ABSTRACT
Monovalent bispecific antibodies (BsAbs) are projected to have broad clinical applications due to their ability to bind two different targets simultaneously. Although they can be produced using recombinant technologies, the correct pairing of heavy and light chains is a significant manufacturing problem. Various approaches exploit mutations or linkers to favor the formation of the desired BsAb, but a format using a single common light chain has the advantage that no other modification to the antibody is required. This strategy reduces the number of formed molecules to three (the BsAb and the two parent mAbs), but the separation of the BsAb from the two monovalent parent molecules still poses a potentially difficult purification challenge. Current methods employ ion exchange chromatography and linear salt gradients, but are only successful if the difference in the observed isoelectric points (pIs) of two parent molecules is relatively large. Here, we describe the use of highly linear pH gradients for the facile purification of common light chain BsAbs. The method is effective at separating molecules with differences in pI as little as 0.10, and differing in their sequence by only a single charged amino acid. We also demonstrate that purification resins validated for manufacturing are compatible with this approach.
KEYWORDS: Bispecific antibody, common light chain, isoelectric point, ion exchange chromatography, linear pH gradient, purification
Abbreviations
- BsAbs
bispecific antibodies
- CDRs
complementarity-determining regions
- CEX
cation exchange chromatography
- DVD-IgG
dual-variable-domain IgG
- HCs
heavy chains
- IgG
Immunoglobulin G
- IEX
ion exchange chromatography
- LCs
light chains
- mAbs
monoclonal antibodies
- MACS
magnetic assisted cell sorting
- pI
isoelectric point
- scFv
single-chain variable fragment
Introduction
Monoclonal antibodies (mAbs) are widely used to treat a variety of human diseases. Classic IgGs contain two identical antigen-binding regions and therefore bind monospecifically and bivalently. For certain applications, however, it can be advantageous to target two pathological factors or pathways simultaneously, which has led to increased interest in the development of bispecific molecules.1,2 The production of these asymmetric molecules in sufficient quantity and purity poses a challenge because it involves the heterodimerization of two different heavy chains (HCs) and correct pairing of the respective light chains (LCs) with their cognate HCs. Purification of the small percentage of correctly assembled molecules from the large number of almost identical potential byproducts is essentially impossible. To overcome this issue, bispecific antibodies (BsAbs) have been developed in a large variety of formats, each with their own sets of advantages but also potential shortcomings. Many early formats consist of single-chain-variable fragment (scFv) domains or other antibody fragments with various linkers or proteins to connect them.3-8 However, these non-IgG-like molecules can suffer from issues with manufacturability, stability, immunogenicity and rapid clearance in vivo. Other structures such as dual-variable-domain IgG (DVD-IgG),9 or chemically crosslinked antibodies10,11 are bivalent for each antigen and therefore bispecific and tetravalent. This can be desirable in some applications, but it precludes them from being used in applications where receptor homodimerization is undesirable, or where the avidity for either antigen could lead to non-target toxicity issues. A similar potential issue needs to be considered in the case of dual-targeting or so-called two-in-1 antibodies.12-14 Additionally, this latter approach requires extensive variable-region engineering for each new antigen pair, making it difficult to use universally. Heterodimeric IgG-like bispecifics therefore have emerged as an advantageous format for monovalent bispecific mAbs. Multiple protein engineering efforts have been reported to overcome the main issues with their production, namely effective heterodimerization of the two different HCs and correct formation of the two light-chain/heavy-chain interactions. Some approaches rely on the mixing of two antibodies under reducing conditions, followed by removal of the reductant and preferred assembly of heterodimers due to mutations in the Fc- and hinge domains.15,16 This annealing method, however, involves additional process development to an already complex manufacturing process. Other designs use non-native arrangement of domains,17 newly created disulfide bridges,18 linkers that need to be removed by several consecutive protease steps,19 or large numbers of mutations to favor correct assembly of the desired bispecific molecule.20,21 All of these approaches potentially lead to developability issues due to risk for misfolding, aggregation, disulfide scrambling, instability, reduced titers or additional product related impurities. Fischer and coworkers recently described the isolation of a common heavy-chain bispecific with kappa and lambda LCs using kappa and lambda specific resins.22 While this elegantly solves the purification problem, it also limits the library diversity to only the LCs, while additionally having the restriction that one LC needs to be kappa and the other one lambda.
Given the potential shortcomings of each of the current approaches, the simplest (and perhaps therefore lowest risk) bispecific format for therapeutic use would be an unmodified human IgG. This format would combine the already established manufacturing processes and validated properties of therapeutic mAbs with the expanded modes of action of bispecifics. It is generally accepted that the specificity of an antibody is predominantly defined by the complementarity-determining regions (CDRs) residing in the HC.23 As such, an obvious approach to reduce the chain association issue is the use of a common LC. Such antibodies are very readily isolated from libraries that have vast HC repertoires and a unique or very few LCs.24 This method has been combined with mutations driving heterodimeric heavy-chain assembly,25,26 but variable purity of the heterodimer product and potential immunogenicity of the introduced mutations engineered into the HC introduce new risks for this heterodimerization approach.
With a focus on reduced potential immunogenicity and good manufacturability, Sampei and coworkers recently demonstrated the identification and multidimensional optimization of a common light chain bispecific mimicking the function of factor VIII cofactor activity.27 Their impressive engineering tour de force has led to a heterodimeric molecule that, based on large differences in heavy-chain isoelectric points (pI), can be isolated from residual homodimers using cation exchange chromatography (CEX) and a salt gradient. However, the amount of isoelectric point engineering necessary to generate the bispecific makes this approach not generally applicable.
In order to devise a more generally applicable purification method, we attempted to find a way to remove contaminating HC homodimers using liquid chromatography without having to change any part of the HC. The current standard for removal of homodimer HC contaminants from heterodimer is ion exchange chromatography (IEX) employing a salt gradient, as demonstrated by Sampei and colleagues.27 This method, however, does not have very high resolution capabilities, forcing one to rely on large differences in pI of the two HCs. Higher resolution methods used for analytical separation of antibodies make use of relatively simple pH gradients,28-30 but these approaches have been hampered by difficulties with forming controllable, linear pH gradients over a broad pH range. Furthermore, the resins used in these high performance liquid chromatography (HPLC)-based methods are not suitable for preparative scale purifications.
Using an established in silico buffer optimization tool, Kröner and Hubbuch recently used a systematic approach to generate buffer compositions for pH gradient anion- and cation-exchange chromatography, providing a broad pH range with low ionic strength.31 Their system overcomes the previously described need to compensate gradient non-linearities with a software-enabled, algorithmic control of the gradient mixing,32 making it simpler and easier to use and implement. Here, we describe the use of this buffer system for the purification of BsAb heterodimers from homodimers on a preparative scale. We also show that homodimers with computed pI differences as little as 0.10 pH units and differing in only one charged amino acid can be separated using commercially scalable ion exchange resins.
