Abstract
Canonical mutations in IDH1 and IDH2 produce high levels of the R-enantiomer of 2-hydroxyglutarate (R-2HG), which is a competitive inhibitor of αKG-dependent enzymes and a putative oncometabolite. Mutant IDH1 collaborates with HoxA9 to induce monocytic leukemia in vivo. We employed two mouse models and a patient derived AML xenotransplantation (PDX) model to evaluate the in vivo transforming potential of R-2HG, S-2HG, and αKG independent of the mutant IDH1 protein. We show that R-2HG, but not S-2HG or αKG, is an oncometabolite in vivo that does not require the mutant IDH1 protein to induce hyperleukocytosis and to accelerate the onset of murine and human leukemia. Thus, circulating R-2HG acts in a paracrine fashion and can drive the expansion of many different leukemic and preleukemic clones that may express wildtype IDH1, and therefore can be a driver of clonal evolution and diversity. In addition we show that the mutant IDH1 protein is a stronger oncogene than R-2HG alone when comparable intracellular R-2HG levels are achieved. We therefore propose R-2HG independent oncogenic functions of mutant IDH1 that may need to be targeted in addition to R-2HG production to exploit the full therapeutic potential of IDH1 inhibition.
Introduction
Mutations in the metabolic enzymes isocitrate dehydrogenase 1 (IDH1) and isocitrate dehydrogenase 2 (IDH2) are frequently found in several tumors, including acute myeloid leukemia (AML), glioma, chondrosarcoma and intrahepatic cholangiocarcinoma. (1–4) Mutations in IDH1 and IDH2 occur almost exclusively at conserved amino acids (5–7) that affect the active sites where IDH1/2 substrates isocitrate and NADP+ bind. (8–10) The conversion of isocitrate to αKG is impaired in the mutant proteins, resulting in reduced production of αKG and NADPH. (11) Furthermore, mutant IDH1 proteins gain a neomorphic ability to convert αKG to R-2-hydroxyglutarate (R-2HG, also known as D-2HG). (8–10) Almost all patients with canonical IDH1/2 mutations express high levels of intracellular R-2HG, (8) while an increase of the S-enantiomer of 2HG has never been described in AML, glioma, chondrosarcoma or intrahepatic cholangiocarcinoma patients. While αKG is a cofactor for many dioxygenases (12) involved in diverse processes like hypoxic response, nucleic acid repair and modification, fatty acid metabolism, and chromatin modification, (13) it was shown that 2HG is an inhibitor of these dioxygenases. (14) Surprisingly, S-2HG inhibits most dioxygenases more potently than R-2HG. (14–16) Therefore, the pathophysiologic role of R-2HG remains unclear. (17) As was shown previously, mutant IDH1 is not sufficient to cause leukemia as a single oncogenic hit. (18, 19) We found that mutant IDH1 is frequently associated with high expression of HOXA and HOXB cluster genes in AML patients and co-expression of mutant IDH1 with HoxA9 in mouse bone marrow cells induced a rapid monocytic leukemia in mice. (19)
Whether oncometabolites (e.g. R-2HG and/or S-2HG) have a causative function in leukemogenesis/carcinogenesis or rather are only biomarkers for oncogenic IDH remains to be demonstrated. To test this, we performed daily administration of R-2HG, S-2HG and αKG in vivo in the HoxA9 mouse model and additional models containing only wild-type IDH1.
Materials and methods
Retroviral vectors and infection of primary bone marrow cells
Retroviral vectors MSCV-HoxA9-PGKneo, pSF91-IRES-eGFP, pSF91-IDH1mut-IRES-eGFP, pSF91-IDH1wt-IRES-eGFP have been described previously, (19) and MSCV-MLL-AF9-IRES-eGFP was a kind gift from Dr. Florian Kuchenbauer. Primary mouse bone marrow cells from C57BL/6J mice were transduced with helper-free recombinant retrovirus as previously described.(19) In brief, bone marrow cells were harvested from 5-FU (Medac, Hamburg, Germany) treated mice, and were infected first by cocultivation with a HoxA9 viral producer cell line followed by either control (CTL) vector or IDH1wt or IDH1mut viral producer GP+E86 cells. Alternatively, prestimulated bone marrow cells were transduced with MLL-AF9. Cells were then sorted for GFP expression and maintained in 6 ng/ml mIL-3, 10 ng/ml hIL-6, and 20 ng/ml mSCF (Peprotech, Hamburg, Germany).
Bone marrow transplantation, treatment and monitoring of mice
6-8 weeks old female C57BL/6J mice were purchased from Charles River, Sulzfeld, Germany and NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) were purchased from the animal facility of Hannover Medical School and kept in pathogen free conditions at the central animal laboratory of Hannover Medical School. Neither randomization, nor blinding was used since all animal experiments were performed with homogeneous strain, age, and similar variance.
Equal numbers of GFP-expressing, sorted bone marrow cells from C57BL/6J mice were injected intravenously in lethally irradiated syngeneic recipient mice that were irradiated with 800 cGy, accompanied by a life-sparing dose of 1x105 freshly isolated bone marrow cells from syngeneic mice. NSG mice were irradiated with 3 Gy and transplanted with IDH1 wildtype, NPM1 mutated human AML patient cells (PDX-AML). After injection of 1 million cells the mice developed leukemia within 90 days with greater than 95% chimerism, which could engraft secondary recipients. Untransplanted C57BL/6J mice, and mice receiving transplants of HoxA9+CTL or MLL-AF9 transduced bone marrow cells or cells from an AML patient (PDX-AML) were treated once daily with R-2HG disodium salt (H8378, Sigma-Aldrich, Steinheim, Germany), S-2HG disodium salt (90790, Sigma-Aldrich), or α-KG (K-1128, Sigma-Aldrich) intraperitoneally at a dose of 1 mg/mouse or 100 μl of phosphate buffered saline (PBS).
