ABSTRACT
Enterovirus 71 (EV71) causes outbreaks of hand, foot and mouth disease (HFMD), primarily in the Asia-Pacific area, that are often associated with complications of severe to fatal neurological symptoms. There are currently no anti-viral therapies or vaccines available for the treatment of EV71 infection. Illustrating human antibody responses neutralizing EV71 infection could potentially provide valuable information for the development of effective therapies and vaccines. Here, we constructed a comprehensive phage display library based on peripheral blood of eight EV71-infected donors and identified 27 EV71-specific human antibodies, of which four have neutralizing activity in in vitro experiments. Deep sequencing analysis of the antibody heavy chains at the transcript level of another three independent EV71-infected donors and three controls demonstrates that heavy chains of the EV71-specific antibodies are conserved among EV71-infected individuals but absent in controls, suggesting convergent evolution of human antibodies against EV71.
KEYWORDS: Deep sequencing; EV71; human antibody; phage display
Introduction
Enterovirus 71 is a virus of the Enterovirus genus in the Picornaviridae family that was first isolated from patients suffering from neurological disease in California in 1969.1 EV71 is one of the major causative agents for hand, foot, and mouth disease (HFMD) in children, often associated with complications of severe to fatal neurological symptoms.2
Large outbreaks have recently occurred in Southeast Asia, e.g., in Singapore in 2008,3 Fuyang, China, in 2008,4 and Guangdong, China, in 2009.5 In 2010 and 2011, a total of 3,394,375 cases of HFMD were recorded in Mainland China, accounting for 1,414 deaths.6 However, the underlying mechanism of EV71 pathogenesis remains unclear, and effective vaccines or specific antiviral drugs are not available.
The importance of an antibody response to EV71 infection has been demonstrated in animal experiments.7 However, most antibody research on EV71 is currently focused on murine antibodies. Reports about human antibodies against EV71 are very rare, although high-quality human antibodies with high specificity, avidity and neutralizing activity might be a viable treatment option for EV71 infection in humans.
Substantial advances have been made and much is known about the diversity, affinity, and specificity of human antibodies against various antigens,5,8 but critical questions about EV71 infection are still unanswered, including: 1) the antibody repertoire variance between patient and healthy people; 2) the antibody repertoire variance among individuals infected by the same virus; and 3) the abundance distribution of virus-specific antibodies in the antibody repertoires. Preparation of EV71-specific human antibodies may provide valuable tools to seek answers to these questions, which would further help understanding of the human antibody responses to EV71 infection.
Here, we constructed a comprehensive phage display library based on peripheral blood of eight EV71-infected donors and identified 27 EV71-specific human antibodies. Further evaluation suggests that four have neutralizing activity at least in in vitro experiments. Deep sequencing analysis of the antibody heavy chains at the transcript level of another three independent EV71-infected donors demonstrates that heavy chains of the EV71-specific antibodies are conserved among EV71-infected individuals but absent in healthy controls. These observations suggest convergent evolution of human antibodies against EV71 infection, providing valuable clues for the development of effective antiviral therapies and vaccines.
Results
Human Fab antibodies against EV71 identified by phage display
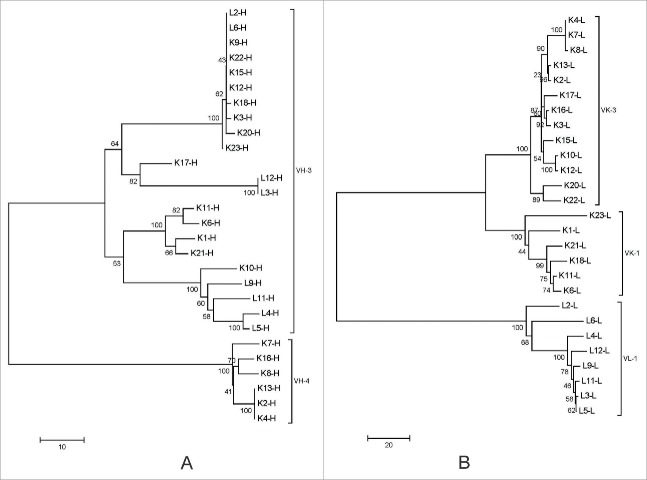
We established antigen-binding fragment (Fab) phage display libraries, one each for λ and κ light chains, from peripheral blood mononuclear cells (PBMCs) of eight volunteers infected with EV71 (Strain Shzh-98, confirmed by PCR). These two phage libraries contained 1.2 × 108 and 1.7 × 108 independent clones. After three rounds of panning, 1920 randomly picked colonies were screened by enzyme-linked immunosorbent assay (ELISA) for binding activity to inactivated EV71 virions, and 413 positive Fab clones were finally identified. Sequence analysis of all the 413 positive Fab clones revealed the presence of 27 unique clones. These 27 unique Fabs were then analyzed for their variable immunoglobulin gene segments using VBASE2 (www.vbase2.org), and were named L n or K n according to the types of their light chains (Table 1). There are 17 unique VH genes in these 27 Fabs; 13 are derived from the families VH3, and 4 are derived from the families VH4 (Fig. 1A). The light chains of these Fabs are derived from the lambda gene family Vλ1 and the kappa gene families Vκ1 and Vκ3 (Fig. 1B).
