Abstract
Mandibular torus (MT) is a common intraoral osseous outgrowth located on the lingual surface of the mandible. Histologic features include hyperplastic bone consisting of mature cortical and trabecular bone. Some theories on the etiology of MT have been postulated, such as genetic factors, masticatory hyperfunction, trauma, and continued growth, but the underlying mechanism remains largely unknown. In this study, we investigated the potential role of mesenchymal stem cells (MSCs) derived from human MT in the pathogenesis of bone outgrowth. We demonstrated that MT harbored a distinct subpopulation of MSCs, with enhanced osteogenic and decreased adipogenic differentiation capacities, as compared with their counterparts from normal jaw bone. The increased osteogenic differentiation of mandibular torus MSCs was associated with the suppression of Notch3 signaling and its downstream target genes, Jag1 and Hey1, and a reciprocal increase in the transcriptional activation of ATF4 and NFATc1 genes. Targeted knockdown of Notch3 expression by transient siRNA transfection promoted the expression of osteogenic transcription factors in normal jaw bone MSCs. Our data suggest that the loss of Notch3 signaling may contribute partly to bone outgrowth in MT, as mediated by enhanced MSC-driven osteogenic differentiation in the jaw bone.
Keywords: cell signaling, jaw bone anomalies, bone formation, gene expression, oral and maxillofacial surgery, adult stem cell(s)
Introduction
Oral torus is an exostosis, or benign bony outgrowth, and mandibular torus (MT) is the most common torus on the lingual aspect, above the mylohyoid ridge (Choi et al. 2012). Usually, MT presents as a very slow and progressive growth that can stop spontaneously (Komori and Takato 1998). The prevalence of MT ranges from 0.54% to 64.4%, and varies among ethnic groups (Sirirungrojying and Kerdpon 1999; García-García et al. 2010).
To date, the etiology of MT is still unknown. Several mechanisms have been proposed, including genetics, trauma, and dietary habits (Ladizinski and Lee 2014). Despite the most widely accepted theory of genetic etiology, torus has not always been reported as autosomal dominant (Eggen 1989; Bruce et al. 2004). Mechanical stress, especially occlusal force, is another important factor of MT formation. It has been reported that bone response and remodeling happen under functional stress (Frost 2004; Cortes et al. 2014), so MT is frequently observed in patients who have powerful masticatory muscles and parafunctional habits (Eggen 1989; Sirirungrojying and Kerdpon 1999; Sonnier et al. 1999). Since MT often is asymptomatic, the treatment is not necessary except for prosthetic needs, autologous bone graft, and functional problems, such as food retention or phonetic disturbances and cancer-phobic patients (Barker et al. 2001; Proussaefs 2006).
Bone formation and resorption are coupled events during bone growth and remodeling (Zuo et al. 2012). Despite the fact that they originate from different cell linages and possess opposite functions, osteoblasts, osteoclasts, and osteocytes interact and establish microscopic basic multicellular units to orchestrate bone remodeling (Zanotti and Canalis 2012; Zuo et al. 2012). At the molecular level, several major signals and transcription factors—including Hedgehog, wingless-type MMTV integration site family member (Wnt), bone morphogenetic protein (BMP), fibroblast growth factor (FGF), and Notch signals—have been implicated in the regulation of differentiation and function of bone cells (Long 2012). Evolutionarily conserved Notch signaling plays a critical role during embryonic development and tissue renewal. There are 4 receptors (Notch 1 to 4) and 5 Delta/Serrate/Lag-2 ligands (Jagged 1 [JAG1] and Jagged 2; Delta-like 1, 3, and 4). In the canonical signaling pathway, Notch receptors are cleaved by γ-secretase complex, leading to the release of the Notch intracellular domain (NICD), which regulates the downstream gene expression (Kopan and Ilagan 2009; Zanotti and Canalis 2012). The Notch3 gene was initially reported as being expressed in proliferating neuroepithelium, while targeted deletion of murine Notch3 does not lead to embryonic lethality possibly due to its more restricted tissue distribution (Bellavia et al. 2008). Numerous studies have shown that deregulated Notch3 signaling is closely linked to the pathogenesis of several diseases, such as heart failure (Ragot et al. 2016), pulmonary arterial hypertension (Li et al. 2009), renal disease (El Machhour et al. 2015), and tumorigenesis (Serafin et al. 2011). Furthermore, several studies have shown that Notch canonical signaling plays a critical role in prenatal skeletal development and postnatal bone remodeling by regulating the proliferation and differentiation of bone cells (Engin and Lee 2010). Dysregulation of Notch signaling is associated with numerous developmental and postnatal skeletal diseases, such as Alagille syndrome, brachydactyly and spondylocostal dysostosis, and Hajdu-Cheney syndrome (Zanotti and Canalis 2012). Previous studies have also shown that Notch-Hey1 or Notch-Hes signaling contributes to impaired osteoblast differentiation/maturation or osteogenesis of bone marrow mesenchymal stem cells (MSCs) (Zamurovic et al. 2004; Hilton et al. 2008; Fei et al. 2015).
