Abstract
The 7B-1 tomato (Solanum lycopersicum L. cv Rutgers) is a male-sterile mutant with enhanced tolerance to abiotic stress, which makes it a potential candidate for hybrid seed breeding and stress engineering. To underline the molecular mechanism regulating the male-sterility in 7B-1, transcriptomic profiles of the 7B-1 male-sterile and wild type (WT) anthers were studied using mRNA sequencing (RNA-Seq). In total, 768 differentially expressed genes (DEGs) were identified, including 132 up-regulated and 636 down-regulated transcripts. Gene ontology (GO) enrichment analysis of DEGs suggested a general impact of the 7B-1 mutation on metabolic processes, such as proteolysis and carbohydrate catabolic process. Sixteen candidates with key roles in regulation of anther development were subjected to further analysis using qRT-PCR and in situ hybridization. Cytological studies showed several defects associated with anther development in the 7B-1 mutant, including unsynchronized anther maturation, dysfunctional meiosis, arrested microspores, defect in callose degradation and abnormal tapetum development. TUNEL assay showed a defect in programmed cell death (PCD) of tapetal cells in 7B-1 anthers. The present study provides insights into the transcriptome of the 7B-1 mutant. We identified several genes with altered expression level in 7B-1 (including beta-1,3 glucanase, GA2oxs, cystatin, cysteine protease, pectinesterase, TA29, and actin) that could potentially regulate anther developmental processes, such as meiosis, tapetum development, and cell-wall formation/degradation.
Introduction
In flowering plants, male-fertility is a highly regulated process, which requires proper cellular differentiation in anthers and timely regulation of microsporogenesis. Male-sterility on the other hand has potential application in hybrid seed breeding and understanding its molecular mechanism is currently an important research topic in plant science [1]. Large number of male-sterile tomato mutants have been identified, however, in most cases the mutant gene(s) have not been precisely identified and often mapped only to a large genomic region [1,2,3]. A polygalacturonase gene is the only well characterized gene known so far, which is responsible for male-sterile phenotype of ps-2 tomato mutant [1]. Male-sterile tomato mutants with desired agricultural traits are advantageous for hybrid seed breeding. Male-sterile mutants in tomato have been classified into functional, structural, and sporogenous classes [4]. For example, positional sterile-2 (ps-2) tomato is a functional male-sterile mutant with defected pollen dehiscence [1]. Stamenless-2 (sl-2) tomato is a structural mutant, which produces abnormal stamens with aborted microspores [5]. In sporogenous mutants, microsporogenesis could break down during meiosis, formation of tetrads or separation of microspores. In male-sterile (ms) 3 and ms15 tomato mutants, pollen mother cells (PMC) collapse in pre-meiotic anthers [6]. In ms5 and ms1035 (allelic to ms10) tomato mutants, microsporogenesis beaks down at meiosis due to aberrant regulation of tapetal cells [4,7].
Several genes with key roles in anther development have been characterized in Arabidopsis, among those, SPL/NZZ, EMS1/EXS, and TPD1 are essential for differentiation of anther wall cells [8–11], and MS1 and MS2 are required for pollen wall formation [11,12]. In rice, GAMYB [13], MYB33/MYB65 [14], DYT1 [15], TDF1 [16], AMS [17,18], MS1 [19], PTC1 [20], TDR-2 [21], UDT1 [22], TDR [23], and EAT1 [24] play key roles in tapetum development and regulation of microsporogenesis. Studies in tomato and rapeseed suggest that male-sterility is, in part, a manifestation of hormonal imbalance in flowers, particularly in stamens [25–27]. Male-sterility is also known to be regulated by environmental factors, i.e., temperature, and photoperiod [28,29], and it has been suggested that the effects of these external agents are mediated through hormonal changes [26].
In most angiosperms, the anther consists of four lobes, each containing four highly specialized layers (from outer to inner: epidermis, endothecium, middle layer and tapetum), which houses the reproductive cells [30]. The tapetal cells play an important physiological role as all nutritional materials entering the sporogenous cells either passes through or originates from the tapetum [31]. In addition, the tapetum produces callase, an enzyme which removes the callose around tetrads. Aberrant regulation of tapetum development has been often associated with male-sterile anther phenotypes [32]. Tapetum degeneration is proposed to be triggered by PCD processes during the late stage of pollen development, which in turn provide cellular contents supporting pollen wall formation and maturation. Rice TDR mutant exhibits delayed tapetal PCD and retarded degeneration, resulting in male-sterility [32].
The 7B-1 tomato mutant line (Solanum lycopersicum L. cv. Rutgers) was previously described as a photoperiod-dependent male-sterile line [33,34]. In long days (LD), the 7B-1 flowers are male-sterile, which produce shrunken stamens with no viable microspores, while in short days (SD), flowers are fertile, stamens are intact and produce viable pollen. Compared to the WT, the mutant shows reduced de-etiolation, has higher content of endogenous Abscisic acid (ABA), but less gibberellins (GAs), indole-3-acetic acid (IAA), and cytokinins (CKs), and is hypersensitive to exogenous ABA [35–37]. Seed germination and hypocotyl growth in 7B-1 mutant are more tolerant to various abiotic stresses, especially under blue light [36]. Molecular studies showed defects in blue light perception and hormonal balance in the 7B-1 mutant, associated with a large number of proteins being differentially expressed between 7B-1 and WT anthers [36,38]. A recent study by Omidvar and Fellner [39] showed distinct DNA methylation dynamics and transcriptional regulation in response to different light qualities and abiotic stresses between 7B-1 and WT seedlings. Several microRNAs (miRNAs) with key roles in regulation of anther development, male-sterility and stress-response in 7B-1 have been identified and characterized [40,41]. With primary effect of the 7B-1 mutation yet unknown, studies indicate that modulation of the 7B-1 mutation and its effect on the gene expression is coordinated through a complex interplay between light signalling components, hormonal balance and their crosstalk with miRNAs and DNA methylation programming, which all collectively tune the downstream gene expression associated with anther development and male-sterility in 7B-1 anthers.
The aim of our study is to gain a deeper insight into the molecular mechanism of male-sterility and transcriptional regulation of anther developmental processes in 7B-1 anthers. Using RNA-Seq, we identified a number of genes with potential key roles in regulation of anther development and microsporogenesis, which were differentially expressed between WT and 7B-1 anthers. Expression profiles of these candidate genes were further investigated at different developmental stages of 7B-1 anthers using qRT-PCR and in situ hybridization. Cytological studies showed differences between WT and 7B-1 anthers, including anther structure, callose deposition and tapetum development.
Materials and methods
Plant materials
7B-1 mutant and WT seedlings (Solanum lycopersicum L., cv. Rutgers) were grown in long days (16/8 h light/dark) in temperature controlled growth chamber. Flower buds at different developmental stages, including buds smaller than 4–5 mm (pre-meiotic anthers; referred to as S1), equal to 4–5 mm (meiotic anthers; S2) and bigger than 4–5 mm (post-meiotic anthers; S3) were collected and anthers were dissected under a stereomicroscope. Stages of flower buds were selected according to Sheoran et al. [38] and confirmed by analysis of anther squashes. Gibberelic acid treatment was carried out by spraying (0.1 mM GA3) directly onto the 7B-1 buds at the panicle primordium stage and repeated once a week until the buds reached the length of ≥ 5 mm.