Table 1.
EV71-specific human antibodies identified by phage display.
| Neutralization | ||||||
|---|---|---|---|---|---|---|
| Fabs | VL | LCDR3 | VH | HCDR3 | ELISA | titers |
| L2 | 1–40*01 | QSYDSSLSGSGV | 3–11*01 | VRGTLYYYDMSGQLHDWYFDL | 0.8750 | |
| L3 | 1–51*01 | GTWDSSLSVWV | 3–21*02 | GLVFAYYHAPGRSFHYGMDV | 1.2780 | |
| L4 | 1–51*01 | GTWDSSLSSVV | 3–30–3*01 | APVWFGAMDV | 1.6250 | 1:16 |
| L5 | 1–51*01 | GTWDSSLSWV | 3–30–3*01 | APVWFGAMDV | 0.8540 | |
| L6 | 1–44*01 | AVWDDGLIGPV | 3–11*01 | VRGTLYYYDMSGQLHDWYFDL | 1.3550 | 1:8 |
| L9 | 1–51*01 | GTWDSRLSAGV | 3–30*18 | SPVWFGPLDY | 0.9610 | |
| L11 | 1–51*01 | GTWDSSLSAWV | 3–30–3*01 | DPMAGYYGSGALNY | 1.5800 | 1:8 |
| L12 | 1–51*01 | GTWDGSLTAWV | 3–21*02 | GLVFAYYHAPGRSFHYGMDV | 0.6150 | |
| K1 | 1D-12*02 | QQANSFPWT | 3–23*04 | VEAVAGRGVFDY | 1.5910 | |
| K2 | 3–20*01 | QQYGSSHRT | 4-b*02 | DSSYEGIIY | 1.3090 | |
| K3 | 3–20*01 | QQYGSSPPRLT | 3–11*01 | VRGTLYYYDMSGQLHDWYFDL | 0.5490 | |
| K4 | 3–20*01 | QQYATSLRT | 4-b*02 | DSSYEGIIY | 1.1920 | |
| K6 | 1D-39*01 | QQSYSTPLT | 3–23*04 | GAAAGRRSWFDP | 1.1370 | |
| K7 | 3–20*01 | QQYATSLRT | 4-b*02 | DSSYNGFDP | 1.5680 | 1:16 |
| K8 | 3–20*01 | QQYATSLRT | 4-b*02 | DLPYGGIDY | 1.7910 | |
| K9 | 1D-39*01 | QQSYSTPYT | 3–11*01 | VRGTLYYYDMSGQLHDWYFDL | 1.3940 | |
| K10 | 3–20*01 | QQYGSSPIT | 3–33*05 | DRGTWIQLRGSTGRYFQH | 0.3840 | |
| K12 | 3–20*01 | QQYGSSPSIT | 3–11*01 | VRGTLYYYDMSGQLHDWYFDL | 1.0690 | |
| K13 | 3–20*01 | QQYGSSQRT | 4-b*02 | DSSYEGIIY | 1.0470 | |
| K15 | 3–20*01 | QQYGSSPRNT | 3–11*01 | VRGTLYYYDMSGQLHDWYFDL | 1.1800 | |
| K16 | 3–20*01 | QQYGSSLLT | 4-b*02 | DTNFWSALDY | 1.4640 | |
| K17 | 3–20*01 | QQYGSPRT | 3–21*02 | DWGYEALFDY | 1.1790 | |
| K18 | 1D-39*01 | QQSYTTPRAVT | 3–11*01 | VRGTLYYYDMSGQLHDWYFDL | 1.0800 | |
| K20 | 3–11*01 | QQRSNFPIT | 3–11*01 | VRGTLYYYDMSGQLHDWYFDL | 1.5250 | |
| K21 | 1D-39*01 | QQSYSTPIT | 3–23*04 | GAAAGRQAYFDY | 1.5050 | |
| K22 | 3–11*01 | QQRSNWPPL | 3–11*01 | VRGTLYYYDMSGQLHDWYFDL | 1.3860 | |
| K23 | 1D-33*01 | QQYENLSLVT | 3–11*01 | VRGTLYYYDMSGQLHDWYFDL | 0.7080 |
Abbreviations: VL: V gene segments of the light chain; LCDR3: amino acid sequences of the CDR3 of the light chain; VH: V gene segments of the heavy chain; HCDR3: amino acid sequences of the CDR3 of the heavy chain.
Figure 1.
Phylogenetic analysis of the heavy and light chains of EV71-specific human Fab antibodies identified in this study. The phylogenetic trees were constructed by the minimum evolution method implemented in the software MEGA5. (A) the heavy chains; (B) the light chains. The bars indicate the distance scales in the trees measured by the number of sequence differences at the nucleic acid level.
These Fab antibodies demonstrate high avidity to EV71 but do not react with the control virus
According to the reactivity of Fabs to EV71, the 27 Fabs can be divided into three classes. class I (OD450<0.5), the lowest activating to EV71, only includes Fab K10. class II (OD450>1.5), the highest activating to EV71, includes Fabs L4, L11, K1, K7, K8, K20 and K21. class III (0.5<OD450<1.5) includes all the other Fabs (Fig. 2A). The immune specificity of all Fabs was tested by ELISA in 96-well micro-plates coated with inactivated enteroviruses (EV71, CVA4, CVA16, CVB3, CVB5, Echo25 and Echo30). A serum sample from an EV71-infected patient reacting with all of the above enteroviruses was set as the positive control. Of the 27 Fab clones we selected, 26 reacted with only the inactivated EV71, and did not react with the inactivated CVA4, CVA16, CVB3, CVB5, Echo25 and Echo30. K10 also reacted with CVA4 and CVB5 (Fig. 2B).