Here, we reported the isolation and characterization of MSCs derived from human mandibular torus (T-MSCs) and normal jaw bone (JB-MSCs) and explored their potential role and mechanisms in the pathogenesis of MT. In addition, we demonstrated the functional role of Notch3 signaling in the regulation of osteogenesis of T-MSCs and JB-MSCs as a potential mechanism of bone overgrowth in MT.
Materials and Methods
Subjects and Isolation and Culture of Cells
All tissues were collected from generally healthy patients (48 to 65 y of age) after informed consents were obtained following the protocol approved by the Institutional Review Board of University of Pennsylvania. Normal jaw bone tissues (n = 5) were collected from patients undergoing extraction of impacted third molars. MT tissues (n = 5) were obtained during surgical removal of MT for prosthodontic restoration. Bone tissues were treated aseptically, cut into small pieces, and digested at 37 °C for 2 h in sterile 1× phosphate-buffered solution (PBS) containing 4 mg/mL of collagenase IV (Gibco). The dissociated cell suspension was filtered through a 70-μm cell strainer (Falcon) and plated on nontreated 10-cm Petri dishes in α-Minimum Essential Medium (α-MEM; Gibco) containing 10% fetal bovine serum (FBS; Gibco), 100 U/mL of penicillin, 100 µg/mL of streptomycin, 2 mM of L-glutamine, 100 mM of nonessential amino acid, and 550 μM of 2-mercaptoethanol (Sigma-Aldrich). Nonadherent cells were removed after 3 d, and the cells were washed with PBS. The medium was exchanged twice weekly. At 70% to 80% confluence, the fibroblast-like colonies were harvested with 0.05% trypsin–EDTA (Gibco). The subcultures were continuously maintained at 37 °C in a 5% CO2 humidified atmosphere.
Immunophenotyping of MSCs by Flow Cytometric Analysis
JB-MSCs and T-MSCs (1 × 105) were immunolabeled with specific mouse monoclonal antibodies for human CD29, CD44, CD73, CD90, CD105, stro-1 (BioLegend), CD14, CD34, CD146 (BD Biosciences), and stage-specific embryonic antigen (SSEA)–4 (R&D) at 4 °C for 1 h. Mouse isotype antibodies served as controls (BD Biosciences). After washing with PBS, cells were incubated with fluorescein-isothiocyanate-conjugated secondary antibodies for 30 min in the dark. Cells were analyzed on a BD LSR II flow cytometer (BD Bioscience).
In Vitro Multipotent Differentiation Assay
Osteogenic Differentiation
JB-MSCs and T-MSCs were plated at 5 × 105 cells per well in 6-well plates in MSC growth medium and allowed to adhere. When 100% confluency was achieved, osteogenic inductive medium (α-MEM supplemented with 10% FBS, 50 U/mL of penicillin, 50 µg/mL of streptomycin, 100 nM of dexamethasone, 100 µM of L-ascorbic acid 2-phosphate, and 10 mM of β-glycerophosphate) was replaced and changed every 3 d. After differentiation periods (9 d and 3 wk), cell cultures were fixed and assayed for mineralized deposits of calcium by alizarin red S staining (Fisher Scientific). Cells in negative control groups were grown in α-MEM with 10% FBS for 9 d.
Adipogenic Differentiation
JB-MSCs and T-MSCs (5 × 105 cells respectively) were seeded in 6-well plates in MSC growth medium and allowed to adhere. When 80% to 90% confluency was reached, adipogenic inductive medium (α-MEM supplemented with 10% FBS, 50 U/mL of penicillin, 50 µg/mL of streptomycin, 100 mM of L-ascorbate-2-phosphate, 100 nM of dexamethasone, 0.45 mM of 3-isobutyl-1-methylxanthine, and 60 mM of indomethacin) was replaced and changed every 3 d for 2 wk. Oil red O staining (Sigma-Aldrich) was performed to detect intracellular lipid vacuoles characteristic of adipocytes.
Western Blotting
Equal amounts of protein extracts (30 mg) were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) with 10% polyacrylamide gels and transferred to nitrocellulose membrane. Bands were detected immunologically with polyclonal antibodies against Notch1, Notch2, Notch3, or Notch4 (1:1000; Cell Signaling). Monoclonal antibody against mouse β-actin (Santa Cruz) was used as a loading control. The binding of all antibodies was detected with an enhanced chemiluminescence (ECL) detection system (Thermo Scientific).
Quantitative Real-time Polymerase Chain Reaction
Total RNA was isolated from cultured cells with the RNeasy Mini kit (Qiagen). cDNA was synthesized with Maxima First Strand cDNA Synthesis Kit (Thermo Scientific) according to the instructions of the manufacturer. Quantitative real-time polymerase chain reaction was performed with the ABI Prism 7900 HT Sequence Detection System. All reactions were performed in triplicate. The lists of all primers are summarized in the Appendix Table. All primers were synthesized by Integrated DNA Technologies.