Results
Discovery of IgG-like common light chain BsAbs
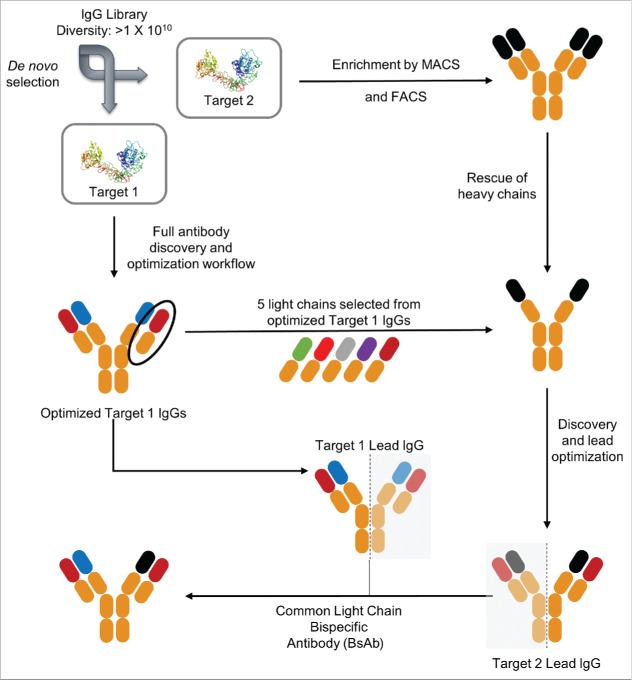
Common LC BsAbs were discovered according to a workflow depicted in Fig. 1. Monospecific IgGs against target 1 were isolated from a full-length human IgG antibody library using an in vitro yeast selection system.33 To further optimize binding characteristics of the isolated antibodies, they were then affinity matured as described in Materials and Methods. For target 2, the same antibody library was enriched in clones binding to the antigen using magnetic assisted cell sorting (MACS). The HC plasmids were then recovered from the enriched population and combined with 5 LCs selected from optimized IgGs specific to target 1 (isolated in step 1). The resulting library was then used to isolate and affinity mature IgGs against target 2. In the final step, individual HCs from optimized IgGs against targets 1 and 2 were combined with common LCs, expressed in HEK293 cells and BsAbs were purified according to the scheme in Fig. S1.
Figure 1.
Schematic representation of discovery of IgG-like common light chain BsAbs. The first step was a full antibody discovery and optimization workflow from large yeast naïve libraries (1×1010) for target 1. Step 2 was the enrichment of a target 2 binding population from the same libraries using magnetic assisted cell sorting (MACS) and fluorescence activated cell sorting (FACS). This was followed by rescue of heavy chains from the enriched target 2 binding population and pairing with 5 selected light chains of optimized target 1 IgGs (discovered in step 1). Target 1 and target 2 lead heavy chains as well as the common light chains from affinity matured leads were then cloned into mammalian expression vectors to yield IgG-like common light chain BsAbs.
Ion exchange chromatography using highly linear pH gradients provides superior results to using salt gradients for purification of bispecific antibodies with common light chains
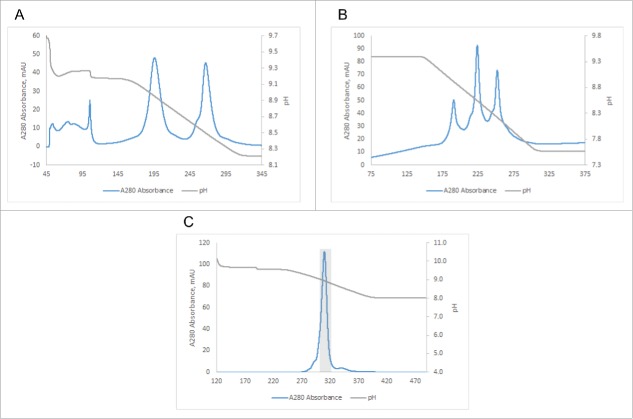
The current state of the art for purification of heterodimeric BsAbs from contaminating homodimers is cation exchange chromatography (CEX) employing linear salt gradients.15,19,27,34 In order to determine how separation using a highly linear pH gradient compares to this method, we expressed seven different common LC BsAbs in HEK293 cells as a test panel and purified the mixtures of BsAb and homodimeric byproducts using Protein A chromatography to remove host cell proteins and other product-related impurities (see schematic in Fig. S1). The individual BsAbs in the panel had been chosen to span differences in calculated pI between the parent antibody pairs from as large as 1.33 pH units to as little as 0.10 pH unit (see Table 1). We then chose two of the mixtures, BsAb#1 and BsAb#5, with each to be separated by CEX using a MonoS 5/50 GL column and either a 1 M NaCl gradient or a linear pH gradient from pH 4 to pH 11. Fig. S2A shows that the full pH gradient can achieve baseline resolution of the BsAb#1 mixture, whereas the 1 M salt gradient failed to do so (Fig. S2B). For the BsAb#5 mixture, in which the parent antibodies are only separated by a pI difference of 0.26, resolution of three distinct peaks can be achieved with the full pH gradient (Fig. S2D), whereas only a single peak is observed in the case of the 1 M salt gradient (Fig. S2E). Only when the salt gradient is optimized, i.e., made more shallow (in our case a gradient from 0 to 0.25 M NaCl), resolution similar to the non-optimized, full pH gradient can be observed (Figs. S2C and 2F).
Table 1.
Bispecific antibodies used in this study.
| Designation | pI mAb1 | pI mAb2 | pI BsAb | ΔpI mAb1/mAb2 |
|---|---|---|---|---|
| BsAb#1 | 9.32 | 7.99 | 8.94 | 1.33 |
| BsAb#2 | 8.95 | 8.36 | 8.73 | 0.59 |
| BsAb#3 | 9.19 | 8.71 | 9.01 | 0.48 |
| BsAb#4 | 9.08 | 8.7 | 8.92 | 0.38 |
| BsAb#5 | 9.19 | 8.93 | 9.08 | 0.26 |
| BsAb#6 | 9.19 | 9.08 | 9.14 | 0.11 |
| BsAb#7 | 9.19 | 9.09 | 9.14 | 0.10 |
pI = isoelectric point (calculated); BsAb, bispecific antibody; mAb, monoclonal antibody.
pH gradient-based IEX purification can be used for separation of BsAb mixtures of widely varying pI differences
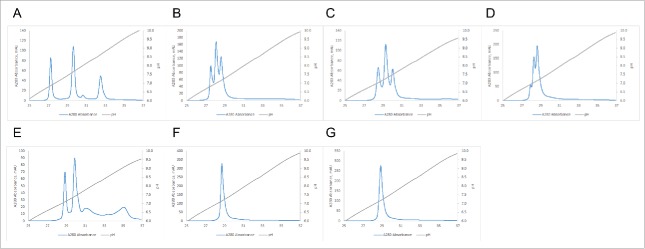
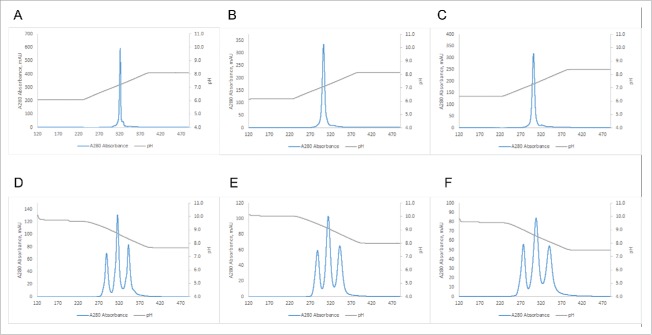
In order to test the useful range of the method, we purified common LC BsAbs with varying pI differences between their individual monospecific antibodies using a Mono S 5/50 column and a linear pH gradient. We routinely use the small MonoS and MonoQ 5/50 GL columns with a full pH gradient for screening purposes to determine the apparent pI of the molecules and whether AEX or CEX provides the best resolution. Using the apparent pI, we then choose the appropriate pH range for the shallower gradients used for the larger, preparative MonoS and MonoQ 10/100 GL columns. As is evident from Fig. 2, the difference in apparent pI, and the resolution achieved using the pH gradient, correlates very well with the difference in calculated pI of the components of the mixture to be separated. Fig. 2A demonstrates that, even with the small screening column and the full pH gradient, the BsAb#1 mixture that has the largest difference in calculated pI between the parent antibodies can be readily resolved. As the pI difference between the two parent antibodies for the bispecific gets smaller (BsAb#2 to BsAb#5 in Figs. 2B to 2E), the individual peaks are less resolved. Once this difference is as small as 0.10 (BsAb#6 and BsAb#7 in Figs. 2F and 2G), no separation between the two homodimers and the heterodimer in the mixture can be achieved under the chosen purification conditions.
