Abstract
Temporomandibular disorder (TMD) is a musculoskeletal condition characterized by pain and reduced function in the temporomandibular joint and/or associated masticatory musculature. Prevalence in the United States is 5% and twice as high among women as men. We conducted a discovery genome-wide association study (GWAS) of TMD in 10,153 participants (769 cases, 9,384 controls) of the US Hispanic Community Health Study/Study of Latinos (HCHS/SOL). The most promising single-nucleotide polymorphisms (SNPs) were tested in meta-analysis of 4 independent cohorts. One replication cohort was from the United States, and the others were from Germany, Finland, and Brazil, totaling 1,911 TMD cases and 6,903 controls. A locus near the sarcoglycan alpha (SGCA), rs4794106, was suggestive in the discovery analysis (P = 2.6 × 106) and replicated (i.e., 1-tailed P = 0.016) in the Brazilian cohort. In the discovery cohort, sex-stratified analysis identified 2 additional genome-wide significant loci in females. One lying upstream of the relaxin/insulin-like family peptide receptor 2 (RXP2) (chromosome 13, rs60249166, odds ratio [OR] = 0.65, P = 3.6 × 10−8) was replicated among females in the meta-analysis (1-tailed P = 0.052). The other (chromosome 17, rs1531554, OR = 0.68, P = 2.9 × 10−8) was replicated among females (1-tailed P = 0.002), as well as replicated in meta-analysis of both sexes (1-tailed P = 0.021). A novel locus at genome-wide level of significance (rs73460075, OR = 0.56, P = 3.8 × 10−8) in the intron of the dystrophin gene DMD (X chromosome), and a suggestive locus on chromosome 7 (rs73271865, P = 2.9 × 10−7) upstream of the Sp4 Transcription Factor (SP4) gene were identified in the discovery cohort, but neither of these was replicated. The SGCA gene encodes SGCA, which is involved in the cellular structure of muscle fibers and, along with DMD, forms part of the dystrophin-glycoprotein complex. Functional annotation suggested that several of these variants reside in loci that regulate processes relevant to TMD pathobiologic processes.
Keywords: epidemiology, functional annotation, population, genetics, Hispanic Americans, musculoskeletal pain
Introduction
Chronic pain is pain that persists beyond the normal healing time (Bonica 1953) and no longer serves the adaptive warning function of nociception. In the United States, 25 million adults report chronic pain on a daily basis (Nahin 2015), and prevalence of pain disorders increased, including facial pain, from 1997–1999 to 2011–2013 (Case and Deaton 2015). Because mechanisms underlying chronic pain are largely unknown, treatment in general has low effectiveness. For example, management with nonsteroidal anti-inflammatory drugs has limited efficacy (da Costa et al. 2016).
Much of the burden of chronic pain is attributable to tension headache, low back pain, neck pain, fibromyalgia, and face pain. One such condition, temporomandibular disorder (TMD), is a musculoskeletal condition characterized by nonodontogenic pain and loss of function in the region innervated by the trigeminal nerve. The condition is diagnosed clinically by assessing palpation tenderness and functional pain in the masseter and temporalis masticatory muscles (myalgia) and/or the temporomandibular joint/s (arthralgia). In contrast, most population-based surveys classify the condition using screening questions. For example, in the 2014 US National Health Interview Survey, 4.6% of adults reported pain in the face or jaw in the previous 3 mo, with prevalence being higher among women (5.8%) than men (3.4%) (Blackwell et al. 2014).
TMD pain is not attributable to any known tissue damage, and its pathophysiology is unclear. Multiple clinical and biopsychosocial risk factors are implicated in its sensory and affective dimensions. Among these, anxiety, depression and somatosensory amplification (Von Korff and Simon 1996; Manfredini et al. 2004), and sleep disturbance (Edwards et al. 2009; Smith et al. 2009) have undergone intensive investigation. Frequently, TMD overlaps with other pain disorders, including headache, irritable bowel syndrome, low back pain, and chronic widespread pain. In a study that asked 200 women with persistent TMD to indicate the anatomic locations of their painful body sites on manikins, only 19% indicated sites limited to the trigeminal system (Türp et al. 1998). Of the 5% of adults with TMD pain in 2000 to 2006 National Health Interview Surveys, 53% reported concomitant headache/migraine and 64% reported concomitant low back pain (Plesh et al. 2011). Prevalence was similar in Sweden, affecting 5% of women and 2% of men (Lövgren et al. 2016), but higher prevalence was reported in other population-based studies, including Germany (Gesch et al. 2004) and Italy (Ciancaglini and Radaelli 2001). Genetic studies are beginning to shed light onto the molecular basis for pain perception and to identify putative genes associated with various chronic pain disorders. Although these associations have not been consistently replicated (Foulkes and Wood 2008; Mogil 2012), a genetic basis for TMD pain seems plausible. The largest twin study of 1,236 monozygotic and 570 dizygotic female twin pairs estimated that the heredity of TMD pain was 27% (95% confidence limit [CL], 15 to 38) of the population variance in TMD pain (Plesh et al. 2012).
