Abstract
The high morbidity and mortality rate of bloodstream infections involving antibiotic-resistant bacteria necessitate a rapid identification of the infectious organism and its resistance profile. Traditional methods based on culturing the blood typically require at least 24 h, and genetic amplification by PCR in the presence of blood components has been problematic. The rapid separation of bacteria from blood would facilitate their genetic identification by PCR or other methods so that the proper antibiotic regimen can quickly be selected for the septic patient. Microfluidic systems that separate bacteria from whole blood have been developed, but these are designed to process only microliter quantities of whole blood or only highly diluted blood. However, symptoms of clinical blood infections can be manifest with bacterial burdens perhaps as low as 10 CFU/mL, and thus milliliter quantities of blood must be processed to collect enough bacteria for reliable genetic analysis. This review considers the advantages and shortcomings of various methods to separate bacteria from blood, with emphasis on techniques that can be done in less than 10 min on milliliter-quantities of whole blood. These techniques include filtration, screening, centrifugation, sedimentation, hydrodynamic focusing, chemical capture on surfaces or beads, field-flow fractionation, and dielectrophoresis. Techniques with the most promise include screening, sedimentation, and magnetic bead capture, as they allow large quantities of blood to be processed quickly. Some microfluidic techniques can be scaled up.
Keywords: bacterial bloodstream infection, rapid identification, filtration, centrifugation, sedimentation, hydrodynamic focusing, chemical binding
Introduction
The rising prevalence of antibiotic resistance in bacterial pathogens is a significant threat to global health.1 Life-threatening bloodstream infections caused by antimicrobial-resistant bacteria are a tremendous burden on healthcare and lead to much higher patient treatment costs, increased length of hospitalization, and most importantly, higher rates of morbidity and mortality.2–4 Of particular concern are gram-negative pathogens, given their ease of resistance-gene acquisition. Furthermore, few new antibiotics are in development to treat these pathogens, leaving clinicians with an ever-diminished arsenal against these increasingly resistant opponents. 4–6 The most concerning relevant gram-negative pathogens include Klebsiella pneumoniae, Escherichia coli, and members of the genus Enterobacter. The carbapenem class of antibiotics has been used as a last-resort against members of the Enterobacteriaceae, inevitably leading to the generation and spread of resistant strains, called carbapenem-resistant Enterobacteriaceae (CRE).7, 8 Infections with these agents have produced mortality rates as high as 50%,7, 9 and they have been associated with significant outbreaks.10 Of the gram-positive pathogens, Staphylococcus epidermidis and Staphylococcus aureus are often associated with bacteremia and bloodstream sepsis, but usually have lower mortality rates.11–13
The gold standard of profiling antibiotic resistance has long been based on phenotypic assays; however, these tests require a relatively long turnaround time (at least 12–24 h) between sample acquisition and assay reporting.14 Even recently approved molecular techniques for bloodstream infections, designed to diagnose the organism and its antibiotic resistance profile, require a positive blood culture before the assay can be performed.15, 16 As would be expected, many studies have suggested an association between appropriate, timely administration of effective antibiotic regimens and decreased human mortality from bloodstream infections. 13, 17–20 However, without detailed information on both the infecting bacterium and its resistance profile, clinicians cannot reliably determine the most effective antimicrobial treatment. Instituting empiric therapy based upon initial clinical impressions delays effective treatment, and may exacerbate the emergence of resistant organisms if the administered antibiotic is not well-matched to the yet-unknown pathogen. Rapid acquisition of accurate information with an assay that requires no pretest blood culture, and simultaneously identifies both the bacterial species and its resistance profile, would enable the modulation of empirical regimens with targeted therapy shortly after clinical suspicion of infection, leading to improved clinical outcomes.
The need to rapidly identify and initiate treatment of bloodstream infection becomes critical if the infecting organism is resistant to the initial antibiotic regimen. Studies have shown that empirical treatment decisions of bacteremia are incorrect as much as 25 to 33% of the time,21–23 and in these cases, the infection will likely progress to serious and perhaps fatal consequences because of inappropriate treatment. Survival rates drop by as much as 7.9% per hour as effective treatment is delayed, giving urgency to early identification of antimicrobial susceptibility profiles.24, 25 Nearly half of all hospital deaths involve sepsis, helping make septicemia the 11th leading cause of death in U.S. in 2011, killing over 35,000 individuals.26, 27 The exact threshold of bacterial colony forming units per milliliter (CFU/mL) that produces clinical symptoms is unclear and may depend on species, but certainly at levels as low as 10 CFU/mL, sepsis becomes a threat.22, 28–30 The current clinical identification process employed for suspected septicemia typically requires at least 24 h, as a sample must be grown in culture before additional evaluation can identify the infecting species and determine its antibiotic resistance profile.31 Such evaluation is performed by a variety of techniques, including phenotypic assays, fluorescence in situ hybridization, mass spectrometry, and polymerase chain reaction (PCR). These FDA-approved methods determine either the identity of the pathogen alone, or both its identity and drug-susceptibility.32 PCR methods carry the possibility of broadly multiplexed assays and high levels of sensitivity, allowing rapid pathogen detection and characterization.33 However, blood has proven to be a particularly difficult medium for PCR analysis, as blood contains endogenous inhibitors that decrease the efficiency of the amplification step through interference with the polymerase or interaction with the DNA itself.34 Furthermore, PCR cannot discriminate between live and dead cells and thus cannot quantify a virulent load. The two currently approved PCR assays for analyzing gram-negative bloodstream infections require an initial culture step adding 18 to 24 h to the time to diagnosis, and also report lower limits of detection for one assay just below 106 CFU/mL.15, 16 Other techniques have been developed that have much lower limits of detection, such as fluorescent in situ hybridization and mass spectroscopy,35–37 but they require a positive blood culture which usually entails 24 h of growth of a blood sample. The design of a more effective, rapid nucleic acid-based test must include increased sensitivity over current approaches. For example, laser-based diagnostics are acquiring the sensitivity to detect single polynucleotide strands for molecular-based detection of pathogens.38
The anticipated arrival of single molecule detection of polynucleotides will enable rapid genomic identification of the bacterial species and its antibiotic resistance genes. Thus, the infectious agent and its resistance profile could be ascertained within perhaps an hour, since there would be no need for genome amplification by culture or by PCR. However, there remains a need to separate the bacteria from the blood since blood cells and proteins may interfere with the sensitivity of some molecular detection techniques, through light scattering (cells) and competitive binding or blocking (proteins).
The objective of this review is to analyze the possible methods by which bacteria have been or possibly could be separated from blood. Because our interest is in rapidly identifying blood pathogens, the emphasis of this review will be on techniques of high throughput (rapid processing) and efficient recovery of bacteria. Since blood samples from infected patients are often obtained in 7 to 10 mL increments in vacutainer tubes, this provides a logical target volume: 7 mL of blood with a concentration of only 10 CFU/mL, or only 70 bacteria to separate from billions of blood cells. Bacterial separation should be done in 10 min or less to provide time to subsequently lyse the bacteria, release, and label genomic DNA, and then molecularly identify the species and resistance profile.
Perhaps the largest challenge in separating bacteria from blood is the extremely high concentration of red cells, which can outnumber the bacteria by nearly one billion to one. Platelets are another complication as they are very easily activated to aggregate to each other or to adhere to any surface. Even when the blood is anticoagulated with calcium chelators such as citrate or ethylenediaminetetraacetic acid (EDTA), the platelets still tend to stick to some surfaces. In addition, anticoagulants such as heparin and EDTA interfere with PCR by interacting with the polymerase enzyme or with the DNA itself.39 Table 1 presents some basic facts about blood components that are relevant to this review.
Table 1.
Some Properties of Human Blood Components
| Component | Concentration (#/mL) | Size (µm) | Density (g/mL) | Comments | Ref |
|---|---|---|---|---|---|
| Red blood cell | 4.0–5.7 × 109 | 6.2–8.2 | 1.086–1.122 | Usually enucleate; generally no DNA; flexible biconcave discoid shape. |
40 |
| White blood cell | 4–11 × 106 | 7–30 | 1.057-1.092 | DNA in every cell; spherical with many different sizes; monocytes can be as large as 30 µm. |
41,42 |
| Platelets | 1.3–4.0 × 108 | 2.0–4.0 | 1.072–1.077 | No DNA; easily activated to aggregate and adhere to surfaces; biconcave discoid when not activated. |
41 |
| Plasma | – | – | 1.024 | Contains high concentrations of proteins and other biomolecules. |
43 |
While this article focuses on data pertaining to the separation of bacteria from blood, we will also present a few other processes employed to separate blood components from each other, as they might also be extended as possible methods to separate bacteria. This review is organized in general by the speed of blood processing, from techniques that can quickly process large volumes of blood to those with very slow process times. We will begin by reviewing mechanical separation by filtration, screening, and sedimentation, all of which usually employ larger volumes of blood. These will be followed by reviews of microfluidic techniques involving hydrodynamic focusing, chemical capture, field-flow fractionation, and dielectrophoresis—analytical techniques that most often employ very small samples or very slow flow rates.
Mechanical Filtration and Screening
Qualitative filtration principles
Separation by mechanical filtration is primarily based upon the size difference between the suspended particles and the size of the passageways (pores) through the filter. However, other properties can have a significant influence, such as the aggregation of particles, adhesion to the filter and its pores, how particles pack with each other in a filter cake, their mechanical compliance (shape change under pressure), their interaction with the suspending liquid, and the viscosity of the suspending liquid.
As suspended particles are convected towards the filter, a first layer is deposited on the filter surface and subsequent particles are deposited upon them, forming what is called a filter cake.44 Rigid particles deposited randomly leave open spaces between them, through which fluid continues to pass. The open space, or void volume fraction (ε) is one of the defining characteristics of the filter cake and the pressure drop through the cake. The pressure drop per thickness of cake is proportional to (1 − ε)2/ε3, 44 so a small void fraction creates tremendous pressure drop. In filtration at constant flow rate, the cake thickens linearly with time or flow volume, causing the required pressure to increase linearly with time. However, if particles in the cake settle, pack, or deform over time, the pressure required to maintain the constant flow increases even faster as the void fraction quickly decreases.
Most filtration processes occur at a constant pressure differential. In this case, the filter cake thickness increases, and the flow rate decreases, with a decaying exponential function of time; i.e., Q = Q0 exp [−k((1 − ε)2/ε3)t], where Q0 is the initial flow rate, t is time, and k is related to fluid viscosity, particle size, and other factors. Again, a small void fraction or deformable particles quickly shuts down the flow through the filter.
One possible means to separate suspended bacteria from blood is to use a filter with pores that allow bacteria to pass, but prevent the passage of blood cells. Using a sequence of filters is called screening; smaller particles pass through the filter pores while larger particles are retained by the filter. Screening is only slightly more complicated than filtration, in that the particles to be passed (the bacteria) must be sufficiently smaller than the retained particles (blood cells and platelets) so that the particles to be passed (in this case the bacteria) can fit through the pores and channels of the filter cake. In the theoretical case of rigid spherical particles packing randomly into a filter cake, a simple geometric calculation shows that the ratio of small particle diameter to large particle diameter must be less than 0.155 (=2/√3 – 1) to guarantee that all small particles could possibly pass through the smallest pores of a filter cake of large spheres.
Two aspects of bacteria suspended in blood make separation by screening very challenging. First, the particle sizes are not greatly different, particularly because the length of a bacterium is about the same as the diameter of a platelet. Assuredly, the size ratio of bacteria to RBCs is greater than 0.155. But the real “show stopper” is that the red cells are so flexible that they pack together in a filter cake with nearly zero porosity, and thus provide no connected pores sufficiently large to allow the passage of a bacterium. Hence bacteria will be terminally trapped in the blood cell filter cake, and only those bacteria close to the pores on the filter surface at the commencement of filtration will be passed. The filter cake will trap all other bacteria and shut down further flow. With a blood hematocrit of ~45, this stoppage occurs very rapidly. Diluting the blood slows the build-up of the filter cake, but also dilutes the bacteria, and theoretically provides no gain in the recovery of bacteria through the filter, except for the case of motile bacteria in a very dilute and extremely slow flowing filtration process, neither of which is practical for the desired separation process at hand.
An alternative to dead-end filtration is a cross-flow screening process in which a fluid flow parallel to the surface, carries the large particles along, and keeps them from forming a filter cake, while there is a small velocity component perpendicular to the filter surface that carries the small particles (and the large particles) toward the surface.44 In theory, large rigid particles would rebound from the surface, while the smaller particles would eventually flow through the pores in the filter. This might be feasible if the particles were rigid and large dilution volumes and process times were allowed. Furthermore, the pliable nature of platelets and some small red blood cells allows them to squeeze into the 2 to 3 µm pores, a size that would be required to freely pass rod-shaped enteric bacteria. Thus even a cross-flow filtration would be very challenging owing to the flexible nature of RBCs and the small size of platelets. Cross-flow filtration is used on highly anticoagulated blood,45 but we have found no reports of separating bacteria from blood by cross-flow filtration.
Filtration applications to separate bacteria from blood
A search of the literature revealed no publications in which bacteria were directly separated from blood by filtration processes. However, there were several studies describing the successful separation of blood components from each other, including some good reviews.46, 47 Creative study of these reports might provide some guidance (or sound rejection) of a filtration process to isolate bacteria from blood.
