Abstract
Diaminopimelate epimerase (DapF) is one of the crucial enzymes involved in l-lysine biosynthesis, where it converts l,l-diaminopimelate (l,l-DAP) into d,l-DAP. DapF is also considered as an attractive target for the development of antibacterial drugs. Here, we report the crystal structure of DapF from Corynebacterium glutamicum (CgDapF). Structures of CgDapF obtained under both oxidized and reduced conditions reveal that the function of CgDapF is regulated by redox-switch modulation via reversible disulfide bond formation between two catalytic cysteine residues. Under oxidized condition, two catalytic cysteine residues form a disulfide bond; these same cysteine residues exist in reduced form under reduced condition. Disulfide bond formation also induces a subsequent structural change in the dynamic catalytic loop at the active site, which results in open/closed conformational change at the active site. We also determined the crystal structure of CgDapF in complex with its product d,l-DAP, and elucidated how the enzyme recognizes its substrate l,l-DAP as a substrate. Moreover, the structure in complex with the d,l-DAP product reveals that CgDapF undergoes a large open/closed domain movement upon substrate binding, resulting in a completely buried active site with the substrate bound.
Corynebacterium glutamicum is a Gram-negative bacterium widely utilized for biotechnological production of various amino acids, such as l-glutamate and l-lysine1,2. As an essential building block for protein synthesis, l-lysine is an industrially important substance for animal feed and dietary supplements. Owing to global increases in meat consumption, l-lysine is particularly required for the animal feed market; the global market for l-lysine is expected to reach USD 6.96 billion by 20203. Furthermore, meso-diaminopimelate (d,l-DAP; mDAP), the intermediate precursor of l-lysine, is involved in cross-linking of the peptidoglycan cell wall of certain species of Gram-negative bacteria4. Because l-lysine biosynthesis is unavailable in animals, members of l-lysine biosynthetic pathway are regarded as attractive targets for antibacterial drug development5,6.
In C. glutamicum, l-lysine is synthesized from aspartate7,8,9. The aspartate is converted into l-aspartate semialdehyde (ASA) by the consecutive reaction of two enzymes. ASA is a precursor for the biosynthesis of various amino acids such as l-threonine, l-isoleucine, l-methionine, and l-lysine. On the l-lysine biosynthetic pathway, ASA is condensed with pyruvate to generate dihydrodipicolinate (DHDP). DHDP reductase reduces DHDP to produce tetrahydrodipicolinate (THDP)10. Currently, four different pathways for the biosynthesis of l-lysine that branch out from THDP have been reported in bacteria11,12: the succinylase pathway, the acetylase pathway, the mDAP dehydrogenase pathway, and the recently discovered aminotransferase pathway13,14. Although most bacteria possess only one of these pathways, C. glutamicum utilizes both the succinylase and mDAP dehydrogenase pathways15. In the succinylase pathway, l,l-diaminopimelate (L,L-DAP) is synthesized from THDP by succinylation, transamination, and desuccinylation steps; L,L-DAP is converted to d,l-DAP by the action of DAP epimerase (DapF). In contrast, in the mDAP dehydrogenase pathway, DAP dehydrogenase converts THDP into d,l-DAP in one step; DAP decarboxylase subsequently catalyzes the decarboxylation of d,l-DAP to form l-lysine.
DapF (EC 5.1.1.7), a pyridoxal 5-phosphate-independent amino acid racemase, catalyzes the stereochemical inversion between l,l-DAP and d,l-DAP. DapF catalyzes the final step of the succinylase pathway. DapF plays an important role in the biosynthesis of l-lysine because DAP decarboxylase only recognizes d,l-DAP. Previous structural and functional studies of DapFs from several organisms such as Haemophilus influenzae16,17,18,19,20,21, Mycobacterium tuberculosis22, Escherichia coli23, and Arabidopsis thaliana24 have reported that the DapFs consist of two structurally similar α/β domains and each domain provides one of the two key active site cysteine residues. In addition, DapFs utilizes two cysteine residues for catalysis: one cysteine residue acts as a base and abstracts a proton from L,L-DAP, whereas the other acts as an acid and re-protonates the molecule to form d,l-DAP. Despite the importance of C. glutamicum in the production of l-lysine, detailed structural and biochemical studies had not been reported on CgDapF prior to this study.
In this study, we determined the crystal structures of CgDapF in both the oxidized and the reduced form. We elucidated that CgDapF is regulated by redox-switch modulation via reversible disulfide bond formation between two catalytic cysteine residues, depending on the environmental redox state. We also report that CgDapF undergoes a large open/closed domain movement upon substrate binding.
Results and Discussion
Overall structure of CgDapF
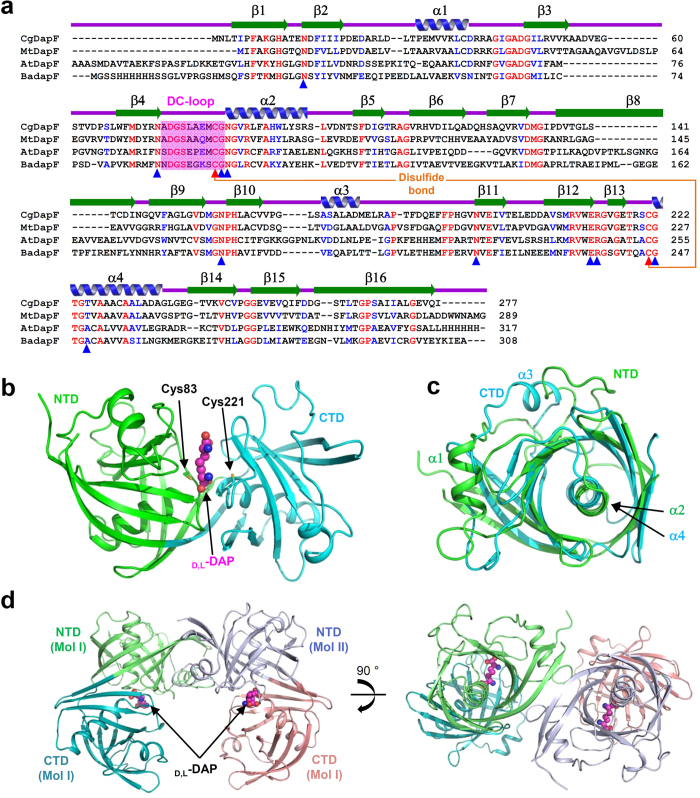
To reveal the molecular mechanism of CgDapF, we determined its crystal structure (2.0 Å resolution) via the single-wavelength anomalous dispersion method, using a selenium-substituted crystal (Table 1). The structure of CgDapF adopts an overall fold similar to those of diaminopimelate epimerases from Mycobacterium tuberculosis (MtDapF; PDB code 3FVE)22, Arabidopsis thaliana (AtDapF; PDB code 3EJX)24, and Bacillus anthracis (BaDapF; PDB code 2OTN) (Fig. 1a). Each CgDapF monomer consists of two distinct domains: an N-terminal domain (NTD; Met1–Asp131 & Gly268–Ile277) and a C-terminal domain (CTD; Met132–Thr267). Each domain contains a set of five-stranded and three-stranded antiparallel β-sheets and two α-helices (Fig. 1b). One α-helix of each domain (α2 in the NTD and α4 in the CTD) is sandwiched between the five-stranded and three-stranded β-sheets, whereas the other helix lies on the surface of the protein. The NTD and the CTD are structurally homologous to each other, with a root-mean-square deviation (RMSD) of 3.4 Å. However, there is one striking structural difference between the two domains; namely, the position of the α-helix that lies on the surface (Fig. 1c). Two catalytic cysteine residues are positioned at the active site, which is located at a cleft between the two domains (Fig. 1b). CgDapF functions as a dimer and the asymmetric unit contains a CgDapF dimer (Fig. 1d), which is consistent with our size-exclusion chromatography results under both the oxidized and the reduced conditions (data not shown). The dimerization interface is mainly constituted by the contacts between β16 from both monomers, and contact between two β-strands connects the two β-sheets of the NTDs from both monomers (Fig. 1d). Contacts between the connecting loops (α1–β3) from both monomers also mediate dimerization of the protein (Fig. 1d). The PISA25 server was used to calculate the buried interface area (854.9 Å2) and the percentage of participating residues (8.3%).