Transfection with siRNA
Twenty-four hours before siRNA transfection, JB-MSCs were seeded in 6-well plates containing fresh medium without antibiotics. The cells were then transfected with siRNAs against human Notch3 gene and a nontarget siRNA control with Lipofectamine RNAi MAX (Invitrogen), as described in the manufacturer’s instruction. The sequences for downregulating Notch3 in JB-MSCs were as follows: Notch3 siRNA-1, 5′-GAGCCAAUAAGGACAUGC A-3′; Notch3 siRNA-2, 5′-U AUAGGUGUUGACGCCAUCCACGCA-3′; nontarget control siRNA, 5′-UUCUCCGAACGUGUCACGU-3′. The cells were harvested at 48 h posttransfection for further analysis.
Statistical Analysis
Data obtained from 3 independent experiments were presented as mean ± SD and analyzed by unpaired t test. P values <0.05 were considered statistically significant.
Results
Characterization of MSCs from Human Normal Jaw Bone and MT
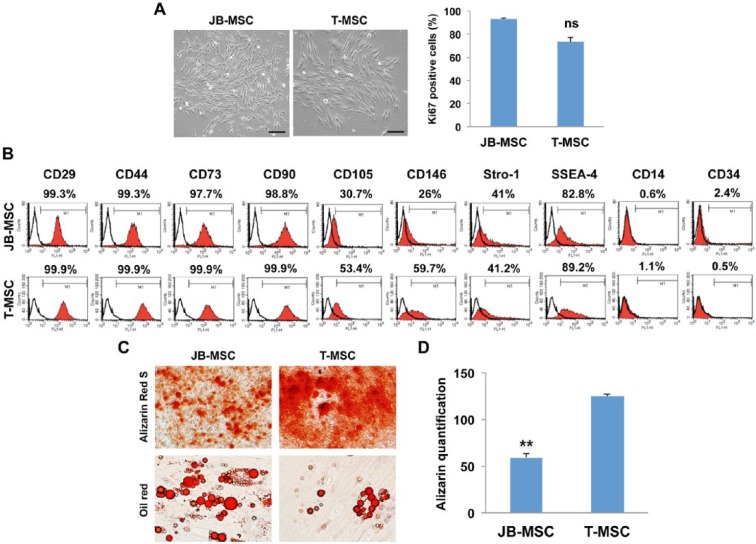
After culture for 7 to 10 d, the adherent MSCs from normal jaw bone (JB-MSCs) and mandibular torus (T-MSCs) formed colonies and displayed fibroblast-like morphologies (Fig. 1A). In all MT samples examined, the derived JB-MSCs and T-MSCs showed similar proliferative activities by immunostaining of nuclear Ki67 expression (Fig. 1A). Flow cytometric analysis of representative JB-MSCs and T-MSCs showed that both types of cells were negative or very low for CD14 and CD34 (hematopoietic stem cell markers) but positive for CD29, CD44, CD73, and CD90 (positive markers of MSCs) and stem cell–related markers Stro-1 and SSEA-4 (Fig. 1B). Both CD105 and CD146 were relatively more abundant in T-MSCs; however, their significance remains unknown. Interestingly, in vitro differentiation assays showed that T-MSCs exhibited increased osteogenic differentiation capabilities and decreased potentials for adipogenic differentiation as compared with JB-MSCs (Fig. 1C, D). These results suggest that normal jaw bone and MT harbor distinct subpopulation of MSCs (i.e., JB-MSCs and T-MSCs, respectively) that possess different differentiation capabilities.
Figure 1.
Characteristics of mesenchymal stem cells from jaw bone (JB-MSCs) and mandibular torus (T-MSCs). (A) After culture for 7 to 10 d, the adherent cells from normal jaw bone and mandibular torus formed colonies and displayed fibroblast-like morphologies and similar proliferative activities as determined by immunostaining of Ki67 expression. (B) The expression of MSC-associated cell surface markers was determined by flow cytometric analysis. (C, D) In vitro osteogenic and adipogenic differentiation of JB-MSCs and T-MSCs were determined by alizarin red S and oil red O staining, respectively. Scale bars, 50 µm. The data are expressed as the mean ± SD of 3 independent experiments (**P < 0.01). ns, no significant difference.
Downregulation of Notch3 Protein Expression in T-MSCs
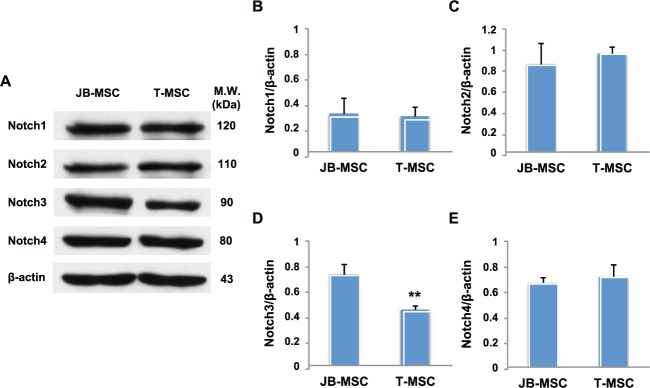
Previous studies have shown that Notch plays a central role in skeletal development and bone remodeling and that dysregulation of Notch signaling is associated with developmental and postnatal skeletal diseases (Zanotti and Canalis 2012). We next examined the expression of Notch family members in JB-MSCs and T-MSCs. Using Western blot analysis, we showed that the baseline expression of Notch1, Notch2, and Notch4 was similar in JB-MSCs and T-MSCs (Fig. 2A–C, E). However, in comparison with JB-MSCs, T-MSCs expressed a significantly lower level of Notch3 protein (41.66% of decrement; Fig. 2A, D). These results suggest that suppressed Notch3 may play a role in the biological function of T-MSCs.