Figure 2.
Purification of BsAbs with varying differences in pI using a linear pH gradient. Panels (A) to (G) correspond to BsAbs#1 to 7 respectively (ΔpI from 1.33 to 0.10, see Table 1 for details). pH gradient was from pH 4.0 to pH 11.0 (20 CV) on a Mono S 5/50GL column.
Parameters for pH-gradient-based purification of BsAbs can quickly be optimized to allow for separation of homodimers differing in calculated pI as little as 0.10 pH units
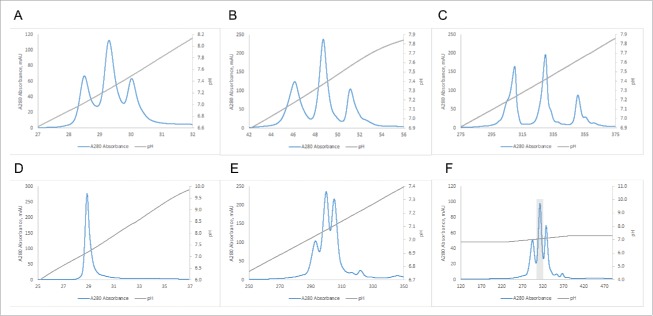
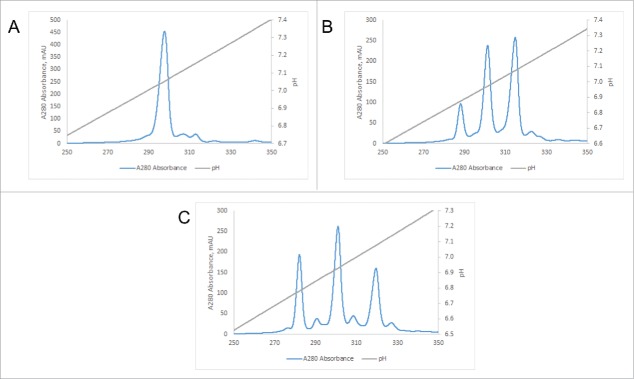
One of the cumbersome characteristics of salt elution-based IEX is the fact that the optimal loading pH needs to be determined individually for each target protein, which is usually done by measuring the effect of pH on selectivity in batch mode, followed by optimization in dynamic binding mode in packed columns.35 This is not necessary when using a pH gradient-based elution method for BsAbs. Fig. 3 shows two examples of the optimization of elution parameters for the purification of BsAbs. In Fig. 3A, the BsAb#2 mixture of homodimeric antibodies with a difference in pI of 0.59 pH units and the corresponding heterodimer was purified using a MonoS 5/50 GL column and a full pH gradient (from pH 4.0 to 11.0). The elution pHs of the two homodimers as determined by the screening run are then used to choose the appropriate gradient for the preparative runs (usually 0.5 pH units above and below homodimer peaks, for example pH 6.5 to 8.0 as shown in Fig. 3B). As expected, the shallower gradient leads to better separation of homodimeric and heterodimeric species as judged by the chromatogram. This separation can further be improved by performing the separation on a larger MonoS 10/100 GL column using the same gradient as shown in Fig. 3C. Consequently, we were able to recover pure heterodimer (100% purity by mass spec; see Fig. S3A) in several of the fractions from this preparative size column. In the second example, the BsAb#7 mixture of homodimeric antibodies with a difference in pI of only 0.10 pH units and the corresponding heterodimer were run on the small screening column using a full pH gradient (from pH 4.0 to 11.0). Fig. 3D shows that resolution of the individual species is not possible under these conditions. However, when a shallower pH gradient and a longer column are employed, separation of the 3 species is observed (Fig. 3E). Already at this stage, we were able to isolate about 95% pure (as judged by mass spectrometry) heterodimer from 3 fractions of the central peak. From 7.57 mg of total mixture, we recovered 2.28 mg, equivalent to a yield of 30%. As shown in Fig. 3F, further reducing the slope of the gradient (0.40 pH units overall), leads to even better separation and recovery of 3.73 mg of about 95% pure heterodimer (see mass spectrum in Fig. S3B) from a total mixture of 6.69 mg for an essentially quantitative yield.
Figure 3.
Improvement in resolution by narrowing gradient and increasing residence time. (A) BsAb#2 run on Mono S 5/50GL with pH gradient from 4.0 to 11.0. (B) BsAb#2 run on Mono S 5/50GL with pH gradient from 6.5 to 8.0. (Note: the load amount was 2 mg) (C) BsAb#2 run on Mono S 10/100GL with pH gradient from 6.5 to 8.0. (D) BsAb#7 run on Mono S 5/50GL with pH gradient from 4.0 to 11.0. (E) BsAb#7 run on Mono S 10/100GL with pH gradient from 6.65 to 7.65. (F) BsAb#7 run on Mono S 10/100GL with pH gradient from 6.87 to 7.27.
pH gradient-based elution can be used in conjunction with anion or cation exchange chromatography
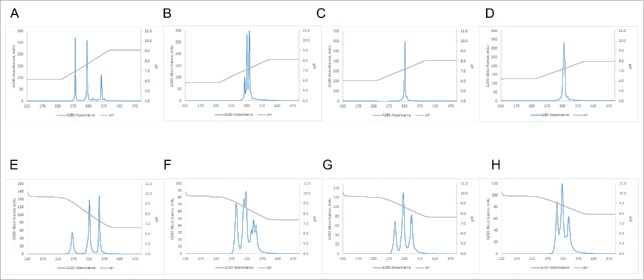
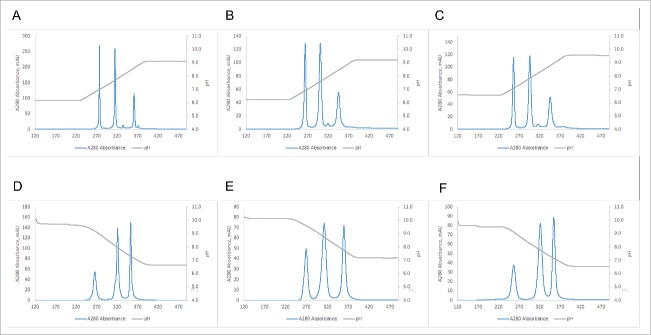
A standard purification scheme for mAbs usually contains a Protein A capture step followed by AEX or CEX or a combination of both.36 We therefore also tested the possibility of using an AEX chromatography resin with the linear pH gradient elution. Fig. 4 shows a comparison of purifications of BsAbs#1, 5, 6 and 7 using either a MonoS 10/100 GL CEX or a MonoQ 10/100 GL AEX column. Comparison of Figs. 4A and 4E shows that in the case of BsAb#1 the superior resolution can be achieved using the CEX resin. In contrast, BsAb#5 shows better resolution when separated over the AEX column (Fig. 4B vs Fig. 4F). Here, even charge-related subspecies can be resolved as the additional peaks in Fig. 4F indicate. The same increased performance of AEX over CEX is true for BsAbs#6 and 7 (compare Fig. 4C with 4G and Fig. 4D with 4H). It is noteworthy that in the case of BsAb#6, where no acceptable resolution between the three species can be achieved using the CEX resin, almost baseline resolution can be accomplished using the AEX resin.
