-P. Protein design in systems metabolic engineering for industrial strain development. Biotechnol. J. 8, 523–33 (2013). [DOI] [PubMed] [Google Scholar]
- Menzel K. & Zeng A. Kinetic,dynamic, and pathway studies of glycerol metabolism by Klebsiella pneumoniae in anaerobic continuous culture: I. The phenomena and characterization of oscillation. Biotechnol.Bioeng 52, 549–560 (1996). [DOI] [PubMed] [Google Scholar]
- Tang X., Tan Y., Zhu H., Zhao K. & Shen W. Microbial conversion of glycerol to 1,3-propanediol by an engineered strain of Escherichia coli. Appl. Environ. Microbiol. 75, 1628–1634 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachovchin W. W., Eagar R. G., Moore K. W. & Richards J. H. Mechanism of action of adenosylcobalamin: glycerol and other substrate analogues as substrates and inactivators for propanediol dehydratase--kinetics, stereospecificity, and mechanism. Biochemistry 16, 1082–1092 (1977). [DOI] [PubMed] [Google Scholar]
- Lim H. G., Noh M. H., Jeong J. H., Park S. & Jung G. Y. Optimum rebalancing of the 3-hydroxypropionic acid production pathway from glycerol in Esherichia coli. ACS Synth. Biol., doi: 10.1021/acssynbio.5b00303 (2016). [DOI] [PubMed] [Google Scholar]
- Baumgart M. et al. Construction of a prophage-free variant of Corynebacterium glutamicum ATCC 13032 for use as a platform strain for basic research and industrial biotechnology. Appl. Environ. Microbiol. 79, 6006–15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauch A. et al. Efficient electrotransformation of corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr. Microbiol. 45, 362–7 (2002). [DOI] [PubMed] [Google Scholar]
- Gibson D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–5 (2009). [DOI] [PubMed] [Google Scholar]
- Rest M. van der, Lange C. & Molenaar D. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52, 541–545 (1999). [DOI] [PubMed] [Google Scholar]
- Toraya T., Ushio K., Fukui S. & Studies H. P. on the mechanism of the adenosylcobalamin-dependent diol dehydrase reaction by the use of analogs of the coenzyme. J Biol Chem 252, 963–970 (1977). [PubMed] [Google Scholar]
- Forage R. G. & Foster M. A. Glycerol fermentation in Klebsiella pneumoniae: Functions of the coenzyme B12-dependent glycerol and diol dehydratases. J. Bacteriol. 149, 413–419 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. et al. Improved production of propionic acid in Propionibacterium jensenii via combinational overexpression of glycerol dehydrogenase and malate dehydrogenase from Klebsiella pneumoniae. Appl. Environ. Microbiol. 2256–2264, doi: 10.1128/AEM.03572-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., He Z., Shi Z. & Enock M. Improving the stability of glutamate fermentation by Corynebacterium glutamicum via supplementing sorbitol or glycerol. Bioresour. Bioprocess. 2, 9 (2015). [Google Scholar]
- Krauter H., Willke T. & Vorlop K. Production of high amounts of 3-hydroxypropionaldehyde from glycerol by Lactobacillus reuteri with strongly increased biocatalyst lifetime and productivity. N. Biotechnol. 29, 211–217 (2012). [DOI] [PubMed] [Google Scholar]
- Hall R. H. & Stern E. S. Acid-catalysed hydration of acraldehyde. Kinetics of the reaction and isolation of beta-hydroxypropaldehyde. J. Chem. Soc. 1950, 490–498 (1950). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.