For example, a series of chambers separated by filters of decreasing pore sizes was used by Liu et al. to separate white blood cells (WBCs) from red blood cells (RBCs) and platelets in diluted (1:20) blood.48 There are a few articles that describe using arrays of posts and channels with gradients in size or spacing to separate WBCs from blood.49–51 In these devices, the RBCs, platelets, and plasma flow through. While these may be useful in separating a minority member (many fewer WBCs than RBCs) that is larger in size, it may be difficult to design arrays that retain RBCs while passing bacteria, again because of the overwhelming number of RBCs and the issue of forming a cell pack. In theory and practice, it is possible to separate 0.9 µm and 1.0 µm hard spherical particles from each other in dilute flow,52 but this has not been done with bacteria and platelets that have a similar size ratio.
Centrifugation and Sedimentation Techniques
Centrifugation and sedimentation principles
We define centrifugation as the process of rotating a sample to generate a centrifugal force that operates on the particles and liquid. In contrast, sedimentation is the movement of particles through a liquid under the influence of an external field, such as a gravitational field, centrifugal field, electric field, etc. In the laboratory or clinic, centrifugation is a convenient method to produce sedimentation. Most importantly, because particles may sediment at different velocities, they may be separable from each other during a sedimentation process.
Sedimentation occurs in a gravity or centrifugal field because the gravity or centrifugal force that moves a particle is balanced by the buoyancy force and drag force on the particle. The buoyancy and gravity forces are constant, and their imbalance creates a net acceleration on the particle; the acceleration creates movement, and this movement creates a drag force that increases as the particle velocity increases. A centrifugal force is slightly different in that it increases with rotational velocity and distance from the axis of rotation; but at constant rotation over a short distance centrifugal force can be considered constant. In either case, within a fraction of a second the particle reaches a velocity at which its gravity (or centrifugal accelerating force) is exactly balanced by the drag and buoyancy forces. That velocity remains constant (or increases slowly during centrifugation) until the particle hits an immobile interface (wall or packed particles), it enters a fluid of the same or greater density, or the separation driving force is stopped. The very small particles in blood sediment in the Stokes flow regime, as they have Reynolds numbers less than 0.1. (, where Dp is the particle diameter, ρf is the fluid density, and µ is the fluid viscosity.) In theory, for rigid spheres in Newtonian fluids under Stokes flow, the sedimentation velocity νs is given by
| (1) |
where ρp is the particle density, R is the rotational radius, g is the rotational angular velocity, and g is the gravitational constant.53 In practice, the actual sedimentation velocity is often less because of interparticle interactions (collisions) and nonspherical shapes. For blood and bacteria, the velocities are slower than predicted by Eq. (1) because RBCs and most enteric bacteria are not spherical, blood is not a Newtonian fluid, and the particles interact with each other. Nevertheless, Eq. (1) gives a fairly good first-order estimate of sedimentation velocities, as the correction factors for spheroids are not large,53 and blood plasma is nearly Newtonian at the low shear rates generated during sedimentation.43
Centrifugation processes have been used for more than a century to separate blood into its various components (cells, platelets, and plasma). In such cases, the centrifugation time is sufficiently long that the particles sediment until they are separated into layers according to their density. We will refer to this as isopycnic (equal density) or equilibrium centrifugation in which all dynamic sedimentation movement has stopped either because there is no more density difference to drive particles to move, or because the particles have come to an impenetrable barrier. At the end of the process, all the particles will be separated sequentially according to their density differences.
Referring to Table 2, we see that isopycnic centrifugation of septic blood would not separate the bacteria from the red cells, as the range of bacterial density overlaps the range of RBC density. But Table 2 also shows that the sedimentation velocities of the various blood components are different— and in some cases very different—from bacteria. Therefore, the principles of sedimentation velocity may be used to separate bacteria. Specifically, the nominal νs of WBCs is about 96 times faster than E. coli, and that of RBCs is 30 times faster. In the time that it take RBCs to move to the end of a centrifuge tube, the bacteria have only moved about 1/30 of that distance, and only about 3.3% of randomly dispersed bacteria will have encountered the end of the tube (ignoring all other cells in the way). Quickly collecting the plasma (which will have been clarified of RBCs and WBCs) could theoretically recover about 97% of the bacteria. However, the actual amount will be less because a fraction of bacteria may become trapped in the cell pack. Another deficiency is that there will also be many platelets in this plasma because their sedimentation velocity is similar to bacteria, but at least the RBCs and WBCs will be separated from these smaller components.
Table 2.
Relative Sedimentation Properties of Blood Components and Bacteria
| Component | Density Range (g/cm3) | Density used for Calculation (g/cm3) |
Nominal Size used for Calculation (µm) |
Relative Velocity* |
|---|---|---|---|---|
| Red blood cell | 1.086–1.122 | 1.098 | 8 | 30 |
| White blood cell | 1.057-1.092 | 1.092 | 15 | 96 |
| Platelet | 1.072–1.077 | 1.077 | 3 | 3 |
| E. coli | 1.08–1.10 | 1.095 | 1.5 | 1 |
Relative to E. coli (modeled as 1.095 g/cm3, 1.5 µm sphere) using the nominal values in the table.
Examples of centrifugation of macroscopic quantities of blood
Blood collected clinically in blood tubes is often separated into components for subsequent analysis. We have found no reports of separating bacteria from blood by isopycnic centrifugation of blood in tubes.
However, we present an intriguing possibility that may be worthy of pursuit. The use of a separator tube might be possible, although we have found no such reports for bacterial separation. A separator tube contains a gel that has a density between that of plasma and cellular components. This gel has thixotropic viscosity, which means that when it experiences shear stress during centrifugation, the viscosity decreases (in a time-dependent manner) and then stiffens again when the shear stress abates. During centrifugation, the gel, which is originally at the bottom or sides of the centrifuge tube, slowly migrates upward through the cell pack while the cells sediment downward.54 It stops moving when it encounters clarified plasma of lower density than itself; then it spreads sideways, coalescing to form a horizontal plug between the cell pack and the plasma, which keeps the cells from mixing with plasma after centrifugation is stopped. The thixotropic nature of the gel is designed so that it migrates slowly, giving the cells time to sediment before the plug forms, because once the plug forms it becomes stiff enough that an individual bacterial cell cannot penetrate it, as the stress imposed by a single sedimenting bacterium is on the order of 10−6 Pa and the yield stress of the gel is usually greater than 20 Pa.55
To trap most of the bacteria in the upper plasma phase, one would have to design the viscosity, density, and volume of the thixotropic gel such that it moves upward and forms at a precise rate such that the last red cells are below the gel when it completes the plug formation. But because of the differential in sedimentation velocity between cells and bacteria, the plug formation could be completed before an appreciable fraction of bacteria has sedimented down with the cells and is trapped under the separating gel. Unfortunately, there would also be a large fraction of platelets remaining in the plasma phase. Once the gel has formed, the remaining platelets and sedimenting bacteria may collect on the gel surface, but will not be able to penetrate, since a single platelet or bacterium cannot generate enough shear stress to penetrate the shear-thinning gel. Cessation of centrifugation would need to be performed quickly and at a precise time—quickly, so the bacteria do not collect in a mass of platelets on the surface of the plug and precisely when the gel forms a plug to recover as many bacteria as possible. The limitations of this technique are that less than 100% of bacteria would be trapped in the upper phase of plasma, and further separation from the platelets would be required. As mentioned, such a system has not been studied experimentally or theoretically, but it may be possible to confine a fraction of the bacteria to the upper phase, which could be collected by decanting the plasma and scraping the upper surface of the separating gel.
Centrifugation of microscopic quantities of blood
Traditionally blood is collected by venipuncture into vacutainer “blood tubes” ranging in size from 2 to 10 mL. However, there is impetus to reduce the required volume of blood to microliter sizes that can be obtained by a finger or earlobe prick in a clinician’s office, and then separate the blood for rapid diagnosis on site.
There are several reports of centrifugal disk separation processes in which microliter quantities of blood are placed within a chamber built into a disk the size of a compact disk (CD) or digital video disc; then the disk is spun.56–58 In most cases, after equilibrium centrifugation has thoroughly separated plasma from the cellular components, the separated plasma flows further (downstream on the disk) toward a diagnostic process. To our knowledge, these centrifugal microfluidic processes have not yet been used to differentially separate bacteria from blood, but it may be very possible to do so.
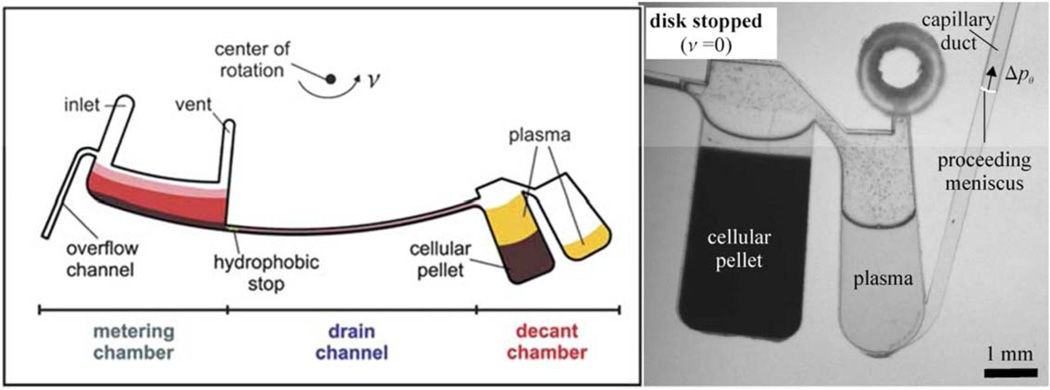
An example of such a device was developed by Haeberle et al. in which whole blood was deposited into a 5 µL metered chamber (containing an overflow drain so not too much blood was deposited).56 When the disk was spun, the centrifugal force pushed the blood past a hydrophobic stop and down a long microfluidic channel whose cross-section and length control the time to empty the metered chamber (see Figure 1). Sedimentation commences in the metering chamber, so the first components to travel down the drain channel are red cells, which then collect in a carefully sized chamber. Figure 4 of the Haeberle et al.’s article show that separation by sedimentation also occurs in the drain channel. 56 The cellular pack enters the first chamber and fills it, with some plasma on top; then eventually the plasma spills over into the plasma chamber, but cannot escape through the capillary duct until the rotation stops.
Figure 1.
Centrifugal separation device adapted from Ref. 56, with permission from Royal Society of Chemistry. Left: schematic representation of microfluidic channels on a rotating disk. Right: photograph of blood cell pellet and separated plasma after rotation has stopped, and plasma is being drained through a capillary duct.
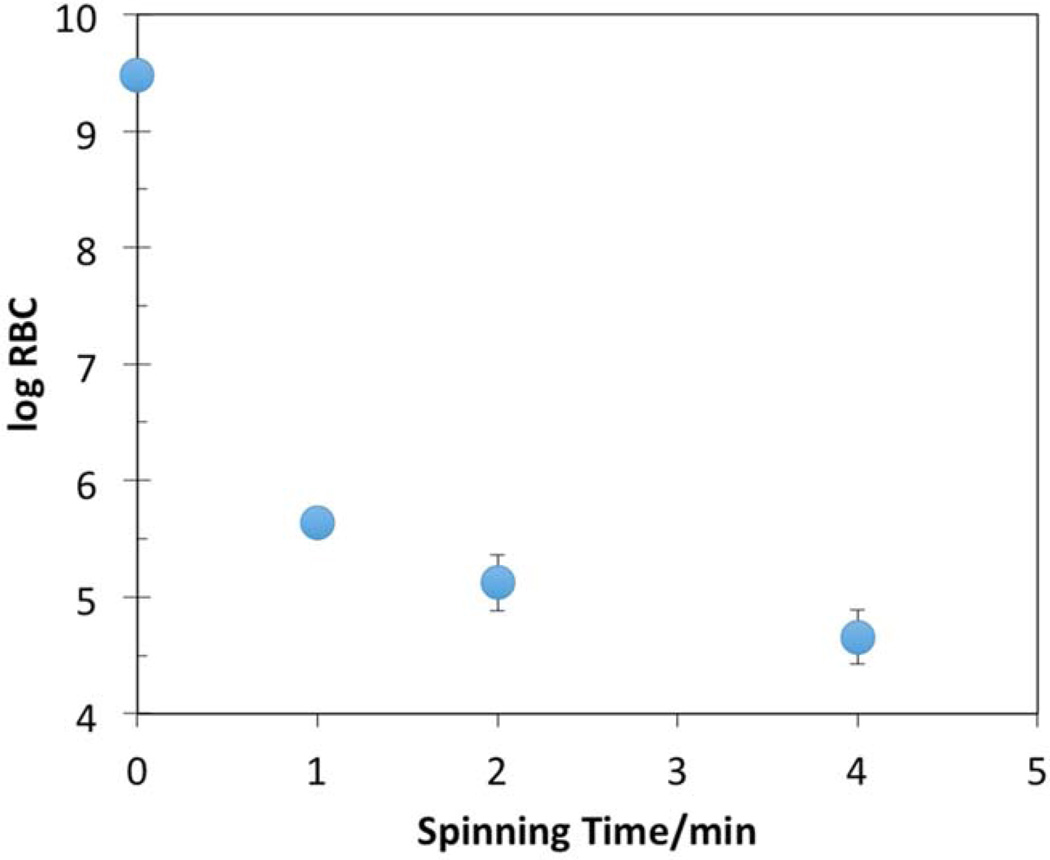
Figure 4.