The discovery phase of this study sought to identify genetic loci associated with painful TMD via a genome-wide association scan of 10,153 men and women of Hispanic/Latino ancestry. The top signals were tested for replication in a meta-analysis using data from independent studies that conducted genome-wide association scans of painful TMD. We also conducted functional genomic analysis of significant loci to assess how the associated alleles could modulate gene or protein function.
Materials and Methods
Ethical Statement
This study was conducted according to the principles expressed in the Declaration of Helsinki. This study was approved by the institutional review boards (IRBs) of the 4 Hispanic Community Health Study/Study of Latinos (HCHS/SOL) field centers affiliated with San Diego State University, Northwestern University in Chicago, Albert Einstein College of Medicine in the Bronx area of New York, and the University of Miami. The Orofacial Pain: Prospective Evaluation and Risk Assessment (OPPERA) study was approved by the IRBs at the 4 study sites at the University of North Carolina at Chapel Hill, University of Maryland at Baltimore, University of Buffalo, University of Florida at Gainesville, and the data coordinating center Battelle Memorial Institute. The Study of Health in Pomerania (SHIP) was approved by the IRB affiliated with study at the University of Greifswald, Germany. The Northern Finland Birth Cohort (NFBC) study was approved by the human study ethical committees of the University of Oulu and the Northern Ostrobothnia Hospital District. The Brazilian study was approved by the IRB affiliated with the University of Campinas, Piracicaba, São Paulo in Brazil.
This study conformed with Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines for observational studies.
HCHS/SOL
The HCHS/SOL is a population-based cohort in the United States. Between 2008 and 2011, the HCHS/SOL enrolled 16,415 study participants who were recruited as self-identified Hispanic/Latino participants aged 18–74 y at each of 4 US communities located in the Bronx, New York; San Diego, California; Miami, Florida; and Chicago, Illinois. The stratified multistage area probability sample of the 4 study sites was purposefully designed to have high concentrations of specific Hispanic/Latino backgrounds, allowing estimation of prevalence rates of diseases and risk factors for each background. Details of the design, recruitment, and implementation of HCHS/SOL have been published (Lavange et al. 2010; Sorlie et al. 2010). The present study reports cross-sectional analysis of baseline data.
TMD Phenotype in the Discovery HCHS/SOL Cohort
The HCHS/SOL Oral Health Questionnaire asked participants, “In the past 12 mo have you had or do you currently have pain in your face?” They were also asked, “In the past 12 mo have you had or do you currently have pain in your jaw joint?” Response options for both questions were yes/no. To be classified as a TMD case, participants had to report having had pain in both their face and in their jaw joint. The requirement of pain in both face and jaw joint is a stringent case definition. The rationale was to reduce misclassification bias due to other causes underlying one or the other pain locations and limit phenotypic heterogeneity that could arise from the presence of pain in either the muscle or the joint. However, no information was available on the duration of symptoms; thus, TMD cases may include individuals with acute (i.e., self-limited short-term pain) or chronic (i.e., persistent) TMD.
Descriptive univariate multivariate analyses were conducted on all 15,344 HCHS/SOL participants who provided information about TMD symptoms. These analyses were weighted to account for the stratified multistage area probability sampling and to at least partially adjust for any bias effects due to differential nonresponse in the selected sample at the household and person levels. The adjusted weights were also trimmed to limit precision losses due to the variability of the adjusted weights and calibrated to the 2010 Census characteristics by age, sex, and Hispanic background in each field site’s target population. All analyses also account for cluster sampling and the use of stratification in sample selection.
Genome-Wide Association Study Quality Control
In the HCHS/SOL, DNA was extracted from blood samples according to standard protocols. Participants were genotyped on the HCHS Custom 15041502 B3 array (Illumina Omni2.5M+custom content). Quality control was conducted as previously described (Laurie et al. 2010; Conomos et al. 2016). In brief, DNA samples were checked for annotated versus genetic sex discrepancies, gross chromosomal anomalies, missing call rates, contaminations, and batch effects. Single-nucleotide polymorphism (SNP) quality metrics included Illumina/LA Biomed assay failure indicator, missing call rates, deviation from Hardy-Weinberg equilibrium, Mendelian errors, and duplicate sample discordance. After quality control, there were about 1.7 million genotyped SNPs and 10.3 million imputed SNPs available for analysis.
Genotypes were prephased using SHAPEIT2 and imputed using IMPUTE2 into the 1000 Genomes Phase 1 reference panel. For imputed SNPs, we calculated the imputation quality scores “info” and “oevar” (ratio of observed to expected variance of imputed dosage) for each SNP and excluded SNPs with oevar <0.3 (Li et al. 2010). Finally, SNPs with 50 or fewer counts of the expected or effective number of copies (effN) of the minor allele were excluded from analysis (Cade et al. 2016). A variant’s effN is approximately its minor allele count and was estimated as 2 × MAF × (1−MAF) × N × oevar, where MAF is the frequency of the minor allele, N is the number of participants, and oevar is set to 1 for genotyped variants. After quality control, there were about 1.4 million genotyped SNPs and 7 million imputed SNPs available for analysis.