Figure 2.
Expression of Notch signals in mesenchymal stem cells from jaw bone (JB-MSCs) and mandibular torus (T-MSCs). (A) The expression of Notch1, Notch2, Notch3, and Notch4 proteins in JB-MSCs and T-MSCs was determined by Western blot analysis. (B–E) The ratio of Notch1, 2, 3, 4/β-actin expression in JB-MSCs and T-MSCs was determined through densitometric analysis. The data are expressed as the mean ± SD of 3 independent experiments (**P < 0.01).
Downregulation of Notch3 Signals Is Conversely Correlated with Osteogenic Gene Expressions in T-MSCs and JB-MSCs
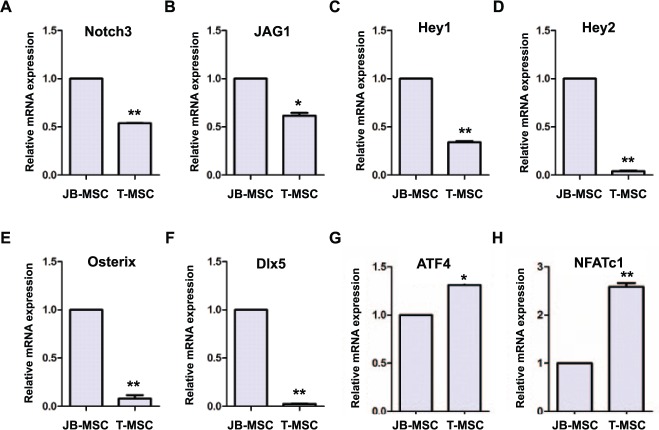
To explore the relationship between Notch3 signaling and osteogenic differentiation potentials of JB-MSCs and T-MSCs, we examined the mRNA expression of Jag1, a surface ligand of Notch receptors; Hey1 and Hey2, 2 family members of bHLH transcription factors of Notch signaling; Osterix, Dlx5, activating transcription factor 4 (ATF4), and nuclear factor of activated T-cell (NFATc), genes that play an important role in the regulation of different stages of osteogenesis (Deng et al. 2008; Samee et al. 2008; Long 2012). As shown in Figure 3, a reduced expression of Notch signals, including Notch3, Jag1, Hey1, and Hey2, was consistently observed in T-MSCs in comparison with JB-MSCs (Fig. 3A–D). In accordance, the downregulation of Notch signals in T-MSCs was coupled with a decreased expression of Osterix and Dlx5 genes (Fig. 3E, F), 2 transcription factors that play an important role in the early formation and proliferation of premature and immature osteoblasts (Komori and Takato 1998). In contrast, the expression of ATF4 and NFATc1 genes—2 transcription factors that play a critical role in the late differentiation and maturation of osteoblasts (Komori and Takato 1998; Long 2012)—was significantly increased in T-MSCs versus JB-MSCs (Fig. 3G, H). These data suggest that the suppressed Notch3 signaling might play a role in augmenting the osteogenic capability of T-MSCs, at least in part, by promoting the expression of ATF4 and NFATc1 genes.
Figure 3.
Osteogenic gene expressions in mesenchymal stem cells from jaw bone (JB-MSCs) and mandibular torus (T-MSCs). (A–H) The transcriptional expression of Notch3, Jag1, Hey1, Hey2, Osterix, Dlx5, ATF4, and NFATc1 genes was determined by quantitative real-time polymerase chain reaction. The data are expressed as the mean ± SD of 3 independent experiments (*P < 0.05, **P < 0.01).
