Abstract
The ability to suppress host macrophage apoptosis is essential for M. tuberculosis (Mtb) to replicate intracellularly while protecting it from antibiotic treatment. We recently described that Mtb infection upregulated expression of the host phosphatase PPM1A, which impairs the antibacterial response of macrophages. Here we establish PPM1A as a checkpoint target used by Mtb to suppress macrophage apoptosis. Overproduction of PPM1A suppressed apoptosis of Mtb-infected macrophages by a mechanism that involves inactivation of the c-Jun N-terminal kinase (JNK). Targeted depletion of PPM1A by shRNA or inhibition of PPM1A activity by sanguinarine restored JNK activation, resulting in increased apoptosis of Mtb-infected macrophages. We also demonstrate that activation of JNK by subtoxic concentrations of anisomycin induced selective apoptotic killing of Mtb-infected human macrophages, which was completely blocked in the presence of a specific JNK inhibitor. Finally, selective killing of Mtb-infected macrophages and subsequent bacterial release enabled rifampicin to effectively kill Mtb at concentrations that were insufficient to act against intracellular Mtb, providing proof of principle for the efficacy of a “release and kill” strategy. Taken together, these findings suggest that drug-induced selective apoptosis of Mtb-infected macrophages is achievable.
Apoptosis, the process of programmed cell death, is fundamental for the proper maintenance of many biological processes including embryonic development, cell differentiation, and immune system development and regulation1. In the context of infectious diseases, both pathogen-induced apoptosis of host cells and the ability of pathogens to suppress host cell apoptosis play an important role in determining disease progression.
This is the case in Mycobacterium tuberculosis (Mtb) infections, where the ability of the pathogen to control the timing and mode of host cell death plays a pivotal role for Mtb persistence and replication2,3. It is well established that Mtb infection suppresses host cell apoptosis to replicate inside the phagosome of infected macrophages3,4. On the host cell side, apoptosis of infected macrophages has been shown to facilitate intracellular bacterial killing, priming of cell mediated immunity, and limits unnecessary tissue inflammation2,3,5,6,7. Apoptotic bodies containing Mtb are scavenged by activated macrophages and taken up by dendritic cells to facilitate the priming of antigen specific T cells to stimulate adaptive immunity8,9,10. In contrast, the loss of membrane integrity that defines necrosis is used by Mtb to exit macrophages, to evade the host immune defenses, and to disseminate3,11. The ability to prevent macrophage apoptosis is thus essential for the ability of Mtb to replicate and persist in its human host. In extension, the ability to modulate cell death could have immense therapeutic potential for the treatment of Mtb infections12,13. A clear mechanistic understanding of the host signaling pathways exploited by Mtb to inhibit macrophage apoptosis would allow for the development of targeted therapeutics aimed to restore the ability of macrophages to undergo apoptosis, leading to selective elimination of Mtb-infected macrophages.
The majority of previous studies investigating the regulation of host cell death in response to Mtb infection have focused on mycobacterial proteins, which resulted in the identification of multiple virulence factors (nuoG14, SecA215, pknE16, ndkA17, cpnT11,18) that interfere with macrophage cell death19. However, research from the host cell perspective is lacking despite the knowledge that Mtb infection can regulate apoptosis through both extrinsic and intrinsic pathways by release of cytokines or modulation of the mitochondrial membrane permeability20. Evidence has been accumulating that host eicosanoids play an important role in the regulation of Mtb-mediated macrophage cell death as it was found that Mtb infection induces lipoxin A4 expression, which downregulates the pro-apoptotic and necrosis-protecting prostaglandin E29,10. While these pathways are known to affect cell fate, the upstream signals and molecular regulators that control these processes in the context of Mtb infection remain largely unknown.
We here demonstrate that the Protein Phosphatase, Mg2+/Mn2+-dependent 1A (PPM1A), which we recently identified as a key regulator of the innate antibacterial and antiviral response in macrophages21, is targeted by Mtb to prevent host macrophage apoptosis.
Host serine/threonine phosphatases are known to play important roles for regulation of cellular apoptosis22,23, and this has been extensively described as potential drug targets in the cancer field24,25. However, phosphatases have not received much attention in the context of infectious diseases or more specifically pathogen-mediated host cell apoptosis. Kinome analysis provided us with a basic understanding of the protein-protein interaction network governed by PPM1A and allowed us to identify pharmacologically addressable targets to bring proof of principle that therapeutic restoration of the ability of macrophages to undergo apoptosis in response to Mtb infection can be achieved. Beyond this, we demonstrate that selective killing of Mtb-infected macrophages increased the efficacy of the first-line anti-tuberculosis drug rifampicin, which has been reported to be less effective against intracellular Mtb26,27 due to inefficient penetration into cells28,29. As such, a “release and kill” strategy to deprive the replicative niche of Mtb by inducing Mtb-infected macrophage cell death would be a means to more efficiently expose the bacteria to already existing Mtb drugs, thereby shortening the currently long treatment times.
Results
PPM1A inhibits the macrophage apoptosis pathways
Apoptosis regulation is a critical component of the antibacterial response that has clear implications on pathogen clearance, stimulation of cell mediated immunity, and ultimately disease progression2,3,5,6. Kinome analysis of persistently Mtb-infected THP-1 macrophages that had guided our previous research on the effect of PPM1A expression on the innate antibacterial response of macrophages21, also suggested a possible link of Mtb-induced upregulation of PPM1A with the downregulation of a number of proteins with functions in the apoptosis pathways21. Indeed, there is precedence that phosphatases play important roles in the regulation of cell death22,23,30.
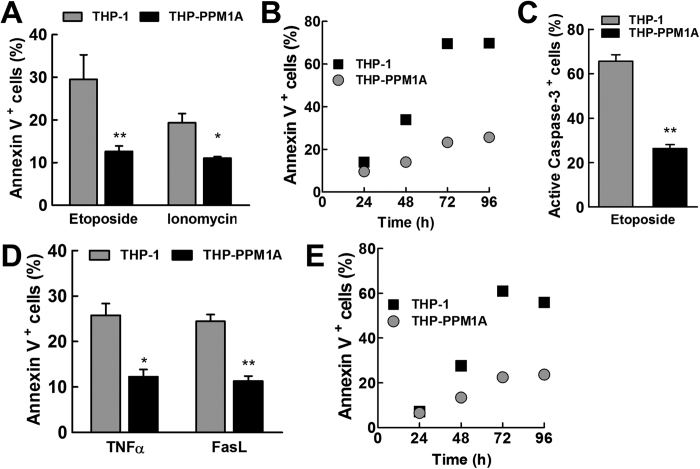
Cellular apoptosis can be induced by intrinsic (cell stress events leading to collapse of mitochondria membrane potential) and extrinsic (death ligand mediated) signals, leading to the eventual cleavage and activation of caspase 3, the executioner caspase31. As THP-1 cells are the most commonly used human monocyte/macrophage model, including frequent use for Mtb infection experiments32,33, we addressed the question whether upregulation of PPM1A, as observed in Mtb infection21, could prevent macrophage apoptosis through either of these apoptosis pathways using genetically manipulated THP-1 cells. To induce the intrinsic apoptotic death pathway, we used etoposide, a topoisomerase II inhibitor34, and ionomycin, a calcium ionophore35. A single addition of etoposide at 0.3 μM induced 30% apoptosis in THP-1 cells after 48 h, but only 13% in THP-1 cells overexpressing PPM1A (THP-PPM1A), as measured by Annexin V assays (Fig. 1A). Etoposide-induced apoptosis levels increased as a function of time. After 96 h, etoposide induced apoptosis in >60% of the THP-1 cells, but THP-PPM1A cells remained protected (<25% apoptotic cells) (Fig. 1B). A single ionomycin treatment at 10 μM for 24 h induced 20% apoptosis in THP-1 cells, but only 11% in THP-PPM1A cells (Fig. 1A). As Annexin V staining is mostly suitable for the detection of early apoptotic events, we also stained for active caspase-3 as an alternative indicator for apoptosis using the Fluorochrome Inhibitor of Caspases (FLICA) method36. When apoptosis levels were measured by FLICA caspase-3 assays, cells stimulated with 1 μM etoposide for 24 h showed that 66% of THP-1 cells stained positive for active caspase-3, while only 26% of THP-PPM1A cells stained positive for active caspase-3 (Fig. 1C), confirming the previous results.
Figure 1. PPM1A overexpression inhibits intrinsic and extrinsic apoptotic pathways.
(A) THP-1 or THP-PPM1A cells were stimulated with 300 nM etoposide or 10 μM ionomycin for 48 h and 24 h, respectively. Then, cells were stained with Annexin V and analyzed by flow cytometry to quantify the amount of apoptotic cells. (B) THP-1 or THP-PPM1A cells were stimulated with 300 nM etoposide for a time course of 24–96 h. Samples were stained every 24 h with Annexin V and analyzed by flow cytometry. (C) The amount of apoptotic cells was determined following treatment with 1 μM etoposide for 24 h by the FLICA caspase-3 assay and flow cytometry. (D) THP-1 or THP-PPM1A cells were stimulated with 100 ng/ml TNFα or 1 μg/ml FasL for 48 h, and thereafter stained with Annexin V and analyzed by flow cytometry. (E) Cells were stimulated with 200 ng/ml TNFα and analyzed as in (B). Data in (A,C,D) represent the means ± S.D. of three independent experiments. *p < 0.05; **p < 0.01 relative to THP-1 control cells.
We found PPM1A to also be involved in the control of the extrinsic apoptosis pathway. TNFα and Fas Ligand (FasL) are two death ligands that induce apoptosis through the extrinsic pathway. A single addition of TNFα (100 ng/ml) or FasL (1 μg/ml) for 48 h induced apoptosis in ~25% of THP-1 cells, but in less than 15% of THP-PPM1A cells (Fig. 1D). Again, in TNFα-treated THP-1 cells, we observed a time-dependent increase in the frequency of apoptotic events to >60%, whereas THP-PPM1A cells were mostly apoptosis-resistant with the percentage of apoptotic cells increasing to only ~20% (Fig. 1E). Thus, PPM1A appears to play an important role in the control of macrophage apoptosis as its expression directly inhibited both intrinsic and extrinsic apoptotic pathways.
