Abstract
Synovial sarcoma (SS) is an aggressive soft tissue sarcoma (STS) that typically occurs in the extremities near a joint. Metastatic disease is common and usually occurs in the lungs and lymph nodes. Surgical management is the mainstay of treatment with chemotherapy and radiation typically used as adjuvant treatment. Although chemotherapy has a positive impact on survival, the prognosis is poor if metastatic disease occurs. The biology of sarcoma invasion and metastasis remain poorly understood. Chromosomal translocation with fusion of the SYT and SSX genes has been described and is currently used as a diagnostic marker, although the full impact of the fusion is unknown. Multiple biomarkers have been found to be associated with SS and are currently under investigation regarding their pathways and mechanisms of action. Further research is needed in order to develop better diagnostic screening tools and understanding of tumor behavior. Development of targeted therapies that reduce metastatic events in SS, would dramatically improve patient prognosis.
Keywords: SYT-SSX, CDCA2, KIF14, IGFBP7, Secernin-1, E2H2, MMPs, NY-ESO-1, CXCR4
Background
Synovial sarcoma (SS) is the fourth most common type of soft tissue sarcoma (STS) and accounts for 5–10% of all soft tissue sarcomas [1]. SS was originally described by Simon in 1865, and given the name “synovial sarcoma” by Sabrazes et al. in 1934 based on a similar appearance to developing synovial tissue under light microscopy [2]. SS has a tendency to arise in the soft tissue surrounding larger joints, but it does not have a synovial cell origin [3]. While the exact cellular origin is an ongoing topic of investigation, it is likely to arise from undifferentiated mesenchymal stem cells [4]. Compared with other soft tissue sarcomas, SS occurs predominantly in younger adults with a median age of diagnosis of 35 years [5]. The most common tumor location is in the extremities, where approximately 70% of these tumors develop [6], and these patients have significantly better long term survival outcomes than those with non-extremity involvement [7]. In patients with localized disease, 10 year survival outcomes vary from 8 to 88% depending on tumor size and location [8]. Standard treatment for synovial sarcoma is tumor resection frequently accompanied by radiotherapy and/or chemotherapy, although some data suggests that therapies in addition to surgery substantially increase long-term metastatic risks [7].
Metastasis negatively impacts patient prognosis and significantly reduces survival outcomes [9]. For patients who present with or develop metastatic tumors median survival outcomes vary from 7 to 37 months depending on lymph node involvement and metastatic location [10]. SS can evolve slowly and there is a high incidence of late metastasis which occurs in about 50% of all cases [7]. Most metastatic tumors develop in the lungs (80%), although bone (9.9%) and liver (4.5%) are the next most frequent locations [9]. Synovial sarcoma can metastasize through the lymph nodes with clinically detectable disease found in 15–20% of newly diagnosed patients [11]. Histologically, synovial sarcoma is classified into four subtypes consisting of a biphasic (BPSS), monophasic fibrous or monophasic epithelial (MPSS), and poorly differentiated (PDSS, round cell) tumor cells [1]. While these histological subtypes do not seem to be associated with metastatic events [7, 12], they have been linked to survival. A study of 3756 SS patients registered in the National Cancer Data Base from 1998 to 2010 showed a significant difference in average 5-year survival numbers among patients with biphasic (65%), monophasic (56%) and undifferentiated (52%) tumors [13].
While many factors that influence synovial sarcoma patient outcome have been identified, tumor behavior remains highly unpredictable [14]. An extremely high level of metastasis means that further studies are needed to characterize the mechanisms that influence tumor action. Ultimately, the identification of highly relevant molecular biomarkers associated with metastatic outcomes and patient prognosis establishes the foundation for development of better therapeutic strategies to target these oncogenic factors. Several metastatic biomarkers that have been described to date are described in the following sections and summarized in Fig. 1.
Fig. 1.
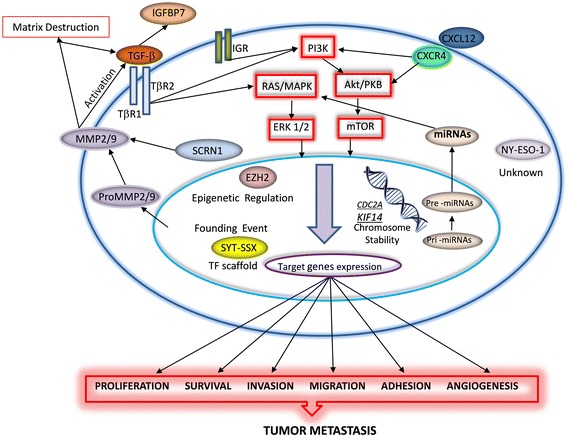
Schematic diagram summarizing functional relevance of metastatic signals in synovial sarcoma. SYT-SSX fusion is a founding event in the development of this cancer which frequently results in the production of molecular signals that promote tumor metastasis. Abbreviations: Akt (Serine/threonine kinase); CDCA2 (Cell division cycle A2); CXCR (Chemokine receptor); ERK1/2 (Extracellular signal-regulated kinase 1/2); EZH2 (Enhancer of zeste homologue 2); IGFBP7 (Insulin-like growth factor-binding protein-7); IGR (Insulin like growth factor receptor); KIF14 (Kinesin family member 14); MAPK (Mitogen-activated protein kinases); MicroRNAs (miRNAs); MMPs (Matrix metalloproteinases); mTOR (Mammalian target of rapamycin); NY-ESO-1 (New York esophageal squamous cell carcinoma 1); PI3K (Phosphatidylinositol-3-kinase); pri-miRNA (primary microRNA); pre-miRNA (precursor microRNA); PKB (Protein kinase B); RAS (Ras GTPase); SCRN1 (Secernin-1); TGF-β (Transforming growth factor beta); TβR (TGF-beta receptor); TF (transcription factor)
Significance of the syt-ssx fusion gene
