Abstract
Primula veris L. is an important medicinal plant with documented use for the treatment of gout, headache and migraine reaching back to the Middle Ages. Triterpenoid saponins from roots and flowers are used in up-to-date phytotherapeutic treatment of bronchitis and colds due to their expectorant and secretolytic effects. In addition to the wild type plants with yellow petals, a red variant and an intermediate orange form of Primula veris L. have recently been found in a natural habitat. The secondary metabolite profiles of roots, leaves and flowers of these rare variants were investigated and compared with the wild type metabolome. Two flavonoids, six flavonoid glycosides, four novel methylated flavonoid glycosides, five anthocyanins and three triterpenoid saponins were identified in alcoholic extracts from the petals, leaves and roots of the three variants by high performance liquid chromatography (HPLC)-diode array detection (DAD)/mass spectrometry (MSn) analyses. Anthocyanins were detected in the petals of the red and orange variety, but not in the wild type. No other effects on the metabolite profiles of the three varieties have been observed. The possibility is discussed that a regulatory step of the anthocyanin biosynthetic pathway may have been affected by mutation thus triggering color polymorphism in the petals.
Keywords: Primula veris L., primrose, HPLC-DAD-MSn, triterpenoid saponins, flavonoids, anthocyanins, color polymorphism
1. Introduction
Primula veris L. (Primulaceae), also known under the common names cowslip or cowslip primrose, is native to Europe and Siberia, as well as West and Central Asia. The perennial plant with a short rhizome grows in warm, sunny and dry habitats, especially on dry meadows and light-flooded deciduous forests [1]. Various subspecies and varieties are differentiated, however, the species is also known to hybridize, for instance, with Primula elatior L. [2]. Besides the yellow wild type of P. veris, a report of a natural orange variety [3] exists, and more recently a red flowered type has been offered commercially in garden plant markets. From a natural habitat in Southern Germany, a population of yellow, red and intermediate orange plants has been known for more than 10 years. This recently offered the possibility to analyze the phytochemistry of these rare varieties, particularly with respect to similarity or differences in pharmacologically interesting compounds known from the wild type.
The metabolome of the wild type of P. veris has been well described in the scientific literature. Beside the triterpenoid saponins and carotenoids, essential oils are also present in P. veris petals [4], whereby the carotenoids provide the typically yellow flower color. However, phenolic compounds belonging to the flavones and flavonols are the main components of the yellow petals. Already in 1927, the phenolic compound patterns of various Primula species have been characterized and used for species identification [5]. Especially for P. veris, the flavonoid glycosides quercetin-3-O-rutinoside (rutin) as well as kaempferol-dirhamnoside were identified [6]. The discovery of the floral pigments gossypetin and kaempferol-3-O-gentiotrioside has also been reported [7]. Furthermore, leucocyanidin, leucodelphinidin and the flavonols myricetin, quercetin, kaempferol and their derivatives have been detected in the leaf extract of the wild type [8,9]. Recent studies have demonstrated the habitats and the respective abiotic factors to influence the flavonoid glycoside pattern in the petals and leaves of P. veris [10]. However, only the petals and roots are used pharmaceutically, based on the expectorant and secretolytic effects of the triterpenoid saponins like priverosaponin B-22-acetate, primula acid I and primula acid II [11,12]. Phenolic glycosides, mainly primeverin and primulaverin, are also characteristic compounds of Primula roots and have been suggested as indicators of the age of the plant material [12]. Moreover, Mueller et al. [12] described the first liquid chromatographic method suitable for the characterization of bioactive compounds, i.e., saponins and phenolic glycosides, present in P. veris and P. elatior. A more comprehensive characterization of the metabolite profile with focus on triterpenoid saponins and phenolic compounds (flavonoid glycosides including anthocyanins) in the petals, leaves and roots has not been reported so far. Therefore, we have developed a high performance liquid chromatography-diode array detection/mass spectrometry (HPLC-DAD-MSn) method to analyze and compare the secondary metabolite profile of the different plant parts. The study presented here is also focused on the identification of secondary compounds, especially anthocyanins in the Primula petals in order to unravel the chemical background of the different Primula color variants. Furthermore, the possible correlation between the variations in the flower colors caused by the pigment change and the variation in the metabolite profile of P. veris was also investigated. The phytochemical characterization of the Primula variants and the different plant shall complement the literature data of the therapeutically used Primula species.
2. Results
2.1. Color Variants of Primula veris
Three noticeable color variants of P. veris were found in their natural habitat in southern Germany (Figure 1). Besides the typical wild type with yellow petals and orange spots at the base (Figure 1(A1,A2)), a form with deeply red petals around the yellow central part (Figure 1(C1,C2)) occurred. A third variety (Figure 1(B1,B2)) appeared intermediate to the wild type and red type in that the petal color shifted from yellow in the center to red towards the tips, thus imparting the flower an orange appeal. Light microscopy of the different petals revealed the yellow wild type to be devoid of reddish vacuoles, which were found in the epidermal papilla of the orange and red forms, most likely due to the accumulation of anthocyanins (Figure 1(A3,B3,C3–A4,B4,C4)). However, all three variants showed yellow chromoplasts in the epidermal cells colored by carotenoids. This suggested that in the red colored areas, anthocyanins in the papillate cells completely cover the chromoplasts of the basal epidermal cells and that the orange variety represents only an attenuated form, where the yellow pigments are still visible. Besides that, the color varieties shared all other phenotypical features and could not be differentiated from the wild type by distinctive morphological characters. The flower stalks reached 20–25 cm in length harboring 10–30 blossoms. The flowering stage was uniformly reached in April to May.
Figure 1.
Primula veris L. with different petal colors from the botanical garden of Hohenheim University, May 2015. Overview of P. veris with yellow petals (A1), orange petals (B1) and red petals (C1). Close-up view of the yellow petals (A2), orange petals (B2) and red petals (C2). Light microscopic images of the differently colored petals in supervision (A3,B3,C3) and in cross-section (A4,B4,C4). Yellow chromoplasts (arrows) are shown in the papilliform epidermis cells of the yellow petals. The vacuoles of the cells are colorless (A3,A4). The orange colored petals also have yellow chromoplasts. Papillate epidermal cells are characterized by the occurrence of anthocyanins in vacuoles (B3,B4). Some epidermis cells include anthocyanins (*) in their vacuoles, thus exhibiting red color. All papilliform epidermis cells of the red-colored petals have anthocyanin-containing vacuoles. Yellow chromoplasts (arrows) exist, but are masked by the content of the vacuoles (C3,C4).
