Abstract
MicroRNAs (miRNAs) have within the past decade emerged as key regulators of metabolic homoeostasis. Major tissues in intermediary metabolism important during development of the metabolic syndrome, such as β‐cells, liver, skeletal and heart muscle as well as adipose tissue, have all been shown to be affected by miRNAs. In the pancreatic β‐cell, a number of miRNAs are important in maintaining the balance between differentiation and proliferation (miR‐200 and miR‐29 families) and insulin exocytosis in the differentiated state is controlled by miR‐7, miR‐375 and miR‐335. MiR‐33a and MiR‐33b play crucial roles in cholesterol and lipid metabolism, whereas miR‐103 and miR‐107 regulates hepatic insulin sensitivity. In muscle tissue, a defined number of miRNAs (miR‐1, miR‐133, miR‐206) control myofibre type switch and induce myogenic differentiation programmes. Similarly, in adipose tissue, a defined number of miRNAs control white to brown adipocyte conversion or differentiation (miR‐365, miR‐133, miR‐455). The discovery of circulating miRNAs in exosomes emphasizes their importance as both endocrine signalling molecules and potentially disease markers. Their dysregulation in metabolic diseases, such as obesity, type 2 diabetes and atherosclerosis stresses their potential as therapeutic targets. This review emphasizes current ideas and controversies within miRNA research in metabolism.
Keywords: adipocytes, metabolism, microRNA, non‐alcoholic hepato‐steatosis, type 2 diabetes mellitus, β‐cells
For the past 50 years, the term ‘gene’ has been synonymous with regions of the genome encoding mRNAs that are translated into protein. However, the past decade's explosion of large‐scale genome sequencing has revealed that opposed to the original expectation that more complex organisms would have a greater number of genes, it is now clear that human and mice shares a similar number of protein encoding genes as the round worm C. elegans. A possible explanation for this paradox comes from the insight that biological complexity generally correlates with proportion of the genome which is non‐protein‐coding (Taft et al. 2007). The majority of this non‐protein‐coding region is transcribed into long non‐coding RNA or small non‐coding RNA, which orchestrate the regulation of protein expression both at the transcriptional and translational level. A class of small non‐coding RNAs termed microRNAs (miRNAs) was discovered in 1993 by Lee, Feinbaum and Ambros (Lee et al. 1993). MiRNAs consist of approx. 22 nucleotides and regulate gene expression by binding to their complementary sites within the 3′‐untranslated regions (3′UTRs) of target mRNAs (Lagos‐Quintana et al. 2001) resulting in mRNA translational repression or transcript degradation. The degree of miRNA‐target base‐pairing complementarity determines the fate of the target transcript. Perfect complementarity leads to target cleavage and degradation. In contrast, imperfect complementarity triggers mRNA silencing by distinct mechanisms which may involve translational repression, slicer‐independent mRNA degradation and/or sequestration in cytoplasmic processing bodies (Roberts 2015) (Fig. 1).
Figure 1.
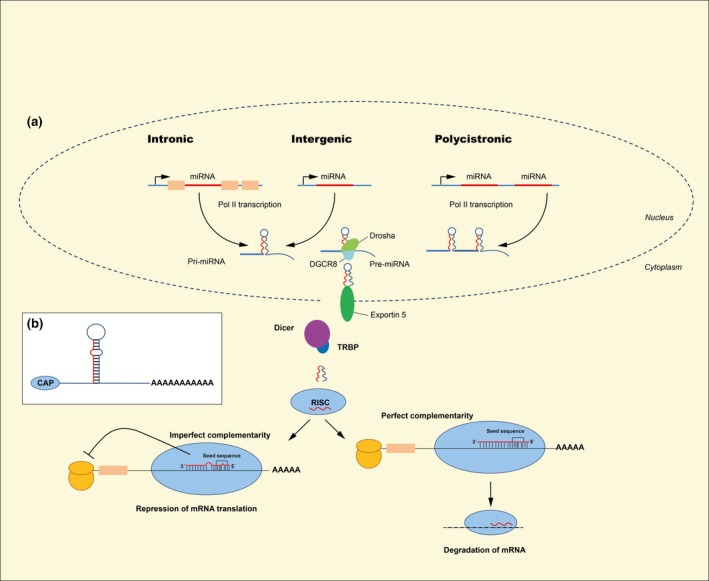
The canonical miRNA biogenesis pathway (a) and the average precursor miRNA (b). (a): The miRNA genes lies either intronic, intergenic or polycistronic. The primary miRNA (Pri‐miRNA) is transcribed by polymerase II (or polymerase III). The Pri‐miRNA is cleaved by the microprocessor complex Drosha‐DGCR8 in the nucleus. The precursor miRNA (pre‐miRNA) is transported out in the cytoplasm by Exportin 5. In the cytoplasm, the pre‐miRNA is further cleaved to its mature length (approx. 22 nt) by the RNase III Dicer in complex with the double‐stranded RNA‐binding protein TRBP. Argonaute (AGO2) proteins unwind the miRNA duplex and facilitate incorporation of the guide strand (red) into the RNA‐induced silencing complex (RISC). AGO2 then guides the RISC miRNA assembly to target mRNAs, whereas the passenger strand (blue) is degraded. Some miRNA bind mRNA with perfect complementarity and induce degradation of mRNA. miRNA also bind to targets with imperfect complementarity and block translation. (b): An average precursor miRNA with a hairpin stem of 33 base pairs, a terminal loop and two flanking region, where 5′ end are capped and a polyadenylated 3′ end.
Each miRNA may have hundreds of mRNA targets, just as well as a single mRNA may be regulated by several distinct miRNAs adding to the layer of complexity of protein expression. MiRNAs are encoded in diverse regions of the genome including non‐coding regions (intronic or intergenic) as well as protein coding (in exons). The canonical biogenesis of the mature functional miRNAs involves multiple processing steps, described in Figure 1. Each processing step contains another layer of regulation and therefore adds to the complexity of gene expression.
The aim of this review is to highlight the recent progress and challenges within research of miRNAs in metabolism and metabolic disease, with a special emphasis on specific and major tissues and cell types important for development of the metabolic syndrome, obesity and type 2 diabetes: β‐cells, liver, skeletal and heart muscle and adipose tissue. Although organ to organ crosstalk greatly impacts metabolism, it is not the scope of this review. Furthermore, we also describe recent progress on development of miRNA therapeutics and biomarkers, as well as challenges in quantification of miRNAs.
Pancreatic islets and β‐cells
The islet of Langerhans constitutes an important node of control for maintaining normoglycaemia, as sufficient β‐cell insulin secretion is needed for proper peripheral glucose uptake, and α‐cell glucagon secretion is important for hepatic glucose production. The importance of β‐cells for glucose homoeostasis is underlined by the observation that type 2 diabetes mellitus only develops in the context of β‐cell failure. Moreover, genetic studies reveal that amongst the more than 75 genetic loci associating with type 2 diabetes, the largest proportion harbours transcripts important for β‐cell function or proliferation (Rutter 2014).
The level of gene expression or function is regulated at many levels, and miRNA networks constitute control points for integration of environmental and genetic factors influencing physiological responses of the β‐cell. In general, most genetics studies with miRNA mutants display no obvious phenotype unless the animal is confronted by physiological stressors. For β‐cells, increased stress occurs for example as their workload increases due to peripheral insulin resistance, which may result in progression to type 2 diabetes (Halban et al. 2014).
Evidence to support an important role of miRNAs in β‐cells has been obtained from cre‐mediated deletion of Dicer1 in different pancreatic lineages. Pdx1‐cre‐mediated Dicer1 deletion shows that developmentally expressed miRNAs are important for proper islet and β‐cell development (Lynn et al. 2007) and both induced and constitutive Dicer1 deletion in β‐cells results in impaired insulin secretion and diabetes (Kalis et al. 2011, Melkman‐Zehavi et al. 2011, Martinez‐Sanchez et al. 2015) with impaired glucose‐stimulated insulin secretion (GSIS) preceding changes in insulin content or β‐cell mass.
Another important role of miRNAs is thought to be through selective repression of mRNAs, whose expression is detrimental to correct functioning of specific cell types, termed ‘disallowed’ genes. For β‐cells Slc16a1 (pyruvate transporter), Ldha (lactate dehydrogenase A), Fcgrt1 (Neonatal Fc receptor), Pdfgra (PDGF receptor, alpha‐type) and Oat (ornithine aminotransferase) are selectively repressed by miRNAs, as these mRNAs are de‐repressed in dicer1 KO islets and their 3′UTRs confer increased reporter‐gene activity in dicer1 depleted islet cells (Pullen et al. 2011, Martinez‐Sanchez et al. 2015). The Argonaute 2 (Ago2) component of the RISC complex controls compensatory β‐cell proliferation and is under the control of miR‐184, which is negatively regulated by blood glucose levels, thus providing a systemic feedback to the β‐cells reflecting systemic insulin resistance (Tattikota et al. 2014, 2015).
Although very few miRNAs are tissue specific, a number of miRNAs can be designated as being either enriched in endocrine, neuro‐endocrine or epithelial tissues, where the miR‐375 belongs to the endocrine enriched miRNAs (10% of β‐cell microRNA is miR‐375) (Poy et al. 2004, van de Bunt et al. 2013), the miR‐7 family to the neuro‐endocrine, and the 200‐family is expressed in epithelial tissues from which islet cells originate. The phenotype of the global miR‐375 knockout (KO) (Poy et al. 2009) shows progressive hyperglycaemia with lower numbers of β‐cells and impaired compensatory β‐cell proliferation as well as effects on Na+ channel inactivation properties (Salunkhe et al. 2015a). Thus, the miR‐375 deficient β‐cells' adaptation to stress and insulin resistance resembles a phenocopy of Dicer1‐KO β‐cells (Lynn et al. 2007). Conversely, specific β‐cell re‐expression of miR‐375 in the KO mouse normalizes glucose tolerance. Although miR‐375 is the most highly expressed miRNA in pancreatic β‐cells, under normal physiological conditions, only 1% of plasma miR‐375 is derived from β‐cells, which only doubles after streptozotocin induced diabetes (Latreille et al. 2015). Thus, these findings challenge the use of miR‐375 as a circulating biomarker for β‐cell injury (Erener et al. 2013).