PPM1A expression inhibits apoptosis of Mtb-infected macrophages
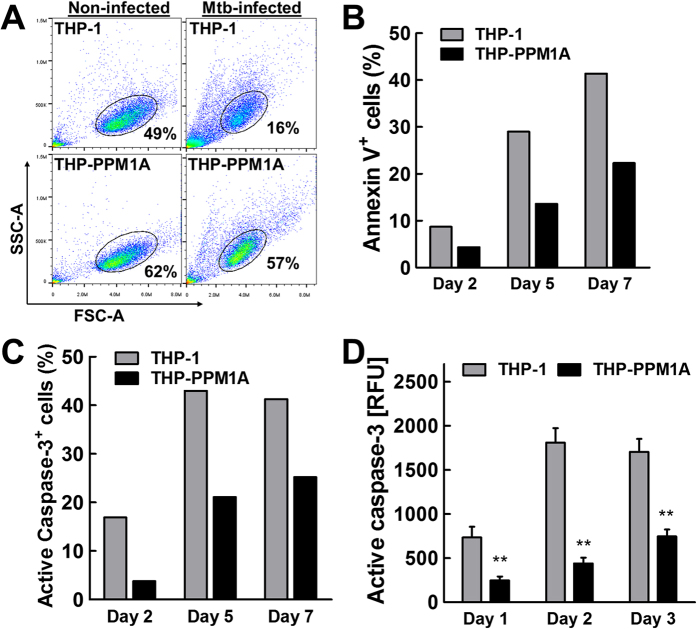
We next examined whether PPM1A plays a role during Mtb infection to control macrophage apoptosis. As it has been previously shown that a higher multiplicity of infection (MOI) induces macrophage apoptosis37, we infected THP-PPM1A cells with Mtb at an MOI of 20. In this scenario, PPM1A overexpressing cells retained higher cell viability relative to parental THP-1 cells (57% vs. 16%), as assessed by flow cytometry on day 2 post infection (Fig. 2A). These results show that increased PPM1A expression levels have a clear impact on the survival of THP-1 cells during Mtb infection.
Figure 2. PPM1A inhibits Mtb-infection induced apoptosis.
(A) THP-1 or PPM1A overexpressing THP-1 (THP-PPM1A) cells were infected with Mtb at an MOI of 20. 48 h post-infection, cells were harvested for flow cytometry analysis using forward (FSC-A) and side scatter (SSC-A) parameters. Elliptical gate shows percentage of viable cells as determined by scattering profile. (B,C) THP-1 or THP-PPM1A cells were infected with Mtb at an MOI of 20 to induce apoptosis. At 2, 5 and 7 days post-infection, cells were harvested and stained with (B) Annexin V or (C) FLICA capsase-3 and analyzed by flow cytometry to measure the amount of apoptotic cells. Results in (B,C) are representative of three independent experiments. (D) THP-1 and THP-PPM1A cells were infected with Mtb as in (B,C) and the Apo-ONE assay was used to quantify relative levels of apoptosis. Relative fluorescence units (RFU) correlate to amount of caspase-3/7 activity in each sample, and represent the means ± S.D. of three individual wells. **p < 0.01 relative to THP-1 control cells.
To detail this finding, we next addressed whether the observed Mtb-induced cell death was indeed caused by apoptosis. THP-1 or THP-PPM1A cells were infected with Mtb at an MOI of 20 and cells were stained for Annexin V at 2, 5, and 7 days post-infection. Over this period of time, apoptosis levels, as determined by Annexin V stains, increased continuously until 40% of THP-1 cells exhibited an apoptotic phenotype on day 7 post-infection (Fig. 2B). In contrast, only ~ 25% of THP-PPM1A cells underwent apoptosis during the same period (Fig. 2B). FLICA caspase-3 staining showed a similar reduction in apoptosis of Mtb-infected THP-PPM1A cells in comparison to parental THP-1 cells at the same time points post infection (Fig. 2C). These data show that increased PPM1A levels suppress apoptosis of Mtb-infected macrophages. While PPM1A has a major impact on macrophage apoptosis, it does not completely abrogate the apoptotic response, which would be consistent with reports for the involvement of other host-derived or bacterial factors that act outside of the PPM1A signaling axis14,38.
Measuring apoptosis over time by single cell staining techniques (FLICA, Annexin V) does not provide information on accumulative effects, as cells that underwent apoptosis at earlier time points disintegrate and are not accounted for at later time points. Since Mtb infection can also induce macrophage necrosis11,18, which could result in false positive Annexin V signals or the loss of active caspase-3 signal, both arising from plasma membrane destruction and eventual cellular breakdown, we wanted to confirm our findings with another method for the detection of apoptosis. We thus utilized the Apo-ONE assay39, which measures caspase-3/7 activity in a homogenous format following lysis of cells without the need to remove culture media that may already contain released caspase-3/7. The results using this method confirmed that PPM1A overexpression indeed inhibited Mtb-induced THP-1 cell apoptosis, as indicated by 4-fold reduction in active caspase-3/7 in the samples from THP-PPM1A cells on day 2 post-infection (Fig. 2D). These data collectively show that elevated PPM1A levels, as observed during persistent Mtb infections21, inhibit macrophage apoptosis.
PPM1A controls macrophage apoptosis through JNK activation
Our results clearly demonstrate that PPM1A controls both the extrinsic and the intrinsic apoptotic pathways of macrophages. However, PPM1A has not been previously linked to macrophage apoptosis, which suggests that PPM1A must control downstream effectors that directly mediate the apoptotic response. Given the diversity of proteins that have been reported to be involved in regulation of apoptosis, we used kinome analysis to identify a possible link between PPM1A and proteins known to be involved in the control of apoptosis pathways. The utilized Kinexus antibody array provide information on changes in the expression or phosphorylation state of 309 protein kinases, 38 protein phosphatases, 37 stress response proteins, 24 transcription factors, and 109 proteins involved in other signaling pathways.
Kinome analysis of THP-PPM1A cells in comparison to THP-1 cells revealed a total of 131 statistically relevant signals, which indicate up- or down-regulated protein expression or phosphorylation events. Of these, 78 (59%; p = 1.06e-35) proteins were associated with the regulation of apoptotic processes as indicated by Gene Ontology (GO) process analysis (Table 1). Using the proteins identified from this analysis as seed nodes, we generated a protein-protein interaction network (PIN) using Metacore and identified several nodes that could potentially function as downstream signaling effectors of PPM1A (Supplemental Fig. 1). This analysis identified several highly linked nodes in the JNK-AP1 pathway (c-Jun N-terminal kinase) (Supplemental Fig. 1). As JNK had been previously reported to be a substrate of PPM1A activity40 and is known to play a central role in the control of the cellular extrinsic and intrinsic apoptosis pathways41, it made for a particularly interesting candidate that could act as a mediator of PPM1A signaling into the apoptotic pathway.
Table 1. Differentially expressed proteins in THP-PPM1A cells involved in the regulation of apoptotic processes.