Figure 4.
Comparison of performance of CEX (A-D) (10/100GL Mono S column) vs AEX (E-F) (10/100GL Mono Q column). (A) BsAb#1 using pH gradient from 6.19 to 9.19. (B) BsAb#5 using pH gradient from 5.85 to 8.10. (C) BsAb#6 using pH gradient from 6.1 to 8.1. (D) BsAb#7 using pH gradient from 6.26 to 8.01. (E) BsAb#1 using pH gradient from 9.65 to 6.65. (F) BsAb#5 using pH gradient from 9.68 to 7.43. (G) BsAb#6 using pH gradient from 9.66 to 7.66. (H) BsAb#7 using pH gradient from 9.74 to 7.99.
Up to this point in the project, for simplicity sake, we had used the buffer system described by Kröner and colleagues31 for CEX also for our AEX-based purifications by simply switching the start and end buffers (CEX buffer gradient from pH 4.0 to pH 11.0 for CEX resin-based purifications and CEX buffer gradient from pH 11.0 to pH 4.0 for AEX resin-based purifications). However, when we attempted to use the CEX buffer system on the preparative AEX column for the purification of BsAb#6, we noticed the presence of homodimer 1 in all three peaks although the chromatogram had suggested acceptable separation (see chromatogram in Fig. 5A and the mass spectrum of the heterodimer pool in Fig. S4A). To determine whether this phenomenon was related to the buffer system used, we ran the same sample on the same column with the AEX buffer system suggested by Kröner and colleagues. Although the chromatogram does not show increased resolution (compare Figs. 5A and 5B), the heterodimer isolated in the middle peak is 100% pure by mass spectrometry (see Fig. S4B). We are currently investigating whether this is a sample specific effect, or an inherent characteristic of the two buffer systems.
Figure 5.
Purification using AEX of BsAbs that could not be purified to homogeneity using CEX. (A) Purification of BsAb#6 using a Mono Q 10/100GL column and the CEX buffer system with a pH gradient from pH 9.17 to 8.21 (B) Purification of BsAb#6 using a Mono Q 10/100GL column and the AEX buffer system with a pH gradient from pH 9.34 to 7.51 (C) Secondary purification of the fractions of BsAb#7 highlighted in Fig. 3F using a Mono Q 10/100GL column and a pH gradient from pH 9.57 to 8.07 (Note: the load amount was 4.4 mg).
We also used AEX (this time again using the CEX buffer system) to further purify the 95% pure fractions from the CEX purification of BsAb#7 from the example shown in Fig. 3F above. The chromatogram in Fig. 5C shows that the remaining homodimers appear as two very small shoulders that can easily be removed by peak cutting (indicated by the shaded rectangle). The mass spectrum in Fig. S4C of a representative fraction demonstrates that the method efficiently removes the last remaining homodimer impurities to below the level of detection.
pH gradient-based purification is sensitive to single amino acid differences
In the course of antibody discovery, affinity maturation, and optimization, small changes in the primary antibody sequence often lead to accompanying shifts in calculated antibody pI. For example, four of the antibodies in our test panel, BsAbs#3, 5, 6 and 7, had been determined to differ in only four residues in one of their HCs (see Table 2). The chromatograms in Fig. 6 demonstrate that the replacement of a single charged amino acid (in our case lysine in BsAb#6 and BsAb#7) with either the amide version (i.e., glutamine in BsAb#5) or one of the opposite charge (i.e., glutamic acid in BsAb#3) can lead to dramatic differences in separation when using IEX combined with linear pH gradient elution. As can be seen in the elution profile for one of the lysine-containing clones BsAb#6 in Fig. 6A, the homodimers and heterodimer cannot be resolved under the chosen purification conditions. Mutation of the lysine to glutamine increases the difference in pI of the two homodimers from 0.11 pH units to 0.26 pH units and results in resolution of the three species (see Fig. 6B). A mutation of the glutamine to glutamic acid results in a further increase of pI difference to 0.48 pH units and not only in baseline resolution of heterodimer from the homodimers, but also resolution of apparent charge isoforms (see small peaks in Fig. 6C). It should be noted that these mutations arose naturally during selection for desirable affinity and polyspecificity properties, and were not introduced specifically to assist in IEX separation.
Table 2.
Sequence details for BsAbs#3 and 5 through 7.
| Designation | VH mAb1 | pI mAb1 | VH mAb2* | pI mAb2 | ΔpI mAb1/mAb2 |
|---|---|---|---|---|---|
| BsAb#3 | Same | 9.19 | I A E Y | 8.71 | 0.48 |
| BsAb#5 | I A Q Y | 8.93 | 0.26 | ||
| BsAb#6 | I S K Y | 9.08 | 0.11 | ||
| BsAb#7 | V A K H | 9.09 | 0.10 |
Four positions in which the VHs differ.
Figure 6.
Separation of BsAbs with highly similar sequences. (A) BsAb#6 run on 10/100GL Mono S column using pH gradient from 6.65 to 7.65. (B) BsAb#5 run on 10/100GL Mono S column using pH gradient from 6.43 to 7.65. (C) BsAb#3 run on 10/100GL Mono S column using pH gradient from 6.33 to 7.66.
Process scale CEX and AEX resins are compatible with pH gradient-based elution
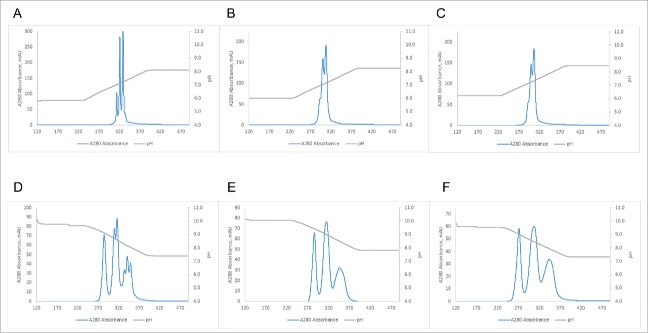
While MonoS or MonoQ resins can be employed to purify BsAbs at scales up to several milligrams, their small bead size precludes them from also being used in commercial scale settings. To determine whether process scale resins with bead sizes ranging from 30 to 90 µm show acceptable resolution when used with a pH gradient, we compared several commercial resins in small scale scouting experiments using BsAb#1 (ΔpI = 1.33) and BsAb#5 (ΔpI = 0.26) (for details on the resins compared, see Table 3). When BsAb#1 was purified using various CEX resins, several of the smaller bead size resins showed almost baseline resolution (see Fig. S5). Similar resolution was also observed for most of the AEX resins (see Fig. S6). For BsAb#5, however, none of the CEX resins could separate heterodimer from the two homodimers under the scouting type purification conditions (results not shown). On the other hand, all of the tested AEX resins showed at least some resolution between the three species using the small scale scouting columns (see Fig. S7). To determine the resolution limits of the resins and compare them to MonoS and MonoQ, the two best candidates each for CEX and AEX were scaled up to larger 8 ml columns and shallower gradients were employed. For these experiments, we used BsAbs#1, 5, and 6 with pI differences of as little as 0.11 pH units. The two CEX resins with the best resolution as determined by scouting runs were Source 30S and SP Sepharose High Performance, and the two AEX resins with the best resolution as determined by scouting runs were Source 30Q and Q Sepharose High Performance. Fig. 7 shows that for BsAb#1 baseline resolution is achievable using either the process scale CEX resins or AEX resins. For BsAb#5, both CEX resins show slightly lower resolution than MonoS, whereas the two AEX resins show similar resolution as MonoQ when regarding separation of homodimers from heterodimer (see Fig. 8). It is noteworthy, however, that the process scale resins are not able to resolve charge-related subspecies to the same level as MonoQ (see Fig. 8D). In the case of BsAb#6 with a ΔpI of only 0.11, neither MonoS nor the two process scale CEX resins provide any detectable resolution under the chosen conditions, whereas MonoQ and the two process scale AEX resins are all able to achieve almost baseline resolution (see Fig. 9).