The chromosomal translocation t(X;18)(p11.2;q11.2) fuses the SS18 (SYT) gene to the SSX gene (predominantly SSX1 or SSX2) and is regarded as a founding event in the oncogenic development of synovial sarcoma [15]. Yet, the exact transformative event of the chimeric SYT-SSX gene product has not been fully elucidated. It has been shown that SYT-SSX can interfere with assembly of BAF (BRG-/BRM-associated factor) complexes affecting the integration of a tumor suppressor component and consequent SRY (sex-determining region Y)-box 2 (SOX2) activation which stimulates cell proliferation [16]. Over 95% of SS can be characterized by expression of the SYT-SSX gene and it is used as a routine diagnostic marker for this type of cancer [15]. Although both SYT and SSX proteins contain transcriptional regulation domains, they lack any DNA binding regions, so their regulatory effects are surmised to occur through interactions with other proteins [17]. Accordingly, the SYT-SSX fusion oncogene has been shown to interact with several major epigenetic regulators as well as other DNA binding proteins and exert both direct and indirect effects on transcript regulation [17, 18]. Of note, SYT-SSX has been shown to act as a scaffold linking two master transcription regulators TLE1 (transducin like enhancer of split 1) and ATF2 (activating transcription factor 2) such that TLE1 acts as a repressor of ATF2 target genes regulating cell cycle, apoptosis, and more, demonstrating that the SYT-SSX/TLE1/ATF2 complex is important not only to oncogenic transformation but also tumor cell survival [19]. TLE has emerged as a useful diagnostic marker of SS with robust expression detected in approximately 95% of these tumors however a low level of positive staining in non-SS tumors indicates that this antibody should only be used in a panel with other antibodies to confirm diagnosis [20]. Recently, a highly specific assay to detect the association of SYT-SSX and TLE in either localized or metastatic SS tumors was developed and shown to be useful in drug discovery assays seeking to disrupt this interaction [21].
A number of studies have investigated the association between SYT-SSX gene fusion type and metastatic risk. Some have reported that the SYT-SSX1 fusion approximately doubles the risk of metastatic tumor development compared with SYT-SSX2 [22, 23]. Intriguingly, a similar pattern was evident in a survival study demonstrating a median survival of 6.1 years for SS patients with an SYT-SSX1 fusion gene compared with 13.7 years for those with SYT-SSX2 [24]. In contrast, several other studies report no significant associations between SYT-SSX fusion type and metastasis or survival [7, 25]. These conflicting data appear to suggest that the influence of unspecified additional factors have a much larger role in determining metastatic outcomes. These clinicopathological characteristics must be better defined to understand in which context SYT-SSX has an important versus an inconsequential role.
Circulating biomarkers
Development of a diagnostic blood test to detect SS and assess metastatic risk would be a valuable tool for the management of this disease. A whole blood microRNA signature shows that SS patients demonstrate significant upregulation of seven microRNAs (miR-99a-5p, miR-146b-5p, miR-148b-3p, miR-195-5p, miR-223-3p, miR-500b-3p and miR-505-3p) compared with patients in remission and healthy controls [26]. Furthermore, expression levels of these seven microRNAs are significantly reduced by 5–50 fold increments after local tumor resection, but once again dramatically upregulated if a patient develops recurrent local or metastatic disease [26]. Thus, since a clearly defined microRNA expression panel can be used to distinguish SS from other malignancies as well as characterize metastatic and recurrent tumor status, this method could be useful in therapeutic monitoring of patients [26]. miRNAs have a critical role in activation of the Ras GTPase/Mitogen-activated protein kinases (Ras/MAPK) pathway necessary for tumor development [27], and miRNA suppression significantly inhibits SS cell proliferation in vitro [28].
Detection of a circulating SYT-SSX fusion gene product could be another important method for assessing metastatic risk in patients with SS. In culture, it has been shown that SS cells produce microvesicles containing the SYT-SSX fusion gene, yet this research group was unable to detect the biomarker in microvesicles or PBMCs (peripheral blood mononuclear cell) obtained from patient blood samples regardless of their metastatic status [29]. In contrast, the SYT-SSX gene was detected in peripheral blood in a case study of a single SS patient who developed multiple lung metastasis 2 months after local tumor resection from the thigh [30]. The presence of circulating tumor cells (CTC) could be indicative of a patient’s metastatic risk given their potential to extravasate and form tumors in new locations [31], and CTC abundance may account for discrepancies in SYT-SSX detection abilities. CTCs can be readily isolated from patient blood by size exclusion, and CTC quantities vary considerably among different SS patients [32].
Cinsarc signature genes
Gene expression profiling of soft tissue sarcomas revealed a prognostic panel of 67 genes (CINSARC, complexity index in sarcomas) with functional roles in mitosis and chromosome management that are also highly predictive of metastatic risk [33]. While many CINSARC genes have been identified as molecular markers associated with metastasis in other types of cancers, they are usually ascribed a proliferative function, although they may actually have a greater role in chromosome instability [33]. When the CINSARC classification criteria were used to stratify SS tumor specimens into two prognostic groups, a highly significant difference was observed in metastatic outcomes among these patients [34]. However, since the CINSARC profile emerged from an analysis of multiple types of soft tissue sarcomas, these authors sought to determine if a better metastatic prognostic profile could be developed for SS [34]. Comparison of whole genome expression in 51 metastatic and 49 non-metastatic SS tumors revealed significant upregulation of 59 genes in metastatic specimens, 24% of which were common to the CINSARC classification panel [34]. Importantly, singular expression of the 2 most differentially regulated genes, CDCA2 (cell division cycle A2) and KIF14 (kinesin family member 14), could better predict metastatic outcomes than the overall CINSARC score in this patient cohort [34]. Functionally, both of these genes code for proteins that help to maintain chromosome integrity and are essential for the completion of cytokinesis. KIF14 is localizes to the central spindle during late phase mitosis and its inhibition in tumor cells results in cell cycle arrest and the formation of binucleated cells [35]. KIF14 was also found to regulate adhesive components on the tumor cell surface influencing migratory and invasive properties that promote cell motility during metastasis [36]. CDCA2 has critical catalytic and structural functions during late mitosis that help coordinate chromosome segregation and nuclear envelope reformation after division of nuclear contents [37].