RNA-seq analysis
Total RNA was extracted from WT and 7B-1 anthers at different stages using the RNeasy Plant Mini Kit (Qiagen). Samples were pooled separately in equimolar ratio and used for construction of sequencing libraries using the Truseq™ RNA Sample Prep Kit (Illumina, San Diego, CA, USA). Sequencing was carried out on the Illumina HiSeq™ 2000 platform. Short reads and low quality bases were trimmed using Trimmomatic [42]. The remaining reads were mapped to the ribosome RNA database [43] using bowtie [44], allowing up to 3 mismatches and rRNA-mapping reads were subsequently filtered out. The cleaned reads were then mapped (allowing 2 mismatches) to the tomato reference genome ITAG v2.5 release using TopHat2 [45]. Read counts were normalized using the FPKM (fragments per kilobase per million) approach [46]. Differential expression analysis was carried out using NOISeq [47] and presented as offset fold change (OFC), with an offset of 20 as described by Mohorianu et al. [48]. Genes with log2 (OFC) ≥ 1.5 and probabilities > 0.95 were identified as DEGs. Gene ontologies were assigned using the Blast2go tool (http://www.blast2go.com/b2ghome). Enrichment analysis was carried out using PANTHER [49].
Quantitative PCR
qRT-PCR experiments were carried out using the SensiFAST SYBR Lo-ROX kit (Bioline). First-strand cDNAs were synthesized using the PrimeScript First Strand cDNA Synthesis kit (Takara). Gene-specific primers are listed in S2 Table. Data normalization was carried out using the CAC and α-tubulin housekeeping genes (data were shown only for CAC). PCR thermal cycles were set for initial denaturation at 95°C for 2 min, 40 cycles of 95°C for 5 s, followed by annealing/extension at 60°C for 20 s. Differential expression values were calculated as normalized fold changes of expression using the ΔΔCT method [50].
Light microscopy
Cryosections were prepared as described previously [40]. In brief, flower buds were embedded in Paraplast® PlusTM and transversal sections of 8 μM thickness were cut using a Leica Ultracut R ultramicrotome (Leica Bensheim, Germany). Callose was detected by staining the tissue sections with 0.05% (w/v) aniline blue and visualized with fluorescence microscopy (λexc = 330-385nm, λem = 480nm; Olympus BX60). In situ hybridization assay was carried out as previously described [40]. Oligo-probes (S3 Table) with sequences complementary to the candidate genes and murine miR122a (as a negative control) were synthesized and DIG-labelled at 5'-end by Eastport (Eastport, Czech Republic). Probe concentration and hybridization temperature were experimentally optimized to 10 nM and 50°C, respectively. In situ localization signals were detected using light microscopy in a colorimetric-based reaction using DIG-specific antibodies coupled to alkaline phosphatase.
TUNEL assay
Anther sections were washed in PBS (160 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4) for 5 min and incubated in 20 mg/mL proteinase K in proteinase K buffer (100 mM Tris-HCl, pH 8.0, and 50 mM EDTA) for 20 min at 37°C in a humid chamber. Sections were washed in PBS for 5 min and fixed in 4% (w/v) paraformaldehyde in PBS for 10 min. PBS wash was repeated twice, each for 5 min. Detection of nuclear DNA fragmentation was performed using Terminal deoxynucleotidyl transferase (TdT) dUTP Nick-End Labeling (TUNEL) assay (DeadEnd Fluorometric TUNEL system, Promega) according to the manufacturer’s instructions. Fluorescence signal in samples was analyzed by fluorescence microscopy (wavelength of 520 ± 20nm; Olympus BX60).
Experimental design and statistical analysis
Experiments were conducted in three biological replicates and arranged in a completely randomized design. Analysis of the variance (ANOVA) and mean comparison using duncan new multiple range test (DNMRT p = 0.05) were carried out using the SAS software version 9.2.
Results
Callose degradation is perturbed in 7B-1 anthers
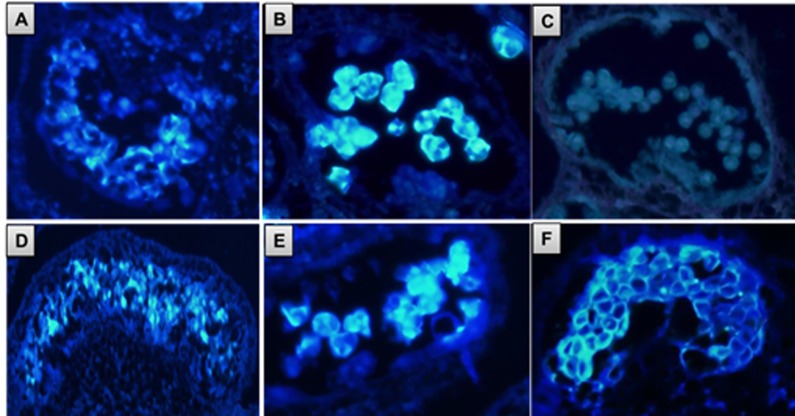
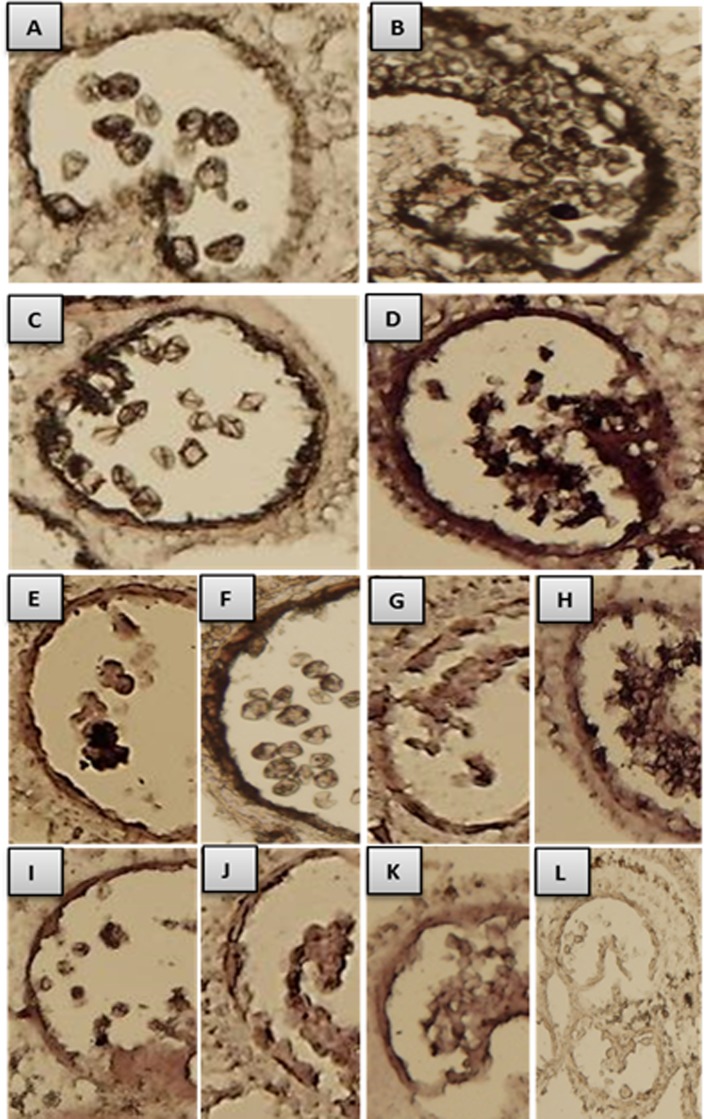
We have previously showed that anther maturation in 7B-1 was not synchronized and microsporogenesis was impaired partially in some anthers/lobes as evidenced by arrested microspores. In addition, some anthers had abnormal tapetum phenotype, where the tapetal cells were vacuolated and failed to degenerate [41]. In this study, callose localization was examined in WT and 7B-1 anthers during meiosis (Fig 1). At the early PMC stage, callose was detected around the PMCs in both WT and 7B-1 anthers (Fig 1A and 1D). Callose was also detected in WT and 7B-1 meiotic anthers around the tetrads (Fig 1B and 1E). With release of microspores from the tetrads in WT anthers, callose was completely degraded as evidenced by lack of the signal, while it persisted around the arrested microspores in 7B-1 anthers (Fig 1C and 1F). This result showed that callose degradation was perturbed in 7B-1 anthers at the end of meiosis, resulting in the arrested microspore phenotype.
Fig 1.