Statistical Analysis in HCHS/SOL
To account for population structure and relatedness, we applied a logistic mixed model using GMMAT (Chen et al. 2016) to test for a SNP genotype and case versus control status. The model adjusted for fixed effects of age, sex, recruitment site, log sampling weight, 5 principal components of ancestry, and 6 genetic analysis groups. We provide 95% CLs obtained from the replication cohort but not the discovery cohort; confidence intervals from the discovery study underestimate the plausible range of values for the measure of association (Zhong and Prentice 2008). We provide 95% CLs obtained from the replication cohort, as these are expected to have the correct 95% coverage, while the confidence limits from the discovery study underestimate the plausible range of values for the measure of association.
The genetic analysis groups are based on self-reported ethnicity and genetic similarity between participants, and it was shown by Conomos et al. (2016) that it is advantageous to adjust for this variable, compared to self-reported ethnicity.
Meta-Analysis
We looked for replication of the top findings in 4 independent cohorts of examiner-determined TMD. Meta-analysis was used to combine the effects of all replication cohorts, where possible, as a primary test of replication. For SNPs that were not found in multiple replication cohorts, evidence from the single available cohort was used to assess replication. Random-effects meta-analysis was performed using PLINK 1.07 software (Purcell et al. 2007). Allelic odds ratios (ORs), 95% CLs, and P values were calculated separately for each genome-wide association study (GWAS) included in the meta-analysis. Study-specific SNP filters for imputation quality and minor allele frequency (MAF) were imposed, as described for each study. All SNPs passing quality assurance (QA)/quality control (QC) filters in at least 2 cohorts were included in the meta-analysis. The forward strand allele (using reference assemble GRCh37) was used as the reference to standardize direction of ORs. As replication implicitly requires the same direction of effect as the original finding, P values were calculated for one-sided tests; P < 0.05 in a 1-sided test was considered the threshold for a replicated association. The Cochrane’s Q statistic P value was used as a measure of heterogeneity between studies (Xu et al. 2008).
Replication Cohorts
Details of the clinical examination used to determine TMD case classification are described in the online Appendix. Here we describe the TMD phenotype.
OPPERA
The OPPERA case-control cohort included 3,030 participants, of whom 999 were TMD cases and 2,031 TMD-free controls. Examiners determined classification of TMD according to the Research Diagnostic Criteria for TMD (Dworkin and LeResche 1992). As reported previously (Ohrbach et al. 2011), cases met all 3 of the following criteria: 1) pain reported with sufficient frequency in the cheeks, jaw muscles, temples, or jaw joints during the preceding 6 mo; 2) pain reported in the examiner-defined orofacial region for at least 5 d out of the prior 30 d; and 3) pain reported in at least 3 masticatory muscles or at least 1 temporomandibular joint in response to palpation of the orofacial muscles or maneuver of the jaw.
SHIP
The German cohort was derived from a large cross-sectional survey of a representative sample of Pomerania, Germany, the SHIP study (Bernhardt et al. 2004). Participants were aged 20 to 81 y and included 51% females. Participants reported symptoms by questionnaire regarding pain in the temporomandibular joint and facial muscles; presence and frequency of pain were assessed. During a clinical exam, the examiner inquired about pain or discomfort upon palpation of masticatory tissues, including temporomandibular joints (dorsocranial and lateral) at 2 kg/cm2 and masseter, temporalis, and medial pterygoid at 1 kg/cm2.
NFBC
The NFBC is a cohort study of all births in 1966 in the Oulu and Lapland provinces of northern Finland. An assessment for TMD was performed at the 46-y follow-up time point. Participants (52% female) reported symptoms by responding to a questionnaire with the following questions: 1) “Do you experience temple, temporomandibular joint, face, or jaw pain once a week or more often?” 2) “Do you experience pain once a week or more often while opening your mouth wide?” A clinical exam determined the presence of examiner-evoked pain in 3 or more temporomandibular muscles and/or joints.
Brazilian Cohort
The Brazilian participants were enrolled in a community-based case-control study in Piracicaba, São Paulo, Brazil, and included females between the ages of 18 and 44 y. Pain history was determined by asking participants the following question: “Have you had pain in your head, face, jaw, or in front of the ears in the last 30 d?” The examiner manually palpated lateral and posterior temporomandibular joints (0.45 kg) and asked participants to report yes or no responses to the presence of pain.
Results
Descriptive univariate associations between study participant characteristics and TMD in the full HCHS/SOL cohort are reported in Appendix Table 1 along with recruitment site-adjusted OR and 95% CL. Prevalence of TMD was 5.1%. Females had 2-fold greater odds of TMD than males. Odds of TMD declined after age 55 y. Adults who did not graduate high school had 20% greater odds of TMD than those with education beyond high school education. Current smokers had 30% greater odds than lifetime nonsmokers. Depressive symptoms, anxiety, and poor mental and physical health were all associated with increased odds of TMD. Odds of TMD did not differ among Hispanic/Latino groups or differ on the basis of nativity, body mass index, or health insurance status.