For secondary transplantation, 1 million cells from the bone marrow of primary transplanted and R-2HG, S-2HG, α-KG or PBS treated mice were injected intravenously in lethally irradiated syngeneic recipient mice and subsequently treated with the corresponding metabolite as described above. Engraftment was monitored by tail vein bleeds and FACS analysis of GFP-expressing cells every four weeks as previously described.(19, 20) Blood counts with differential WBC analysis were performed using an ABC Vet Automated Blood counter (Scil animal care company GmbH, Viernheim, Germany). Lineage distribution was determined by FACS analysis (FACS Calibur, Becton Dickinson, Heidelberg, Germany) as previously described. (21)
Antibodies for FACS staining and morphologic analysis
Monoclonal antibodies for lineage staining used were Gr1-PE (clone-RB6-8CS), B220-PE (RA3-6B2), Sca-1-PE (D7), ckit-APC (2B8) and CD45-FITC (HI30) from BD Biosciences, CD11b-APC (M 1/70) and F4/80-PE (BM8) from eBioscience. Antibodies used for the lineage cocktail were PerCP Cy5.5 labelled CD4 (GK1.5) and Ter119 (Ter-119) from Biolegend, B220 (RA3-6B2) and CD3 (145-2C11) from BD Biosciences and Gr-1 (RB6-8C5C), CD8 (53-6.7), IL7R (A7R34), CD11b (M1/70) from eBioscience. Antibodies used for SLAM HSC staining were CD48-APC (HM48-1), CD150-PECy7 (TC-15-12F12.2) from Biolegend and CD244-PE (2B4) from eBioscience. Antibodies used for progenitor staining were CD16/32-PE (93), CD34-biotin (RAM34), Sca1-PECy7 (D7) from eBioscience, cKit-APC (2B8) and Streptavidin-APC-Cy7 from BD Biosciences. Cytospin preparations were stained with Wright-Giemsa stain. The morphology of bone marrow was assessed with an Olympus BX51 (Olympus, Tokyo, Japan) microscope and a 40x/0.75 numerical aperture objective, or a 100x/1.3 numerical aperture objective with Zeiss immersol medium (Zeiss, Jena, Germany). OlympusXC50 (Olympus) and analySIS software (Soft Imaging System, Stuttgart, Germany) were used to capture images.
Patient samples
Diagnostic bone marrow, peripheral blood or serum samples collected from AML patients registered for the multicenter treatment trial AML-SHG 01/99 or 02/95 were analyzed for mutations in IDH1 and 2HG levels as previously described. (6, 19) Five million cells were used per sample for 2HG quantification. Written informed consent was obtained according to the Declaration of Helsinki, and the study was approved by the institutional review board of Hannover Medical School.
2-hydroxyglutarate quantification
Bone marrow cells or serum were isolated from mice either transplanted with IDH1mut expressing cells or treated with metabolites for four weeks. To determine intracellular R- and S-2HG levels, either 10μl serum was directly used or 5- million cells were sonicated for 10 minutes at intervals of 30 seconds on, 30 seconds off at 4°C using the Bioruptor (Diagenode, Liege, Belgium) followed by two rounds of freeze thaw cycles. All analyses were performed on an AB Sciex 4000 Q-trap triple quadruple mass spectrometer (Applied Biosystems, Darmstadt, Germany). LC was performed on a Waters Acquity UPLC BEH C18 analytical column [100 × 2.1 mm (i.d.); 1.7-μm bead size with water–acetonitrile (96.5:3.5 by volume) containing 125 mg/L ammonium formate (pH adjusted to 3.6 by addition of formic acid) as mobile phase as described. (22)
Immunoblotting
Cellular lysates were prepared and immunoblotting was performed as described previously. (19, 20) Primary antibodies used were polyclonal goat anti-HoxA9 (N-20) from Santa Cruz Biotechnology (Heidelberg, Germany) and monoclonal mouse-anti-β-actin (AC-15) from Sigma-Aldrich (Hannover, Germany). Secondary horseradish peroxidase-conjugated antibodies used were bovine-anti-goat (Santa Cruz Biotechnology, Heidelberg, Germany), and goat-anti-mouse (Beckman Coulter, Fullerton, CA, USA).
Cell cycle analysis
For cell cycle analysis mice were injected i.p. with 100 μl BrdU (1mM) at 36, 24 and 12 hours before harvest of cells. 1x107 bone marrow cells per mouse were stained with antibodies specific for cell surface antigens followed by permeabilization, fixation, and staining with an anti-BrdU antibody according to the manufacturers protocol (BD Pharmingen Cat no. 559619).
Gene expression profiling
GFP positive cells were sorted from mouse bone marrow cells four weeks after transplantation/start of treatment and were hybridized to Affymetrix Mouse Genome 430 2.0 GeneChips as previously described. (19) Data were analyzed using R and Bioconductor. Microarray quality was assessed using the ArrayQualityMetrics package.(23) Arrays were background corrected, normalized and summarized using RMA.(24) For unsupervised hierarchical clustering the expression set was filtered based on a log2 (interquartile range) IQR.(25) LIMMA was used to detect differentially expressed probe sets applying Benjamini-Hochberg step up multiple testing correction considering FDR of <0.05 significant.(26) Genome-wide gene expression data from R-2HG treated HoxA9 CTL, IDH1wt and IDH1mut cells were used for gene set enrichment analysis.(27) The Broad Institute GSEA software package was employed for gene set enrichment analysis using gene ontology gene sets from the Molecular Signatures Database (http://www.broad.mit.edu/gsea/msigdb/). Gene expression profiling data can be found at the gene expression omnibus database under accession number GSE77594.