Amount of RBCs (log10 of RBC/mL) from porcine blood remaining in the plasma recovered from the spinning hollow disk apparatus. Spinning was done at 3,000 rpm. Error bars indicate the standard deviation of the mean of repeated experiments (n = 3). Where no error bars are evident, the range of standard deviation is less than the symbol size.
Because bacteria sediment so much more slowly than RBCs and WBCs, it would be possible to control the timing of filling the pellet chamber (by designing the length and cross-section of the drain channel) such that not all of the bacteria will have sedimented into the cellular pellet when the upper layer plasma starts to overflow into the plasma chamber. Again, it would be difficult to separate platelets from bacteria with this system unless other steps were taken.
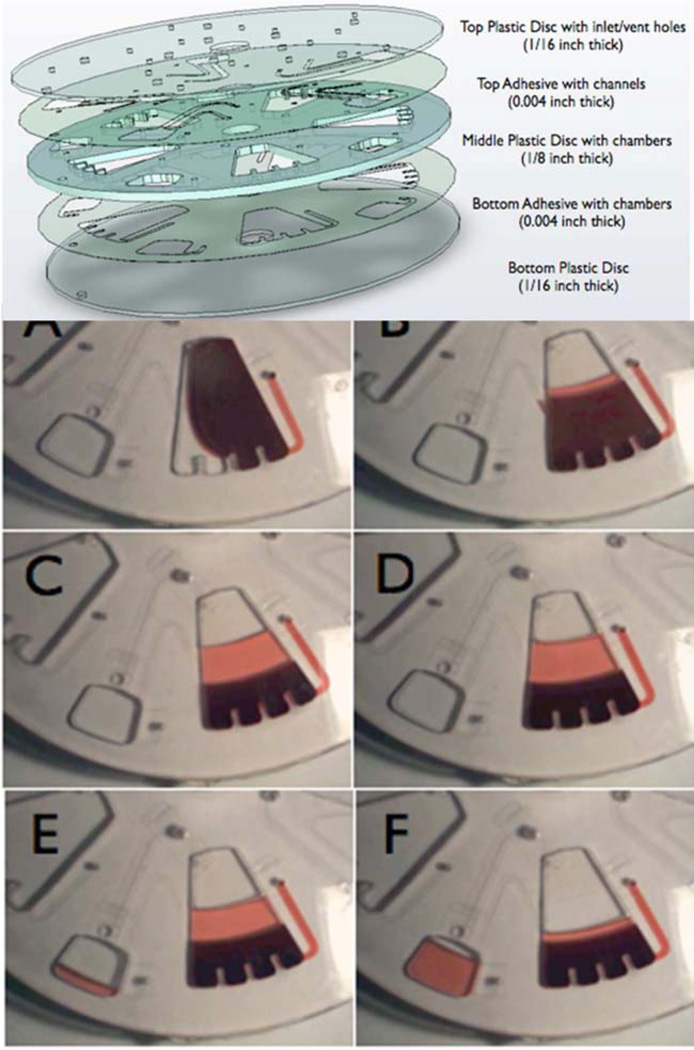
Amasia and Madou57 have designed a similar system on a spinning disk format that also separates larger quantities of blood (Figure 2). In this device, there is no drain channel, but there is a capillary duct drain channel that empties the plasma into a collection chamber (on the left in panel F of Figure 2).57 Again, by stopping the centrifugation just as the RBCs and WBCs have sedimented to the bottom, the majority of bacteria (and platelets) will still be in the plasma and could be recovered. One chamber of the device shown in Figure 2 is loaded with 2.0 mL whole blood, and so a disk with five chambers could process 10 mL of blood.
Figure 2.
Centrifugal separation device adapted from Ref. 57, with permission from Bioanalysis as agreed by Future Science Ltd. This compact disk device has a separation chamber and a capillary connection to a plasma collection chamber (bottom left of panels E and F) that drains the plasma when centrifugation is stopped.
Gorkin et al. have written a good review of other blood separation devices on a CD size scale.58
There are pros and cons to these small centrifugal devices for separating bacteria. Advantages include the lack of complexity required to control pressure, in that there are no pumps nor control valves. With the correct timing, the plasma can be collected before all the bacteria sediment out. One disadvantage is that platelets are not automatically separated. Another is that the volumes are very small for the chamber as presently designed on these compact disks. One chamber of the Haeberle et al. design processes 0.005 mL of blood, while the Amasia and Madou design processes 2.0 mL.57 Of course, several chambers could be placed on one CD, and the disks could be made thicker. While the Amasia and Madou design could process 7 mL of blood, it might be problematic to process this volume with the Haeberle et al. design. It would be necessary to process this much blood, since the bacterial load may be only 10 CFU/mL, and in a single collection chamber of the Haeberle et al. design, the probability of finding even one CFU would be on the order of 0.05 (only 1 out of 20 collection chambers would contain a single CFU).
Differential sedimentation of macroscopic quantities of blood
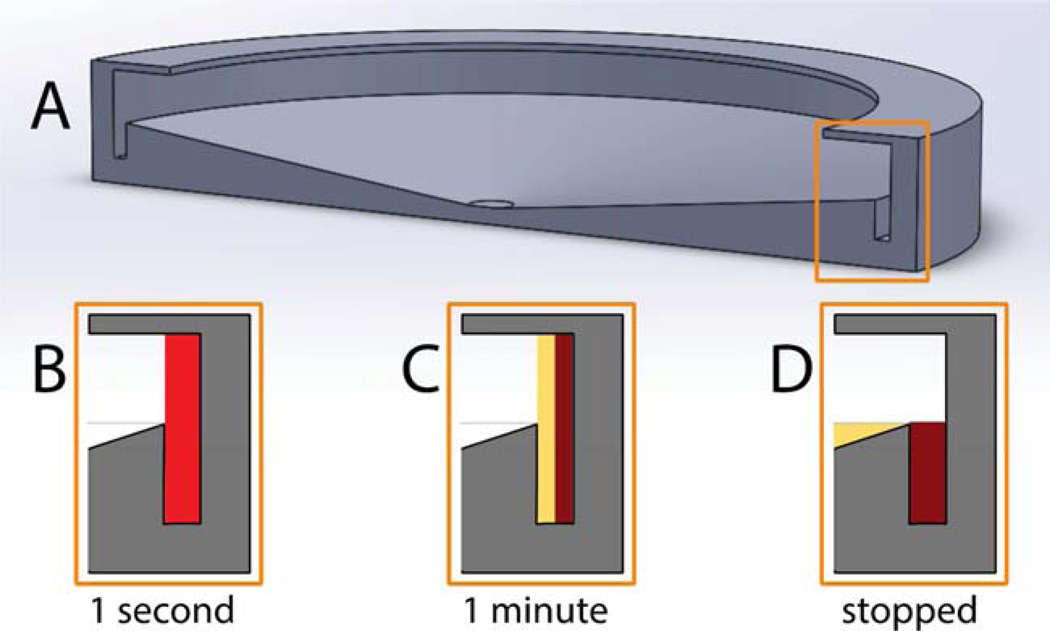
Our lab has explored the possibility of separating E. coli bacteria from blood using the spinning disk design shown in Figure 3. A 12-cm diameter hollow disk was built of photopolymerizable acrylates using rapid-prototyping technology. This disk clips onto a CD spinner in a CD player. The disk has a channel at the periphery that holds 3.5 mL, and a centrally open lid placed at a height such that 7.0 mL of blood occupies the annular volume from the channel up to the lid. The surface of the disk outside the channel is slanted toward the center of the disk. When blood is pipetted into the disk through the open lid, it drains toward the center. When spinning starts, the blood is flung to the inside surface of the wall of the disk. The thickness of the blood is 2 mm. Sedimentation of WBCs and RBCs creates a clear plasma layer within seconds that grows in thickness with spinning time. After a predetermined time, the disk is slowly decelerated (at ~33 rpm/s, to prevent mixing at the plasma-cell interface), and finally the red cells slide down the wall and are trapped in a well, while the plasma spills over the edge of the channel and flows to the center where it is collected with a pipette. In most experiments, the disk is spun at about 3,000 rpm, as measured by a tachometer.
Figure 3.
(A) Drawing (to scale) of rotating hollow disk for sedimentation-based separation of bacteria from blood. (B) Schematic showing that initially 7 mL of bacteria-spiked blood are placed in the disk. Upon rotation at 3,000 rpm, the blood is spun to the inside surface of the wall of the hollow disk and the blood components start to sediment toward the wall. (C) After about 1 min there is a layer of clear plasma which still contains the bacteria because they sediment much more slowly than RBCs and WBCs. (D) When the rotation stops, the packed cells slough down and are trapped in the well at the base of the channel, while the plasma containing bacteria drains off and is collected.
For preliminary experiments, we collected porcine blood at a local abattoir from clinically normal swine via exsanguination and immediately anticoagulated with heparin. Then 7.0 mL of this blood was spun at 3,000 rpm for various periods of time and the plasma collected. The concentration of RBCs in the blood was counted with a hemocytometer before spinning and in the collected plasma after spinning. Figure 4 shows the results of these experiments. The concentration of porcine RBCs decreased rapidly during the first minute of spinning, and then continued to decrease slowly after that. With only 1 min of spinning, the concentration of RBCs decreased by 3½ orders of magnitude, but there were always some RBCs remaining in the plasma. This was attributed to a similar distribution in the density of porcine RBCs to that found in human blood. Some RBCs that are less dense will sediment at a lower velocity, thus remaining in the plasma for longer times.
Blood was collected from human volunteers into anticoagulant-containing vacutainer tubes (sodium citrate, BD #369714; EDTA, BD #366643; sodium heparin, BD #367874; Becton Dickinson, Franklin Lakes, NJ) according to an IRB-approved protocol at Brigham Young University. The blood was processed on the same day as collection. Just before the sedimentation, the blood was spiked with E. coli in PBS (1 mL in 7 mL blood) and vortexed to produce a concentration of about 3 × 105 CFU/mL. Then 7 mL of this suspension was pipetted through the open top of the hollow disk and the disk was spun at 3,000 rpm for 1 min. The plasma was collected and the amount of bacteria recovered was measured by viable plate counting. Recovery was quantified by dividing the number of recovered bacteria by the original amount in the blood delivered to the disk.
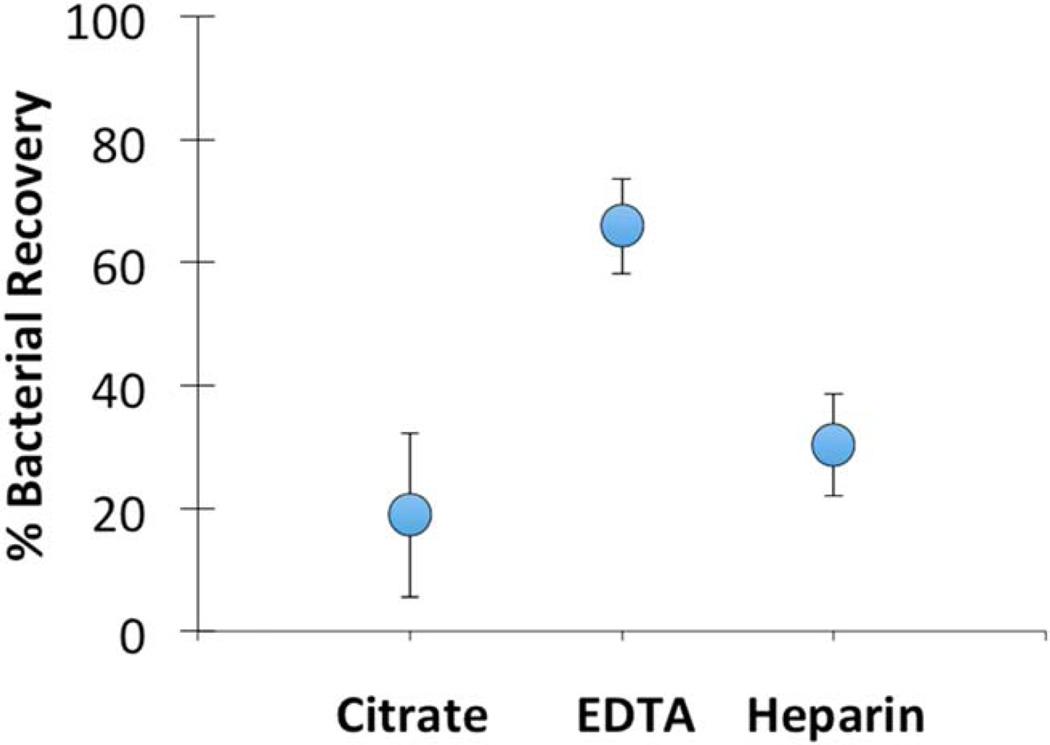
Figure 5 shows the bacterial recovery as a function of the type of anticoagulant used in the vacutainer tubes. While these preliminary results do not show significant differences between the types of anticoagulants used, it appears that the plasma recovered from EDTA-treated blood had on average more bacteria than the others. Using hemocytometry, we observed that plasma from blood collected in EDTA vacutainer tubes contained large numbers of platelets, while that from the other types of tubes had very few and sometimes no platelets. We are still exploring the relationship between platelet recovery and bacterial recovery.
Figure 5.
Preliminary results of recovery of E. coli spiked into fresh anticoagulant-treated human blood. Blood was collected from human volunteers into vacutainer tubes containing anticoagulants of either citrate, EDTA, or heparin. Error bars represent the standard error of the mean from repeated experiments, n ≥ 4.