Table 1. Data collection and refinement statistics of CgDapF.
| PDB code | CgDapF_Oxidized (SeMet) | CgDapF_Reduced (SeMet) | CgDapF in complex with d,l-DAP |
|---|---|---|---|
| 5H2G | 5H2Y | 5M47 | |
| Data collection | |||
| Wavelength (Å) | 0.97934 | 0.97934 | 0.97934 |
| Cell dimensions (a, b, c; α,β,γ) (Å; °) | 101.7, 119.1, 155.6; 90.0, 90.0 90.0 | 101.8, 118.9, 153.3; 90.0, 90.0 90.0 | 155.7 155.7 155.7; 90.0, 90.0, 90.0 |
| Wavelength (Å) | 0.97918 | 0.97924 | 0.97934 |
| Space group | I222 | I222 | P4332 |
| Resolution range (Å) | 50.00–2.00 (2.03–2.0) | 50.00–2.00 (2.03–2.00) | 50.00–2.60 (2.64–2.60) |
| Rsym or Rmerge (%) | 8.2 (40.5) | 8.3 (41.4) | 11.5 (34.4) |
| I/σI | 46.3 (7.4) | 22.5 (2.4) | 21.1 (4.0) |
| Completeness (%) | 99.8 (100.0) | 96.0 (91.8) | 97.4 (93.4) |
| Redundancy | 8.1 (8.1) | 4.5 (2.7) | 12.0 (4.5) |
| Refinement | |||
| Resolution (Å) | 50.0–2.0 | 50.0–2.0 | 50.0–2.6 |
| No. reflections | 59911 | 58955 | 19071 |
| Rwork/Rfree | 18.9/21.9 | 20.7/25.2 | 19.1/22.9 |
| No. atoms | 4563 | 4273 | 2154 |
| Protein | 4126 | 4088 | 2073 |
| Ligand/ion | 11 | — | 13 |
| Water | 426 | 185 | 68 |
| B-factors | 37.0 | 43.0 | 34.0 |
| Protein | 36.0 | 43.1 | 34.3 |
| Ligand/ion | 37.9 | — | 30.8 |
| Water | 45.8 | 44.7 | 27.8 |
| B from Wilson plot (Å2) | 29.2 | 28.5 | 27.6 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.023 | 0.022 | 0.018 |
| Bond angles (°) | 2.137 | 2.180 | 2.103 |
*Mean B value is for both protein atoms and the solvent molecules.
Figure 1. Overall structure of CgDapF.
(a) Amino acid sequence alignment of the DapF proteins. Identical and highly conserved residues are presented in red and blue colored characters, respectively. Secondary structure elements are shown and labeled based on the structure of CgDapF. The DC-loop is shown in a magenta-colored box. Two catalytic cysteine residues, Cys83 and Cys221, involved in the disulfide bond formation are distinguished with orange-colored line and marked with red colored triangles. Residues involved in the substrate binding are marked with blue colored triangles. Cg, Mt, At, and Ba represent DapF from Corynebaterium glutamicum, Mycobacterium tuberculosis, Arabidopsis thaliana, and Bacillus anthracis, respectively. (b) Monomeric structure of CgDapF. A monomeric CgDapF is shown as a cartoon diagram. NTD and CTD are distinguished with green and cyan colors, respectively. d,l-DAP molecule bound in the enzyme is shown as a sphere model with a magenta color. (c) Superimposition of structures of NTD and CTD. The NTD and CTD structure are presented with a color scheme in (b). (d) Dimeric structure of CgDapF. The dimeric structure of CgDapF is presented as a cartoon diagram with one monomer with a color scheme in (b) and the other in light-blue and salmon for the NTD and CTD, respectively. The bound d,l-DAP is shown as a sphere model with a magenta color. The right-side figure is a 90° rotation in the vertical direction from the left-side figure.
Reversible disulfide bond formation at the active site of CgDapF
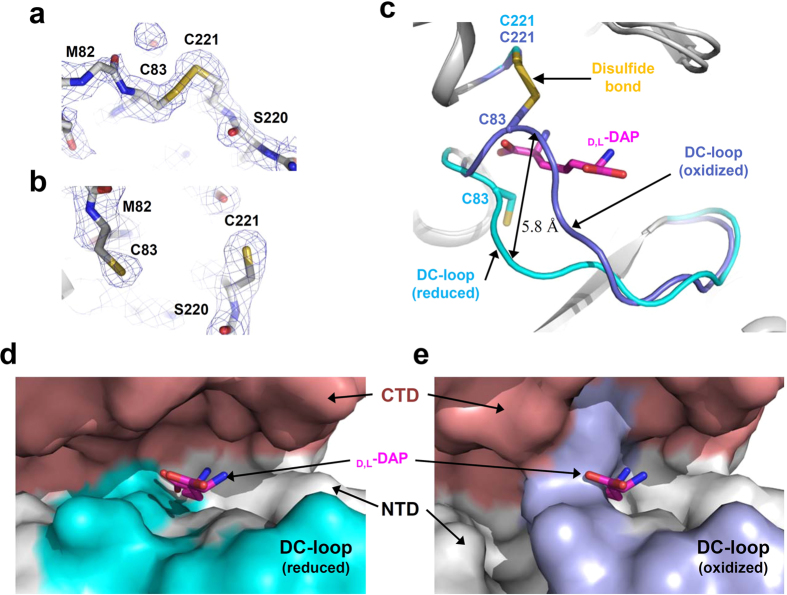
Although both monomers adopt an almost identical overall shape with respect to each other (RMSD = 0.31 Å), an interesting structural difference was observed between the active sites of each monomer. In one monomer, two catalytic cysteine residues (Cys83 and Cys221) were oxidized, forming a disulfide bond with each other; in contrast, these residues were reduced in the other monomer (Fig. 2a,b). The formation of this disulfide bond also caused a large conformational change in the loop that contains Cys83 (Ala75–Gly84) (Fig. 2c). In the reduced form, the active site exhibits an open conformation that is freely accessible to the substrate (Fig. 2d). In contrast, in the oxidized form of CgDapF, the loop was positioned 5.8 Å closer to the substrate binding site, compared to the reduced form (Fig. 2c), resulting in closure of the substrate binding site (Fig. 2e). Based on these observations, we speculate that the activity of CgDapF is regulated by the environmental redox potential via the formation of a reversible disulfide bond and by the subsequent open/closed conformational change at the active site. Because the loop conformation seems to influence the activity of CgDapF directly, we will henceforth refer to this loop as a ‘dynamic catalytic loop’ (DC-loop).
Figure 2. Reversible disulfide bond formation of CgDapF.