2.2. Metabolite Profiles of the Primula Variants
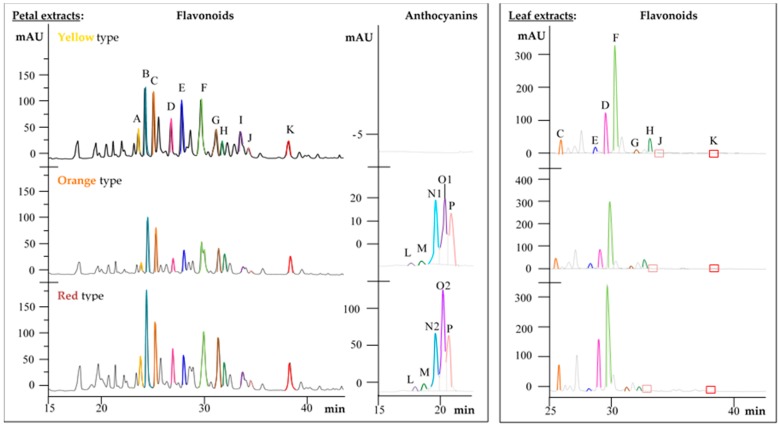
HPLC-DAD-MSn analyses of petal extracts of all three Primula varieties showed a broad spectrum of flavonoids and their glycosylated or methylated derivatives (Figure 2). By means of mass spectrometric data and database comparison, the following known compounds were assigned for all Primula variants: (+)-catechin, kaempferol-3-O-galactoside-rhamnoside-7-O-rhamnoside, kaempferol-3-O-rutinoside, kaempferol-3-O-diglucoside-7-O-glucoside, quercetin-3-O-rutinoside, quercetin-3-O-gentiobioside, quercetin-trihexoside and primula acid I. Four methylated flavonoid derivatives tentatively assigned to methyl-quercetin-dihexoside, methyl-quercetin-rhamnoside-hexoside, quercetin-methylether-rhamnoside-glucoside-rhamnoside and methyl-myricetin-trihexoside were found for the first time and characterized according to mass spectrometric data reported in the literature. The only difference between the three Primula samples was found in isorhamnetin (data not shown), which occurred in very small amounts in the orange and red varieties, but was not detected in the yellow wild type petals.
Figure 2.
Comparison of high performance liquid chromatograms of Primula veris L. petal and leaf extracts covering flavonoid glycosides at 280 nm and anthocyanins at 520 nm. Identical compounds are marked with the same color and the letters A to P: A: (+)-Catechin (yellow); B: Methyl-myricetin-trihexoside (light blue); C: Quercetin-trihexoside (orange); D: Kaempferol-3-O-diglucoside-7-O-glucoside (pink); E: Quercetin-3-O-gentiobioside (blue); F: Kaempferol-3-O-galactoside-rhamnoside-7-O-rhamnoside (light green); G: Quercetin-methylether-rhamnoside-glucoside-rhamnoside (brown); H: Quercetin-3-O-rutinoside (grey); I: Methyl-quercetin-dihexoside (purple); J: Kaempferol-3-O-rutinoside (light red); K: Methyl-quercetin-rhamnoside-hexoside (red); L: Cyanidin-hexoside (light purple); M: Cyanidin-glucoside (dark green); N1: Coelution of peonidin-dihexoside and peonidin-hexoside (aqua); N2: Coelution of peonidin-hexoside and malvidin-hexoside (aqua); O1: Coelution of malvidin-hexoside and peonidin-hexoside (purple); O2: Peonidin-dihexoside (purple); P: Malvidin-hexoside (apricot).
In addition, the petal extracts contained the saponin primula acid I in all three varieties. The metabolic difference between the color varieties was found in the HPLC patterns of methanol-water extracts of the petals, which allowed the determination of anthocyanins (Figure 2, central chromatograms). While the orange and red variety both contained cyanidin-hexoside, cyanidin-3-O-glucoside, peonidin-hexoside, peonidin-dihexoside and malvidin-hexoside, these compounds were not detected in the yellow wild type plants. All other identified flavonoids, flavonoid glycosides and triterpenoid saponins were uniformly found in the petals of all three color variants with only marginal quantitative deviations (Figure 2, left chromatograms).
Leaf extracts of the three Primula samples were characterized by very similar flavonoid and saponin profiles compared to the petals; however, individual compounds were present in different relative quantities (Figure 2, right chromatograms). Kaempferol-3-O-galactoside-rhamnoside-7-O-rhamnoside (peak F in Figure 2) was the most prominent compound in all three varieties. In the red colored type, kaempferol-3-O-diglucoside-7-O-glucoside and quercetin-trihexoside appeared slightly increased in their contents when compared with the wild type, and the orange variety contained somewhat smaller amounts of all compounds. In those parts of the chromatograms exhibiting more lipophilic compounds, trace amounts of kaempferol-3-O-rutinoside (light red box J) and methyl-quercetin-rahmnoside-hexoside (red box K) were detected and their occurrence verified by mass spectrometry (MS) data.
The root extracts of all three Primula samples provided identical compound profiles, which consisted of triterpenoid saponins, but were devoid of the flavonoids and anthocyanins detected in the above ground plant organs (Table 1). Besides, primula acid I, primula acid II and priverosaponin B-22-acetate were found in roots, thus supporting previous findings for P. veris [12,13,14]. All identified compounds with their characteristic spectrometric data and their occurrence in petals, leaves and roots of the three Primula variants are summarized in Table 1.
Table 1.