Quantitative RT-PCR
RNA was extracted and reverse transcribed as previously described. (28) Quantitative reverse-transcriptase polymerase chain reaction (RT-PCR) was performed as previously described using SYBR green (Life Technologies, Darmstadt, Germany) for quantification of double stranded DNA on a StepOne Plus cycler (Life Technologies, Darmstadt, Germany). (29) Relative expression was determined with the 2-ΔΔCT method, (30) and the housekeeping gene transcript Abl1 was used to normalize the results.
Reduced representation bisulfite sequencing and analysis of methylome data (RRBS-Seq)
DNA from HoxA9 cells expressing CTL vector, IDH1wt or IDH1mut proteins was used for RRBS library preparation using a published protocol with minor modifications. (31) BSMAP aligner (32) and Lister protocol was used for data analysis. (33) Detailed methods for RRBS and its analysis are provided in the data supplement. RRBS data can be found at the gene expression omnibus database under accession number GSE77828.
Statistical analysis
Pairwise comparisons were performed by Student t test for continuous variables and by chi-squared test for categorical variables. The 2-sided level of significance was set at P less than 0.05. Statistical analyses were performed with Microsoft Excel (Microsoft, Munich, Germany) or GraphPad Prism 5 (GraphPad Software, La Jolla, CA). Graphical representation was prepared using Adobe illustrator CS5.1 (Adobe systems GmbH, Munich, Germany).
The size of animal cohorts was based on our previous study. (19)
Results
2HG, but not S-2HG, collaborates with HoxA9 to induce monocytic leukemia in vivo
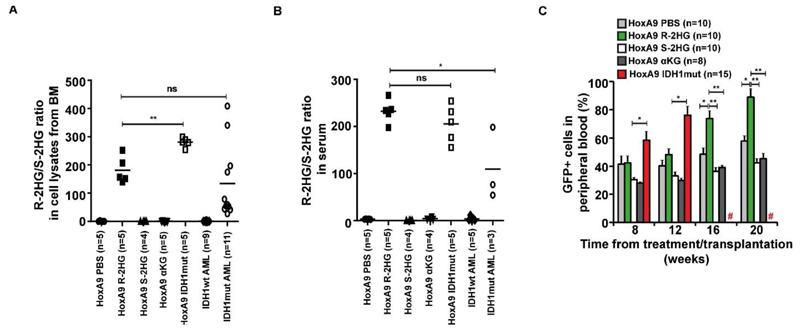
To evaluate the oncogenic properties of 2HG in vivo, mouse bone marrow cells were transduced with HoxA9 (Supplementary Figure S1), selected for GFP expression, and transplanted in lethally irradiated mice (Supplementary Figure S2). We treated these mice once daily with intraperitoneal injections of 1 mg of R-2HG, S-2HG, αKG or 100 μl of PBS. As a control HoxA9+IDH1mut expressing cells were transplanted in recipient mice and were kept without further treatment. At 4 weeks some mice were sacrificed to measure intracellular 2HG levels in bone marrow cells. The ratio of the R- to S-enantiomer is the most reliable measure of R-2HG in cells as it is independent of cell number. This ratio was similar in R-2HG treated mice as in primary human AML cells (Figure 1A and Supplementary Table S1) and slightly higher in serum of mice treated with R-2HG than in serum of AML patients (Figure 1B). The higher concentration in the serum of R-2HG treated mice is not surprising as R-2HG is provided from the extracellular space, while the origin of R-2HG in AML patients is intracellular. S-2HG was increased in cells and serum of S-2HG treated mice (Supplementary Figure S3 and Supplementary Table S1).
Figure 1. R-2HG/S-2HG levels in bone marrow cells of treated mice and engraftment of transduced cells in peripheral blood.
A) Ratio of R-2HG to S-2HG in mouse bone marrow cells transduced with HoxA9 and a control vector or mutant IDH1 and primary AML cells at diagnosis with wildtype or mutant IDH1. Control vector transduced cells were either treated with PBS, R-2HG, S-2HG or αKG (1 mg intraperitoneally per day). Cells were harvested from bone marrow four weeks after treatment or transplantation (HoxA9 IDH1mut). Bars represent mean ratios.
B) Ratio of R-2HG to S-2HG in serum of mice as explained in (B). Bars represent mean ratios.
C) Engraftment of transduced cells in peripheral blood at different time points after start of treatment/transplantation (mean ± SEM).
# indicates all mice are dead, * indicates P<.05, ** indicates P<.01, ns, not significant
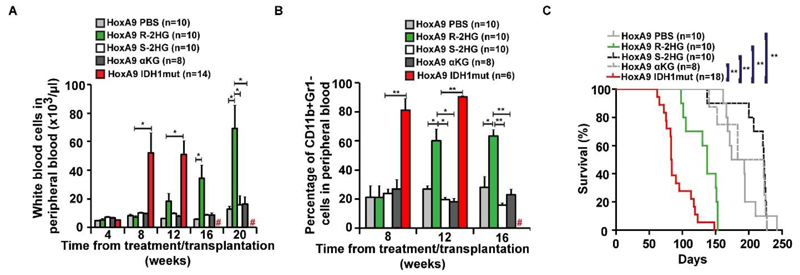
Monitoring of leukemic onset showed that R-2HG treated mice had significantly higher engraftment levels in peripheral blood (Figure 1C), developed striking leucocytosis from 12 weeks onwards (Figure 2A), and had declining hemoglobin and platelets compared to S-2HG, αKG or PBS treated mice (Supplementary Figure S4A and S4B).