Figure 2.
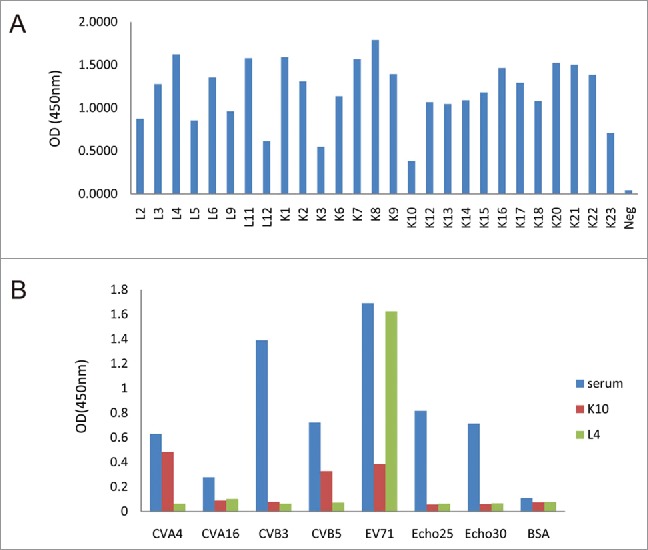
Characterization of specificity of Fab antibodies (A) reactivity of the identified Fabs to EV71 strain Shzh-98 at the concentrations of 100 ng/ml to the indicated virus measured by ELISA; (B) reactivity of the identified Fabs K10 and L4 to EV71 and other enteroviruses (CVA4, CVA16, CVB3, CVB5, Echo25 and Echo30) at the concentrations of 100 ng/ml to the indicated virus measured by ELISA. The reactivity profiles of other Fabs against all the enteroviruses are similar to L4 (positive to EV71 and negative to others).
Four of these Fab antibodies demonstrate neutralizing activity
To evaluate the neutralizing activities of the 27 Fabs, the neutralizing assays were performed using the EV71 virus strains Shzh-98 and Fuyang-0805. Two-fold serial dilutions of each of the purified Fabs were incubated together with 100TCID50 EV71 and rhabdomyosarcoma (RD) cells, and the cytopathic effects (CPE) of cells was observed 4 d after infection. Serum from a convalescent EV71-infected patient was used as a positive control. Among the 27 Fabs, 23 did not have any neutralizing activity. In contrast, 4 of the Fabs (L4, L6, L11 and K7) protected RD cells from EV71-induced CPE at antibody concentrations ranging from 2.2–7.8 ug/ml (Fig. 3). In the western blot experiments, 4 of the neutralizing Fabs did not demonstrate reactivity against EV71, indicating that these Fabs likely bind to conformational epitopes on EV71 (data not show).
Figure 3.
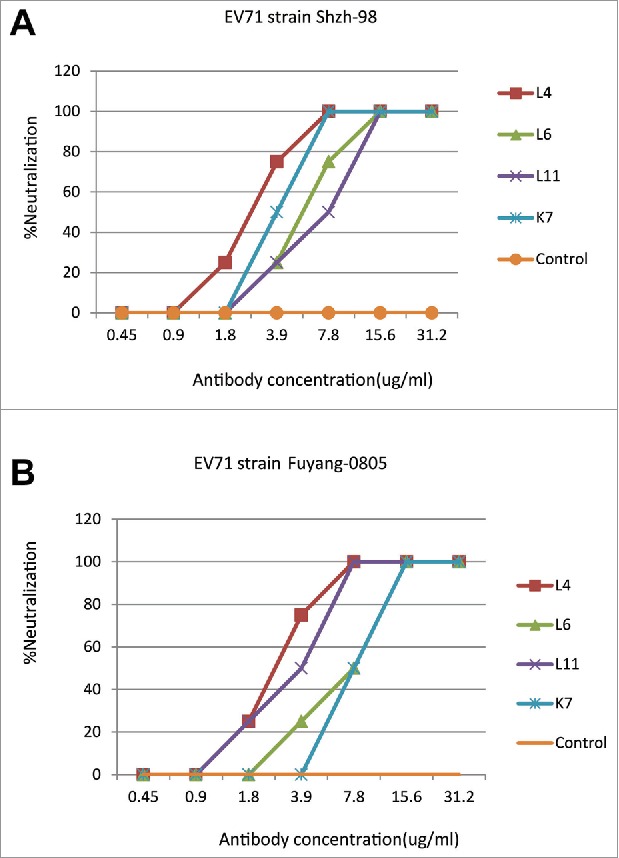
Neutralization activities of Fab antibodies to the EV71 strain Shzh-98 (A) and Fuyang-0805 (B). In vitro neutralizing assays of the positive Fabs L4, L6, L11, K7 and control IgG (anti-Flu) were illustrated. Fabs (ug/ml) were serially 2-fold diluted. Neutralization activities were evaluated by CPE assays using RD cells. The neutralizing antibody titer was defined as the highest dilution of Fabs that completely suppressed the virus-induced CPE in 50% of the wells.