While the average overall bacterial recovery from these experiments was only about 40%, this preliminary observation suggests that it may be possible to quickly separate bacteria from nondiluted blood using this sedimentation technique.
Microfluidic Techniques
Basic principles of microfluidic flow
Laminar Flow in Microfluidic devices
Given that in perhaps all microfluidic devices described below for processing blood, the flow is laminar and is in rectangular channels, this section briefly reviews flow rates and pressure drops in circular and rectangular channels for Newtonian fluids (constant viscosity fluids).
For flows in circular channels (tubes), the Hagen-Poiseuille equation gives the relationship between volumetric flow rate (Q), tube radius (R), viscosity (µ), and pressure drop per length (ΔP/L): .
However, circular channels are difficult to build in microfluidic devices. For rectangular channels, the pressure drop for a channel of height a and width b is given by , where ϕ is a correction factor that depends on the aspect ratio of the channel.59 For a square channel, ϕ =56.91, and for rectangular channels, ϕ can be estimated using the relationship ln ϕ = 4.0038−0:1957ln (a/b). Whether circular or rectangular, small channels create large pressure drops, even for short channels.
Other important aspects to consider in microfluidic systems are that pressure drops are additive when flow is in series, and pressure drops are equivalent when flow is in parallel, thus allowing scale-up through multiple parallel channels. Also spontaneous capillary flow can occur when the channel walls are wettable, and flow can initially be halted (at least at low pressures) by a hydrophobic (nonwettable) surface in a channel.
There are many interesting forces and phenomena acting on particles flowing in microchannels, both active and passive, which can be exploited to “focus” the particles in a particular portion of the channel to concentrate them or to separate them from each other based on size, density, and deformability. Details can be found in excellent books and review articles of the separation of particles from fluids;46, 47, 60–68 however, only a few of these mention the separation of bacteria in blood.60, 62 To understand these separation methods, we briefly mention herein the basic hydrodynamic principles that were exploited to separate bacteria from blood in the examples given in the next section. We will first mention fluid velocity distributions and then the forces acting on particles.
The local fluid velocity is determined by the transport of momentum in the channels and the resulting velocities and velocity gradients. While the velocity profiles of pure fluids are easily calculated, the presence of particles perturb the local velocities, which leads to a very complex fluid flow problem to model, particularly as the particle density increases (such as in blood). In straight channels, velocities are parallel with the channel walls, unless forces move particles away or toward a wall, which in turn generate fluid flows toward or away from the walls. Some secondary transverse flows occur in curved channels—or even a straight channel with a non-Newtonian fluid. Dean flow69 is one such secondary flow in curved rectangular channels that has been exploited to separate bacteria from blood cells,70 as will be discussed later.
Focusing and separating particles in microchannels is accomplished by manipulating several forces that change the inertial momentum of the particle and control its final trajectory. These include buoyancy, body, and lift forces. Buoyancy forces always act on a particle when its density is different from the surrounding fluid, but often these forces are negligibly small. Body forces that are applied directly to the particle include gravity force, centrifugal force, and magnetic force. The latter two can be fairly strong forces and can be used in bacterial separation processes.
There is a set of lift forces that act on particles that cause displacement in the lateral direction in microchannels. The first is called a “wall lift” force, and moves particles from the wall toward the center of the channel, and which decreases in magnitude with distance from the wall. It arises from the asymmetric wake of a particle near the wall that produces a lift force away from the wall.68, 71 In channels with fluid shear gradients there is a lift force acting on particles down the velocity gradient, pushing toward the wall.68, 72 The combination of these opposing forces is called “inertial lift” and results in annular focusing in a circular channel; however, in rectangular channels, the particles are focused at a small distance perpendicular to the center of each of the four walls. Still another force at play is a centric force arising from the flexibility of the particles, with more deformable particles moving further from the wall.62 Thus, larger particles or more flexible particles are focused further from the wall than are smaller or more rigid particles. In small channels carrying blood, these forces cause the red blood cells to move away from the wall to produce a margin or layer that is fairly free of RBCs.73, 74 This is called “margination,” as it leaves a margin near the wall with a lower RBC concentration. Many interesting rheological phenomena have been attributed to the migration of RBCs away from the wall, such as the Fahraeus effect (the hematocrit in very small capillaries is less than that of the supply reservoir) 75 and the Fahraeus-Lindqvist effect (the apparent viscosity of blood decreases as the capillary diameter decreases).76, 77 The other cellular components of blood do not migrate from the walls to the same extent as the RBCs, and this becomes a point of exploitation for separation of blood components in narrow channels. It appears that both the discoidal shape and the flexibility of the red cells are involved in their migration away from the channel wall.73, 74 Flexible spheres or rigid discoids do not migrate to the same extent.
A very significant hydrodynamic force, drag force, is always present when other forces impart a particle trajectory that is different from the local fluid flow. This drag force points opposite to the particle trajectory and balances with the other forces in creating a final trajectory, velocity, and eventual focused position in a channel. Still other forces can cause particle focusing, such as the collision of a particle with a solid wall in pinched flow fractionation, which leads to separation of particles based on their diameter.62
Bacterial separation in microfluidic devices
There are a few examples in the literature of bacterial separation from blood using the concepts of red cell migration and particle focusing. All the following reports involve microfluidic devices, and most are batch or semibatch processes because finite blood samples were used in “proof-of-principle” experiments.
Hou et al. took advantage of RBC margination and migration away from the walls in small rectangular channels to preferentially concentrate bacteria near the walls using a suspension of bacteria in washed blood cells, resuspended at a blood hematocrit of 45.78 After flowing down a 5 mm length for margination to occur, the square channel of 20 µm × 20 µm was slowly expanded to 200 µm × 20 µm and two side-stream bifurcations drew off some fluid from the RBC-depleted region at the periphery of the wide channel. The central section of blood was directed to a second 20 µm × 20 µm channel, marginated again, expanded (200 µm × 20 µm) and the RBC-depleted layer skimmed a second time. A demonstration of this technique, using washed human blood spiked with E. coli or S. cerevisiae, showed that the microbial separation efficiency was about 80% for the two-stage process. However, about 80% of the WBC and platelets were also recovered with the bacteria. Thus a second separation process would be needed to remove the WBCs if subsequent analysis required no contamination human DNA from WBCs. The report indicated that there were problems with large pressure drops and leakage at flow rates greater than 15 µL/min.
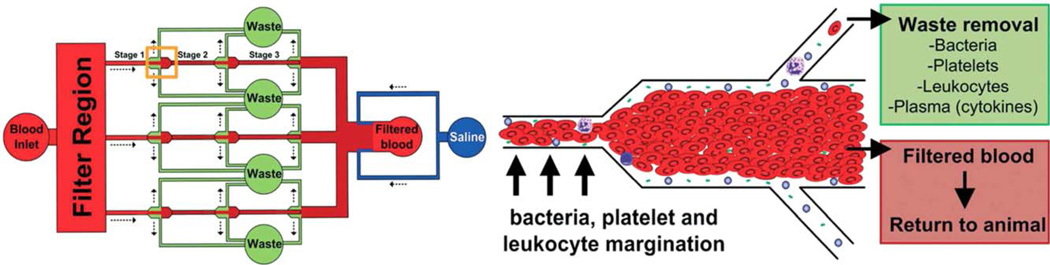
Later, the same group validated this technique in a mouse model.79 A device with three parallel channels (see Figure 6), 20 µm × 40 µm (WxH) with three skimming stages, was fed from the carotid artery of a mouse (via a peristaltic pump); the skimmed plasma (with bacteria) was collected and the concentrated red cells returned to the mouse via a catheter to the jugular vein. The mouse had received a cecal ligation and puncture wound 24 h before the procedure and had septicemia. Mouse blood was processed at 30 µL/min for 25 min. The bacterial count in the mouse blood was reduced by about 50% during this procedure. To demonstrate that this concept could be scaled further for human application, they built and tested (in vitro) a 16-channel platform that processed 1.5 mL/min and reduced bacterial concentration by >50% in 1 h.79
Figure 6.
Illustration of the three-stage separation device by which bacteria were separated from RBCs in whole mouse blood (above left). The bacteria and plasma are skimmed from the cell-free layer near the wall (above right), and the core of red cells is returned to the mouse. Taken from Ref. 79, with permission from Royal Society of Chemistry.
Mach and Di Carlo80 capitalized on inertial lift forces in a high aspect ratio rectangular channel (60 µm × 20 µm) to focus dilute RBCs in a suspension of bacteria. After 4 mm of flow length, the channel was expanded gradually to 160 µm × 20 µm and the RBCs remained focused near the wall. The RBCs were then drawn off through side channels leaving the bacterial suspension in the middle of the channel. Drawing off the RBCs removed about 88% of the RBCs (in the first stage), but also removed some bacteria. As in the first example, two stages of separation were employed, and this device was built into a massively parallel system with forty 2-stage channels that could process 8 mL/min of diluted blood. However, the processed blood was diluted 1:200 with sodium chloride solution containing E. coli, so this represents only about 40 µL/min of whole blood in 40 channels, or about 1 µL/min of whole blood per channel. The bacterial concentration was rather high, at 108 CFU/mL, but 80% of the bacteria were recovered. While this is a substantial recovery, the processed volume is tremendously large (7 mL blood would become 1.4 L) and it would require 25 of these 40-channel devices to process 7 mL of blood in 7 min.
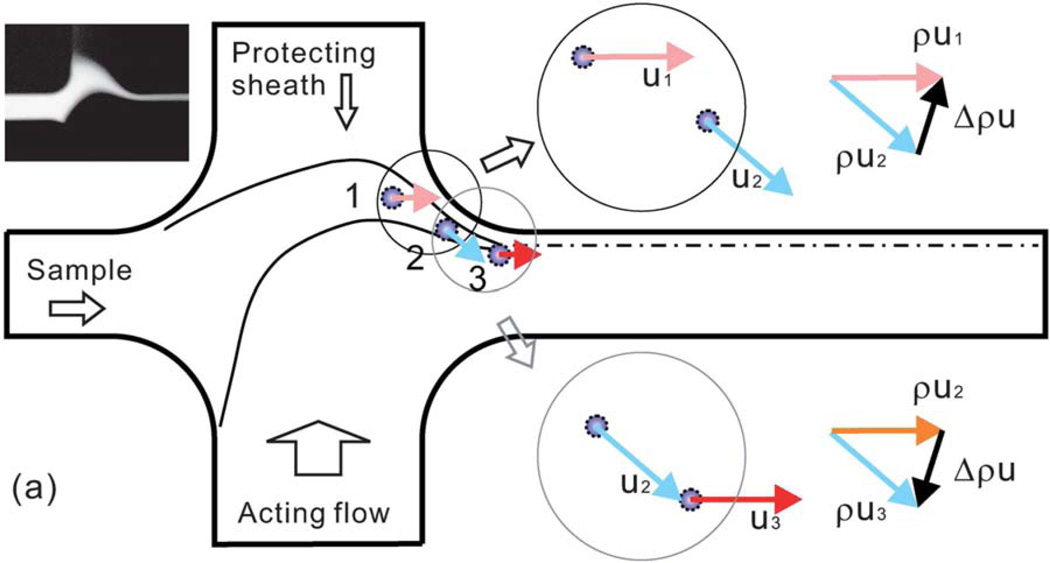
In a very creative application of inertial microfluidics, Wu et al. separated E. coli mixed into human blood using a flow system in which the diluted blood was sheathed with another flow (to protect cells from hitting the wall) and then deflected by an “acting” flow.81 These impinging flows changed the fluid and particle momentum twice. The more massive blood cells were deflected further than bacteria, leading to a separation and collection of the bacteria. While the separation was not perfect (some bacteria were in the RBCs and vice versa), the bacteria were focused and collected at a 300-fold enriched concentration (see Figure 7). There was no mention of where platelets or WBCs were focused. While this technique has great potential for small flows on a chip, the flow rate was only 18 µL/min, and there are large volumes of fluid involved. For example, in these experiments the final dilution of the blood was from 1:500 to 1:1,000, mostly due to the “sheath” and “acting” flows that produced the focusing of the bacteria. While this process had good separation and produced actual concentration of bacteria, it might be impractical to use this process to treat 7 mL in less than 10 min.
Figure 7.
Illustration of the concept of inertial microfluidic focusing by which bacteria were separated from RBCs in highly diluted blood. The bacteria and red cells are deflected by the acting flow and protected by the sheath flow, and finally focused at a particular location in the exit channel. Reproduced from Ref. 81, with permission from Royal Society of Chemistry.