Callose deposition in WT (A, B, C) and 7B-1 (D, E, F) anthers. A, D: PMCs at early stage of meiosis. B, E: tetrad stage. C, F: microspores release stage.
Aberrant regulation of tapetum PCD in 7B-1 anthers
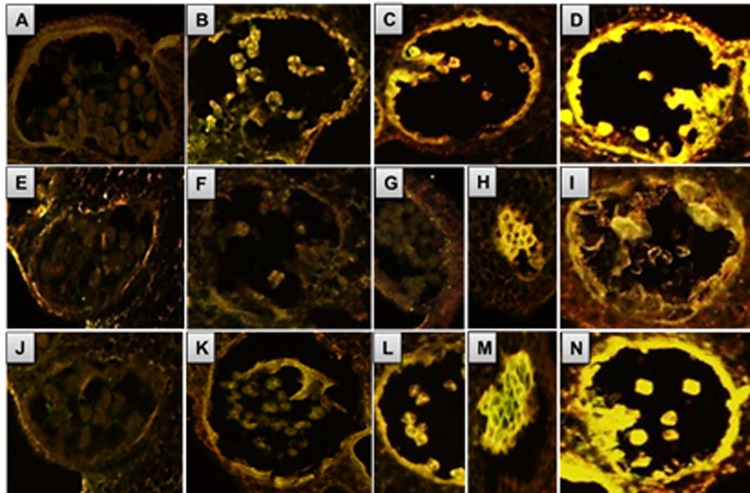
The PCD in tapetal cells is characterized by cleavage of the nuclear DNA. To test if 7B-1 anthers are defective in PCD, we performed the TUNEL (terminal deoxynucleotidyl transferase–mediated dUTP nick-end labeling) assay (Fig 2). The assay measures nuclear DNA fragmentation, which can be visualized directly by fluorescence microscopy. Both WT and 7B-1 anthers undergoing meiosis showed TUNEL-negative signal (Fig 2A and 2E), indicating a lack of DNA fragmentation of nuclei at the PMC stage. At the tetrad stage, the TUNEL-positive signal was marginally detectable in WT tapetal cells, but not in 7B-1, suggesting the onset of PCD in WT tapetum (Fig 2B and 2F). At the binucleate microspore stage, strong TUNEL-positive signal was detected in WT tapetal cells (Fig 2C), while a lack of the signal in 7B-1 tapetum indicated a delay or failure of PCD in these cells (Fig 2G and 2H). At the mature pollen stage, TUNEL-positive signal was detected in WT anthers in fully degenerated tapetal cells (Fig 2D), while a weak signal observed in 7B-1 anthers in the vacuolated tapetal cells and collapsed microspores (Fig 2I). These observations demonstrated that PCD in WT tapetum has commenced at the tetrad stage, while in 7B-1 anthers the tapetum was failed to degenerate due to retardation or defect of PCD.
Fig 2. TUNEL assay in WT and 7B-1 anthers.
Panels A, B, C, D: WT anthers at PMCs, tetrads, free binucleate microspores, and mature pollens stages, respectively. Panels E, F, G, H, I: 7B-1 anthers at PMCs, tetrads, free binucleate microspores, arrested binucleate microspores and mature pollens stages, respectively. Panels J, K, L, M, N: GA-treated 7B-1 anthers at the same stages as E-I.
As mentioned earlier, free microspores could be marginally formed in very few of the 7B-1 anthers/lobes, while in most of them, they were arrested and lysed. Strong TUNEL-positive signal was detected in the arrested microspores, but not in the tapetal cells of either free or arrested microspores phenotypes (Fig 2G and 2H). To test if GA3 could restore the timely PCD in 7B-1 tapetal cells, 7B-1 buds were treated with GA3 at the panicle primordium stage. GA3 restored the PCD of tapetal cells in anthers/lobes, which produced free microspores, but not in those showing arrested microspores (Fig 2L and 2M). These observations confirmed that GA is essential for triggering of PCD in 7B-1 tapetal cells.
Expression profiling revealed genes associated with male-sterile phenotype of 7B-1 anthers
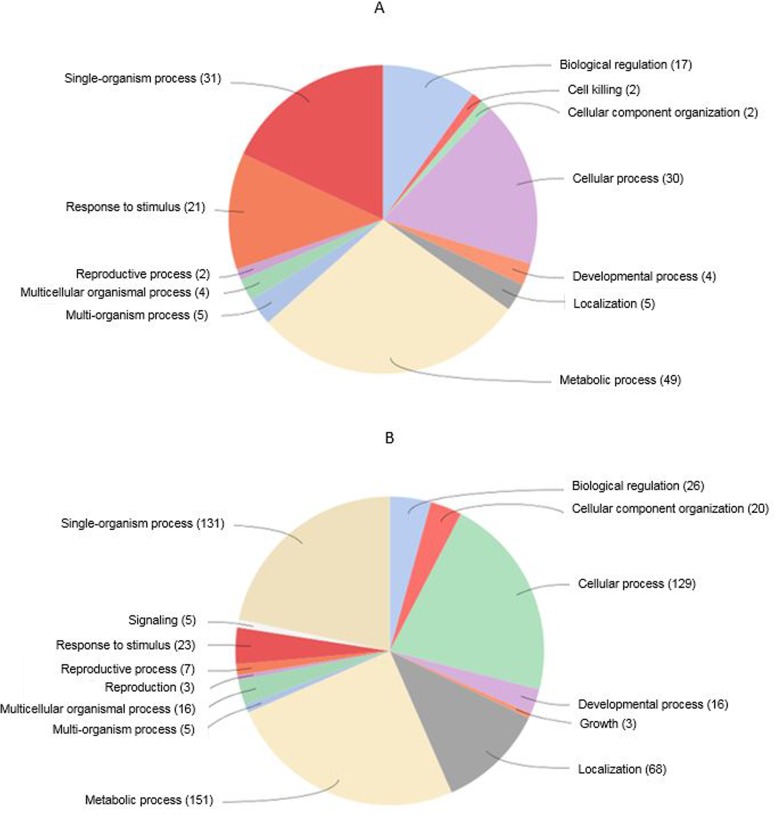
Total RNA from anthers at three developmental stages of pre-meiosis, meiosis, and post-meiosis (designated as S1, S2, and S3) were pooled with equimolar ratio and used for construction of RNA-Seq libraries. Total of 14.1 and 13.9 million raw reads were sequenced for WT and 7B-1 libraries, respectively. After removal of short reads and rRNA matching reads, the clean reads were mapped (allowing 2 mismatches) to the tomato (cv. Heinz) reference genome ITAG v2.5. Read statistics are shown in Table 1. We identified 768 DEGs, including 132 up-regulated and 636 down-regulated genes (S1 Table). To gain insight into functional categories of DEGs, gene ontologies were assigned based on the biological processes using BLAST2GO (Fig 3). The majority of both up- and down-regulated genes corresponded to three major biological classes, including metabolic process, single-organism process and cellular process.
Table 1. Read statistics in WT and 7B-1 libraries.
| Sample | Total | Adaptor trimming | Removal of rRNA-matching reads | Genome-matching reads | ||||
|---|---|---|---|---|---|---|---|---|
| Clean | %Clean | rRNA-matching | Clean | %Clean | Mapped | %Mapped | ||
| WT | 14,116,742 | 12,882,309 | 91.26 | 370,592 | 12,511,717 | 97.12 | 10,370,234 | 82.88 |
| 7B-1 | 13,927,913 | 12,696,565 | 91.16 | 351,302 | 12,345,263 | 97.23 | 10,319,965 | 83.59 |
Fig 3. Gene ontology of DEGs.
Up-regulated (A) and down-regulated (B) genes were categorized into different biological classes and numbers in the parenthesis indicate the frequency of members in each category.