Whereas miR‐375 expression is necessary for correct β‐cell function and proliferation, targeted deletion of either miR‐7 or miR‐200 family members display improved β‐cell function in mice fed a normal diet indicating that the role of these miRNAs are to constitutively repress β‐cell function (Latreille et al. 2014, Belgardt et al. 2015). Conditional β‐cell KO of all three miRNAs of the miR‐7 family preferentially changed the abundance of synaptic proteins involved in exocytosis, for which target gene de‐repression was observed and insulin exocytosis was increased (Latreille et al. 2014). Whether the miR‐7 family is regulated the same way in other tissues with high rate of exocytosis still needs to be examined. The miR‐200 family consists of miR‐200a/141 and miR‐200b/miR‐200c/miR‐429 clusters. These miRNAs are generally dysregulated in cancers, but miRNAs belonging to these subfamilies are also very abundant in β‐cells and amount up to 2/3 of all miRNAs in β‐cells. They are decreased by high fat diet and upregulated in diabetic db/db (BKS background) about threefold. Furthermore, forced expression of the miR‐141/200a cluster by just fivefold in β‐cells results in overt diabetes and subsequent death of the mice and is accompanied by massive β‐cell apoptosis (Belgardt et al. 2015). The double KO of miR‐141/200a and 200b/c/429 clusters protects against β‐cell ER‐stress in the Akita mouse model, which as misfolding of insulin, as well as in both multiple low‐dose streptozotocin and single‐dose streptozotocin induced β‐cell damage. The β‐cell protection is mediated through regulation of Tp53 activity. Thus, two large families of miRNAs negatively control β‐cell function and survival. One could ask why the pancreatic β‐cells are under such heavy negative control. In the setting of limited access to nutrients, having excessive insulin production and secretion would be detrimental to an individual due to the risk of hypoglycaemia. Consequently, there will be a high selective pressure to develop mechanisms keeping β‐cells under control.
However, the environment to which most subjects are exposed is not one of famine but rather one of feast. Therefore, it is important to study β‐cell failure in type 2 diabetes mellitus and the impact of environment on islet response and impaired insulin secretion. Insulin exocytosis is a very specialized feature of β‐cells. It is affected by several diabetes associated gene variants, but the expression of exocytotic genes has been shown to be reduced islets from patients with type 2 diabetes, which is not due to genetic variation in the vicinity of these genes. Epigenetic variation, such as DNA methylation (Dayeh et al. 2014) or miRNAs, may mediate some of the expression changes. In an attempt to identify miRNAs involved in β‐cell decompensation, islets were isolated from Goto Kakizaki (GK)‐rats, a model of type 2 diabetes with β‐cell dysfunction. miR‐335, miR‐152 and miR‐130a/b were found to be increased (Esguerra et al. 2011) and computational analysis showed that miR‐335 targeted transcripts encoding exocytotic proteins (Stxbp1, Syt11, Snap25). Moreover, overexpression of miR‐335 leads to decreased GSIS as well as decreased depolarization‐induced insulin exocytosis (Salunkhe et al. 2015b). Additionally, the poor insulin‐secretor cell line INS‐832/2 had increased levels of miR‐152 and miR‐130a/b compared with INS‐832/13 cells, which maintain a high GSIS (Hohmeier et al. 2000, Ofori et al. 2014). In line with this, knockdown of miR‐152 leads to increased GSIS, whereas overexpression impaired GSIS in the INS‐832/13 cells, with a concomitant decrease in insulin content (Ofori et al. 2014). Moreover, miR‐187 was shown to be increased in human type 2 diabetic islets, negatively correlating with GSIS in islets from normoglycaemic donors, and forced expression in rat islets and INS‐1 cells reduced GSIS (Locke et al. 2014). Thus, multiple miRNAs participate in and control insulin exocytosis, are dysregulated in type 2 diabetic islets and constitute natural nodes in cellular interaction networks regulating insulin exocytosis (Eliasson 2014).
β‐cells have low proliferation rates, which decrease even further as an animal age (Wang et al. 2015b). However, mature β‐cells release more insulin in response to glucose than β‐cells from young animals (Jacovetti et al. 2015), making it attractive to understand how a β‐cell can be kept ‘young’ in terms of proliferative capacity and ‘old/mature’ in terms of insulin secretion. When rat pup islets from 10 days of age (D10) are compared with adult islets, proliferation rate is higher and the level of GSIS is lower, while insulin content is unchanged. The transition was found to occur 2–5 days following weaning, and the associated change in nutrients is thought to induce the shift between proliferation rate and GSIS. Rats prematurely weaned showed the same metabolic shift and maturation, further indicating that it is a change in nutrients that drive the shift. Interestingly, several miRNAs were altered in the young vs. the mature islets. For example, were the miR‐29 family, miR‐204 and miR‐129 upregulated, whereas the miR‐17‐92 cluster, miR‐181b and miR‐215 were suppressed more than twofold from young (D10) to adult islets. The same expression pattern was present in prematurely weaned pups. Surprisingly, when pups were fed a high fat diet the transition into high GSIS did not occur and this was associated with an extended immature miRNA profile. To investigate the contribution of single miRNAs to this phenotype, D10 isolated islets were dissociated into single islet cells, transfected using antisense oligonucleotides targeting miRNAs or miRNA mimetics and GSIS and β‐cell proliferation were determined. Using this approach, both the miR‐17‐92 cluster and miR‐181b were found to regulate β‐cell replication as well as GSIS. Using luciferase reporter assays, the miR‐17‐92 cluster was shown to target 3′UTRs of Pfkp (phosphofructokinase, platelet), Tgfbr2 (transforming growth factor beta receptor II) and Pten (phosphatase and tensin homolog), while miR‐181b targeted Gpd2 (glycerol‐3‐phosphate dehydrogenase 2), Mdh1 (malate dehydrogenase 1) and Sirt1 (sirtuin 1). Of note, the miR‐17‐92 cluster is also a known transcriptional target of E2F and regulates cMyc levels in other cell types (Aguda et al. 2008). It seems likely that a similar pathway could control the decrease in β‐cell proliferation occurring with weaning and maturation. β‐cell proliferation is decreased by ageing, which is partly due to decreased amounts of Pdgfra, mediated by an ageing‐induced increase in miR‐34a (Tugay et al. 2015). From these studies, it is clear that nutritional state, malnutrition and foetal programming affect the maturity state of the β‐cells. However, how this translates into human subjects and human nutrition, and for example, the role of infant formula compared to breast milk feeding is not known and will need to be addressed.
Liver metabolism regulated by miRNAs
The liver plays a major role in energy metabolism as it is a main contributor to absorptive glucose storage and post‐absorptive glucose release, amino acid metabolism and is the main regulator of lipoprotein metabolism. Although functions of miRNAs were first described for the regulation of proper development of C. elegans (Lee et al. 1993), several studies have revealed that miRNAs play a pivotal role for controlling metabolic homoeostasis. miR‐122 is the most abundant miRNA in liver and has been shown to be involved in hepatic cholesterol and lipid metabolism (Krützfeldt et al. 2005). Two studies have shown that antisense targeting miR‐122 results in significant reduction in plasma cholesterol levels (Krützfeldt et al. 2005, Esau et al. 2006), but the effect on cholesterol metabolism by miR‐122 seems to be indirect, as the exact targets of this particular miRNA are still unclear (Fernandez‐Hernando et al. 2013). Another miRNA implicated in hepatic cholesterol metabolism is miR‐33 originating from two intronic miRNAs, miR‐33a and miR‐33b, which are encoded within the introns of Srebf2 and Srebf1 genes respectively (Najafi‐Shoushtari et al. 2010). Both miRNAs are co‐transcribed with their host genes and under their regulation. MiR‐33a directly targets the cholesterol transporters Abca1 and Abcg1, which are responsible for the efflux of cholesterol from the cell, suggesting the importance of this miRNA in cholesterol metabolism. In agreement with this, the miR‐33a KO mouse has shown an increase in Abca1 expression and plasma HDL levels (Horie et al. 2010). The data are supported by three independent studies using different strategies to inhibit endogenous miR‐33a, which also increase plasma HDL levels (Marquart et al. 2010, Najafi‐Shoushtari et al. 2010, Rayner et al. 2010). Besides cholesterol metabolism miR‐33b has also been shown to be implicated in fatty acid β‐oxidation, as carnitine palmitoyltransferase (Cpt1a) is regulated by miR‐33b. Interestingly, miR‐33 is also highly abundant in brain (Rayner et al. 2010), and it has previously been shown that the cholesterol metabolism in brain of diabetic animals is indeed compromised (Suzuki et al. 2010), suggesting a potential role for miR‐33 in brain cholesterol metabolism.
In an effort to systematically identify miRNAs that regulate cholesterol metabolism through the low‐density lipoprotein receptor (LDLR), Goedeke et al. (2015) developed a high‐throughput screen to monitor the effect of miRNAs to induce cellular low‐density lipoprotein (LDL) uptake. During this screen, miR‐148a was discovered as a top candidate for LDLR regulation. And indeed, when miR‐148a is suppressed, LDL levels decrease, whereas HDL levels increase, highlighting the therapeutic potential for the treatment of atherosclerosis and related dyslipidaemias of this particular miRNA.
Several other miRNAs have been shown to be implicated in liver metabolism and glucose homoeostasis. miR‐143 (Jordan et al. 2011), miR‐181a (Zhou et al. 2012), miR‐103 and miR‐107 (Trajkovski et al. 2011) have all been shown to affect hepatic insulin sensitivity, and more recently, miR‐802 has been shown to be increased with obesity and that its reduction improves glucose tolerance and insulin action (Kornfeld et al. 2013). From the above studies, it is clear that miRNAs play a central role in regulation of liver metabolism and most likely more metabolically important miRNAs will be discovered in the future and will serve as potential targets for treatment of metabolic disorders. However, using miRNAs as therapeutics faces numerous challenges, which will be discussed in the section of ‘miRNAs as therapeutics’.
Muscle and heart miRNAs
Skeletal muscle accounts for more than 40% of the body weight of a normal healthy person and is by far the largest organ of the body. Furthermore, impaired insulin‐stimulated muscle glucose disposal is the primary defect in the insulin resistant state during a hyperinsulinaemic–euglycaemic clamp (DeFronzo et al. 1985), highlighting the importance of skeletal muscle in glucose homoeostasis. Besides their importance in whole‐body glucose homoeostasis, skeletal muscles also play a pivotal role in healthy ageing, and muscle wasting is present in many diseases, such as sarcopenia, HIV–AIDS, cancer cachexia, renal failure and bed rest (Bodine 2013, Polge et al. 2013), showing the importance of development and maintenance of muscle mass. MiRNAs have been shown to be necessary for appropriate muscle development, as muscle specific dicer1 knockout mice have marked dysregulation of muscle development resulting in embryonic lethality (O'Rourke et al. 2007). In addition, miRNAs are involved in control of muscle fibre type (Van Rooij et al. 2009), namely miR‐208a, miR‐208b and miR‐499, with each of these miRNAs encoded in the myosin heavy chain (MHC) genes (McCarthy et al. 2009) signifying their importance in muscle phenotype. In agreement, miR‐208b and miR‐499 are decreased concomitant with the expression of the respective MHC genes, inversely correlated with myostatin in human skeletal muscle after spinal cord injury, and thereby linked to the regulation of muscle mass (Boon et al. 2015). In line with this, age‐related changes in miR‐143 have recently been shown to affect muscle regeneration in vitro (Soriano‐Arroquia et al. 2016).