IGFBP7
Insulin-like growth factor-binding protein-7 (IGFBP7,) also termed IGFBP-related protein-1 (IGFBP-rP1), is a secreted 31-kDa protein belonging to the IGFBP family [38]. In various cancer types including hepatocellular carcinoma [39], breast [40], brain [41], and colon [42], IGFBP7 can function as a tumor suppressor and have the ability to suppress proliferation, adhesion, angiogenesis, survival, or induce apoptosis and senescence [43–45]. Yet the role of this protein in tumor behavior is complex, as it undergoes extensive proteolytic processing that reverses its cellular function and influence over cell proliferation and adhesion activities [46]. In the tumor stroma microenvironment, IGFBP7 expression is closely related with the transforming growth factor beta (TGF-β secretion [47], which is a potent factor promoting tumor cell invasiveness and metastasis [48, 49]. TGF-β upregulates expression of IGFBP7 and angiogenic capacity of tumor cells [50]. Recently, it was shown that IGFBP7 expression in SS was higher that other types of STSs and significantly associated with metastatic events [51]. In addition, nuclear expression of another IGFBP family protein, insulin like growth factor 1 receptor (IGF1R),, was significantly related to poor survival in SS patients who did not receive adjuvant chemotherapy [52]. In sarcoma, IGF1R activation is known to activate the phosphatidylinositol-3-kinase/serine/threonine kinase Akt/Mammalian target of rapamycin (PI3K/Akt/mTOR) pathway which promotes cancer progression and metastasis [53]. These studies, highlight the potential for proteins from the IGF family to be used as prognostic biomarkers to help guide treatment decisions in SS.
MMPs
The matrix metalloproteinases (MMPs), a family of zinc and calcium dependent proteolytic enzymes, are involved in the degradation extracellular matrix (ECM) components and play key roles in tumor cell invasion and metastasis [54]. Notably, the proteolytic activity of MMPs can help release inactive TGFB in the extracellular space so that it can bind to its receptors and activate downstream pathways such as PI3k/Akt and MAPK which are critical to the epithelial mesenchymal transition (EMT) process underlying metastasis [55]. Benassi et al. demonstrated that high levels of MMP2 and MMP9 in biopsied tissue from patients with SS, was significantly associated with metastasis (P = 0.008 and P = 0.005, respectively) [56]. In addition, lack of expression the tissue inhibitors of metalloproteinases (TIMP) was found to be a poor prognostic factor for disease-free survival in synovial sarcoma (P = 0.009) [56]. The proteolytic activity of MMPs and their activation process can be inhibited by the TIMPs [57]. The presence of TIMPs can suppress metastasis by preserving ECM integrity [58]. It has been shown that a decrease in TIMP-1 correlated with a poor outcome in high-risk STS [59] and colorectal cancer patients [60]. Both MMPs and TIMPs can further be evaluated as biological markers for predicting progression, metastasis and prognosis of human SS.
Secernin-1
Secernin-1 (SCRN1), a 50-kDa cytosolic protein, is a member of the secernin gene family that regulates exocytosis in mast cells through a mechanism that has not been well defined [61]. Exocytosis is a process by which cells transport and release secretory products through the cytoplasm to the cell membrane and several studies have described that this promotes tumor growth, metastasis and invasion [62, 63]. In colon cancer, SCRN1 expression promoted exocytosis secretion of MMP2 and MMP, while silencing this gene reduced MMP2 secretion, inhibited cell proliferation and decreased invasion capability [63]. The poor prognostic significance of SCRN1 expression in colon cancer [63, 64], is contrary to that reported in synovial sarcoma [65]. A proteomics analysis of tumor specimens collected from 13 SS patients, identified SCRN1 as positive prognostic factor with significantly higher expression among patients who were alive and disease free for at least 5 years [65]. Further analysis of SCRN1 expression in 45 SS tumor specimens revealed a 5-year survival rate was 77.6 and 21.8% for patients with secernin-1 positive and negative primary tumors, respectively (p = 0.0015), and significantly associated with metastatic outcomes [65]. Metastasis-free survival was significantly higher (62.8% vs. 16.7%) in the SS patient group with SCRN1 positive tumors compared to that with SCRN1 negative tumors (p = 0.0012) [65].
EZH2
Enhancer of zeste homologue 2 (EZH2) is a member of the polycomb group (PcG) protein family, which is composed of epigenetic transcriptional regulators that participate in cell cycle regulation, DNA damage repair, cell differentiation, senescence, and apoptosis [66]. In cancer, EZH2 expression is associated with a worse prognosis and required for promotion of metastasis [67]. In SS, overexpression of EZH2 helps to distinguish PDSS, which is defined by high cellularity, high nuclear grade, high mitotic activity and an agresssive clinical course that tends to include early recurrence and metastasis, from the MPSS and BPSS histological subtypes [68]. EZH2 overexpression in SS is correlated with high H3K27 trimethylation, which facilitates chromatin compaction and gene silencing [68]. Importantly, high expression of EZH2 is predictive of developing distant metastasis even in the better-differentiated MPSS and BPSS subtypes [68]. In a recent preclinical study, the anti-tumor effect of EZH2 inhibition was evidenced in human SS models in vitro, as well as xenograft and patient-derived xenograft (PDX) models in vivo [69]. Moreover, Ramagila et al. found high EZH2 expression to be correlated with metastatic disease in pediatric soft tissue sarcomas [70]. Low expression of EZH2 restricts cell proliferation and induces cell cycle arrest at the G2 phase, whereas the overexpression of EZH2 can shorten the G1 phase of the cell cycle and lead to cell accumulation in the S phase [71, 72]. Moore et al. showed that EZH2 knockdown is sufficient to reduce distant metastasis in vivo [73]. EZH2-specific inhibition is an active area of researcher, with several human phase 1 and 2 trials now underway, such as an ongoing phase II, multicenter study of the EZH2 inhibitor tazemetostat in adult subjects with INI1-negative tumors or relapsed/refractory SS (https://clinicaltrials.gov/ct2/show/NCT02601950 ).