Effect of Loss of Notch3 Signals on Osteogenic Differentiation of JB-MSCs
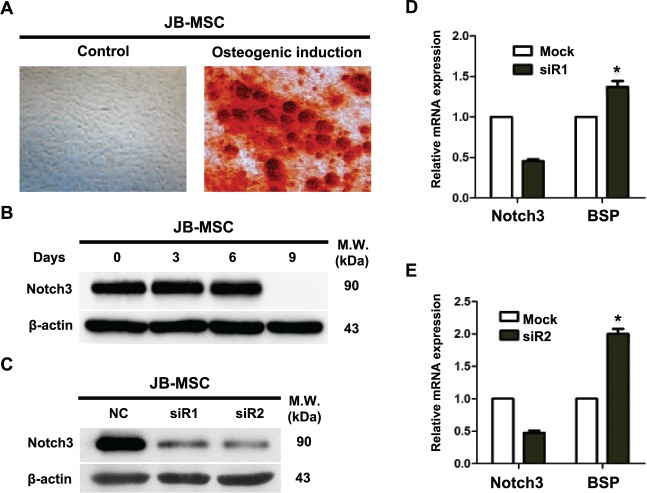
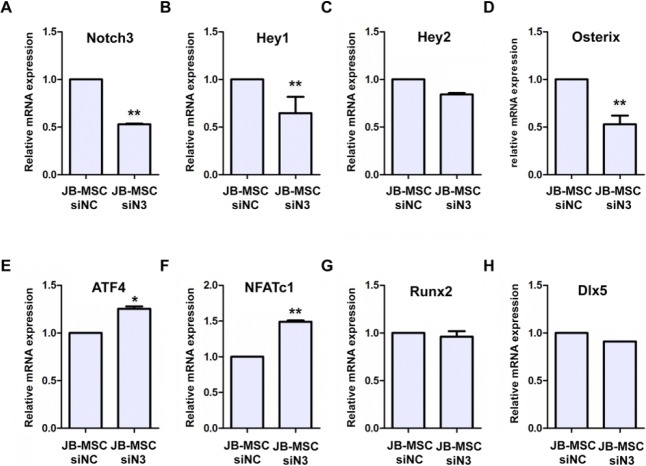
Using in vitro osteogenic assay, we showed that upon exposure to osteogenic induction condition, JB-MSCs gradually lost Notch3 signals; the decrease in Notch3 is associated with the appearance of mineralization on day 9 (Fig. 4A, B). To confirm the direct contribution of Notch3 in osteogenic differentiation, we performed transient loss of function of Notch3 in JB-MSCs by using specific siRNAs. Our results showed that transfection of 2 Notch3 siRNAs efficiently knocked down Notch3 expression at the mRNA and protein levels in JB-MSCs (Fig. 4C, D, E), which correlated with a significant increase in the mRNA expression of bone sialoprotein, a marker of bone formation (Fig. 4D, E). Mechanistically, knockdown of Notch3 expression in JB-MSCs led to a significantly reduced expression of the downstream transcriptional factor Hey1 but not Hey2 (Fig. 5A–C) and, correspondingly, the expression of Osterix (Fig. 5D). On the contrary, knockdown of Notch3 expression simultaneously led to an increased expression of AFT4 and NFATc1 (Fig. 5E, F) but had no obvious effects on the expression of Runx2 and Dlx5 (Fig. 5G, H). These results further support the notion that loss of function of Notch3 signals may lead to enhanced osteogenic differentiation of JB-MSCs, a characteristic phenotype conferred by T-MSCs.
Figure 4.
Correlation between Notch3 expression and osteogenic differentiation of mesenchymal stem cells from jaw bone (JB-MSCs). (A) JB-MSCs were cultured under osteogenic induction condition for 9 d, and the mineralization was determined by alizarin red S staining. (B) JB-MSCs were cultured under osteogenic induction condition for different periods, and the expression of Notch3 protein was determined by Western blot analysis. (C) Notch3 expression was knocked down by siRNA transfection (siR1, siR2) in JB-MSCs for 48 h, while transfection with a nonspecific siRNA (NC) was used as a control. (D, E) JB-MSCs were transfected with siRNAs against Notch3 (siR1 and siR2) for 48 h, and the expression of bone sialoprotein (BSP) mRNA was determined by quantitative real-time polymerase chain reaction. The data are expressed as the mean ± SD of 3 independent experiments (*P < 0.05).
Figure 5.
Effect of Notch3 gene expression on osteogenic markers in mesenchymal stem cells from jaw bone (JB-MSCs). JB-MSCs were transfected with siRNAs against Notch3 (siN3) for 48 h, while transfection with a nonspecific siRNA (siNC) was used as a control. (A–H) The expression of Notch3, Hey1, Hey2, Osterix, ATF4, NFATc1, Runx2, and Dlx5 mRNA was determined by quantitative real-time polymerase chain reaction. The data are expressed as the mean ± SD of 3 independent experiments (*P < 0.05, **P < 0.01).
Discussion
MSCs represent a unique subpopulation of stromal progenitor cells with multipotent and self-renewing capabilities, which exist not only in bone marrow but in almost all nonskeletal tissues. Beyond the potential use of MSCs in tissue regeneration and cell-based therapy of autoimmune and inflammatory diseases, dysregulated phenotypes and functions of MSCs may play an important role in the pathogenesis of skeletal and nonskeletal diseases. Most recently, we identified a subpopulation of MSCs from keloids, a benign skin tumor (Zhang et al. 2009). These MSCs possess distinctive properties in comparison to normal MSCs and may play an important role in the pathogenesis of these benign tumors.
MT constitutes the most common oral exostoses that do not have cartilage involvement, owing to their anatomic location. Even though several hypotheses, including genetic and environmental causes, have been proposed for the etiology of this benign bony outgrowth, there is still a lack of basic knowledge about the cellular and molecular mechanisms underlying its pathogenesis. In the present study, we have demonstrated for the first time, to our knowledge, that MT harbors a distinct subpopulation of MSCs (T-MSCs) that display enhanced osteogenic and decreased adipogenic differentiation capabilities as compared with their counterparts derived from normal jaw bone (JB-MSCs), suggesting that dysregulated functions of MSCs may contribute to the formation of MT.
