With structural glycoscience finally gaining popularity, the need for a clear way of depicting glycans and their interactions in three dimensions is more pressing than ever. Here the Glycoblocks representation is introduced, which combines a simplified bonding-network depiction with the familiar two-dimensional glycan notation brought into three-dimensions.
Keywords: Glycoblocks, molecular graphics, glycans, interactions, CCP4mg, Privateer, carbohydrates, three-dimensional representations
Abstract
The close-range interactions provided by covalently linked glycans are essential for the correct folding of glycoproteins and also play a pivotal role in recognition processes. Being able to visualise protein–glycan and glycan–glycan contacts in a clear way is thus of great importance for the understanding of these biological processes. In structural terms, glycosylation sugars glue the protein together via hydrogen bonds, whereas non-covalently bound glycans frequently harness additional stacking interactions. Finding an unobscured molecular view of these multipartite scenarios is usually far from trivial; in addition to the need to show the interacting protein residues, glycans may contain many branched sugars, each composed of more than ten non-H atoms and offering more than three potential bonding partners. With structural glycoscience finally gaining popularity and steadily increasing the deposition rate of three-dimensional structures of glycoproteins, the need for a clear way of depicting these interactions is more pressing than ever. Here a schematic representation, named Glycoblocks, is introduced which combines a simplified bonding-network depiction (covering hydrogen bonds and stacking interactions) with the familiar two-dimensional glycan notation used by the glycobiology community, brought into three dimensions by the CCP4 molecular graphics project (CCP4mg).
1. Introduction
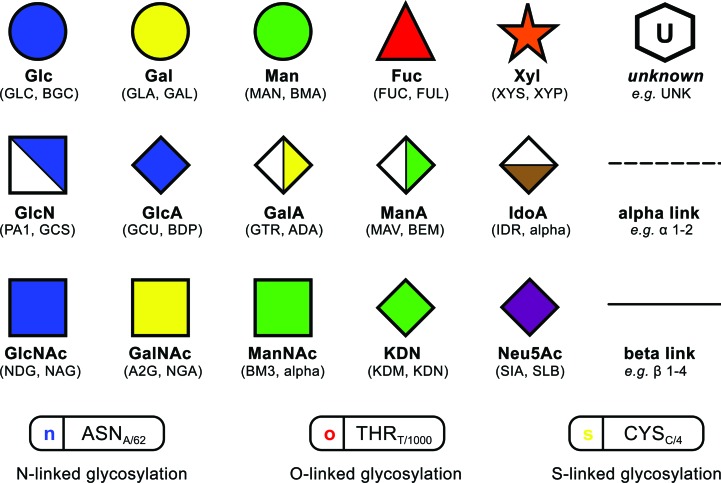
Unlike proteins or nucleic acids, polysaccharides are frequently branched and in addition have two alternative configurations in their glycosidic linkages. While this imposes considerable restrictions on their three-dimensional conformations, it is precisely this nonlinear nature of glycans that poses a challenge in terms of two-dimensional representation. A number of sequence formats (e.g. LINUCS, Bohne-Lang et al., 2001 ▸; GLYCO-CT, Herget et al., 2008 ▸) have been developed for creating textual renditions of branched polysaccharides, each with their respective strengths and pitfalls. Many of the more complex formats are particularly well suited for conveying identification results from techniques such as mass spectrometry, and have been successfully used for mapping glycan sequences to proteoglycan structure entries in the Protein Data Bank (Berman et al., 2003 ▸), effectively bridging the gap between two- and three-dimensional information (Campbell et al., 2014 ▸). While most sequence formats provide machine-readable, univocal descriptions of glycan sequences, graphical conventions are better suited for human interaction and visualization. The graphical convention first introduced by Kornfeld et al. (1978 ▸) gained widespread popularity after being standardised (Varki et al., 1999 ▸) and perfected (Varki et al., 2009 ▸, 2015 ▸) to match the needs of the glycobiology community. This convention (hereafter termed the ‘Essentials’ notation) assigns a colour to the different stereochemistries occurring in glycans (e.g. blue, glucose; green, mannose; yellow, galactose; see Fig. 1 ▸) and identifies the different sugar types by altering each block’s shape (e.g. square, amino sugars; diamond, acidic sugars; or a white hexagon for unknown sugars; see Fig. 1 ▸), with α and β bonds being depicted as dashed and continuous lines respectively, a feature adopted from Oxford nomenclature (Harvey et al., 2009 ▸). Also, connecting lines may be oriented according to the ring position where the linkage starts (e.g. a 45° rotation for a 1–6 link). Nowadays, it is even possible to employ the Essentials convention to search databases, such as UniCarbKB (Campbell et al., 2014 ▸) or glycosciences.de (Loss & Lütteke, 2015 ▸), for particular glycans through the use of graphical tools such as GlycanBuilder (Damerell et al., 2012 ▸).
Figure 1.
Legend to the two-dimensional representation as drawn by Privateer, and its correspondence to three-letter codes from the PDB Chemical Component Dictionary. The current version of Privateer adopts all features of the Essentials notation except the bond–angle relation, which will be available in a forthcoming update to the software. Those sugars typically found in both anomeric forms in covalently bound glycans have both three-letter codes assigned to the same shape, e.g. GLC (α-d-glucopyranose) and BGC (β-d-glucopyranose) to a blue circle. The anomeric form is mentioned explicitly for those cases where just one form is present in the PDB. As the SVG file format supports tooltips (messages that get displayed when the mouse hovers a graphical component), all information related to the linkages is displayed there in order to keep the diagrams minimal. This figure features all three-letter codes recognised by Glycoblocks up to the date of this publication.