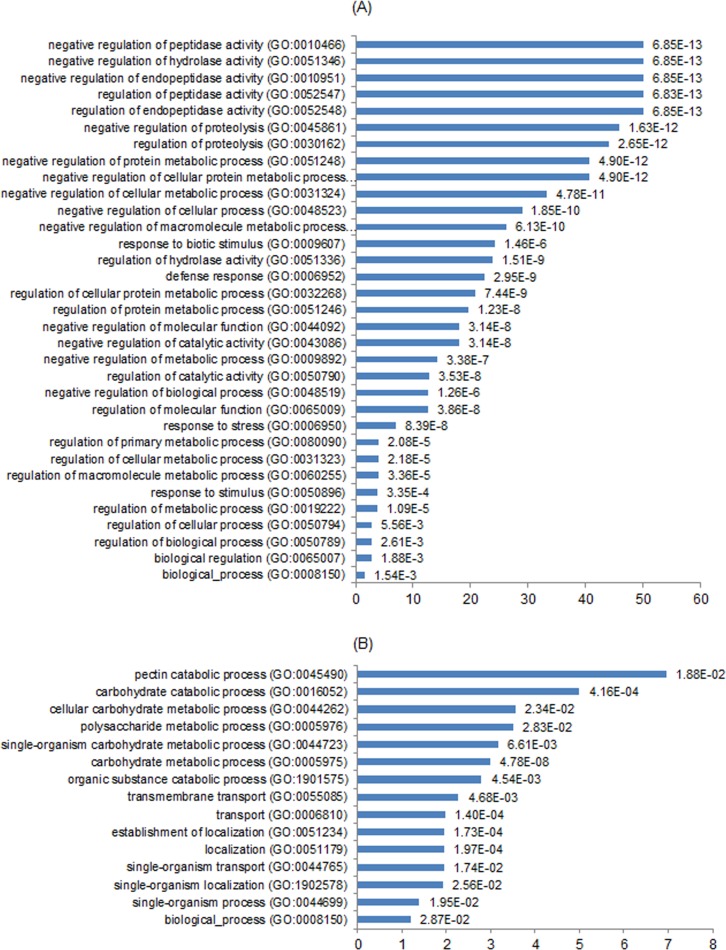
GO enrichment analysis was carried out in order to identify the major biological processes affected by the 7B-1 mutation. Thirty three and fifteen GO terms were over-represented (p<0.05) among up- and down-regulated DEGs, respectively (Fig 4). This indicates the broad effect of the 7B-1 mutation on transcriptional regulation of anther development, affecting diverse biological processes from regulation of proteolysis, defense response, response to stress to pectin catabolic and carbohydrate metabolic processes. Although several biological processes were enriched, nonetheless it was difficult to point a direct link between any of the enriched terms (with exception of the pectin catabolic process) and the male-sterile phenotype of 7B-1 anthers. Therefore, we focused our attention to DEGs with putative roles in regulation of anther development in 7B-1 mutant based on their expression, annotation and literature search. Sixteen candidates (Table 2) with key roles in regulation of meiosis, tapetum development, and cell-wall formation/degradation were further examined using qRT-PCR and in situ hybridization.
Fig 4.
GO enrichment analysis of up- (A) and down-regulated (B) DEGs. Biological processes are listed on the Y-axis with their enrichment folds against all tomato genes (reference) presented on the X-axis. P-values are indicated for each GO term.
Table 2. List of DEGs with potential roles in anther development in 7B-1 mutant.
| GeneID | Normalized reads | Statistics | Annotation | ||
|---|---|---|---|---|---|
| WT | 7B-1 | DE | P-value | ||
| Solyc10g079860.1.1 | 3.73 | 34.55 | 3.21 | 0.99 | Beta-1,3-glucanase |
| Solyc04g005610.2.1 | 11.45 | 54.70 | 2.26 | 0.98 | NAC transcription factor |
| Solyc00g071180.2.1 | 239.56 | 891.96 | 1.90 | 0.97 | Cystatin |
| Solyc01g079200.2.1 | 32.79 | 152.49 | 1.70 | 0.98 | Gibberellin 2-oxidase |
| Solyc05g052110.2.1 | 60.99 | 9.35 | -2.74 | 0.99 | Pectinesterase |
| Solyc06g008530.1.1 | 23.17 | 0.87 | -4.64 | 0.99 | Myosin XI |
| Solyc07g044870.2.1 | 358.24 | 13.27 | -4.64 | 1.00 | Polygalacturonase |
| Solyc12g098930.1.1 | 24.02 | 0.79 | -5.06 | 0.99 | Pyruvate dehydrogenase kinase |
| Solyc05g051250.2.1 | 271.66 | 8.71 | -5.06 | 1.00 | Glutamine synthetase |
| Solyc02g078370.1.1 | 307.04 | 9.23 | -5.06 | 1.00 | Anther-specific protein TA29 |
| Solyc10g086460.1.1 | 291.50 | 9.83 | -5.06 | 1.00 | Actin |
| Solyc01g111540.2.1 | 172.06 | 5.29 | -5.06 | 1.00 | Beta-galactosidase |
| Solyc07g053460.2.1 | 75.02 | 1.37 | -5.64 | 1.00 | Cysteine proteinase |
| Solyc06g005180.1.1 | 33.48 | 0.63 | -5.64 | 0.99 | Zinc finger transcription factor |
| Solyc06g059970.2.1 | 204.97 | 2.67 | -6.64 | 1.00 | MADS-box transcription factor |
| Solyc06g059820.1.1 | 29.45 | 0.35 | -6.64 | 0.99 | F-box transcription factor |
DE is differential expression values, which were calculated as log2-fold changes of the expression. Positive and negative values mean up- and down-regulation of expression in 7B-1, respectively.
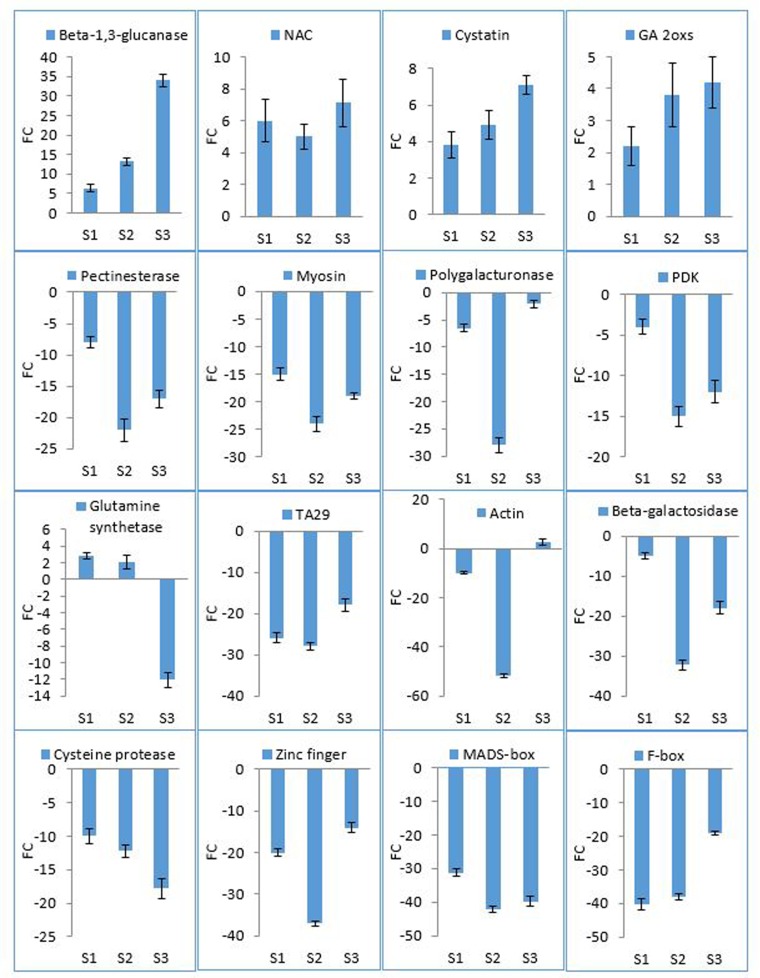
Candidate DEGs were validated using qRT-PCR at different developmental stages of 7B-1 anthers (Fig 5). Despite some quantitative differences in the expression levels, qRT-PCR results showed the same expression pattern as RNA-seq data. Beta-1,3-glucanase was up-regulated in S1, S2, and more strongly in S3. NAC was up-regulated in all stages. Cystatin and gibberellin 2-oxidases (GA2ox) were up-regulated with an increasing pattern during anther maturation. Pectinesterase, myosin, polygalacturonase, pyruvate dehydrogenase kinase (PDK), beta-galactosidase, and zinc finger were down-regulated in S1, S3, and more strongly in S2. Glutamine synthetase (GS1) was slightly up-regulated in S1 and S2, but strongly down-regulated in S3. TA29 and F-box were down-regulated in S1 and S2, more strongly compared to S3. Actin was down-regulated in S1, very strongly in S2, but slightly up-regulated in S3. Cysteine protease was down-regulated S1, S2 and more strongly in S3. MADS-box was down-regulated more strongly in S2 and S3 compared to S1.