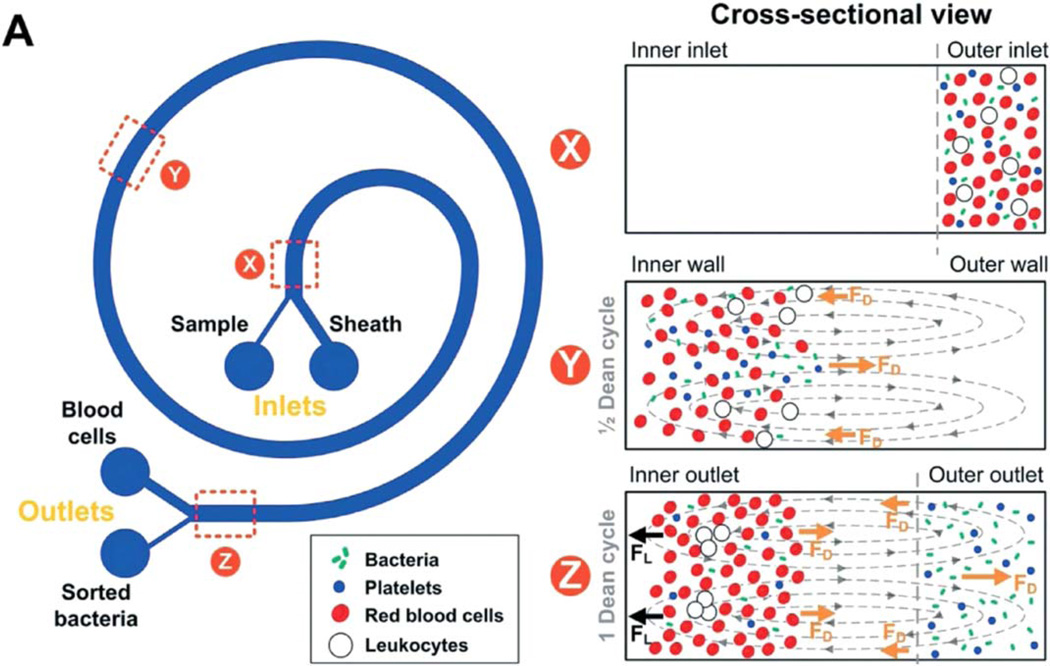
By employing Dean secondary flows,69, 82 Hou et al. demonstrated that E. coli bacteria could be separated from blood cells in threefold diluted whole blood, even when the bacteria were present at a physiologically low value of only 10 to 50 CFU/mL.70 As mentioned, Dean secondary flow is a circulation perpendicular to the axial flow that occurs in curved channels. Because the drag forces on bacteria are different than on the larger red and white cells, the bacteria were isolated on one side of the channel while the larger cells were isolated on the other side (Figure 8). The bacteria were collected by splitting the flow, and platelets were collected along with the bacteria. For loadings greater than 100 CFU/ mL, bacterial recovery was greater than 65%. At loadings between 10 and 50 CFU/mL, bacteria were also recovered, but the efficiency was not reported. The drawbacks to this approach is that there is a sheathing flow of buffer required for the separation, so each milliliter of whole blood requires 33 mL of process fluid to be disposed of. The flow rate for optimal separation is only about 50 µL of whole blood per minute, and the authors indicated that this could be scaled up by building devices with parallel flow. Thus an array of 20 systems could process 7 mL of blood in 7 min.
Figure 8.
Illustration of the concept of Dean flow focusing by which bacteria and platelets were separated from RBCs in slightly diluted blood. The Dean flow circulation cycles the bacteria and platelets from the outer wall to the inner wall and back to the outer wall while the RBCs get trapped near the inner wall of the curved channel. Reproduced from Ref. 70, with permission from Royal Society of Chemistry.
To date only Hou et al.79 have presented a fluid-mechanics-based separation of bacteria from milliliter-quantities of blood in many parallel channels in a practical time scale. This is promising, but the process also diluted the blood and so there would be copious amounts of waste fluid to deal with.
There are commercial blood analysis instruments in clinics that process milliliter-quantities of blood, and then proceed to quantitatively count the cellular components of the sample; but even in these cases, the actual counting is usually done by passing a fraction of the blood sample at very low flow rates (but high velocities) through a very small capillary channel that allows only one cellular component at a time to flow past a detector. Appropriate chemical labels and various markers of size and complexity are used to differentiate RBCs, platelets, and the various types of WBCs. To our knowledge, the identification of bacterial species and their number is not a routine capability of these instruments. However, with the rising prevalence of antibiotic-resistant blood infections, we would not be surprised to see the introduction of bacterial typing into blood analysis instruments.
Chemical Capture of Bacteria from Blood
Chemical and physical principles
The governing principle for chemical capture of a bacterium is that a physical or chemical interaction attaches the bacterium to a particle that is more easily separated from the blood than the bacterium alone could be separated. In theory, any of the separation principles described above could be used to separate the capturing particle from the blood. In current practice, magnetic separation is most prevalent, but sedimentation or filtration could also be used.
One of the stringent requirements for chemical capture of a yet-unidentified bacterium is that the capturing agent must be very general in attachment to every kind of bacterium, but have no or very minimal interaction with the human blood components, including blood proteins that may block the bacterial binding sites. While certain species or classes of bacteria may have specific antigens that could be bound by complementary antibodies, what is needed is a chemical group that is present on all bacteria, including gram-negative, gram-positive, and acid-fast, and yet is absent from all mammalian cells. Although no such chemical group is yet known, humans possess an immune system protein called mannose binding lectin in their blood that binds to the surface of all of the clinical pathogens of interest in antibiotic resistance situations, including Enterococcus ssp., Klebsiella spp., E. coli, S. aureus, S. pneumonia, S. pyogenes, P. aeruginosa, and many other bacteria, viruses, fungi, and some protozoa.83–85 This lectin and parts thereof have been used to capture and separate bacteria from blood.86, 87
Another molecule of recent interest is Zn-dipicolylamine, an organic molecule much smaller than a lectin, which is reported to bind to many species of bacteria.88–90 It displays minimal binding to mammalian cells90 and thus has been used to capture bacteria from blood.91 It appears to become associated with anionic phospholipids on the surfaces of gram-negative and gram-positive bacteria.
Surprisingly the biopolymer heparin sulfate (HS) appears to bind some bacteria. HS is structurally related to the heparin biopolymer that is often employed as an anticoagulant for blood. HS is found on the surface of many mammalian cells, and some bacteria have developed a binding affinity for HS, including some species of Heliobacter, Staphylococcus, Streptococcus, Pseudomonas, Escherichia, and Mycobacterium. 92 Some types of antibiotics (e.g., vancomycin, daptomycin, polymixin B) are used for bacterial capture, but it remains unproven if these antibiotics have general affinity for all bacteria.93, 94
Of course, antibodies that bind bacteria could be used as ligands. However, antibodies are usually very specific, and unless one already knows the species of the blood pathogen, this would probably be an inefficient approach as many and various types of antibodies would need to be simultaneously employed to evaluate an unknown clinical blood infection. One possibility is to use antibodies to the lipopolysaccharides found on the outer membrane of gram-negative bacteria.95
An often-overlooked aspect of using ligands to generically capture bacteria is that tight binding is often desired for more rapid association and/or adhesion. However, if the captured bacterium needs to be released at a later time for analysis, there must be a method to easily release the bacteria from its binding particle. Thus high binding affinity may be a conflicting requirement in the optimization of a separation and analytical system.
Another significant aspect of capture by ligands is that the bacterium must come in close enough contact with the ligand to be captured. If there is a requirement for a certain steric approach, then the ligand and bacterium must spend some time in the same vicinity to increase the probability of capture. Furthermore, if binding is not irreversible, the bacterium may be released and need to be captured again. These concepts lead to a discussion of the time-course, or kinetics of capture. Theoretical and experimental analysis of bacterial capture by lectin-coated beads has been presented previously.87 Basically, the bacterium must move to the capture surface, or the surface (such as a bead) must move to the bacterium, or both. These processes take time, and the time to capture decreases as the concentration of binding surface (concentration of beads, or surface area of a packed column) increases. In the case of capturing 10 CFU from 1 mL of blood in 1 min, the blood volume would need to be loaded with billions of beads/mL,87 and then one would have to capture all the beads and look for very few beads that were successful in capturing a bacterium. To make this more complicated, there will be many RBCs and platelets that get in the way (sterically interfere) with the approach of a bacterium to a bead in whole blood.
Examples of capture on magnetic beads
Superparamagnetic (SPM) beads have a useful magnetic property in that they are negligibly magnetic in the absence of a strong magnetic field (so they do not magnetically stick to each other), but become magnetic in the presence of a strong field and are easily captured by a magnet. SPM beads are usually made of iron or manganese oxides, and are quite small, often having a size on the order of 10 to 100 nm. They can be purchased with “ready-to-conjugate” surface chemistry, so they can be easily attached to biomolecules.96 One challenge with magnetic beads is that a single bead, while superparamagnetic may still not generate sufficient force in the magnetic field to quickly move to a capture surface or region because of the very strong viscous drag forces.97 Thus hundreds of magnetic particles may be required to “pull down” the bacteria in the face of strong hydrodynamic forces.86 Other important issues involve the sensitivity to detect attachment of single magnetic beads.98, 99
One of the most interesting applications of bacterial capture and separation is the “artificial spleen” developed by Kang et al.86, 87, 100 Genetic manipulation produced a molecule with the binding end of mannose binding lectin attached to the Fc fragment of IgG. The Fc end was bound to 250-nm SPM beads. When mixed with septic blood, E. coli and S. aureus were surrounded by these beads and magnetically pulled from whole blood through a porous barrier and then discarded, while the whole blood was returned to the rat. A clearance of about 90% from whole blood was reported when the septic load was about 104 CFU/mL. Using some mathematical modeling they backed out binding parameters that showed that the beads bound to bacteria about 3 orders of magnitude less in whole blood than in buffer. They attributed low binding in whole blood to interference with proteins and blood cells.87
Sen et al. present an example of soil-derived E. coli capture by magnetic beads coated with binding ligands, which might be applied to capture from blood.101 However, the fluorescent detection method employed had a limit of detection of 106 CFU/mL, using 3 × 108 beads/mL. A more sensitive detection system would be required to detect the low concentrations found in clinical blood samples.
In an in vitro model using bovine blood spiked with 1 × 107 CFU/mL of E. coli, Lee et al. used SPM beads coated with bis-Zn-dipicolylamine.91 In a nonflowing system, a first separation removed 70% of the bacteria from diluted blood (1:50), and a second separation was reported to remove all the remaining bacteria. Using whole blood in a flow system at 1 mL/min, about 80% of the E. coli was cleared in one pass, and a second pass boosted the clearance to 90 to 95%. The authors suggested that there was some nonspecific binding of RBCs to the SPM beads.
Examples of capture on column support or other surfaces
Polymixin B binds to the lipopolysaccharide of gram-negative bacteria, and passing septic blood through a column of fibers coated with polymixin B has been used clinically in Japan to treat septic shock.102, 103 While it appears that the column binds the endotoxins released into blood, there was no evidence that whole bacteria were bound to the column.
Similarly, a heparin-coated fiber column was used to remove proinflammatory cytokines from septic blood.104 Heparin-coated beads in a column were successfully used to remove about 60% of S. aureus from spiked blood in vitro.105 The adsorbed bacteria were then eluted with a 2M NaCl solution.
Wang et al. used various antibodies attached to glass to capture E. coli from whole blood.95 The use of an antibody to lipopolysaccharide binding protein, along with the lipopolysaccharide binding protein (which bound to the E. coli), was best able to capture E. coli on a glass surface. Up to 70% binding efficiency was measured, and limits of detection were down to 50 CFU/mL. However, this was a batch process that required at least 3 h to bind the E. coli and then wash out the blood.
Capture by particles that differentially sediment
The observation that bacteria can be captured from whole or diluted blood by coated beads leads to a proposed combination of sedimentation and bead capture. We have found no reports of such a system in the literature, but it may be possible to capture bacteria on small nondense beads, such as plastic beads or hollow glass spheres. In a sedimentation process these may have an even slower sedimentation velocity than platelets, allowing for the separation of the bacteria from all the interfering blood components. Although it may be a challenge to find all the bacteria in concentrations as low as 10 CFU/mL, these possibilities are worth investigating.
Capture by particles that can be filtered
Another possibility may be to capture the bacteria from whole blood onto particles that are much larger than WBCs. These particles might be mixed vigorously with the blood for adequate time to collect the bacteria on their surfaces. Then this suspension could be filtered, allowing the plasma and cellular components to pass through the open filter cake of packed beads. The beads could be washed and then the bacteria could be lysed to release their DNA or eluted from the beads and collected in another filtration process.
Other Methods
Field-flow fractionation
Field-flow fractionation (FFF) is usually considered an analytical technique in which suspended particles flow laminarly through a chamber and are subjected to some kind of force transverse to the flow direction. This causes differential transverse migration in the channel and a separation from the other particles and the fluid. Common forces are crossflow fluid movement causing drag force, sedimentation force, thermophoretic force, electrical, magnetic, and dielectric forces.106 The cross-flow filtration discussed in Qualitative Filtration Principles section is a type of drag-force-FFF. Various types of FFF have been used to separate blood components, 107–109 and separate bacteria from each other,110–114 but we have found no reports of its use in separation of bacteria from blood. There are a few reports of separating blood-dwelling parasites from blood by FFF methods, but these reports take advantage of the much larger size and motility of filamentous parasites compared to RBCs.115, 116 There is a report of FFF separation of a small spherical parasite (Toxoplasma gondii) from mouse blood, and in this case, the parasite was larger (10.3 vs. 4.6 µm diameter) and less dense (1.056 vs. 1.085 g/mL) than the mouse RBCs.117 As sedimentation forces can dynamically separate blood cells from smaller bacteria, it may be possible to apply sedimentation-FFF to remove RBCs and WBCs from bacteria; however, bacterial separation from platelets may be more challenging. Furthermore, in most cases of FFF involving blood, the blood is diluted with saline or a buffer. This may create a challenge for managing the volume to be processed and the short time allowed. Commercial FFF equipment is available, but is expensive and not miniaturized, and our future vision of routine septic blood analysis may require disposable or “easily sterilized” equipment.