With most of the biotechnological interest in glycosylation of proteins focused on how and where ligand carbohydrates appear in a protein–sugar complex structure, the important interactions provided by covalently attached glycans are often overlooked. These contacts have clear implications on correct glycoprotein folding and on protein–glycan and glycan–glycan recognition, for example having a clear impact on the therapeutic effects of antibodies. Their importance has been evident for several years (Sinclair & Elliott, 2005 ▸), as they can show how and why the glycans are required. To some extent, it is possible to visualise them in two dimensions by using ligand-focused software such as Ligplot+ (Laskowski & Swindells, 2011 ▸), or by accessing the online facilities provided by the PDB (Stierand & Rarey, 2010 ▸). Nevertheless, the number of contacts provided by glycosylation trees (which can be composed of more than ten monosaccharide units and may establish contacts with other equally complex glycans) limits this in practice to simple cases. As further a complication, glycans may also bind to aromatic residues (reviewed in Hudson et al., 2015 ▸), which frequently line active sites in carbohydrate-active enzymes (Lombard et al., 2014 ▸). Such well defined stacking interactions are visually evident but rarely identified in graphics programs, requiring scientists to follow complicated bespoke protocols for their depiction (e.g. creating two dummy atoms at the centre of each ring system and drawing a line between them).
Extracting visual information from densely populated scenarios requires simplification, i.e. not all information is relevant at the same time. Conceptualization in three dimensions has been successfully implemented already with the ribbon diagram (Richardson, 1985 ▸), which is the representation of choice for proteins, and has been implemented with minor local variations in all major structure visualization software. Perhaps unsurprisingly, carbohydrates have not been so fortunate in this regard and up until now only a handful of programs have introduced ad-hoc solutions that simplify sugar representation. For example, SweetUnityMol (Pérez, Tubiana et al., 2015 ▸) converts monosaccharides into textured hexagonal shapes, which are then coloured based on an expansion of the Essentials colour scheme, while the Azahar plugin for PyMOL (Schrodinger, 2015 ▸) also produces hexagonal shapes that can be coloured based on a set of predefined choices. However, to the best of our knowledge none of these programs are able to produce shapes that match those specified in the standard Essentials notation, nor have they extended functionality for simplifying complex interactions.
Here, we introduce a schematic three-dimensional representation that minimises graphic complexity while retaining the visual identification, spatial orientation and branching structure of the monosaccharides. This feature, named Glycoblocks, is available as part of CCP4mg, the CCP4 Molecular Graphics program (McNicholas et al., 2011 ▸), which is distributed as part of the CCP4 suite (Winn et al., 2011 ▸). CCP4mg, incorporating Privateer (Agirre, Iglesias-Fernández et al., 2015 ▸), a required component, is alternatively available as a standalone program (http://www.ccp4.ac.uk/MG). Glycoblocks aims to provide a simplified view for glycans, similar to what the ribbon diagram achieved for proteins, reducing each entity and interaction to an easily identifiable three-dimensional sketch.
2. Materials and methods
2.1. Graphic conventions
An analysis of N- and O-glycans in the PDB (Agirre, Davies et al., 2015 ▸) performed with the CCP4 program Privateer (Agirre, Iglesias-Fernández et al., 2015 ▸) identified which sugars were present in the deposited structures, generating a list of three-letter codes from the PDB’s chemical component dictionary. Privateer’s most recent version (MKIII) introduced a Python scripting interface for seamless integration into other programs and pipelines (e.g. CCP4mg and CCP4i2, which handle most of their logic in Python code). The functions in this interface produce validation data in extensible markup language (XML) format, with scalable vector graphics (SVG) two-dimensional diagrams of the glycan structures being embedded in the XML output. These diagrams are encoded according to the most recent Essentials notation (Varki et al., 2015 ▸), with dashed lines for α-bonds and continuous lines for β-bonds, a feature that the Essentials system has recently adopted from the Oxford nomenclature (Harvey et al., 2009 ▸). For added interactivity, they are annotated with all the validation information produced by Privateer (checks on stereo- and regiochemistry, ring puckering and conformation, and linkage torsions, all available as a tooltip), and with HTML links containing MMDB (the CCP4 coordinate library; Krissinel et al., 2004 ▸) residue selections, which CCP4mg is able to process in order to focus on the selected sugar upon clicking on a two-dimensional shape (sugar, amino acid or link). These diagrams can be shown by choosing Glycan viewer from the CCP4mg menu.
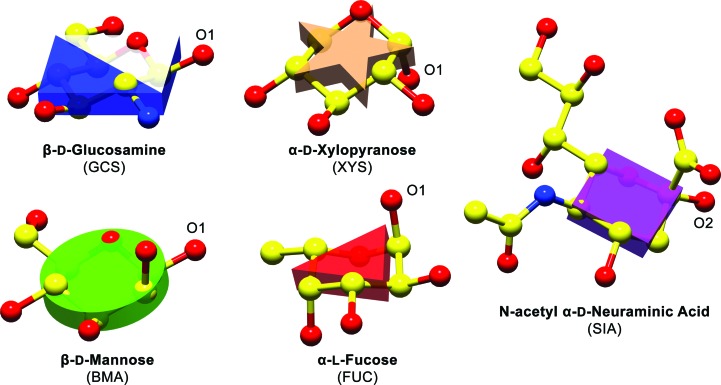
In Glycoblocks the Essentials notation (Varki et al., 2009 ▸, 2015 ▸) has been translated into three-dimensional solids, matching each shape, sugar name and colour with the corresponding three-letter codes recognised by Privateer (Fig. 1 ▸). Each Glycoblock is a vertical extension of the original two-dimensional shape, producing a triangular prism for fucose, a rectangular prism for N-acetylgalactosamine or a cylinder for mannose (Fig. 2 ▸). In order to provide a notion of the particular orientation of the sugars, the Glycoblocks are oriented according to the mean ring plane, defined in the following section.