Notch signaling governs cell fate determination, proliferation, differentiation, and apoptosis (Long 2012; Chen et al. 2014). In recent years, accumulating evidence has implicated the critical role of Notch-mediated signaling in normal bone remodeling and homeostasis. It has been reported that osteo- and chondroprogenitors require Notch signaling to maintain multipotency, proliferation, and hypertrophy and to suppress premature and terminal differentiation into osteoblasts and chondrocytes (Chen et al. 2014). Several lines of evidence have demonstrated the expression of Notch1 and Notch2 by skeletal cells, which appear to mediate the effects of Notch signaling on bone microarchitecture and homeostasis (Engin et al. 2008; Hilton et al. 2008; Zanotti and Canalis 2014). It has been shown that activation of Notch signaling in the early phases of osteoblastic differentiation leads to reduce cancellous bone volume and disorganized deposition of woven bone (Engin et al. 2008; Zanotti et al. 2008; Tao et al. 2010) and elevated Notch-Hey1 or Notch-Hes signals result in impaired osteogenesis of bone marrow MSCs (Zamurovic et al. 2004; Hilton et al. 2008; Fei et al. 2015). In addition, a persistent Notch activation in MSCs has been shown to contribute to decreased osteoblast differentiation associated with rheumatoid arthritis (Zhang et al. 2014), and galectin-3 secreted in the bone microenvironment niche inhibits osteoblast differentiation through Notch1 signaling (Nakajima et al. 2014). However, inactivation of Notch signaling in osteoblasts results in increased trabecular bone volume due to enhanced osteoblastogenesis (Engin et al. 2008).
In the present study, we demonstrated that JB-MSCs and T-MSCs expressed similar baseline levels of Notch1, Notch2, and Notch4 signals; however, a relatively lower level of Notch3 signal was expressed in T-MSCs. Furthermore, we showed that transient knockdown of Notch3 expression increased the expression of osteogenic differentiation markers in JB-MSCs, suggesting that loss of function of Notch3 signals may play an important role in torus formation through promoting osteogenic differentiation of T-MSCs. Collectively, these data highlight the significance of Notch signaling pathways in the physiology and pathology of human bones. Of note, several lines of evidence have shown that causative mutations in exons 3 and 4 of the Notch3 gene result in a gain or loss of a cysteine residue within the extracellular domain, which leads to an inherited disorder: cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (Mosca et al. 2014). It would be worthwhile to determine in a future study whether the lower/absent Notch3 signal in MT is caused by a pathogenic polymorphism or a mutation in the Notch3 gene.
Binding of Notch receptors with cognate ligands leads to the proteolytic cleavage of the NICD, which translocates to the nucleus and transcriptionally activates the downstream target genes, including those encoding for the transcriptional repressors hairy enhancer of split (Hes) and Hes related with YRPW motif (Hey; Kopan and Ilagan 2009). To date, it is still unknown how Notch signaling pathways are involved in the regulation of osteoblast differentiation and bone formation. Runt-related transcription factor 2 (Runx2), one of the major osteogenic transcription factors, is essential for early but not the late stage of osteoblast differentiation, and it needs to be suppressed for the terminal differentiation of osteoblast into mature osteocytes (Liu and Lee 2013). Previous studies have shown that NICD and Hey1 can bind and inhibit the transactivation of Runx2, thus impairing osteoblast differentiation (Long 2012). However, in the present study we found that T-MSCs expressed a level of Runx2 gene similar to that in JB-MSCs (data not shown) and that knockdown of Notch3 gene expression had no obvious effects on Runx2 expression in JB-MSCs. There is also evidence showing that Notch suppresses Wnt signaling that is required for osteoblastogenesis (Hilton et al. 2008; Zanotti et al. 2008). ATF4, another osteogenic transcription factor, plays an important role in the differentiation of more mature osteoblast cells. It can directly regulate the expression of osteocalcin and receptor activator of nuclear factor-κB ligand (RANKL) and promote amino acid import to ensure proper protein synthesis by osteoblast (Long 2012). NFATc1, another key osteogenic transcription factor, governs the differentiation and function of osteoblasts, whereby the osteogenic differentiation is inhibited in Nfatc1- and Nfatc2-deficient cells (Koga et al. 2005). Later studies indicated that NFATc1nuc mice display massive osteoblast overgrowth and enhanced osteoblast proliferation and that viable NFATc1-deficient mice have defects in skull bone formation (Winslow et al. 2006). Furthermore, recent studies have shown that Notch inhibited ATF4 transactivation and NFATc1 transcription, while NFATc1 reciprocally inhibited the transactivation of Notch target genes, suggesting that Notch and NFATc1 form a regulatory network to regulate the differentiation of osteoblasts (Zanotti et al. 2011). Herein, we showed that T-MSCs constitutively express a higher level of ATF4 and NFATc1 than JB-MSCs, which is conversely correlated with the decreased expression of Notch3 in T-MSCs; knockdown of Notch3 expression led to increased expression of ATF4 and NFATc1 in JB-MSCs.