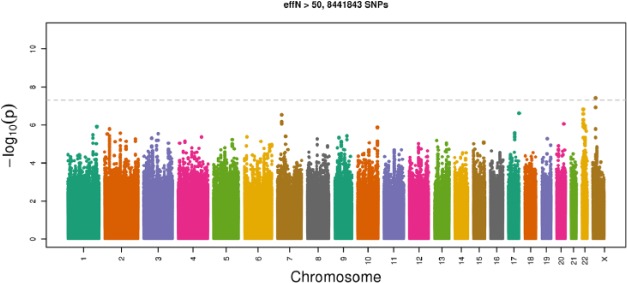
The genome-wide scan of the HCHS/SOL cohort used DNA from whole blood collected at baseline from 10,153 study participants (769 TMD cases and 9,384 controls). Excluded from analysis were 2,603 participants with missing information for the TMD trait and an additional 63 participants with missing information for genetic subgroup. A quantile-quantile (Q-Q) plot analysis was conducted of the observed versus expected P values for all 8,441,843 SNPs that passed quality filters (effN > 50 in both cases and controls) and had a MAF of >3% (Appendix Fig. 1). Departures in the extreme tail of the distribution of test statistics are due to regions with a strong signal for association. The genomic inflation factor (lambda statistic 1.007) shows no evidence of genomic inflation.
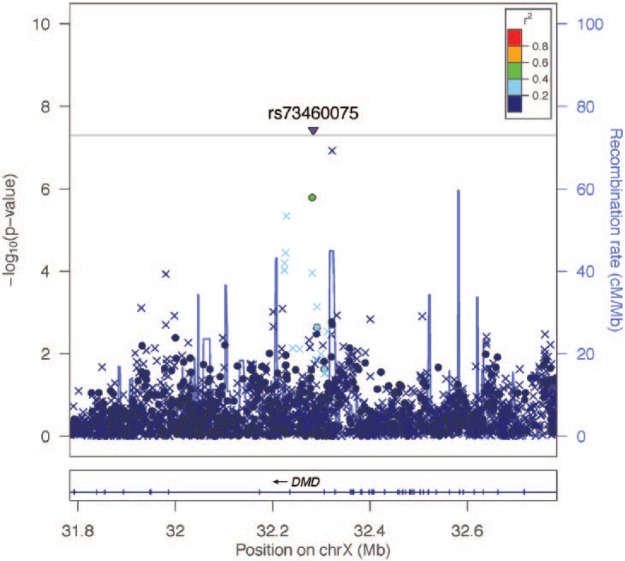
The discovery GWAS (Table 1) revealed a novel locus that exceeded the genome-wide threshold for significance on the X chromosome, rs73460075 (Manhattan plot, Fig. 1), OR = 0.56, P = 3.8 × 10−8. A regional association plot (Fig. 2) shows all SNPs in this region plotted according to the significance of their association with TMD and color-coded according to their linkage disequilibrium (r2) with the most significant SNP. This locus is rare in European populations (MAF < 0.001) but is common in African populations (MAF = 0.146), according to the 1000 Genomes Project Phase 1. This SNP lies in the intron of the dystrophin-encoding DMD gene responsible for Duchenne muscular dystrophies. In addition, we found suggestive evidence of an association with TMD for 2 loci (Appendix Fig. 4A, B). One was rs4794106 near the sarcoglycan alpha (SGCA) gene (OR = 1.30, P = 2.6 × 10−6), and the other was rs73271865 upstream of the Sp4 transcription factor (SP4) gene (OR = 0.56, P = 2.9 × 10−7) (Appendix Fig. 2A, B).
Table 1.
Summary of the Association Results of the Discovery GWAS in the HCHS/SOL.
| Analysis | Chromosome | Position | rsID | Nearest Gene | A1/A2 | A1.freq | n | OR | P Value |
|---|---|---|---|---|---|---|---|---|---|
| All | 23 | 32283492 | rs73460075 | DMD | G/C | 0.964 | 10,142 | 0.563 | 3.79E-08 |
| All | 17 | 48238294 | rs4794106 | SGCA | T/C | 0.549 | 10,153 | 1.296 | 2.60E-06 |
| All | 7 | 21399327 | rs73271865 | SP4 | C/T | 0.959 | 10,153 | 0.559 | 2.91E-07 |
| Females | 13 | 32084901 | rs60249166 | RXP2 | C/T | 0.812 | 5,820 | 0.645 | 3.57E-08 |
| Females | 17 | 79380547 | rs1531554 | BAHCC1 | T/C | 0.476 | 5,820 | 0.678 | 2.92E-08 |
A1, effect allele, A1.freq, its frequency; A2, other allele; GWAS, genome-wide association study; HCHS/SOL, Hispanic Community Health Study/Study of Latinos; OR, odds ratio; rsID, reference SNP cluster ID.
Figure 1.