Figure 2. R-2HG, but not S-2HG, collaborates with HoxA9 to induce monocytic leukemia.
A) White blood cell count in peripheral blood at different time points after the start of treatment/transplantation (mean ± SEM).
B) Percentage of immature myeloid cells in peripheral blood at different time points after the start of treatment/transplantation (mean ± SEM).
C) Survival of mice in the different treatment and transplantation groups.
# indicates all mice are dead, * indicates P<.05, ** indicates P<.01, ns, not significant
Peripheral blood from R-2HG treated mice revealed significantly more immature CD11b+Gr1- and less mature CD11b+Gr1+ myeloid cells at 12 and 16 weeks after treatment than S-2HG, αKG and PBS treated mice (Figure 2B and Supplementary Figure S4C). Interestingly, the R-2HG treated mice died significantly earlier with a median latency of 137 days post transplantation from monocytic leukemia than mice treated with S2HG, αKG and PBS (median latency of 223, 202 and 184 days respectively; P<.001, Figure 2C; Supplementary Figure S4D shows high engraftment of HoxA9 cells in bone marrow, spleen and peripheral blood at death; Supplementary Figure S4E shows monoblastic cells in bone marrow). As expected, these cells were readily transplantable in secondary animals (Supplementary Figure S5A and S5B).
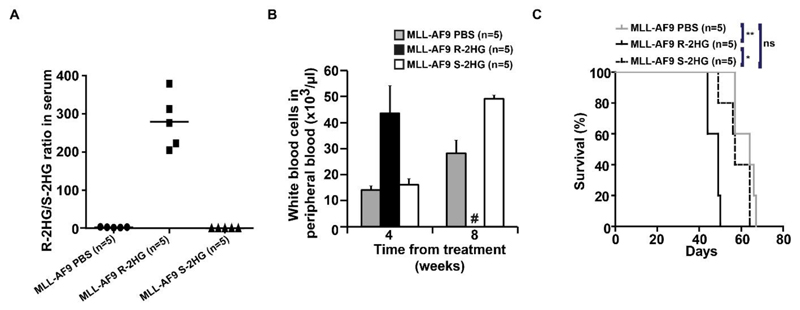
R-2HG, but not S-2HG, accelerates leukemic onset of MLL-AF9 leukemia in vivo
To investigate whether R-2HG is an oncometabolite in vivo independent of HoxA9, we next treated mice transplanted with MLL-AF9 transduced mouse bone marrow cells. R-2HG treated mice had a high R-2HG/S-2HG ratio in serum (Figure 3A). Similarly, S-2HG treated mice had a high S-2HG/R-2HG ratio in serum (Supplementary Figure S6A). R-2HG treatment induced the same effects as in the HoxA9 model with severe leukocytosis and thrombocytopenia and shorter disease latency compared to S-2HG or PBS treated mice (Figure 3B, Supplementary Figure S6B and 3C), whereas hemoglobin levels were similar between the treatment groups (Supplementary Figure S6C). Peripheral blood from R-2HG treated mice had significantly more immature F4/80+CD11b- and less mature F4/80+CD11b+ cells than S-2HG and PBS treated mice at four weeks after treatment (Supplementary Figure S6D). This confirms the oncometabolic potential of R-2HG in an independent murine leukemia model.
Figure 3. R-2HG, but not S-2HG, collaborates with MLL-AF9 to accelerate leukemia onset.
(A) Ratio of R-2HG to S-2HG in serum of mice transplanted with mouse bone marrow cells transduced with MLL-AF9 and either treated with PBS, R-2HG or S-2HG (1 mg intraperitoneally per day). Bars represent mean ratios.
(B) White blood cell count in peripheral blood at different time points after the start of treatment (mean ± SEM).
(C) Survival of mice in the different treatment groups.
# indicates all mice are dead, * indicates P<.05, ** indicates P<.01, ns, not significant
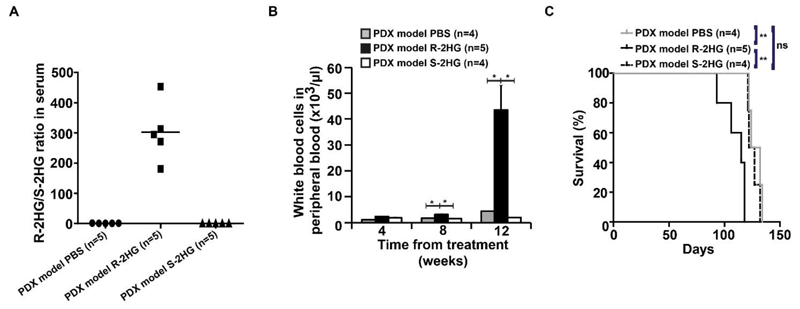
R-2HG, but not S-2HG, accelerates primary human IDH1 wildtype AML in vivo
We next confirmed the oncometabolic role of R-2HG in a patient derived xenotransplantation (PDX) model of human AML. This PDX model was established from an IDH1 wildtype/NPM1 mutant patient. R-2HG treated mice had a high R-2HG/S-2HG ratio in serum (Figure 4A). S-2HG treated mice had a high S-2HG/R-2HG ratio in serum (Supplementary Figure S7A). Again, R-2HG treatment induced significantly higher engraftment of human CD45+ cells and white blood cells at 12 weeks and significantly lower platelet counts at 4, 8 and 12 weeks compared to S-2HG and PBS treated mice (Supplementary Figure S7B, Figure 4B and Supplementary Figure S7C), while hemoglobin levels were similar between the treatment groups (Supplementary Figure S7D). R-2HG treated mice died significantly earlier than S-2HG and PBS treated mice (Figure 4C), confirming the leukemogenic function of R-2HG in primary human cells in vivo.