NY-ESO-1
New York esophageal squamous cell carcinoma 1 (NY-ESO-1), encoded by the CTAG1B gene, is a 22 kD hydrophobic protein cancer-testis antigen [74]. NY-ESO-1 is expressed in many cancers, associated with poor prognosis, and elevated metastatic risk [75]. The function of NY-ESO-1 is still unknown, but is of particular interest to researchers because it is highly immunogenic eliciting both cellular and humoral responses, and a large number of major histocompatibility complex (MHC) class I- and class II-restricted NY-ESO-1 epitopes have been identified [76]. Furthermore, NY-ESO-1 protein expression is significantly higher in metastatic versus primary tumors [75, 77, 78]. NY-ESO-1 is an attractive target for SS treatment because chemotherapy has a limited durable efficacy in relapsed or metastatic SS demonstrating the need for novel more effective therapies. NY-ESO-1 is expressed in approximately 80% of patients with SS which could be useful for distinguishing this cancer from other types of mesenchymal tumors, and identifying patients who would benefit from NY-ESO-1 targeted therapies [79]. In a clinical trial using genetically engineered T cells reactive with NY-ESO-1 in patients with metastatic synovial cell sarcoma, tumor regression was achieved in 67% of the patients [80]. Several clinical trials employing NY-ESO-1 are currently under way in patients with SS (https://clinicaltrials.gov/ct2/show/NCT01343043, NCT02609984).
CXCR4
Chemokine receptor 4 (CXCR4), is a 352-amino acid seven-transmembrane G protein-coupled cell surface receptor with an important role in homing of hematopoietic stem cells and lymphocyte trafficking that has been found to promote cell migration, invasion and angiogenesis [52, 81]. The CXCR4 pathway has been shown to be associated with tumor progression and poor prognosis in many types of cancer including breast [82], lung [83], colon [84], melanoma [85] and soft tissue sarcomas [86]. CXCR4 promotes metastasis by activating activating extracellular signal-regulated kinase 1/2 (ERK1/2), Akt/PKB and Nuclear factor-κB (NF-κB), which increases the adhesion and invasive ability of cancer cells in part by the activity of MMP2 and MMP9 [87–89]. CXCR4 has a pivotal role in the migration of cancer cells between the primary and the metastatic site in synovial sarcoma [52, 90]. Tumor cells expressing CXCR4 that detach from the primary tumor and enter the circulatory system can migrate toward organs that express its ligand CXCL12 [91]. Lung, lymph node, bone marrow, and liver, the most frequent metastatic locations in SS [9], all express very high levels of CXCL12 [92]. A study of SS patients found that 5-year overall survival (OS) rates were 47% for those with positive CXCR4 staining, and 86% (P = 0.0003) for those with negative CXCR4 staining [52]. A second study, reported that 5 year survival outcomes for SS patients with positive CXCR4 staining was less than 30% [4]. Importantly, it was found that SS cultures contain a subpopulation of cells expressing high levels of CXCR4 that also express high levels stem cell markers (NANOG, OCT4, SOX2), and these cells have an increased tumor initiating capacity in xenographic mouse models [4]. Although no group has directly measured the metastatic risk of CXCRX4 expression in SS, perhaps due to limited patient numbers, expression of this marker appears to be a key factor both for cell migration and tumor propagation at a distal site. Perhaps the use of CXCR4 antagonists, many of which are already in various stages of clinical development [93], and shown to significantly reduce lung metastasis in mouse models of osteosarcoma [94], would be a beneficial treatment option for patients with metastatic SS, particularly in cases where tumors are unresectable.
Conclusions
SS is a rare, aggressive subtype of soft tissue sarcoma. It has a predilection for metastases to multiple organs, including the lungs, lymph nodes, and bone. The ability of this tumor to metastasize to multiple organs demonstrates that the tumor can interact and invade multiple environments. The overall prognosis is poor given the high rate of metastatic disease and lack of effective therapeutic agents. Nearly all mortality in patients with SS is caused by metastatic disease, yet the biological cause of these events has not been well characterized. The full transformation that occurs with the X;18 chromosomal translocation has not been completely elucidated at this time. Several oncogenic biomarkers have been found to be elevated in SS. As summarized in Fig. 1, many of these biomarkers may be used to evaluate for recurrence and metastatic disease and also help determine prognosis. The lack of the full understanding of how the translocation and elevated biomarkers interact with the host is a significant limitation in our ability to effectively treat SS. Further research should be done to help develop a greater understanding of these interactions and the downstream effects that occur in SS, with an emphasis on preventing metastatic disease. This will not only enable patients to be monitored for progression of disease and allow for counseling regarding prognosis, but also be used to develop better treatments for this subtype of soft tissue sarcoma.
Acknowledgements
We would like to thank Loma Linda University School of Medicine Dean’s Office & Department of Orthopedic Surgery for the Grants to Promote Collaborative and Translational Research (GCAT 2016, #681163) for supporting synovial sarcoma research and providing professional materials relevant to this review topic.
Funding
All costs associated with the study design, collection, analysis, interpretation of data, and writing the manuscript were funded by the Loma Linda University Cancer Center.
Availability of data and materials
This is a review article and the specific datasets supporting the conclusions of this article were generated by other research groups. Our supporting data was obtained from the referenced publications.