Fig 5. qRT-PCR analysis of DEGs in 7B-1 anthers.
Expression changes are presented as normalized fold changes (FC) between 7B-1 and WT reference tissue. Positive and negative values indicate up- and down-regulation of the expression, respectively. Two-fold threshold was considered as a cutoff value for significant changes in the expression. Error bars represent standard errors of three biological replicates.
Localization profile of DEGs in 7B-1 anthers
Fig 6 shows in situ localization of beta-1,3 glucanase, GA2oxs, TA29, and pectinesterase in WT and 7B-1 anthers. Beta-1,3 glucanase and GA2oxs were expressed in WT tapetum and binucleate microspores (Fig 6A and 6C), and more strongly in 7B-1 vacuolated tapetum and arrested microspores (Fig 6B and 6D). In WT anthers, TA29 transcripts were localized in the tapetum, tetrads (Fig 6E), and the binucleate microspores (Fig 6F), while in 7B-1 anthers, they were localized in the tapetum, tetrads (Fig 6G), and the arrested microspores (Fig 6H). Pectinesterase transcripts were localized in the tapetum and the tetrads in both WT and 7B-1 anthers (Fig 6I and 6J) as well as in the arrested binucleate microspores in 7B-1 anthers (Fig 6K). The murine miR122a probe was used as negative control, which did not produce any hybridization signal (Fig 6L).
Fig 6. In situ localization of beta-1,3-glucanase, GA2oxs, TA29 and pectinesterase.
A and B: localization of beta-1, 3-glucanase in WT and 7B-1 anthers respectively at binucleate microspores stage. C and D: GA2ox in WT and 7B-1 anthers at binucleate microspores stage, respectively. E and F: TA29 in WT anthers at tetrads and binucleate microspores stages, respectively. G and H: TA29 in 7B-1 anthers at tetrads and arrested binucleate microspores stages, respectively. I, J, K: pectinesterase in WT anthers at tetrads, in 7B-1 anthers at tetrads, and in 7B-1 anthers at arrested binucleate microspores stages, respectively. L: negative control, where a murine miR122a-specific probe was used to ensure that the experimental staining is not an artifact.
Discussion
Despite the importance of male-sterility in hybrid seed breeding, the physiological mechanisms, i.e. nutritional, hormonal and environmental, which regulate the male-sterility are not yet fully understood. Until now, only a small number of genes have been identified that are specifically involved in this developmental process and the molecular mechanism of genetic male-sterility is still largely unknown. The transcriptomic profiling in our study showed differential expression of a large number of genes between WT and 7B-1 anthers. Majority of DEGs belonged to three major biological classes, including metabolic process, single-organism process and cellular process. This indicates that diverse gene regulation pathways are affected by or involved in the regulation of male-sterility in 7B-1 anthers. Further examination of GO terms showed enrichment of several biological processes, including those of special interest related to protein and carbohydrate metabolic processes. Several pectinesterase and pectate lyase-related genes were enriched within down-regulated DEGs, which were further characterized. Enrichment analysis suggested a broad impact of 7B-1 mutation primarily on the metabolism. Sixteen candidates were identified with potential roles in regulation of anther development and male-sterility in 7B-1 anthers and further characterized in different developmental stages between WT and 7B-1 anthers. These DEGs and their roles are discussed below.
During meiosis, tapetal cells undergo PCD and release beta-1,3-glucanase, which hydrolyses the callose from tetrads [51]. Persistent callose or delay in its dissolution could result in collapse of the developing microspores [52]. While callose was no longer detectable in the early microspore stage in WT anthers, it persisted around the tetrads and newly formed microspores in 7B-1 anthers, resulting in an arrested-microspore phenotype. A similar phenotype was observed in male-sterile anthers of Brassica napus, where callose was persistent around the tetrads [53]. qRT-PCR analysis showed up-regulation of beta-1,3-glucanase in 7B-1 anthers and in situ hybridization showed the prominent expression of this enzyme in 7B-1 tapetum at late stage of meiosis, where tapetal cells were vacuolated but not degenerated. Delay of tapetum degeneration in 7B-1 anthers could have led to beta-1,3-glucanase build-up level in these cells as detected by qRT-PCR and in situ hybridization signal, while callose around the newly formed microspores was not degraded, probably due to lack of the acting enzyme.
Several pectinesterase and pectate lyase-related genes were enriched within down-regulated DEGs. In addition to pectinesterase, several other cell wall modifying enzymes, including beta-galactosidase, a cellulose-modifying enzyme, and polygalacturonase which is a pectin-modifying enzyme [54,55] were strongly down-regulated in 7B-1 meiotic anthers. In qrt1 and qrt2 mutants of Arabidopsis thaliana, microspores were arrested as pectin was not degraded in primary cell walls around tetrads [56]. Pectinesterase transcripts were localized in tapetum, tetrads and arrested binucleate microspores in 7B-1 anthers. Suppression of the pectin-modifying enzymes in 7B-1 anthers were more pronounced during meiosis (stage S2), which could have impaired enzymatic degradation of cell wall pectin around tetrads, resulting in an arrested-microspores phenotype, similar to those observed in qrt mutants.
Previously, we found that cystatin and cysteine protease were up- and down-regulated in 7B-1 anthers, respectively with a pattern correlated to tapetum degeneration during anther development [41]. Similar results were observed using mRNA-seq and qRT-PCR in the present study. TUNEL assay showed a delay of PCD in 7B-1 tapetal cells. There results strongly suggest that suppression of cysteine protease could have caused a delay or defect of PCD in tapetal cells. GA plays an important role in floral organ growth, especially anther development. Tapetum is an important source of bioactive gibberellins in anthers [57], and alteration of GA level is often associated with abnormalities in anther development and male-sterility. GA-deficient mutants of tomato, rice and Arabidopsis exhibited common defects in PCD of tapetal cells, resulting in a post-meiotic arrest in male-sterile stamens [13,58,59]. In sl-2 tomato mutant GA3 could restore the male-fertility [5,60]. Application of GA3 also partially restored the male-fertility in 7B-1 anthers (Omidvar et al., unpublished data). GA2oxs regulates the GA level through inactivation of endogenous bioactive GAs [61]. 7B-1 seedlings have a lower GA level compared to WT. Up-regulation of GA2oxs in 7B-1 anthers could have decreased the GA level in 7B-1 anthers, resulting in a defect in PCD of tapetal cells. Using TUNEL assay, we showed that application of GA3 restored the PCD of tapetal cells in 7B-1 anthers similar to those of WT, which suggests that GA3 is likely to regulate the initiation of PCD in tapetal cells.
