Abstract
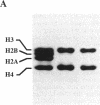
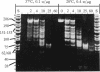
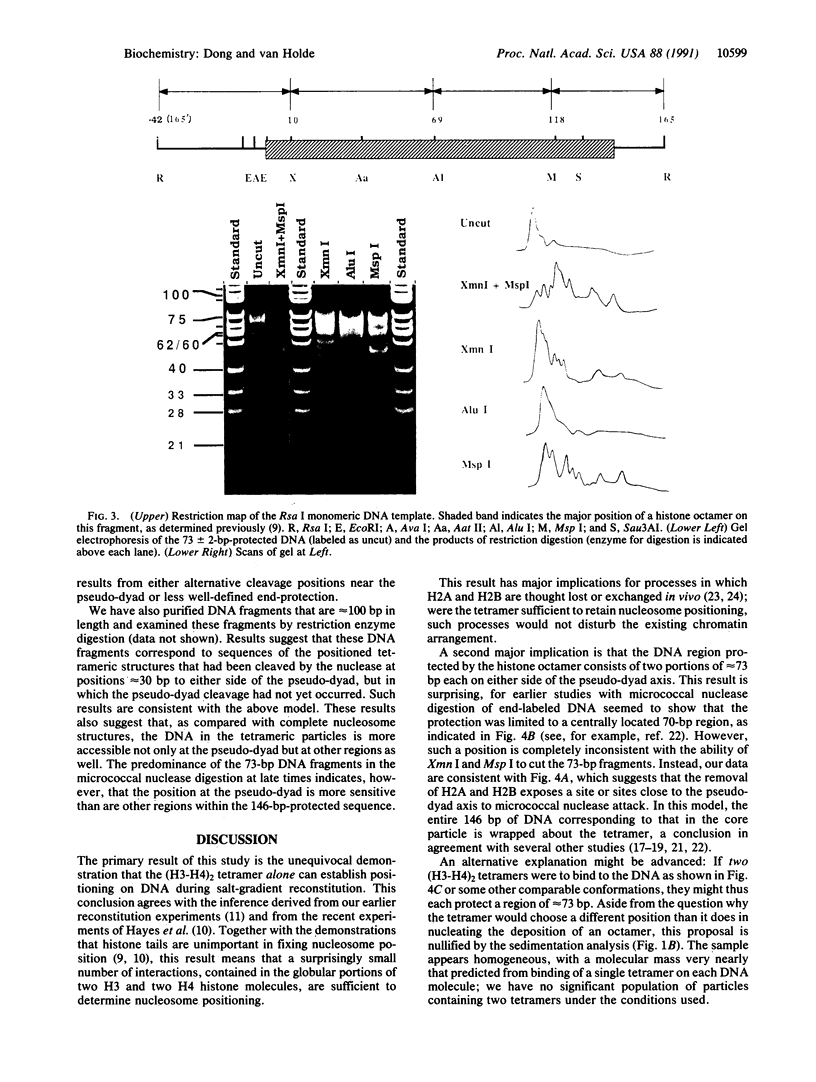
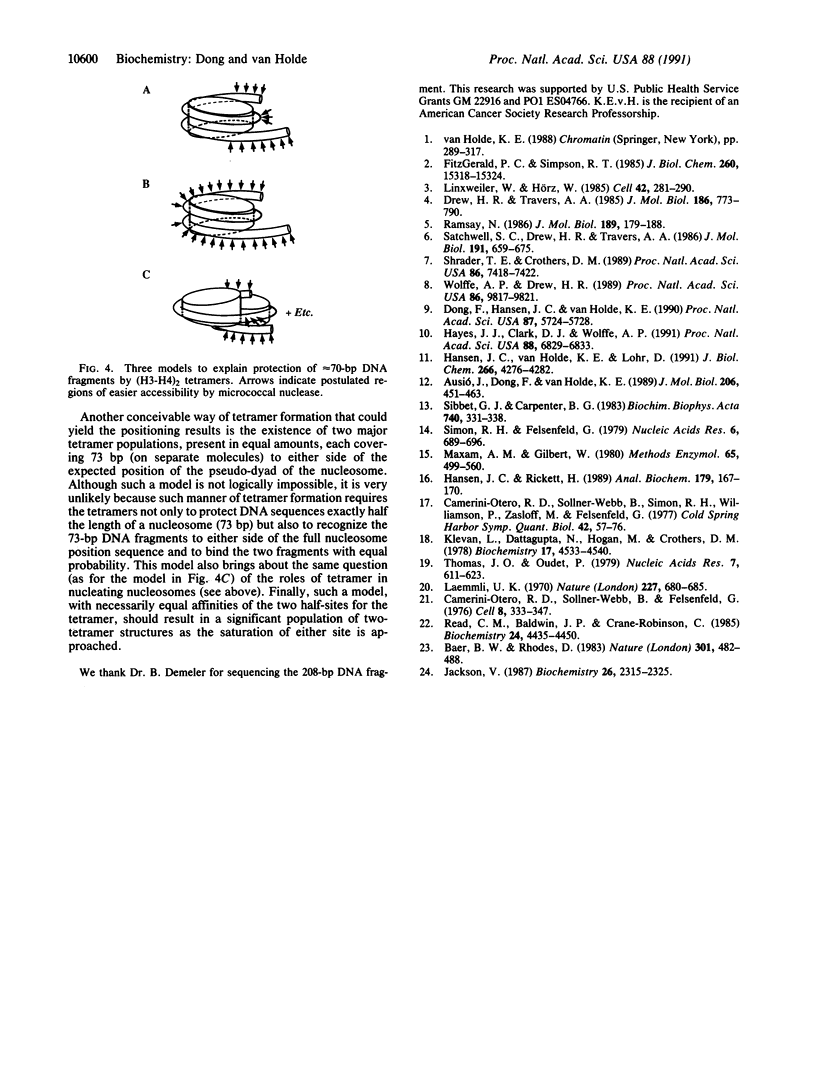
It is demonstrated that the histone (H3-H4)2 tetramer can find specific positions on DNA, even in the absence of other histones. Purified histone (H3-H4)2 tetramers were reconstituted onto 208-base-pair (bp) DNA molecules containing a nucleosome-positioning sequence by using salt-gradient dialysis. The stoichiometry of histone tetramer to DNA was shown to be 1:1. Digestion with micrococcal nuclease led to formation of protected DNA fragments of approximately 73 bp. Cleavage of the 73-bp DNA with restriction enzymes produced a small set of defined bands, demonstrating positioning of the (H3-H4)2 tetramer on DNA. Analysis of the restriction digests shows that the 73-bp DNA corresponds mainly to two fragments, one lying on either side of the pseudo-dyad axis of the major position adopted by complete histone octamers on this DNA. This result means that a single (H3-H4)2 histone tetramer can fold approximately 146 bp of DNA with the same positioning as the complete octamer but that a region near the pseudo-dyad is only weakly protected against micrococcal nuclease attack in the absence of histones H2A and H2B.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ausio J., Dong F., van Holde K. E. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone "tails" in the stabilization of the nucleosome. J Mol Biol. 1989 Apr 5;206(3):451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- Baer B. W., Rhodes D. Eukaryotic RNA polymerase II binds to nucleosome cores from transcribed genes. Nature. 1983 Feb 10;301(5900):482–488. doi: 10.1038/301482a0. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976 Jul;8(3):333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Simon R. H., Williamson P., Zasloff M., Felsenfeld G. Nucleosome structure, DNA folding, and gene activity. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):57–75. doi: 10.1101/sqb.1978.042.01.008. [DOI] [PubMed] [Google Scholar]
- Dong F., Hansen J. C., van Holde K. E. DNA and protein determinants of nucleosome positioning on sea urchin 5S rRNA gene sequences in vitro. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5724–5728. doi: 10.1073/pnas.87.15.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- FitzGerald P. C., Simpson R. T. Effects of sequence alterations in a DNA segment containing the 5 S RNA gene from Lytechinus variegatus on positioning of a nucleosome core particle in vitro. J Biol Chem. 1985 Dec 5;260(28):15318–15324. [PubMed] [Google Scholar]
- Hansen J. C., Rickett H. Large-scale purification of plasmid insert DNA sequences using low-percentage agarose exclusion chromatography. Anal Biochem. 1989 May 15;179(1):167–170. doi: 10.1016/0003-2697(89)90219-4. [DOI] [PubMed] [Google Scholar]