Figure 4. R-2HG, but not S-2HG accelerates leukemia onset in a primary human AML PDX model.
(A) Ratio of R-2HG to S-2HG in serum of mice transplanted with human IDH1wt patient cells and either treated with PBS, R-2HG or S-2HG (1 mg intraperitoneally per day). Bars represent mean ratios.
(B) White blood cell count in peripheral blood at different time points after the start of treatment (mean ± SEM).
(C) Survival of mice in the different treatment groups.
* indicates P<.05, ** indicates P<.01, ns, not significant
Mutant IDH1 has additional oncogenic potency and function beyond R-2HG
A question of current investigation is whether R-2HG alone is responsible for the oncogenic effects of mutant IDH1 and IDH2 or if the mutant protein contributes additional oncogenic functions. Both cohorts, HoxA9+IDH1mut and HoxA9 treated with R-2HG, developed monocytic leukemia, albeit with different kinetics. The Hoxa9+IDH1mut mice died with a median latency of 83 days while the R-2HG cohort died with a median latency of 137 days post transplantation (P<0.001, Figure 2C). Engraftment in peripheral blood was significantly higher in IDH1mutant than in R-2HG treated mice at 8 and 12 weeks (Figure 1C). In addition, severe leukocytosis, anemia and thrombocytopenia developed earlier in IDH1 mutant mice than in R-2HG treated mice (Figure 2A and Supplementary Figure S4A and S4B). No difference was seen for the frequency of ckit+/Sca1- cells between the treatment groups and mutant IDH1 (Supplementary Figure S8).
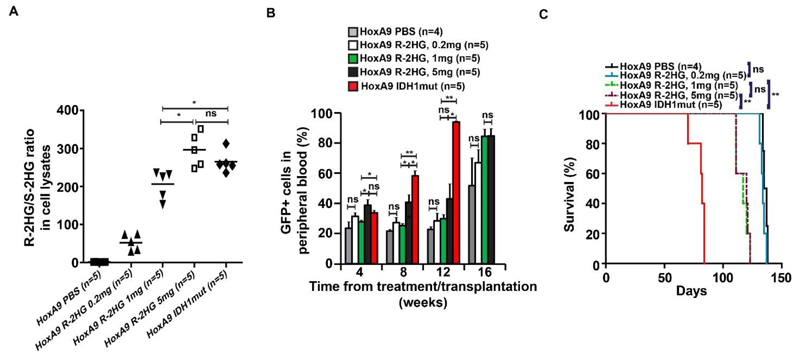
Therefore, we examined whether the faster disease kinetics of mutant IDH1 compared to R-2HG was due to a lower concentration of R-2HG in the treated mice, and would be overcome if the R-2HG dose was increased. Mice transplanted with HoxA9 expressing cells were treated with 0.2, 1 and 5 mg of R-2HG. We observed a dose dependent increase of R-2HG in bone marrow cells of treated mice, while the dose of 5 mg R-2HG per day even slightly exceeded R-2HG of cells expressing mutant IDH1 (Figure 5A). Despite unequal intracellular R-2HG levels, mice treated with 1 or 5 mg of R-2HG had similar engraftment levels and died with an identical latency, while IDH1 mutant mice died significantly earlier and lastly mice treated with 0.2 mg R-2HG died with the same latency as the PBS cohort (Figure 5B and Figure 5C). Thus, we have established non-oncogenic R-2HG levels and identified a ceiling effect for R-2HG beyond which no further oncogenicity is seen. The full oncogenic potential of R-2HG was seen at a dose of 1 mg/day R-2HG, corresponding to an R-2HG/S-2HG ratio of 200 and an R-2HG concentration of 300 μM. Strikingly, at comparable intracellular R-2HG levels IDH1 mutant mice died significantly earlier than R-2HG treated mice strongly suggesting oncogenic functions of mutant IDH1 beyond R-2HG.
Figure 5. Dose dependent leukemogenic effects of R-2HG.
(A) Ratio of R-2HG to S-2HG in mouse bone marrow cells that carried HoxA9 and a control vector or mutant IDH1. Control vector transduced cells were treated with 0.2mg, 1mg or 5mg of R-2HG every day. Bars represent mean ratios.
(B) Engraftment of HoxA9-transduced cells in peripheral blood at different time points after the start of treatment or transplantation (IDH1 mutant cells) (mean ± SEM)
(C) Survival of mice treated with different doses of R-2HG or transplanted with IDH1mut cells (mean ± SEM).