Several miRNAs are enriched in muscle and heart (Sempere et al. 2004), and especially, miR‐1, miR‐133 and miR‐206 are defined as myogenic miRNAs capable of inducing skeletal muscle differentiation in murine models (Chen et al. 2006, Dey et al. 2011). To address the miRNA network of human skeletal muscle cell differentiation, Sjögren and colleagues performed a time course analysis from day 0 (myoblast) to day 10 (myotube) coupled to microarray analyses to identify differential miRNA and mRNA expression (Sjogren et al. 2015). Confirming data from murine muscle cells largest fold change in miRNA expression were indeed observed for miR‐1, miR‐133a, miR‐133b and miR‐206. Integrating the miRNAs and mRNAs that were differentially expressed in a network analysis pointed to nodes of miRNA regulation containing genes such as HEYL, NR4A2, NR4A3, PAX7 and PHIP. Interestingly, these are all annotated as regulators of muscle developments and differentiation. Moreover, functional studies with overexpression of miR‐30b, also differentially expressed during muscle differentiation, showed that not all in‐silico predicted gene targets were necessarily regulated by manipulation of this particular miRNA. Despite the integration of simultaneous miRNA and mRNA, expression data with target prediction algorithms and network this study clearly show that bioinformatics‐based deductions cannot substitute for experimental validation of miRNA function (Sjogren et al. 2015). Therefore, hypothesis‐driven functional studies of miRNA and target genes within skeletal muscle development, as well as other biological settings, still needs to be addressed.
The muscular walls of the heart (the myocardium) are responsible for pumping blood through the lungs and the rest of the body, and it is clearly important that the muscles in the heart maintain their activity under all conditions. The increased risk of premature death with type 2 diabetes is not because of diabetes per se, but the cardiovascular diseases and their comorbidities which follow with the development of diabetes and obesity. Microvascular complications occur as long‐term sequelae to poorly controlled type 2 diabetes and include diabetic retinopathy, neuropathy and impaired wound healing as well as nephropathy. The role of miRNAs and their impact on microvascular complications has been excellently covered elsewhere (Moura et al. 2014, Banerjee & Sen 2015, Bhatt et al. 2016).
Several miRNAs are enriched in the heart (Sempere et al. 2004), and heart‐specific dicer1 KO mice die young by severe heart failure mainly characterized by hypertrophic growth (Chen et al. 2008) demonstrating the importance for Dicer in cardiac contraction and indicate that miRNAs play a key role in proper heart function. In humans, cardiac hypertrophy is the main risk factor for the development of heart failure and lethal arrhythmias (Towbin & Bowles 2002) and is usually linked to hypertension and macrovascular complications. The development of cardiac hypertrophy is linked to aberrant reactivation of embryonic gene programmes, which is still not completely understood. It is known, however, that transcription factors sharing a basic helix‐loop‐helix domain are important for determination and differentiation of various cell types, including cardiomyocytes. Elegantly, Dirkx and colleagues (Dirkx et al. 2013) showed that such a transcription factor, heart and neural crest derivatives expressed transcript (Hand2) which is required for proper heart development (Hutson & Kirby 2007, Snider et al. 2007), is re‐activated in the failing heart, where it drives the induction of a gene network controlling cardiac growth, dilation and dysfunction. Interestingly, Hand2 is inversely expressed with miR‐1, miR‐92a, miR‐92b and miR‐25 in experimental models of heart disease and in vivo inhibition of miR‐25 resulted in cardiac dysfunction in a Hand2 dependent manner (Dirkx et al. 2013). Several other miRNAs have been shown to be implicated in pathological cardiac hypertrophy including miR‐133 (Care et al. 2007), miR‐199b (Da Costa Martins et al. 2010) and miR‐378 (Ganesan et al. 2013) signifying the potential for targeting specific miRNAs with RNA based therapeutics.
MiRNAs in adipose tissue and adipocyte differentiation
The traditional view of adipose tissues as biologically inactive lipid storage depots has changed over the course of the last 30 years. It is now clear that the adipose tissues are highly responsive endocrine organs that influence metabolic homoeostasis and inflammation (Rosen & Spiegelman 2006). The importance of adipose tissue function in health and disease is revealed by the range of diseases previously associated with ageing that are more prevalent amongst overweight and obese individuals. The discovery of an age‐induced decline in miRNA processing, specifically in the adipose tissue, underlines the importance of this tissue. The abundance of the key miRNA processing enzyme, Dicer, is reduced in white adipose tissue (inguinal and perigonadal) by ageing and followed by a coordinated decline in levels of multiple miRNAs, an observation conserved between mice, human and nematodes. In mice subjected to calorie restriction, Dicer levels do not decline (Mori et al. 2012b). Knockdown of Dicer in cells results in premature senescence, and mice deficient of Dicer in adipose tissue develop lipodystrophic loss of intra‐abdominal and subcutaneous white fat, severe insulin resistance and enlargement and ‘whitening’ of intrascapular brown fat (Mori et al. 2012b, 2014), and miR‐365 was identified as a miRNA that partially can explain the ‘whitening’ phenotype.
Brown adipose tissue (BAT) has an obvious therapeutic potential, and many studies highlight the importance of miRNAs in the formation of BAT, such as miR‐193b/‐365 (Sun et al. 2011, Feuermann et al. 2013), miR‐196a (Mori et al. 2012a), miR‐155 (Chen et al. 2013) and miR‐133a/b (Trajkovski et al. 2012). Recently, Zhang et al. showed by combining miRNA and mRNA microarray data that miR‐455 plays a crucial role in brown adipogenesis. MiR‐455, a BMP7‐induced miRNA, targets several key adipogenic regulators, such as Necdin and Runx1t1, which are important adipogenic suppressors gating an adipocyte differentiation programme. In addition, miR‐455 targets hypoxia‐inducible factor 1a inhibitor (HIF1an), a hydroxylase that normally inhibits AMPK activity by hydroxylation, which leads to AMPK activation. Thus, miR‐455 suppresses Necdin and Runx1t1 to initiate adipogenic programme and suppresses HIF1an to activate AMPK which, in turn, acts as a metabolic trigger to induce a brown adipogenesis (Zhang et al. 2015). It is indeed interesting to observe that BAT expressed miRNAs to a certain degree reflect the myogenic lineage of these cells in that BAT shares important functional miRNAs with skeletal muscle (Table 1).
Table 1.
MiRNAs of importance for metabolically relevant tissues
| β‐cells | Liver | Sk. Muscle | Heart | WAT | BAT |
|---|---|---|---|---|---|
| miR‐375 | miR‐122 | miR‐1 | miR‐25 | miR‐365 | miR‐365 |
| miR‐200 family | miR‐33a/b | miR‐206 | miR‐199a‐214 | miR‐193b | miR‐193b |
| miR‐7 family | miR‐148a | miR‐208b | miR‐155 | miR‐126 | miR‐155 |
| miR‐335 | miR‐143 | miR‐133a/b | miR‐92a | miR‐133a/b | |
| miR‐152 | miR‐103/107 | miR‐499 | miR‐196a | ||
| miR‐29 family | miR‐802 | miR‐30b | miR‐455 | ||
| miR‐181b | miR‐181a | miR‐143 | |||
| miR‐184 | |||||
| miR‐187 | |||||
| miR‐204 | |||||
| miR‐17‐92 | |||||
| miR‐129 | |||||
| miR‐34a | |||||
| miR‐215 | |||||
| miR‐130a/b |
MiRNAs in this table are all cited in the text and is not an exhaustive list of all miRNAs shown to be important for the function of these tissues. MiRNAs are given by number only, for identity of 5′ or 3′ mature species please refer to the original publication.
Several studies have compared miRNA expression profiles in obese and lean white adipose tissue from mice and from humans. In fat cells from mice with diet‐induced obesity, 35 of the 574 detected miRNAs were differentially expressed (Xie et al. 2009). Microarray screening of human WAT tissues identified a number of miRNAs that are potentially dysregulated in patients with obesity, both in those with concurrent type 2 diabetes mellitus and in glucose tolerant subjects (Heneghan et al. 2011, Keller et al. 2011). However, it has proven difficult to validate reported miRNAs in other cohorts, as well as the direction of expression of the miRNA between lean and obese groups, all of which is excellently reviewed in Arner & Kulyte (2015) (Arner & Kulyte 2015). This highlights the challenge that many miRNA studies are difficult to reproduce. As possible sources of variation, one has to consider the sample origin, which tissue or cell type is examined, the gender of test subjects and disease state. Furthermore, many microarray studies are often underpowered, which also contributes to ambiguous results (Witwer 2013), not to mention which platform was used to detect the miRNAs (Mestdagh et al. 2014).
MiRNAs often assert their action in families and thus work in networks. Arner et al. (2012) found that 11 miRNAs were downregulated in obese subjects, all of which target the expression of CCL2 (C‐C motif ligand 2, also known as MCP1 (macrophage chemoattractant protein 1) (Arner et al. 2012). The chemokine CCL2 is suspected to initiate inflammation in the adipose tissue, which in turn is could be a driver of whole‐body insulin resistance. Also, it is known that obese individuals have an increased secretion of CCL2 compared to lean controls (Arner et al. 2012). A subnetwork involving miR‐126, miR‐193b and miR‐92a was shown to associate inversely with transcription factors controlling inflammation in the adipose tissue and that the regulation of these miRNAs had an additive effect on CCL2 secretion (Kulyte et al. 2014). Even though there is conflicting data, especially in the context of WAT and miRNAs, accumulating data indicate that miRNAs are central modulators of normal WAT and BAT differentiation and biology (Rottiers & Naar 2012).
MiRNAs as therapeutics
The growing knowledge about miRNAs and their molecular actions gives rise to innovative industrial applications for this class of molecules (Fig. 2a). Amongst the most promising perspectives are the usage of miRNAs in medical therapy and their potential as new biomarkers (Hayes et al. 2014). Both topics will be discussed in the following section with focus on current challenges, existing solution strategies and future perspectives.
Figure 2.