Figure 2.
Orientation of Glycoblocks with respect to the atomic models they represent. All monosaccharides have been oriented with the oxygen linked to the anomeric carbon (see annotations on the picture) on the right. Despite the d- and l-sugars showing 4C1 and 1C4 conformations, respectively, the orientation of the block remains representative, providing a clear hint at the stereochemistry. For clarity, object outlines and H atoms have been omitted.
As Privateer is able to detect covalently linked N-, O- and S-glycans, CCP4mg shows them automatically in the Glycoblocks representation upon loading the structure of a glycoprotein. Although less frequently used, the representation may also be selected when examining ligand mono- and polysaccharides, provided that they are identified using monosaccharide three-letter codes, i.e. two β1,4-linked glucoses (BGC) and not a single cellobiose entity (CBI). This requirement is not expected to have a negative impact, as the majority of sugar structures have been deposited using the monosaccharide codes. Nevertheless, common di- and oligosaccharides will be added to the Glycoblocks representation in a forthcoming update. While the Essentials notation covers most of the N- and O-glycan-forming carbohydrates, unknown sugars can be depicted as white/grey hexagons (Fig. 1 ▸), with the first letter of the three-letter code being shown in the two-dimensional diagram for quick identification. In CCP4mg, these are shown as grey hexagonal prisms.
2.2. Computing interactions
Hydrogen bonds are depicted using a dashed line from each block’s centre to the Cα of the amino acid with which it interacts in the protein backbone (Figs. 3 to 7), the hydrogen bonds being computed internally by CCP4mg (Potterton et al., 2002 ▸, 2004 ▸; McNicholas et al., 2011 ▸). Covalent bonds, including the protein–glycan ones such as AsnND2–GlcNAcC1, are depicted as solid lines, with each linkage arising from the projected side of the glycoblock. Stacking interactions are computed according to the criterion defined by Hudson et al. (2015 ▸), whereby interaction distances must fall within a 4 Å limit and the angle formed by vectors orthogonal to the aromatic and mean carbohydrate planes must not exceed 30°. These are depicted as red dashed lines between each ring’s centre of mass.
Linkages are determined by chemistry perception instead of relying on the deposited LINK records, as it has been reported that many structures contain wrongly specified links (Lütteke et al., 2004 ▸; Lütteke & Lieth, 2009 ▸). Distances in Å, corresponding to the actual distance between the two atoms forming the bond, and residue numbers can be optionally annotated adjacent to each line, providing quantitative details on the interactions. Thickness and size may be changed for bond cartoons and blocks, with the default values having been optimized for close-up views. For reasons of clarity, the bonding network is not shown by default and has to be activated from the Preferences menu.
2.3. Orientation of the blocks
Let i, j and k represent three consecutive atoms in a sugar ring of size s and 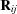 the position vector that goes from atom i to atom j, identifying the bond between both atoms. A vector
the position vector that goes from atom i to atom j, identifying the bond between both atoms. A vector  is then calculated
is then calculated
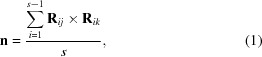 |
with  normal to the plane that will define the block’s orientation.
normal to the plane that will define the block’s orientation.
2.4. Figure preparation
All three-dimensional figures have been produced with CCP4mg (McNicholas et al., 2011 ▸) and all two-dimensional vector diagrams with Privateer (Agirre, Iglesias-Fernández et al., 2015 ▸), as CCP4mg shows interactive versions of these on-screen, but does not offer an option to save them to disk. The Shadows, Occlusion and Object outlines options were activated under the Lighting menu in CCP4mg, and labels were added using the same program. Interactions were represented as dashed cylinders.
3. Discussion
The Glycoblocks representation was originally developed to simplify the visualization of the complex interactions that can be found in heavily glycosylated structures, such as those in Agirre et al. (2016 ▸). Reducing a 12-atom-plus entity to a single block while retaining its overall orientation and link cardinality greatly helps uncluttering the view of a glycan. In addition, depicting covalent, stacking and close-range electrostatic interactions as lines between the blocks and the Cα from the linked residue, allows for the removal of the side chains from complex interaction scenarios. The integration of Glycoblocks into CCP4mg makes it instantly possible to represent interactions between monomers related by crystallographic symmetry, to create movies that can be integrated in slide shows, or to generate stereoviews with an enhanced sense of depth (Agirre et al., 2016 ▸).
The representation has been tested in most practical scenarios with positive results, summarised as follows.