(a,b) Electron density map of the two cysteine residues in the oxidized (a) and the reduced (b) condition. The 2Fo-Fc electron density map of the reversible disulfide bond in the oxidized form and a breakage of the disulfide bond in the reduced form of CgDapF are shown as a blue mesh and contoured at 1.2 σ. (c) Disulfide bond-mediated conformational change of DC-loop. The DC-loop in the oxidized form and the reduced form is presented as a cartoon diagram with blue and cyan colors, respectively. Two catalytic cysteine residues involved in the disulfide bond formation are shown as a stick model. The bound d,l-DAP is shown as a stick model with a magenta color. (d,e) A surface model of the active site of CgDapF. The CgDapF structures in reduced and oxidized form are presented with a surface model with colors of white and salmon for the NTD and CTD, respectively. DC-loops in reduced and oxidized form are shown with the same color scheme as in (c).
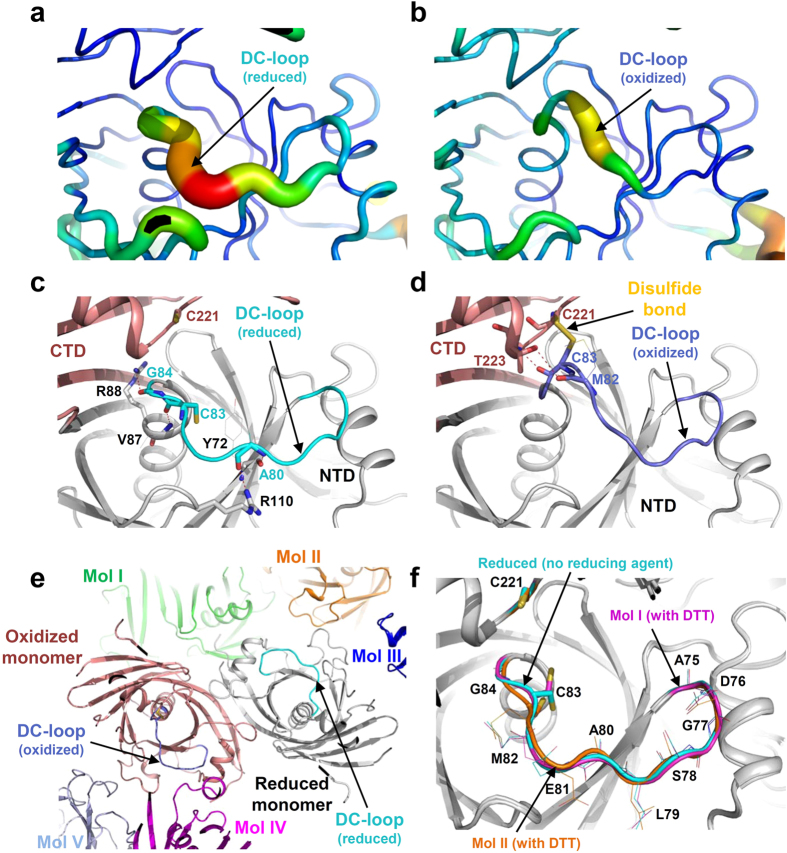
The conformational change of the DC-loop seems to result from the high flexibility of the loop. The DC-loop shows much higher B-factors in both the oxidized and the reduced forms, as compared with the rest of the protein, indicating that the DC-loop region is highly flexible (Fig. 3a,b). The stabilization modes of the loop are also quite different from each other. In the reduced form of CgDapF, the loop is mainly stabilized by interactions with the NTD. The main chain portion of Ala80 forms direct hydrogen bonds with the main chain portion of Tyr72 and the side chain of Arg110, and the main chain oxygen atoms of Cys83 and Gly84 formed hydrogen bonds with the main chain nitrogen atom of Val87 and the side chain of Arg88 (Fig. 3c). In contrast, in the oxidized form of CgDapF, the DC-loop interacts with the CTD instead. The disulfide bond between Cys83 and Cys221 is the main contributor to the stability of the loop, and the side chain of Thr223 forms hydrogen bonds with the main chain portions of Met82 and Cys83 (Fig. 3d).
Figure 3. Disulfide bond-mediated conformational change in CgDapF.
(a,b) B-factor presentation of CgDapF structures in the reduced (a) and oxidized (b) form. Structures in (a) and (b) are drawn in a same orientation. (c,d) Stabilization mode of the DC-loop in the reduced (c) and oxidized (d) form. The DC-loops in the reduced and oxidized form are presented as cartoon diagrams with cyan and blue colors, respectively. Side-chains involved in the stabilization of the DC-loop are shown as stick models. Main-chains involved in the stabilization of the DC-loop are shown as stick models and their side-chains are as line models. Hydrogen bonds are presented as red-colored dotted lines. Structures in (c) and (d) are drawn in a same orientation. (e) Different contacts of CgDapF monomer with the neighboring molecules in the crystal. Oxidized or reduced monomers are shown as a cartoon diagram with salmon and white color, respectively. The DC-loops are distinguished with the same color scheme in (a,b). The molecules near each monomer are shown as cartoon models with different colors. (f) Superposition of reduced monomers in CgDapF. Monomers are shown as a cartoon diagram and distinguished by the color of DC-loop. DC-loop in reduced monomer without reducing agent is colored with cyan. DC-loops from the reduced monomer I and II with DTT are presented as magenta and orange colors, respectively. The catalytic cysteine residues and other residues in DC-loop are presented as a stick and line model, respectively.
In fact, no reducing agent was added, neither to the reservoir nor to the protein solution used for crystallization, indicating that the protein was crystallized under somewhat oxidized condition. Consequently, we sought to answer why each monomer adopted a different conformation and why only one monomer contained a disulfide bond under identical redox condition. When we generated I222 symmetry operations, it became clear that the contacts of the two monomers with neighboring molecules completely differ from each other. In particular, the DC-loop of the oxidized monomer makes a close contact with one neighboring molecule, whereas that of the reduced monomer makes no contact with any molecule (Fig. 3e). These observations suggested the possibility that disulfide bond formation at the active site may have been an artifact caused by different crystal contacts, rather that constituting an authentic structural feature of the protein. We then determined a crystal structure of CgDapF in the presence of 1 mM DTT, to investigate whether reversible disulfide bond formation and the associated conformational change we had observed indeed constitute authentic structural features of the protein (Table 1). The structure determined in the presence of 1 mM DTT belonged to space group I222, the same group as that of the oxidized structure, and thus identical crystal contacts were formed as when the protein had been crystallized without a reducing agent. Interestingly, under reduced condition the structures of both monomers adopted the same conformation that had been adopted in the ‘reduced monomer’ in the earlier structure that had been determined in the absence of a reducing agent: no disulfide bond was observed in either monomer and the DC-loops showed open conformations (Fig. 3f). These results demonstrate that, in the earlier structure that was determined in the absence of a reducing agent, the pattern of crystal contacts was not responsible for the formation of disulfide bond and the closed conformation. Rather, these features were derived from somewhat oxidized condition during crystallization. Taken together, we propose that CgDapF regulates its function by redox-switch modulation via reversible disulfide bond formation and the subsequent open/closed conformational change.