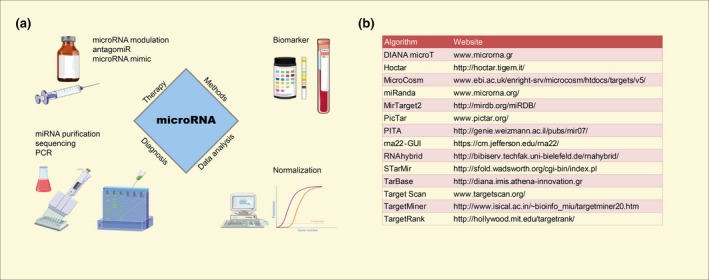
(a) Translational aspects of miRNAs. For therapy, miRNAs may be used as antagonists (antagomiR) or agonists (microRNA mimics). Specific methods, such as next‐generation sequencing of small RNAs and qPCR‐based miRNA arrays are used to identify possible miRNA biomarkers from patient specimens, but may also be used to monitor therapeutic effects or unintended side effects by treatment. Integral to the translational use of miRNAs are improved and consistent data analysis strategies of qPCR results with special focus on using optimal data normalization and identification of proper reference genes or miRNAs. For these purposes, current techniques and analysis strategies need to be adjusted and optimized. The figure was assembled with the help of Servier Medical Art (http://www.servier.com/Powerpoint-image-bank). (b) Table of Web‐tools commonly used for prediction of miRNA‐target mRNAs.
Given that miRNAs are involved in regulation of a magnitude of cellular processes (Esau et al. 2006, Trajkovski et al. 2011, Rottiers & Naar 2012, Dumortier et al. 2013), it is not surprising that miRNA expression patterns change in obesity (Ortega et al. 2013) and diabetes (Kong et al. 2010, Hayes et al. 2014, Yang et al. 2014). Pathophysiologically, upregulated miRNAs can be manipulated by complementary nucleotide analogues to decrease their effective activity (Stenvang et al. 2012). These ‘antagomiRs’ often have a modified nucleotide architecture, where, for example, the nucleic acid ribose moieties have been replaced by the high‐affinity RNA analogue Locked Nucleic Acid™ (LNA™, Exiqon). AntagomiRs bind to mature and precursor miRNAs and thus effectively remove them from the biologically available pool (Elmen et al. 2007, Gebert et al. 2014). MiRNA sponges constitute an alternative inhibition strategy. Acting after a similar principle, miRNA sponges contain multiple miRNA‐binding sites in their sequence and thus compete for miRNAs (Zhang et al. 2013). Modified oligo‐ribonucleotides may be used for miRNA overexpression and in parallel with inhibitors these are convenient tools for modifying cellular miRNA levels in in vitro models (Stenvang & Kauppinen 2008, Chen et al. 2015).
The Miravirsen antagomiR targeting the liver‐specific miR‐122 (Roche Innovation Center Copenhagen, previously Santaris Pharma) shows feasibility of miRNA inhibition and is a promising translation of basic microRNA research into therapeutic context (Janssen et al. 2013, Ottosen et al. 2015). MiR‐122 is required for hepatitis C virus (HCV) replication (Jopling et al. 2005, 2008, Henke et al. 2008), and Miravirsen administration is able to suppress miR‐122 expression and thus prevent HCV replication (Krützfeldt et al. 2005, Ottosen et al. 2015).
Within metabolism, only few miRNA inhibitors are in development, one of which the N‐acetylgalactosamine (GalNAc)‐conjugated anti‐miR‐103/107 RG‐125(AZD4076), being developed by Regulus Therapeutics and AstraZeneca for the treatment of non‐alcoholic steatohepatitis (NASH) in patients with type 2 diabetes/pre‐diabetes (RegulusTherapeutics 2014, 2015). The current treatment of NASH with thiazolidinediones is commonly accompanied with undesirable weight gain (Musso et al. 2012), and there is an unmet need for improved therapy for this disorder. Treatment with RG‐125(AZD4076) is based on its ability to inhibit the activity of miR‐103/107, whose hepatic upregulation causes insulin resistance (Trajkovski et al. 2011). AntagomiR‐based silencing of miR‐103/107 in mice was followed by decreased liver triglyceride content and improved insulin sensitivity. RG‐125 (AZD4076) has therefore the potential to acts as an efficacious insulin sensitizer (RegulusTherapeutics 2015).
RG‐125 (AZD4076) is modified by addition of an N‐acetylgalactosamine (RegulusTherapeutics 2015), which targets the oligonucleotide preferentially to hepatocytes via the binding to the liver enriched ASGR1 (asialoglycoprotein receptor 1). This conjugation enhances potency and aids in avoiding cross‐reactivity with similar miRNA families like miR‐15/16, circumventing two of the challenges facing miRNA‐based pharmaceutical agents; delivery and specificity for intended miRNAs. Currently, AstraZeneca initiated dosing in a first‐in‐human Phase I clinical study of RG‐125(AZD4076) at the end of 2015.
MiRNA‐based therapies offer some distinct advantages over other nucleic acid directed therapies: MiRNAs are efficient silencers and in contrast to plasmid DNA or synthetic oligonucleotides, miRNAs occur naturally in the blood stream. As they target multiple mRNAs, resulting synergistic effects could be positive for therapy (Chen et al. 2015) and increase the barriers for formation of resistance (Janssen et al. 2013, Wang et al. 2015a). A low toxicity and good tolerance in antagomiR‐treated patients support the beneficial role of miRNAs in therapy (Janssen et al. 2013, Van Der Ree et al. 2014).
However, there are multiple important challenges facing miRNA‐mediated treatments: unmodified miRNAs are rapidly degraded (Chen et al. 2015), emphasizing the requirement for chemically modified derivatives or encapsulation. Moreover, activation of the innate immunity or neurotoxicity is potential and important side effects, and miRNA inhibitors are restricted in their actions by cellular uptake and incorporation into RISC, and improper dosing can lead to inhibition of unintended targets causing side effects (Lindow et al. 2012). Currently, two of the major challenges with miRNA‐based drugs are delivery (Wang et al. 2015a) and low tissue specificity (Kwekkeboom et al. 2016). Administration of miRNA‐based drug candidates is mostly carried out by injection either intravenously or locally (Obad et al. 2011, Shu et al. 2015, Wang et al. 2015a), but alternative approaches include oral administration, enema formulation followed by gut delivery, topical application and intra‐ocular delivery (Stenvang & Kauppinen 2008, Kwekkeboom et al. 2016). Nevertheless, improved strategies for precise and efficient tissue‐delivery are needed.
Thus, miRNA‐based therapy approaches have potential as new and innovative tools in various diseases, but there is currently only few in development for metabolic disease most likely due to the promiscuous nature of the miRNA targets as well as the difficulties of obtaining tissue specificity.
Biomarkers
MiRNAs are found in bio fluids, such as blood, urine, plasma and saliva (Javidi et al. 2014, Arrese et al. 2015). Although pure RNA is prone to rapid degradation, miRNAs from bio fluids show a remarkable stability (Mitchell et al. 2008). The common explanation is that these miRNAs are contained in exosomes, which are small membrane vesicles of 40–100 nm size (Chevillet et al. 2014), which contain DNA, mRNA and proteins in addition to miRNAs (Ban et al. 2015). Surrounded by such membrane structures, miRNAs are protected from RNases. The process of miRNA‐secretion in exosomes is still largely unknown. It is unclear, which tissues contribute to miRNA secretion and whether it is the result of active sorting. Furthermore, it is an open question, whether secreted miRNAs have a biological function. But given the low abundancy of miRNAs in exosomes, which might be as low as one copy per exosome (Chevillet et al. 2014), a communication function seems unlikely. Moreover, miRNAs in bio fluids may also be stabilized via binding to Ago2 or lipoproteins (Arroyo et al. 2011).
Despite these unclear issues, it is well described that the miRNA patterns of bio fluids change under pathological conditions (Mitchell et al. 2008, Ortega et al. 2013, Yang et al. 2014). MiRNA signatures show promise for use in diagnostic or prognostic tests in a variety of diseases, for example cancer (Javidi et al. 2014), polycystic ovary syndrome (Sørensen et al. 2014) and liver diseases (Arrese et al. 2015) amongst others. Advantages of using miRNAs as biomarkers include sensitivity of detection and possibility of multiplexing analyses for increased specificity. A major challenge for the analysis of miRNAs as biomarkers from bio fluids is to establish consequent and robust protocols for pre‐analytical sample handling, miRNA extraction and measurement, which are all important for reliable results (Blondal et al. 2013). All taken together, miRNAs can be used as diagnostic tools, but studies for the future should include investigations of large, population based cohorts to establish base‐line values and degree of between subject variation to enable the use of miRNAs for minimal to non‐invasive biomarkers.
Quantification of miRNA
Even though RNA molecules have been studied for decades, the adaption of standard laboratory techniques to miRNA research is challenging. While both column‐ and chemical‐based RNA isolation techniques are successfully used for mRNA isolation, these have limitations with regard to miRNA (Trevorlstokes, 2012, McAlexander et al. 2013, Moldovan et al. 2014). For analysis of miRNA levels, two commonly used approaches are platforms based on sequencing and quantitative PCR (qPCR). During the miRNA quality control study (MiRQC), commercial platforms from 12 different providers were assessed and compared by different quality metrics (Mestdagh et al. 2014). It was investigated how well platforms perform when faced with challenges such as discrimination between highly similar miRNA sequences or low abundant miRNAs. As it turned out, each platform has specific strengths and weaknesses. The choice of an optimal approach is therefore highly dependent on the experimental setup and goal.
Integration of mRNA and miRNA expression data based on micro‐array analysis or next‐generation sequencing is a convenient method to investigate possible regulatory mechanisms of miRNAs. However, it is important to realize that miRNA targets preferentially repressed at the translational level will not be detected using this approach. Thus, using mRNA arrays to characterize miRNA‐target regulation is likely to miss true targets regulated by translational inhibition, and therefore, this approach is more likely to report false negative findings. Using Ago2 immunoprecipitation to enrich for mRNAs incorporated into RISC, and thus targeted by a miRNA, is an alternative approach to identify miRNA targets enabling identification of targets regulated at the translational level as well.
Analysis, quantification and transparency of miRNA data have become critical steps. A particular drawback is the absence of a standard reference for normalization. The use of snoRNAs, such as U6, is undesirable, because of different stability and biogenesis compared with miRNAs (Vandesompele 2013, Hellemans & Vandesompele 2014). Other strategies, including global mean normalization (Zhao et al. 2010) or several stable reference genes (Bustin et al. 2009, Mestdagh et al. 2009), should be used. To increase analysis transparency, detailed information regarding miRNA sequence and name should be explicitly noted in resulting publications (Van Peer et al. 2014), for example using the miR‐tracker software (Van Peer et al. 2014).