Dielectrophoresis
Dielectrophoretic separation occurs when polarized particles (no net charge) placed in a nonuniform electric field gain an electrokinetic force that creates net movement through the surrounding fluid, which in turn creates a drag force opposing the movement. One can exploit the differences in size, shape, density, and dielectric properties of various particles to create a separation. There are many types of dielectrophoretic separation of small particles,118 and in general this is an analytical technique employing very small sample volumes (µL size scale).
For example, Cheng et al. used a quadruple electrode array to separate bacteria (S. aureus and P. aeruginosa) from blood cells by causing the bacteria to migrate and become trapped at the center of the quadruple electrode while the blood cells migrated away from the center.119 The bacterial species was identified by also collecting Ag nanoparticles with the bacteria at the center, and then identifying the species by surface enhanced Raman spectroscopy. The challenge with this separation technique is that in order to provide sufficient signal-to-noise for detection the blood was highly diluted (to 107 “cells”/mL) and the bacteria were very concentrated (107 CFU/mL). This bacterial load is much higher than would be found in a living patient with blood sepsis. In a later study, Cheng et al. separated blood cells and bacteria using a flow-through system that operated at 1 µL/min and used a bacterial concentration of 107 CFU/mL (still unrealistically high).120
A combination of dielectrophoresis and FFF was employed by Piacentini et al. to separate platelets from RBCs in diluted (about 1:10) blood using an alternating array of electrodes in a microfluidic channel.108 It might be possible to do the same to separate out the bacteria, or perhaps to collect bacteria along with the platelets.
Analysis and Outlook
As we have shown above, there are numerous techniques that have been used or might be used to separate bacteria from blood. Many of these have been used on bacteria of interest in blood sepsis.
When we examine these in light of the requirements for a rapid identification technique using septic blood with a loading of only 10 CFU/mL, we realize that volumetric throughput and bacterial recovery efficiency are key distinguishing criteria. With only 10 CFU/mL, one needs 7 to 10 mL (or more) to collect enough bacteria for sufficient signal-to-noise to provide molecular identification, even with laser-based single molecule diagnostics.
Examples of techniques that have been proven to separate bacteria from blood are presented in Table 3. Those with the highest flow rate of whole blood are centrifugal sedimentation in a disk (7 mL/min), lectin on magnetic beads (8.9 mL/ min),86, 87 and parallel skimming channels (1.5 mL/min).79 The others are in the µL/min range. Those with the highest recovery efficiency are lectin on magnetic beads (90%), and those using margination and hydrodynamic focusing in straight channels (80%). Four of those techniques in Table 3 can process whole blood, although this is not a stringent requirement. While those techniques with low flow rates in single microfluidic channels can be manufactured in parallel configuration, these often have very high pressure requirements, which could complicate production of reliable leak-proof systems.
Table 3.
Summary of Sampling of Successful Techniques used to Separate Bacteria from Blood
| Description and/or Principle | Whole Blood Flow Rate |
Dilution | Recovery Efficiency (%) |
Comments | Reference | Manufacturing Costs and Considerations |
Time Efficiency per mL of Blood |
Possible Points of Application |
|---|---|---|---|---|---|---|---|---|
| Centrifugal sedimentation in hollow disk | 7,000 µL/min | Undiluted | 25–65 | Recovery dependent on anticoagulant |
This paper | Low cost, single unit | Quick for large volumes |
Hospital, point of care |
| RBC margination in straight channels | 15 µL/min | Undiluted | 80 | High pressures, low flow rates |
78,79 | Low single channel cost; demonstrated parallel channels |
Slow unless massively parallel |
Hospital, point of care |
| Hydrodynamic focusing: channel flow | 1 µL/min | 1:200 | 80 | High pressures, low flow rates |
80 | Low single channel cost; but costly parallel channel manufacturing |
Slow unless massively parallel |
Hospital, point of care |
| Hydrodynamic focusing: sheath flow | 18 µL/min | 1:1000 | Not reported | Large volumes of fluid |
81 | Low single channel cost; but costly parallel channel manufacturing |
Slow unless massively parallel |
Hospital, point of care |
| Hydrodynamic focusing: Dean circulation |
50 µL/min | 1:3 | 65 | Low dilution and good recovery |
70 | Low single channel cost; but costly parallel channel manufacturing |
Moderate to fast depending on parallel channels |
Hospital, point of care |
| Lectin on magnetic beads | 8,900 µL/min | Undiluted | 90 | Done with in vivo mouse model |
86 | Costly beads and lectin coat; simple flow system |
Quick for large volumes |
Hospital, point of care, field |
| Bis-Zn-dipicolylamine on magnetic beads |
1,000 µL/min | Undiluted | 80 | Two passes produce ~95% recovery |
91 | Costly beads; simple flow system |
Quick for small volumes |
Hospital, point of care, field |
| Dielectrophoresis | 1 µL/min | ~1:500 | Not reported | May be difficult or costly for parallel systems |
119 | Expensive equipment | Slow unless massively parallel |
Hospital |
Thus far we have not discussed the cost and manufacturing challenges associated with these devices. Microfluidic devices can be manufactured in high quantities using technologies gleaned from the semiconductor industry. As the production quantity increases, the cost per item will decrease. The three columns on the right side of Table 3 are some estimated figures of merit that should be considered when comparing the possible utility of these technologies in rapidly separating bacteria from blood. When considering the merit of production and use costs, production of a single microchannel is fairly low, as the manufacturing technology is readily available. However, most of the microchannel techniques would require parallel devices, or even massively parallel devices to process 1 mL of blood in 1 min. The technology and costs of assembling parallel flow channels that never leak under very high pressures may be a challenge but can probably be accomplished. The hollow disk could be made for low cost using current plastic molding technology. The costs of the magnetic beads may be high, but this may be offset by the simple flow system associated with simply mixing and then magnetically separating the beads; thus capital costs may be low in such systems. The rapid processing time may also give advantage to this magnetic technology.
Our first estimates of possible application locations for the technologies listed in Table 3 are given in the right-most column. These are based on the roughly estimated size and costs of the processing equipment, and their sensitivity to environmental jostling. Of course, they all will work well in the spacious and stable environment of a hospital laboratory. The dielectrophoretic technology may be too specialized and expensive to place routinely in physician’s clinics. As to which might be used in the field (ambulance, portable clinic, battlefield, etc.), the separation process would need to be insensitive to jostling, and the equipment should not be massive. The magnetic bead technologies would be amenable to field use, as mixing is a hardy process and simple magnets could be used to collect the beads. Possibly some of the microfluidic devices could be portable if the associated systems can be made compact and low weight. In our lab we find that the centrifugal sedimentation system is currently sensitive to movement (jostling) during the processing; but this might be ameliorated with further development.
Future considerations
We see two directions that rapid identification of resistant organisms in blood sepsis can take. One direction is the construction of large analytical machines that take whole blood and process it through microfluidic systems to identify the bacteria in blood, like blood analysis devices presently used to identify and count white cells, cancer stem cells, and other blood components. The microfluidic devices and perhaps magnetic bead capture could be employed in these systems. We envision this as expensive nondisposable hardware for a hospital lab that can be used for tens of thousands of assays with sterilization between each assay. An alternative direction is to develop small inexpensive disposable devices that separate the bacteria and pass it on to another analytical device designed specifically to identify the bacteria or its DNA. Filtration and centrifugal sedimentation can process large quantities of whole (or slightly diluted) blood quickly, and the device with residual blood components can be disposed of after each assay. However, these devices might have lower bacterial recovery. Both directions appear to be feasible, and much research and development needs to be done. As the mortality rate from antibiotic-resistant blood infections continues to increase, there is an essential need and an increasing demand for this technology.
Acknowledgments
Support is gratefully acknowledged from the National Institutes of Health grant R01AI116989.
Contributor Information
William G. Pitt, Dept. of Chemical Engineering, Brigham Young University, Provo, UT
Mahsa Alizadeh, Dept. of Chemical Engineering, Brigham Young University, Provo, UT.
Ghaleb A. Husseini, Dept. of Chemical Engineering, American University of Sharjah, Sharjah, UAE
Daniel S. McClellan, Dept. of Chemical Engineering, Brigham Young University, Provo, UT
Clara M. Buchanan, Dept. of Chemical Engineering, Brigham Young University, Provo, UT
Colin G. Bledsoe, Dept. of Chemical Engineering, Brigham Young University, Provo, UT
Richard A. Robison, Dept. of Microbiology and Molecular Biology, Brigham Young University, Provo, UT
Rae Blanco, Dept. of Chemical Engineering, Brigham Young University, Provo, UT.
Beverly L. Roeder, Dept. of Biology, Brigham Young University, Provo, UT
Madison Melville, Dept. of Chemical Engineering, Brigham Young University, Provo, UT.
Alex K. Hunter, Dept. of Chemical Engineering, Brigham Young University, Provo, UT
Literature Cited
- 1.CDC. [Accessed May 18, 2016];Antibiotic Resistance Threats in the United States, 2013. 2013 http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf.
- 2.Lodise TP, McKinnon PS, Swiderski L, Rybak MJ. Outcomes analysis of delayed antibiotic treatment for hospital-acquired Staphylococcus aureus bacteremia. Clin Infect Dis. 2003;36:1418–1423. doi: 10.1086/375057. [DOI] [PubMed] [Google Scholar]
- 3.Malik S, Ravishekhar K. Significance of coagulase negative Staphylococcus species in blood culture. J Clin Diagn Res. 2012;6:632–635. [Google Scholar]
- 4.Howell L. Global Risks 2013: An Initiative of the Risk Response Network. New York, NY: World Economics Forum; 2013. [Google Scholar]
- 5.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 6.Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. Reply. N Engl J Med. 2010;363:1483–1484. doi: 10.1056/NEJMc1006641. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Vital signs: carbapenem-resistant enterobacteriaceae. MMWR Morbid Mortal Wkly Rep. 2013;62:165–170. [PMC free article] [PubMed] [Google Scholar]
- 8.Schwaber MJ, Carmeli Y. Carbapenem-resistant Enterobacteriaceae: a potential threat. JAMA. 2008;300:2911–2913. doi: 10.1001/jama.2008.896. [DOI] [PubMed] [Google Scholar]
- 9.Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol. 2008;29:1099–1106. doi: 10.1086/592412. [DOI] [PubMed] [Google Scholar]
- 10.Cooke J, Ellis J. Healthcare-associated infections. [Accessed May 18, 2016];Clinical Lab Products Magazine. 2015 http://www.clpmag.com/2015/09/health-care-associated-infections/, 2016. [Google Scholar]
- 11.Kleinschmidt S, Huygens F, Faoagali J, Rathnayake IU, Hafner LM. Staphylococcus epidermidis as a cause of bacteremia. Fut Microbiol. 2015;10:1859–1879. doi: 10.2217/fmb.15.98. [DOI] [PubMed] [Google Scholar]
- 12.Lv H, Ning B. Pathogenesis of bloodstream infection in children with blood cancer. Exp Ther Med. 2013;5:201–204. doi: 10.3892/etm.2012.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duerden B, Fry C, Johnson AP, Wilcox MH. The control of methicillin-resistant Staphylococcus aureus blood stream infections in England. Open Forum Infect Dis. 2015;2 doi: 10.1093/ofid/ofv035. :ofv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diekema DJ, Pfaller MA. Rapid detection of antibiotic-resistant organism carriage for infection prevention. Clin Infect Dis. 2013;56:1614–1620. doi: 10.1093/cid/cit038. [DOI] [PubMed] [Google Scholar]
- 15.USFDA. [Accessed May 18, 2016];Film Array Blood Culture Identification Panel Kit. 2013 http://www.accessdata.fda.gov/cdrh_docs/reviews/k130914.pdf.
- 16.USFDA. [Accessed May 18, 2016];Verigene Gram Negative Blood Culture Nucleic Acid Test. 2014 http://www.accessdata.fda.gov/cdrh_docs/pdf13/ k130914.pdf.