| Target Protein Name | Phospho Site (Human) | Globally Normalized -THP-1 | Globally Normalized -THP-PPM1A | Z-ratio (THP-PPM1A/THP-1) | Uniprot Link |
|---|---|---|---|---|---|
| JNK1/2/3 | Pan-specific | 8182 | 1720 | −3.80 | P45983 |
| MEK1 (MAP2K1) | Pan-specific | 1911 | 521 | −3.39 | Q02750 |
| ErbB2 (HER2) | Pan-specific | 7069 | 1809 | −3.35 | P04626 |
| KHS (MAP4K5) | Pan-specific | 2960 | 923 | −3.01 | Q9Y4K4 |
| PKR1 | Pan-specific | 2953 | 1097 | −2.59 | P19525 |
| Lck | Pan-specific | 861 | 353 | −2.54 | P06239 |
| PP2A/Ca | Pan-specific | 599 | 252 | −2.53 | P67775 |
| Hsp105 | Pan-specific | 9909 | 3539 | −2.52 | Q92598 |
| Hpk1 (MAP4K1) | Pan-specific | 13234 | 4744 | −2.47 | Q92918 |
| Hsp27 | S82 | 4186 | 1617 | −2.45 | P04792 |
| Mcl1 | Pan-specific | 8514 | 3241 | −2.39 | Q07820 |
| Plk2 | Pan-specific | 5303 | 2089 | −2.37 | Q9NYY3 |
| PP2A B’ (B56) | Pan-specific | 12073 | 4679 | −2.29 | Q15172 |
| LAR | Pan-specific | 1450 | 648 | −2.26 | P10586 |
| Fos | Pan-specific | 15992 | 6482 | −2.15 | P01100 |
| Jun | Pan-specific | 981 | 481 | −2.09 | P05412 |
| COX2 | Pan-specific | 783 | 399 | −2.04 | P35354 |
| JNK1/2/3 | Pan-specific | 731 | 383 | −1.98 | P45983 |
| Hsc70 | Pan-specific | 7101 | 3320 | −1.93 | P11142 |
| PKCm (PKD) | Pan-specific | 2313 | 1167 | −1.91 | Q15139 |
| PKBb (Akt2) | Pan-specific | 4790 | 2369 | −1.85 | P31751 |
| CDK1 (CDC2) | Pan-specific | 6759 | 3391 | −1.77 | P06493 |
| PKBa (Akt1) | Pan-specific | 13995 | 6781 | −1.75 | P31749 |
| HO1 | Pan-specific | 15873 | 7684 | −1.73 | P09601 |
| Hsp27 | S78 | 14306 | 7090 | −1.69 | P04792 |
| eIF2a | S52 | 2743 | 1506 | −1.68 | P05198 |
| Hsp90a/b | Pan-specific | 11594 | 5938 | −1.64 | P07900 |
| Lck | Pan-specific | 6160 | 3315 | −1.61 | P06239 |
| Catenin b1 | Pan-specific | 18725 | 9440 | −1.61 | P35222 |
| IRAK1 | Pan-specific | 8278 | 4572 | −1.51 | P51617 |
| CDK1 (CDC2) | Pan-specific | 7669 | 4303 | −1.48 | P06493 |
| FRS2 | Y348 | 5084 | 2931 | −1.48 | Q8WU20 |
| PDK1 | Pan-specific | 578 | 382 | −1.46 | O15530 |
| PKCh | Pan-specific | 6323 | 3633 | −1.46 | P24723 |
| PKR1 | T446 | 2498 | 1535 | −1.43 | P19525 |
| PKCm (PKD) | S738 + S742 | 765 | 511 | −1.40 | Q15139 |
| Hsc70 | Pan-specific | 23652 | 12908 | −1.39 | P11142 |
| CASP1 | Pan-specific | 6292 | 3775 | −1.35 | P29466 |
| Hsp60 | Pan-specific | 27986 | 15480 | −1.34 | P10809 |
| PKBb (Akt2) | Pan-specific | 5431 | 3325 | −1.33 | P31751 |
| Lyn | Pan-specific | 9390 | 5674 | −1.28 | P07948 |
| CDK1 (CDC2) | Pan-specific | 11030 | 6640 | −1.27 | P06493 |
| Abl | Pan-specific | 1510 | 1048 | −1.21 | P00519 |
| Hsp60 | Pan-specific | 10753 | 6639 | −1.21 | P10809 |
| STAT1a | Pan-specific | 421 | 886 | 1.23 | P42224 |
| FAK | Y576/Y577 | 2946 | 5519 | 1.23 | Q05397 |
| Hsp27 | S15 | 480 | 1020 | 1.27 | P04792 |
| MEK1 (MAP2K1) | T292 | 1028 | 2108 | 1.29 | Q02750 |
| Jun | S73 | 5622 | 10590 | 1.33 | P05412 |
| Hsp90a/b | Pan-specific | 1144 | 2394 | 1.36 | P07900 |
| GSK3b | Pan-specific | 350 | 787 | 1.36 | P49841 |
| IRAK2 | Pan-specific | 3275 | 6557 | 1.40 | O43187 |
| CDK5 | Pan-specific | 674 | 1509 | 1.44 | Q00535 |
| Grp94 | Pan-specific | 407 | 949 | 1.46 | P14625 |
| ErbB2 (HER2) | Pan-specific | 226 | 548 | 1.48 | P04626 |
| IGF1R | Pan-specific | 244 | 592 | 1.48 | P08069 |
| GSK3a | Y279 | 624 | 1430 | 1.48 | P49840 |
| HDAC4 | Pan-specific | 719 | 1649 | 1.51 | P56524 |
| GSK3b | Pan-specific | 453 | 1074 | 1.52 | P49841 |
| NR1 | S896 | 2774 | 5896 | 1.52 | Q05586 |
| CAMK2d | Pan-specific | 398 | 951 | 1.52 | Q13557 |
| MEK2 (MAP2K2) | Pan-specific | 412 | 989 | 1.53 | P36507 |
| NFkappaB p65 | S529 | 871 | 2018 | 1.56 | Q04206 |
| PKCd | S645 | 834 | 1957 | 1.58 | Q05655 |
| GSK3a | Pan-specific | 420 | 1044 | 1.62 | P49840 |
| Hsp90a/b | Pan-specific | 591 | 1445 | 1.63 | P07900 |
| TAO3 (JIK) | Pan-specific | 488 | 1228 | 1.67 | Q9H2K8 |
| Jun | S243 | 697 | 1719 | 1.68 | P05412 |
| GroEL | Pan-specific | 4427 | 9914 | 1.71 | P10809 |
| CREB1 | S133 | 1042 | 2583 | 1.74 | P16220 |
| Nek4 | Pan-specific | 277 | 753 | 1.77 | P51957 |
| NFkappaB p65 | S536 | 2603 | 6280 | 1.81 | Q04206 |
| GNB2L1 | Pan-specific | 600 | 1616 | 1.86 | P63244 |
| PKBa (Akt1) | S473 | 1014 | 2770 | 1.97 | P31749 |
| CDK5 | Pan-specific | 1175 | 3245 | 2.02 | Q00535 |
| FAK | S732 | 1081 | 3471 | 2.36 | Q05397 |
| FAK | S843 | 933 | 3371 | 2.62 | Q05397 |
| NFkappaB p65 | S276 | 590 | 2483 | 2.91 | Q04206 |
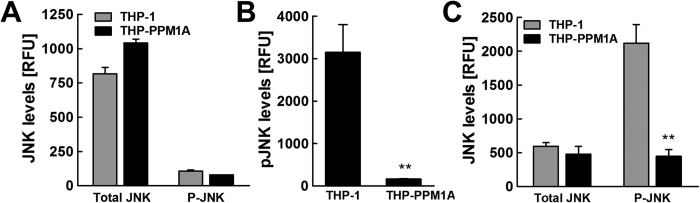
Using a multiplex bead assay with specific antibodies to total or phosphorylated JNK (Thr183/Tyr185), our data show similar baseline protein levels of JNK and no significant activation (phosphorylation) in either resting THP-1 or THP-PPM1A cells (Fig. 3A). Treatment of THP-1 cells with anisomycin, a potent JNK activator42, induced activation (phosphorylation) of JNK, whereas JNK activation was completely abrogated in THP-PPM1A cells (Fig. 3B). Importantly, following Mtb infection of THP-1 and THP-PPM1A cells, while total JNK protein levels remained similar, Mtb infection induced JNK activation (phosphorylation) in THP-1 cells, but not in THP-PPM1A cells (Fig. 3C). As PPM1A has also been reported to inactivate p38 MAPK40, we examined total and phosphorylated p38 levels in resting and Mtb-infected THP-1 macrophages. We found that p38 levels were unchanged at baseline and after Mtb-infection in THP-1 and THP-PPM1A macrophages (Supplemental Fig. 2), suggesting that p38 does not play a role in mediating cell death in our system. These data are consistent with the known role of JNK phosphorylation in pro-apoptotic signaling41, and thus link PPM1A to apoptosis control at the level of JNK phosphorylation.
Figure 3. PPM1A control of apoptosis is mediated through inactivation of JNK.
(A) Resting THP-1 or THP-PPM1A cells were lysed and the amount of total and activated JNK (phosphorylated; T183/Y185) were measured using a 2-plex Milliplex assay kit. RFU indicates relative levels of total or phosphorylated JNK in these cells at baseline. (B) THP-1 or THP-PPM1A cells were stimulated with 1 μM anisomycin for 2 h to induce activation of JNK. Thereafter, cells were lysed and levels of phosphorylated JNK were measured by the Milliplex assay. (C) THP-1 and THP-PPM1A cells were infected with Mtb at an MOI of 20 for 48 h. Then, cells were lysed and the amount of total and activated JNK were measured using the Milliplex assay. Data in this figure represent the means ± S.D. of three independent experiments. **p < 0.01 relative to THP-1 control cells.
Depletion of PPM1A promotes selective apoptosis of Mtb-infected macrophages
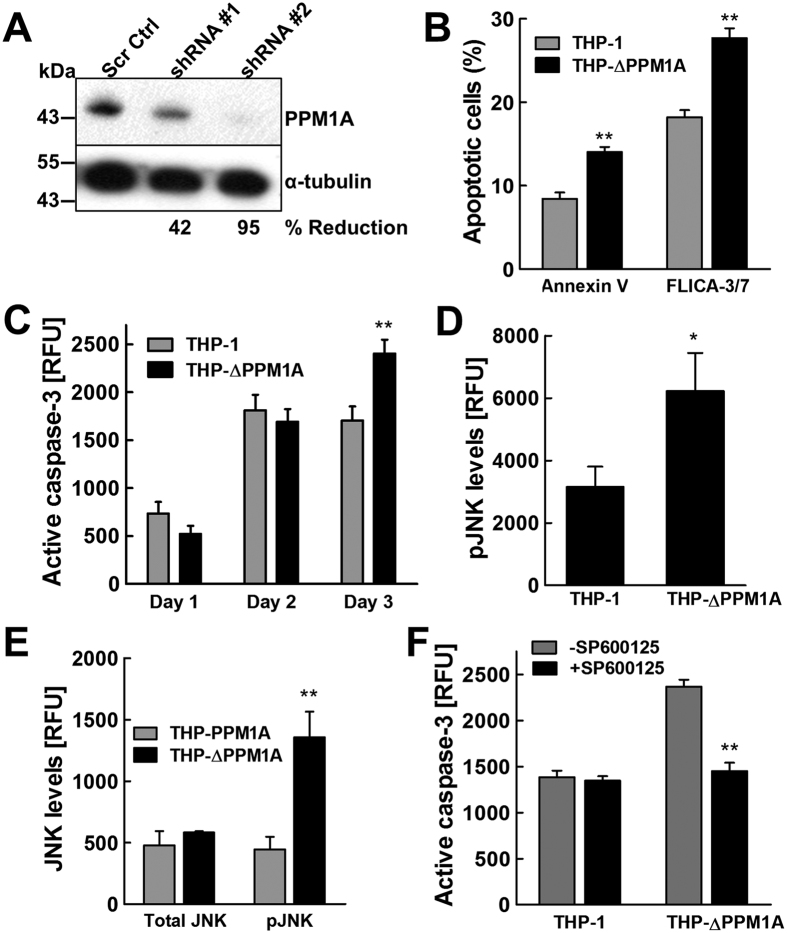
Since Mtb infection induces elevated levels of PPM1A expression, which in turn inhibits macrophage apoptosis in response to pathogen invasion, inhibition of PPM1A signaling may be an attractive therapeutic strategy to selectively kill Mtb-infected macrophages by inducing infected macrophages to undergo apoptosis. To address this possibility, we generated PPM1A knockdown cells. Transduction of THP-1 cells with a PPM1A-specific shRNA vector produced a cell population (THP-ΔPPM1A cells) in which PPM1A protein levels were reduced to ~10% of the protein levels in parental THP-1 cells (Fig. 4A).
Figure 4. Depletion of PPM1A restores the ability of macrophages to undergo Mtb-induced apoptosis.