Manhattan plot showing association between the tested single-nucleotide polymorphisms and painful temporomandibular disorder for men and women combined in the Hispanic Community Health Study/Study of Latinos discovery genome-wide association analysis. The X axis shows the chromosomal position, and the Y axis shows the significance level with the horizontal dashed line indicating the genome-wide significance threshold (P = 5 × 10−8).
Figure 2.
Regional association plot (LocusZoom) for the DMD-region (X chromosome) significantly associated with painful temporomandibular disorder in the Hispanic Community Health Study/Study of Latinos discovery cohort, men and women combined. The –log10 of P values are plotted against the single-nucleotide polymorphism (SNP) genomic position based on NCBI build 37; the estimated recombination rate from the 1000 Genomes Project is on the right vertical axis and is plotted in blue. SNPs are colored to reflect correlation with the most significant SNP. The inverted triangle indicates that the genome-wide significant SNP, rs73460075, was imputed. Other x-symbols denote imputed SNPs, and circles denote genotyped SNPs.
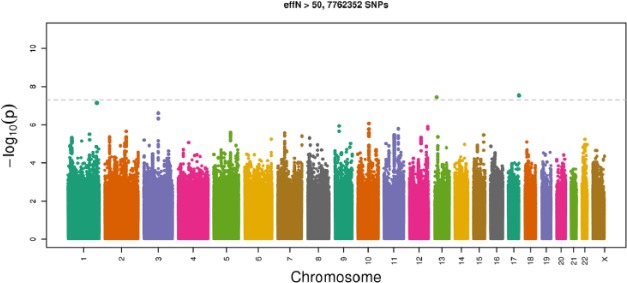
The Q-Q plots for sex-stratified genome-wide association analyses did not indicate any evidence for systematic increase in false positives as the observed distribution did not deviate from the expected distribution (Appendix Fig. 3). The sex-stratified genome-wide association in females identified 2 additional significant loci shown in the Manhattan plot (Fig. 3). The first, rs60249166 on chromosome 13 (P = 3.6 × 10−8), lies upstream of the RXP2 gene (relaxin/insulin-like family peptide receptor 2, Appendix Fig. 4A). The second, rs1531554 on chromosome 17 (P = 2.9 × 10−8), is located in the first exon of the BAHCC1 gene (BAH domain and coiled-coil containing 1, Appendix Fig. 4B).
Figure 3.
Manhattan plot showing the chromosomal position and –log10 (P value) of the single-nucleotide polymorphisms (SNPs) among females in the Hispanic Community Health Study/Study of Latinos discovery genome-wide association analysis (P < 5 × 10−8 indicated by the dotted black line). Two SNPs exceed the genome-wide significance threshold.
The 3 SNPs associated with TMD among all adults (rs73460075, rs4794106, and rs73271865) and the 2 SNPs associated with TMD among females only (rs60249166, rs1531554) in the HCHS/SOL discovery phase were further evaluated in 4 independent cohorts (Appendix Table 2), as well as in a meta-analysis of their combined evidence of associations (Table 2). These independent studies are OPPERA (conducted in the United States), SHIP (conducted in Germany), NFBC, and a case-control study of TMD conducted in Piracicaba, São Paulo, Brazil.
Table 2.
Summary of the Association Results of the Meta-analysis.
| Included | rsID | Chromosome | Base Pair | Closest Gene | Coded/Noncoded Allele | No. of Studies in Meta-analysis | OR (95% CL) | P | Q | H |
|---|---|---|---|---|---|---|---|---|---|---|
| All | rs73460075 | 23 | 32283492 | DMD | G/C | 1 | NA | NA | NA | NA |
| All | rs4794106 | 17 | 48238294 | SGCA | T/C | 4 | 1.03 (0.94 to 1.11) | 0.357 | 0.085 | 54.74 |
| All | rs73271865 | 7 | 21399327 | SP4 | C/T | 1 | NA | NA | NA | NA |
| Females | rs60249166 | 13 | 32084901 | RXP2 | C/T | 2 | 0.87 (0.74 to 1.03) | 0.052 | 0.854 | 0 |
| Females | rs1531554 | 17 | 79380547 | BAHCC1 | T/C | 2 | 0.83 (0.73 to 0.95) | 0.002 | 0.869 | 0 |
CL, confidence limits; OR, odds ratio from random-effects meta-analysis; H, index of heterogeneity; NA, not applicable; Q, Cochrane’s Q statistic.
The SNPs in the DMD (rs73460075) and SP4 (rs73271865) loci were only available in the OPPERA cohort, and neither was significantly associated with TMD (P = 0.28 and 0.36, respectively). However, the SNP near the SGCA locus (rs4794106) was found in all 4 replication cohorts. Although the combined evidence from the 4 cohorts was not significant (meta-analysis P = 0.40), the association observed in the cohort from Brazil supported the original finding (P = 0.02). To confirm the female-specific findings, we used the results of sex-stratified analyses in the replication cohorts. There was evidence from the meta-analysis of 2 studies to support the association near the RXP2 locus (rs60249166, P = 0.05), although it was not significant in either the OPPERA study (P = 0.10) or SHIP study (P = 0.11) alone. Finally, the SNP in the BAHCC1 locus was significant in both OPPERA (P = 0.001) and SHIP (P = 0.03), as well as in the meta-analysis of both cohorts (P = 0.002).