3.1. High-mannose N-glycans
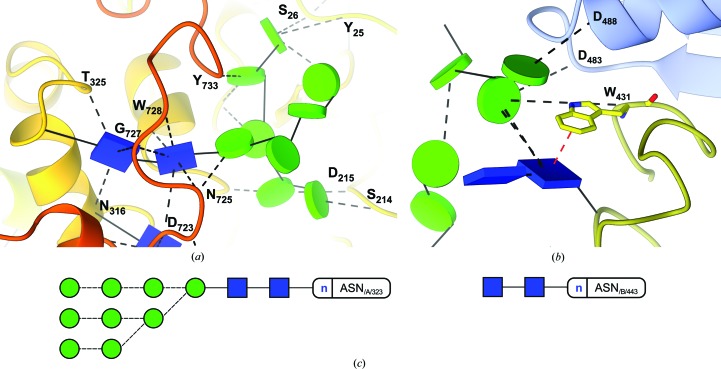
Of simple composition (9 × Man and 2 × GlcNAc, arranged in up to three branches), high-mannose glycans can establish hydrogen bonds with other domains or even chains of the protein to which they are attached, as distant from the original glycosylation point as 30 Å (Fig. 3a ▸). These glycans are typically seen as structural reinforcement for glycoproteins, with mostly intact trees being found linked to asparagine residues in the core of the protein, and shorter trees or even single GlcNAc monosaccharides (possibly a result of the action of endoglycosidases) appearing more frequently towards the surface (Agirre et al., 2016 ▸). While they are far more frequent in binding sites (Hudson et al., 2015 ▸), stacking interactions may play a role on the conformation of glycans too; particular linkage conformations can be enforced when aromatic residues are in the neighbourhood (Fig. 3b ▸). These are depicted in the Glycoblocks representation as a red dashed line, which can be traced to the centre of mass of the aromatic residue or to the Cα, similar to how hydrogen bonds are shown.
Figure 3.
Visualizing interactions with Glycoblocks. In the figure, the structure of a heavily glycosylated fungal glycosylhydrolase (PDB code 5fjj), reported by Agirre et al. (2016 ▸). (a) View of the interactions of a high-mannose tree. The glycan connected to Asn323 is perhaps the only example of a three-dimensional structure of a complete high-mannose tree in the PDB. As the protein part has been coloured in rainbow style, it can immediately be seen that the glycan establishes hydrogen bonds across multiple domains and with other glycans which, in turn, interact with other parts of the protein. (b) Visualizing stacking interactions. The first GlcNAc sugar is linked in a flipped conformation to Asn443 due to the stacking interaction with Trp431 (W431 in the picture). These interactions are depicted in red. (c) Two-dimensional representation by Privateer. Dashed lines indicate an alpha link.
3.2. Plant glycans
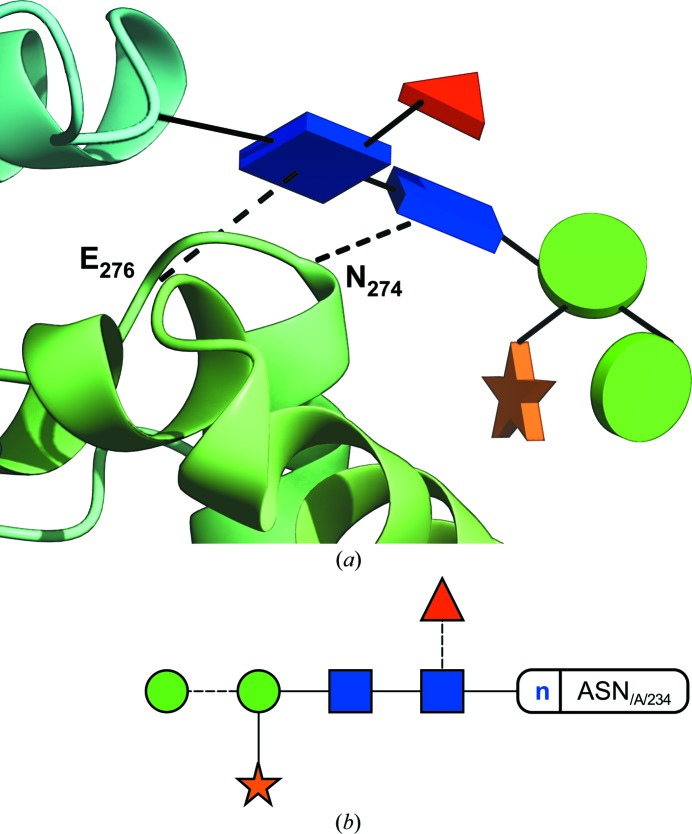
Common plant complex N-glycans include core α1,3-fucose (not to be confused with the core α1,6 linkage found in mammalians) and β1,2-linked xylose saccharides (Fig. 4 ▸), which are added during the final stages of processing in the Golgi apparatus (Strasser, 2014 ▸). Their function remains uncertain, although their tendency to show up on the protein’s surface hints at their potential implications in recognition processes.
Figure 4.
(a) Glycoblocks representation of a plant N-glycan and its interactions (PDB code 5aog). The structure depicted is a cationic class III peroxidase purified from Sorghum bicolor (Nnamchi et al., 2016 ▸), which shows the typical core α1,3-fucosylated glycans covalently attached to it. The two core GlcNAc sugars establish two hydrogen bonds (dashed lines in the three-dimensional view), respectively, to one end of a neighbouring α-helix. (b) Two-dimensional representation produced by Privateer. Dashed lines indicate an α-link.