The heavy chains of these antibodies are conserved among EV71-infected patients
To further evaluate whether the EV71-specific human antibodies are conserved between EV71-infected patients, we sequenced the B-cell transcriptomes of another three independent EV71-infected patients using next-generation sequencing technology. We first amplified by reverse transcription polymerase chain reaction (RT-PCR) the full-length variable regions of human antibody heavy chains in the PBMCs extracted two (Time point One, T1) and twelve (Time point Two, T2) days after hospitalization from three EV71-infected patients with severe HFMD symptoms. Then, all the amplicons were sequenced on the Roche 454 Genome Sequencer FLX System. A total of 274055, 247219 and 149739 reads were obtained for the three patients at T1 and 301633, 172610 and 248467 reads were obtained at T2. After filtering those sequencing reads with errors, we evaluated the abundance of the EV71-specific antibody heavy chains in each individual.
Because some of the 27 EV71-specific antibodies share the same heavy chains, we clustered EV71-specific antibody heavy chains into 17 unique groups (G1: K9, K12, K15, K18, K22, K23, L2 and L6; G2: K2, K4 and K13; G3: L3 and L12; G4: K7; G5: K8; G6: K20; G7: K1; G8: K16; G9: K21; G10: L11; G11: K17; G12: K6; G13: L9; G14: K3; G15: L4; G16: K10; G17: L5). We then clustered the sequencing reads with these 17 unique antibody heavy chains after trimming the 5′ and 3′ end irregularity (see the Materials and Methods section for details). We found that all the 17 unique heavy chains except G11, G16 and G17 were found in all three of the EV71-infected patients with more than 99% sequence identity (Fig. 4). G11 was found in two patients. G17 was found only in one patient, and G16 was not found in either patient. A total of 14, 9 and 8 unique heavy chains were found with more than 10 reads in Patients 1, 2 and 3, respectively, while 6, 3 and 3 unique heavy chains were found with more than 100 reads in Patients 1, 2 and 3, respectively.
Figure 4.
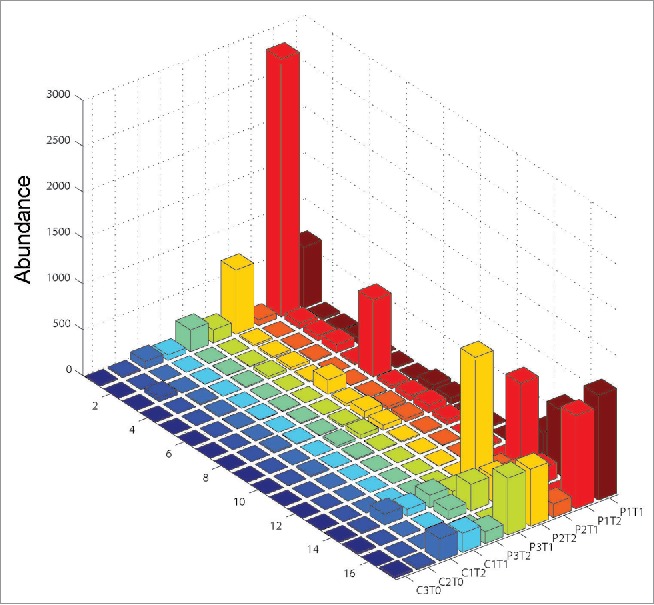
Number of sequencing reads (after normalization of the total read numbers to 100000) of the 17 unique heavy chain groups of the 27 EV71-specific human antibodies in three EV71-infected patients (P1, P2 and P3), one CVA4-infected control (C1) and two healthy controls without known EV71-infection history (C2 and C3). G1: K9, K12, K15, K18, K22, K23, L2 and L6; G2: K2, K4 and K13; G3: L3 and L12; G4: K7; G5: K8; G6: K20; G7: K1; G8: K16; G9: K21; G10: L11; G11: K17; G12: K6; G13: L9; G14: K3; G15: L4; G16: K10; G17: L5. T0: sampling time points for healthy controls; T1: 2 d after hospitalization; T2: 12 d after hospitalization.
As controls, we sequenced the B-cell transcriptomes of another three individuals. One was identified as infected by coxsackievirus A4 (CVA4); this patient's PBMCs were sampled two and 12 d after hospitalization. The other two controls were healthy children with no known history of EV71 infection (serum-negative to EV71). In total, 200665, 223768, 238336 and 217903 sequencing reads were obtained for each control. Twelve unique heavy chains were found in the CVA4-infected control, among which 5 unique heavy chains had more than 10 reads and only 1 heavy chain has more than 100 reads. Twelve and 10 unique heavy chains were found in the healthy controls, respectively; 4 and 3 unique heavy chains had more than ten reads, respectively. However, no individual had unique heavy chains with more than 100 reads.
We further analyzed whether the abundance of these EV71-specific antibody heavy chains can statistically distinguish the EV71-infected patients from the controls by Wilcoxon rank sum test. Although the small sample size provides quite limited statistical power, with none of the 17 unique heavy chains showing statistical significance, 9 unique heavy chains demonstrated small p-values (0.1).