* indicates P<.05, ** indicates P<.01, ns, not significant
R-2HG alone is insufficient to induce myeloproliferation or leukemia in vivo
In order to assess whether R-2HG alone was sufficient as a single hit to induce myeloproliferation or leukemia, normal C57BL/6 mice (without HoxA9) were treated with R-2HG, S-2HG or PBS for eight months. Three mice were sacrificed at 4 weeks of treatment and 2HG levels were quantified in bone marrow cells. High levels of R-2HG were measured in R-2HG treated mice, while high levels of S-2HG were measured in S-2HG treated mice (Supplementary Figure S9A and S9B). No differences were observed for survival (Supplementary Figure S9C), spleen weight (Supplementary Figure S9D), blood counts (Supplementary Figures S9E, S9F and S9G), and immunophenotype in peripheral blood (Supplementary Figure S9H) and bone marrow (Supplementary Figure S10) after 8 months of treatment. We found comparable frequencies of stem and progenitor cells (lin-ckit+sca1+, CMP, GMP and MEP) in bone marrow between treatment and control groups at 4 months of treatment (Supplementary Figure S9I) and a similar cell cycle distribution between these cell types (Supplementary Figure S11). This data shows that the metabolite R-2HG, but not S-2HG, has oncogenic potential in vivo in cooperation with HoxA9 like the IDH1 mutant protein but is insufficient to induce leukemia by itself.
Mutant IDH1 and R-2HG have similar and distinct effects on gene expression and DNA methylation
To identify similarities and potential differences between IDH1 mutant and R-2HG-treated HoxA9 cells, we compared gene expression and DNA methylation profiles of bone marrow cells harvested 4 weeks after transplantation/treatment. Based on the gene expression data R-2HG treated cells clustered with IDH1 mutant cells, while IDH1 wildtype cells clustered with HoxA9 control cells (Figure 6A, Supplementary Table S2). However, many genes that were overexpressed in IDH1 mutant cells were not upregulated in R-2HG treated cells and principal component analysis showed that R-2HG treated cells were at an intermediate stage between control/IDH1 wildtype cells on the one hand and IDH1 mutant cells on the other hand (Figure 6A and Supplementary Figure S12). By gene set enrichment analysis using gene ontology gene sets we found that 16% of the enriched gene ontology terms were shared between IDH1 mutant and R-2HG treated cells (Figure 6B and Supplementary Table S3). We selected genes that had a role in myeloid differentiation and were downregulated in R-2HG treated and IDH1mut cells, and validated their expression by real time RT-PCR (Figure 6C). Expression of Myadm and Ccl6 were most strongly downregulated, confirming a differentiation inhibiting effect of R-2HG.
Figure 6. Gene expression profiles suggest similarities of the mutant IDH1 protein compared to the R-2HG oncometabolite.
A) The Top 100 significantly differentially expressed genes between IDH1 mutant and IDH wiltype were used for hierarchical clustering of control, IDH1 wildtype, IDH mutant or R-2HG treated cells.
B) Overlap of top 100 enriched gene ontology gene sets from gene set enrichment analysis in IDH1 mutant and R-2HG treated cells.
C) Gene expression levels determined by RT-PCR relative to the housekeeping gene Abl and was normalized to gene expression in HoxA9 CTL cells (mean ± SEM).
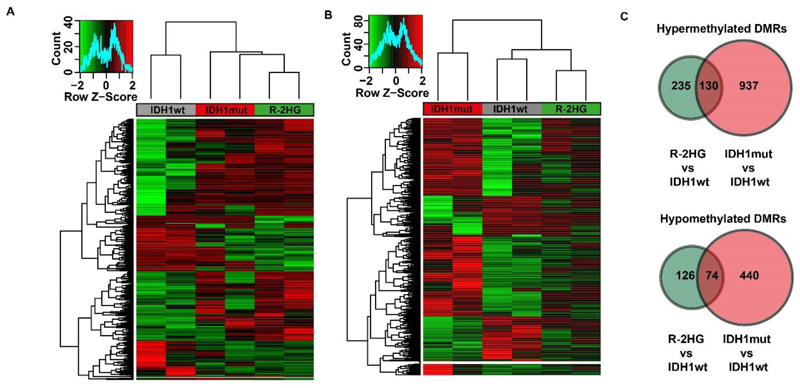
Next, we compared DNA methylation in IDH1 wildtype, mutant and R-2HG treated cells by reduced representation bisulfite sequencing using bone marrow cells harvested 4 weeks after transplantation/treatment that were sorted for transgene expression (Supplementary Figure S13). The differentially methylated regions (DMRs) between R-2HG treated cells and IDH1 wildtype cells were well represented in IDH1 mutant cells (IDH1 mutant cells cluster together with R-2HG treated cells, Figure 7A and Supplementary Table S4). However, DMRs between IDH1 mutant and IDH1 wildtype cells were not well represented in R-2HG treated cells and these cells clustered with IDH1 wildtype and not IDH1 mutant cells (Figure 7B and Supplementary Table S5). The higher number of hyper- and hypomethylated regions in IDH1 mutant cells compared to R-2HG treated cells as shown in Figure 7C, supports an additional function of the mutant protein on transcriptional regulation and DNA methylation beyond the function of the metabolite R-2HG and may explain the earlier disease onset of IDH1 mutant cells compared to R-2HG treated cells.
Figure 7. Methylation profiles suggest additional functions of the mutant IDH1 protein compared to the R-2HG oncometabolite.
A) Heat map based on differentially methylated CpGs between R-2HG treated and IDH1 wildtype cells showing the clustering of IDH1 mutant cells with R-2HG samples.
B) Heat map based on differentially methylated CpGs between IDH1 mutant and wildtype cells showing the clustering of R-2HG treated cells with the IDH wildtype samples.