(A) THP-1 cells were transduced with lentiviral vectors expressing shRNA targeting PPM1A, or scrambled shRNA (Scr Ctrl). Cell lysates were prepared and PPM1A protein levels were analyzed by Western blotting. Densitometry analysis was performed by ImageJ to quantify PPM1A band intensities as normalized to α-tubulin, and % reduction of PPM1A levels are expressed relative to the control THP-1 cells. (B) THP-1 and THP-ΔPPM1A cells were infected with Mtb at an MOI of 5. At 3 days post-infection, cells were stained with Annexin V and FLICA capsase-3 and analyzed by flow cytometry to measure the amount of apoptotic cells. (C) THP-1 and THP-ΔPPM1A cells were infected with Mtb (MOI 5) and the Apo-ONE assay was used to quantify relative levels of apoptosis. Relative fluorescence units (RFU) correlate to amount of caspase-3/7 activity in each sample, and represent the means ± S.D. of three individual wells. (D) THP-1 or THP-ΔPPM1A cells were stimulated with 1 μM anisomycin for 2 h to induce activation of JNK. Thereafter, cells were lysed and levels of phosphorylated JNK were measured by the Milliplex assay. (E) THP-PPM1A and THP-ΔPPM1A cells were infected with Mtb at an MOI of 20 for 48 h. Then, cells were lysed and the amount of total and activated JNK were measured using the Milliplex assay. (F) THP-1 and THP-ΔPPM1A cells were infected with Mtb (MOI of 5) and at the same time mock treated or treated with 20 μM SP600125 to block JNK activation. At 3 days post infection, relative levels of apoptosis were quantified with the Apo-ONE assay, which measures the amount of caspase-3/7 activity. Data in this figure represent the means ± S.D. of three independent experiments. *p < 0.05; **p < 0.01 relative to THP-1, THP-PPM1A control cells, or non-drug treated cells.
If PPM1A inhibitors, as hypothesized, would trigger selective apoptosis of Mtb-infected macrophages, we would expect Mtb infection to trigger increased apoptosis in THP-ΔPPM1A cells compared to parental THP-1 cells. To observe the effect of PPM1A depletion on apoptosis of Mtb-infected macrophages, we used an MOI of 5, a physiologically representative scenario where Mtb infection would naturally suppress host macrophage apoptosis. THP-1 and THP-ΔPPM1A cells were infected with Mtb and relative levels of apoptosis were measured by Annexin V, FLICA caspase-3 staining, and Apo-ONE assay. In line with our predictions, using three different methods, we indeed observed a 67% (Annexin V), 52% (FLICA) or 40% (Apo-ONE) increase in Mtb-infection induced apoptosis in THP-ΔPPM1A cells, relative to the parental THP-1 cells (Fig. 4B,C).
Our data would suggest that the underlying cause for this increased ability of Mtb-infected THP-ΔPPM1A cells to undergo apoptosis would be the restoration of the JNK pathway to respond to the incoming infection. Consistent with our data that PPM1A overexpression would abrogate anisomycin induced JNK activation (Fig. 3B), THP-ΔPPM1A cells stimulated with 1 μM anisomycin showed a 2-fold increase in JNK phosphorylation compared to THP-1 cells (Fig. 4D), adding further evidence that PPM1A expression levels effectively control JNK activation in macrophages. Importantly, different from THP-PPM1A cells, where Mtb infection did not induce JNK activation (Fig. 3C), Mtb infection induced potent JNK activation in THP-ΔPPM1A cells (Fig. 4E).
To conclusively demonstrate that JNK functions in the PPM1A signaling pathway in the context of Mtb infection, we examined whether increased apoptosis associated with the absence of PPM1A (Fig. 4B,C) would actually depend on JNK activation. To this end, we treated Mtb-infected THP-ΔPPM1A macrophages with the specific JNK inhibitor SP600125, which has 300-fold selectivity over p38 and no effect on p38 phosphorylation at the concentration used in our experiement43. Consistent with our data showing increased JNK phosphorylation in Mtb-infected THP-ΔPPM1A macrophages, inhibition of JNK activity by SP600125 abrogated the ability of THP-ΔPPM1A macrophages to undergo apoptosis in response to Mtb infection (Fig. 4F), thereby providing further evidence for the involvement of JNK in the PPM1A-mediated block of macrophage apoptosis.
These data demonstrate that modulation of PPM1A levels controls apoptosis of Mtb-infected macrophages and suggest that PPM1A could be an attractive drug target to eliminate persistently Mtb-infected macrophages.
Inhibition of PPM1A activity increases JNK activation during Mtb infection
Given that genetic manipulations of PPM1A expression showed that it controlled JNK activation as a means to inhibit apoptosis of Mtb-infected macrophages, we next sought to test whether PPM1A phosphatase activity was required to inactivate JNK signaling. This would also show whether pharmacological perturbations targeting PPM1A could restore proper JNK activation during Mtb infection.
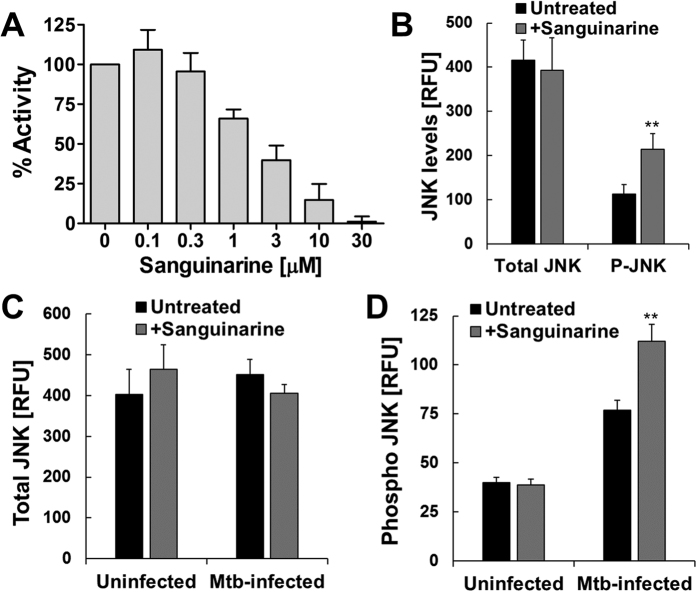
Consistent with a previous report that showed the plant product sanguinarine as a potent and selective inhibitor of PPM1A activity44, we demonstrated that sanguinarine can block PPM1A phosphatase activity (Fig. 5A). Using sanguinarine as a chemical probe, we treated primary human monocyte derived macrophages (hMDMs) for 24 h, and then induced JNK activation with anisomycin. As we would predict, inhibition of PPM1A activity by sanguinarine increased anisomycin induced phosphorylation of JNK by ~2-fold (Fig. 5B). Importantly, sanguinarine treated hMDMs infected with Mtb showed ~45% increase in JNK activation, while levels of total JNK remained similar (Fig. 5C,D). These data show that PPM1A activity is essential for inactivation of JNK during Mtb infection and suggests that targeting the PPM1A-JNK signaling axis could restore the proper apoptotic response in Mtb-infected macrophages.
Figure 5. Inhibition of PPM1A activity induces JNK activation.
(A) Enzymatic activity of PPM1A in the presence of increasing concentrations of sanguinarine was measured by pNPP assay and normalized as 100% in the absence of sanguinarine. (B) Primary human monocyte derived macrophages (hMDMs) were treated with 1 μM sanguinarine for 24 h, and then stimulated for 2 h with 100 nM anisomycin. Thereafter, total and phosphorylated JNK levels were measured using the bead-based Milliplex assay. (C) Total and (D) phosphorylated JNK levels were measured by the Milliplex assay in uninfected and Mtb-infected hMDMs (MOI 5; 24 h) that were mock treated or pre-treated with 1 μM sanguinarine for 24 h prior to infection. Data in this figure represent the means ± S.D of three independent experiments. **p < 0.01 relative to untreated macrophages.
Anisomycin induces selective cell death of Mtb-infected primary human macrophages
While we had shown that PPM1A in THP-1 cells and primary human macrophages were similarly upregulated in response to Mtb-infection, resulting in a severe impairment of the innate immune response21, we would have to verify that this similarity would extend to the ability of PPM1A to control apoptosis of primary macrophages in particular as a function of the identified PPM1A-JNK signaling axis.
Differentiated primary macrophages are highly adherent, which requires aggressive manipulations to de-adhere the cells for downstream single cell analysis. Also, these methods are not conducive to study the kinetic effects of drugs on cell viability. To enable direct kinetic measurements of the effect of chemical research probes on the viability of the Mtb-infected primary macrophages, we used Real Time Cell Analysis (RTCA), a method that is frequently used to kinetically determine viability of adherent cells45,46,47. The RTCA system measures changes in impedance between electrodes at the bottom of E-well plates, which is then translated into a Cell Index (CI) measurement, a dimensionless value. An increase in the CI reflects an increase in macrophage adherence (differentiation), whereas a decrease in the CI reflects the loss of macrophage viability as they detach from the bottom of the well (Supplemental Fig. 3). Suspension cells trigger no measurable signal in this system.