- 17.Vergara-Lopez S, Dominguez MC, Conejo MC, Pascual A, Rodriguez-Bano J. Lessons from an outbreak of metallo-betalactamase-producing Klebsiella oxytoca in an intensive care unit: the importance of time at risk and combination therapy. J Hosp Infect. 2015;89:123–131. doi: 10.1016/j.jhin.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Daikos GL, Petrikkos P, Psichogiou M, Kosmidis C, Vryonis E, Skoutelis A, Georgousi K, Tzouvelekis LS, Tassios PT, Bamia C, Petrikkos G. Prospective observational study of the impact of VIM-1 metallo-beta-lactamase on the outcome of patients with Klebsiella pneumoniae bloodstream infections. Antimicrob Agents Chemother. 2009;53:1868–1873. doi: 10.1128/AAC.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zarkotou O, Pournaras S, Tselioti P, Dragoumanos V, Pitiriga V, Ranellou K, Prekates A, Themeli-Digalaki K, Tsakris A. Predictors of mortality in patients with bloodstream infections caused by KPC-producing Klebsiella pneumoniae and impact of appropriate antimicrobial treatment. Clin Microbiol Infect. 2011;17:1798–1803. doi: 10.1111/j.1469-0691.2011.03514.x. [DOI] [PubMed] [Google Scholar]
- 20.Kang CI, Kim SH, Park WB, Lee KD, Kim HB, Kim EC, Oh MD, Choe KW. Bloodstream infections caused by antibiotic-resistant Gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob Agents Chemother. 2005;49:760–766. doi: 10.1128/AAC.49.2.760-766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Lilienfeld-Toal M, Lehmann LE, Raadts AD, Hahn-Ast C, Orlopp KS, Marklein G, Purr I, Cook G, Hoeft A, Glasmacher A, Stïuber F. Utility of a commercially available multiplex real-time PCR assay to detect bacterial and fungal pathogens in febrile neutropenia. J Clin Microbiol. 2009;47:2405–2410. doi: 10.1128/JCM.00491-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehmann LE, Hunfeld KP, Emrich T, Haberhausen G, Wissing H, Hoeft A, Stïuber F. A multiplex real-time PCR assay for rapid detection and differentiation of 25 bacterial and fungal pathogens from whole blood samples. Med Microbiol Immunol. 2008;197:313–324. doi: 10.1007/s00430-007-0063-0. [DOI] [PubMed] [Google Scholar]
- 23.Bloos F, Hinder F, Becker K, Sachse S, Mekontso Dessap A, Straube E, Cattoir V, Brun-Buisson C, Reinhart K, Peters G, Bauer M. A multicenter trial to compare blood culture with polymerase chain reaction in severe human sepsis. Intensive Care Med. 2010;36:241–247. doi: 10.1007/s00134-009-1705-z. [DOI] [PubMed] [Google Scholar]
- 24.Kumar A, Roberts D, Wood KE. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Ann Intern Med. 2007;147:413–413. doi: 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- 25.Schweizer ML, Furuno JP, Harris AD, Johnson JK, Shardell MD, McGregor JC, Thom KA, Cosgrove SE, Sakoulas G, Perencevich EN. Comparative effectiveness of nafcillin or cefazolin versus vancomycin in methicillin-susceptible Staphylococcus aureus bacteremia. BMC Infect Dis. 2011;11:279. doi: 10.1186/1471-2334-11-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu V, Escobar GJ, Greene JD, Soule J, Whippy A, Angus DC, Iwashyna TJ. Hospital deaths in patients with sepsis from 2 independent cohorts. JAMA. 2014;312:90–92. doi: 10.1001/jama.2014.5804. [DOI] [PubMed] [Google Scholar]
- 27.Hoyert DL, Xu J. Deaths: preliminary data for 2011. Natl Vit Stat Rep. 2012;61:1–51. [PubMed] [Google Scholar]
- 28.Kreger BE, Craven DE, Carling PC, McCabe WR. Gram-negative bacteremia. III. Reassessment of etiology, epidemiology and ecology in 612 patients. Am J Med. 1980 Mar;68:332–343. doi: 10.1016/0002-9343(80)90101-1. [DOI] [PubMed] [Google Scholar]
- 29.Leggieri N, Rida A, Francois P, Schrenzel J. Molecular diagnosis of bloodstream infections: planning to (physically) reach the bedside. Curr Opin Infect Dis. 2010;23:311–319. doi: 10.1097/QCO.0b013e32833bfc44. [DOI] [PubMed] [Google Scholar]
- 30.Yagupsky P, Nolte FS. Quantitative aspects of septicemia. Clin Microbiol Rev. 1990 Jul;3:269–279. doi: 10.1128/cmr.3.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price C, Douglas I, Tuttle E, Shorr A, Mensack M, Bessesen M, Miquirray S, Overdier K, Duarte N, Shamsheyeva A, Gamage D, Allers E, Hance K, Michel C, Turng B, Metzger S. ePoster presented at ECCMID. Copenhagen, Denmark: 2015. Rapid identification and antimicrobial susceptibility testing of bacteria in bloodstream infections using the Accelerate ID/AST technology. [Google Scholar]
- 32.Kothari A, Morgan M, Haake D. Emerging technologies for rapid identification of bloodstream pathogens. Clin Infect Dis. 2014;59:272–278. doi: 10.1093/cid/ciu292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruimy R, Dos-Santos M, Raskine L, Bert F, Masson R, Elbaz S, Bonnal C, Lucet JC, Lefort A, Fantin B, Wolff M, Hornstein M, Andremont A. Accuracy and potential usefulness of triplex real-time PCR for improving antibiotic treatment of patients with blood cultures showing clustered gram-positive cocci on direct smears. J Clin Microbiol. 2008;46:2045–2051. doi: 10.1128/JCM.02250-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hedman J, Radstrom P. Overcoming inhibition in real-time diagnostic PCR. Methods Mol Biol. 2013;943:17–48. doi: 10.1007/978-1-60327-353-4_2. [DOI] [PubMed] [Google Scholar]
- 35.Peters RP, van Agtmael MA, Danner SA, Savelkoul PH, Vandenbroucke-Grauls CM. New developments in the diagnosis of bloodstream infections. Lancet Infect Dis. 2004;4:751–760. doi: 10.1016/S1473-3099(04)01205-8. [DOI] [PubMed] [Google Scholar]
- 36.Makristathis A, Riss S, Hirschl AM. A novel fluorescence in situ hybridization test for rapid pathogen identification in positive blood cultures. Clin Microbiol Infect. 2014;20:O760–O763. doi: 10.1111/1469-0691.12561. [DOI] [PubMed] [Google Scholar]
- 37.Calderaro A, Martinelli M, Motta F, Larini S, Arcangeletti MC, Medici MC, Chezzi C, De Conto F. Comparison of peptide nucleic acid fluorescence in situ hybridization assays with culture-based matrix-assisted laser desorption/ionization-time of flight mass spectrometry for the identification of bacteria and yeasts from blood cultures and cerebrospinal fluid cultures. Clin Microbiol Infect. 2014;20:O468–O475. doi: 10.1111/1469-0691.12490. [DOI] [PubMed] [Google Scholar]
- 38.Cai H, Parks JW, Wall TA, Stott MA, Stambaugh A, Alfson K, Griffiths A, Mathies RA, Carrion R, Patterson JL, Hawkins AR, Schmidt H. Optofluidic analysis system for amplification-free, direct detection of Ebola infection. Sci Rep. 2015;5:14494. doi: 10.1038/srep14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hedman J, Radstrom P. Overcoming inhibition in real-time diagnostic PCR. In: Wilkes M, editor. PCR Detection of Microbioal Pathogens, Methods Mol Biol. Vol. 943. Springer; 2013. pp. 17–48. [DOI] [PubMed] [Google Scholar]
- 40.Danon D, Marikovsky Y. Determination of density distribution of red cell population. J Lab Clin Med. 1964;64:668. [PubMed] [Google Scholar]
- 41.Turgeon ML. Clinical Hematology: Theory and Procedures. 4th. Philadelphia: Lippincott Williams & Wilkins; 2004. [Google Scholar]
- 42.Zucker RM, Cassen B. Separation of normal human leukocytes by density and classification by size. J Hematol. 1969;34:591. [PubMed] [Google Scholar]
- 43.Cooney DO. Biomedical Engineering Principles. Vol. 2. New York: Marcel Dekker; 1976. [Google Scholar]
- 44.McCabe WL, Smith JC, Harriot P. Unit Operations of Chemical Engineering. 7th. New York: McGraw Hill; 2005. [Google Scholar]
- 45.Beaudoin G, Jaffrin MY. Plasma filtration in couette-flow membrane devices. Artif Organs. 1989;13:43–51. doi: 10.1111/j.1525-1594.1989.tb02831.x. [DOI] [PubMed] [Google Scholar]
- 46.Toner M, Irimia D. Blood-on-a-chip. Annu Rev Biomed Eng. 2005;7:77–103. doi: 10.1146/annurev.bioeng.7.011205.135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hou HW, Bhagat AAS, Lee WC, Huang S, Han J, Lim CT. Microfluidic devices for blood fractionation. Micromachines (Basel) 2011;2:319–343. [Google Scholar]
- 48.Liu V, Patel M, Lee A. A microfluidic device for blood cell sorting and morphology analysis. 17th International Conference on Miniaturized Systems for Chemistry and Life Sicences; October, 2013; Freiburg, Germany. pp. 27–31. [Google Scholar]
- 49.Wilding P, Kricka LJ, Cheng J, Hvichia G, Shoffner MA, Fortina P. Integrated cell isolation and polymerase chain reaction analysis using silicon microfilter chambers. Anal Biochem. 1998 Mar 15;257:95–100. doi: 10.1006/abio.1997.2530. [DOI] [PubMed] [Google Scholar]
- 50.Mohamed H, McCurdy LD, Szarowski DH, Duva S, Turner JN, Caggana M. Development of a rare cell fractionation device: application for cancer detection. IEEE T Nanobiosci. 2004 Dec;3:251–256. doi: 10.1109/tnb.2004.837903. [DOI] [PubMed] [Google Scholar]
- 51.Carlson RH, Gabel CV, Chan SS, Austin RH, Brody JP, Winkelman JW. Self-sorting of white blood cells in a lattice. Phys Rev Lett. 1997;79:2149–2152. [Google Scholar]
- 52.Huang LR, Cox EC, Austin RH, Sturm JC. Continuous particle separation through deterministic lateral displacement. Science. 2004;304:987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 53.Hiemenz PC, Rajagopalan R. Principles of Colloid and Surface Chemistry. 3rd. New York: Marcel Dekker; 1997. [Google Scholar]
- 54.Lin FC, Cohen R, Losada R, Bush V. Cellular sedimentation and barrier formation under centrifugal force in blood collection tubes. Lab Med. 2001;32:588. [Google Scholar]
- 55.Kessler SB. Assembly, Composition and Method for Separating Blood. 4,350,593 assigned to Becton Dickinson. US Patent Office: Washington DC; US Patent. 1979
- 56.Haeberle S, Brenner T, Zengerle R, Ducree J. Centrifugal extraction of plasma from whole blood on a rotating disk. Lab Chip. 2006;6:776–781. doi: 10.1039/b604145k. [DOI] [PubMed] [Google Scholar]
- 57.Amasia M, Madou M. Large-volume centrifugal microfluidic device for blood plasma separation. Bioanalysis. 2010;2:1701–1710. doi: 10.4155/bio.10.140. [DOI] [PubMed] [Google Scholar]
- 58.Gorkin R, Park J, Siegrist J, Amasia M, Lee BS, Park JM, Kim J, Kim H, Madou M, Cho YK. Centrifugal microfluidics for biomedical applications. Lab Chip. 2010;10:1758–1773. doi: 10.1039/b924109d. [DOI] [PubMed] [Google Scholar]
- 59.White FM. Fluid Mechanics. 6th. New York: McGraw-Hill; 2008. [Google Scholar]
- 60.Lenshof A, Laurell T. Continuous separation of cells and particles in microfluidic systems. Chem Soc Rev. 2010;39:1203–1217. doi: 10.1039/b915999c. [DOI] [PubMed] [Google Scholar]
- 61.Pamme N. Continuous flow separations in microfluidic devices. Lab Chip. 2007;7:1644–1659. doi: 10.1039/b712784g. [DOI] [PubMed] [Google Scholar]
- 62.Sajeesh P, Sen AK. Particle separation and sorting in microfluidic devices: a review. Microfluid Nanofluid. 2014;17:1–52. [Google Scholar]
- 63.Zhu G, Nguyen N-T. Particle Sorting and Microfluidic Systems. Micro Nanosyst. 2010;2:202–216. [Google Scholar]
- 64.Kumar CS. Microfluidic Devices in Nanotechnology. Fundamental Concepts. Hoboken, New Jersey: Wiley; 2010. [Google Scholar]
- 65.Kirby BJ. Micro- and Nanoscale Fluid Mechanics: Transport in Microfluidic Devices. New York: Cambrige University Press; 2010. [Google Scholar]
- 66.Gossett DR, Weaver WM, Mach AJ, Hur SC, Tse HT, Lee W, Amini H, Di Carlo D. Label-free cell separation and sorting in microfluidic systems. Anal Bioanal Chem. 2010;397:3249–3267. doi: 10.1007/s00216-010-3721-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Di Carlo D. Inertial microfluidics. Lab Chip. 2009;9:3038–3046. doi: 10.1039/b912547g. [DOI] [PubMed] [Google Scholar]
- 68.Di Carlo D, Irimia D, Tompkins RG, Toner M. Continuous inertial focusing, ordering, and separation of particles in microchannels. Proc Natl Acad Sci USA. 2007;104:18892–18897. doi: 10.1073/pnas.0704958104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bhagat AA, Kuntaegowdanahalli SS, Papautsky I. Continuous particle separation in spiral microchannels using Dean flows and differential migration. Lab Chip. 2008;8:1906–1914. doi: 10.1039/b807107a. [DOI] [PubMed] [Google Scholar]