The redox sensitivity of CgDapF
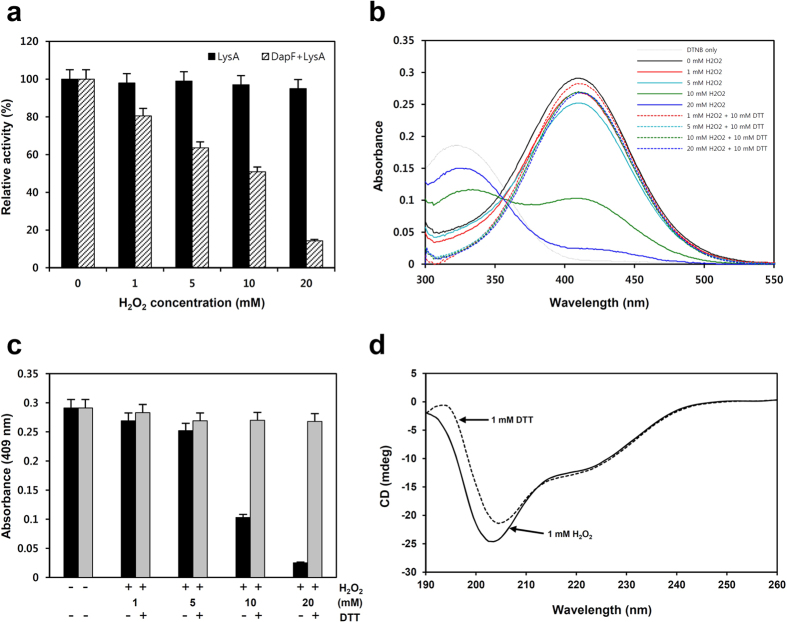
In general, redox-mediated modification of cellular proteins confers a respose to changes in the environmental redox potential26,27,28. Our crystal structures of CgDapF in both oxidized and reduced forms suggest that CgDapF is regulated by redox-switch modulation, via reversible disulfide bond formation. To investigate the redox sensitivity of CgDapF, we examined the susceptibility of CgDapF to hydrogen peroxide (H2O2). When CgDapF was treated with various concentrations of H2O2, the enzyme showed a concentration-dependent loss of activity (Fig. 4a), indicating the enzyme is indeed sensitive to oxidative environment. To verify whether disulfide bond formation was indeed reversible, we performed an activity recovery test on CgDapF using an Ellman assay, which measures the amount of free thiol groups in a protein. CgDapF was pre-incubated with various concentrations of H2O2, and the concentration of free thiol groups in the enzyme was incrementally decreased (Fig. 4b,c). Then, when the environment was switched to reduced condition by removing H2O2 and adding 10 mM DTT; the oxidized protein indeed recovered and free thiol groups were measured (Fig. 4b,c). In highly oxidized condition, cysteine thiol groups can form sulfinic or sulfonic acids, which cannot be reduced back to thiol groups by DTT. However, the recovery of thiol groups in our Ellman assay results indicate that CgDapF formed disulfide bonds when oxidized by H2O2 and that the thiol groups of the enzyme were recovered through breakage of disulfide bonds by DTT. Our proposal of a reversible disulfide bond formation that accompanies the structural change was investigated by circular dichroism spectroscopy (CD). CD measurements were recorded over the wavelengths 190–260 nm on the CgDapF protein, which had been treated with either 1 mM H2O2 or 1 mM DTT (Fig. 4d). The CD results demonstrate that there are notable differences in the ellipticity values recorded from CgDapF that depending on whether it had been treated with DTT or H2O2, indicating that CgDapF undergoes substantial redox-dependent structural changes. The secondary structure elements calculated using the CD data were quite similar to the structural analysis from the crystal structures, and there was no significant difference between the oxidized and the reduced conditions. Taking our structural and biochemical observations on CgDapF together, we propose that the activity of CgDapF is regulated by redox-switch modulation via the reversible disulfide bond formation in response to environmental redox changes. We further propose that the open/closed conformational change caused by the reversible disulfide bond formation also contributes to the redox sensitive regulation of the protein.
Figure 4. Redox sensitivity of CgDapF.
(a) Susceptibility of CgDapF to hydrogen peroxide (H2O2). The CgDapF protein was treated with various concentration of H2O2 and DapF activity was measured. To test the effect of H2O2 on LysA, LysA activity with various concentrations of H2O2 were also measured. (b) UV/Vis spectra of CgDapF depending on redox state. The filled-lines indicate the UV/Vis spectra of CgDapF with various concentration of H2O2, and the dotted-lines indicate those after switch of the redox states to the reduced condition by addition of 10 mM DTT. (c) Absorbance at 409 nm of CgDapF depending on redox state. The absorbance values at 409 nm from the Uv/vis spectra of (b) are compared. (d) The far-UV CD spectra of CgDapF in different redox conditions. Structural changes of CgDapF depending on different redox conditions are measured using the far-UV CD spectra. For the reduced and the oxidized condition, 1 mM each of DTT and H2O2 are added to the CgDapF protein, respectively.
Comparison of CgDapF with other DapFs
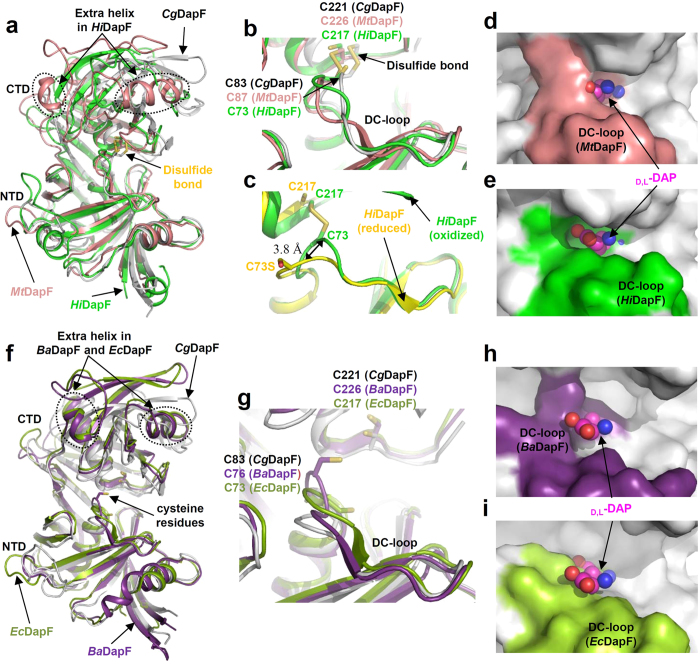
The structures of DapF from several organisms have been determined in both oxidized and reduced conditions. To investigate the differences between DapFs, we compared the oxidized form of CgDapF with MtDapF and DapF from Haemophilus influenzae (HiDapF; PDB code 1BWZ and 2Q9H)16,21 and the reduced form of CgDapF is compared with BaDapF and DapF from Escherichia coli (EcDapF; PDB code 4IJZ)23. The overall structure is almost conserved in DapFs and the quaternary structure of DapFs is dimer. In both oxidized and reduced forms of CgDapF, PISA25 server compute that the oligomeric status is dimer and other DapFs show same results. The dimeric interface of DapFs is also conserved. The two β-strands from both monomers mainly contribute to the dimerization of DapFs. The buried interface areas of MtDapF, HiDapF, BaDapF and EcDapF were calculated by PISA25 server and the values are 687.7 Å, 1084.6 Å, 854.6 Å, and 771.8 Å, respectively. Similarities in the overall structure extend to the active site residues. In DapF structures, the residues involved in the substrate binding are almost identical. Although Thr223 in CgDapF that stabilizes the carboxyl group of d,l-DAP on the side bearing the d-amino moiety was substituted for Ser219 in HiDapF and EcDapF, the role of the residues seems to be similar each other.