Discussion
This GWAS in the HCHS/SOL cohort revealed several promising association signals for painful TMD, with supporting evidence from both biological relevance to nociception as well as replication in a meta-analysis of 4 independent cohorts. Two prominent associations (near DMD and SGCA) implicate the same dystrophin-glycoprotein pathway, suggesting that biomechanical properties of muscle fibers contribute to orofacial pain. Although we were not able to replicate the association at the DMD locus, the association at the SGCA locus was significantly replicated in 1 independent cohort and was consistent in direction in the meta-analysis of 4 cohorts. The SGCA gene encodes SGCA, which is involved in the cellular structure of muscle fibers and, along with DMD, forms part of the dystrophin-glycoprotein complex. Likewise, we were limited in our ability to confirm the suggestive association at the SP4 locus, but the effect direction was the same in the 1 independent cohort with data available. As SP4 is a transcription factor for the transient receptor potential vanilloid 1 (TRPV1), which is upregulated in chronic pain states, this finding should be investigated further. The biological relevance to nociception of the 2 loci discovered in the female-only GWAS analyses (near RXP2 and BAHCC1) is not as easily explained, but these associations were consistently supported by the replication cohorts and may be important novel discoveries.
A potential limitation of the HCHS/SOL discovery cohort was its dependence on self-reported facial pain, which is known to have good specificity (95%) but poor sensitivity (43%) compared to examiner-determined TMD (Janal et al. 2008).
However, in most situations, misclassification that is nondifferential with respect to the genotype will bias estimates toward the null, assuming that the misclassification of TMD pain is independent of other errors. Based on this assumption, our estimates of association in the discovery cohort are conservative and reduce the risk of false-positive genetic associations. A related point is that 3 of the 4 replication cohorts used examiner-determined case classifications for TMD, which means that data from those studies do not satisfy requirements for “exact replication” of the HCHS/SOL discoveries (Ioannidis et al. 2009). This is a common problem for GWAS replication studies where at least some variation in phenotype definitions has been described as “unavoidable” (Kraft et al. 2009). The replication studies also varied in sampling methods, study population, and demographic characteristics. We therefore interpret the current findings of consistency among cohorts as evidence that the associations are robust and generalizable to different study populations.
Overall, we discovered and replicated SNP associations with TMD in multiple loci. For 2 loci, we propose plausible mechanism of regulation and potential causal SNPs via comprehensive functional annotation of processes relevant to TMD pathology.
Author Contributions
A.E. Sanders, contributed to conception, design, data analysis, and interpretation, drafted and critically revised the manuscript; D. Jain, T. Sofer, contributed to data analysis and interpretation, drafted and critically revised the manuscript; K.F. Kerr, C.C. Laurie, J.R. Shaffer, M.L. Marazita, contributed to data interpretation, critically revised the manuscript; L.M. Kaste, contributed to data acquisition, critically revised the manuscript; G.D. Slade, C. Schwahn, contributed to data analysis and interpretation, critically revised the manuscript; R.B. Fillingim, R. Ohrbach, W. Maixner, T. Kocher, O. Bernhardt, A. Teumer, K. Sipilä, R. Lähdesmäki, M. Männikkö, P. Pesonen, M. Järvelin, C.M. Rizzatti-Barbosa, C.B. Meloto, M. Ribeiro-Dasilva, L. Diatchenko, P. Serrano, contributed to data acquisition and interpretation, critically revised the manuscript; S.B. Smith, contributed to design, data analysis, and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplementary Material
Acknowledgments
We thank the participants and staff of the Hispanic Community Health Study/Study of Latinos (HCHS/SOL).
Footnotes
The baseline examination of HCHS/SOL was supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (grant number N01-HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC 65236), and San Diego State University (N01-HC65237). The National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research (NIDCR), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Neurological Disorders and Stroke, and National Institutes of Health (NIH) Office of Dietary Supplements also contributed funding to HCHS/SOL. The Genetic Analysis Center at the University of Washington was supported by NHLBI and NIDCR contracts (grant numbers HHSN268201300005C AM03 and MOD03). Additional analysis support was provided by 1R01DK101855-01 and 13GRNT1 6490017. Genotyping efforts were supported by NHLBI HSN 26220/20054C, NCATS CTSI grant UL1TR000124, and NIDDK Diabetes Research Center (DRC) (grant DK063491). Additional support for rs117672662 functional studies was provided by grant R01 DK072193. Data from HCHS/SOL are available through the NIH database of Genotypes and Phenotypes (dbGaP): phs000810.v1.p1.