Spectroscopic data (UV, MS) and HPLC retention times (tR) of secondary metabolites from petal color variants of Primula veris L.
| Compound | tR (min) | λmax (nm) | m/z | HPLC-ESI-MS/MS Fragments, m/z | Plant Variants | Reference | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yellow Variant | Orange Variant | Red Variant | ||||||||||||
| p | l | r | p | l | r | p | l | r | ||||||
| Flavonoids | [M-H]− | |||||||||||||
| (+)-Catechin (A) | 23.7 | 228, 280 | 289 | 245, 203, 186 | + | − | − | + | − | − | + | − | − | st |
| Isorhamnetin | 50.3 | 254, 368 | 315 | 300, 271, 151 | − | − | − | + | − | − | + | − | − | st |
| Flavonoid glycosides | [M-H]− | |||||||||||||
| Quercetin-3-O-rutinoside (H) | 32.1 | 256, 356 | 609 | 301, 179, 151 | + | + | − | + | + | − | + | + | − | st |
| Quercetin-3-gentiobioside (E) | 28.3 | 256, 354 | 625 | 463, 301, 179, 151 | + | + | − | + | + | − | + | + | − | [15,16] |
| Quercetin-trihexoside (C) | 25.5 | 256, 358 | 787 | 625, 463, 301, 179 | + | + | − | + | + | − | + | + | − | [16] |
| Kaempferol-3-O-diglucoside-7-O-glucoside (D) | 29.4 | 270, 350 | 771 | 609, 285, 257, 151 | + | + | − | + | + | − | + | + | − | [17] |
| Kaempferol-3-O-rutinoside (J) | 34.4 | 264, 350 | 593 | 447, 285, 257, 229 | + | + | − | + | + | − | + | + | − | [15] |
| Kaempferol-3-O-galactoside-rhamnoside-7-O-rhamnoside (F) | 30.1 | 254, 356 | 739 | 575, 429, 282, 255 | + | + | − | + | + | − | + | + | − | [18] |
| Methylated flavonoid glycosides | [M-H]− | |||||||||||||
| Quercetin-methylether-rhamnoside-glucoside-rhamnoside (G) | 31.5 | 254, 354 | 769 | 737, 623, 605, 315, 300, 271 | + | + | − | + | + | − | + | + | − | [13] |
| Methyl-quercetin-dihexoside (I) | 33.2 | 254, 356 | 639 | 477, 315, 300, 271 | + | − | − | + | − | − | + | − | − | |
| Methyl-quercetin-rhamnoside-hexoside (K) | 37.6 | 254, 356 | 623 | 477, 315, 300, 271 | + | + | − | + | + | − | + | + | − | |
| Methyl-myricetin-trihexoside (B) | 24.3 | 278, 342 | 817 | 655, 493, 331, 316, 271 | + | − | − | + | − | − | + | − | − | |
| Anthocyanins | [M]+ | |||||||||||||
| Cyanidin-3-O-glucoside (M) | 18.1 | 280, 520 | 449 | 287, 231, 213 | − | * | * | + | * | * | + | * | * | st |
| Cyanidin-hexoside (L) | 17.3 | 280, 520 | 449 | 287, 231, 213 | − | * | * | + | * | * | + | * | * | [19] |
| Peonidin-hexoside (O1) | 19.6 | 280, 520 | 463 | 301, 286, 258 | − | * | * | + | * | * | + | * | * | [19] |
| Peonidin-dihexoside (O2) | 19.2 | 280, 520 | 625 | 301, 286, 258 | − | * | * | + | * | * | + | * | * | [19] |
| Malvidin-hexoside (P) | 20.2 | 260, 520 | 493 | 331, 315, 298, 281 | − | * | * | + | * | * | + | * | * | [19] |
| Triterpenoid saponins | [M-H]− | |||||||||||||
| Primula acid I | 53.1 | − | 1104 | 924, 465, 447, 246 | + | + | + | + | + | + | + | + | + | st |
| Primula acid II | 52.4 | − | 1236 | 924, 465 | − | − | + | − | − | + | − | − | + | [12] |
| Priverosaponin B-22-acetate | 52.1 | − | 1162 | 982, 465 | − | − | + | − | − | + | − | − | + | [12] |
p, petals; l, leaves; r, roots; +, detected; −, not detected; * not analyzed; (A to P), compounds are shown in Figure 2; st, reference standard used to verify retention time and/or UV spectrum and/or MS fragmentation pattern.
2.3. Comparison of Metabolite Profiles between Plant Parts
The characterization of different plant parts with regard to their secondary metabolite profile revealed that eight out of ten flavonoid glycosides identified in the petals were also detected in the leaves. Of the methylated flavonoids, only quercetin-methylether-rhamnoside-glucoside-rhamnoside and methyl-quercetin-rhamnoside-hexoside were detected in leaves, whereas methyl-quercetin-dihexoside and methyl-myricetin-trihexoside were missing or below the limit of detection. A range of yet unknown peaks in a retention time range of 50 to 60 min were detected only in leaf extracts but not in petal extracts, however, the yields of these components were insufficient for structure identification (data not shown). Additionally, the main triterpenoid saponin primula acid I occurred in each of the different plant parts. In contrast, primula acid II and priverosaponin B-22-acetate were exclusively detected in Primula roots (Table 1).
2.4. Identification of Novel Compounds in the Primula Variants
Three novel methylated quercetin derivatives were detected in the petals or leaf extracts for the first time. The fragment ions at m/z 315 and 300 were important for their identification and revealed the presence of an aromatic methoxyl group. Tentative structure assignment was performed as follows. Methyl-quercetin-dihexoside was identified in petal extracts from a peak with a retention time of 33.2 min by means of mass spectra revealing a characteristic [M-H]− ion at m/z 639 and fragment ions at m/z 477 and 315 ([M-H162162]−). Methyl-quercetin-rhamnoside-hexoside was detected in petals as well as leaves. The fragmentation in the MS2 experiment revealed the loss of a rhamnose moiety ([M-H146]−) with a fragment ion at m/z 477, followed by an O-hexose cleavage ([M-H-146-162]−) resulting in the corresponding fragment ion at m/z 315. Due to the lack of literature data and insufficient sample amounts for NMR investigations, further characterization of the two methyl-quercetin derivatives was not performed. Based on the study of Pemp and Krenn [13], the quercetin-methylether-rhamnoside-glucoside-rhamnoside could be characterized in more detail. Compound assignment was based on its UV spectrum (maxima at 254 nm and 354 nm) as well as mass spectrometric data, which revealed a base peak at m/z 769 and a fragment ion at m/z 737 indicating methylether cleavage by releasing methanol. The loss of rhamnose [M-H-146]− and glucose [M-H-162]− moieties were found in further fragmentation steps.