When we superimposed the NTD of the oxidized form of CgDapF with those of MtDapF and HiDapF, NTD structures matched each other closely, whereas the CTDs from three oxidized DapFs show somewhat different conformations (Fig. 5a). HiDapF contains two additional helices; one helix is inserted at the entrance of CTD and the other is inserted into the connecting loop between β10 and β11 (Fig. 5a). The catalytic cysteine residues from CgDapF and HiDapF form a disulfide bond, whereas those from MtDapF exist in mixed states of the oxidized and the reduced conformations (Fig. 5b). As we mentioned above, the disulfide bond between two catalytic cysteine residues results in the conformational change in the DC-loop. Although the disulfide bond mediated conformational change is also observed in HiDapF, the movement of the DC-loop is less dynamic compared with CgDapF; the loop containing Cys73 (Val70-Gly76) in HiDapF was moved only by 3.8 Å (Fig. 5c). The DC-loop of MtDapF (Asn78-Gly90) is also located at the similar position as that of HiDapF (Fig. 5b). The oxidized conformations also exhibit the closed conformation at the substrate binding site in both MtDapF and HiDapF (Fig. 5d,e). When we compared the reduced form of CgDapF with BaDapF and EcDapF, the conformations of both NTD and CTD are almost identical (Fig. 5e). Compared to the reduced form of CgDapF, BaDapF and EcDapF also contain two additional helices at the similar position to HiDapF (Fig. 5f). The catalytic cysteine residues are all reduced (Fig. 5g) and the DC-loops show the open conformation (Fig. 5h,i). Based on these structural comparisons, we suggest that the activities of DapF proteins might be regulated by redox-switch modulation via the reversible disulfide bond formation to respond the environmental redox changes. We also measured the kinetic parameters of CgDapF and the Km and kcat values of CgDapF were 1.86 mM and 58 sec-1, respectively. Comparing with the kinetic parameters of other DapFs29,30, the enzyme activity of CgDapF seems to be somewhat lower than other DapFs.
Figure 5. Structural comparison of CgDapF with other DapFs.
(a) Superimposition of the oxidized form of DapF structures. Structures of the oxidized forms of CgDapF, MtDapF, and HiDapF are superimposed and shown as a cartoon diagram. CgDapF, MtDapF, and HiDapF are distinguished with different colors of gray, salmon, and green, respectively. The disulfide bond are presented as a stick model and labeled. (b) Comparison of the DC-loops of DapFs. Structures of CgDapF, MtDapF, and HiDapF are superposed and shown with a color scheme same in (a). (c) Disulfide bond-mediated conformational change of DC-loop of HiDapF. The DC-loops in the oxidized form and the reduced form are presented as a cartoon diagram with green and yellow colors, respectively. Two catalytic cysteine residues involved in the disulfide bond formation are shown as a stick model. (d,e) A surface model of the active site of MtDapF and HiDapF. The MtDapF (d) and HiDapF (e) structures in oxidized form are presented with a surface model. The DC-loops in MtDapF and HiDapF are distinguished with colors of salmon and green, respectively. (f) Superimposition of the reduced form of DapF structures. Structures of the reduced form of CgDapF, BaDapF, and EcDapF are superimposed and shown as a cartoon diagram. CgDapF, BaDapF, and EcDapF are distinguished with different colors of gray, purple, and lime green, respectively. The catalytic cysteine residues are presented as a stick model and labeled. (g) Comparison of the DC-loop of DapFs. Structures of CgDapF, BaDapF, and EcDapF are superposed and shown with a color scheme same in (f). (h,i) A surface model of the active site of BaDapF and EcDapF. The BaDapF (h) and EcDapF (i) structures in reduced form are presented with a surface model. DC-loops in BaDapF and EcDapF are distinguished with colors of purple and lime green, respectively.
Substrate binding mode of CgDapF
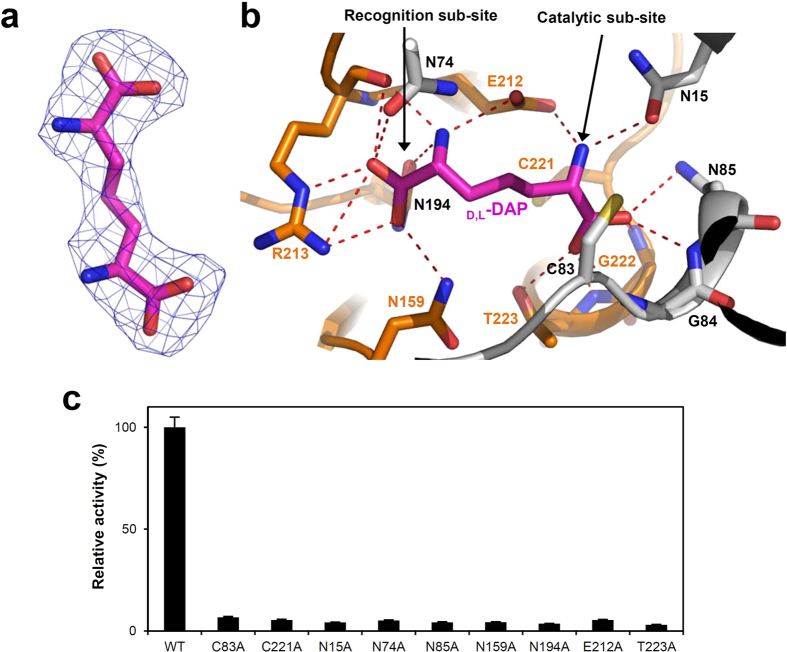
To investigate the substrate binding mode of CgDapF, we determined the crystal structure of the protein in complex with the d,l-DAP product (Table 1). In fact, although we added a mixture of the three DAP enantiomers (l,l-DAP, d,d-DAP, d,l-DAP) into the crystallization solution, we only observed density consistent with d,l-DAP in the electron density map (Fig. 6a). We suspect that the l,l-DAP substrate was converted to the d,l-DAP product during the crystallization procedure because we used the wild-type protein. The substrate binding site is located at a deep cleft formed between the two domains. It can be divided into two sub-sites, a catalytic sub-site and a recognition sub-site, where the d- and l-amino moieties of d,l-DAP are positioned, respectively. The two catalytic cysteine residues are located at the catalytic sub-site, and the amino group in the d-amino moiety is hydrogen bonded to the carboxyl group of Glu212 (Fig. 6b). The carboxyl group of d,l-DAP on the side bearing the d-amino moiety, is stabilized through hydrogen bonds with the main chain nitrogen atoms of Gly84, Asn85, Gly222, and Thr223, and the side chains of Asn85 and Thr223 (Fig. 6b). In the recognition sub-site, the carboxyl group of d,l-DAP on the side bearing the l-amino moiety is stabilized by direct interactions with the main chain of Arg213 and the side chains of Asn74, Asn159, Asn194, and Arg213 (Fig. 6b). The l-amino group of d,l-DAP forms hydrogen bonds with the main chain oxygen atom of Arg213 and side chain of Glu212 (Fig. 6b). In our structure, the stabilization mode of l-amino moiety of d,l-DAP provides a clear explanation of the means by which the enzyme utilizes l,l-DAP as a substrate.
Figure 6. Substrate binding mode of CgDapF.
(a) Electron density map of d,l-DAP. The Fo-Fc electron density map of the bound d,l-DAP is shown as a blue mesh and contoured at 3.5 σ. (b) Substrate binding mode of CgDapF. The bound d,l-DAP is presented in a stick model with a magenta color and labeled. NTD and CTD are distinguished by white and orange colors. Residues involved in the d,l-DAP stabilization are shown as stick models. Hydrogen bonds formed between d,l-DAP and neighboring residues are presented with red dotted lines. (c) Site directed mutagenesis of CgDapF. Residues involved in substrate binding are replaced by alanine residues. The relative activity of recombinant mutant proteins are measured and compared with that of wild-type CgDapF. Each experiment was performed in triplicate.