The abundance distribution of the heavy chains is not even, but it is similar between EV71-infected patients
We noticed that the abundance of each unique heavy chain was not evenly distributed. For EV71-infected Patient 1, G7 had the highest abundance (1143 reads) and G1 had the second highest abundance (713 reads). G11 was the detectable unique heavy chain with the lowest abundance (4 reads). The ratio between the highest and lowest abundance values thus reaches 285-fold. Similarly, the ratios between the highest and lowest abundance values in Patients 2 and 3 reach 73-fold and 619-fold, respectively.
However, the abundance distributions were similar between EV71-infected patients (Fig. 5). We calculated the Spearman's correlation coefficients between the three distribution and observed that the correlation coefficients were as high as 0.9. But the abundance distributions between EV71-specific patients and the controls were not similar, with the Spearman's correlation coefficients as small as 0.3.
Figure 5.
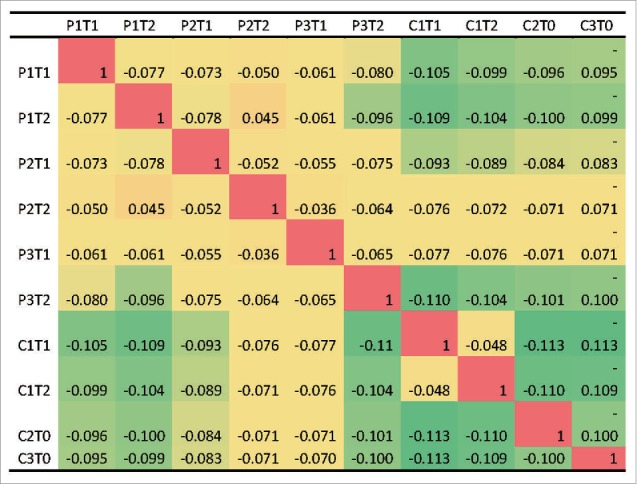
Similarities of abundance distributions among the EV71-infected patients and the controls measured by the Spearman correlation coefficients. P1, P2 and P3: EV71-infected patients; C1: CVA4-infected control; C2 and C3: two healthy controls without known EV71-infection history. T0: sampling time points for healthy controls; T1: 2 d after hospitalization; T2: 12 d after hospitalization.
Discussion
In this study, we prepared human antibodies specifically against EV71 for the first time, to the best of our knowledge, and identified four with neutralizing activity. These EV71-specific human antibodies may provide valuable information and serve as tools for the development of effective antiviral therapies and vaccines against EV71 infection.
Using next-generation sequencing technology, we also examined the distribution of the heavy chains of EV71-specific human antibodies among EV71-infected patients and three representative controls, including one patient infected by another enterovirus and two healthy individuals. This technology allows repertoire-wide mapping of the human antibodyome at the transcript level and has been widely used. The successful application of next-generation technology to sequencing of the antibody repertoires of humans and model organisms has been used to improve understanding of the composition and variation of human humoral immune responses.4,9-12 However, the abundance and distribution of virus-specific antibody heavy chains have not been reported until now, to our best knowledge. The conservation of EV71-specific human antibodies among EV71-infected patients with regards to both the sequences and the abundance distribution is expected to provide important insights into the human humoral immune responses against viral infection.
The identification of epitopes for these EV71-specific human antibodies is important for future studies. We applied western blotting and peptide arrays to screen the possible linear epitopes. However, in the western blot, the neutralizing Fabs did not react with denatured EV71 virions, and only the neutralizing Fab K7 had weak activity with some synthesized peptides (VP1, amino-acid residues 202–254), in which some linear epitopes (VP1 208–222 and 240–260) had been reported as neutralizing epitopes.13,14 These results suggest that these Fabs are likely to bind to conformational epitopes on live EV71 virions.
It is impossible that the apparent conservation was caused by nucleic contamination because the experiments of EV71-infected patients and the controls were conducted at the same time. The few EV71-specific antibody heavy chains in the healthy controls may not imply the existence of EV71-specific antibodies because the status of the corresponding light chains is still unknown. The relatively more abundant EV71-specific antibody heavy chains appearing in the coxsackievirus-A4-infected control may indicate that antibodies against EV71 and CVA4 might share similar heavy chains. However, many more experiments should to be done to clarify these phenomena.
In contrast to many of the human antibody repertoire studies which only analyzed complementarity-determining region (CDR)3,11,15-17 we sequenced the full length antibody variable regions. One benefit to our approach is that we can depict more finely and accurately the whole variations of human antibodies. Because of the ignorance of variations induced by CDR1 and CDR2, antibodies with the same CDR3 but different CDR1 and CDR2 may be merged together, and thus the true antibody abundance distribution may be distorted.
We also sequenced the repertoires in sufficient depth to capture those EV71-specific antibodies with low abundance. For example, we obtained 274055 raw 454 reads for the heavy chains for patient 1, and identified 16 of the total 17 unique heavy chains of the EV71-specific antibodies. If the sequencing depth was only 10% of the current depth (∼27405 reads), two (average on 100 bootstrap data sets) types of the unique heavy chains would not been detected. If the sequencing depth became only 5% (∼13703 reads), only nine unique heavy chains of the EV71-specific antibodies would be detected (average on 100 bootstrap data sets).