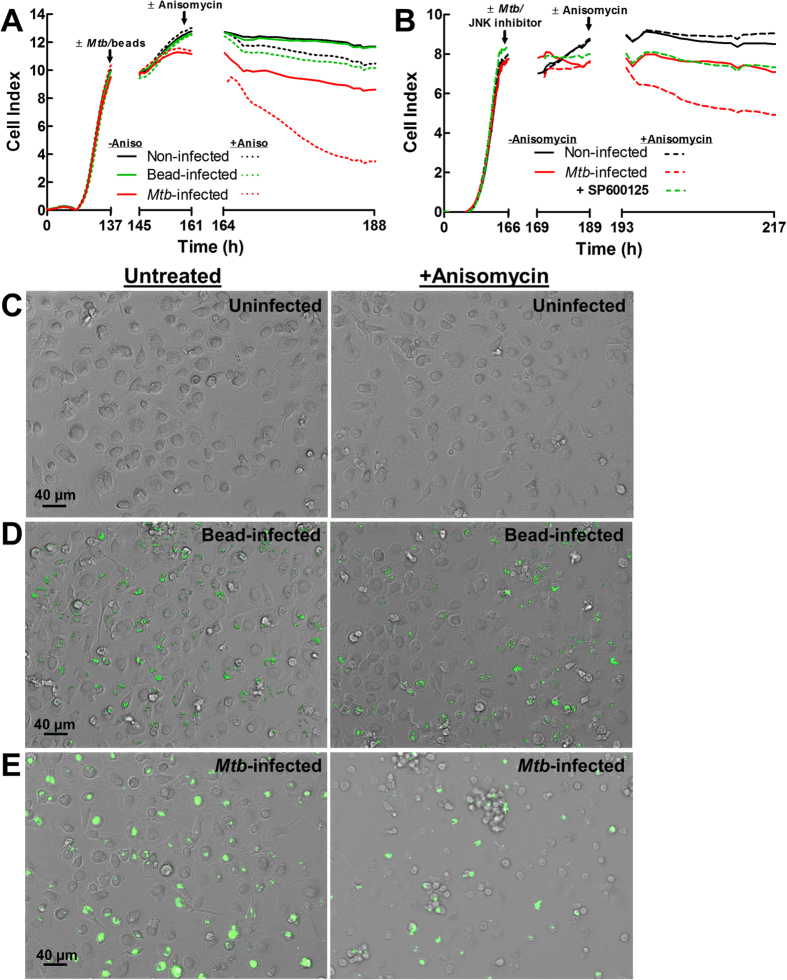
To address the question whether interference with the PPM1A-JNK axis would provide a means to selectively kill Mtb-infected macrophages, we infected hMDM with Mtb, and 24 h post infection, treated the macrophages with or without anisomycin, and then used changes in the CI to kinetically quantify compound effects. Briefly, monocytes were seeded in 96 E-well plates and monitored for GM-CSF induced differentiation. As indicated by the increase in the cell index, primary human monocytes attached to the E-well plate over the first ~140 h (6 days) in response to GM-CSF and differentiated into hMDMs. The hMDMs were left untreated, were exposed to 3 μm inert latex bead particles (phagocytosis control) or infected with Mtb. Wells for each condition were then mock treated or treated with anisomycin (75 nM) and macrophage viability (adherence) was followed kinetically by RTCA for the next 24 h to record changes in the CI.
While the CI remained stable for all control conditions (no anisomycin), and for pseudo-infected MDMs that had phagocytosed the inert latex beads, we observed a selective and rapid decrease in CI in anisomycin treated Mtb-infected hMDMs (Fig. 6A). The selective killing of Mtb-infected macrophages was indicated by the decrease in CI from 11 to 4, while the CI of untreated Mtb-infected macrophages remained above 10 (Fig. 6A). The RTCA experiment demonstrates that Mtb-infected primary macrophages were efficiently and selectively killed by the use of anisomycin as a JNK agonist. To unequivocally show that anisomycin induced selective killing of Mtb-infected macrophages is not mediated through a p38 dependent mechanism and that JNK activation is absolutely required in this process, we used a specific JNK inhibitor, SP60012543 to probe the system. Using RTCA as described above, we show that the presence of SP600125 completely blocked the ability of anisomycin to induce selective apoptotic killing of Mtb-infected human macrophages (Fig. 6B). This result demonstrates that anisomycin induced selective cell death can be reversed by directly inhibiting JNK.
Figure 6. Anisomycin induces selective cell death of Mtb-infected primary human macrophages.
(A) Human monocytes were seeded into E-well plates and GM-CSF (5 ng/ml) differentiation into MDMs was monitored by RTCA to measure cellular adherence as indicated by the Cell Index. Following differentiation (~140 h), MDMs were infected with Mtb (MOI 5) or 3 μm latex bead for 24 h, at which point 75 nM anisomycin was added to corresponding wells (~161 h), and changes in the Cell Index was followed for the next 24 h. (B) hMDMs were differentiated as in (A) and left uninfected or infected with Mtb (MOI 5) in the absence or presence of 20 μM SP600125, a specific JNK inhibitor, for 24 h. Thereafter, 75 nM anisomycin was added and changes in the Cell Index was followed for 24 h. CelI Index measurements in (A,B) were made every 30 min and represent the average of 3 independent wells. (C) Representative bright field images of human MDMs mock treated or treated with 75 nM anisomycin for 24 h. (D,E) Representative merged bright field and GFP channel images of (D) fluorescent 3 μm latex beads or (E) Mtb-GFP (MOI 5) infected MDMs following 24 h treatment with or without 75 nM anisomycin.
The RTCA data were confirmed by fluorescence microscopy analysis in an independent experiment. On day 6 post GM-CSF differentiation, macrophage differentiation was indicated by a strongly adherent, flattened, pancake-like morphology indicative of M1 polarization48, which was not altered by the addition of anisomycin (Fig. 6C). Similarly, no influence of anisomycin on the viability or phenotype of hMDMs loaded with 3 μm inert latex bead particles was observed (Fig. 6D). However, anisomycin treatment of Mtb-infected macrophages resulted in extensive cell death compared to Mtb-infected macrophages in the absence of anisomycin (Fig. 6E). While untreated Mtb-infected macrophages maintained the flattened, pancake-like morphology, anisomycin treated Mtb-infected macrophages exhibited features of cell shrinkage, detachment, and formation of apoptotic bodies with some extracellular bacteria, typical features of apoptotic cells (Fig. 6E).
Taken together, these data suggest that selective killing of Mtb-infected macrophages can be achieved by pharmacological perturbation and that targeting the PPM1A-JNK signaling network is a promising therapeutic strategy to induce selective elimination of persistently Mtb-infected macrophages.
Selective killing of Mtb-infected macrophages enhances antimicrobial chemotherapy to kill Mtb
Apoptosis of infected macrophages stimulates the antibacterial response5 and plays a significant role in promoting adaptive immunity3. Alternatively, given reports that several key TB drugs such as rifampicin26,27, streptomycin, and fluoroquinolones have decreased efficacy against intracellular Mtb likely due to penetration properties29, induction of apoptosis may also improve the ability of existing antimicrobial chemotherapy against Mtb.
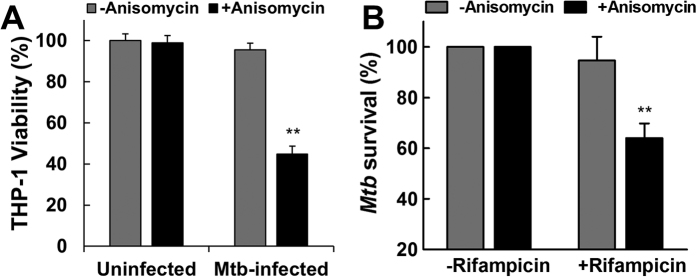
To test this hypothesis, Mtb-infected THP-1 macrophages were treated with anisomycin to induce apoptosis-mediated bacterial release, and subsequently treated with rifampicin at a concentration below the minimal inhibitory concentration (MIC) necessary to kill intracellular Mtb.
Consistent with our data using primary human macrophages, we show that anisomycin treatment induces selective killing of Mtb-infected THP-1 macrophages (Fig. 7A). Subsequently, to measure Mtb viability, we used a well-characterized luciferase reporter system in Mtb49,50, which has been shown to correlate well with the standard colony unit formation plating method51. Our experiments show that while anisomycin or rifampicin at the used concentration had little to no effect on the killing of intracellular Mtb, combination treatment decreased the survival of Mtb by ~40% as measured by luciferase assays (Fig. 7B). This experiment suggests that selective induction of Mtb-infected macrophage apoptosis improves standard antimicrobial chemotherapy and should provide an attractive treatment strategy that has the potential to shorten the currently lengthy treatment period.
Figure 7. Selective killing of Mtb-infected macrophages by anisomycin enhances killing of Mtb by rifampicin.
(A) THP-1 macrophages were uninfected or infected with Mtb (MOI 5) for 24 h, and subsequently mock treated or treated with 100 nM anisomycin for 72 h. THP-1 macrophage viability was then measured using CellTiter-Glo luminescent assay and normalized to uninfected, untreated cells as 100% viability. (B) Mtb-infected THP-1 macrophages (MOI 5) were treated with or without 100 nM anisomycin for 3 days. Then cells were treated with or without 0.1μg/ml rifampicin for 1 day. The amount of viable Mtb in each condition was quantified using the luciferase activity assay. Using relative luminescence units, data were normalized to control conditions (without rifampicin) as 100% Mtb survival. Results in this figure represent the means ± S.D. of three independent experiments. **p < 0.01 relative to untreated macrophages.
To explore the possible effect of such a “release and kill” strategy on TB treatment kinetics, we used a simple mathematical model that assumes a biphasic decline of the bacterial load in patients following onset of chemotherapy (e.g. rifampicin treatment) (Supplemental Fig. 4). Based on a series of data published by other groups52,53,54, we assumed that during phase I (initial kill), therapy would eliminate extracellular Mtb and intracellular Mtb in macrophages located in tissues where sufficiently high drug concentrations can be easily achieved. During this phase, a reduction of the bacterial burden by ~70% would be achieved within a month of standard treatment. Phase II (slow kill) would be characterized by a much slower decline of the residual bacterial burden, which under standard treatment would be completely eliminated after ~6 months (24 weeks) of treatment (Supplemental Fig. 4; black curve). This slow decline would be representative of the effect of drug treatment against intracellular Mtb, possibly in a persistent and therefore more therapy resistant form and in tissue locations that are poorly penetrated by drugs. The model provided us with the ability to predict various theoretical treatment scenarios, in which Mtb-release drugs that trigger selective apoptosis of Mtb-infected macrophages would be added as adjunctive therapy to the ongoing standard Mtb treatment regimen.