OPPERA was supported by NIDCR (grant number U01DE017018). The OPPERA program also acknowledges resources specifically provided for this project by the respective host universities: University at Buffalo, University of Florida, University of Maryland–Baltimore, and University of North Carolina–Chapel Hill. Funding for genotyping was provided by NIDCR through a contract to the Center for Inherited Disease Research at Johns Hopkins University (HHSN268201200008I). Data from the OPPERA study are available through the NIH dbGaP: phs000796.v1.p1, phs000761.v1.p1.
SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, by the Federal Ministry of Education and Research (grant numbers 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs, and the Social Ministry of the Federal State of Mecklenburg–West Pomerania and the network “Greifswald Approach to Individualized Medicine (GANI_MED),” funded by the Federal Ministry of Education and Research (grant number 03IS2061A).
Genome-wide data have been supported by the Federal Ministry of Education and Research (grant number 03ZIK012) and a joint grant from Siemens Healthcare (Erlangen, Germany) and the Federal State of Mecklenburg–West Pomerania. The University of Greifswald is a member of the Caché Campus program of the InterSystems GmbH.
The Northern Finland Birth Cohort 1966 has received financial support from the Academy of Finland (project grants 104781, 120315, 129269, 1114194, 24300796, Center of Excellence in Complex Disease Genetics and SALVE), University Hospital Oulu, Biocenter, University of Oulu (75617), NHLBI grant 5R01HL087679-02 through the STAMPEED program (1RL1MH083268-01), NIH/National Institute of Mental Health (NIMH) (5R01MH63706:02), ENGAGE project and grant agreement HEALTH-F4-2007-201413, EU FP7 EurHEALTH Ageing-277849, the Medical Research Council (G0500539, G0600705, G1002319, PrevMetSyn/SALVE), and the MRC, Centenary Early Career Award. The program is currently being funded by the H2020-633595 DynaHEALTH action and Academy of Finland EGEA-project (285547). The DNA extractions, sample quality controls, biobank upkeeping, and aliquoting were performed in the National Public Health Institute, Biomedicum Helsinki and supported financially by the Academy of Finland and Biocentrum Helsinki. We thank the late Professor Paula Rantakallio (launch of NFBCs) and Ms Outi Tornwall and Ms Minttu Jussila (DNA biobanking). The authors would like to acknowledge the contribution of the late Academian of Science Leena Peltonen. The study has been financially supported by the Academy of Finland, the European Commission (EURO-BLCS, Framework 5 award QLG1-CT-2000-01643), the Sigrid Jusélius Foundation, and US National Institute of Mental Health (5R01MH 63706:02).
The Brazilian cohort has been funded by the São Paulo Research Foundation (grant numbers 2006/56019-8R and 2009/02520-6), and genotyping was funded by the Canadian Excellence Research Chairs (CERC) Program (grant CERC09).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
A supplemental appendix to this article is available online.
References
- Bernhardt O, Gesch D, Schwahn C, Bitter K, Mundt T, Mack F, Kocher T, Meyer G, Hensel E, John U. 2004. Signs of temporomandibular disorders in tinnitus patients and in a population-based group of volunteers: results of the Study of Health in Pomerania. J Oral Rehabil. 31(4):311–319. [DOI] [PubMed] [Google Scholar]
- Blackwell DL, Lucas JW, Clarke TC. 2014. Summary health statistics for U.S. adults: national health interview survey, 2012. Vital Health Stat. 10(260):1–161. [PubMed] [Google Scholar]
- Bonica JJ. 1953. The management of pain. Philadelphia (PA): Lea & Febiger. [Google Scholar]
- Cade BE, Chen H, Stilp AM, Gleason KJ, Sofer T, Ancoli-Israel S, Arens R, Bell GI, Below JE, Bjonnes AC, et al. 2016. Genetic associations with obstructive sleep apnea traits in Hispanic/Latino Americans. Am J Respir Crit Care Med. 194(7):886–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case A, Deaton A. 2015. Rising morbidity and mortality in midlife among white non-Hispanic Americans in the 21st century. Proc Natl Acad Sci U S A. 112(49):15078–15083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wang C, Conomos MP, Stilp AM, Li Z, Sofer T, Szpiro AA, Chen W, Brehm JM, Celedon JC, et al. 2016. Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am J Hum Genet. 98(4):653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciancaglini R, Radaelli G. 2001. The relationship between headache and symptoms of temporomandibular disorder in the general population. J Dent. 29(2):93–98. [DOI] [PubMed] [Google Scholar]
- Conomos MP, Laurie CA, Stilp AM, Gogarten SM, McHugh CP, Nelson SC, Sofer T, Fernandez-Rhodes L, Justice AE, Graff M, et al. 2016. Genetic diversity and association studies in US Hispanic/Latino populations: applications in the Hispanic community health study/study of Latinos. Am J Hum Genet. 98(1):165–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa BR, Reichenbach S, Keller N, Nartey L, Wandel S, Juni P, Trelle S. 2016. Effectiveness of non-steroidal anti-inflammatory drugs for the treatment of pain in knee and hip osteoarthritis: a network meta-analysis. Lancet. 387(10033):2093–2105. [DOI] [PubMed] [Google Scholar]