Table 3.
Characteristics of tested ion exchange resins.
| Resin | Type | Matrix | Particle size [µm] | Functional group | Process scale? |
|---|---|---|---|---|---|
| Mono S1 | Cation | Polystyrene / divinyl benzene | 10 | Methyl sulfonate | N |
| SP Sepharose Fast Flow1 | Cation | 6% crosslinked agarose | 90 | Sulfopropyl | Y |
| Macro-Prep High S2 | Cation | Methacrylate copolymer bead | 50 | Sulfonate | Y |
| Poros XS3 | Cation | Polystyrene / divinyl benzene | 50 | Sulfopropyl | Y |
| Poros HS3 | Cation | Polystyrene / divinyl benzene | 50 | Sulfopropyl | Y |
| Capto SP ImpRes1 | Cation | High flow agarose | 40 | Sulfonate | Y |
| SP Sepharose High Performance1 | Cation | 6% highly crosslinked spherical agarose | 34 | Sulfopropyl | Y |
| SOURCE 30S1 | Cation | Polystyrene / divinyl benzene | 30 | Sulfonate | Y |
| Capto MMC1 | Multimodal | Highly crosslinked agarose | 75 | Multimodal | Y |
| Mono Q1 | Anion | Polystyrene / divinyl benzene | 10 | Quaternary amine | N |
| Poros XQ3 | Anion | Crosslinked poly[styrene divinylbenzene] | 50 | Quaternary amine | Y |
| Poros HQ3 | Anion | Crosslinked poly[styrene divinylbenzene] | 50 | Quaternized polyethyleneimine | Y |
| Capto Q ImpRes1 | Anion | High flow agarose | 40 | Quaternary amine | Y |
| Q HP1 | Anion | Crosslinked agarose | 34 | Quaternary amine | Y |
| SOURCE 30Q1 | Anion | Polystyrene / divinyl benzene | 30 | Quaternary amine | Y |
GE Healthcare Life Sciences.
Bio-Rad.
Life Technologies.
Figure 7.
Performance of the best 3 tested CEX and AEX resins using 3 pH unit gradients for BsAb#1. Resins: (A) MonoS (gradient from pH 6.19 to 9.19), (B) SOURCE 30S (gradient from pH 6.22 to 9.22), (C) SP Sepharose High Performance (gradient from pH 6.53 to 9.53), (D) MonoQ (gradient from pH 9.65 to 6.65), (E) SOURCE 30Q (gradient from pH 10.21 to 7.21), (F) Q Sepharose High Performance (gradient from pH 9.56 to 6.56).
Figure 8.
Performance of the best 3 tested CEX and AEX resins using 2.25 pH unit gradients for BsAb#5. Resins: (A) MonoS (gradient from pH 5.85 to 8.10), (B) SOURCE 30S (gradient from pH 6.01 to 8.26), (C) SP Sepharose High Performance (gradient from pH 6.23 to 8.48), (D) MonoQ (gradient from pH 9.68 to 7.43), (E) SOURCE 30Q (gradient from pH 10.09 to 7.84), (F) Q Sepharose High Performance (gradient from pH 9.57 to 7.32).
Figure 9.
Performance of the best 3 tested CEX and AEX resins using 2.0 pH unit gradients for BsAb#6. Resins: (A) MonoS (gradient from pH 6.10 to 8.10), (B) SOURCE 30S (gradient from pH 6.15 to 8.15), (C) SP Sepharose High Performance (gradient from pH 6.38 to 8.38), (D) MonoQ (gradient from pH 9.66 to 7.66), (E) SOURCE 30Q (gradient from pH 10.09 to 8.09), (F) Q Sepharose High Performance (gradient from pH 9.52 to 7.52).
Discussion
With the recent rise in BsAb formats and applications, the problem of purifying the desired bispecific from undesired homodimers and other product-related impurities has become an important factor in the commercial development of these potential drugs. The current state-of-the-art downstream process for purification of bispecifics remains IEX using a salt gradient. Due to the low resolution that is achievable with this method, a significant amount of engineering of the bispecific molecule needs to be accomplished before purification can be pursued at a commercial scale. If this is not done, removal of some impurities is difficult, if not impossible. Sampei and colleagues used isoelectric point engineering of the HC variable regions to increase the difference in pI of the homodimers from almost zero to over 2 pH units and thereby facilitated purification of the target BsAb.27 This difference in pI of the homodimers then had to be painstakingly maintained throughout the remaining optimization for improved solubility, removal of deamidation sites and deimmunization.
It has been shown that for analytical characterization of mAbs the use of pH gradients results in higher resolution than salt gradients,28-30 making this method the current state-of-the-art for profiling charge heterogeneity of these molecules. Kluters and coworkers recently demonstrated the feasibility of using CEX with pH gradient elution on a preparative scale.37 In their application, they added polyethylene glycol as a mobile phase modifier to remove high molecular weight and low molecular weight impurities from a “CrossMab.”17 Although their main focus was the removal of impurities of different molecular weight, their results suggest that IEX combined with pH gradient elution could become a valuable tool for the purification of bispecifics.
Common LC BsAbs normally do not contain large amounts of high or low molecular weight impurities. Nevertheless, in addition to mass spectrometry, we routinely also use size exclusion chromatography (SEC) to determine the amount of aggregates or fragments in the various fractions. As suggested by the work of Kluters, we find the purified samples to be highly homogeneous and usually in the range of 99% pure by SEC (results not shown).
Here, we show that either AEX or CEX in combination with a pH gradient is able to separate homodimers that differ in their computed pI by as little as 0.10 pH units from the desired heterodimer. Since the same buffer system is used for both techniques, very little optimization of purification parameters needs to be done, nor is there a need for specialized software to control the formation of the gradient. Because the effective charge of the protein is often not equal to the calculated net charge due to charge patches on the surface, the apparent pI of the protein usually needs to be determined by short screening runs using AEX and CEX. This also determines which of the two techniques shows the best separation for a given molecule. The apparent pI is then used to pick the starting and ending pH for the elution gradient that is used for the preparative runs. Readily available MonoS and MonoQ 10/100 GL columns can be used for purifications of up to several milligrams of protein.
While it is convenient to use the buffer system that originally had been designed for CEX also for AEX, one needs to consider that in the latter case the buffer components will interact with the resin. The AEX chromatography in this case is not only using a pH gradient, but can also have a displacement component. While this did not result in reduced performance in most cases we tested, we saw one instance where a switch to the designated AEX buffer was necessary to achieve the required resolution.
Several other considerations regarding the pH gradient buffer system in general are its range, complexity, and availability of its components in good manufacturing practices (GMP) grade. Since the primary purpose of our study was to demonstrate the superiority of linear pH gradients over salt gradients in general, we picked the buffer system with the most consistent linearity over the greatest pH range irrespective of its complexity. Because the pIs of BsAbs and mAbs are usually in a much narrower range, in a manufacturing setting one would probably choose a more simple mixture with fewer toxic and difficult to source components. Examples of such buffer systems have recently been described by a group specializing in the modeling of pH gradients for IEX.38,39 An additional consideration for preparative scale purifications is the stability of the protein at the starting pH of the separation, since the protein mixture is usually being loaded onto the column in a buffer of the same pH. Although we did not encounter any stability issues at the high end of the pH range (the loading pH for AEX), we found that, especially for BsAbs made up of IgG4 parents, a starting pH of 4.0 led to significant amounts of aggregation (results not shown). In these cases we therefore set the starting pH to 6.0, where negligible amounts of aggregation were observed.