In Supplemental Fig. 4, several scenarios are visualized. In a scenario where release drugs are not added to the regimen during phase I or their addition would not increase phase I treatment efficacy, this model would predict that a weekly application schedule of Mtb-release drugs that would result in a 10%, 20%, or 40% reduction in the bacterial burden (based on what can be achieved in Fig. 7B) would lead to a reduction in treatment times from 24 weeks to 15, 12, or 9 weeks, respectively. If Mtb-release drugs were to improve phase I treatment efficacy by 10% or 20%, and subsequently reduce the Mtb burden by even only 10%/week during phase II, treatment times would be reduced to 13 or 9 weeks, respectively. While these are only possible predictions based on simple, but realistic assumptions, these calculations suggest that a release and kill strategy-based adjunctive therapy to the standard TB treatment regimen could lead to a major reduction in treatment time.
Discussion
Mtb is a prototypical intracellular bacterial pathogen that replicates efficiently in the phagosome of macrophages55. We recently demonstrated that to enable this process, Mtb alters the kinase and phosphatase signaling networks within the infected macrophage resulting in an environment conducive to the establishment of a persistent infection. We found that Mtb-induced upregulation of PPM1A controls several aspects of the macrophage innate antibacterial response21.
In the current study, we demonstrate that PPM1A is an apoptosis checkpoint that controls both the intrinsic and extrinsic apoptosis pathways (Fig. 1), a critical innate immune response with major implications for intracellular pathogens. Kinome analysis enabled the identification of the JNK/AP-1 signaling pathway as a downstream effector through which PPM1A exerts its control on the apoptotic pathway in macrophages (Supplemental Fig. 1 and Table 1), which was confirmed by a series of experiments using PPM1A overexpressing and knockdown cells (Figs 3 and 4). These experiments revealed that the ability to control PPM1A expression provided Mtb with a means to hijack macrophages by suppressing host cell apoptosis. Suppression of apoptosis, which is otherwise an innate defense mechanism against intracellular pathogens, provides Mtb with a replicative niche within which it can persist for long periods. Indeed, apoptosis of infected macrophages has been shown to facilitate intracellular bacterial killing, priming of cell mediated immunity, and limits unnecessary tissue inflammation2,3,5,6,7. Suppression of apoptosis is also essential for eventual Mtb dissemination, which is achieved by Mtb-induced host cell necrosis. PPM1A upregulation thus seems to be a key factor for Mtb to establish a persistent infection in macrophages, by hijacking the innate immune response of its host cells, and disabling the natural ability of macrophages to undergo apoptosis in response to pathogen infection.
Our finding that a host phosphatase (PPM1A) controls both the extrinsic and intrinsic apoptotic pathways, and more importantly inhibits apoptosis of Mtb-infected macrophages corroborates with research on other members of the PPM (monomeric, metal-dependent) family of serine/threonine phosphatases56, which have well described functions in the regulation of cell death23,30,57. In addition, it was recently demonstrated that the Mtb phosphatase PtpA could dephosphorylate host Glycogen Synthase Kinase-3 (GSK3α), thereby inhibiting apoptosis of Mtb-infected macrophages38. Consistent with this, kinome analysis from persistently Mtb-infected THP-1 cells21 and that of PPM1A overexpressing cells (Supplemental Fig. 1) both show dysregulation of GSK3. There is also evidence showing that Mtb PtpA exploits host ubiquitin to inactivate JNK and p38, thereby suppressing innate immunity58. Our finding that PPM1A controls the innate immune response21 and apoptosis during Mtb infection may suggest the existence of redundant mechanisms (via host and bacterial phosphatases) used by Mtb to ensure survival of the host macrophage. Such functional redundancy would also explain why knockdown of PPM1A in our experiments induced apoptosis in a large proportion of, but not all Mtb-infected macrophages.
As the PPM1A-JNK signaling axis presented itself as an attractive target to restore the ability of Mtb-infected macrophages to undergo apoptosis, we also address the important question whether this signaling network could be targeted to selectively kill Mtb-infected macrophages. Such a therapeutic intervention would (i) disrupt the intracellular replication cycle of Mtb, (ii) boost the adaptive immune response to Mtb8, and (iii) increase the susceptibility of Mtb to existing drug treatments through induction of macrophage apoptosis given that common anti-tuberculosis drugs (e.g. rifampicin) have reduced efficacy against intracellular bacteria26,27.
Given that we had identified JNK as a key downstream signaling effector of PPM1A, it was logical to target JNK to deliver proof of concept that selective apoptosis induction of Mtb-infected macrophages can be achieved. Indeed, anisomycin, a reported JNK phosphorylation agonist could counteract the PPM1A upregulation enforced suppression of JNK phosphorylation following Mtb-infection, and restored the ability of macrophages to undergo apoptosis in response to Mtb infection (Fig. 6).
Drugs that could selectively induce apoptosis of Mtb-infected macrophages are highly desirable as this would restore a key innate immune response that is actively blocked by Mtb infection, which will have therapeutic benefits through the generation of an improved adaptive immune response and the ability to potentiate vaccines. Beyond this, drug-induced selective killing of Mtb-infected macrophages could likely increase the efficacy of existing anti-Mtb drugs as they gain better access to the bacteria. This approach would be particularly useful as adjunctive TB therapy since the commonly used anti-Mtb drugs rifampicin26,27, streptomycin, and fluoroquinolones having decreased efficacy against intracellular Mtb likely due to penetration properties29. Indeed, in a process we term “release and kill”, our results show that anisomycin induced killing of Mtb-infected macrophages removes the protective niche of the bacteria and increases the efficacy of rifampicin to kill Mtb at concentrations that are normally inactive against intracellular Mtb.
To examine the possible impact of the addition of Mtb-release drugs to standard TB treatment regimens, a simple mathematic model that allowed for some basic predictions suggested that a shortening of the treatment course by 35–60% seemed rather realistic. As clinical anti-tuberculosis drug treatment, particularly in resource poor settings, is primarily hampered by long treatment periods lasting over 6 months59 leading to patient non-adherence, any significant shortening of the treatment period would be highly desirable.
Methods
Cell culture, reagents, and antibodies
THP-1 monocytes (ATCC TIB-202) and primary monocytes were maintained in RPMI 1640 medium supplemented with 2 mM L-glutamine and 10% heat-inactivated fetal bovine serum (FBS) at 37 °C in a humidified atmosphere of 5% CO2. THP-PPM1A cells were generated previously21. Human PBMCs were isolated from buffy coats by the Ficoll-Paque density centrifugation method. Monocytes were enriched by positive selection using anti-CD14 mAb-coated microbeads from Miltenyi Biotec (San Diego, CA) according to manufacturer’s protocol. Monocytes were then differentiated with 5 ng/ml GM-CSF (R&D Systems, Minneapolis, MN) for 5–7 days to obtain monocyte derived macrophages (MDM). Fetal bovine serum was obtained from Life Technologies (Grand Island, NY). Phorbol ester 13-phorbol-12-myristate acetate (PMA), puromycin, etoposide, kanamycin and anisomycin were purchased from Sigma (St. Louis, MO). Sanguinarine and SP600125 were purchased from Fisher Scientific (Pittsburgh, PA). CellTiter-Glo luminescent cell viability assay kit was purchased from Promega (Madison, WI). Polyclonal PPM1A antibodies were purchased from Thermo Scientific (Rockford, IL). Monoclonal mouse antibodies to GAPDH and α-tubulin were purchased from Santa Cruz (Dallas, Texas) and Cell Signaling (Danvers, MA), respectively.
Bacteria and plasmids
The M. tuberculosis H37Rv derived auxotroph strain mc26206 was grown in Middlebrook 7H9 medium (Difco) supplemented with 0.2% glycerol, 0.02% Tyloxapol and 10% OADC (Remel) or on Middlebrook 7H10 plates supplemented with 0.5% glycerol and 10% OADC (Remel). Growth media of the auxotrophic M. tuberculosis strain was supplemented with 24 μg/ml pantothenate and 50 μg/ml L-leucine60. Hygromycin B was purchased from Calbiochem. gfp expressing M. tuberculosis mc26206 was generated previously21. M. tuberculosis mc26206 expressing luciferase was generated by transformation of the pJAK.1-luciferase plasmid49 (generous gift from Dr. Hmama). Escherichia coli strain DH5α was used for plasmid propagation, and was routinely grown in Luria-Bertani broth at 37 °C. Lentiviral vectors expressing shRNA targeting human PPM1A were generated by Genecopoeia (Rockville, MD) using the sequences: shRNA #1: 5′-GTACCTGGAATGCAGAGTA and shRNA #2: 5′- GTCGACACCTGTTTGTATA. THP-ΔPPM1A cells generated using shRNA #2 showed the best knockdown and was used for all subsequent experiments.
Generation of THP-ΔPPM1A cells
HEK 293T cells seeded at 50% confluency were transfected with lentiviral plasmids expressing shRNA targeting human PPM1A or scrambled control using FuGENE (Promega, Madison, WI). Culture supernatants were harvested after 48 h and 72 h, aliquoted, and stored at −80 °C. The supernatants containing lentiviral particles were used to transduce THP-1 cells. Cells were selected by puromycin and analyzed by Western blot to verify knockdown of PPM1A protein levels.
Bacterial infection
M. tuberculosis mc26206 growing in log-phase was quantified by optical density measurement at 600 nm using the conversion of 3 × 108 (Mtb) bacteria per ml for OD 1.0. The amount of bacteria required for various MOIs were washed and resuspended in RPMI 1640 cell culture media without antibiotics. For infection of THP-1 monocytes, the prepared bacteria were added to the THP-1 cells and resuspended together to ensure even exposure of the monocytes to bacteria. For infection of differentiated THP-1 macrophages and primary human MDMs, bacteria were added to the cells and incubated at 37 °C for 4 h. Extracellular, non-phagocytosed bacteria were removed by 3 washes and infection was continued 37 °C for the desired time.
Survival of Mtb during macrophage infection
Entire wells of Mtb-infected macrophages were harvested by collecting the cells in the supernatant and cells that were still attached to the well bottoms 3 days post infection. These fractions were combined and macrophages were lysed in Glo Lysis Buffer (Promega) to measure the amount of viable Mtb in each well. Luciferase activity, proportional to bacterial load, was determined by using the BrightGlo Luciferase Assay System (Promega) according to the manufacturer’s protocol. Resultant luminescence was measured with the Cytation 3 Multi-Mode Reader (BioTek, Winooski, VT) using 96-well solid white plates (Corning, Corning, NY) and an integration time of 1 s per well.