Based on our CgDapF structure in complex with the d,l-DAP product, we performed site-directed mutagenesis experiments to verify residues inferred to be involved in catalysis and substrate binding. We compared the enzyme activities of the resulting mutants to the activity of the wild-type protein. To test their importance as catalytic residues, Cys83 and Cys221 were mutated to alanine; the C83A and C221A mutants almost completely lost their activity (Fig. 6c). This suggests that CgDapF uses both of these residues for catalysis, and the enzymatic reaction mechanism is similar to that of other DapF proteins. In addition, we mutated residues structurally inferred to be involved in the stabilization of the substrate to alanine, generating N15A, N74A, N85A, N159A, N194A, E212A, R213A, and T223A. As anticipated, the enzymatic activities of all these mutants were almost completely abolished, as compared with that of the wild-type enzyme (Fig. 6c).
Domain movement in CgDapF
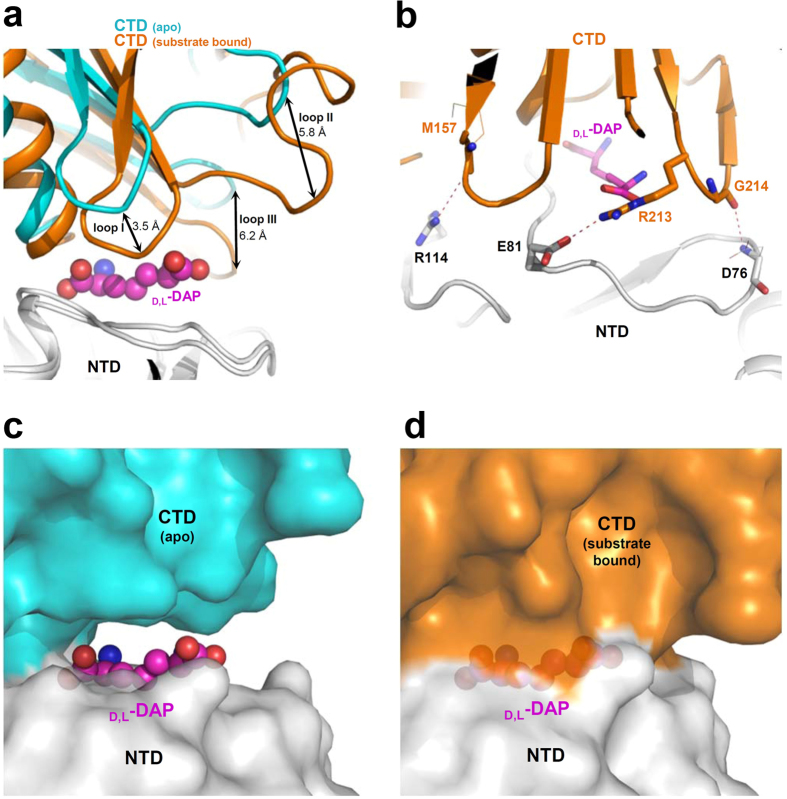
When we superimposed the apo-structure of CgDapF with that of the complex with d,l-DAP, it became evident that substrate binding had induced the CTD to move towards the NTD (Fig. 7a). In particular, three loops (Loop I: Met157–Asn159; Loop II: Met177–Val193; Loop III: Arg213–Gly214) move by 3.5 Å, 5.8 Å, and 6.2 Å, respectively (Fig. 7a). For the stabilization of reorganized CTD, residues from NTD interact with the CTD through hydrogen bonds: Met157 from Loop I, and Arg213 and Gly214 from Loop III interact with the Arg114, Glu71 and Asp76 of the NTD, respectively (Fig. 7b). In addition, the interactions between the reorganized CTD and the residues located at the entrance of the active site cleft result in a completely buried substrate binding site (Fig. 7c,d). These structural observations clearly show that CgDapF undergoes an open/closed conformational change during the entrance of the substrate and the release of the product.
Figure 7. Domain movement of CgDapF.
(a) Domain movement of CgDapF. CgDapF structure in the apo-form and in complex with D,L-DAP are superimposed in a cartoon diagram. In the apo-form, the NTD is shown in white and CTD in cyan colors. In the complex structure, NTD is also presented in a white and CTD in an orange color. The d,l-DAP product is shown as a sphere in a magenta color. Three loops and the distances between structures are labeled. (b) Stabilization mode of the reorganized CTD in CgDapF in complex with d,l-DAP. CgDapF is shown as a cartoon diagram with an orange color. Residues involved in the inter-domain interactions are presented as a stick model and labeled. Side-chains involved in the stabilization of the reorganized CTD are shown as stick models. Main-chains involved in the stabilization the reorganized CTD are shown as stick models and their side-chains are as line models. Hydrogen bonds are presented as red-colored dotted lines. (c,d) Active site closure of CgDapF. CgDapF structures in the apo-form and in complex with d,l-DAP are presented with a surface model with same color scheme in (a).
In this report, we have revealed that the function of CgDapF is regulated by redox-switch modulation via the reversible disulfide bond formation in the response to the environmental redox conditions. This reversible disulfide bond formation also induces structural change in the DC-loop of the active site. Under oxidized condition, the position of the DC-loop occludes the active site, which inactivates the enzyme. However, under reduced condition, the DC-loop undergoes a conformational change that opens the active site, restoring enzymatic activity. Because redox-switch modulation is one of the key regulatory mechanisms used to control the function of enzymes in response to changes in the environmental redox state, we suspect that cellular redox changes might significantly influence l-lysine biosynthesis. Further investigations of the relationship between the cellular redox state and l-lysine biosynthesis in C. glutamicum seem to be needed.
Methods
Production of CgDapF
The CgDapF gene was amplified by polymerase chain reaction (PCR) using genomic DNA from C. glutamicum strain ATCC 13032 as a template. The PCR product was then subcloned into pET30a (Life Science Research), and the resulting expression vector pET30a:CgdapF was transformed into the E. coli strain B834(DE3), which was grown in 1 L of LB medium containing kanamycin at 37 °C. After induction by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), the culture medium was maintained for a further 20 h at 18 °C. The culture was then harvested by centrifugation at 4,000 × g for 20 min at 4 °C. The cell pellet was resuspended in buffer A (40 mM Tris-HCl, pH 8.0) and then disrupted by ultrasonication. The cell debris was removed by centrifugation at 13,500 × g for 30 min and the lysate was applied to an Ni-NTA agarose column (Qiagen). After washing with buffer A containing 30 mM imidazole, the bound proteins were eluted with 300 mM imidazole in buffer A. Finally, trace amounts of contaminants were removed by size-exclusion chromatography by using a Superdex 200 prep-grade column (320 mL, GE Healthcare) equilibrated with buffer A. All purification experiments were performed at 4 °C and SDS-polyacrylamide gel electrophoresis analysis of the purified proteins showed a single polypeptide of 29.9 kDa that corresponded to the estimated molecular weight of the CgDapF monomer. The purified protein was concentrated to 40 mg/mL in 40 mM Tris-HCl, pH 8.0. Site-directed mutagenesis experiments were performed using the Quick Change site-directed mutagenesis kit (Stratagene). The production and purification of the CgDapF mutants were carried out by the same procedure employed for the wild-type protein.