Authors’ contributions
RN prepared the manuscript draft, LZ contributed information and important written content, SK critically reviewed the literature, HRM and CSC provided important material and suggestions, SM designed the study, prepared the figure, and revised the manuscript. All authors read and approved the final manuscript draft.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- Akt
Serine/threonine kinase
- ATF2
Activating transcription factor 2
- BAF
BRG-/BRM-associated factor
- BPSS
Biphasic synovial sarcoma
- CDCA2
Cell division cycle A2
- CTC
Circulating tumor cells
- C-X-C motif
Chemokine
- CXCL12
ligand 12
- CXCR4
Chemokine receptor 4
- ECM
extracellular matrix
- EMT
Epithelial mesenchymal transition
- ERK1/2
extracellular signal-regulated kinase 1/2
- EZH2
Enhancer of zeste homologue 2
- IGF1R
Insulin like growth factor 1 receptor
- IGFBP7
Insulin-like growth factor-binding protein-7
- IGFBP-rP1
IGFBP-related protein-1
- KIF14
Kinesin family member 14
- MAPK
Mitogen-activated protein kinases
- MMPs
Matrix metalloproteinases
- MPSS
Monophasic synovial sarcoma
- mTOR
Mammalian target of rapamycin
- NF-κB
Nuclear factor-κB
- NY-ESO-1
New York esophageal squamous cell carcinoma 1
- PBMCs
Peripheral blood mononuclear cell
- PcG
Polycomb group
- PDSS
Poorly differentiated synovial sarcoma
- PDX
Patient-derived xenograft
- PI3K
Phosphatidylinositol-3-kinase
- PKB
Protein kinase B
- RAS
Ras GTPase
- SCRN1
Secernin-1
- SOX2
SRY (sex-determining region Y)-box 2
- SS
Synovial sarcoma
- STS
Soft tissue sarcoma
- TGF-β
Transforming growth factor beta
- TIMP
Tissue inhibitors of metalloproteinases
- TLE1
Transducin like enhancer of split 1
References
- 1.Rajwanshi A, Srinivas R, Upasana G. Malignant small round cell tumors. J Cytol. 2009;26(1):1–10. doi: 10.4103/0970-9371.54861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rong R, et al. Metastatic poorly differentiated monophasic synovial sarcoma to lung with unknown primary: a molecular genetic analysis. Int J Clin Exp Pathol. 2009;3(2):217–21. [PMC free article] [PubMed] [Google Scholar]
- 3.Thway K, Fisher C. Synovial sarcoma: defining features and diagnostic evolution. Ann Diagn Pathol. 2014;18(6):369–80. doi: 10.1016/j.anndiagpath.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Kimura T, et al. Identification and analysis of CXCR4-positive synovial sarcoma-initiating cells. Oncogene. 2016;35(30):3932–43. doi: 10.1038/onc.2015.461. [DOI] [PubMed] [Google Scholar]
- 5.Eilber FC, Dry SM. Diagnosis and management of synovial sarcoma. J Surg Oncol. 2008;97(4):314–20. doi: 10.1002/jso.20974. [DOI] [PubMed] [Google Scholar]
- 6.Sultan I, et al. Comparing children and adults with synovial sarcoma in the Surveillance, Epidemiology, and End Results program, 1983 to 2005: an analysis of 1268 patients. Cancer. 2009;115(15):3537–47. doi: 10.1002/cncr.24424. [DOI] [PubMed] [Google Scholar]
- 7.Krieg AH, et al. Synovial sarcomas usually metastasize after >5 years: a multicenter retrospective analysis with minimum follow-up of 10 years for survivors. Ann Oncol. 2011;22(2):458–67. doi: 10.1093/annonc/mdq394. [DOI] [PubMed] [Google Scholar]
- 8.Deshmukh R, Mankin HJ, Singer S. Synovial sarcoma: the importance of size and location for survival. Clin Orthop Relat Res. 2004;419:155–61. doi: 10.1097/00003086-200402000-00025. [DOI] [PubMed] [Google Scholar]
- 9.Vlenterie M, et al. Outcome of chemotherapy in advanced synovial sarcoma patients: Review of 15 clinical trials from the European Organisation for Research and Treatment of Cancer Soft Tissue and Bone Sarcoma Group; setting a new landmark for studies in this entity. Eur J Cancer. 2016;58:62–72. doi: 10.1016/j.ejca.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Salah S, et al. Factors influencing survival in metastatic synovial sarcoma: importance of patterns of metastases and the first-line chemotherapy regimen. Med Oncol. 2013;30(3):639. doi: 10.1007/s12032-013-0639-z. [DOI] [PubMed] [Google Scholar]
- 11.Amankwah EK, Conley AP, Reed DR. Epidemiology and therapies for metastatic sarcoma. Clin Epidemiol. 2013;5:147–62. doi: 10.2147/CLEP.S28390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spurrell EL, et al. Prognostic factors in advanced synovial sarcoma: an analysis of 104 patients treated at the Royal Marsden Hospital. Ann Oncol. 2005;16(3):437–44. doi: 10.1093/annonc/mdi082. [DOI] [PubMed] [Google Scholar]
- 13.Corey RM, Swett K, Ward WG. Epidemiology and survivorship of soft tissue sarcomas in adults: a national cancer database report. Cancer Med. 2014;3(5):1404–15. doi: 10.1002/cam4.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vlenterie M, Jones RL, van der Graaf WT. Synovial sarcoma diagnosis and management in the era of targeted therapies. Curr Opin Oncol. 2015;27(4):316–22. doi: 10.1097/CCO.0000000000000198. [DOI] [PubMed] [Google Scholar]
- 15.Kubo T, et al. Prognostic value of SS18-SSX fusion type in synovial sarcoma; systematic review and meta-analysis. Springerplus. 2015;4:375. doi: 10.1186/s40064-015-1168-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell. 2013;153(1):71–85. doi: 10.1016/j.cell.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Przybyl J, et al. Downstream and intermediate interactions of synovial sarcoma-associated fusion oncoproteins and their implication for targeted therapy. Sarcoma. 2012;2012:249219. doi: 10.1155/2012/249219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia CB, Shaffer CM, Eid JE. Genome-wide recruitment to Polycomb-modified chromatin and activity regulation of the synovial sarcoma oncogene SYT-SSX2. BMC Genomics. 2012;13:189. doi: 10.1186/1471-2164-13-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su L, et al. Deconstruction of the SS18-SSX fusion oncoprotein complex: insights into disease etiology and therapeutics. Cancer Cell. 2012;21(3):333–47. doi: 10.1016/j.ccr.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rekhi B, et al. Immunohistochemical validation of TLE1, a novel marker, for synovial sarcomas. Indian J Med Res. 2012;136(5):766–75. [PMC free article] [PubMed] [Google Scholar]