Apoptosis assays
Cells were stained with Annexin V conjugated to AlexaFluor 488 or APC (Life Technologies) according to manufacturer’s protocol. FAM FLICA™ (green) or FLICA™ 660 (far red) caspase-3/7 staining kits were used to stain for active caspase-3/7 in cells, and were used according to manufacturer’s protocol (ImmunoChemistry Technologies, Bloomington, MN). Apo-ONE homogenous caspase-3/7 detection kit was purchased from Promega, and used according to manufacturer’s protocol.
Flow cytometry
Cells following Annexin V or FLICA staining were analyzed by flow cytometric analysis (FCM). FCM analysis was performed on a Guava EasyCyte (Guava Technologies Inc., Billerica, MA). Data analysis was performed using Guava Express software (Guava Technologies Inc.) or FlowJo V10 software (Ashland, OR).
Milliplex assays
Cells following various treatments or Mtb infections were lysed according to Milliplex kit protocol (Millipore, Billerica, MA). Total and phosphorylated JNK (T183/Y185) or p38 (T180/Y182) protein levels were measured using separate 2-plex Milliplex kits. Experiments were performed according to manufacturer’s protocol and read-out was performed using a Bio-Plex 200 instrument. Data analysis was performed using Bio-Plex manager 6.0 software (Bio-Rad, Hercules, CA).
PPM1A activity assay
The enzymatic activity of PPM1A was measured using the pNPP phosphatase assay kit (BioAssay Systems, Hayward, CA). In brief, 10 μg/ml recombinant PPM1A (Abnova, Walnut, CA) was prepared in reaction buffer (25 mM HEPES, 100 mM NaCl, 2 mM MnCl2, pH 7.2) in the absence or presence of 0–30 μM sanguinarine for 15 min at room temperature. The resulting reaction was mixed with pNPP substrate mix according to manufacturer’s protocol and incubated at 37 °C for 15 min. The reaction was then stopped using the kit provided stop solution and absorbance was measured in a plate reader at 405 nm.
Real-time cell analyzer assay (RTCA)
Macrophage adhesion was measured in specialized 96-well plates (E-plate 96) with the xCELLigence Real-time Cell Analyzer (RTCA) SP apparatus (ACEA Biosciences, San Diego, CA). Data was quantified by measuring impedance changes between the sensing electrodes located in the well-bottom, which changes as a function of the adhesion of cells to the surface of the plate. A dimensionless value, the Cell Index (CI), is representative of these impedance changes. Using this system, macrophage adhesion and therefore viability was monitored in real-time. Plates were removed at various time points for the addition of anisomycin or Mtb for infection. After each manipulation, plates were placed back in the RTCA apparatus for kinetic monitoring. The CI at every time point represents the mean of three independent measurements. A schematic description of this system is shown in Supplemental Fig. 3.
Western blot
Cells were harvested by centrifugation, washed once with PBS, and lysed in RIPA buffer (Cell Signaling) according to the manufacturer’s instructions. Protein concentration of the lysates was determined by the bicinchoninic acid (BCA) method according to the manufacturer’s recommendations (Thermo Scientific). About 10 to 20 μg of protein per sample was separated on 10% Mini-Protean TGX gels (Bio-Rad) and subsequently transferred to a polyvinylidene difluoride (PVDF) membrane using an iBlot gel transfer system (Life Technologies). Western blot analysis was performed according to standard protocols. Total PPM1A, α-tubulin, or GAPDH proteins were detected with specific antibodies (see antibody section). A horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse polyclonal antibody (Santa Cruz) was used as the secondary antibody. The blot was developed using the Western Lightning Ultra chemiluminescent substrate from Perkin Elmer, Inc., and detected in an EpiChemi3 darkroom (UVP BioImaging Systems).
Kinex™ Antibody Microarray-based analysis
Kinex™ antibody array analysis was performed as before21. Briefly, 50 μg of lysate from each sample were covalently labeled with a proprietary fluorescent dye according to the manufacturer’s instructions (Kinexus, Canada). After blocking non-specific binding sites on the array, an incubation chamber was mounted onto the microarray to permit the loading of one control (THP-1) and one experiment (THP-PPM1A) sample side-by-side on the same chip. Following sample incubation, unbound proteins were washed away. KAM-850 chips are spotted in duplicates with over 850 antibodies, including 517 pan-specific antibodies and 337 phosphosite-specific antibodies. By this means, the microarrays provide information about the expression and phosphorylation levels of these target proteins.
Each array produces a pair of 16-bit images captured with a Perkin-Elmer ScanArray Reader laser array scanner (Waltham, MA). Signal quantification was performed with ImaGene 8.0 from BioDiscovery (El Segundo, CA) with predetermined settings for spot segmentation and background correction. The background-corrected raw intensity data are logarithmically transformed with base 2. Z-scores are calculated by subtracting the overall average intensity of all spots within a sample from the raw intensity for each spot, and dividing it by the standard deviations (SD) of all of the measured intensities within each sample61. Z-ratios are further calculated by taking the difference between the averages of the observed protein Z-scores and dividing by the SD of all of the differences for that particular comparison. Calculated Z-ratios have the advantage that they can be used in multiple comparisons without further reference to the individual conditional standard deviations by which they were derived.
Statistical analysis
Data are expressed as the mean ± the standard deviation of three independent experiments. Statistical analysis was performed using the Student t test. Values of p < 0.05 were considered to be significant.
Additional Information
How to cite this article: Schaaf, K. et al. Mycobacterium tuberculosis exploits the PPM1A signaling pathway to block host macrophage apoptosis. Sci. Rep. 7, 42101; doi: 10.1038/srep42101 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Dr. Zakaria Hmama for the generous gift of the pJAK.1-luciferase plasmid. This work was funded in part by NIH grants R01-AI104499, R56-AI122842 and R21-AI116188 to OK and in part by R01-AI104952 to FW. Parts of the work were performed in the UAB CFAR BSL-3 facilities and by the UAB CFAR Flow Cytometry Core/Joint UAB Flow Cytometry Core, which are funded by NIH/NIAID P30 AI027767 and by NIH 5P30 AR048311.
Footnotes
The authors declare no competing financial interests.
Author Contributions K.S., F.W., A.O.W., O.K. and J.S. conceived and designed experiments and analyzed data; K.S., S.S., A.D., F.W. and J.S. performed experiments; O.K. and J.S. wrote and edited the manuscript.
References
- Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol 35, 495–516, doi: 10.1080/01926230701320337 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar S. M., Divangahi M. & Remold H. G. Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy? Nat Rev Microbiol 8, 668–674, doi: 10.1038/nrmicro2387 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraco A. H. & Kornfeld H. Cell death and autophagy in TB. Semin Immunol 26, 497–511, doi: 10.1016/j.smim.2014.10.001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Hartman M. & Kornfeld H. Macrophage apoptosis in tuberculosis. Yonsei Med J 50, 1–11, doi: 10.3349/ymj.2009.50.1.1 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberdein J. D., Cole J., Bewley M. A., Marriott H. M. & Dockrell D. H. Alveolar macrophages in pulmonary host defence the unrecognized role of apoptosis as a mechanism of intracellular bacterial killing. Clin Exp Immunol 174, 193–202, doi: 10.1111/cei.12170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst S., Schaible U. E. & Schneider B. E. Interferon gamma activated macrophages kill mycobacteria by nitric oxide induced apoptosis. PLoS One 6, e19105, doi: 10.1371/journal.pone.0019105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sly L. M., Hingley-Wilson S. M., Reiner N. E. & McMaster W. R. Survival of Mycobacterium tuberculosis in host macrophages involves resistance to apoptosis dependent upon induction of antiapoptotic Bcl-2 family member Mcl-1. J Immunol 170, 430–437 (2003). [DOI] [PubMed] [Google Scholar]
- Behar S. M., Martin C. J., Nunes-Alves C., Divangahi M. & Remold H. G. Lipids, apoptosis, and cross-presentation: links in the chain of host defense against Mycobacterium tuberculosis. Microbes Infect 13, 749–756, doi: 10.1016/j.micinf.2011.03.002 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divangahi M. et al. Mycobacterium tuberculosis evades macrophage defenses by inhibiting plasma membrane repair. Nat Immunol 10, 899–906, doi: 10.1038/ni.1758 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. et al. Lipid mediators in innate immunity against tuberculosis: opposing roles of PGE2 and LXA4 in the induction of macrophage death. J Exp Med 205, 2791–2801, doi: 10.1084/jem.20080767 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilchanka O. et al. An outer membrane channel protein of Mycobacterium tuberculosis with exotoxin activity. Proc Natl Acad Sci USA 111, 6750–6755, doi: 10.1073/pnas.1400136111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H. et al. Cell death and infection: a double-edged sword for host and pathogen survival. J Cell Biol 195, 931–942, doi: 10.1083/jcb.201108081 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber G. N. Host defense, viruses and apoptosis. Cell Death Differ 8, 113–126, doi: 10.1038/sj.cdd.4400823 (2001). [DOI] [PubMed] [Google Scholar]
- Velmurugan K. et al. Mycobacterium tuberculosis nuoG is a virulence gene that inhibits apoptosis of infected host cells. PLoS Pathog 3, e110, doi: 10.1371/journal.ppat.0030110 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchey J. et al. Enhanced priming of adaptive immunity by a proapoptotic mutant of Mycobacterium tuberculosis. J Clin Invest 117, 2279–2288, doi: 10.1172/JCI31947 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakumar D., Jacobs W. R. Jr. & Narayanan S. Protein kinase E of Mycobacterium tuberculosis has a role in the nitric oxide stress response and apoptosis in a human macrophage model of infection. Cell Microbiol 10, 365–374, doi: 10.1111/j.1462-5822.2007.01049.x (2008). [DOI] [PubMed] [Google Scholar]