C) Venn diagram of hypermethylated DMRs (upper panel) and hypomethylated (lower panel) between R-2HG treated and IDH1 mutant cells.
Discussion
We show that R-2HG, but not S-2HG, promotes leukemogenesis in murine and human pre-transformed cells in vivo. Previously, it has been shown that R-2HG can recapitulate properties of mutant IDH1 in vitro like enhanced proliferation, blocked differentiation and histone and DNA methylation, (14, 18, 34, 35) but the unique oncogenic potential of R-2HG compared to S-2HG had not been evaluated in vivo. It has been puzzling why all 2HG-producing tumors expressed R-2HG but not S-2HG. Our findings offer an explanation to this phenomenon as R-2HG treatment provided a competitive advantage to immortalized myeloid progenitor cells in vivo. Our findings also show that R-2HG not only acts in an autocrine fashion on the cells which contain a mutated IDH1 protein and therefore produce R-2HG, but also in a paracrine fashion on cells that are susceptible to transformation by R-2HG. This suggests that high R-2HG levels also affect the growth of IDH1 wildtype clones that may be present in IDH1 mutant tumors and may enhance clonal diversity and evolution.
2HG inhibits many αKG-dependent dioxygenases, many of which are more strongly inhibited by S-2HG than by R-2HG.(14–16) Our data suggests that dioxygenases that are equally or more efficiently inhibited by R-2HG than S-2HG are most relevant for leukemogenesis like the H3K4 and H3K36 demethylases KDM4A and KDM4C and the mitochondrial electron transport component cytochrome c oxidase (COX).(15, 34, 36) On the other hand, enzymes that are more potently inhibited by S-2HG than by R-2HG like TET2 or BBOX1 (14, 15) seem to play a minor role in IDH1-induced transformation. In contrast to R-2HG being oncogenic, Losman et al. have postulated that S-2HG can antagonize the transforming effects of TET2 knockdown in TF1 cells, (35) thus assigning a tumor suppressive role to S-2HG. However, in the context of HoxA9, MLL-AF9 and primary human AML cells, S-2HG treatment neither accelerated nor delayed disease onset in our in vivo model, suggesting that S-2HG is neutral towards leukemogenesis.
Our data confirm previous observations that mutant IDH1 requires a collaborating oncogene to induce leukemia or glioma (18, 19, 37), as R-2HG treatment had no measurable effect in normal mice during the observation period of 8 months. The low oncogenic potential of R-2HG in normal tissues is further supported by the observation that children with hydroxyglutaric aciduria never develop tumors or leukemia. (38) Hydroxyglutaric aciduria is a metabolic disorder, which presents with neurologic symptoms like seizures, developmental retardation, ataxia and others in young children. (38) The limited survival of these patients may not provide the time frame that is required to develop mutations which can synergize with high R-2HG levels.
The mutant IDH1 protein induced leukemia more rapidly than R-2HG in HoxA9 cells, even at comparable doses of R-2HG. We show that the R-2HG-induced gene expression and methylation changes are mostly preserved in mutant IDH1 cells, while the mutant IDH1 protein induces broader gene expression and methylation changes. Gene ontology gene sets that were shared between mutant IDH1 and R-2HG fell into the category of signalling, suggesting that R-2HG has a critical effect on signal transduction. In R-2HG treated cells gene sets related to ion transport were uniquely enriched, while transcriptional regulation was uniquely enriched in IDH1 mutant cells. This suggests that mutant IDH1 contributes to leukemogenesis with additional mechanisms than R-2HG production and that full inhibition of mutant IDH1 may require more than just inhibition of R-2HG production.
In summary, we show that R-2HG, but not S-2HG, is an oncometabolite that does not require the mutant IDH1 protein to induce hyperleukocytosis and to accelerate disease onset in vivo. Thus, circulating R-2HG acts in a paracrine fashion and can drive the expansion of many different leukemic and preleukemic clones that may even express wildtype IDH1, and therefore can be a source of clonal evolution and diversity.
Supplementary Material
Acknowledgement
We acknowledge assistance of the Cell Sorting Core Facility of Hannover Medical School supported in part by the Braukmann-Wittenberg-Herz-Stiftung and the Deutsche Forschungsgemeinschaft. We would like to thank the staff of the Central Animal Facility of Hannover Medical School, Vishwas Sharma, Razif Gabdoulline, Michael Morgan, Silke Glowotz, Nicole Ernst and Martin Wichmann for their support on this project. We would also like to thank Dr. Florian Kuchenbauer for providing the MLL-AF9 construct. This study was supported by an ERC grant under the European Union’s Horizon 2020 research and innovation programme (No. 638035), by grants 110284, 110287, 110292 and 111267 from Deutsche Krebshilfe; grant DJCLS R13/14 from the Deutsche José Carreras Leukämie-Stiftung e.V; the German Federal Ministry of Education and Research grant 01EO0802 (IFB-Tx); DFG grant HE 5240/5-1 and HE 5240/6-1; grants from Dieter-Schlag Stiftung, and a HiLF grant from Hannover Medical School awarded to AC.
Footnotes
Author contributions
A.C. and M.H. designed the research; A.C., M.M.A.C., N.J., A.Sh., R.Go., A.Sc., K.G., R.S., E.S., E.J., C.R., C.M., R.Ge., and G.G., performed the research; F.T., A.G., and M.H. contributed patient samples and clinical data; A.C., M.M.A.C., A.Sc., C.R., R.Ge., and M.H. analyzed the data. A.C. and M.H. wrote the manuscript.