In regards to the choice of an appropriate separation matrix, we showed that several bulk resins, which can be used for purifications on a large commercial scale, have resolution capabilities similar to the Mono resins. Although our maximum protein load amounts were chosen to allow for maximum resolution, one also needs to consider that our method is envisioned to be used as a secondary purification or polishing step where dynamic binding capacity is a secondary consideration.40 This combination of IEX and linear pH gradient elution can thus be used where the lower resolution salt gradients result in insufficient separation of heterodimer from homodimers and other impurities. Although we demonstrated here the application of our method for the purification of common LC bispecifics, it can also be used for the purification of other bispecific formats as long as the impurities to be removed have a sufficiently different apparent pI. These include BsAbs incorporating Fc heterodimerization techniques, such as electrostatic steering21 or the more widely used “knobs-into-holes.”25 Furthermore, the technique can also be combined with continuous purification procedures,41 as well as purification schemes specifically designed for common LC BsAbs.34 As such, the combination of IEX and linear pH gradient elution has the potential to become a state-of-the-art method for the purification and polishing of BsAbs in general.
Materials and methods
Discovery of common light chain antibodies
Common LC antibodies were isolated from a full-length human IgG1 antibody library using an in vitro yeast selection system and associated methods. Target-binding mAbs were enriched by incubating biotin-labeled antigens with antibody-expressing yeast cells at different concentrations, followed by magnetic bead selection (Miltenyi, Biotec) and fluorescence-activated cell sorting (FACS) on a FACSAria II cell sorter (BD Biosciences) employing streptavidin-labeled secondary reagents in several successive selection rounds. After the last round of enrichment, yeast cells were sorted and plated onto agar plates, clones were analyzed by DNA sequencing and used for IgG production. Optimization of antibodies for higher affinity was performed in successive cycles of selection rounds using lower concentrations of antigen baits with sub-libraries generated by LC shuffling, targeted mutagenesis of CDR1 and CDR2 of the HCs and ePCR of the variable region of the heavy or light chain.
Expression and purification of antibodies
BsAbs were expressed in HEK293 cells grown in shake flasks. After six days of growth, the cell culture supernatant was harvested by centrifugation and passed over Protein A agarose (MabSelect SuRe™ from GE Healthcare Life Sciences). The bound antibodies were then washed with phosphate-buffered saline and eluted with buffer consisting of 200 mM acetic acid and 50 mM NaCl at pH3.5 into 1/8th volume 2 M HEPES, pH 8.0.
Chromatographic separation of bispecific antibodies
All chromatographic separations were performed on a computer controlled ÄKTA avant 150 preparative chromatography system (GE Healthcare Life Sciences) equipped with an integrated pH electrode, enabling in-line pH monitoring during the run. The columns Mono S 5/50 GL, Mono Q 5/50 GL, Mono S 10/100 GL, and Mono Q 10/100 GL were purchased from GE Healthcare Life Sciences. The CEX buffer was composed of 15.6 mM CAPS (Sigma), 9.4 mM CHES (Sigma), 4.6 mM TAPS (Sigma), 9.9 mM HEPPSO (VWR/MP Biomedicals), 8.7 mM MOPSO (Sigma), 11.0 mM MES (Sigma), 13.0 mM Acetate (BDH), 9.9 mM formate (EMD), 10 mM NaCl (VWR/BDH), and the pH was adjusted up to 4.0 or 11.0 using NaOH. The AEX buffer was composed of 9.8 mM methylamine (Sigma), 9.1 mM 1,2-ethanediamine (Sigma), 6.4 mM 1-methylpiperazine (Sigma), 13.7 mM 1,4-dimethylpiperazine (Sigma), 5.8 mM bis-Tris (Affymetrix), 7.7 mM hydroxylamine (Sigma), 10 mM NaCl (VWR/BDH), and the pH was adjusted to 10.5 or 3.5 using HCl. The pH gradient forming solutions were freshly prepared before each experiment by dissolving the buffering species in water and dividing the solution into two equal parts. One half was then adjusted to pH 4.0 (buffer A) using sodium hydroxide, while the other half was adjusted to pH 11.0 (buffer B) also using sodium hydroxide. Unless otherwise indicated, the following procedure was used to perform all separations. 0.5 mg (for the 5/50 columns) or 10 mg (for the 10/100 column) (for exceptions, please see figure legends 3B and 5C) of the common LC BsAbs mixture to be separated were buffer exchanged into the starting pH buffer and filtered through a 0.2 µm filter. Before each separation, the column was equilibrated with 10 column volumes of starting buffer (either buffer A, buffer B, or the appropriate mixture of buffer A and buffer B). The protein mixture was then loaded onto the column via a capillary loop, and the column was washed with another 10 column volumes of starting buffer to remove the unbound material. Subsequently, a linear pH gradient of 20 column volumes of the appropriate mixtures of buffer A and buffer B was used for separation of the common LC BsAbs mixture.
For salt gradient-based separations, the buffer was composed of 20 mM MES pH 6.0 and the appropriate concentration of NaCl (0 or 1 M).
Confirmation of heterodimer purity by mass spectrometry
To remove heterogeneity introduced by Fc glycans, F(ab’)2 fragments of antibodies were generated by cleavage with IdeS (FabRICATOR®, Genovis) according to the manufacturer's protocol.
Digested samples were injected onto an Agilent 1100 series HPLC with an Applied Biosystems POROS® R2 10 µm column, (2.1×30 mm, 0.1 mL) maintained at 65°C. Mobile phase A was 0.1% formic acid in H20 and mobile phase B was 0.1% formic acid in acetonitrile. After injection of 25 µL of IdeS digested sample, a 2.1 minute LC gradient with a flow rate of 2 mL/min was used to elute the sample from the column (0.0 min, 2% B; 0.2 min, 35% B; 0.21 min, 95% B; 1.4 min, 95% B; 1.41 min, 2% B; 2.1 min, 5% B).
The Bruker maXis 4G mass spectrometer was run in positive ion mode with detection in the range of 700 to 2500 m/z. The remaining source parameters were set as follows; the capillary was set at 5500V, the nebulizer at 4.0 Bar, dry gas at 4.0 l/min, and dry temp set at 200°C. The resulting spectra were analyzed with Bruker Compass Data Analysis version 4.1.
Detection of intact F(ab’)2 heterodimer and homodimer species were confirmed based on mass measurement as compared with the theoretical sequence. Relative quantitation for each of the heterodimer and homodimer species was calculated based on the intensities of the peaks with respect to the sum of all the heterodimer and homodimer peak intensities.
Computation of theoretical pIs
Theoretical pIs of mAbs were computed based on their protein sequence using the Henderson-Hasselback equation with the known pKa according to EMBOSS.42
Supplementary Material
Disclosure of potential conflicts of interest
All authors are employees and shareholders of Adimab, LLC.
Acknowledgments
We acknowledge valuable discussions with Tillman Gerngross, Eric Krauland, Michael Ruse, William Roach, Max Vásquez, and Yingda Xu. We thank the Core facility of Adimab for plasmid construction and preparation.