Identifying target mRNAs of a given miRNA is crucial in understanding the biological context of miRNAs, but nevertheless remains a complex issue. The imperfect base pairing of miRNA‐mRNA duplexes is challenging for software algorithms (Witkos et al. 2011). Consequently, most algorithms struggle with a high number of false‐positive results and low accuracy (Ekimler et al. 2014). Current prediction algorithms are available as online tools (Fig. 2b) or source code. The starting point is usually the ‘seed region’ from nucleotide 2–7 of miRNAs (Mazière & Enright 2007, Vlachos & Hatzigeorgiou 2013, Peterson et al. 2014), which used by RISC to bind mRNAs by Watson‐Crick base pairing to the miRNA (Saito & Sætrom 2010, Peterson et al. 2014). Depending on the algorithm, other features are also taken into account: the location of the binding site on the mRNA, sequence conservation across species, free energy of the formed duplex, accessibility of the mRNA binding site, surrounding miRNA‐recognition sites and miRNA expression profiles in the investigated tissue (Saito & Sætrom 2010, Vlachos & Hatzigeorgiou 2013, Ekimler et al. 2014, Peterson et al. 2014). Different balances of these features can cause discrepancies in the results. Preferably, several algorithms can be combined in the analysis (Witkos et al. 2011) followed by experimental validation. Even though bioinformatic target prediction methods have their limits, they can supplement experimental approaches, giving rise to a more efficient identification of miRNA targets. In conclusion, both the analysis and target prediction of miRNAs provides new challenges for wet and dry laboratory.
Conclusions and perspectives
A large number of miRNAs have been implicated in different facets of the metabolic syndrome and diabetes mellitus, and currently, there are no well‐established or unifying sets of miRNAs characterizing the various subphenotypes of metabolic disease. However, a number of miRNAs appear to affect the function or differentiated state of the pancreatic β‐cell, whereas miRNAs in skeletal muscle, liver and adipose tissue constitute different and almost non‐overlapping sets of miRNAs (Table 1). The field of miRNA research is developing rapidly with new tools and models arising. This will enable the further development of miRNA‐based therapeutics for the treatment of metabolic diseases for which there is an unmet need worldwide. Moreover, seeing the impact of regulation of miRNA processing in adipose tissue, it could be highly useful to identify endogenous as well as small molecule regulators of miRNA processing with the potential use of modifying adipose tissue metabolism for treatment of metabolic disease.
A general conclusion is that with many contrasting studies regarding expression regulation and function of miRNAs in different cell types suggests that there is a genuine need for more studies. As many miRNAs appear to have roles in stress response modulation, whereas they are dispensable in the unperturbed state, it will be necessary to investigate the proper models of metabolic stress in order to elucidate their functions. Moreover, because miRNAs often exist in families, an increasingly important approach will be to modulate levels of entire miRNA families or coregulated miRNAs together as well as separately in order to establish their roles in the intact tissue or organ.
This work, and the Symposium on MicroRNAs in Metabolism, from which this review originates, was sponsored by the Danish Diabetes Academy supported by the Novo Nordisk Foundation. We are very grateful for the support from the Invited Speakers of this Symposium for sharing their view on MicroRNA research in Metabolism and for their constructive comments to this review. A full list of Speakers at the Symposium on MicroRNAs in Metabolism can be found in Appendix S1. The authors would like to thank Lena Eliasson, Romano Regazzi, Marcelo Mori, Brendan Egan, Peter Mouritzen, Bader Zarrouki and Hongbin Zhang for critical revision and their constructive comments on this manuscript. Funding: This work is supported by the Danish Diabetes Academy supported by the Novo Nordisk Foundation, Roskilde University and the FSS, DFF¦Danish Medical Research Council 1331‐00033 to L.T.D. The funders had no role in decision to publish or preparation of the manuscript.
Conflicts of interest
JG, SM and LTD have nothing to declare. SV is currently employed by Novo Nordisk A/S, a pharmaceutical company selling products for treatment of diabetes mellitus.
Supporting information
Appendix S1 Speakers at the microRNAs in metabolism symposium, who all contributed to the manuscript.
References
- Aguda, B.D. , Kim, Y. , Piper‐Hunter, M.G. , Friedman, A. & Marsh, C.B. 2008. MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR‐17‐92, E2F, and Myc. Proc Natl Acad Sci USA 105, 19678–19683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner, P. & Kulyte, A. 2015. MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol 11, 276–288. [DOI] [PubMed] [Google Scholar]
- Arner, E. , Mejhert, N. , Kulyte, A. , Balwierz, P.J. , Pachkov, M. , Cormont, M. , Lorente‐Cebrian, S. , Ehrlund, A. , Laurencikiene, J. , Heden, P. et al 2012. Adipose tissue microRNAs as regulators of CCL2 production in human obesity. Diabetes 61, 1986–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese, M. , Eguchi, A. & Feldstein, A.E. 2015. Circulating microRNAs: emerging biomarkers of liver disease. Semin Liver Dis 35, 43–54. [DOI] [PubMed] [Google Scholar]
- Arroyo, J.D. , Chevillet, J.R. , Kroh, E.M. , Ruf, I.K. , Pritchard, C.C. , Gibson, D.F. , Mitchell, P.S. , Bennett, C.F. , Pogosova‐Agadjanyan, E.L. , Stirewalt, D.L. , Tait, J.F. & Tewari, M. 2011. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA 108, 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban, J.‐J. , Lee, M. , Im, W. & Kim, M. 2015. Low pH increases the yield of exosome isolation. Biochem Biophys Res Commun 461, 76–79. [DOI] [PubMed] [Google Scholar]
- Banerjee, J. & Sen, C.K. 2015. microRNA and wound healing. Adv Exp Med Biol 888, 291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgardt, B.F. , Ahmed, K. , Spranger, M. , Latreille, M. , Denzler, R. , Kondratiuk, N. , von Meyenn, F. , Villena, F.N. , Herrmanns, K. , Bosco, D. , Kerr‐Conte, J. , Pattou, F. , Rulicke, T. & Stoffel, M. 2015. The microRNA‐200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat Med 21, 619–627. [DOI] [PubMed] [Google Scholar]
- Bhatt, K. , Kato, M. & Natarajan, R. 2016. Mini‐review: emerging roles of microRNAs in the pathophysiology of renal diseases. Am J Physiol Renal Physiol 310, F109–F118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondal, T. , Jensby Nielsen, S. , Baker, A. , Andreasen, D. , Mouritzen, P. , Wrang Teilum, M. & Dahlsveen, I.K. 2013. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 59, S1–S6. [DOI] [PubMed] [Google Scholar]
- Bodine, S.C. 2013. Disuse‐induced muscle wasting. Int J Biochem Cell Biol 45, 2200–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon, H. , Sjogren, R.J. , Massart, J. , Egan, B. , Kostovski, E. , Iversen, P.O. , Hjeltnes, N. , Chibalin, A.V. , Widegren, U. & Zierath, J.R. 2015. MicroRNA‐208b progressively declines after spinal cord injury in humans and is inversely related to myostatin expression. Physiol Rep 3, e12622, doi: 10.14814/phy2.12622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Bunt, M. , Gaulton, K.J. , Parts, L. , Moran, I. , Johnson, P.R. , Lindgren, C.M. , Ferrer, J. , Gloyn, A.L. & McCarthy, M.I. 2013. The miRNA profile of human pancreatic islets and beta‐cells and relationship to type 2 diabetes pathogenesis. PLoS ONE 8, e55272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin, S.A. , Benes, V. , Garson, J.A. , Hellemans, J. , Huggett, J. , Kubista, M. , Mueller, R. , Nolan, T. , Pfaffl, M.W. , Shipley, G.L. , Vandesompele, J. & Wittwer, C.T. 2009. The MIQE guidelines: minimum information for publication of quantitative real‐time PCR experiments. Clin Chem 55, 611–622. [DOI] [PubMed] [Google Scholar]
- Care, A. , Catalucci, D. , Felicetti, F. , Bonci, D. , Addario, A. , Gallo, P. , Bang, M.L. , Segnalini, P. , Gu, Y. , Dalton, N.D. et al 2007. MicroRNA‐133 controls cardiac hypertrophy. Nat Med 13, 613–618. [DOI] [PubMed] [Google Scholar]
- Chen, J.F. , Mandel, E.M. , Thomson, J.M. , Wu, Q. , Callis, T.E. , Hammond, S.M. , Conlon, F.L. & Wang, D.Z. 2006. The role of microRNA‐1 and microRNA‐133 in skeletal muscle proliferation and differentiation. Nat Genet 38, 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.F. , Murchison, E.P. , Tang, R. , Callis, T.E. , Tatsuguchi, M. , Deng, Z. , Rojas, M. , Hammond, S.M. , Schneider, M.D. , Selzman, C.H. , Meissner, G. , Patterson, C. , Hannon, G.J. & Wang, D.Z. 2008. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci USA 105, 2111–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Siegel, F. , Kipschull, S. , Haas, B. , Frohlich, H. , Meister, G. & Pfeifer, A. 2013. miR‐155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat Commun 4, 1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Gao, D.Y. & Huang, L. 2015. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv Drug Deliv Rev 81, 128–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevillet, J.R. , Kang, Q. , Ruf, I.K. , Briggs, H.A. , Vojtech, L.N. , Hughes, S.M. , Cheng, H.H. , Arroyo, J.D. , Meredith, E.K. , Gallichotte, E.N. et al 2014. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci USA 111, 14888–14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa Martins, P.A. , Salic, K. , Gladka, M.M. , Armand, A.S. , Leptidis, S. , El Azzouzi, H. , Hansen, A. , Coenen‐De Roo, C.J. , Bierhuizen, M.F. , Van Der Nagel, R. et al 2010. MicroRNA‐199b targets the nuclear kinase Dyrk1a in an auto‐amplification loop promoting calcineurin/NFAT signalling. Nat Cell Biol 12, 1220–1227. [DOI] [PubMed] [Google Scholar]