- 21.Laporte AN et al. Identification of cytotoxic agents disrupting synovial sarcoma oncoprotein interactions by proximity ligation assay. Oncotarget, 2016;7(23):34384–94. [DOI] [PMC free article] [PubMed]
- 22.Panagopoulos I, et al. Clinical impact of molecular and cytogenetic findings in synovial sarcoma. Genes Chromosomes Cancer. 2001;31(4):362–72. doi: 10.1002/gcc.1155. [DOI] [PubMed] [Google Scholar]
- 23.Sun Y, et al. Prognostic implication of SYT-SSX fusion type and clinicopathological parameters for tumor-related death, recurrence, and metastasis in synovial sarcoma. Cancer Sci. 2009;100(6):1018–25. doi: 10.1111/j.1349-7006.2009.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ladanyi M, et al. Impact of SYT-SSX fusion type on the clinical behavior of synovial sarcoma: a multi-institutional retrospective study of 243 patients. Cancer Res. 2002;62(1):135–40. [PubMed] [Google Scholar]
- 25.Takenaka S, et al. Prognostic implication of SYT-SSX fusion type in synovial sarcoma: a multi-institutional retrospective analysis in Japan. Oncol Rep. 2008;19(2):467–76. [PubMed] [Google Scholar]
- 26.Fricke A, et al. Identification of a blood-borne miRNA signature of synovial sarcoma. Mol Cancer. 2015;14:151. doi: 10.1186/s12943-015-0424-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masliah-Planchon J, Garinet S, Pasmant E. RAS-MAPK pathway epigenetic activation in cancer: miRNAs in action. Oncotarget. 2016;7(25):38892–907. doi: 10.18632/oncotarget.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hisaoka M, et al. Identification of altered MicroRNA expression patterns in synovial sarcoma. Genes Chromosomes Cancer. 2011;50(3):137–45. doi: 10.1002/gcc.20837. [DOI] [PubMed] [Google Scholar]
- 29.Fricke A, et al. Synovial Sarcoma Microvesicles Harbor the SYT-SSX Fusion Gene Transcript: Comparison of Different Methods of Detection and Implications in Biomarker Research. Stem Cells Int. 2016;2016:6146047. doi: 10.1155/2016/6146047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hashimoto N, et al. Detection of SYT-SSX fusion gene in peripheral blood from a patient with synovial sarcoma. Am J Surg Pathol. 2001;25(3):406–10. doi: 10.1097/00000478-200103000-00017. [DOI] [PubMed] [Google Scholar]
- 31.Chang L, et al. Circulating tumor cells in sarcomas: a brief review. Med Oncol. 2015;32(1):430. doi: 10.1007/s12032-014-0430-9. [DOI] [PubMed] [Google Scholar]
- 32.Chinen LT, et al. Isolation, detection, and immunomorphological characterization of circulating tumor cells (CTCs) from patients with different types of sarcoma using isolation by size of tumor cells: a window on sarcoma-cell invasion. Onco Targets Ther. 2014;7:1609–17. doi: 10.2147/OTT.S62349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chibon F, et al. Validated prediction of clinical outcome in sarcomas and multiple types of cancer on the basis of a gene expression signature related to genome complexity. Nat Med. 2010;16(7):781–7. doi: 10.1038/nm.2174. [DOI] [PubMed] [Google Scholar]
- 34.Lagarde P, et al. Chromosome instability accounts for reverse metastatic outcomes of pediatric and adult synovial sarcomas. J Clin Oncol. 2013;31(5):608–15. doi: 10.1200/JCO.2012.46.0147. [DOI] [PubMed] [Google Scholar]
- 35.Gruneberg U, et al. KIF14 and citron kinase act together to promote efficient cytokinesis. J Cell Biol. 2006;172(3):363–72. doi: 10.1083/jcb.200511061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmed SM, et al. KIF14 negatively regulates Rap1a-Radil signaling during breast cancer progression. J Cell Biol. 2012;199(6):951–67. doi: 10.1083/jcb.201206051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vagnarelli P, et al. Repo-Man coordinates chromosomal reorganization with nuclear envelope reassembly during mitotic exit. Dev Cell. 2011;21(2):328–42. doi: 10.1016/j.devcel.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwa V, Oh Y, Rosenfeld RG. The insulin-like growth factor-binding protein (IGFBP) superfamily. Endocr Rev. 1999;20(6):761–87. doi: 10.1210/edrv.20.6.0382. [DOI] [PubMed] [Google Scholar]
- 39.Chen D, et al. Insulin-like growth factor-binding protein-7 functions as a potential tumor suppressor in hepatocellular carcinoma. Clin Cancer Res. 2011;17(21):6693–701. doi: 10.1158/1078-0432.CCR-10-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burger AM, et al. Essential roles of IGFBP-3 and IGFBP-rP1 in breast cancer. Eur J Cancer. 2005;41(11):1515–27. doi: 10.1016/j.ejca.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 41.Jiang W, et al. Insulin-like growth factor binding protein 7 mediates glioma cell growth and migration. Neoplasia. 2008;10(12):1335–42. doi: 10.1593/neo.08694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shao L, et al. Detection of the differentially expressed gene IGF-binding protein-related protein-1 and analysis of its relationship to fasting glucose in Chinese colorectal cancer patients. Endocr Relat Cancer. 2004;11(1):141–8. doi: 10.1677/erc.0.0110141. [DOI] [PubMed] [Google Scholar]