3.3. Antibodies
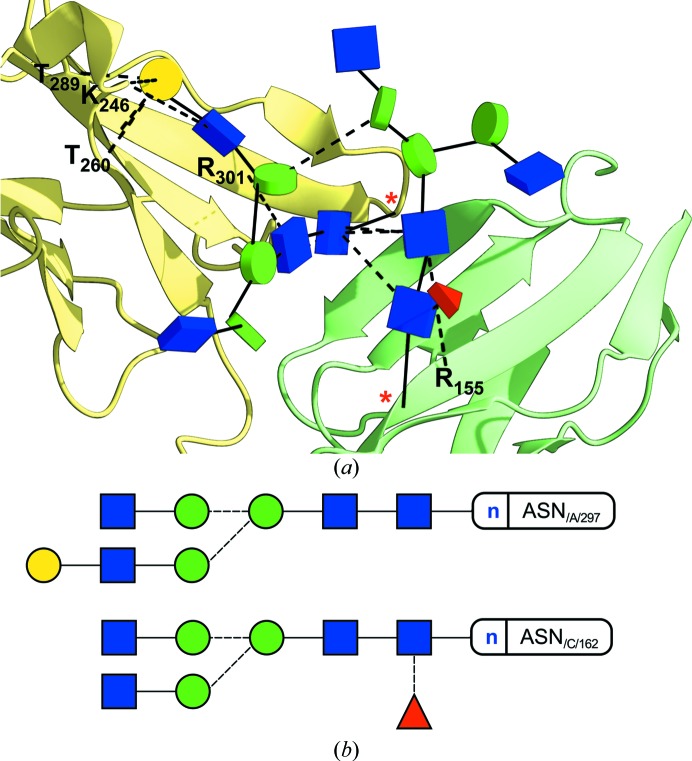
N-glycosylation is a key functional part of antibodies, vital for their structure and interactions and hence for their therapeutic effectiveness. In Fig. 5 ▸, a glyco-engineered Fc fragment lacking core-linked fucose binds to the human Fc γ receptor IIIa (FcγRIIIa) through a network of hydrogen bonds (Mizushima et al., 2011 ▸). This non-fucosylated variant was shown to have stronger affinity to FcγRIIIa because of reduced steric hindrance in the region where the core α1,6-fucose would be found in the fucosylated form. A second example using antibodies (PDB code 4byh; Crispin et al., 2013 ▸) can be found in the supplementary video protocol.
Figure 5.
Glycan–glycan and glycan–protein contacts in an antibody–Fc γ receptor IIIa (FcγRIIIa) complex (PDB code 4a5t). (a) Glycoblocks representation. The non-fucosylated Fc fragment has been coloured in yellow, FcγRIIIa is in green. Most of the contacts that bind the two structures together occur between the glycans themselves. The missing fucose residue would have appeared at the interface between both chains, causing steric hindrance according to the authors (Mizushima et al., 2011 ▸). The glycosylation points (asparagine residues 297 and 162) have been marked with a red asterisk. (b) Two-dimensional representation produced by Privateer. Dashed lines indicate an α-link.
3.4. O-GalNAc glycans
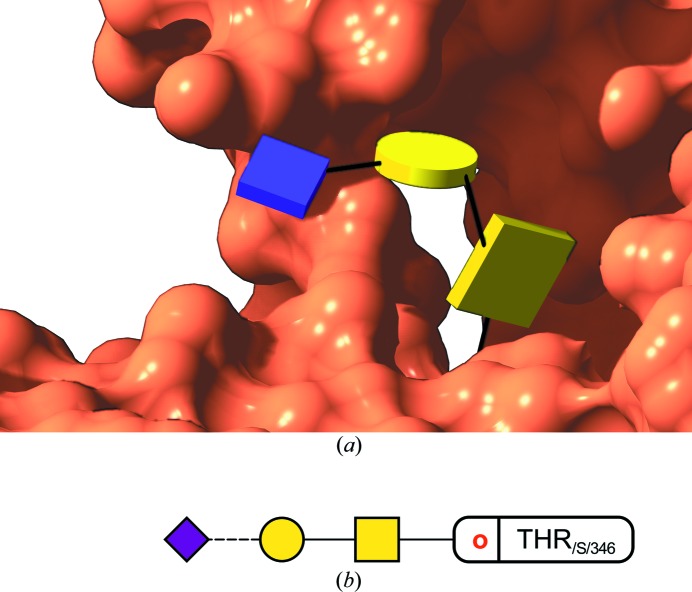
O-glycosylation, only known to be present in eukaryotes, groups the covalent modification of a serine or threonine residue with, most frequently, N-acetylgalactosamine (GalNAc, yellow square in Fig. 1 ▸), although other modifications do exist, such as O-linked mannose, fucose, xylose, galactose, glucose or, notably, the intracellular O-GlcNAc modification with N-acetylglucosamine. O-GalNAc glycans, usually found linked to mucins, have implications in many signalling and communication processes occurring for instance in cancer, including metastasis formation (Pinho & Reis, 2015 ▸). In addition to over- or under-expression, structural changes in O-GalNAc glycans can be associated with certain types of cancer and, therefore, be used as biomarkers for diagnosis (Tuccillo et al., 2014 ▸). A partial O-GalNAc glycan can be seen attached to human native plasminogen (PDB code 4a5t) in Fig. 6 ▸, with sialic acid (Neu5Ac) at its terminus. This structure was determined at low resolution (3.49 Å), and contains modelling errors such as a wrong GalNAc–Thr linkage, which must be α. These problems become apparent in the resulting two-dimensional diagrams, which incorporate anomeric information on the linkages (dashed versus continuous line).
Figure 6.
(a) View of an O-glycan. This shows one of the rare examples of O-glycosylation found in the PDB (code 4a5t, solved to 3.49 Å resolution), reported by Xue et al. (2012 ▸). As can be seen from the two-dimensional diagram (see b), the GalNAc–Thr linkage was originally modelled as β whilst in reality it had to be α. It is only by using all available knowledge of glycochemistry that these mistakes can be avoided, as the fit to a featureless map must always be tightly restrained to what is known in terms of link distances, angles and torsions. (b) Two-dimensional representation by Privateer. The dashed line indicates an α-link.