Another flavonoid glycoside, which has not been described in P. veris before, was detected in petals. MS data of the compound with a retention time of 24.3 min suggested the occurrence of a methyl-myricetin-trihexoside with characteristic fragment ions at m/z 817, 655 and 493. The MS2 experiments revealed the losses of three O-hexose moieties. The assignment of the flavonol aglycone myricetin was based on identical MS data reported by Kim et al. [14].
The anthocyanins, exclusively found in orange and red Primula petals, were also assigned based on UV and mass spectrometric measurements. Cyanidin-3-O-glucoside showed identical HPLC behavior (retention time 18.1 min) and spectroscopic characteristics (UVmax 280 and 520 nm) compared to the corresponding reference standard. The fragment ions at m/z 449 and 287 ([M-162]+) reported in the literature were confirmed. A second cyanidin-hexoside was detected exhibiting a retention time of 17.3 min. Additionally, peonidin and malvidin derivatives were identified and confirmed by comparison with literature data of Wu et al. [19]. Peaks N1,2 and O1,2 (Figure 2, central chromatograms) appeared to be coelutions of several compounds, which could only partly be resolved and indicated most likely quantitative differences between the orange and red color variants. A mixture of peonidin-dihexoside and peonidin-hexoside (m/z 625 and 463, respectively) and the typical fragment ion at m/z 301 for the peonidin aglycone at a retention time of 19.3 min was detected in the extract of the orange petals. Peak N2 in the red variety contained a substance mixture of malvidin-hexoside, peonidin-dihexoside and peonidin-hexoside. Malvidin-hexoside exhibited absorption maxima at 260 and 520 nm and a molecular ion [M]+ at m/z 493 as well as a fragment ion at m/z 331, thus indicating the loss of a hexose moiety. Furthermore, the peak at a retention time of 20.0 min revealed the presence of malvidin-hexoside and peonidin-hexoside in the orange petal extract. The peak with the same retention time in the red petal extract revealed only the occurrence of peonidin-hexoside. Finally, malvidin-hexoside was re-assigned to the peak with a retention time of 20.5 min in both orange and red petals. A tailing or fronting might be the reason for this multiple assignment of probably the same peaks. Consequently, this coelution aggravated unambiguous compound assignment, which is assumed to be as follows: N: Peonidin-dihexoside, O: Peonidin-hexoside and P: Malvidin-hexoside. All identified substances with their characteristic spectrometric data and their occurrence in the petals, leaves and roots of the three Primula variants are summarized in Table 1.
3. Discussion
Within the past century phytochemical studies of 100 species from 25 different genera of Primulaceae have been conducted [7]. Flavonoids and saponins were among the predominant compounds characteristic for this family. However, significant diversity was found with respect to specific compounds or derivatives and in their occurrence in different plant organs. This is particularly relevant for the pharmacological use of Primula species, among which P. veris plays the most important role.
Previous studies reported the petal pigment kaempferol-3-gentiotrioside from P. veris and 39 further Primula species [7]. Furthermore, the leaf pigment quercetin-3-O-rutinoside was also found in P. veris before [7]. The occurrence of these two flavonoid glycosides was confirmed in the present work, and both were found in petals as well as leaves. Further quercetin derivatives have already been identified in previous studies of P. veris and P. elatior and were used for analytical differentiation between these two medicinal plants [20]. Quercetin-3-gentiobioside, quercetin-3-O-gentiotrioside, kaempferol-3-O-rutinoside and kaempferol-3-O-robinobioside were isolated from petals of the wild type P. veris [7] and were also detected in flowers and leaves of the current study.
In addition, we were able to identify three methylated quercetin-derivatives and methyl-myricetin-trihexoside in the petal extracts, which, to the best of our knowledge, have not yet been reported in the species before. Mono-, di-, tri- and penta-methoxyflavones have been identified and characterized during the last years by liquid chromatography and mass spectrometry in flowers of P. veris and have long been recognized to possess antiallergic, antiviral, anti-inflammatory activities. They also play an important part in the biochemistry and physiology of plants, for example they act as antioxidants, enzyme inhibitors and precursors of toxic substances [21,22,23,24]. However, glycosylated methoxyflavones for P. veris are unknown [23]. The exact structural identification of the novel methylated quercetin- and myricetin-derivatives was not possible due to lack of sufficient amounts required for extensive 2D-NMR experiments. However, the structural elements identified from the MS/MS measurements suggested similarity to compounds recently described from phytochemical studies of other plant species known for pharmacological use. Thus, the methyl-quercetin-rhamnoside-hexoside from the petals and leaves of the yellow, orange and red variants resembles 4′-O-methylquercetin-3-rutinoside recently extracted from Albizzia amara leaves [25]. The methyl-quercetin-dihexoside with the molecular weight of 640 displayed characteristics of tamarixetin-3,7-diglucoside, a compound found in the aerial parts of Echium longifolium and Heliotropium digynum [26]. A compound structurally similar to the methyl-myricetin-trihexoside from P. veris was found in Artemisia annua [27].
In contrast to the flavonoids quercetin and kaempferol, myricetin occurs very rarely in the family Primulaceae [7] and has not been found in P. veris so far [28]. Among the flavonoid aglyca, (+)-catechin was also identified for the first time in the petals of this species. Isorhamnetin was found only in trace amounts in petal extracts of the orange and red variants. The absence of this compound in the wild type is most likely attributed to the extremely low compound concentration. This assumption is supported by the fact that Karl et al. [20] have previously reported the extraction of isorhamnetin and several of its glycosylated derivatives from the petals of Primula officinalis Hill. (a synonym for P. veris L.). Strong variation in the compound concentration may also account for the absence of the flavonoids gossypetin, luteolin and apigenin in our samples, which have been reported for P. veris before [20].