- 43.Gambaro K, et al. Low levels of IGFBP7 expression in high-grade serous ovarian carcinoma is associated with patient outcome. BMC Cancer. 2015;15:135. doi: 10.1186/s12885-015-1138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nousbeck J, et al. Insulin-like growth factor-binding protein 7 regulates keratinocyte proliferation, differentiation and apoptosis. J Invest Dermatol. 2010;130(2):378–87. doi: 10.1038/jid.2009.265. [DOI] [PubMed] [Google Scholar]
- 45.Wajapeyee N, et al. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132(3):363–74. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmed S, et al. Proteolytic processing of IGFBP-related protein-1 (TAF/angiomodulin/mac25) modulates its biological activity. Biochem Biophys Res Commun. 2003;310(2):612–8. doi: 10.1016/j.bbrc.2003.09.058. [DOI] [PubMed] [Google Scholar]
- 47.Rao C, et al. High expression of IGFBP7 in fibroblasts induced by colorectal cancer cells is co-regulated by TGF-beta and Wnt signaling in a Smad2/3-Dvl2/3-dependent manner. PLoS One. 2014;9(1):e85340. doi: 10.1371/journal.pone.0085340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Padua D, Massague J. Roles of TGFbeta in metastasis. Cell Res. 2009;19(1):89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 49.Massague J. TGFbeta in Cancer. Cell. 2008;134(2):215–30. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pen A, et al. Glioblastoma-secreted factors induce IGFBP7 and angiogenesis by modulating Smad-2-dependent TGF-beta signaling. Oncogene. 2008;27(54):6834–44. doi: 10.1038/onc.2008.287. [DOI] [PubMed] [Google Scholar]
- 51.Benassi MS, et al. Tissue and serum IGFBP7 protein as biomarker in high-grade soft tissue sarcoma. Am J Cancer Res. 2015;5(11):3446–54. [PMC free article] [PubMed] [Google Scholar]
- 52.Palmerini E, et al. Prognostic and predictive role of CXCR4, IGF-1R and Ezrin expression in localized synovial sarcoma: is chemotaxis important to tumor response? Orphanet J Rare Dis. 2015;10:6. doi: 10.1186/s13023-014-0222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wan X, Helman LJ. The biology behind mTOR inhibition in sarcoma. Oncologist. 2007;12(8):1007–18. doi: 10.1634/theoncologist.12-8-1007. [DOI] [PubMed] [Google Scholar]
- 54.Roomi MW, et al. Modulation of u-PA, MMPs and their inhibitors by a novel nutrient mixture in adult human sarcoma cell lines. Int J Oncol. 2013;43(1):39–49. doi: 10.3892/ijo.2013.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willis BC, Borok Z. TGF-beta-induced EMT: mechanisms and implications for fibrotic lung disease. Am J Physiol Lung Cell Mol Physiol. 2007;293(3):L525–34. doi: 10.1152/ajplung.00163.2007. [DOI] [PubMed] [Google Scholar]
- 56.Benassi MS, et al. Metalloproteinase expression and prognosis in soft tissue sarcomas. Ann Oncol. 2001;12(1):75–80. doi: 10.1023/A:1008318614461. [DOI] [PubMed] [Google Scholar]
- 57.Hidalgo M, Eckhardt SG. Development of matrix metalloproteinase inhibitors in cancer therapy. J Natl Cancer Inst. 2001;93(3):178–93. doi: 10.1093/jnci/93.3.178. [DOI] [PubMed] [Google Scholar]
- 58.Ferrari C, et al. Role of MMP-9 and its tissue inhibitor TIMP-1 in human osteosarcoma: findings in 42 patients followed for 1–16 years. Acta Orthop Scand. 2004;75(4):487–91. doi: 10.1080/00016470410001295-1. [DOI] [PubMed] [Google Scholar]
- 59.Benassi MS, et al. Tissue and serum loss of metalloproteinase inhibitors in high grade soft tissue sarcomas. Histol Histopathol. 2003;18(4):1035–40. doi: 10.14670/HH-18.1035. [DOI] [PubMed] [Google Scholar]
- 60.Moran A, et al. Clinical relevance of MMP-9, MMP-2, TIMP-1 and TIMP-2 in colorectal cancer. Oncol Rep. 2005;13(1):115–20. [PubMed] [Google Scholar]
- 61.Way G, et al. Purification and identification of secernin, a novel cytosolic protein that regulates exocytosis in mast cells. Mol Biol Cell. 2002;13(9):3344–54. doi: 10.1091/mbc.E01-10-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hendrix A, et al. The tumor ecosystem regulates the roads for invasion and metastasis. Clin Res Hepatol Gastroenterol. 2011;35(11):714–9. doi: 10.1016/j.clinre.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 63.Lin S, et al. Secernin-1 contributes to colon cancer progression through enhancing matrix metalloproteinase-2/9 exocytosis. Dis Markers. 2015;2015:230703. doi: 10.1155/2015/230703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miyoshi N, et al. SCRN1 is a novel marker for prognosis in colorectal cancer. J Surg Oncol. 2010;101(2):156–9. doi: 10.1002/jso.21459. [DOI] [PubMed] [Google Scholar]
- 65.Suehara Y, et al. Secernin-1 as a novel prognostic biomarker candidate of synovial sarcoma revealed by proteomics. J Proteomics. 2011;74(6):829–42. doi: 10.1016/j.jprot.2011.02.033. [DOI] [PubMed] [Google Scholar]
- 66.Wang W, et al. Polycomb Group (PcG) Proteins and Human Cancers: Multifaceted Functions and Therapeutic Implications. Med Res Rev. 2015;35(6):1220–67. doi: 10.1002/med.21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Volkel P, et al. Diverse involvement of EZH2 in cancer epigenetics. Am J Transl Res. 2015;7(2):175–93. [PMC free article] [PubMed] [Google Scholar]