- Sun J. et al. Mycobacterium tuberculosis nucleoside diphosphate kinase inactivates small GTPases leading to evasion of innate immunity. PLoS Pathog 9, e1003499, doi: 10.1371/journal.ppat.1003499 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. et al. The tuberculosis necrotizing toxin kills macrophages by hydrolyzing NAD. Nat Struct Mol Biol, doi: 10.1038/nsmb.3064 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parandhaman D. K. & Narayanan S. Cell death paradigms in the pathogenesis of Mycobacterium tuberculosis infection. Front Cell Infect Microbiol 4, 31, doi: 10.3389/fcimb.2014.00031 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar S. M. et al. Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal Immunol 4, 279–287, doi: 10.1038/mi.2011.3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. et al. Protein Phosphatase, Mg2+/Mn2+-dependent 1A controls the innate antiviral and antibacterial response of macrophages during HIV-1 and Mycobacterium tuberculosis infection. Oncotarget, doi: 10.18632/oncotarget.8190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia A. et al. Serine/threonine protein phosphatases PP1 and PP2A are key players in apoptosis. Biochimie 85, 721–726 (2003). [DOI] [PubMed] [Google Scholar]
- Chen W. et al. Ppm1b negatively regulates necroptosis through dephosphorylating Rip3. Nat Cell Biol 17, 434–444, doi: 10.1038/ncb3120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo J., da Cruz E. S. O. A. & Fardilha M. Protein phosphatase 1 and its complexes in carcinogenesis. Curr Cancer Drug Targets 14, 2–29 (2014). [DOI] [PubMed] [Google Scholar]
- Kiely M. & Kiely P. A. PP2A: The Wolf in Sheep’s Clothing? Cancers (Basel) 7, 648–669, doi: 10.3390/cancers7020648 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophe T. et al. High content screening identifies decaprenyl-phosphoribose 2′ epimerase as a target for intracellular antimycobacterial inhibitors. PLoS Pathog 5, e1000645, doi: 10.1371/journal.ppat.1000645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartkoorn R. C. et al. Differential drug susceptibility of intracellular and extracellular tuberculosis, and the impact of P-glycoprotein. Tuberculosis (Edinb) 87, 248–255, doi: 10.1016/j.tube.2006.12.001 (2007). [DOI] [PubMed] [Google Scholar]
- Donald P. R. Cerebrospinal fluid concentrations of antituberculosis agents in adults and children. Tuberculosis (Edinb) 90, 279–292, doi: 10.1016/j.tube.2010.07.002 (2010). [DOI] [PubMed] [Google Scholar]
- Schaaf K. et al. A Macrophage Infection Model to Predict Drug Efficacy Against Mycobacterium Tuberculosis. Assay Drug Dev Technol 14, 345–354, doi: 10.1089/adt.2016.717 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers T. et al. Role of PP2Calpha in cell growth, in radio- and chemosensitivity, and in tumorigenicity. Mol Cancer 6, 65, doi: 10.1186/1476-4598-6-65 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain D. R., Berger T. & Mak T. W. Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol 5, a008656, doi: 10.1101/cshperspect.a008656 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes R. W. & Doxsee D. The receptor-mediated uptake, survival, replication, and drug sensitivity of Mycobacterium tuberculosis within the macrophage-like cell line THP-1: a comparison with human monocyte-derived macrophages. Cell Immunol 197, 1–9, doi: 10.1006/cimm.1999.1554 (1999). [DOI] [PubMed] [Google Scholar]
- Theus S. A., Cave M. D. & Eisenach K. D. Activated THP-1 cells: an attractive model for the assessment of intracellular growth rates of Mycobacterium tuberculosis isolates. Infect Immun 72, 1169–1173 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J. D., Gogvadze V., Zhivotovsky B. & Orrenius S. Distinct pathways for stimulation of cytochrome c release by etoposide. J Biol Chem 275, 32438–32443, doi: 10.1074/jbc.C000518200 (2000). [DOI] [PubMed] [Google Scholar]
- Gil-Parrado S. et al. Ionomycin-activated calpain triggers apoptosis. A probable role for Bcl-2 family members. J Biol Chem 277, 27217–27226, doi: 10.1074/jbc.M202945200 (2002). [DOI] [PubMed] [Google Scholar]
- Grabarek J. & Darzynkiewicz Z. In situ activation of caspases and serine proteases during apoptosis detected by affinity labeling their enzyme active centers with fluorochrome-tagged inhibitors. Exp Hematol 30, 982–989 (2002). [DOI] [PubMed] [Google Scholar]
- Lee J., Remold H. G., Ieong M. H. & Kornfeld H. Macrophage apoptosis in response to high intracellular burden of Mycobacterium tuberculosis is mediated by a novel caspase-independent pathway. J Immunol 176, 4267–4274 (2006). [DOI] [PubMed] [Google Scholar]
- Poirier V., Bach H. & Av-Gay Y. Mycobacterium tuberculosis promotes anti-apoptotic activity of the macrophage by PtpA protein-dependent dephosphorylation of host GSK3alpha. J Biol Chem 289, 29376–29385, doi: 10.1074/jbc.M114.582502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miret S., De Groene E. M. & Klaffke W. Comparison of in vitro assays of cellular toxicity in the human hepatic cell line HepG2. J Biomol Screen 11, 184–193, doi: 10.1177/1087057105283787 (2006). [DOI] [PubMed] [Google Scholar]
- Takekawa M., Maeda T. & Saito H. Protein phosphatase 2Calpha inhibits the human stress-responsive p38 and JNK MAPK pathways. EMBO J 17, 4744–4752, doi: 10.1093/emboj/17.16.4744 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanasekaran D. N. & Reddy E. P. JNK signaling in apoptosis. Oncogene 27, 6245–6251, doi: 10.1038/onc.2008.301 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazzalin C. A., Le Panse R., Cano E. & Mahadevan L. C. Anisomycin selectively desensitizes signalling components involved in stress kinase activation and fos and jun induction. Mol Cell Biol 18, 1844–1854 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B. L. et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc Natl Acad Sci USA 98, 13681–13686, doi: 10.1073/pnas.251194298 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aburai N., Yoshida M., Ohnishi M. & Kimura K. Sanguinarine as a potent and specific inhibitor of protein phosphatase 2C in vitro and induces apoptosis via phosphorylation of p38 in HL60 cells. Biosci Biotechnol Biochem 74, 548–552, doi: 10.1271/bbb.90735 (2010). [DOI] [PubMed] [Google Scholar]
- Atienza J. M. et al. Dynamic and label-free cell-based assays using the real-time cell electronic sensing system. Assay Drug Dev Technol 4, 597–607, doi: 10.1089/adt.2006.4.597 (2006). [DOI] [PubMed] [Google Scholar]
- Limame R. et al. Comparative analysis of dynamic cell viability, migration and invasion assessments by novel real-time technology and classic endpoint assays. PLoS One 7, e46536, doi: 10.1371/journal.pone.0046536 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Serra J. et al. xCELLigence system for real-time label-free monitoring of growth and viability of cell lines from hematological malignancies. Onco Targets Ther 7, 985–994, doi: 10.2147/OTT.S62887 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhorter F. Y., Wang T., Nguyen P., Chung T. & Liu W. F. Modulation of macrophage phenotype by cell shape. Proc Natl Acad Sci USA 110, 17253–17258, doi: 10.1073/pnas.1308887110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. et al. A broad-range of recombination cloning vectors in mycobacteria. Plasmid 62, 158–165, doi: 10.1016/j.plasmid.2009.07.003 (2009). [DOI] [PubMed] [Google Scholar]
- Sorrentino F. et al. Development of an intracellular screen for new compounds able to inhibit Mycobacterium tuberculosis growth in human macrophages. Antimicrob Agents Chemother, doi: 10.1128/AAC.01920-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snewin V. A. et al. Assessment of immunity to mycobacterial infection with luciferase reporter constructs. Infect Immun 67, 4586–4593 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeyborne I. et al. The molecular bacterial load assay replaces solid culture for measuring early bactericidal response to antituberculosis treatment. J Clin Microbiol 52, 3064–3067, doi: 10.1128/JCM.01128-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram R. et al. Pharmacokinetics-pharmacodynamics of rifampin in an aerosol infection model of tuberculosis. Antimicrob Agents Chemother 47, 2118–2124 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutelle S., Bourguignon L., Jelliffe R. W., Conte J. E. Jr. & Maire P. Mathematical modeling of pulmonary tuberculosis therapy: Insights from a prototype model with rifampin. J Theor Biol 282, 80–92, doi: 10.1016/j.jtbi.2011.05.013 (2011). [DOI] [PubMed] [Google Scholar]
- Stanley S. A. & Cox J. S. Host-pathogen interactions during Mycobacterium tuberculosis infections. Curr Top Microbiol Immunol 374, 211–241, doi: 10.1007/82_2013_332 (2013). [DOI] [PubMed] [Google Scholar]
- Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484, doi: 10.1016/j.cell.2009.10.006 (2009). [DOI] [PubMed] [Google Scholar]
- Le Guezennec X. & Bulavin D. V. WIP1 phosphatase at the crossroads of cancer and aging. Trends Biochem Sci 35, 109–114, doi: 10.1016/j.tibs.2009.09.005 (2010). [DOI] [PubMed] [Google Scholar]
- Wang J. et al. Mycobacterium tuberculosis suppresses innate immunity by coopting the host ubiquitin system. Nat Immunol 16, 237–245, doi: 10.1038/ni.3096 (2015). [DOI] [PubMed] [Google Scholar]
- Zumla A. et al. Tuberculosis treatment and management–an update on treatment regimens, trials, new drugs, and adjunct therapies. Lancet Respir Med 3, 220–234, doi: 10.1016/S2213-2600(15)00063-6 (2015). [DOI] [PubMed] [Google Scholar]
- Sampson S. L. et al. Protection elicited by a double leucine and pantothenate auxotroph of Mycobacterium tuberculosis in guinea pigs. Infect Immun 72, 3031–3037 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheadle C., Vawter M. P., Freed W. J. & Becker K. G. Analysis of microarray data using Z score transformation. J Mol Diagn 5, 73–81, doi: 10.1016/S1525-1578(10)60455-2 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.