- Dayeh, T. , Volkov, P. , Salo, S. , Hall, E. , Nilsson, E. , Olsson, A.H. , Kirkpatrick, C.L. , Wollheim, C.B. , Eliasson, L. , Ronn, T. , Bacos, K. & Ling, C. 2014. Genome‐wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non‐diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet 10, e1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFronzo, R.A. , Gunnarsson, R. , Bjorkman, O. , Olsson, M. & Wahren, J. 1985. Effects of insulin on peripheral and splanchnic glucose metabolism in noninsulin‐dependent (type II) diabetes mellitus. J Clin Invest 76, 149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey, B.K. , Gagan, J. & Dutta, A. 2011. miR‐206 and ‐486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol 31, 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirkx, E. , Gladka, M.M. , Philippen, L.E. , Armand, A.S. , Kinet, V. , Leptidis, S. , El Azzouzi, H. , Salic, K. , Bourajjaj, M. , Da Silva, G.J. et al 2013. Nfat and miR‐25 cooperate to reactivate the transcription factor Hand2 in heart failure. Nat Cell Biol 15, 1282–1293. [DOI] [PubMed] [Google Scholar]
- Dumortier, O. , Hinault, C. & van Obberghen, E. 2013. MicroRNAs and metabolism crosstalk in energy homeostasis. Cell Metab 18, 312–324. [DOI] [PubMed] [Google Scholar]
- Ekimler, S. , Sahin, K. , Watanabe, Y. , Tomita, M. & Kanai, A. 2014. Computational methods for microRNA target prediction. Methods Enzymol 5, 671–683. [DOI] [PubMed] [Google Scholar]
- Eliasson, L. 2014. The exocytotic machinery. Acta Physiol (Oxf) 210, 455–457. [DOI] [PubMed] [Google Scholar]
- Elmen, J. , Lindow, M. , Silahtaroglu, A. , Bak, M. , Christensen, M. , Lind‐Thomsen, A. , Hedtjarn, M. , Hansen, J.B. , Hansen, H.F. , Straarup, E.M. , McCullagh, K. , Kearney, P. & Kauppinen, S. 2007. Antagonism of microRNA‐122 in mice by systemically administered LNA‐antimiR leads to up‐regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res 36, 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erener, S. , Mojibian, M. , Fox, J.K. , Denroche, H.C. & Kieffer, T.J. 2013. Circulating miR‐375 as a biomarker of beta‐cell death and diabetes in mice. Endocrinology 154, 603–608. [DOI] [PubMed] [Google Scholar]
- Esau, C. , Davis, S. , Murray, S.F. , Yu, X.X. , Pandey, S.K. , Pear, M. , Watts, L. , Booten, S.L. , Graham, M. , McKay, R. et al 2006. miR‐122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3, 87–98. [DOI] [PubMed] [Google Scholar]
- Esguerra, J.L. , Bolmeson, C. , Cilio, C.M. & Eliasson, L. 2011. Differential glucose‐regulation of microRNAs in pancreatic islets of non‐obese type 2 diabetes model Goto‐Kakizaki rat. PLoS ONE 6, e18613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez‐Hernando, C. , Ramirez, C.M. , Goedeke, L. & Suarez, Y. 2013. MicroRNAs in metabolic disease. Arterioscler Thromb Vasc Biol 33, 178–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuermann, Y. , Kang, K. , Gavrilova, O. , Haetscher, N. , Jang, S.J. , Yoo, K.H. , Jiang, C. , Gonzalez, F.J. , Robinson, G.W. & Hennighausen, L. 2013. MiR‐193b and miR‐365‐1 are not required for the development and function of brown fat in the mouse. RNA Biol 10, 1807–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan, J. , Ramanujam, D. , Sassi, Y. , Ahles, A. , Jentzsch, C. , Werfel, S. , Leierseder, S. , Loyer, X. , Giacca, M. , Zentilin, L. , Thum, T. , Laggerbauer, B. & Engelhardt, S. 2013. MiR‐378 controls cardiac hypertrophy by combined repression of mitogen‐activated protein kinase pathway factors. Circulation 127, 2097–2106. [DOI] [PubMed] [Google Scholar]
- Gebert, L.F.R. , Rebhan, M.A.E. , Crivelli, S.E.M. , Denzler, R. , Stoffel, M. & Hall, J. 2014. Miravirsen (SPC3649) can inhibit the biogenesis of miR‐122. Nucleic Acids Res 42, 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedeke, L. , Rotllan, N. , Canfran‐Duque, A. , Aranda, J.F. , Ramirez, C.M. , Araldi, E. , Lin, C.S. , Anderson, N.N. , Wagschal, A. , de Cabo, R. , Horton, J.D. , Lasuncion, M.A. , Naar, A.M. , Suarez, Y. & Fernandez‐Hernando, C. 2015. MicroRNA‐148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat Med 21, 1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halban, P.A. , Polonsky, K.S. , Bowden, D.W. , Hawkins, M.A. , Ling, C. , Mather, K.J. , Powers, A.C. , Rhodes, C.J. , Sussel, L. & Weir, G.C. 2014. Beta‐cell failure in type 2 diabetes: postulated mechanisms and prospects for prevention and treatment. J Clin Endocrinol Metab 99, 1983–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, J. , Peruzzi, P.P. & Lawler, S. 2014. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 20, 460–469. [DOI] [PubMed] [Google Scholar]
- Hellemans, J. & Vandesompele, J. 2014. Selection of reliable reference genes for RT‐qPCR analysis. Methods Mol Biol 1160, 19–26. [DOI] [PubMed] [Google Scholar]
- Heneghan, H.M. , Miller, N. , McAnena, O.J. , O'Brien, T. & Kerin, M.J. 2011. Differential miRNA expression in omental adipose tissue and in the circulation of obese patients identifies novel metabolic biomarkers. J Clin Endocrinol Metab 96, E846–E850. [DOI] [PubMed] [Google Scholar]
- Henke, J.I. , Goergen, D. , Zheng, J. , Song, Y. , Schüttler, C.G. , Fehr, C. , Jünemann, C. & Niepmann, M. 2008. microRNA‐122 stimulates translation of hepatitis C virus RNA. EMBO J 27, 3300–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmeier, H.E. , Mulder, H. , Chen, G. , Henkel‐Rieger, R. , Prentki, M. & Newgard, C.B. 2000. Isolation of INS‐1‐derived cell lines with robust ATP‐sensitive K+ channel‐dependent and ‐independent glucose‐stimulated insulin secretion. Diabetes 49, 424–430. [DOI] [PubMed] [Google Scholar]
- Horie, T. , Ono, K. , Horiguchi, M. , Nishi, H. , Nakamura, T. , Nagao, K. , Kinoshita, M. , Kuwabara, Y. , Marusawa, H. , Iwanaga, Y. , Hasegawa, K. , Yokode, M. , Kimura, T. & Kita, T. 2010. MicroRNA‐33 encoded by an intron of sterol regulatory element‐binding protein 2 (Srebp2) regulates HDL in vivo . Proc Natl Acad Sci USA 107, 17321–17326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson, M.R. & Kirby, M.L. 2007. Model systems for the study of heart development and disease. Cardiac neural crest and conotruncal malformations. Semin Cell Dev Biol 18, 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacovetti, C. , Matkovich, S.J. , Rodriguez‐Trejo, A. , Guay, C. & Regazzi, R. 2015. Postnatal beta‐cell maturation is associated with islet‐specific microRNA changes induced by nutrient shifts at weaning. Nat Commun 6, 8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen, H.L.A. , Reesink, H.W. , Lawitz, E.J. , Zeuzem, S. , Rodriguez‐Torres, M. , Patel, K. , van der Meer, A.J. , Patick, A.K. , Chen, A. , Zhou, Y. , Persson, R. , King, B.D. , Kauppinen, S. , Levin, A.A. & Hodges, M.R. 2013. Treatment of HCV infection by targeting microRNA. N Engl J Med 368, 1685–1694. [DOI] [PubMed] [Google Scholar]
- Javidi, M.A. , Ahmadi, A.H. , Bakhshinejad, B. , Nouraee, N. , Babashah, S. & Sadeghizadeh, M. 2014. Cell‐free microRNAs as cancer biomarkers: the odyssey of miRNAs through body fluids. Med Oncol 31, 295. [DOI] [PubMed] [Google Scholar]
- Jopling, C.L. , Yi, M. , Lancaster, A.M. , Lemon, S.M. & Sarnow, P. 2005. Modulation of hepatitis C virus RNA abundance by a liver‐specific microRNA. Science 309, 1577–1581. [DOI] [PubMed] [Google Scholar]
- Jopling, C.L. , Schütz, S. & Sarnow, P. 2008. Position‐dependent function for a tandem microRNA miR‐122‐binding site located in the hepatitis C virus RNA genome. Cell Host Microbe 4, 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, S.D. , Kruger, M. , Willmes, D.M. , Redemann, N. , Wunderlich, F.T. , Bronneke, H.S. , Merkwirth, C. , Kashkar, H. , Olkkonen, V.M. , Bottger, T. , Braun, T. , Seibler, J. & Bruning, J.C. 2011. Obesity‐induced overexpression of miRNA‐143 inhibits insulin‐stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol 13, 434–446. [DOI] [PubMed] [Google Scholar]
- Kalis, M. , Bolmeson, C. , Esguerra, J.L. , Gupta, S. , Edlund, A. , Tormo‐Badia, N. , Speidel, D. , Holmberg, D. , Mayans, S. , Khoo, N.K. , Wendt, A. , Eliasson, L. & Cilio, C.M. 2011. Beta‐cell specific deletion of Dicer1 leads to defective insulin secretion and diabetes mellitus. PLoS ONE 6, e29166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, P. , Gburcik, V. , Petrovic, N. , Gallagher, I.J. , Nedergaard, J. , Cannon, B. & Timmons, J.A. 2011. Gene‐chip studies of adipogenesis‐regulated microRNAs in mouse primary adipocytes and human obesity. BMC Endocr Disord 11, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, L. , Zhu, J. , Han, W. , Jiang, X. , Xu, M. , Zhao, Y. , Dong, Q. , Pang, Z. , Guan, Q. , Gao, L. , Zhao, J. & Zhao, L. 2010. Significance of serum microRNAs in pre‐diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol 48, 61–69. [DOI] [PubMed] [Google Scholar]
- Kornfeld, J.W. , Baitzel, C. , Konner, A.C. , Nicholls, H.T. , Vogt, M.C. , Herrmanns, K. , Scheja, L. , Haumaitre, C. , Wolf, A.M. , Knippschild, U. , Seibler, J. , Cereghini, S. , Heeren, J. , Stoffel, M. & Bruning, J.C. 2013. Obesity‐induced overexpression of miR‐802 impairs glucose metabolism through silencing of Hnf1b. Nature 494, 111–115. [DOI] [PubMed] [Google Scholar]
- Krützfeldt, J. , Rajewsky, N. , Braich, R. , Rajeev, K.G. , Tuschl, T. , Manoharan, M. & Stoffel, M. 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438, 685–689. [DOI] [PubMed] [Google Scholar]