References
- 1.Spiess C, Zhai Q, Carter PJ. Alternative molecular formats and therapeutic applications for bispecific antibodies. Mol Immunol 2015; 67:95-106; PMID:25637431; http://dx.doi.org/ 10.1016/j.molimm.2015.01.003 [DOI] [PubMed] [Google Scholar]
- 2.Kontermann RE. Dual targeting strategies with bispecific antibodies. mAbs 2012; 4:182-97; PMID:22453100; http://dx.doi.org/ 10.4161/mabs.4.2.19000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonagh CF, Huhalov A, Harms BD, Adams S, Paragas V, Oyama S, Zhang B, Luus L, Overland R, Nguyen S, et al.. Antitumor activity of a novel bispecific antibody that targets the ErbB2/ErbB3 oncogenic unit and inhibits heregulin-induced activation of ErbB3. Mol Cancer Ther 2012; 11:582-93; PMID:22248472; http://dx.doi.org/ 10.1158/1535-7163.MCT-11-0820 [DOI] [PubMed] [Google Scholar]
- 4.Michaelson JS, Demarest SJ, Miller B, Amatucci A, Snyder WB, Wu X, Huang F, Phan S, Gao S, Doern A, et al.. Anti-tumor activity of stability-engineered IgG-like bispecific antibodies targeting TRAIL-R2 and LTbetaR. mAbs 2009; 1:128-41; PMID:20061822; http://dx.doi.org/ 10.4161/mabs.1.2.7631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson S, Burke S, Huang L, Gorlatov S, Li H, Wang W, Zhang W, Tuaillon N, Rainey J, Barat B, et al.. Effector cell recruitment with novel Fv-based dual-affinity re-targeting protein leads to potent tumor cytolysis and in vivo B-cell depletion. J Mol Biol 2010; 399:436-49; PMID:20382161; http://dx.doi.org/ 10.1016/j.jmb.2010.04.001 [DOI] [PubMed] [Google Scholar]
- 6.Baeuerle PA, Reinhardt C. Bispecific T-cell engaging antibodies for cancer therapy. Cancer Res 2009; 69:4941-4; PMID:19509221; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-0547 [DOI] [PubMed] [Google Scholar]
- 7.Kelton C, Wesolowski JS, Soloviev M, Schweickhardt R, Fischer D, Kurosawa E, McKenna SD, Gross AW. Anti-EGFR biparatopic-SEED antibody has enhanced combination-activity in a single molecule. Arch Biochem Biophys 2012; 526:219-25; PMID:22426455; http://dx.doi.org/ 10.1016/j.abb.2012.03.005 [DOI] [PubMed] [Google Scholar]
- 8.Mabry R, Lewis KE, Moore M, McKernan PA, Bukowski TR, Bontadelli K, Brender T, Okada S, Lum K, West J, et al.. Engineering of stable bispecific antibodies targeting IL-17A and IL-23. Protein Eng Des Sel 2010; 23:115-27; PMID:20022918; http://dx.doi.org/ 10.1093/protein/gzp073 [DOI] [PubMed] [Google Scholar]
- 9.Wu C, Ying H, Grinnell C, Bryant S, Miller R, Clabbers A, Bose S, McCarthy D, Zhu RR, Santora L, et al.. Simultaneous targeting of multiple disease mediators by a dual-variable-domain immunoglobulin. Nat Biotechnol 2007; 25:1290-7; PMID:17934452; http://dx.doi.org/ 10.1038/nbt1345 [DOI] [PubMed] [Google Scholar]
- 10.Yankelevich M, Kondadasula SV, Thakur A, Buck S, Cheung NK, Lum LG. Anti-CD3 × Anti-GD2 bispecific antibody redirects T-cell cytolytic activity to neuroblastoma targets. Pediatr Blood Cancer 2012: 59:1198-1205; PMID:22707078; http://dx.doi.org/ 10.1002/pbc.24237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doppalapudi VR, Huang J, Liu D, Jin P, Liu B, Li L, Desharnais J, Hagen C, Levin NJ, Shields MJ, et al.. Chemical generation of bispecific antibodies. Proc Natl Acad Sci 2010; 107:22611-6; PMID:21149738; http://dx.doi.org/ 10.1073/pnas.1016478108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bostrom J, Yu SF, Kan D, Appleton BA, Lee CV, Billeci K, Man W, Peale F, Ross S, Wiesmann C, Fuh G. Variants of the antibody Herceptin that interact with HER2 and VEGF at the antigen binding site. Science 2009; 323:1610-4; PMID:19299620; http://dx.doi.org/ 10.1126/science.1165480 [DOI] [PubMed] [Google Scholar]
- 13.Schaefer G, Haber L, Crocker LM, Shia S, Shao L, Dowbenko D, Totpal K, Wong A, Lee CV, Stawicki S, et al.. A two-in-one antibody against HER3 and EGFR has superior inhibitory activity compared with monospecific antibodies. Cancer Cell 2011; 20:472-86; PMID:22014573; http://dx.doi.org/ 10.1016/j.ccr.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, Yun S, Batuwangala TD, Steward M, Holmes SD, Pan L, Tighiouart M, Shin HJ, Koenig L, Park W, et al.. A dual-targeting antibody against EGFR-VEGF for lung and head and neck cancer treatment. Int J Cancer 2012; 131:956-69; PMID:21918971; http://dx.doi.org/ 10.1002/ijc.26427 [DOI] [PubMed] [Google Scholar]
- 15.Strop P, Ho WH, Boustany LM, Abdiche YN, Lindquist KC, Farias SE, Rickert M, Appah CT, Pascua E, Radcliffe T, et al.. Generating bispecific human IgG1 and IgG2 antibodies from any antibody pair. J Mol Biol 2012; 420:204-19; PMID:22543237; http://dx.doi.org/ 10.1016/j.jmb.2012.04.020 [DOI] [PubMed] [Google Scholar]
- 16.Labrijn AF, Meesters JI, de Goeij BE, van den Bremer ET, Neijssen J, van Kampen MD, Strumane K, Verploegen S, Kundu A, Gramer MJ, et al.. Efficient generation of stable bispecific IgG1 by controlled Fab-arm exchange. Proc Natl Acad Sci 2013; 110:5145-50; PMID:23479652; http://dx.doi.org/ 10.1073/pnas.1220145110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaefer W, Regula JT, Bähner M, Schanzer J, Croasdale R, Dürr H, Gassner C, Georges G, Kettenberger H, Imhof-Jung S, et al.. Immunoglobulin domain crossover as a generic approach for the production of bispecific IgG antibodies. Proc Natl Acad Sci 2011; 108:11187-92; PMID:21690412; http://dx.doi.org/ 10.1073/pnas.1019002108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazor Y, Oganesyan V, Yang C, Hansen A, Wang J, Liu H, Sachsenmeier K, Carlson M, Gadre DV, Borrok MJ, et al.. Improving target cell specificity using a novel monovalent bispecific IgG design. mAbs 2015; 7:377-89; PMID:25621507; http://dx.doi.org/ 10.1080/19420862.2015.1007816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wranik BJ, Christensen EL, Schaefer G, Jackman JK, Vendel AC, Eaton D. LUZ-Y, a novel platform for the mammalian cell production of full-length IgG-bispecific antibodies. J Biol Chem 2012; 287:43331-9; PMID:23118228; http://dx.doi.org/ 10.1074/jbc.M112.397869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis SM, Wu X, Pustilnik A, Sereno A, Huang F, Rick HL, Guntas G, Leaver-Fay A, Smith EM, Ho C, et al.. Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface. Nat Biotechnol 2014; 32:191-8; PMID:24463572; http://dx.doi.org/ 10.1038/nbt.2797 [DOI] [PubMed] [Google Scholar]