3.5. Ligand glycans
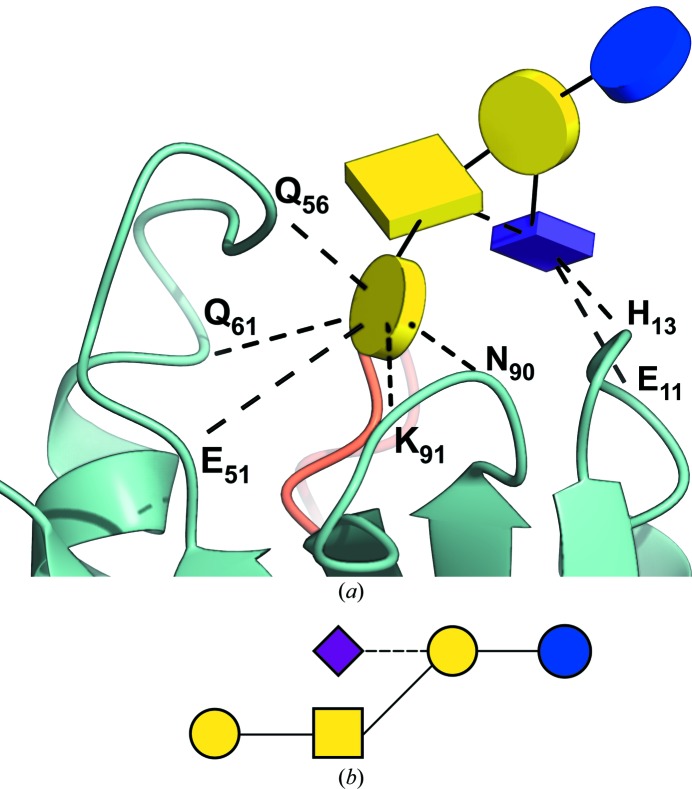
The GM1/choleratoxin B-pentamer complex (Merritt et al., 1994 ▸, 1998 ▸) is a classic example of an intricate bonding network between a ligand glycan and a protein. Despite having waters removed from their original three-dimensional stereographic depiction, the interaction network proved visually challenging to interpret, and had to be explained in an expanded, cleverly drawn planar diagram (Merritt et al., 1994 ▸). In Fig. 7 ▸, a Glycoblocks three-dimensional interpretation of this scenario provides a simplified way of looking at the same interactions, reducing the number of atoms and dashed lines to a minimum and eliminating the need for a stereo figure.
Figure 7.
Visualizing ligand glycans. (a) A simplified three-dimensional view of the interactions between the GM1 pentasaccharide and the subunit B5 of the choleratoxin pentamer (PDB code 3chb), reported in Merritt et al. (1994 ▸) and re-refined in Merritt et al. (1998 ▸). Only direct hydrogen bonds are shown, as waters have been omitted from the picture. The protein part has been coloured by chain. There is an unlabelled hydrogen bond between the GalNAc and Neu5Ac monosaccharides, also drawn as a dashed line. All the depicted interactions, computed on the fly by CCP4mg, match those manually determined in the original research (Merritt et al., 1994 ▸). (b) Two-dimensional representation by Privateer. The dashed line indicates an α-link.
3.6. Analysing NMR structures
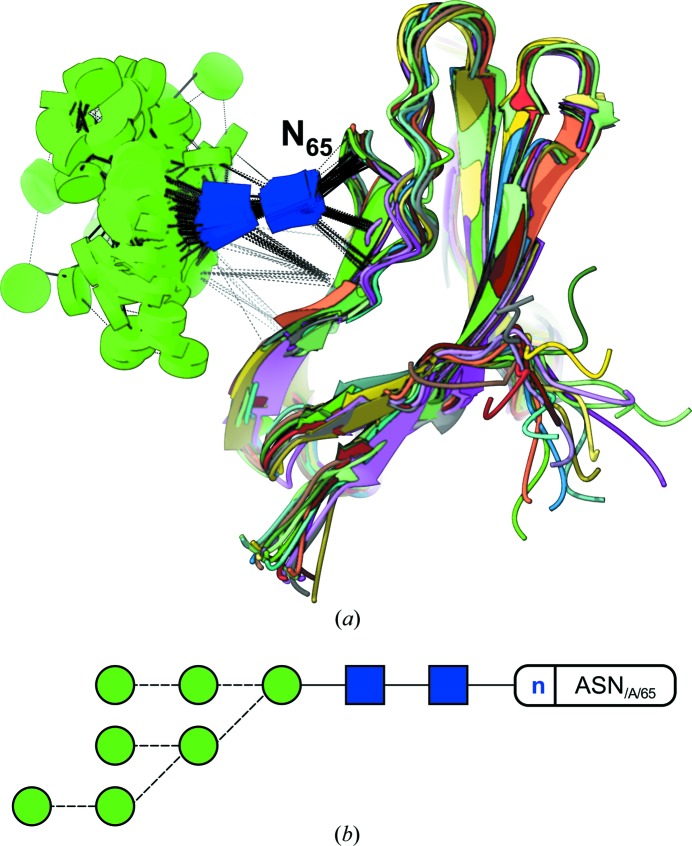
Removing glycans from the surface of a glycoprotein enzymatically (e.g. using EndoH) has become standard practice in X-ray crystallography whenever the first crystallization trials fail. Other techniques, such as NMR, are able to cope with the glycans’ conformational variability and thus represent a suitable alternative for those cases when the external glycans are not an obstacle but the very target of the study, e.g. in those cases where terminal sugars play a central role in molecular recognition (Ardá et al., 2013 ▸; Canales et al., 2013 ▸). An example is shown in Fig. 8 ▸, where the glycan takes part in counterbalancing the positive charge density near the glycosylation point (N65 in the figure) in the adhesion domain of human CD2 (Wyss et al., 1995 ▸).