Triterpenoid saponins are among the pharmacologically most interesting compounds of Primula species. Primula acid I was detected as major triterpenoid saponin in all three plant organs, thus confirming former reports where the compound was isolated from P. veris, P. elatior and Primula vulgaris [29]. The saponins primula acid II and priverosaponin B-22-acetate, which were exclusively found in P. veris roots, are also secondary metabolites of P. elatior. According to literature data, primeverin is the main glycoside of P. veris roots [12]. Interestingly, we were not able to detect primeverin in the roots of any of the three P. veris variants considered in this study.
The substance identification of the anthocyanins is unambiguous. Extracts of the yellow petals were devoid of anthocyanins. This coincided with the lack of red colored vacuoles in the papillate epidermal cells visualized by light microscopy. For the wild type, this result is confirmed by earlier studies, where P. veris, P. vulgaris and P. elatior were characterized by the lack of anthocyanins, whereas carotenoids were detected in all three Primula species [7]. The orange and red Primula types analyzed in our study contained cyanidin-3-O-glucoside, cyanidin-hexoside, peonidin-hexoside, peonidin-dihexoside and malvidin-hexoside. These water-soluble vacuolar pigments have not been described in P. veris before. However, anthocyanins like malvidin glycosides and their derivatives are very widespread in some other European Primula species [7] and were also detected in the Japanese ornamental plant Primula sieboldii [30]. Peonidin, delphinidin, cyanidin and rosinidin glycosides occur occasionally [7].
In conclusion, the comparison of the plant secondary metabolites demonstrated that the three P. veris color types only differ in the occurrence and profile of the petal anthocyanins. Other compounds detected in the present study were not found to be affected by the mutation. This accounts particularly for saponins, which, due to their expectorant and secretolytic effects, are most relevant for the pharmacological use of P. veris. Currently, only the roots and petals of the wild type are used as a mild expectorant for the treatment of coughs and bronchitis; however, our study demonstrates that saponins are also present in the leaves. This confirms earlier studies, which also reported the extraction of saponins from Primula leaves [11], whereas their concentrations are lower in leaves as compared to petals [12].
The color polymorphism of P. veris is still not completely understood, but also known in P. vulgaris with petal colors ranging from white, pink and red to violet [3]. In Western Europe, P. vulgaris shows a uniform appearance with pale-yellow petals. However, in Eastern Europe (Caucasus, Greece, Turkey and Iran), common primrose is known with pale-yellow, pink, violet and purple petals side by side in one population. Morphological investigations of plants found along the northeast coast of the Black Sea (250 km) showed that the petal color is the only phenotypic difference between the varieties. A classification into different subtypes was therefore found unnecessary [31]. Recent molecular analyses based on chloroplast DNA and internal transcribed spacer markers demonstrated the color polymorphism not to be related to general phylogenetical trends in the population of P. vulgaris. Rather, the color polymorphism of P. vulgaris was suggested to reflect the postglacial distribution from different refugia and is assumed to be originated by the cross territorial distribution [32]. Molecular investigations aiming at the explanation of color polymorphism of P. veris are still pending. Genetic modifications of the anthocyanin biosynthetic pathway may be one option for the occurrence of color variations, whereby a change in gene regulation of the pathway is more likely than a mutation in one of the biosynthetically active enzymes. The ability to synthesize anthocyanins is most likely also present in the yellow wild type, but is not activated in the yellow parts of wild type petals. Observations in the progeny of our plants indicate an intermediate inheritance of the red color phenotype (data not shown). Mutations in the regulation of flavanone-3β-hydroxylase (F3H), a key enzyme of the flavonoid biosynthetic pathway, have the consequence that neither flavonols nor anthocyanins are synthesized, as it was demonstrated for Ipomoea purpurea (Convolvulaceae) with a modified pale-yellow phenotype [33]. In the case of P. veris, quercetin- as well as kaempferol-derivatives are synthesized. Consequently, mutation in the regulation downstream of F3H, especially the dihydroflavonol 4-reductase (DFR), or in transcription regulator gene(s) were suggested to be reasons for color polymorphism and result in modified phenotypes [34].
Differences in the gene regulation of P. veris are already known to be responsible for variation in the development of reproductive structures. Primula plants produce two morphological types of flowers: the long-styled flower type forms anthers attached midway along the floral tube (L-morph) and the short-styled flower forms anthers at the top of the floral tubes (S-morph). The flower phenotype is genetically associated with heterostyly genes, a system of heteromorphic self-incompatibility [35]. Recent studies identified 113 candidate heterostyly genes that revealed significant morph-specific differential expression in P. veris. One of these candidate genes has been duplicated in P. veris and is completely silenced in the long-styled flower type [36]. Similarly, a mutation in a regulatory step of the anthocyanin biosynthetic pathway might be the trigger of color polymorphism in petals.
4. Materials and Methods
4.1. Plant Material
Primula veris L. with different petal color variants from a natural habitat (Vaihingen Enz, Baden-Wuerttemberg/Germany) were cultivated in the Botanical Garden of Hohenheim University. Plant samples were harvested at flowering stage in April and May. Prior to analysis, petals, leaves and roots were manually separated and kept at −20 °C. Specimens were deposited at the herbarium of Hohenheim University (HOH-014883 to HOH-014985).
4.2. Chemicals
Reference standards quercetin-3-O-rutinoside (rutin), (+)-catechin, luteolin, quercetin and apigenin, kaempferol, isorhamnetin and cyanidin-3-O-glucoside were obtained from Carl Roth GmbH (Karlsruhe, Germany). Primeverin and primula acid I were purchased from Phytolab GmbH (Vestenbergsgreuth, Germany).
4.3. Sample Preparation
The sample composition of the plant parts composes 1 g of four individuals of the same Primula type. Petals, leaves and roots (4 g fresh weight) of the different P. veris types were cut into small pieces and extracted with 40 mL of methanol at room temperature for 30 min. The extraction with methanol was repeated and intensified by 30 min incubation in an ultrasonic bath (VWR, Darmstadt, Germany) at 22 °C. Subsequently, the extracts were decanted, filtered, transferred into a round-bottomed flask and kept at 8 °C until HPLC analysis. Petal extracts were also used for anthocyanin analyses. For this purpose, the manually separated petals were extracted with chloroform at room temperature for 30 min plus 30 min under ultrasonication to remove carotenoids from the plant material. The supernatant was removed, subsequently, petals were extracted with 40 mL of methanol/water (8:2 v/v, pH 3) as described above. All sample preparations were performed in duplicate.