- 68.Changchien YC, et al. Poorly differentiated synovial sarcoma is associated with high expression of enhancer of zeste homologue 2 (EZH2) J Transl Med. 2012;10:216. doi: 10.1186/1479-5876-10-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kawano S, et al. Preclinical Evidence of Anti-Tumor Activity Induced by EZH2 Inhibition in Human Models of Synovial Sarcoma. PLoS One. 2016;11(7):e0158888. doi: 10.1371/journal.pone.0158888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramaglia M, et al. High EZH2 expression is correlated to metastatic disease in pediatric soft tissue sarcomas. Cancer Cell Int. 2016;16:59. doi: 10.1186/s12935-016-0338-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamaguchi H, Hung MC. Regulation and Role of EZH2 in Cancer. Cancer Res Treat. 2014;46(3):209–22. doi: 10.4143/crt.2014.46.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fujii S, et al. Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. J Biol Chem. 2008;283(25):17324–32. doi: 10.1074/jbc.M800224200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moore HM, et al. EZH2 inhibition decreases p38 signaling and suppresses breast cancer motility and metastasis. Breast Cancer Res Treat. 2013;138(3):741–52. doi: 10.1007/s10549-013-2498-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schultz-Thater E, et al. NY-ESO-1 tumour associated antigen is a cytoplasmic protein detectable by specific monoclonal antibodies in cell lines and clinical specimens. Br J Cancer. 2000;83(2):204–8. doi: 10.1054/bjoc.2000.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Park TS, et al. Expression of MAGE-A and NY-ESO-1 in Primary and Metastatic Cancers. J Immunother. 2016;39(1):1–7. doi: 10.1097/CJI.0000000000000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li M, et al. Effective inhibition of melanoma tumorigenesis and growth via a new complex vaccine based on NY-ESO-1-alum-polysaccharide-HH2. Mol Cancer. 2014;13:179. doi: 10.1186/1476-4598-13-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Giesen E, et al. NY-ESO-1 as a potential immunotherapeutic target in renal cell carcinoma. Oncotarget. 2014;5(14):5209–17. doi: 10.18632/oncotarget.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aung PP, et al. Expression of New York esophageal squamous cell carcinoma-1 in primary and metastatic melanoma. Hum Pathol. 2014;45(2):259–67. doi: 10.1016/j.humpath.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lai JP, et al. NY-ESO-1 expression in sarcomas: A diagnostic marker and immunotherapy target. Oncoimmunol. 2012;1(8):1409–10. doi: 10.4161/onci.21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Robbins PF, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol. 2011;29(7):917–24. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu C, et al. CXCR4 in breast cancer: oncogenic role and therapeutic targeting. Drug Des Devel Ther. 2015;9:4953–64. doi: 10.2147/DDDT.S84932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dewan MZ, et al. Stromal cell-derived factor-1 and CXCR4 receptor interaction in tumor growth and metastasis of breast cancer. Biomed Pharmacother. 2006;60(6):273–6. doi: 10.1016/j.biopha.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 83.Gangadhar T, Nandi S, Salgia R. The role of chemokine receptor CXCR4 in lung cancer. Cancer Biol Ther. 2010;9(6):409–16. doi: 10.4161/cbt.9.6.11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lv S, et al. The association of CXCR4 expression with prognosis and clinicopathological indicators in colorectal carcinoma patients: a meta-analysis. Histopathology. 2014;64(5):701–12. doi: 10.1111/his.12321. [DOI] [PubMed] [Google Scholar]
- 85.Scala S, et al. Expression of CXCR4 predicts poor prognosis in patients with malignant melanoma. Clin Cancer Res. 2005;11(5):1835–41. doi: 10.1158/1078-0432.CCR-04-1887. [DOI] [PubMed] [Google Scholar]
- 86.Oda Y, et al. Chemokine receptor CXCR4 expression is correlated with VEGF expression and poor survival in soft-tissue sarcoma. Int J Cancer. 2009;124(8):1852–9. doi: 10.1002/ijc.24128. [DOI] [PubMed] [Google Scholar]
- 87.Yuecheng Y, Xiaoyan X. Stromal-cell derived factor-1 regulates epithelial ovarian cancer cell invasion by activating matrix metalloproteinase-9 and matrix metalloproteinase-2. Eur J Cancer Prev. 2007;16(5):430–5. doi: 10.1097/01.cej.0000236259.88146.a4. [DOI] [PubMed] [Google Scholar]
- 88.Uchida D, et al. Possible role of stromal-cell-derived factor-1/CXCR4 signaling on lymph node metastasis of oral squamous cell carcinoma. Exp Cell Res. 2003;290(2):289–302. doi: 10.1016/S0014-4827(03)00344-6. [DOI] [PubMed] [Google Scholar]
- 89.Guo F, et al. CXCL12/CXCR4: a symbiotic bridge linking cancer cells and their stromal neighbors in oncogenic communication networks. Oncogene. 2016;35(7):816–26. doi: 10.1038/onc.2015.139. [DOI] [PubMed] [Google Scholar]
- 90.Kim RH, Li BD, Chu QD. The role of chemokine receptor CXCR4 in the biologic behavior of human soft tissue sarcoma. Sarcoma. 2011;2011:593708. doi: 10.1155/2011/593708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Murphy PM. Chemokines and the molecular basis of cancer metastasis. N Engl J Med. 2001;345(11):833–5. doi: 10.1056/NEJM200109133451113. [DOI] [PubMed] [Google Scholar]
- 92.Muller A, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410(6824):50–6. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 93.Debnath B, et al. Small molecule inhibitors of CXCR4. Theranostics. 2013;3(1):47–75. doi: 10.7150/thno.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim SY, et al. Inhibition of the CXCR4/CXCL12 chemokine pathway reduces the development of murine pulmonary metastases. Clin Exp Metastasis. 2008;25(3):201–11. doi: 10.1007/s10585-007-9133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This is a review article and the specific datasets supporting the conclusions of this article were generated by other research groups. Our supporting data was obtained from the referenced publications.