Figure 8.
Simplifying NMR model representation. (a) Glycoblocks view of a partial high-mannose glycan N-linked to the adhesion domain of human CD2. This lateral view of the glycoprotein allows for an unobscured way of looking at the contacts that occur between sugars, and sugars and protein. While hydrogen bonds keep the two core GlcNAc sugars tied to the protein, the rest of the glycan shows great conformational variability. The protein part has been coloured by model. (b) Two-dimensional representation by Privateer. Dashed lines indicate an α-link.
3.7. Block orientation and stereochemistry
A decision was made neither to use regular polyhedra nor spheres; instead prisms or cylinders are cut thin by two parallel planes (bases) which are orthogonal to the sides. This has two benefits: the sugar’s orientation can be retained in the block representation; and these occupy a similar volume on-screen. The different orientation between two blocks can hint at their linkage’s torsions, and that way unusual linkage conformations can be ascertained from the pictures (see Fig. 3b ▸).
4. Conclusions
The possibility of detecting glycans in structures will enable databases such as the PDB (Berman et al., 2003 ▸) or Glyco3D (Pérez, Sarkar et al., 2015 ▸) to display images in Glycoblocks format whenever glycans are found in a structure. Embedding validation information in non-intrusive tooltips should encourage users to adopt a critical view on the sometimes subjective and debatable interpretations that can be found in the PDB (Lütteke et al., 2004 ▸; Crispin et al., 2007 ▸; Lütteke & Lieth, 2009 ▸; Agirre, Davies et al., 2015 ▸).
Supplementary Material
Video protocol. DOI: 10.1107/S2059798316013553/ba5253sup1.mp4
Acknowledgments
The authors are indebted to Gideon J. Davies, Keith S. Wilson, Saioa Urresti and Kevin D. Cowtan for insightful discussions and critically reading the manuscript. This work was partly supported by the Biotechnology and Biological Sciences Research Council grant and by the Science and Technologies Facilities Council through CCP4.
References
- Agirre, J., Ariza, A., Offen, W. A., Turkenburg, J. P., Roberts, S. M., McNicholas, S., Harris, P. V., McBrayer, B., Dohnalek, J., Cowtan, K. D., Davies, G. J. & Wilson, K. S. (2016). Acta Cryst. D72, 254–265. [DOI] [PMC free article] [PubMed]
- Agirre, J., Davies, G., Wilson, K. & Cowtan, K. (2015). Nat. Chem. Biol. 11, 303. [DOI] [PubMed]
- Agirre, J., Iglesias-Fernández, J., Rovira, C., Davies, G. J., Wilson, K. S. & Cowtan, K. D. (2015). Nat. Struct. Mol. Biol. 22, 833–834. [DOI] [PubMed]
- Ardá, A., Blasco, P., Varón Silva, D., Schubert, V., André, S., Bruix, M., Cañada, F. J., Gabius, H. J., Unverzagt, C. & Jiménez-Barbero, J. (2013). J. Am. Chem. Soc. 135, 2667–2675. [DOI] [PubMed]
- Berman, H., Henrick, K. & Nakamura, H. (2003). Nat. Struct. Biol. 10, 980. [DOI] [PubMed]
- Bohne-Lang, A., Lang, E., Förster, T. & von der Lieth, C. W. (2001). Carbohydr. Res. 336, 1–11. [DOI] [PubMed]
- Campbell, M. P., Peterson, R., Mariethoz, J., Gasteiger, E., Akune, Y., Aoki-Kinoshita, K. F., Lisacek, F. & Packer, N. H. (2014). Nucleic Acids Res. 42, D215–D221. [DOI] [PMC free article] [PubMed]
- Canales, A., Mallagaray, A., Pérez-Castells, J., Boos, I., Unverzagt, C., André, S., Gabius, H. J., Cañada, F. J. & Jiménez-Barbero, J. (2013). Angew. Chem. Int. Ed. 52, 13789–13793. [DOI] [PubMed]
- Crispin, M., Stuart, D. I. & Jones, E. Y. (2007). Nat. Struct. Mol. Biol. 14, 354–355. [DOI] [PubMed]
- Crispin, M., Yu, X. & Bowden, T. A. (2013). Proc. Natl Acad. Sci. USA, 110, E3544–E3546. [DOI] [PMC free article] [PubMed]
- Damerell, D., Ceroni, A., Maass, K., Ranzinger, R., Dell, A. & Haslam, S. M. (2012). Biol. Chem. 393, 1357–1362. [DOI] [PubMed]
- Harvey, D. J., Merry, A. H., Royle, L., Campbell, M. P., Dwek, R. A. & Rudd, P. M. (2009). Proteomics, 9, 3796–3801. [DOI] [PubMed]
- Herget, S., Ranzinger, R., Maass, K., Lieth, C. W., v, & d, (2008). Carbohydr. Res. 343, 2162–2171. [DOI] [PubMed]