Conflict of interest
All authors read and agreed to the final version of the manuscript and declare no conflict of interest.
References
- 1.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009 Sep 10;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009 Feb 19;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011 Jul;224(3):334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 4.Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17(1):72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thol F, Damm F, Wagner K, Gohring G, Schlegelberger B, Hoelzer D, et al. Prognostic impact of IDH2 mutations in cytogenetically normal acute myeloid leukemia. Blood. 2010 Jul 29;116(4):614–616. doi: 10.1182/blood-2010-03-272146. [DOI] [PubMed] [Google Scholar]
- 6.Wagner K, Damm F, Gohring G, Gorlich K, Heuser M, Schafer I, et al. Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. J Clin Oncol. 2010 May 10;28(14):2356–2364. doi: 10.1200/JCO.2009.27.6899. [DOI] [PubMed] [Google Scholar]
- 7.Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Kronke J, Bullinger L, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010 Aug 1;28(22):3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 8.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010 Mar 16;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2010 Jun 17;465(7300):966. doi: 10.1038/nature09132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010 Feb 15;207(2):339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009 Apr 10;324(5924):261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rose NR, McDonough MA, King ON, Kawamura A, Schofield CJ. Inhibition of 2-oxoglutarate dependent oxygenases. Chem Soc Rev. 2011 Aug;40(8):4364–4397. doi: 10.1039/c0cs00203h. [DOI] [PubMed] [Google Scholar]
- 13.Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat Chem Biol. 2008 Mar;4(3):152–156. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- 14.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011 Jan 18;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011 May;12(5):463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012 Mar 22;483(7390):484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heuser M, Araujo Cruz MM, Goparaju R, Chaturvedi A. Enigmas of IDH mutations in hematology/oncology. Exp Hematol. 2015 May 29; doi: 10.1016/j.exphem.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012 Aug 30;488(7413):656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaturvedi A, Araujo Cruz MM, Jyotsana N, Sharma A, Yun H, Gorlich K, et al. Mutant IDH1 promotes leukemogenesis in vivo and can be specifically targeted in human AML. Blood. 2013 Oct 17;122(16):2877–2887. doi: 10.1182/blood-2013-03-491571. [DOI] [PubMed] [Google Scholar]
- 20.Heuser M, Yun H, Berg T, Yung E, Argiropolous B, Kuchenbauer F, et al. Cell of origin in AML: Susceptibility to MN1-induced transformation is regulated by the MEIS1/AbdB-like HOX protein complex. Cancer Cell. 2011;20(1):39–52. doi: 10.1016/j.ccr.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heuser M, Sly LM, Argiropoulos B, Kuchenbauer F, Lai C, Weng A, et al. Modeling the functional heterogeneity of leukemia stem cells: role of STAT5 in leukemia stem cell self-renewal. Blood. 2009 Nov 5;114(19):3983–3993. doi: 10.1182/blood-2009-06-227603. [DOI] [PubMed] [Google Scholar]
- 22.Struys EA, Jansen EE, Verhoeven NM, Jakobs C. Measurement of urinary D- and L-2-hydroxyglutarate enantiomers by stable-isotope-dilution liquid chromatography-tandem mass spectrometry after derivatization with diacetyl-L-tartaric anhydride. Clin Chem. 2004 Aug;50(8):1391–1395. doi: 10.1373/clinchem.2004.033399. [DOI] [PubMed] [Google Scholar]
- 23.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics--a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009 Feb 1;25(3):415–416. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004 Feb 12;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 26.Smyth GK. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Part V. Springer; New York: 2005. limma: Linear Models for Microarray Data; pp. 397–420. [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005 Oct 25;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heuser M, Beutel G, Krauter J, Dohner K, von Neuhoff N, Schlegelberger B, et al. High meningioma 1 (MN1) expression as a predictor for poor outcome in acute myeloid leukemia with normal cytogenetics. Blood. 2006 Dec 01;108(12):3898–3905. doi: 10.1182/blood-2006-04-014845. [DOI] [PubMed] [Google Scholar]
- 29.Heuser M, Yap DB, Leung M, de Algara TR, Tafech A, McKinney S, et al. Loss of MLL5 results in pleiotropic hematopoietic defects, reduced neutrophil immune function, and extreme sensitivity to DNA demethylation. Blood. 2009 Feb 12;113(7):1432–1443. doi: 10.1182/blood-2008-06-162263. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001 Dec;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Smith ZD, Gu H, Bock C, Gnirke A, Meissner A. High-throughput bisulfite sequencing in mammalian genomes. Methods. 2009 Jul;48(3):226–232. doi: 10.1016/j.ymeth.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009 Nov 19;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012 Mar 22;483(7390):474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013 Mar 29;339(6127):1621–1625. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan SM, Thomas D, Corces-Zimmerman MR, Xavy S, Rastogi S, Hong WJ, et al. Isocitrate dehydrogenase 1 and 2 mutations induce BCL-2 dependence in acute myeloid leukemia. Nat Med. 2015 Feb;21(2):178–184. doi: 10.1038/nm.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kats LM, Reschke M, Taulli R, Pozdnyakova O, Burgess K, Bhargava P, et al. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell. 2014 Mar 6;14(3):329–341. doi: 10.1016/j.stem.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kranendijk M, Struys EA, Salomons GS, Van der Knaap MS, Jakobs C. Progress in understanding 2-hydroxyglutaric acidurias. J Inherit Metab Dis. 2012 Jul;35(4):571–587. doi: 10.1007/s10545-012-9462-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.