- 21.Gunasekaran K, Pentony M, Shen M, Garrett L, Forte C, Woodward A, Ng SB, Born T, Retter M, Manchulenko K, et al.. Enhancing antibody Fc heterodimer formation through electrostatic steering effects. J Biol Chem 2010; 285:19637-46; PMID:20400508; http://dx.doi.org/ 10.1074/jbc.M110.117382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer N, Elson G, Magistrelli G, Dheilly E, Fouque N, Laurendon A, Gueneau F, Ravn U, Depoisier JF, Moine V, et al.. Exploiting light chains for the scalable generation and platform purification of native human bispecific IgG. Nat Commun 2015; 6:1-12; PMID:25672245; http://dx.doi.org/10933393 10.1038/ncomms7113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu JL, Davis MM. Diversity in the CDR3 region of VH is sufficient for most antibody specificities. Immunity 2000; 13:37-45; PMID:10933393; http://dx.doi.org/ 10.1016/S1074-7613(00)00006-6 [DOI] [PubMed] [Google Scholar]
- 24.Carter P. Bispecific human IgG by design. J Immunol Methods 2001; 248:7-15; PMID:11223065; http://dx.doi.org/ 10.1016/S0022-1759(00)00339-2 [DOI] [PubMed] [Google Scholar]
- 25.Merchant AM, Zhu Z, Yuan JQ, Goddard A, Adams CW, Presta LG, Carter P. An efficient route to human bispecific IgG. Nat Biotechnol 1998; 16:677-81; PMID:9661204; http://dx.doi.org/ 10.1038/nbt0798-677 [DOI] [PubMed] [Google Scholar]
- 26.Spreter Von Kreudenstein T, Escobar-Carbrera E, Lario PI, D'Angelo I, Brault K, Kelly J, Durocher Y, Baardsnes J, Woods RJ, Xie MH. Improving biophysical properties of a bispecific antibody scaffold to aid developability. mAbs 2013; 5:646-54; PMID:23924797; http://dx.doi.org/ 10.4161/mabs.25632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sampei Z, Igawa T, Soeda T, Okuyama-Nishida Y, Moriyama C, Wakabayashi T, Tanaka E, Muto A, Kojima T, Kitazawa T. Identification and multidimensional optimization of an asymmetric bispecific IgG antibody mimicking the function of Factor VIII cofactor activity. Plos One 2013; 8:e57479; PMID:23468998; http://dx.doi.org/ 10.1371/journal.pone.0057479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farnan D, Moreno GT. Multiproduct high-resolution monoclonal antibody charge variant separations by pH gradient ion-exchange chromatography. Anal Chem 2009; 81:8846-57; PMID:19795895; http://dx.doi.org/ 10.1021/ac901408j [DOI] [PubMed] [Google Scholar]
- 29.Rea JC, Moreno GT, Lou Y, Farnan D. Validation of a pH gradient-based ion-exchange chromatography method for high-resolution monoclonal antibody charge variant separations. J Pharm Biomed Anal 2011; 54:317-23; PMID:20884149; http://dx.doi.org/ 10.1016/j.jpba.2010.08.030 [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, Patapoff T, Farnan D, Zhang B. Improving pH gradient cation-exchange chromatography of monoclonal antibodies by controlling ionic strength. J Chromatogr A 2013; 1272:56-64; PMID:23253120; http://dx.doi.org/ 10.1016/j.chroma.2012.11.060 [DOI] [PubMed] [Google Scholar]
- 31.Kröner F, Hubbuch J. Systematic generation of buffer systems for pH gradient ion exchange chromatography and their application. J Chromatogr A 2013; 1285:78-87; PMID:23489486; http://dx.doi.org/ 10.1016/j.chroma.2013.02.017 [DOI] [PubMed] [Google Scholar]
- 32.Tsonev LI, Hirsh AG. Theory and applications of a novel ion exchange chromatographic technology using controlled pH gradients for separating proteins on anionic and cationic stationary phases. J Chromatogr A 1200:166-82; PMID:18554604; http://dx.doi.org/ 10.1016/j.chroma.2008.05.076 [DOI] [PubMed] [Google Scholar]
- 33.Xu Y, Roach W, Sun T, Jain T, Prinz B, Yu T-Y, Torrey J, Thomas J, Bobrowicz P, Vásquez M, et al.. Addressing polyspecificity of antibodies selected from an in vitro yeast presentation system: a FACS-based, high-throughput selection and analytical tool. PEDS 2013; 26:663-670; PMID:24046438; https://dx.doi.org/ 10.1093/protein/gzt047 [DOI] [PubMed] [Google Scholar]
- 34.Müller-Späth T, Ulmer N, Aumann L, Ströhlein G, Bavand M, Hendriks LJA, de Kruif J, Throsby M, Bakker ABH. Purifying common light-chain bispecific antibodies. Bioprocess Int 2013; 11:36-45;http://www.bioprocessintl.com/downstream-processing/chromatography/purifying-common-light-chain-bispecific-antibodies-342696/ [Google Scholar]
- 35.Ahamed T, Chilamkurthi S, Nfor BK, Verhaert PDEM, van Dedem GWK, van der Wielen LAM, Eppink MHM, van de Sandt EJAX, Ottens M. Selection of pH-related parameters in ion-exchange chromatography using pH-gradient operations. J Chromatogr A 2008; 1194:22-9; PMID:18154981; http://dx.doi.org/ 10.1016/j.chroma.2007.11.111 [DOI] [PubMed] [Google Scholar]
- 36.Marichal-Gallardo PA, Álvarez MM. State-of-the-art in downstream processing of monoclonal antibodies: process trends in design and validation. Biotechnol Prog 2012; 28:899-916; PMID:22641473; http://dx.doi.org/ 10.1002/btpr.1567 [DOI] [PubMed] [Google Scholar]
- 37.Kluters S, Hafner M, von Hirschheydt T, Frech C. Solvent modulated linear pH gradient elution for the purification of conventional and bispecific antibodies: modeling and application. J Chromatogr A 2015; 1418:119-29; PMID:26431858; http://dx.doi.org/ 10.1016/j.chroma.2015.09.053 [DOI] [PubMed] [Google Scholar]
- 38.Schmidt M, Hafner M, Frech C. Modeling of salt and pH elution in ion-exchange chromatography. J Sep Sci 2014; 37:5-13; PMID:24415551; http://dx.doi.org/ 10.1002/jssc.201301007 [DOI] [PubMed] [Google Scholar]
- 39.Kluters S, Wittkopp F, Jöhnck M, Frech C. Application of linear pH gradients for the modeling of ion exchange chromatography: separation of monoclonal antibody monomer from aggregates. J Sep Sci 2016; 39:663-75; PMID:26549715; http://dx.doi.org/ 10.1002/jssc.201500994 [DOI] [PubMed] [Google Scholar]
- 40.Milne JJ. Scale-up of protein purification: downstream processing issues. Methods Mol Biol 2017; 1485:71-84; PMID:27730549; http://dx.doi.org/27376629 10.1007/978-1-4939-6412-3_5 [DOI] [PubMed] [Google Scholar]
- 41.Steinebach F, Müller-Späth T, Morbidelli M. Continuous counter-current chromatography for capture and polishing steps in biopharmaceutical production. Biotechnol J 2016; 11(9):1126-41; PMID:27376629; http://dx.doi.org/ 10.1002/biot.201500354 [DOI] [PubMed] [Google Scholar]
- 42.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet 2000; 16:276-7; PMID:10827456; http://dx.doi.org/ 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.