- 70.Hou HW, Bhattacharyya RP, Hung DT, Han J. Direct detection and drug-resistance profiling of bacteremias using inertial microfluidics. Lab Chip. 2015;15:2297–2307. doi: 10.1039/c5lc00311c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zeng LY, Balachandar S, Fischer P. Wall-induced forces on a rigid sphere at finite Reynolds number. J Fluid Mech. 2005;536:1–25. [Google Scholar]
- 72.Asmolov ES. The inertial lift on a spherical particle in a plane Poiseuille flow at large channel Reynolds number. J Fluid Mech. 1999;381:63–87. [Google Scholar]
- 73.Mansour MH, Bressloff NW, Shearman CP. Red blood cell migration in microvessels. Biorheology. 2010;47:73–93. doi: 10.3233/BIR-2010-0560. [DOI] [PubMed] [Google Scholar]
- 74.Pranay P, Henriquez-Rivera RG, Graham MD. Depletion layer formation in suspensions of elastic capsules in Newtonian and viscoelastic fluids. Phys Fluids. 2012;24:1–30. [Google Scholar]
- 75.Barbee JH, Cokelet GR. Fahraeus effect. Microvasc Res. 1971;3:6. doi: 10.1016/0026-2862(71)90002-1. [DOI] [PubMed] [Google Scholar]
- 76.Skalak R, Chien S. Handbook of Bioengineering. New York: McGraw Hill; 1986. [Google Scholar]
- 77.Majhi SN, Usha L. Modeling the Fahraeus-Lindqvist effect through fluids of differential type. Int J Eng Sci. 1988;26:503–508. [Google Scholar]
- 78.Hou HW, Gan HY, Bhagat AA, Li LD, Lim CT, Han J. A microfluidics approach towards high-throughput pathogen removal from blood using margination. Biomicrofluidics. 2012;6:24115–2411513. doi: 10.1063/1.4710992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hou HW, Wu L, Amador-Munoz DP, Vera MP, Coronata A, Englert JA, Levy BD, Baron RM, Han J. Broad spectrum immunomodulation using biomimetic blood cell margination for sepsis therapy. Lab Chip. 2016;16:688–699. doi: 10.1039/c5lc01110h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mach AJ, Di Carlo D. Continuous scalable blood filtration device using inertial microfluidics. Biotechnol Bioeng. 2010;107:302–311. doi: 10.1002/bit.22833. [DOI] [PubMed] [Google Scholar]
- 81.Wu ZG, Willing B, Bjerketorp J, Jansson JK, Hjort K. Soft inertial microfluidics for high throughput separation of bacteria from human blood cells. Lab Chip. 2009;9:1193–1199. doi: 10.1039/b817611f. [DOI] [PubMed] [Google Scholar]
- 82.Dean WR. The stream-line motion of a fluid in a curved pipe. Philos Mag. 1928;7:673–695. [Google Scholar]
- 83.Worthley DL, Bardy PG, Mullighan CG. Mannose-binding lectin: biology and clinical implications. Int Med J. 2005;35:548–555. doi: 10.1111/j.1445-5994.2005.00908.x. [DOI] [PubMed] [Google Scholar]
- 84.Neth O, Jack DL, Dodds AW, Holzel H, Klein NJ, Turner MW. Mannose-binding lectin binds to a range of clinically relevant microorganisms and promotes complement deposition. Infect Immun. 2000 Feb;68:688–693. doi: 10.1128/iai.68.2.688-693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Takahashi K, Ip WKE, Michelow IC, Ezekowitz RAB. The mannose-binding lectin: a prototypic pattern recognition molecule. Curr Opin Immunol. 2006;18:16–23. doi: 10.1016/j.coi.2005.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kang JH, Super M, Yung CW, Cooper RM, Domansky K, Graveline AR, Mammoto T, Berthet JB, Tobin H, Cartwright MJ, Watters AL, Rottman M, Waterhouse A, Mammoto A, Gamini N, Rodas MJ, Kole A, Jiang A, Valentin TM, Diaz A, Takahashi K, Ingber DE. An extracorporeal blood-cleansing device for sepsis therapy. Nat Med. 2014;20:1211–1216. doi: 10.1038/nm.3640. [DOI] [PubMed] [Google Scholar]
- 87.Kang JH, Um E, Diaz A, Driscoll H, Rodas MJ, Domansky K, Watters AL, Super M, Stone HA, Ingber DE. Optimization of pathogen capture in flowing fluids with magnetic nanoparticles. Small. 2015;11:5657–5666. doi: 10.1002/smll.201501820. [DOI] [PubMed] [Google Scholar]
- 88.Leevy WM, Johnson JR, Lakshmi C, Morris J, Marquez M, Smith BD. Selective recognition of bacterial membranes by zinc( II)-coordination complexes. Chem Commun. 2006:151595–151597. doi: 10.1039/b517519d. [DOI] [PubMed] [Google Scholar]
- 89.Matosziuk LM, Harney AS, MacRenaris KW, Meade TJ. Synthesis, characterization, and in vitro testing of a bacteria-targeted MR contrast agent. Eur J Inorg Chem. 2012:2099–2107. doi: 10.1002/ejic.201101362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gao M, Hu QL, Feng GX, Tomczak N, Liu R, Xing B, Tang BZ, Liu B. A multifunctional probe with aggregation-induced emission characteristics for selective fluorescence imaging and photodynamic killing of bacteria over mammalian cells. Adv Healthc Mater. 2015;4:659–663. doi: 10.1002/adhm.201400654. [DOI] [PubMed] [Google Scholar]
- 91.Lee JJ, Jeong KJ, Hashimoto M, Kwon AH, Rwei A, Shankarappa SA, Tsui JH, Kohane DS. Synthetic ligand-coated magnetic nanoparticles for microfluidic bacterial separation from blood. Nano Lett. 2014;14:1–5. doi: 10.1021/nl3047305. [DOI] [PubMed] [Google Scholar]
- 92.Wadstrom T, Ljungh A. Glycosaminoglycan-binding microbial proteins in tissue adhesion and invasion: key events in microbial pathogenicity. J Med Microbiol. 1999;48:223–233. doi: 10.1099/00222615-48-3-223. [DOI] [PubMed] [Google Scholar]
- 93.Chu YW, Engebretson DA, Carey JR. Bioconjugated magnetic nanoparticles for the detection of bacteria. J Biomed Nanotechnol. 2013;9:1951–1961. doi: 10.1166/jbn.2013.1701. [DOI] [PubMed] [Google Scholar]
- 94.Herrmann IK, Urner M, Graf S, Schumacher CM, Roth-Z’graggen B, Hasler M, Stark WJ, Beck-Schimmer B. Endo-toxin removal by magnetic separation-based blood purification. Adv Healthc Mater. 2013 Jun;2:829–835. doi: 10.1002/adhm.201200358. [DOI] [PubMed] [Google Scholar]
- 95.Wang S, Inci F, Chaunzwa TL, Ramanujam A, Vasudevan A, Subramanian S, Chi Fai Ip A, Sridharan B, Gurkan UA, Demirci U. Portable microfluidic chip for detection of Escherichia coli in produce and blood. Int J Nanomed. 2012;7:2591–2600. doi: 10.2147/IJN.S29629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Beveridge JS, Stephens JR, Williams ME. The use of magnetic nanoparticles in analytical chemistry. Annu Rev Anal Chem. 2011;4:251–273. doi: 10.1146/annurev-anchem-061010-114041. [DOI] [PubMed] [Google Scholar]
- 97.Eickenberg B, Meyer J, Helmich L, Kappe D, Auge A, Weddemann A, Wittbracht F, Hïutten A. Lab-on-a-chip magneto- immunoassays: how to ensure contact between superparamagnetic beads and the sensor surface. Biosensors. 2013;3:327–340. doi: 10.3390/bios3030327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rife JC, Miller MM, Sheehan PE, Tamanaha CR, Tondra M, Whitman LJ. Design and performance of GMR sensors for the detection of magnetic microbeads in biosensors. Sensor Actuat A. 2003;107:209–218. [Google Scholar]
- 99.Mulvaney SP, Cole CL, Kniller MD, Malito M, Tamanaha CR, Rife JC, Stanton MW, Whitman LJ. Rapid, femtomolar bioassays in complex matrices combining microfluidics and magnetoelectronics. Biosens Bioelectron. 2007;23:191–200. doi: 10.1016/j.bios.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 100.Didar TF, Cartwright MJ, Rottman M, Graveline AR, Gamini N, Watters AL, Leslie DC, Mammoto T, Rodas MJ, Kang JH, Waterhouse A, Seiler BT, Lombardo P, Qendro EI, Super M, Ingber DE. Improved treatment of systemic blood infections using antibiotics with extracorporeal opsonin hemoadsorption. Biomaterials. 2015;67:382–392. doi: 10.1016/j.biomaterials.2015.07.046. [DOI] [PubMed] [Google Scholar]
- 101.Sen A, Harvey T, Clausen J. A microsystem for extraction, capture and detection of E. Coli O157:H7. Biomed Microdevices. 2011;13:705–715. doi: 10.1007/s10544-011-9540-8. [DOI] [PubMed] [Google Scholar]
- 102.Sugiura M, Mitaka C, Haraguchi G, Tomita M, Inase N. Polymyxin B-immobilized fiber column hemoperfusion mainly helps to constrict peripheral blood vessels in treatment for septic shock. J Intensive Care. 2015;3:1–7. doi: 10.1186/s40560-015-0080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hirasawa H. Indications for blood purification in critical care. In: Hiromichi S, Hiroyuki H, editors. Acute Blood Purification. Vol. 166. Basel, Karger: 2010. pp. 21–30. [DOI] [PubMed] [Google Scholar]
- 104.Axelsson J, Ferreira M, Adolfsson L, McCrea K, Ward R, Larm O. Cytokines in blood from septic patients interact with surface-immobilized heparin. ASAIO J. 2010;56:48–51. doi: 10.1097/MAT.0b013e3181c3fec8. [DOI] [PubMed] [Google Scholar]
- 105.Mattsby-Baltzer I, Bergstrom T, McCrea K, Ward R, Adolfsson L, Larm O. Affinity apheresis for treatment of bacteremia caused by Staphylococcus aureus and/or methicillin-resistant S. aureus(MRSA) J Microbiol Biotechnol. 2011;21:659–664. [PubMed] [Google Scholar]
- 106.Reschiglian P, Zattoni A, Roda B, Michelini E, Roda A. Field-flow fractionation and biotechnology. Trends Biotechnol. 2005;23:475–483. doi: 10.1016/j.tibtech.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 107.Caldwell KD, Cheng ZQ, Hradecky P, Giddings JC. Separation of human and animal-cells by steric field-flow fractionation. Cell Biophys. 1984;6:233–251. doi: 10.1007/BF02788630. [DOI] [PubMed] [Google Scholar]
- 108.Piacentini N, Mernier G, Tornay R, Renaud P. Separation of platelets from other blood cells in continuous-flow by dielectrophoresis field-flow-fractionation. Biomicrofluidics. 2011;5:1–9. doi: 10.1063/1.3640045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ibrahim T, Battu S, Cook-Moreau J, Cardot P. Instrumentation of hollow fiber flow field flow fractionation for selective cell elution. J Chromatogr B. 2012;901:59–66. doi: 10.1016/j.jchromb.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 110.Janca J, Halabalova V, Ruzicka J. Role of the shape of various bacteria in their separation by microthermal field-flow fractionation. J Chromatogr A. 2010;1217:8062–8071. doi: 10.1016/j.chroma.2010.10.082. [DOI] [PubMed] [Google Scholar]
- 111.Berg HC, Turner L. Selection of motile nonchemotactic mutants of Escherichia Coli by field-flow fractionation. Proc Natl Acad Sci USA. 1991;88:8145–8148. doi: 10.1073/pnas.88.18.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Saenton S, Lee H, Gao YS, Ranville JF, Williams SKR. Evaluation of different field-flow fractionation techniques for separating bacteria. Sep Sci Technol. 2000;35:1761–1775. [Google Scholar]
- 113.Khoshmanesh A, Sharma R, Beckett R. Biomass of sediment bacteria by sedimentation field-flow fractionation. J Environ Eng. 2001;127:19–25. [Google Scholar]
- 114.Reschiglian P, Zattoni A, Roda B, Cinque L, Melucci D, Min BR, Moon MH. Hyperlayer hollow-fiber flow field-flow fractionation of cells. J Chromatogr A. 2003;985:519–529. doi: 10.1016/s0021-9673(02)01458-9. [DOI] [PubMed] [Google Scholar]
- 115.Gallet S, Metreau JM, Loiseau PM, Bories C, Cardot PJP. Isolation of bloodstream trypanosomes by sedimentation field-flow fractionation. J Microcolumn. 1997;9:469–477. [Google Scholar]
- 116.Merino A, Bories C, Gantier JC, Cardot PJP. Isolation of microfilariae from blood by gravitational field-flow fractionation. J Chromatogr. 1991;572:291–301. doi: 10.1016/0378-4347(91)80493-v. [DOI] [PubMed] [Google Scholar]
- 117.Bouamrane F, Assidjo NE, Bouteille B, Dreyfuss MF, Darde ML, Cardot PJP. Sedimentation field-flow fractionation application to Toxoplasma gondii separation and purification. J Pharmaceut Biomed. 1999;20:503–512. doi: 10.1016/s0731-7085(99)00049-7. [DOI] [PubMed] [Google Scholar]
- 118.Jubery TZ, Srivastava SK, Dutta P. Dielectrophoretic separation of bioparticles in microdevices: a review. Electrophoresis. 2014;35:691–713. doi: 10.1002/elps.201300424. [DOI] [PubMed] [Google Scholar]
- 119.Cheng IF, Chen TY, Lu RJ, Wu HW. Rapid identification of bacteria utilizing amplified dielectrophoretic force-assisted nanoparticle-induced surface-enhanced Raman spectroscopy. Nanoscale Res Lett. 2014;9:1–8. doi: 10.1186/1556-276X-9-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cheng IF, Lin CC, Lin DY, Chang H. A dielectrophoretic chip with a roughened metal surface for on-chip surface-enhanced Raman scattering analysis of bacteria. Biomicrofluidics. 2010;4:1–12. doi: 10.1063/1.3474638. [DOI] [PMC free article] [PubMed] [Google Scholar]