- Kulyte, A. , Belarbi, Y. , Lorente‐Cebrian, S. , Bambace, C. , Arner, E. , Daub, C.O. , Heden, P. , Ryden, M. , Mejhert, N. & Arner, P. 2014. Additive effects of microRNAs and transcription factors on CCL2 production in human white adipose tissue. Diabetes 63, 1248–1258. [DOI] [PubMed] [Google Scholar]
- Kwekkeboom, R.F.J. , Sluijter, J.P.G. , van Middelaar, B.J. , Metz, C.H. , Brans, M.A. , Kamp, O. , Paulus, W.J. & Musters, R.J.P. 2016. Increased local delivery of antagomir therapeutics to the rodent myocardium using ultrasound and microbubbles. J Controlled Release 222, 18–31. [DOI] [PubMed] [Google Scholar]
- Lagos‐Quintana, M. , Rauhut, R. , Lendeckel, W. & Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294, 853–858. [DOI] [PubMed] [Google Scholar]
- Latreille, M. , Hausser, J. , Stutzer, I. , Zhang, Q. , Hastoy, B. , Gargani, S. , Kerr‐Conte, J. , Pattou, F. , Zavolan, M. , Esguerra, J.L. , Eliasson, L. , Rulicke, T. , Rorsman, P. & Stoffel, M. 2014. MicroRNA‐7a regulates pancreatic beta cell function. J Clin Invest 124, 2722–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latreille, M. , Herrmanns, K. , Renwick, N. , Tuschl, T. , Malecki, M.T. , McCarthy, M.I. , Owen, K.R. , Rulicke, T. & Stoffel, M. 2015. miR‐375 gene dosage in pancreatic beta‐cells: implications for regulation of beta‐cell mass and biomarker development. J Mol Med (Berl) 93, 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R.C. , Feinbaum, R.L. & Ambros, V. 1993. The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14. Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- Lindow, M. , Vornlocher, H.‐P. , Riley, D. , Kornbrust, D.J. , Burchard, J. , Whiteley, L.O. , Kamens, J. , Thompson, J.D. , Nochur, S. , Younis, H. , Bartz, S. , Parry, J. , Ferrari, N. , Henry, S.P. & Levin, A.A. 2012. Assessing unintended hybridization‐induced biological effects of oligonucleotides. Nat Biotechnol 30, 920–923. [DOI] [PubMed] [Google Scholar]
- Locke, J.M. , Da, S.X. , Dawe, H.R. , Rutter, G.A. & Harries, L.W. 2014. Increased expression of miR‐187 in human islets from individuals with type 2 diabetes is associated with reduced glucose‐stimulated insulin secretion. Diabetologia 57, 122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn, F.C. , Skewes‐Cox, P. , Kosaka, Y. , McManus, M.T. , Harfe, B.D. & German, M.S. 2007. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes 56, 2938–2945. [DOI] [PubMed] [Google Scholar]
- Marquart, T.J. , Allen, R.M. , Ory, D.S. & Baldan, A. 2010. miR‐33 links SREBP‐2 induction to repression of sterol transporters. Proc Natl Acad Sci USA 107, 12228–12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Sanchez, A. , Nguyen‐Tu, M.S. & Rutter, G.A. 2015. DICER inactivation identifies pancreatic beta‐cell “disallowed” genes targeted by microRNAs. Mol Endocrinol 29, 1067–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazière, P. & Enright, A.J. 2007. Prediction of microRNA targets. Drug Discovery Today 12, 452–458. [DOI] [PubMed] [Google Scholar]
- McAlexander, M.A. , Phillips, M.J. & Witwer, K.W. 2013. Comparison of methods for miRNA extraction from plasma and quantitative recovery of RNA from cerebrospinal fluid. Front Genet 4, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, J.J. , Esser, K.A. , Peterson, C.A. & Dupont‐Versteegden, E.E. 2009. Evidence of MyomiR network regulation of beta‐myosin heavy chain gene expression during skeletal muscle atrophy. Physiol Genomics 39, 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkman‐Zehavi, T. , Oren, R. , Kredo‐Russo, S. , Shapira, T. , Mandelbaum, A.D. , Rivkin, N. , Nir, T. , Lennox, K.A. , Behlke, M.A. , Dor, Y. & Hornstein, E. 2011. miRNAs control insulin content in pancreatic beta‐cells via downregulation of transcriptional repressors. EMBO J 30, 835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh, P. , van Vlierberghe, P. , de Weer, A. , Muth, D. , Westermann, F. , Speleman, F. & Vandesompele, J. 2009. A novel and universal method for microRNA RT‐qPCR data normalization. Genome Biol 10, R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh, P. , Hartmann, N. , Baeriswyl, L. , Andreasen, D. , Bernard, N. , Chen, C. , Cheo, D. , D'Andrade, P. , Demayo, M. , Dennis, L. et al 2014. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods 11, 809–815. [DOI] [PubMed] [Google Scholar]
- Mitchell, P.S. , Parkin, R.K. , Kroh, E.M. , Fritz, B.R. , Wyman, S.K. , Pogosova‐Agadjanyan, E.L. , Peterson, A. , Noteboom, J. , O'Briant, K.C. , Allen, A. et al 2008. Circulating microRNAs as stable blood‐based markers for cancer detection. Proc Natl Acad Sci USA 105, 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan, L. , Batte, K.E. , Trgovcich, J. , Wisler, J. , Marsh, C.B. & Piper, M. 2014. Methodological challenges in utilizing miRNAs as circulating biomarkers. J Cell Mol Med 18, 371–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, M. , Nakagami, H. , Rodriguez‐Araujo, G. , Nimura, K. & Kaneda, Y. 2012a. Essential role for miR‐196a in brown adipogenesis of white fat progenitor cells. PLoS Biol 10, e1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, M.A. , Raghavan, P. , Thomou, T. , Boucher, J. , Robida‐Stubbs, S. , Macotela, Y. , Russell, S.J. , Kirkland, J.L. , Blackwell, T.K. & Kahn, C.R. 2012b. Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab 16, 336–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, M.A. , Thomou, T. , Boucher, J. , Lee, K.Y. , Lallukka, S. , Kim, J.K. , Torriani, M. , Yki‐Jarvinen, H. , Grinspoon, S.K. , Cypess, A.M. & Kahn, C.R. 2014. Altered miRNA processing disrupts brown/white adipocyte determination and associates with lipodystrophy. J Clin Invest 124, 3339–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura, J. , Borsheim, E. & Carvalho, E. 2014. The role of microRNAs in diabetic complications‐special emphasis on wound healing. Genes (Basel) 5, 926–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso, G. , Cassader, M. , Rosina, F. & Gambino, R. 2012. Impact of current treatments on liver disease, glucose metabolism and cardiovascular risk in non‐alcoholic fatty liver disease (NAFLD): a systematic review and meta‐analysis of randomised trials. Diabetologia 55, 885–904. [DOI] [PubMed] [Google Scholar]
- Najafi‐Shoushtari, S.H. , Kristo, F. , Li, Y. , Shioda, T. , Cohen, D.E. , Gerszten, R.E. & Naar, A.M. 2010. MicroRNA‐33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 328, 1566–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obad, S. , Santos, C.O. , Petri, A. , Heidenblad, M. , Broom, O. , Ruse, C. , Fu, C. , Lindow, M. , Stenvang, J. , Straarup, E.M. et al 2011. Silencing of microRNA families by seed‐targeting tiny LNAs. Nat Genet 43, 371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofori, J.K. , Salunkhe, V.A. , Bagge, A. , Wendt, A. , Mulder, H. , Eliasson, L. & Esguerra, J.L.S. 2014. Regulation of Glucose‐Stimulated Insulin Secretion (GSIS) by miR‐130a/b and miR‐152 via pyruvate dehydrogenase E1 component, alpha subunit PDHA1. Diabetologia 57, S170–S170. [Google Scholar]
- O'Rourke, J.R. , Georges, S.A. , Seay, H.R. , Tapscott, S.J. , McManus, M.T. , Goldhamer, D.J. , Swanson, M.S. & Harfe, B.D. 2007. Essential role for Dicer during skeletal muscle development. Dev Biol 311, 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega, F.J. , Mercader, J.M. , Catalan, V. , Moreno‐Navarrete, J.M. , Pueyo, N. , Sabater, M. , Gomez‐Ambrosi, J. , Anglada, R. , Fernandez‐Formoso, J.A. , Ricart, W. , Fruhbeck, G. & Fernandez‐Real, J.M. 2013. Targeting the circulating microRNA signature of obesity. Clin Chem 59, 781–792. [DOI] [PubMed] [Google Scholar]
- Ottosen, S. , Parsley, T.B. , Yang, L. , Zeh, K. , van Doorn, L.‐J. , van der Veer, E. , Raney, A.K. , Hodges, M.R. & Patick, A.K. 2015. In vitro antiviral activity and preclinical and clinical resistance profile of miravirsen, a novel anti‐hepatitis c virus therapeutic targeting the human factor miR‐122. Antimicrob Agents Chemother 59, 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, S.M. , Thompson, J.A. , Ufkin, M.L. , Sathyanarayana, P. , Liaw, L. & Congdon, C.B. 2014. Common features of microRNA target prediction tools. Front Genet 5, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polge, C. , Heng, A.E. , Combaret, L. , Bechet, D. , Taillandier, D. & Attaix, D. 2013. Recent progress in elucidating signalling proteolytic pathways in muscle wasting: potential clinical implications. Nutr Metab Cardiovasc Dis 23(Suppl. 1), S1–S5. [DOI] [PubMed] [Google Scholar]
- Poy, M.N. , Eliasson, L. , Krutzfeldt, J. , Kuwajima, S. , Ma, X. , MacDonald, P.E. , Pfeffer, S. , Tuschl, T. , Rajewsky, N. , Rorsman, P. & Stoffel, M. 2004. A pancreatic islet‐specific microRNA regulates insulin secretion. Nature 432, 226–230. [DOI] [PubMed] [Google Scholar]
- Poy, M.N. , Hausser, J. , Trajkovski, M. , Braun, M. , Collins, S. , Rorsman, P. , Zavolan, M. & Stoffel, M. 2009. miR‐375 maintains normal pancreatic alpha‐ and beta‐cell mass. Proc Natl Acad Sci USA 106, 5813–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullen, T.J. , Da, S.X. , Kelsey, G. & Rutter, G.A. 2011. miR‐29a and miR‐29b contribute to pancreatic beta‐cell‐specific silencing of monocarboxylate transporter 1 (Mct1). Mol Cell Biol 31, 3182–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner, K.J. , Suarez, Y. , Davalos, A. , Parathath, S. , Fitzgerald, M.L. , Tamehiro, N. , Fisher, E.A. , Moore, K.J. & Fernandez‐Hernando, C. 2010. MiR‐33 contributes to the regulation of cholesterol homeostasis. Science 328, 1570–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RegulusTherapeutics, I. 2014. Regulus announces notice of allowance from U.S. patent office related to microRNA‐103/107 Program in Metabolic Disorders. Press release, 1.