4.4. Microscopic Investigations
Fresh petals of the different Primula types were investigated with an Axioplan light microscope (Zeiss, Göttingen, Germany) coupled to a digital camera (Canon Deutschland GmbH, Krefeld, Germany). Petals were fixed in small polystyrene blocks and cut in cross and longitudinal sections.
4.5. HPLC-DAD Analysis
For chromatographic separation, a Thermo Fisher Scientific Dionex Ultimate 3000 RSLC system (Thermo Fisher Scientific GmbH, Dreieich, Germany) equipped with a vacuum degasser, a binary pump, an autosampler, a thermostatic column compartment (25 °C) and a diode array detector (DAD) was used. Gradient binary eluent system (eluent A: 0.1% formic acid (v/v); eluent B: acetonitrile) was applied using a SunFire RP C18 column (150 mm × 2.1 mm, Waters, Wexford, Ireland) at a flow rate of 0.21 mL/min with the following gradient: 0–2 min, 0% B; 2–4 min, 0%–5% B; 4–10 min, 5%–10% B; 10–14 min, 10%–15% B; 14–33 min, 15%–20% B; 33–41 min, 20%–35% B; 41–54 min, 35%–100% B; 54–58 min, 100% B; 58–64 min, 100%–0% B; 64–66 min, 0% B. The detection wavelengths were set at 210, 254, 280, 366 and 520 nm. For HPLC-DAD analyses the samples were diluted with purified water (1:1, v/v). Injection volume was 20 µL (equivalent to 10 mg fresh weight). Chromeleon software (Version 7.2, Dionex, Idstein, Germany) was used for data acquisition and processing.
4.6. LC-MSn Analyses
Mass spectrometric analyses were performed using an Agilent 1200 HPLC system (Agilent, Waldbronn, Germany) connected to an HCT ultra ion trap MS detector interfaced with an ESI ion source (Bruker Daltonik, Bremen, Germany). Negative ionization mode was used for phenolic compound and triterpenoid saponin analysis, whereas anthocyanins were analyzed in the positive ionization mode. The following device parameters were applied: dry gas flow (N2), 8 L/min; nebulizer pressure, 40 psi; capillary temperature, 350 °C; for the negative and positive ionization modes, MS spectra were recorded in a range of m/z 50 to 1500 with a compound stability and trap drive level of 100%. The software Agilent Chemstation (Rev. B.01.03 SR1) (Agilent, Waldbronn, Germany) and Bruker Daltonik esquire control (Version 6.1) (Bruker Daltonik GmbH, Bremen, Germany) were used for data acquisition.
4.7. Characterization of Phenolic Compounds and Triterpenoid Saponins
The identification of compounds previously described in the literature was realized by comparison of retention times, mass spectrometric and UV/Vis data with those of literature reports and reference compounds, respectively.
The flavones (+)-catechin and isorhamnetin were identified by comparison with the corresponding reference standards. However, isorhamnetin was detected only in trace amounts in the orange and red petal extracts, and was not detected in the yellow wild type petal extract and the corresponding leaf extracts as well as root extracts of the three Primula types. The flavonoids apigenin and luteolin were not detected in any of the plant parts of the three Primula types, which is in accordance with previous reports. Kaempferol-3-O-galactoside-rhamnoside-7-O-rhamnoside (m/z 739) was assigned to the peaks with retention times of 29.5 min (petal extracts) and 30.4 min (leaf extracts) and was identified based on MSn spectra with the base peak at m/z 739 as well as comparison with literature data [1]. This flavonoid glycoside revealed fragment ions at m/z 593, 429 and 285, thus indicating the losses of rhamnoside and hexoside moieties and a dehydration reaction. The characteristic fragments of kaempferol (m/z 257, 241, 229, 213, 199, 151) were confirmed by literature data [18]. Kaempferol-3-O-rutinoside was also detected in Primula leaves as well as petals and showed MS base peaks at m/z 593. Fragment ions at m/z 447 and 285 demonstrated the release of hexose and rhamnose moieties. A comparison with literature data of Sánchez-Rabaneda et al. [15] confirmed the compound identification. Kaempferol-3-O-diglucoside-7-O-glucoside showed an [M-H]− ion at m/z 771 and the release of three hexose moieties, thus yielding characteristic fragment ions at m/z 609, 447 and 285 in the MS2 experiments. These data are in agreement with the findings of Gonzales et al. [17] and Harborne [7]. The flavonol quercetin and its glycosides were identified in Primula petals and leaves. For the identification of the respective aglycone quercetin, MSn spectra of Fabre and Rustan [16] were used, which confirmed the main ion at m/z 301 and the characteristic fragment ions at m/z 273 and 257 indicating the losses of CO as well as CO2. Fragment ions at m/z 193 and 179 indicated the fission of the flavonoid B-ring. Quercetin-3-O-rutinoside (rutin) from petal and leaf extracts was identified based on its UV spectral characteristics, retention time and MSn spectra and comparison with the corresponding reference standard. This flavonoid glycoside was not detected in the roots of the different Primula types. Quercetin-3-gentiobioside has been reported in Primula veris L. with yellow petals [37] and was also detected in the present study with a predominant signal at m/z 625. Moreover, a quercetin-trihexoside was determined, exhibiting a signal at m/z 787.
In addition to the phenolic compounds and anthocyanins, triterpenoid saponins were identified in various extracts. The occurrence of primula acid I was confirmed by its mass spectrometric signal at m/z 1104 and its retention time of 53.0 min, which was in accordance with the reference standard. Primula acid II was identified by its [M-H]− ion at m/z 1236 and fragment ions at m/z 924 as well as m/z 465. Priverosaponin B-22-acetate with the [M-H]− ion at m/z 1162 was confirmed by comparison with literature data [12,38].