Another gene which has been differentially expressed between WT and 7B-1 anthers was TA29. It is a tapetal-specific gene in tobacco, and its promoter region has been used for engineering of male-sterility in tobacco as well as other crops [62–65]. Although TA29 is not functionally characterized with respect to regulation of male-sterility, silencing of this gene in tobacco has resulted in male-sterile transgenic plants, where tapetum was prematurely degenerated [65]. In our study TA29 was strongly down-regulated in 7B-1 meiotic anthers, where the TA29 transcripts were predominantly localized in the tapetal cells and tetrads and arrested binucleate microspores. Down-regulation of TA29 in 7B-1 anthers did not result in premature degeneration of tapetum, but it could be associated with the defect of PCD in tapetal cells as it was strongly down-regulated and localized in undegenerated tapetal cells in late meiotic 7B-1 anthers.
Aberrant regulation of actin-, tubulin-, and myosin-related genes could disrupt the organization of actin and microtubules in meiotic cytoskeleton, thus leading to defective cytokinesis in developing pollens and male-sterility in crops [66,67]. In our study actin and myosin were down-regulated in 7B-1 anthers. In addition, actin depolymerizing factors 3/10, and beta-tubulin were also down-regulated in 7B-1 anthers (not validated by qRT-PCR). These observations indicate that the actin cytoskeleton balance may be disturbed in 7B-1 anthers, which could have directly affected the meiosis and pollen cell wall development. A case study showed that suppression of pyruvate dehydrogenase kinase in transgenic tobacco has led to tapetum perturbation and male-sterility [68]. The importance of glutamine synthetase in pollen reproduction has been shown in rice [69], maize [70], and tobacco [71]. Down-regulation of these two enzymes in 7B-1 anthers could also be associated with tapetum perturbation and meiosis break-down. In addition to the above mentioned genes, several transcription factors, including F-box, MADS-box and zinc finger genes were down-regulated, while NAC was up-regulated in 7B-1 anthers. Overexpression of RMF (reduced male fertility) gene, encoding a F-box protein in Arabidopsis caused the delay in tapetum degeneration and male-sterility [72]. Li et al. [73] showed that suppression of a F-box protein-encoding gene, OsADF (anther development F-box), perturbed tapetum degeneration and resulted in male-sterility in rice. MADS-box transcription factors play important roles in floral organ development, anther dehiscence and pollen maturation [74,75]. Arabidopsis MS1 gene encodes PHD-type zinc finger protein, which is redundantly expressed in tapetum and regulates timely PCD in tapetal cells [11,76]. Several NAC transcription factors were differentially expressed between wild type and male-sterile flower buds of Brassica rapa [77]. NACs are key regulators of secondary wall thickening in anther tissue [78]. Although differential expression of these transcription factors in our study could be associated with the 7B-1 mutation and male-sterility phenotype, understating the exact function of these genes require further functional analysis.
A number of genes and transcription factors have been identified that control the tapetum formation and development [16,17,79–82]. However, little is known about the genetic basis regulating the PCD of tapetum during pollen development. In Arabidopsis ms1 and rice tdr male-sterile mutants, tapetum aberrations were associated with failure or delay of PCD [32,76]. TUNEL assay in our study showed a delay of PCD in 7B-1 tapetal cells, where presence of large autophagic vacuolated tapetal cells at this stage suggested the necrotic-based breakdown of cells rather than the normal regulated PCD process. TUNEL-positive signal in arrested 7B-1 microspores was indicative of a PCD-based breakdown, likely as a result of the tapetum aberration. Treatment of GA-deficient male-sterile anthers of rice with GA3, restored the PCD of tapetal cells [13]. GA3 restored the PCD in 7B-1 anthers similar to those in WT, which suggest that GA3 is likely to regulate the PCD onset in 7B-1 anthers.
Conclusions
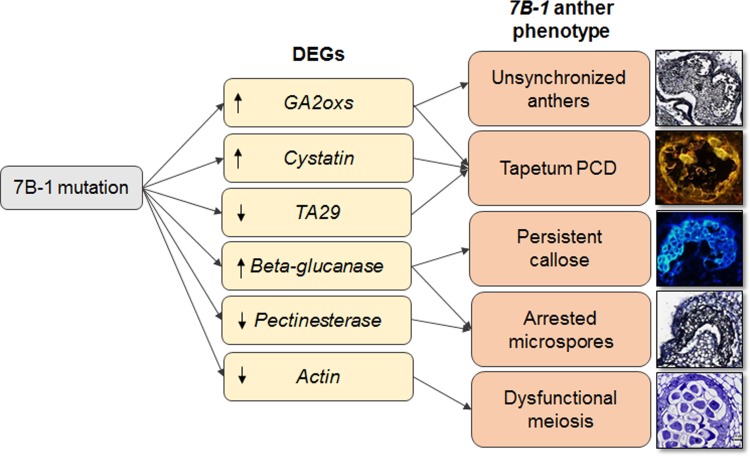
Overall in our study, we found that anther development and microsporogenesis in 7B-1 anthers was perturbed as evidenced by unsynchronized anther growth, dysfunctional meiosis, arrested microspores, defects in callose degradation, retarded PCD and abnormal tapetum profile. In situ localization signals for beta-1,3 glucanase, GA2oxs, TA29, and pectinesterase were coincided with qRT-PCR data, which confirmed the temporal gene expression results, suggesting that these genes could be closely related to tapetum development and regulation of meiosis in 7B-1 anthers. Our findings provide the first insights into the gene regulatory networks underlying the 7B-1 mutation and transcriptome dynamic between WT and 7B-1 anthers (Fig 7). It showed that 7B-1 mutation has predominantly affected genes regulating metabolic processes, and pointed out the distinct gene expression dynamic between 7B-1 and WT anthers. However, there is often a complex interplay of genes, transcription factors, hormonal balance, and environmental stimuli, which collaboratively regulate the male-sterility phenotypes and has to be taken into consideration.
Fig 7. Schematic diagram of transcriptional regulation of male-sterility in 7B-1 anthers.
Supporting information
(DOCX)
(DOCX)
(XLSX)
Acknowledgments
We thank Renata Plotzová and Věra Chytilová for their excellent technical assistance. We thank Vipen K. Sawhney (University of Saskatchewan, Canada) for providing the seeds of 7B-1 mutant. We thank the bioinformatics team at ScienceVision Sdn Bhd (Malaysia) for their technical advises. We thank J. Nauš (Department of Biophysics, Palacky University in Olomouc, Czech Republic) for measurements of the PFD of the lights.
Data availability
The sequencing data are available in the SRA system of the NCBI under accession number GSE85859.