- Hudson, K. L., Bartlett, G. J., Diehl, R. C., Agirre, J., Gallagher, T., Kiessling, L. L. & Woolfson, D. N. (2015). J. Am. Chem. Soc. 137, 15152–15160. [DOI] [PMC free article] [PubMed]
- Kornfeld, S., Li, E. & Tabas, I. (1978). J. Biol. Chem. 253, 7771–7778. [PubMed]
- Krissinel, E. B., Winn, M. D., Ballard, C. C., Ashton, A. W., Patel, P., Potterton, E. A., McNicholas, S. J., Cowtan, K. D. & Emsley, P. (2004). Acta Cryst. D60, 2250–2255. [DOI] [PubMed]
- Laskowski, R. A. & Swindells, M. B. (2011). J. Chem. Inf. Model. 51, 2778–2786. [DOI] [PubMed]
- Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M. & Henrissat, B. (2014). Nucleic Acids Res. 42, D490–D495. [DOI] [PMC free article] [PubMed]
- Loss, A. & Lütteke, T. (2015). Methods Mol. Biol. 1273, 87–95. [DOI] [PubMed]
- Lütteke, T., Frank, M. & von der Lieth, C. W. (2004). Carbohydr. Res. 339, 1015–1020. [DOI] [PubMed]
- Lütteke, T. & von der Lieth, C. W. (2009). Methods Mol. Biol. 534, 293–310. [DOI] [PubMed]
- McNicholas, S., Potterton, E., Wilson, K. S. & Noble, M. E. M. (2011). Acta Cryst. D67, 386–394. [DOI] [PMC free article] [PubMed]
- Merritt, E. A., Kuhn, P., Sarfaty, S., Erbe, J. L., Holmes, R. K. & Hol, W. G. (1998). J. Mol. Biol. 282, 1043–1059. [DOI] [PubMed]
- Merritt, E. A., Sarfaty, S., Akker, F. V. D., L’Hoir, C., Martial, J. A. & Hol, W. G. (1994). Protein Sci. 3, 166–175. [DOI] [PMC free article] [PubMed]
- Mizushima, T., Yagi, H., Takemoto, E., Shibata-Koyama, M., Isoda, Y., Iida, S., Masuda, K., Satoh, M. & Kato, K. (2011). Genes Cells, 16, 1071–1080. [DOI] [PMC free article] [PubMed]
- Nnamchi, C. I., Parkin, G., Efimov, I., Basran, J., Kwon, H., Svistunenko, D. A., Agirre, J., Okolo, B. N., Moneke, A., Nwanguma, B. C., Moody, P. C. & Raven, E. L. (2016). J. Biol. Inorg. Chem. 21, 63–70. [DOI] [PMC free article] [PubMed]
- Pérez, S., Sarkar, A., Rivet, A., Breton, C. & Imberty, A. (2015). Methods Mol. Biol. 1273, 241–258. [DOI] [PubMed]
- Pérez, S., Tubiana, T., Imberty, A. & Baaden, M. (2015). Glycobiology, 25, 483–491. [DOI] [PubMed]
- Pinho, S. S. & Reis, C. A. (2015). Nat. Rev. Cancer, 15, 540–555. [DOI] [PubMed]
- Potterton, E., McNicholas, S., Krissinel, E., Cowtan, K. & Noble, M. (2002). Acta Cryst. D58, 1955–1957. [DOI] [PubMed]
- Potterton, L., McNicholas, S., Krissinel, E., Gruber, J., Cowtan, K., Emsley, P., Murshudov, G. N., Cohen, S., Perrakis, A. & Noble, M. (2004). Acta Cryst. D60, 2288–2294. [DOI] [PubMed]
- Richardson, J. S. (1985). Methods Enzymol. 115, 359–380. [DOI] [PubMed]
- Schrodinger, LLC (2015). The pyMOL Molecular Graphics System, Version 1.8.
- Sinclair, A. M. & Elliott, S. (2005). J. Pharm. Sci. 94, 1626–1635. [DOI] [PubMed]
- Stierand, K. & Rarey, M. (2010). ACS Med. Chem. Lett. 1, 540–545. [DOI] [PMC free article] [PubMed]
- Strasser, R. (2014). Front. Plant Sci. 5, 363. [DOI] [PMC free article] [PubMed]
- Tuccillo, F. M., de Laurentiis, A., Palmieri, C., Fiume, G., Bonelli, P., Borrelli, A., Tassone, P., Scala, I., Buonaguro, F. M., Quinto, I. & Scala, G. (2014). Biomed. Res. Int. 2014, 742831. [DOI] [PMC free article] [PubMed]
- Varki, A., Cummings, R. D., Esko, J. D., Freeze, H. H., Hart, G. W. & Marth, J. D. (1999). Essentials of Glycobiology, 1st ed., edited by A. Varki, R. D. Cummings, J. D. Esko, H. H. Freeze, G. W. Hart & J. D. Marth. Plainview, New York: Cold Spring Harbor Laboratory Press.
- Varki, A., Cummings, R. D., Esko, J. D., Freeze, H. H., Stanley, P., Marth, J. D., Bertozzi, C. R., Hart, G. W. & Etzler, M. E. (2009). Proteomics, 9, 5398–5399. [DOI] [PMC free article] [PubMed]
- Varki, A. et al. (2015). Glycobiology, 25, 1323–1324. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Wyss, D. F., Choi, J. S., Li, J., Knoppers, M. H., Willis, K. J., Arulanandam, A. R., Smolyar, A., Reinherz, E. L. & Wagner, G. (1995). Science, 269, 1273–1278. [DOI] [PubMed]
- Xue, Y., Bodin, C. & Olsson, K. (2012). J. Thromb. Haemost. 10, 1385–1396. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video protocol. DOI: 10.1107/S2059798316013553/ba5253sup1.mp4