- Hansen J. C., van Holde K. E., Lohr D. The mechanism of nucleosome assembly onto oligomers of the sea urchin 5 S DNA positioning sequence. J Biol Chem. 1991 Mar 5;266(7):4276–4282. [PubMed] [Google Scholar]
- Hayes J. J., Clark D. J., Wolffe A. P. Histone contributions to the structure of DNA in the nucleosome. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6829–6833. doi: 10.1073/pnas.88.15.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V. Deposition of newly synthesized histones: new histones H2A and H2B do not deposit in the same nucleosome with new histones H3 and H4. Biochemistry. 1987 Apr 21;26(8):2315–2325. doi: 10.1021/bi00382a037. [DOI] [PubMed] [Google Scholar]
- Klevan L., Dattagupta N., Hogan M., Crothers D. M. Physical studies of nucleosome assemble. Biochemistry. 1978 Oct 17;17(21):4533–4540. doi: 10.1021/bi00614a027. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Linxweller W., Hörz W. Reconstitution experiments show that sequence-specific histone-DNA interactions are the basis for nucleosome phasing on mouse satellite DNA. Cell. 1985 Aug;42(1):281–290. doi: 10.1016/s0092-8674(85)80123-9. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Ramsay N. Deletion analysis of a DNA sequence that positions itself precisely on the nucleosome core. J Mol Biol. 1986 May 5;189(1):179–188. doi: 10.1016/0022-2836(86)90389-x. [DOI] [PubMed] [Google Scholar]
- Read C. M., Baldwin J. P., Crane-Robinson C. Structure of subnucleosomal particles. Tetrameric (H3/H4)2 146 base pair DNA and hexameric (H3/H4)2(H2A/H2B)1 146 base pair DNA complexes. Biochemistry. 1985 Jul 30;24(16):4435–4450. doi: 10.1021/bi00337a027. [DOI] [PubMed] [Google Scholar]
- Satchwell S. C., Drew H. R., Travers A. A. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986 Oct 20;191(4):659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Shrader T. E., Crothers D. M. Artificial nucleosome positioning sequences. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbet G. J., Carpenter B. G. Selective depletion and reconstitution of nucleosome core particles. Biochim Biophys Acta. 1983 Aug 2;740(3):331–338. doi: 10.1016/0167-4781(83)90142-2. [DOI] [PubMed] [Google Scholar]
- Simon R. H., Felsenfeld G. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 1979 Feb;6(2):689–696. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Oudet P. Complexes of the arginine-rich histone tetramer (H3)2(H4)2 with negatively supercoiled DNA: electron microscopy and chemical cross-linking. Nucleic Acids Res. 1979 Oct 10;7(3):611–623. doi: 10.1093/nar/7.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P., Drew H. R. Initiation of transcription on nucleosomal templates. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9817–9821. doi: 10.1073/pnas.86.24.9817. [DOI] [PMC free article] [PubMed] [Google Scholar]