Crystallization of CgDapF
Crystallization of the purified protein was initially performed with commercially available sparse-matrix screens from Rigaku and Molecular Dimensions by using the hanging-drop vapor-diffusion method at 20 °C. Each experiment consisted of mixing 1.0 μL protein solution (40 mg/ml in 40 mM Tris-HCl, pH 8.0) with 1.0 μL reservoir solution and then equilibrating this against 500 μL reservoir solution. The oxidized form of CgDapF crystals were observed in several crystallization screening conditions. After several optimization steps, the best quality crystals appeared in 1.6 M Ammonium sulfate and 0.1 M Bis-Tris, pH 5.0. The reduced form of CgDapF crystals were crystallized in the condition containing 1.4 M Sodium phosphate monobasic/0.9 M Potassium phosphate dibasic, 0.1 M CAPS, pH 10.0, 0.2 M Lithium sulfate, and 1 mM 1,4-Dithiothreitol (DTT). The crystals of CgDapF in complex with D,L-DAP were crystallized in the condition of 1.3 M Sodium citrate and 0.1 M CHES, pH 9.0 and 10 mM DAP isomer.
Data collection and structure determination of CgDapF
The crystals were transferred to cryoprotectant solution composed of the corresponding conditions described above and 30% (v/v) glycerol, fished out with a loop larger than the crystals, and flash-frozen by immersion in liquid nitrogen. All data were collected at the 7 A beamline of the Pohang Accelerator Laboratory (PAL, Pohang, Korea), using a Quantum 270 CCD detector (ADSC, USA). The oxidized form and reduced form of CgDapF crystals diffracted to a resolution 2.3 and 2.0 Å, respectively. The crystals of CgDapF in complex with D,L-DAP diffracted to 2.6 Å resolutions. All data were indexed, integrated, and scaled together using the HKL-2000 software package31. The oxidized form of CgDapF belonged to the space group P32 with the unit cell parameters a = b = 143.584 Å, c = 122.371 Å, α = β = 90.0°, and γ = 120.0°. In order to solve the phase problem for the crystal, we also prepared crystals from a SeMet derivative. SeMet-substituted oxidized form crystals were obtained using the same crystallization condition as used for the native protein crystal. The crystals of SeMet derivative diffracted to a resolution 2.0 Å and belonged to the space group I222 with the unit cell parameters a = 101.74 Å, b = 119.08 Å, c = 155.59 Å, and α = β = γ = 90°. Assuming two molecules of oxidized form of CgDapF (29.2 kDa) per asymmetric unit, the crystal volume per unit of protein mass was approximately 3.94 Å3 Da−1, which corresponded to a solvent content of 68.80%32. The reduced form of CgDapF crystals that substituted with SeMet belonged to the space group I222 with unit cell parameters a = 101.76, b = 118.94 Å, c = 153.33 Å, α = β = γ = 90.0°. With two molecules of CgDapF per asymmetric unit, the crystal volume per unit of protein mass was 3.88 Å3 Da−1, which corresponded to a solvent content of 68.31%32. The crystals of CgDapF in complex with D,L-DAP belonged to space group P4332, with unit cell parameters of parameters a = b = c = 155.7 Å, and α = β = γ = 90.0°. With one molecule of CgDapF in complex with D,L-DAP in an asymmetric unit, the crystal volume per unit of protein mass was 2.63 Å3 Da−1, indicating that the solvent content was approximately 53.26%32. The crystal structures of SeMet-substituted CgDapF in oxidized and reduced form were solved by single-wavelength anomalous dispersion (SAD) using data collected at a determined wavelength (0.97934 Å). Experimentally determined f” and f”’ values by using SOLVE/RESOLVE33. The initial model build was automatically performed using ARP/wARP34, and the final model was built by using the program Wincoot35. The structure of CgDapF in complex with D,L-DAP was determined by molecular replacement with the CCP4 version of MOLREP36 using the refined CgDapF in reduced form structure. Model building was performed manually using the program WinCoot35, and refinement was performed with CCP4 refmac537. The data statistics are summarized in Table 1. The refined models of oxidized and reduced form of CgDapF and CgDapF in complex with D,L-DAP were deposited in the Protein Data Bank with PDB codes of 5H2G, 5H2Y, and 5M47, respectively.
DapF activity assay
The activity of CgDapF was measured using the coupled enzyme assay with DAP decarboxylase from C. glutamicum (CgLysA). The D,L-DAP is generated by the reaction of CgDapF using L,L-DAP and then CgLysA immediately converts D,L-DAP into L-lysine. The production of l-lysine was detected using the lysine oxidase/peroxidase method. Lysine oxidase converts the remaining l-lysine into 6-amino-2-oxohexanoate, NH3, and H2O2, and the H2O2 is then reduced by peroxidase using 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS). Immediately after the addition of CgDapF enzyme and CgLysA into the reaction mixture, equal volume of 2x lysine oxidase/peroxidase solution (0.1 unit ml−1 lysine oxidase, 1 unit ml−1 peroxidase, and 3.6 mM ABTS in 0.1 M potassium phosphate buffer, pH 8.0) was added to the reaction mixture. The amount of oxidized ABTS was detected by measuring absorbance at 412 nm. Activity assays were performed at room temperature and the reaction mixture contains 0.1 M potassium phosphate, pH 8.0, 42.19 μM CgLysA, various concentrations of L,L-DAP, and 3.45 μM purified CgDapF enzyme.
To examine the susceptibility of CgDapF to hydrogen peroxide (H2O2), the CgDapF protein was treated with various concentrations of H2O2 for 1 h and enzyme-buffer assay mixture was added. To switch the redox condition to the reduced state, the protein solutions treated with various concentrations of H2O2 were thoroughly dialyzed against the buffer without H2O2 and 10 mM of DTT was added to the protein samples.
UV/Vis spectroscopy
To quantify the free thiol group, the protein samples were incubated with 5,5-dithiobis(2-nitrobenzoic acid) (DTNB) and the UV/Vis absorbance spectroscopy was performed. In the presence of thiol group compound, the colorless 5,5-dithiobis(2-nitrobenzoic acid) (DTNB) is converted to yellow 2-Nitro-5-mercaptobenzoic acid (TNB), which shows an absorption maximum at 409 nm. The purified CgDapF protein was pre-incubated with various concentration of H2O2 for 1 h and then added into the reaction mixture. To switch the environment to reduced condition, 10 mM DTT was added to H2O2-treated proteins and incubated for 30 min. The reaction mixture contains 0.1 M potassium phosphate, pH 8.0, 10 μM DTNB, and 6.85 μM purified CgDapF enzyme. All incubations and reactions were performed at room temperature.
Circular Dichroism spectroscopy
CD measurements were carried out with a Jasco J-815 CD Spectropolarimeter. Far-UV CD spectra were collected between 190 and 260 nm with a scan speed 20 nm min−1. Spectra of the CgDapF enzymes (33.44 μM in 10 mM potassium phosphate buffer, pH 8.0) reduced by 1 mM DTT or oxidized by 1 mM H2O2 were recorded at 20 °C in a quartz cuvette (0.2-mm pathlength) with a bandwidth 1 nm. The data were expressed in mean residue ellipticity [θ] in degree cm2 dmol−1.
Additional Information
How to cite this article: Sagong, H.-Y. and Kim, K.-J. Structural basis for redox sensitivity in Corynebacterium glutamicum diaminopimelate epimerase: an enzyme involved in l-lysine biosynthesis. Sci. Rep. 7, 42318; doi: 10.1038/srep42318 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
This work was supported by a National Research Foundation of Korea (NRF) grant by C1 Gas Refinery Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2016M3D3A1A01913269), and by the Advanced Biomass R&D Center (ABC) of Global Frontier Project funded by the MEST (NRF-2015M3A6A2053895).