5. Conclusions
In conclusion, the present investigations markedly broadens our knowledge of the phenolic constituents and triterpenoid saponins in P. veris variants with flower color mutations and complement the literature data of the therapeutically used Primula species.
As it is often unclear whether reported chemical diversity within a certain taxon is the result of natural chemotypes used for investigation or just a matter of application of different extraction and identification methods, it was our intention to establish and compare the metabolite profile of different plant organs of P. veris under standardized conditions. Hence, an extraction with methanol and add-on LC-MS analysis in the negative and positive ionization modes was employed and allowed the efficient and comparative phytochemical characterization of phenolic compounds and triterpenoid saponins in flowers, leaves and roots of the plants. Besides the organ-specific occurrence of the investigated metabolites in P. veris, we compared the chemical diversity in prominent secondary metabolites between the mutants and the wild type. In total, two flavonoids, six flavonoid glycosides, four methylated flavonoid glycosides, five anthocyanins and three triterpenoid saponins were identified in the present work. Except for the anthocyanins, significant differences in the saponin and flavonoid profiles of the mutants and the wild type were not observed, thus indicating that flower color mutation does not affect the potential pharmacological use of the respective plant species.
Acknowledgments
We want to thank Torsten Boettcher (Sachsenheim, Germany) for identifying the color variants of P. veris from the natural habitat.
Author Contributions
Otmar Spring, Dietmar R. Kammerer and Florian C. Stintzing conceived and designed the experiments. Lysanne Apel performed the experiments, analyzed the data and wrote the manuscript together with Otmar Spring. All authors contributed to the development of the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Morozowska M., Wesołowska M. In vitro clonal propagation of Primula veris L. and preliminary phytochemical analysis. Acta Biol. Crac. Bot. 2004;46:169–175. [Google Scholar]
- 2.Laenger R., Sankel J. Systematics of Primula veris (Primulaceae) Plant Syst. Evol. 1993;188:31–55. [Google Scholar]
- 3.Köhlein F. Primeln und die Verwandten Gattungen Mannsschild, Heilglöckchen, Götterblume, Troddelblume, Goldprimel. Verlag Eugen Ulmer; Stuttgart, Germany: 1984. pp. 104–108. [Google Scholar]
- 4.HagerRom . Hagers Enzyklopädie der Arzneistoffe und Drogen. Springer; Heidelberg, Germany: 2015. [Google Scholar]
- 5.Karrer P., Widmer R. Über Primelfarbstoffe. Pflanzenfarbstoffe V. Helv. Chim. Acta. 1927;10:758–763. doi: 10.1002/hlca.19270100191. [DOI] [Google Scholar]
- 6.Paris R. On the flavonoids of native species of Primula. Presence of a heteroside of kaempferol in the flowers of Primula officinalis Jacq. Ann. Pharm. Franc. 1959;17:331–335. [PubMed] [Google Scholar]
- 7.Harborne J.B. Comparative biochemistry of the flavonoids—VII, Correlations between flavonoid pigmentation and systematics in the family Primulaceae. Phytochemistry. 1968;7:1215–1230. doi: 10.1016/S0031-9422(00)85616-2. [DOI] [Google Scholar]
- 8.Bat-Smith E.C. The phenolic constituents of plants and their taxonomic significance. I. Dicotyledons. J. Linn. Soc. Bot. 1962;58:95–173. doi: 10.1111/j.1095-8339.1962.tb00890.x. [DOI] [Google Scholar]
- 9.Hegnauer R. Chemotaxonomie der Pflanzen. 5th ed. Birkhäuser Verlag; Basel, Switzerland: 1969. pp. 387–454. [Google Scholar]
- 10.El Morchid E.M., Londono P.T., Papagiannopoulos M., Gobbo-Neto L., Mueller C. Variation in flavonoid pattern in leaves and flowers of Primula veris of different origin and impact of UV-B. Biochem. Syst. Ecol. 2014;53:81–88. doi: 10.1016/j.bse.2013.12.032. [DOI] [Google Scholar]
- 11.Kraft K. Symptomatische Phytotherapie bei Husten. Stellenwert pflanzlicher Antitussiva und Expektorantien. Pharm. Unserer Zeit. 2008;6:478–483. doi: 10.1002/pauz.200800287. [DOI] [PubMed] [Google Scholar]
- 12.Mueller A., Ganzera M., Stuppner H. Analysis of phenolic glycosides and saponins in Primula elatior and Primula veris (primula root) by liquid chromatography, evaporative light scattering detection and mass spectrometry. J. Chromatogr. A. 2006;1112:218–223. doi: 10.1016/j.chroma.2005.10.067. [DOI] [PubMed] [Google Scholar]
- 13.Pemp E., Krenn L. Analytik Pharmazeutisch Relevanter Flavonoide Mit DC und LC/DAD/MS. 1st ed. Südwestdeutscher Verlag für Hochschulschriften AG & Co. KG; Saarbrücken, Germany: 2005. [Google Scholar]
- 14.Kim J., Matsuba Y., Ning J., Schulmiller A.L., Hammar D., Jones A.D., Pickersky E., Last R.L. Analysis of natural and induced variation in tomato glandular trichome flavonoids identifies a gene not present in the reference genome. Plant Cell. 2014;26:3272–3285. doi: 10.1105/tpc.114.129460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sánchez-Rabaneda F., Jáuregui O., Casals I., Andres-Lacueva C., Izquierdo-Pulido M., Lamuela-Raventos R.M. Liquid chromatographic/electrospray ionization tandem mass spectrometric study of the phenolic composition of cocoa (Theobroma cacao) J. Mass Spectrom. 2002;38:35–42. doi: 10.1002/jms.395. [DOI] [PubMed] [Google Scholar]