Funding Statement
This work was supported by the Operational Programs Education for Competitiveness-European Social Fund, project no. CZ.1.07/2.3.00/30.0004 to MF, and by Ministry of Education, Youth and Sports, project no. LO1204. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gorguet B, Schipper D, van Lammeren A, Visser RG, van Heusden AW. ps-2, the gene responsible for functional sterility in tomato, due to non-dehiscent anthers, is the result of a mutation in a novel polygalacturonase gene. Theor Appl Genet. 2009;118:1199–1209 10.1007/s00122-009-0974-9 [DOI] [PubMed] [Google Scholar]
- 2.Emmanuel E, Levy AA. Tomato mutants as tools for functional genomics. Curr Opin Plant Biol. 2002;5:112–117 [DOI] [PubMed] [Google Scholar]
- 3.Lu Q, Li XH, Guo D, Xu CG, Zhang Q. Localization of pms3, a gene for photoperiod-sensitive genic male sterility, to a 284-kb DNA fragment. Mol Genet Genomics. 2005;273:507–511 10.1007/s00438-005-1155-4 [DOI] [PubMed] [Google Scholar]
- 4.Gorman SW, McCormick S. Male sterility in tomato. Crit Rev Plant Sci. 1997;16:31–53 [Google Scholar]
- 5.Sawhney VK, Bhadula SK. Microsporogenesis in the normal and male-sterile stamenless-2 mutant of tomato. Can J Bot. 1988;66:2013–2021 [Google Scholar]
- 6.Rick CM. Genetics and development of nine male-sterile tomato mutants. Hilgardia. 1948;18:599–633 [Google Scholar]
- 7.Jeong HJ, Kang JH, Zhao M, Kwon JK, Choi HS, Hwan J, et al. Tomato Male sterile 1035 is essential for pollen development and meiosis in anthers. J Exp Bot. 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canales C, Bhatt AM, Scott R, Dickinson H. EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr Biol. 2002;12:1718–1727 [DOI] [PubMed] [Google Scholar]
- 9.Zhao DZ, Wang GF, Speal B, Ma H. The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 2002;16:2021–2031 10.1101/gad.997902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito T, Nagata N, Yoshiba Y, Ohme-Takagi M, Ma H, Shinozaki K. Arabidopsis MALE STERILITY1 encodes a PHD-Type transcription factor and regulates pollen and tapetum development. Plant Cell. 2007;19:3549–3562 10.1105/tpc.107.054536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang C, Vizcay-Barrena G, Conner K, Wilson ZA. MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell. 2007;19:3530–3548 10.1105/tpc.107.054981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aarts MG, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, et al. The Arabidopsis male sterility 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 1997;12:615–623 [DOI] [PubMed] [Google Scholar]
- 13.Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M. Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell. 2009;21:1453–1472 10.1105/tpc.108.062935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millar AA, Gubler F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell. 2005;17:705–721 10.1105/tpc.104.027920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H. Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development. 2006;133: 3085–3095 10.1242/dev.02463 [DOI] [PubMed] [Google Scholar]
- 16.Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, et al. Defective in Tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J. 2008;55:266–277 10.1111/j.1365-313X.2008.03500.x [DOI] [PubMed] [Google Scholar]
- 17.Xu J, Yang C, Yuan Z, Zhang D, Gondwe MY, Ding Z, et al. The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell. 2010;22:91–107 10.1105/tpc.109.071803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang X, Wu D, Shi J, He Y, Pinot F, Grausem B, et al. Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J Integr Plant Biol. 2014; [DOI] [PubMed] [Google Scholar]
- 19.Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ. The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 2001;28:27–39 [DOI] [PubMed] [Google Scholar]
- 20.Li H, Yuan Z, Vizcay-Barrena G, Yang C, Liang W, Zong, et al. PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 2011;156(2):615–630 10.1104/pp.111.175760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Z, Yu J, Cheng X, Zong X, Xu J, Chen M, et al. The rice basic helix-loop-helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell 2014;26:1512–1524 10.1105/tpc.114.123745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, et al. Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell. 2005;17:2705–2722 10.1105/tpc.105.034090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang DS, Liang WQ, Yuan Z, Li N, Shi J, Wang J, et al. Tapetum degeneration retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol Plant. 2008;1:599–610 10.1093/mp/ssn028 [DOI] [PubMed] [Google Scholar]
- 24.Niu N, Liang W, Yang X, Jin W, Wilson ZA, Hu J, et al. EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat Commun. 2013;4:1445 10.1038/ncomms2396 [DOI] [PubMed] [Google Scholar]
- 25.Singh S, Sawhney VK. Cytokinins in a normal and the ogura (ogu) cytoplasmic male-sterile line of rapeseed (Brassica napus). Plant Sci. 1992;86:147–154 [Google Scholar]
- 26.Singh S, Sawhney VK, Pearce DW. Temperature effects on endogenous indole-3-acetic acid levels in leaves and stamens of the normal and male sterile ‘stamenless 2’ mutant of tomato. Plant Cell Environ. 1992;15:373–377 [Google Scholar]
- 27.Shukla A, Sawhney VK. Abscisic acid: one of the factors affecting male sterility in Brassica napus. Physiol Plantarum. 1994;91:522–528 [Google Scholar]
- 28.Smith MB, Horner HT, Palmer RG. Temperature and photoperiod effects on sterility in a cytoplasmicmale-sterile soybean. Crop Sci. 2001;41:702–704 [Google Scholar]
- 29.Guo RX, Sun DF, Tan ZB, Rong DF, Li CD. Two recessive genes controlling thermophotoperiod-sensitive male sterility in wheat. Theor Appl Genet. 2006;112:1271–1276 10.1007/s00122-006-0228-z [DOI] [PubMed] [Google Scholar]
- 30.Goldberg RB, Beals TP, Sanders PM. Anther development: basic principles and practical applications. Plant Cell. 1993;5:1217–1229 10.1105/tpc.5.10.1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piffanelli P, Ross JHE, Murphy DJ. Biogenesis and function of the lipidic structures of pollen grains. Sex Plant Reprod. 1998;11:65–80 [Google Scholar]
- 32.Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, et al. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell. 2006;18: 2999–3014 10.1105/tpc.106.044107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawhney VK. Genic male sterility In, Shivanna KR Sawhney VK editors Pollen biotechnology for crop production and improvement. Cambridge, Cambridge University Press; pp. 1997;183–198 [Google Scholar]
- 34.Sawhney VK. Photoperiod-sensitive male-sterile mutant in tomato and its potential use in hybrid seed production. J Hortic Sci Biotech. 2004;79:138–141 [Google Scholar]
- 35.Fellner M, Zhang R, Pharis RP, Sawhney VK. Reduced de-etiolation of hypocotyl growth in a tomato mutant is associated with hypersensitivity to and high endogenous levels of abscisic acid. J Exp Bot. 2001;52:725–738 [DOI] [PubMed] [Google Scholar]
- 36.Fellner M, Sawhney VK. The 7B-1 mutant in tomato shows blue-light-specific resistance to osmotic stress and abscisic acid. Planta. 2002;214:675–682 10.1007/s004250100671 [DOI] [PubMed] [Google Scholar]
- 37.Bergougnoux V, Zalabak D, Jandova M, Novak O, Wiese-Klinkenberg A, Fellner M. Effect of blue light on endogenous isopentenyladenine and endoreduplication during photomorphogenesis and de-etiolation of tomato Solanum lycopersicum L seedlings. PLoS One. 2012;7:e45255 10.1371/journal.pone.0045255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheoran IS, Rossb A, Olsonb D, Sawhney VK. Differential expression of proteins in the wild type and 7B-1 male-sterile mutant anthers of tomato Solanum lycopersicum), A proteomic analysis. J Proteomics. 2009;71:624–636 10.1016/j.jprot.2008.10.006 [DOI] [PubMed] [Google Scholar]
- 39.Omidvar V, Fellner M. DNA methylation and transcriptomic changes in response to different lights and stresses in 7B-1 male-sterile tomato. PLoS ONE. 2015;10: e0121864 10.1371/journal.pone.0121864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Omidvar V, Mohorianu I, Dalmay T, Fellner M. Identification of miRNAs with potential roles in regulation of anther development and male-sterility in 7B-1 male-sterile tomato mutant. BMC Genomics. 2015. a;16:878 10.1186/s12864-015-2077-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Omidvar V, Mohorianu I, Dalmay T, Fellner M. MicroRNA regulation of abiotic stress response in 7B-1 male-sterile tomato mutant. Plant Genome. 2015b; [DOI] [PubMed] [Google Scholar]