In conclusion, we prepared 27 (including four neutralizing) human monoclonal antibodies specifically against EV71 and proposed an elaborate landscape of human antibody repertoires for three EV71-infected patients and three negative controls. The identified EV71-specific antibodies and patterns of human antibody repertoires between EV71-infected patients and controls are expected to provide much help for the diagnosis and therapeutics of EV71 in the future.
Materials and methods
The ethics and human subjects involved in this study
This study was reviewed and approved by the Ethics Committee of the Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College. Peripheral blood samples from HFMD patients and control children were collected by Linyi People's Hospital in Shandong Province, China. Written informed consents from their parents were obtained for the use of peripheral blood samples. The patient information is listed in the supplemental Table 1.
The peripheral blood samples of 11 EV71-infected HFMD children, one coxsackievirus A4 infected child, and two healthy controls were collected by Linyi People's Hospital in Shandong Province, China. HFMD children infected by EV71 and coxsackievirus A4 were selected according to their clinical symptoms (appearing vesicles on hand, foot, buccal mucosa and tongue, with a fever and lymphadenopathy, usually maintained 5 to 6 d before rehabilitation, and with either one of the following severe illnesses: aseptic meningitis, acute hemorrhagic conjunctivitis, acute flaccid paralysis, undifferentiated rash, myocarditis, or neonatal sepsis-like disease), and confirmed by sequencing the amplified VP1 sequences of enterovirus in the corresponding patients.18,19
Cells and viruses involved in phage display study
RD cells (ATCC) were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco) containing 10% fetal bovine serum (FBS; Gibco). The enterovirus used in this study include EV71 strain Shzh-98 (GenBank accession number: AF302996), EV71 strain Fuyang-0805 (GenBank accession number: FJ439769), CoxsackievirusA4 (CVA4,GenBank accession number: hq728260), CoxsackievirusA16, (CVA16, GenBank accession number: AY790926) CoxsackievirusB3, (CVB3, GenBank accession number: JX976770) CoxsackievirusB5, (CVB5,GenBank accession number: jx276378) Echovirus25 (Echo25, GenBank accession number:JX976772) and Echovirus30 (Echo30, GenBank accession number:JX976773) were all isolated from patients with HFMD from China.
Virus titer determination and purification
The enteroviruses (EV71, CVA4, CVA16, CVB3, CVB5, Echo25 and Echo30) titers were determined as the median end-point of the tissue culture infectious dosage (TCID50). Serially diluted virus samples (from 10−2 to10−9) were added to RD cells in 96-well plates (Costar), and eight wells were used at each dilution. The 96-well plates were incubated for 7 d at 37°C, and the TCID50 values were measured by determining CPE. The TCID50 values were calculated by the method of Reed-Muench.20 The virus was inactivated by formalin for 5 d at 44°C, then purified by precipitation with 7% polyethyleneglycol 8000 and then centrifuged onto a 30% sucrose cushion at 25,000 g for 3h.
Generation of recombinant human Fabs to EV71 by phage display
PBMC were isolated from the peripheral blood of eight Chinese volunteers gathered two weeks after hospitalization. The lymphocytes were used for RNA extraction and purification by Qiagen's RNeasy kit following the manufacturer's protocol. The RNAs were on-column treated with DNase I (Qiagen) to eliminate genomic DNA using the provided protocol. cDNA was generated from the total RNA as template using Thermoscript reverse transcriptase (Invitrogen) with oligodT as primer.
The heavy and light chain genes were amplified from the cDNAs by PCR and sequentially cloned into the phagemid vector pComb3H (provided by The Scripps Research Institute). The anti-EV71 phage antibody library was constructed by using primers and methods as previously described.21 The antibody library was screened by panning on purified EV71 strain Shzh-98.22 After three rounds of panning, crude Fab antibody preparations were tested by indirect ELISA using 96-well plates coated with 50 ng per well of purified Shzh-98 virus. Horseradish peroxidase (HRP) conjugated anti-human Fab (Sigma, A0293, 059K4803) was used as the secondary antibody. Sequence analysis of all selected Fab clones with VBASE2 database revealed the presence of 27 unique clones. The 27 human Fabs were purified on an anti-fab affinity chromatography column for further characterization and functional analysis. Purity of human Fabs was confirmed using SDS-PAGE analysis.
ELISA assay
The 96-well plates were coated overnight at 4°C with purified Enterovirus (EV71, CVA4, CVA16, CVB3, CVB5, Echo25 and Echo30) virus per well (100ul, 50ng). After blocking with 2% bovine serum albumin (BSA), the plates were incubated with 100 ng/ml Fabs at the indicated dilutions at 37°C for 1 h and then washed three times with phosphate-buffered saline (PBS) containing 0.1% Tween-20. HRP-conjugated goat anti-human IgG (Fab specific) antibody (Sigma, A0293, 059K4803) was used as secondary antibody at 37°C for 1 h and the reaction was developed by 3,3′,5,5′-tetramethylbenzidine substrate. The absorbance at 450 nm was measured by an ELISA plate reader (Tecan). Each assay was performed three times independently.