Footnotes
The authors declare no competing financial interests.
Author Contributions K.-J.K. designed the project. H.-Y.S. performed the experiments. H.-Y.S. and K.-J.K. wrote the paper.
References
- Kinoshita S., Udaka S. & Shimono M. Studies on the amino acid fermentation. Part 1. Production of L-glutamic acid by various microorganisms. The Journal of general and applied microbiology 50, 331–343 (2004). [PubMed] [Google Scholar]
- Ikeda M. & Takeno S. In Corynebacterium glutamicum 107–147 (Springer, 2013). [Google Scholar]
- Mahmood Z. A. Microbial amino acids production. Microbial Biotechnology: Progress and Trends, 187 (2014). [Google Scholar]
- Pavelka M. & Jacobs W. R. Biosynthesis of diaminopimelate, the precursor of lysine and a component of peptidoglycan, is an essential function of Mycobacterium smegmatis. Journal of bacteriology 178, 6496–6507 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. J. The DAP pathway to lysine as a target for antimicrobial agents. Natural product reports 13, 29–43 (1996). [DOI] [PubMed] [Google Scholar]
- Anishetty S., Pulimi M. & Pennathur G. Potential drug targets in Mycobacterium tuberculosis through metabolic pathway analysis. Computational biology and chemistry 29, 368–378 (2005). [DOI] [PubMed] [Google Scholar]
- Yeh P., Sicard A. M. & Sinskey A. J. General organization of the genes specifically involved in the diaminopimelate-lysine biosynthetic pathway of Corynebacterium glutamicum. Molecular and General Genetics MGG 212, 105–111 (1988). [DOI] [PubMed] [Google Scholar]
- Schrumpf B. et al. A functionally split pathway for lysine synthesis in Corynebacterium glutamicium. Journal of bacteriology 173, 4510–4516 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogovski C. et al. Enzymology of bacterial lysine biosynthesis. (InTech Open Access Publisher, 2012). [Google Scholar]
- Sagong H.-Y. & Kim K.-J. Structural Insight into Dihydrodipicolinate Reductase from Corybebacterium glutamicum for Lysine Biosynthesis. Journal of microbiology and biotechnology 26, 226–232 (2016). [DOI] [PubMed] [Google Scholar]
- Purich D. L. Advances in Enzymology and Related Areas of Molecular Biology, Amino Acid Metabolism. Vol. 72 (John Wiley & Sons, 2009). [Google Scholar]
- Dogovski C. et al. Lysine biosynthesis in bacteria: an unchartered pathway for novel antibiotic design. Encyclopedia of life support systems 11, 116–136 (2009). [Google Scholar]
- McCoy A. J. et al. L, L-diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proceedings of the National Academy of Sciences 103, 17909–17914 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J. et al. L,L-diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proceedings of the National Academy of Sciences of the United States of America 103, 17909–17914, doi: 10.1073/pnas.0608643103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag K., Eggeling L., Degraaf A. A. & Sahm H. Flux Partitioning in the Split Pathway of Lysine Synthesis in Corynebacterium-Glutamicum Quantification by C-13-Nmr and H-1-Nmr Spectroscopy. Eur J Biochem 213, 1325–1331, doi: 10.1111/j.1432-1033.1993.tb17884.x (1993). [DOI] [PubMed] [Google Scholar]
- Cirilli M., Zheng R., Scapin G. & Blanchard J. S. Structural symmetry: the three-dimensional structure of Haemophilus influenzae diaminopimelate epimerase. Biochemistry 37, 16452–16458 (1998). [DOI] [PubMed] [Google Scholar]
- Koo C. W. & Blanchard J. S. Chemical mechanism of Haemophilus influenzae diaminopimelate epimerase. Biochemistry 38, 4416–4422 (1999). [DOI] [PubMed] [Google Scholar]
- Lloyd A. J., Huyton T., Turkenburg J. & Roper D. I. Refinement of Haemophilus influenzae diaminopimelic acid epimerase (DapF) at 1.75 Å resolution suggests a mechanism for stereocontrol during catalysis. Acta Crystallographica Section D: Biological Crystallography 60, 397–400 (2004). [DOI] [PubMed] [Google Scholar]
- Pillai B. et al. Structural insights into stereochemical inversion by diaminopimelate epimerase: an antibacterial drug target. Proceedings of the National Academy of Sciences 103, 8668–8673 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo C. W., Sutherland A., Vederas J. C. & Blanchard J. S. Identification of active site cysteine residues that function as general bases: diaminopimelate epimerase. Journal of the American Chemical Society 122, 6122–6123 (2000). [Google Scholar]
- Pillai B. et al. Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase. Biochemical and biophysical research communications 363, 547–553 (2007). [DOI] [PubMed] [Google Scholar]
- Usha V., Dover L. G., Roper D. I., Fuetterer K. & Besra G. S. Structure of the diaminopimelate epimerase DapF from Mycobacterium tuberculosis. Acta Crystallographica Section D: Biological Crystallography 65, 383–387 (2009). [DOI] [PubMed] [Google Scholar]
- Hor L. et al. Dimerization of bacterial diaminopimelate epimerase is essential for catalysis. Journal of Biological Chemistry 288, 9238–9248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai B. et al. Crystal structure of diaminopimelate epimerase from Arabidopsis thaliana, an amino acid racemase critical for L-lysine biosynthesis. Journal of molecular biology 385, 580–594 (2009). [DOI] [PubMed] [Google Scholar]
- Krissinel E. & Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of molecular biology 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- Barford D. The role of cysteine residues as redox-sensitive regulatory switches. Current opinion in structural biology 14, 679–686 (2004). [DOI] [PubMed] [Google Scholar]
- Choi H.-J. et al. Structural basis of the redox switch in the OxyR transcription factor. Cell 105, 103–113 (2001). [DOI] [PubMed] [Google Scholar]
- Ilbert M. et al. The redox-switch domain of Hsp33 functions as dual stress sensor. Nature structural & molecular biology 14, 556–563 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman J. S. & Nichols J. S. Purification and properties of diaminopimelic acid epimerase from Escherichia coli. Journal of Biological Chemistry 259, 8907–8914 (1984). [PubMed] [Google Scholar]
- Stenta M. et al. Catalytic mechanism of diaminopimelate epimerase: a QM/MM investigation. Journal of chemical theory and computation 5, 1915–1930 (2009). [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. & W. C. C. Jr. Processing of X-ray diffraction data collected in oscillation mode. (1997). [DOI] [PubMed] [Google Scholar]
- Matthews B. W. Solvent content of protein crystals. Journal of molecular biology 33, 491–497 (1968). [DOI] [PubMed] [Google Scholar]
- Wang J. et al. SAD phasing by combination of direct methods with the SOLVE/RESOLVE procedure. Acta Crystallographica Section D: Biological Crystallography 60, 1244–1253 (2004). [DOI] [PubMed] [Google Scholar]
- Langer G., Cohen S. X., Lamzin V. S. & Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nature protocols 3, 1171–1179 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P. & Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D: Biological Crystallography 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- Vagin A. & Teplyakov A. Molecular replacement with MOLREP. Acta Crystallographica Section D: Biological Crystallography 66, 22–25 (2009). [DOI] [PubMed] [Google Scholar]
- Murshudov G. N., Vagin A. A. & Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D: Biological Crystallography 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]