- 16.Fabre N., Rustan I. Determination of flavone, flavonol, and flavanone aglycones by negative ion liquid chromatography electrospray ion trap mass spectrometry. Am. Soc. Mass Spectrom. 2001;12:707–715. doi: 10.1016/S1044-0305(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 17.Gonzales G.B., Raes K., Coleus S., Struijs K., Smagghe G., Camp J.V. Angiotensin converting enzyme inhibitory effects by plant phenolic compounds: A study of structure activity relationships. J. Chromatogr. A. 2013;61:11832–11839. doi: 10.1021/jf404641v. [DOI] [PubMed] [Google Scholar]
- 18.Tsiklauri L., Guohua A., Ruszaj D.M., Alaniyaa M., Kemertelidzea E., Morris M.E. Simultaneous determination of the flavonoids robinin and kaempferol in human breast cancer cells by liquid chromatography-tandem mass spectrometry. J. Pharm. Biomed. 2011;55:109–113. doi: 10.1016/j.jpba.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 19.Wu X., Prior R.L. Systematic identification and characterization of anthocyanins by HPLC-ESI-MS/MS in common foods in the United States: Fruits and berries. J. Agric. Food Chem. 2005;53:2589–2599. doi: 10.1021/jf048068b. [DOI] [PubMed] [Google Scholar]
- 20.Karl C., Mueller G., Pedersen P.A. Die Flavonoide in den Blüten von Primula officinalis. J. Med. Plants Res. 1981;41:96–99. doi: 10.1055/s-2007-971683. [DOI] [PubMed] [Google Scholar]
- 21.Harborne J.B., Mabry T.J. The Flavonoids: Advances in Research. Chapman and Hall; New York, NY, USA: 1982. [Google Scholar]
- 22.McClure J.W. Plant Flavonoids in Biology and Medicine. Alan R. Liss. Inc.; New York, NY, USA: 1986. Biochemical, Pharmacological and Structure—Activity Relationships; pp. 77–135. [Google Scholar]
- 23.Huck C.W., Huber C.G., Ongania K.H., Bonn G.K. Isolation and characterization of methoxylated flavones in the flowers of Primula veris by liquid chromatography and mass spectrometry. J. Chromatogr. A. 2000;870:453–462. doi: 10.1016/S0021-9673(99)00950-4. [DOI] [PubMed] [Google Scholar]
- 24.Huck C.W., Bonn G.K. Evaluation of detection methods for the reversed-phase HPLC determination of 3′,4′,5′-trimethoxyflavone in different phytopharmaceutical products and in human serum. Phytochem. Anal. 2001;12:104–109. doi: 10.1002/pca.547. [DOI] [PubMed] [Google Scholar]
- 25.Sastry C.V., Rukmini C., Ramachandra L. Chemistry of saponins. III. Isolation of new flavonol glycoside, 4′-O-methylquercetin-3-rutinoside, from Albizzia amara. Indian J. Chem. 1967;5:613–615. [Google Scholar]
- 26.Ibrahim L.F., Kawashty S.A., El-Eraky W.I., Shabana M.M., El-Negoumy S.I. A comparative study of flavonoids, pharmacological and antimicrobial effects of Echium longifolium and Heliotropium digynum. Egypt. J. Physiol. Sci. 2002;24:63–82. [Google Scholar]
- 27.Han J., Ye M., Qiao X., Xu M., Wang B.-R., Guo D.-A. Characterization of phenolic compounds in the Chinese herbal drug Artemisia annua by liquid chromatography coupled to electrospray ionization mass spectrometry. J. Pharam. Biomed. Anal. 2008;47:516–525. doi: 10.1016/j.jpba.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Kulevanova S., Stefova M., Panovks T.K., Stafilov T. HPLC identification and determination of myricetin, quercetin, kaempferol and total flavonoids in herbal drugs. Maced. Pharm. Bull. 2003;48:25–30. [Google Scholar]
- 29.Teshesche R., Toja B.T., Wulff G. Über die Sapogenine aus den Wurzeln von Primula veris L. Liebigs Ann. Chem. 1966;696:160–179. doi: 10.1002/jlac.19666960118. [DOI] [Google Scholar]
- 30.Hashimoto N., Ohsawa R., Kitajima J., Iwashina T. New flavonol glycosides from leaves and flowers of Primula sieboldii. Nat. Prod. Commun. 2015;10:421–423. [PubMed] [Google Scholar]
- 31.Shipunov A., Kosenko Y., Volkova P. Floral polymorphism in common primrose (Primula vulgaris Huds., Primulaceae) of the Northeastern Black Sea coast. Plant Syst. Evol. 2011;296:167–178. doi: 10.1007/s00606-011-0484-5. [DOI] [Google Scholar]
- 32.Volkova P.A., Schanzer I.A. Colour polymorphism in common primrose (Primula vulgaris Huds.): Many colours—Many species? Plant Syst. Evol. 2013;299:1057–1087. doi: 10.1007/s00606-013-0780-3. [DOI] [Google Scholar]
- 33.Hoshino A., Abe Y., Saito N., Inagaki Y., Iida S. The gene encoding flavanone 3-hydroxylase is expressed normally in the pale yellow flowers of the Japanese morning glory carrying the speckled mutation which produce neither flavonol nor anthocyanin but accumulate chalcone, aurone and flavanone. Plant Cell Physiol. 1997;38:970–974. doi: 10.1093/oxfordjournals.pcp.a029260. [DOI] [PubMed] [Google Scholar]
- 34.Clegg M., Durbin M.L. Flower color variation: A model for experimental study of evolution. Proc. Natl. Acad. Sci. USA. 2000;97:7016–7023. doi: 10.1073/pnas.97.13.7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCubbin A. Heteromorphic Self-Incompatibility in Primula: Twenty-First Century Tools Promise to Unravel a Classic Nineteeneth Centruy Model System. Springer; Berlin/Heidelberg, Germany: 2008. pp. 289–308. [Google Scholar]
- 36.Nowak M.D., Russo G., Schlapbach R., Huu C.N., Lenhard M., Conti E. The draft genome of Primula veris yields insights into the molecular basis of heterostyly. Genome Biol. 2015;12:1–16. doi: 10.1186/s13059-014-0567-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kofler L. Über das Saponin der Primulawurzel. Arch. Pharm. 1924;262:318–328. doi: 10.1002/ardp.19242620407. [DOI] [Google Scholar]
- 38.Wright A., Sticher O. Triterpene saponins from Primula veris subsp. macrocalyx and Primula elatior subsp. meyeri. J. Nat. Prod. 1992;55:1299–1306. [Google Scholar]