- Dworkin SF, LeResche L. 1992. Research diagnostic criteria for temporomandibular disorders: review, criteria, examinations and specifications, critique. J Craniomandib Disord. 6(4):301–355. [PubMed] [Google Scholar]
- Edwards RR, Grace E, Peterson S, Klick B, Haythornthwaite JA, Smith MT. 2009. Sleep continuity and architecture: associations with pain-inhibitory processes in patients with temporomandibular joint disorder. Eur J Pain. 13(10):1043–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes T, Wood JN. 2008. Pain genes. PLoS Genet. 4(7):e1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesch D, Bernhardt O, Alte D, Schwahn C, Kocher T, John U, Hensel E. 2004. Prevalence of signs and symptoms of temporomandibular disorders in an urban and rural German population: results of a population-based study of health in Pomerania. Quintessence Int. 35(2):143–150. [PubMed] [Google Scholar]
- Ioannidis JP, Thomas G, Daly MJ. 2009. Validating, augmenting and refining genome-wide association signals. Nat Rev Genet. 10(5):318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janal MN, Raphael KG, Nayak S, Klausner J. 2008. Prevalence of myofascial temporomandibular disorder in US community women. J Oral Rehabil. 35(11):801–809. [DOI] [PubMed] [Google Scholar]
- Kraft P, Zeggini E, Ioannidis JP. 2009. Replication in genome-wide association studies. Stat Sci. 24(4):561–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T, Boehm F, Caporaso NE, Cornelis MC, Edenberg HJ, et al. 2010. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol. 34(6):591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavange LM, Kalsbeek WD, Sorlie PD, Aviles-Santa LM, Kaplan RC, Barnhart J, Liu K, Giachello A, Lee DJ, Ryan J, et al. 2010. Sample design and cohort selection in the Hispanic Community Health Study/Study of Latinos. Ann Epidemiol. 20(8):642–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. 2010. Mach: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 34(8):816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lövgren A, Häggman-Henrikson B, Visscher CM, Lobbezoo F, Marklund S, Wänman A. 2016. Temporomandibular pain and jaw dysfunction at different ages covering the lifespan—a population based study. Eur J Pain. 20(4):532–540. [DOI] [PubMed] [Google Scholar]
- Manfredini D, Bandettini di, Poggio A, Cantini E, Dell’Osso L, Bosco M. 2004. Mood and anxiety psychopathology and temporomandibular disorder: a spectrum approach. J Oral Rehabil. 31(10):933–940. [DOI] [PubMed] [Google Scholar]
- Mogil JS. 2012. Pain genetics: past, present and future. Trends Genet. 28(6):258–266. [DOI] [PubMed] [Google Scholar]
- Nahin RL. 2015. Estimates of pain prevalence and severity in adults: United States, 2012. J Pain. 16(8):769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohrbach R, Fillingim RB, Mulkey F, Gonzalez Y, Gordon S, Gremillion H, Lim PF, Ribeiro-Dasilva M, Greenspan JD, Knott C, et al. 2011. Clinical findings and pain symptoms as potential risk factors for chronic TMD: descriptive data and empirically identified domains from the OPPERA case-control study. J Pain. 12(Suppl 11):T27–T45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesh O, Adams SH, Gansky SA. 2011. Temporomandibular joint and muscle disorder-type pain and comorbid pains in a national US sample. J Orofac Pain. 25(3):190–198. [PMC free article] [PubMed] [Google Scholar]
- Plesh O, Noonan C, Buchwald DS, Goldberg J, Afari N. 2012. Temporomandibular disorder-type pain and migraine headache in women: a preliminary twin study. J Orofac Pain. 26(2):91–98. [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. Plink: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MT, Wickwire EM, Grace EG, Edwards RR, Buenaver LF, Peterson S, Klick B, Haythornthwaite JA. 2009. Sleep disorders and their association with laboratory pain sensitivity in temporomandibular joint disorder. Sleep. 32(6):779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie PD, Avilés-Santa LM, Wassertheil-Smoller S, Kaplan RC, Daviglus ML, Giachello AL, Schneiderman N, Raij L, Talavera G, Allison M, et al. 2010. Design and implementation of the Hispanic community health study/study of Latinos. Ann Epidemiol. 20(8):629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Türp JC, Kowalski CJ, O’Leary N, Stohler CS. 1998. Pain maps from facial pain patients indicate a broad pain geography. J Dent Res. 77(6):1465–1472. [DOI] [PubMed] [Google Scholar]
- Von Korff M, Simon G. 1996. The relationship between pain and depression. Br J Psychiatry Suppl. (30): 101–108. [PubMed] [Google Scholar]
- Xu H, Platt RW, Luo ZC, Wei S, Fraser WD. 2008. Exploring heterogeneity in meta-analyses: needs, resources and challenges. Paediatr Perinat Epidemiol. 22(Suppl 1):18–28. [DOI] [PubMed] [Google Scholar]
- Zhong H, Prentice RL. 2008. Bias-reduced estimators and confidence intervals for odds ratios in genome-wide association studies. Biostatistics. 9(4):621–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.