These findings suggest that a dysregulated Notch3-NFATc1 axis may play an important role in the regulation of aberrant osteogenic differentiation of JB-MSCs, thus contributing to the development of MT. However, further studies are warranted to establish stable Notch3-deficient JB-MSCs through appropriate genetic approaches to verify the long-term effects of loss of Notch3 on osteogenesis of JB-MSCs and the molecular mechanisms in detail.
Author Contributions
X.W. Dou, contributed to conception, design, data acquisition, analysis, and interpretation, critically revised the manuscript; W. Park and S. Lee, contributed to data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; Q.Z. Zhang, A.D. Le, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; L.R. Carrasco, contributed to data acquisition, analysis, and interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplementary Material
Footnotes
A supplemental appendix to this article is available online.
This work was supported by Schoenleber funding support (to A.D.L.), the National Institutes of Health (research grant R01DE 019932 to A.D.L.), the Oral and Maxillofacial Surgery Foundation (research grant to Q.Z.Z. and A.D.L.), the Stephen Milam Oral and Maxillofacial Surgery Foundation (research grant to L.R.C.), the Basic National Research Foundation of Korea (NRF-2013R1A1A1009513 to W.P.).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Barker D, Walls AW, Meechan JG. 2001. Ridge augmentation using mandibular tori. Br Dent J. 190(9):474–476. [DOI] [PubMed] [Google Scholar]
- Bellavia D, Checquolo S, Campese AF, Felli MP, Gulino A, Screpanti I. 2008. Notch3: from subtle structural differences to functional diversity. Oncogene. 27(38):5092–5098. [DOI] [PubMed] [Google Scholar]
- Bruce I, Ndanu TA, Addo ME. 2004. Epidemiological aspects of oral tori in a Ghanaian community. Int Dent J. 54(2):78–82. [DOI] [PubMed] [Google Scholar]
- Chen S, Lee BH, Bae Y. 2014. Notch signaling in skeletal stem cells. Calcif Tissue Int. 94(1):68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Park H, Lee JS, Park JC, Kim CS, Choi SH, Cho KS, Chai JK, Jung UW. 2012. Prevalence and anatomic topography of mandibular tori: computed tomographic analysis. J Oral Maxillofac Surg. 70(6):1286–1291. [DOI] [PubMed] [Google Scholar]
- Cortes AR, Jin Z, Morrison MD, Arita ES, Song J, Tamimi F. 2014. Mandibular tori are associated with mechanical stress and mandibular shape. J Oral Maxillofac Surg. 72(11):2115–2125. [DOI] [PubMed] [Google Scholar]
- Deng ZL, Sharff KA, Tang N, Song WX, Luo J, Luo X, Chen J, Bennett E, Reid R, Manning D, et al. 2008. Regulation of osteogenic differentiation during skeletal development. Front Biosci. 13:2001–2021. [DOI] [PubMed] [Google Scholar]
- Eggen S. 1989. Torus mandibularis: an estimation of the degree of genetic determination. Acta Odontol Scand. 47(6):409–415. [DOI] [PubMed] [Google Scholar]
- El Machhour F, Keuylian Z, Kavvadas P, Dussaule JC, Chatziantoniou C. 2015. Activation of Notch3 in glomeruli promotes the development of rapidly progressive renal disease. J Am Soc Nephrol. 26(7):1561–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin F, Lee B. 2010. NOTCHing the bone: insights into multi-functionality. Bone. 46(2):274–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin F, Yao Z, Yang T, Zhou G, Bertin T, Jiang MM, Chen Y, Wang L, Zheng H, Sutton RE, et al. 2008. Dimorphic effects of Notch signaling in bone homeostasis. Nat Med. 14(3):299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei C, Guo J, Zhao Y, Gu S, Zhao S, Li X, Chang C. 2015. Notch-Hes pathway mediates the impaired osteogenic differentiation of bone marrow mesenchymal stromal cells from myelodysplastic syndromes patients through the down-regulation of Runx2. Am J Transl Res. 7(10):1939–1951. [PMC free article] [PubMed] [Google Scholar]
- Frost HM. 2004. A 2003 update of bone physiology and Wolff’s law for clinicians. Angle Orthod. 74(1):3–15. [DOI] [PubMed] [Google Scholar]
- García-García AS, Martínez-González JM, Gómez-Font R, Soto-Rivadeneira A, Oviedo-Roldán L. 2010. Current status of the torus palatinus and torus mandibularis. Med Oral Patol Oral Cir Bucal. 15(2):e353–e360. [PubMed] [Google Scholar]
- Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, Kronenberg HM, Teitelbaum SL, Ross FP, Kopan R, et al. 2008. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med. 14(3):306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga T, Matsui Y, Asagiri M, Kodama T, de Crombrugghe B, Nakashima K, Takayanagi H. 2005. NFAT and Osterix cooperatively regulate bone formation. Nat Med. 11(8):880–885. [DOI] [PubMed] [Google Scholar]