Western blot analysis
Purified EV71 strain Shzh-98 virions suspended in loading buffer were electrophoresed in 12% SDS-PAGE and then transferred to a PVDF membrane. After blocking with PBS containing 1% BSA for 1 h at room temperature, the membrane-immobilized proteins were probed by purified human MAbs (2 ug/ml) or mouse anti-EV71 antibody (2 ug/ml) as positive control overnight at 4°C. Goat anti-human (Sigma, A8667, 060M4795) or anti-mouse antibody conjugated to HRP (Sigma, A4416, 090M6011) was used as secondary antibody. The bands were visualized by diaminobenzidine according to the manufacturer's instructions.
Neutralization assay
Briefly, Fabs were diluted serially 2-fold using DMEM containing 2% FBS, then the 50 ul 100TCID50 EV71 viral samples combined with Fabs were incubated at 37°C for 1 hour. The virus and Fab mixtures (total 100 ul) were inoculated on 70%-confluent RD cells in a 96-wellplate and monitored for the development of characteristic CPE for 4 d. The highest dilution of Fabs that inhibited virus growth was considered the neutralization antibody titer and determined after incubation at 37°C for 4 d.
454 library preparation and sequencing
PBMC were isolated from the peripheral blood of another three Chinese volunteers gathered at 2 and 12 d after hospitalization. The lymphocytes were used for RNA extraction and purification by Qiagen's RNeasy kit following the manufacturer's protocol. The RNAs were on-column treated with DNase I (Qiagen) to eliminate genomic DNA using the provided protocol. cDNA was generated from the total RNA as template using Thermoscript reverse transcriptase (Invitrogen) with oligodT as primer.
Heavy chain fragments were amplified from cDNA obtained above, using human heavy chain constant region primer chg (5- gagttccacgacaccgtcac -3) and separate primers to the variable region of each chain were designed: 6 VH for VH genes. All samples were amplified by PCR with the following conditions: a hotstart at 98°C (90 seconds), followed by 25 cycles of 98°C, (10 seconds); 58°C (30 seconds); 72°C (30 seconds), and 1 cycle of 72°C (5 minutes).
The primer sequences are given in Supplemental Table 2. A single control Ig gene of known sequence was also amplified using the same PCR conditions to evaluate the error rate for the method. PCR Primers were removed from the products by electrophoresis and QIA quick Gel Purification Kit. Prepared libraries were finally sequenced on the Roche 454 GS FLX Titanium platform.4 The raw 454 data were deposited to the NCBI SRA database with accession number SRA061645.
Bioinformatics analysis
A Perl script was developed based on the conserved sequence motifs in the variable region of the human antibody heavy chains in V-Base (http://vbase.mrc-cpe.cam.ac.uk/) to identify full-length antibody heavy chains from 454 reads. The Perl regular expression representation of the conserved motif is CC.22 TCTGG.*GAGTGG.*ACCAT.*TATTACTGT.*TGGGGC. Any read or the corresponding reverse complement containing the motif was extracted as full-length antibody heavy chain after trimming the flanking sequences upstream and downstream of the motif. Sequences that were the same or reversely complemented were clustered to calculate the absolute abundance of a certain unique antibody heavy chain. Sequences with out-of-frame indels were thought to be sequencing errors and were deleted.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the Scripps Research Institute for providing the pComb3H phagemid vector and Linyi People's Hospital in Shandong Province for providing peripheral blood samples.
Funding
This study was supported by the Chinese National Major S & T Project (2013ZX10004–101), the Program for Changjiang Scholars and Innovative Research Team in University of China (IRT13007), the National Science Foundation of China (31500757) from PR China.
References
- 1.Schmidt NJ, Lennette EH, Ho HH. An apparently new enterovirus isolated from patients with disease of the central nervous system. J Infect Dis 1974; 129:304-9; PMID:4361245; http://dx.doi.org/ 10.1093/infdis/129.3.304 [DOI] [PubMed] [Google Scholar]
- 2.McMinn PC. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol Rev 2002; 26:91-107; PMID:12007645; http://dx.doi.org/ 10.1111/j.1574-6976.2002.tb00601.x [DOI] [PubMed] [Google Scholar]
- 3.Wu Y, Yeo A, Phoon MC, Tan EL, Poh CL, Quak SH, Chow VT. The largest outbreak of hand; foot and mouth disease in Singapore in 2008: the role of enterovirus 71 and coxsackievirus A strains. Int J Infect Dis 2010; 14:e1076-81; PMID:20952237; http://dx.doi.org/ 10.1016/j.ijid.2010.07.006 [DOI] [PubMed] [Google Scholar]
- 4.Mao LX, Wu B, Bao WX, FA Han, Xu L, Ge QJ, Yang J, Yuan ZH, Miao CH, Huang XX, et al.. Epidemiology of hand, foot, and mouth disease and genotype characterization of Enterovirus 71 in Jiangsu, China. J clin Virol 2010; 49:100-4; PMID:20719557; http://dx.doi.org/ 10.1016/j.jcv.2010.07.009 [DOI] [PubMed] [Google Scholar]