- RegulusTherapeutics, I. 2015. RG ‐ 125 (AZD4076), a microRNA therapeutic targeting microRNA ‐ 103/107 for the treatment of NASH in patients with type 2 diabetes/Pre – Diabetes, selected as clinical candidate by AstraZeneca. Press release.
- Roberts, T.C. 2015. The microRNA machinery. Adv Exp Med Biol 887, 15–30. [DOI] [PubMed] [Google Scholar]
- Rosen, E.D. & Spiegelman, B.M. 2006. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 444, 847–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottiers, V. & Naar, A.M. 2012. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol 13, 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter, G.A. 2014. Dorothy Hodgkin Lecture 2014. Understanding genes identified by genome‐wide association studies for type 2 diabetes. Diabet Med 31, 1480–1487. [DOI] [PubMed] [Google Scholar]
- Saito, T. & Sætrom, P. 2010. MicroRNAs – targeting and target prediction. New Biotechnol 27, 243–249. [DOI] [PubMed] [Google Scholar]
- Salunkhe, V.A. , Esguerra, J.L. , Ofori, J.K. , Mollet, I.G. , Braun, M. , Stoffel, M. , Wendt, A. & Eliasson, L. 2015a. Modulation of microRNA‐375 expression alters voltage‐gated Na(+) channel properties and exocytosis in insulin‐secreting cells. Acta Physiol (Oxf) 213, 882–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salunkhe, V.A. , Ofori, J. , Gandasi, N.R. , Salo, S.A. , Hansson, S. , Andersson, M.E. , Wendt, A. , Barg, S. , Esguerra, J.L.S. & Eliasson, L. 2015b. MiR‐335 regulates exocytotic proteins and affects glucose‐stimulated insulin secretion through decreased Ca2+‐dependent exocytosis in beta cells. Diabetologia 58, S128–S128. [Google Scholar]
- Sempere, L.F. , Freemantle, S. , Pitha‐Rowe, I. , Moss, E. , Dmitrovsky, E. & Ambros, V. 2004. Expression profiling of mammalian microRNAs uncovers a subset of brain‐expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5, R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, D. , Li, H. , Shu, Y. , Xiong, G. , Carson, W.E. , Haque, F. , Xu, R. & Guo, P. 2015. Systemic delivery of anti‐miRNA for suppression of triple negative breast. ACS Nano 9, 9731–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjogren, R.J. , Egan, B. , Katayama, M. , Zierath, J.R. & Krook, A. 2015. Temporal analysis of reciprocal miRNA‐mRNA expression patterns predicts regulatory networks during differentiation in human skeletal muscle cells. Physiol Genomics 47, 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider, P. , Olaopa, M. , Firulli, A.B. & Conway, S.J. 2007. Cardiovascular development and the colonizing cardiac neural crest lineage. ScientificWorldJournal 7, 1090–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen, A. , Wissing, M. , Salö, S. , Englund, A. & Dalgaard, L. 2014. MicroRNAs Related to Polycystic Ovary Syndrome (PCOS). Genes 5, 684–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano‐Arroquia, A. , McCormick, R. , Molloy, A.P. , McArdle, A. & Goljanek‐Whysall, K. 2016. Age‐related changes in miR‐143‐3p:Igfbp5 interactions affect muscle regeneration. Aging Cell 15, 361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvang, J. & Kauppinen, S. 2008. MicroRNAs as targets for antisense‐based therapeutics. Expert Opin Biol Ther 8, 59–81. [DOI] [PubMed] [Google Scholar]
- Stenvang, J. , Petri, A. , Lindow, M. , Obad, S. & Kauppinen, S. 2012. Inhibition of microRNA function by antimiR oligonucleotides. Silence 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. , Xie, H. , Mori, M.A. , Alexander, R. , Yuan, B. , Hattangadi, S.M. , Liu, Q. , Kahn, C.R. & Lodish, H.F. 2011. Mir193b‐365 is essential for brown fat differentiation. Nat Cell Biol 13, 958–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, R. , Lee, K. , Jing, E. , Biddinger, S.B. , McDonald, J.G. , Montine, T.J. , Craft, S. & Kahn, C.R. 2010. Diabetes and insulin in regulation of brain cholesterol metabolism. Cell Metab 12, 567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft, R.J. , Pheasant, M. & Mattick, J.S. 2007. The relationship between non‐protein‐coding DNA and eukaryotic complexity. BioEssays 29, 288–299. [DOI] [PubMed] [Google Scholar]
- Tattikota, S.G. , Rathjen, T. , McAnulty, S.J. , Wessels, H.H. , Akerman, I. , van de Bunt, M. , Hausser, J. , Esguerra, J.L. , Musahl, A. , Pandey, A.K. et al 2014. Argonaute2 mediates compensatory expansion of the pancreatic beta cell. Cell Metab 19, 122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattikota, S.G. , Rathjen, T. , Hausser, J. , Khedkar, A. , Kabra, U.D. , Pandey, V. , Sury, M. , Wessels, H.H. , Mollet, I.G. , Eliasson, L. et al 2015. miR‐184 regulates pancreatic beta‐cell function according to glucose metabolism. J Biol Chem 290, 20284–20294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin, J.A. & Bowles, N.E. 2002. The failing heart. Nature 415, 227–233. [DOI] [PubMed] [Google Scholar]
- Trajkovski, M. , Hausser, J. , Soutschek, J. , Bhat, B. , Akin, A. , Zavolan, M. , Heim, M.H. & Stoffel, M. 2011. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 474, 649–653. [DOI] [PubMed] [Google Scholar]
- Trajkovski, M. , Ahmed, K. , Esau, C.C. & Stoffel, M. 2012. MyomiR‐133 regulates brown fat differentiation through Prdm16. Nat Cell Biol 14, 1330–1335. [DOI] [PubMed] [Google Scholar]
- Trevorlstokes 2012. Group retracts microRNA paper after realizing reagent was skewing results. Retraction Watch, 1.
- Tugay, K. , Guay, C. , Marques, A.C. , Allagnat, F. , Locke, J.M. , Harries, L.W. , Rutter, G.A. & Regazzi, R. 2015. Role of microRNAs in the age‐associated decline of pancreatic beta cell function in rat islets. Diabetologia 59, 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Ree M.H., van der Meer, A.J. , de Bruijne, J. , Maan, R. , van Vliet, A. , Welzel, T.M. , Zeuzem, S. , Lawitz, E.J. , Rodriguez‐Torres, M. , Kupcova, V. , Wiercinska‐Drapalo, A. , Hodges, M.R. , Janssen, H.L.A. & Reesink, H.W. 2014. Long‐term safety and efficacy of microRNA‐targeted therapy in chronic hepatitis C patients. Antiviral Res 111, 53–59. [DOI] [PubMed] [Google Scholar]
- Van Peer, G. , Lefever, S. , Anckaert, J. , Beckers, A. , Rihani, A. , Van Goethem, A. , Volders, P.J. , Zeka, F. , Ongenaert, M. , Mestdagh, P. & Vandesompele, J. 2014. miRBase tracker: keeping track of microRNA annotation changes. Database (Oxford) pii: bau080. doi: 10.1093/database/bau080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rooij, E. , Quiat, D. , Johnson, B.A. , Sutherland, L.B. , Qi, X. , Richardson, J.A. , Kelm, R.J. Jr & Olson, E.N. 2009. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell 17, 662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele, J. 2013. How to find stably expressed microRNAs | Biogazelle.
- Vlachos, I.S. & Hatzigeorgiou, A.G. 2013. Online resources for miRNA analysis. Clin Biochem 46, 879–890. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Jiang, Y. , Peng, H. , Chen, Y. , Zhu, P. & Huang, Y. 2015a. Recent progress in microRNA delivery for cancer therapy by non‐viral synthetic vectors. Adv Drug Deliv Rev 81, 142–160. [DOI] [PubMed] [Google Scholar]
- Wang, P. , Fiaschi‐Taesch, N.M. , Vasavada, R.C. , Scott, D.K. , Garcia‐Ocana, A. & Stewart, A.F. 2015b. Diabetes mellitus–advances and challenges in human beta‐cell proliferation. Nat Rev Endocrinol 11, 201–212. [DOI] [PubMed] [Google Scholar]
- Witkos, T.M. , Koscianska, E. & Krzyzosiak, W.J. 2011. Practical aspects of microRNA target prediction. Curr Mol Med 11, 93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer, K.W. 2013. Data submission and quality in microarray‐based microRNA profiling. Clin Chem 59, 392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, H. , Lim, B. & Lodish, H.F. 2009. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes 58, 1050–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. , Chen, H. , Si, H. , Li, X. , Ding, X. , Sheng, Q. , Chen, P. & Zhang, H. 2014. Serum miR‐23a, a potential biomarker for diagnosis of pre‐diabetes and type 2 diabetes. Acta Diabetol 51, 823–831. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Wang, Z. & Gemeinhart, R.A. 2013. Progress in microRNA delivery. J Controlled Release 172, 962–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Guan, M. , Townsend, K.L. , Huang, T.L. , An, D. , Yan, X. , Xue, R. , Schulz, T.J. , Winnay, J. , Mori, M. et al 2015. MicroRNA‐455 regulates brown adipogenesis via a novel HIF1an‐AMPK‐PGC1alpha signaling network. EMBO Rep 16, 1378–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Wang, E. , Liu, H. , Rotunno, M. , Koshiol, J. , Marincola, F.M. , Landi, M.T. & McShane, L.M. 2010. Evaluation of normalization methods for two‐channel microRNA microarrays. J Transl Med 8, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B. , Li, C. , Qi, W. , Zhang, Y. , Zhang, F. , Wu, J.X. , Hu, Y.N. , Wu, D.M. , Liu, Y. , Yan, T.T. , Jing, Q. , Liu, M.F. & Zhai, Q.W. 2012. Downregulation of miR‐181a upregulates sirtuin‐1 (SIRT1) and improves hepatic insulin sensitivity. Diabetologia 55, 2032–2043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Speakers at the microRNAs in metabolism symposium, who all contributed to the manuscript.