- Komori T, Takato T. 1998. Time-related changes in a case of torus palatinus. J Oral Maxillofac Surg. 56(4):492–494. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 137(2):216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladizinski B, Lee KC. 2014. A nodular protuberance on the hard palate. JAMA. 311(15):1558–1559. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang X, Leathers R, Makino A, Huang C, Parsa P, Macias J, Yuan JX, Jamieson SW, Thistlethwaite PA. 2009. Notch3 signaling promotes the development of pulmonary arterial hypertension. Nat Med. 15(11):1289–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TM, Lee EH. 2013. Transcriptional regulatory cascades in Runx2-dependent bone development. Tissue Eng Part B Rev. 19(3):254–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F. 2012. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 13(1):27–38. [DOI] [PubMed] [Google Scholar]
- Mosca L, Rivieri F, Tanel R, Bonfante A, Burlina A, Manfredini E, Primignani P, Gesu GP, Marocchi A, Penco S. 2014. Mutational screening of NOTCH3 gene reveals two novel mutations: complexity of CADASIL diagnosis. J Mol Neurosci. 54(4):723–729. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Kho DH, Yanagawa T, Harazono Y, Gao X, Hogan V, Raz A. 2014. Galectin-3 inhibits osteoblast differentiation through notch signaling. Neoplasia. 16(11):939–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proussaefs P. 2006. Clinical and histologic evaluation of the use of mandibular tori as donor site for mandibular block autografts: report of three cases. Int J Periodontics Restorative Dent. 26(1):43–51. [PubMed] [Google Scholar]
- Ragot H, Monfort A, Baudet M, Azibani F, Fazal L, Merval R, Polidano E, Cohen-Solal A, Delcayre C, Vodovar N, et al. 2016. Loss of Notch3 signaling in vascular smooth muscle cells promotes severe heart failure upon hypertension. Hypertension. 68(2):392–400. [DOI] [PubMed] [Google Scholar]
- Samee N, Geoffroy V, Marty C, Schiltz C, Vieux-Rochas M, Levi G, de Vernejoul MC. 2008. Dlx5, a positive regulator of osteoblastogenesis, is essential for osteoblast-osteoclast coupling. Am J Pathol. 173(3):773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafin V, Persano L, Moserle L, Esposito G, Ghisi M, Curtarello M, Bonanno L, Masiero M, Ribatti D, Sturzl M, et al. 2011. Notch3 signalling promotes tumour growth in colorectal cancer. J Pathol. 224(4):448–460. [DOI] [PubMed] [Google Scholar]
- Sirirungrojying S, Kerdpon D. 1999. Relationship between oral tori and temporomandibular disorders. Int Dent J. 49(2):101–104. [DOI] [PubMed] [Google Scholar]
- Sonnier KE, Horning GM, Cohen ME. 1999. Palatal tubercles, palatal tori, and mandibular tori: prevalence and anatomical features in a U.S. population. J Periodontol. 70(3):329–336. [DOI] [PubMed] [Google Scholar]
- Tao J, Chen S, Yang T, Dawson B, Munivez E, Bertin T, Lee B. 2010. Osteosclerosis owing to Notch gain of function is solely Rbpj-dependent. J Bone Miner Res. 25(10):2175–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winslow MM, Pan M, Starbuck M, Gallo EM, Deng L, Karsenty G, Crabtree GR. 2006. Calcineurin/NFAT signaling in osteoblasts regulates bone mass. Dev Cell. 10(6):771–782. [DOI] [PubMed] [Google Scholar]
- Zamurovic N, Cappellen D, Rohner D, Susa M. 2004. Coordinated activation of notch, Wnt, and transforming growth factor-beta signaling pathways in bone morphogenic protein 2-induced osteogenesis: Notch target gene Hey1 inhibits mineralization and Runx2 transcriptional activity. J Biol Chem. 279(36):37704–37715. [DOI] [PubMed] [Google Scholar]
- Zanotti S, Canalis E. 2012. Notch regulation of bone development and remodeling and related skeletal disorders. Calcif Tissue Int. 90(2):69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti S, Canalis E. 2014. Notch1 and Notch2 expression in osteoblast precursors regulates femoral microarchitecture. Bone. 62:22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti S, Smerdel-Ramoya A, Canalis E. 2011. Reciprocal regulation of Notch and nuclear factor of activated T-cells (NFAT) c1 transactivation in osteoblasts. J Biol Chem. 286(6):4576–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti S, Smerdel-Ramoya A, Stadmeyer L, Durant D, Radtke F, Canalis E. 2008. Notch inhibits osteoblast differentiation and causes osteopenia. Endocrinology. 149(8):3890–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hilton MJ, Anolik JH, Welle SL, Zhao C, Yao Z, Li X, Wang Z, Boyce BF, Xing L. 2014. NOTCH inhibits osteoblast formation in inflammatory arthritis via noncanonical NF-kappaB. J Clin Invest. 124(7):3200–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Yamaza T, Kelly AP, Shi S, Wang S, Brown J, Wang L, French SW, Shi S, Le AD. 2009. Tumor-like stem cells derived from human keloid are governed by the inflammatory niche driven by IL-17/IL-6 axis. PLoS One. 4(11):e7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo C, Huang Y, Bajis R, Sahih M, Li YP, Dai K, Zhang X. 2012. Osteoblastogenesis regulation signals in bone remodeling. Osteoporos Int. 23(6):1653–1663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.