- 5.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, et al.. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 2011; 333:1593-602; PMID:21835983; http://dx.doi.org/ 10.1126/science.1207532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang T, Du J, Xue Y, Su H, Yang F, Jin Q. Epidemics and frequent recombination within species in outbreaks of Human Enterovirus B-Associated hand, foot and mouth disease in Shandong China in 2010 and 2011. PLoS One 2013; 8:e67157; PMID:23840610; http://dx.doi.org/ 10.1371/journal.pone.0067157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin YW, Chang KC, Kao CM, Chang SP, Tung YY, Chen SH. Lymphocyte and antibody responses reduce enterovirus 71 lethality in mice by decreasing tissue viral loads. J Virol 2009; 83:6477-83; PMID:19386699; http://dx.doi.org/ 10.1128/JVI.00434-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glanville J, Zhai W, Berka J, Telman D, Huerta G, Mehta GR, Ni I, Mei L, Sundar PD, Day GM, et al.. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Nati Acad Sci USA 2009; 106:20216-21; PMID:19875695; http://dx.doi.org/22200891 10.1073/pnas.0909775106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang N, Weinstein JA, Penland L, White RA, Fisher DS, Quake SR. Determinism and stochasticity during maturation of the zebrafish antibody repertoire. Proc Nati Acad Sci USA 2011; 108:5348-53; PMID:21393572; http://dx.doi.org/22200891 10.1073/pnas.1014277108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prabakaran P, Chen W, Singarayan MG, Stewart CC, Streaker E, Feng Y, Dimitrov DS. Expressed antibody repertoires in human cord blood cells: 454 sequencing and IMGT/HighV-QUEST analysis of germline gene usage, junctional diversity, and somatic mutations. Immunogenetics 2012; 64:337-50; PMID:22200891; http://dx.doi.org/ 10.1007/s00251-011-0595-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnaout R, Lee W, Cahill P, Honan T, Sparrow T, Weiand M, Nusbaum C, Rajewsky K, Koralov SB. High-Resolution Description of Antibody Heavy-Chain Repertoires in Humans. PLoS One 2011; 6:e22365; PMID:21829618; http://dx.doi.org/ 10.1371/journal.pone.0022365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinstein JA, Jiang N, White RA 3rd, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science 2009; 324:807-10; PMID:19423829; http://dx.doi.org/ 10.1126/science.1170020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foo DG, Alonso S, Phoon MC, Ramachandran NP, Chow VT, Poh CL. Identification of neutralizing linear epitopes from the VP1 capsid protein of Enterovirus 71 using synthetic peptides. Virus Res 2007; 125:61-8; PMID:17222936; http://dx.doi.org/ 10.1016/j.virusres.2006.12.005 [DOI] [PubMed] [Google Scholar]
- 14.Chang GH, Luo YJ, Wu XY, Si BY, Lin L, Zhu QY. Monoclonal antibody induced with inactived EV71-Hn2 virus protects mice against lethal EV71-Hn2 virus infection. Virol J 2010; 7:7-106; PMID:20078890; http://dx.doi.org/ 10.1186/1743-422X-7-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung WC, Beausoleil SA, Zhang X, Sato S, Schieferl SM, Wieler JS, Beaudet JG, Ramenani RK, Popova L, Comb MJ, et al.. A proteomics approach for the identification and cloning of monoclonal antibodies from serum. Nat Biotechnol 2012; 30:447-52; PMID:22446692; http://dx.doi.org/ 10.1038/nbt.2167 [DOI] [PubMed] [Google Scholar]
- 16.Reddy ST, Ge X, Miklos AE, Hughes RA, Kang SH, Hoi KH, Chrysostomou C, Hunicke-Smith SP, Iverson BL, Tucker PW, et al.. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat Biotechnol 2010; 28:965-9; PMID:20802495; http://dx.doi.org/ 10.1038/nbt.1673 [DOI] [PubMed] [Google Scholar]
- 17.Racanelli V, Brunetti C, De Re V, Caggiari L, De Zorzi M, Leone P, Perosa F, Vacca A, Dammacco F. Antibody Vh repertoire differences between resolving and chronically evolving Hepatitis C virus infections. PLoS One 2011; 6:e25606; PMID:21980500; http://dx.doi.org/ 10.1371/journal.pone.0025606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iturriza-Gómara M, Megson B, Gray J. Molecular detection and characterization of human enteroviruses directly from clinical samples using RT-PCR and DNA sequencing. J Med Virol 2006; 78:243-53;PMID:16372287; http://dx.doi.org/16891480 10.1002/jmv.20533 [DOI] [PubMed] [Google Scholar]
- 19.Nix WA, Oberste MS, Pallansch MA. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J Clin Microbiol 2006; 44:2698-704; PMID:16891480; http://dx.doi.org/ 10.1128/JCM.00542-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed LJ Mh. A simple method of estimating 50 percent end-points. Am J Hyg 1938; 27:493-7. [Google Scholar]
- 21.Barbas CF 3rd, Kang AS, Lerner RA, Benkovic SJ. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc Natl Acad Sci USA 1991; 88:7978-82; PMID:1896445; http://dx.doi.org/ 10.1073/pnas.88.18.7978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbas CF 3rd, Burton DR. Selection and evolution of high-affinity human anti-viral antibodies. Trends Biotechnol 1996; 14:230-4; PMID:8771795; http://dx.doi.org/ 10.1016/0167-7799(96)10029-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.