- 42.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41: D590–6 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14: R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21: 2213–2223 10.1101/gr.124321.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohorianu I, Schwach F, Jing R, Lopez-Gomollon S, Moxon S, Szittya G, et al. Profiling of short RNAs during fleshy fruit development reveals stage-specific sRNAome expression patterns. Plant J. 2011;67:232–246 10.1111/j.1365-313X.2011.04586.x [DOI] [PubMed] [Google Scholar]
- 49.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protocol. 2013;8:1551–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods. 2001;25:402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 51.Scott RJ, Spielman M, Dickinson HG. Stamen structure and function. Plant Cell. 2004;16(Suppl): S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu P, Maofeng C, Jiange Y, Gang N, Guoliang W, Hong M. The Arabidopsis CALLOSE DEFECTIVE MICROSPORE1 gene is required for male fertility through regulating callose metabolism during microsporogenesis. Plant Physiol. 2014;164:1893–1904 10.1104/pp.113.233387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu Y, Xiaoling D, Zhengfu Z, Shengqian X, Bin Y, Wen J, et al. Separation defect of tapetum cells and microspore mother cells results in male sterility in Brassica napus: the role of abscisic acid in early anther development. Plant Mol Biol. 2010;72:111–123 10.1007/s11103-009-9556-0 [DOI] [PubMed] [Google Scholar]
- 54.Nakamura A, Maeda H, Mizuno M, Koshi Y, Nagamatsu Y. beta-Galactosidase and its significance in ripening of "Saijyo" Japanese Persimmon fruit. Biosci Biotechnol Biochem. 2003;67:68–76 10.1271/bbb.67.68 [DOI] [PubMed] [Google Scholar]
- 55.Lazan H, Ng SY, Goh LY, Ali ZM. Papaya beta-galactosidase/galactanase isoforms in differential cell wall hydrolysis and fruit softening during ripening. Plant Physiol Biochem. 2004;42(11):847–53 10.1016/j.plaphy.2004.10.007 [DOI] [PubMed] [Google Scholar]
- 56.Rhee SY, Somerville CR. Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wall. Plant J. 1998; 15:79–88 [DOI] [PubMed] [Google Scholar]
- 57.Kaneko M, Itoh H, Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Ashikari M, et al. Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants?. Plant J. 2003;35:104–115 [DOI] [PubMed] [Google Scholar]
- 58.Jacobsen SE, Olszewski NE. Characterization of the arrest in anther development associated with gibberellin deficiency of the gib-1 mutant of tomato. Plant Physiol. 1991;97:409–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Plackett ARG, Powers SJ, Fernandez-Garcia N, Urbanova T, Takebayashi Y, Seo M, et al. Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell. 2012;24:941–960 10.1105/tpc.111.095109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rastogi R, Sawhney VK. Flower Culture of a Male Sterile Stamenless-2 Mutant of Tomato (Lycopersicon esculentum). Amer J Bot. 1988; 75:513–518. [Google Scholar]
- 61.Ross JJ, Reid JB, Swain SM, Hasan O, Poole AT, Hedden P, et al. Genetic regulation of gibberellin deactivation in Pisum. Plant J. 1995;7: 513–523 [Google Scholar]
- 62.Koltunow AM, Truettner J, Cox KH, Wallroth M, Goldberg RB. Different temporal and spatial gene expression patterns occur during anther development. Plant Cell. 1990;2:1201–1224 10.1105/tpc.2.12.1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mariani C, De Beuckeleer M, Truettner J, Leemans J, Goldberg RB. lnduction of male sterility in plants by a chimaeric ribonuclease gene. Nature. 1990;347:737–741 [Google Scholar]
- 64.Rong Z, Yu-le L, Feng Z, Sheng-guo L, Liang-yi K, Peng L. Induction of male sterility in oilseed rape by TA29-barnase gene. Acta Botanica Sinica. 1996;38:582–585 [Google Scholar]
- 65.Nawaz-ul-Rehman MS, Mansoor S, Khan AA, Zafar Y, Briddon RW. RNAi-mediated male sterility of tobacco by silencing TA29. Mol Biotechnol. 2007;36(2):159–165 [DOI] [PubMed] [Google Scholar]
- 66.Zhang J, Zhang C, Cheng Y, Qi L, Wang S, Hou X. Microtubule and male sterility in a gene-cytoplasmic male sterile line of non-heading Chinese cabbage. J Sci Food Agric. 2012;92:3046–3054 10.1002/jsfa.5722 [DOI] [PubMed] [Google Scholar]
- 67.Xu C, Liu Z, Zhang L, Zhao C, Yuan S, Zhang F. Organization of actin cytoskeleton during meiosis I in a wheat thermo-sensitive genic male sterile line Protoplasma. 2013;250:415–422 10.1007/s00709-012-0386-6 [DOI] [PubMed] [Google Scholar]
- 68.Yui R, Iketani S, Mikami T, Kubo T. Antisense inhibition of mitochondrial pyruvate dehydrogenase subunit in anther tapetum causes male sterility. Plant J. 2003;34:57–66 [DOI] [PubMed] [Google Scholar]
- 69.Tabuchi M, Sugiyama K, Ishiyama K, Inoue E, Sato T, Takahashi H, et al. Severe reduction in growth rate and grain filling of rice mutants lacking OsGS1;1, a cytosolic glutamine synthetase1;1. Plant J. 2005;42: 641–651 10.1111/j.1365-313X.2005.02406.x [DOI] [PubMed] [Google Scholar]
- 70.Martin A, Lee J, Kichey T, Gerentes D, Zivy M, Tatout C, et al. Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell. 2006;18:3252–3274 10.1105/tpc.106.042689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mamun AN. Reversible male sterility in transgenic tobacco carrying a dominant-negative mutated glutamine synthetase gene under the control of microspore-specific promoter. Indian J Exp Biol. 2007;45:1022–1030 [PubMed] [Google Scholar]
- 72.Kim OK, Jung JH, Park CM. An Arabidopsis F-box protein regulates tapetum degeneration and pollen maturation during anther development. Planta. 2010;232:353–366 10.1007/s00425-010-1178-x [DOI] [PubMed] [Google Scholar]
- 73.Li L, Li Y, Song S, Deng H, Li N, Fu X, et al. An anther development F-box (ADF) protein regulated by tapetum degeneration retardation (TDR) controls rice anther development. Planta. 2015;241:157–166 10.1007/s00425-014-2160-9 [DOI] [PubMed] [Google Scholar]
- 74.Schreiber DN, Bantin J, Dresselhaus T. The MADS box transcription factor ZmMADS2 is required for anther and pollen maturation in maize and accumulates in apoptotic bodies during anther dehiscence. Plant Physiol. 2004;134:1069–1079 10.1104/pp.103.030577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang F, Xu G, Chi Y, Liu H, Xue Q, Zhao T, et al. A soybean MADS-box protein modulates floral organ numbers, petal identity and sterility. BMC Plant Biol. 2014;14:89 10.1186/1471-2229-14-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vizcay-Barrena G, Wilson ZA. Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. J Exp Bot. 2006;57:2709–2717 10.1093/jxb/erl032 [DOI] [PubMed] [Google Scholar]
- 77.Dong X, Feng H, Xu M, Lee J, Kim YK, Lim YP, et al. Comprehensive analysis of genic male sterility-related genes in Brassica rapa using a newly developed Br300K Oligomeric Chip. PLoS ONE. 2013;8(9): e72178 10.1371/journal.pone.0072178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Distelfeld A, Pearce SP, Avni R, Scherer B, Uauy C, Piston F, et al. Divergent functions of orthologous NAC transcription factors in wheat and rice. Plant Mol Biol. 2012;78:515–524 10.1007/s11103-012-9881-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun YJ, Hord CLH, Chen CB, Ma H. Regulation of Arabidopsis early anther development by putative cell-cell signaling molecules and transcriptional regulators J Integr Plant Biol. 2007;49:60–68 [Google Scholar]
- 80.Liu Z, Bao E, Liang W, Yin J, Zhang D. Identification of gamyb-4 and analysis of the regulatory role of GAMYB in rice anther development. J Integr Plant Biol. 2010;52:670–678 10.1111/j.1744-7909.2010.00959.x [DOI] [PubMed] [Google Scholar]
- 81.Zhu J, Lou Y, Xu X, Yang ZN. A genetic pathway for tapetum development and function in Arabidopsis. J Integr Plant Biol. 2011;53:892–900 10.1111/j.1744-7909.2011.01078.x [DOI] [PubMed] [Google Scholar]
- 82.Gu JN, Zhu J, Yu Y, Teng XD, Lou Y, Xu XF. DYT1 directly regulates the expression of TDF1 for tapetum development and pollen wall formation in Arabidopsis. Plant J. 2014;80:1005–1013 10.1111/tpj.12694 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(XLSX)
Data Availability Statement
The sequencing data are available in the SRA system of the NCBI under accession number GSE85859.