Abstract
Lung adenocarcinoma is known for its high rate of somatic mutations and genomic rearrangements. The identification of epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase (ALK) rearrangements that sensitize tumors to specific drugs has changed the therapeutic approach and prognosis in these molecularly-defined subgroups. Several other key genetic alterations have been identified, of which BRAF mutations are found in 4% of non-small cell lung cancer (NSCLC) cases. Targeted drugs against BRAF and downstream MEK were recently approved for the treatment of BRAF-positive melanoma and have entered clinical evaluation in NSCLC. In this review we discuss the latest evidence on the treatment of BRAF-mutated NSCLC, including tumor biology, targeted treatment with BRAF and MEK inhibitors, therapeutic resistance and strategies to overcome resistance.
Keywords: BRAF, NSCLC, TKI
Introduction
State of the art treatment for advanced non-small cell lung cancer (NSCLC) is based on immunohistochemical and molecular tissue characteristics that drive tumorigenesis. In the absence of targetable genetic alterations, first-line therapy generally consists of platinum doublet chemotherapy in which histology directs the choice of the platinum partner drug [Scagliotti et al. 2008]. With the arrival of immunotherapy in the second-line setting, most patients with tumors that do not exhibit a targetable genetic alteration will be treated with an anti-PD-(L1) agent in the second line setting [Borghaei et al. 2015; Brahmer et al. 2015]. On the contrary, patients with a targetable genetic alteration are preferably treated with a targeted strategy, mainly a tyrosine kinase inhibitor (TKI). In phase III trials with patients whose tumors are characterized by a sensitizing epidermal growth factor receptor (EGFR) or anaplastic lymphoma kinase (ALK) alteration, treatment with a TKI results in higher response rates and longer progression free survival (PFS) compared with chemotherapy. These results illustrate the importance of mutational testing to direct the choice of therapy for patients with NSCLC.
Since the discovery of EGFR mutations in 2004 [Lynch et al. 2004; Paez et al. 2004; Pao et al. 2004] and the EML4-ALK translocation in 2007 [Soda et al. 2007], multiple additional genetic alterations that confine targeted drug sensitivity have been discovered in NSCLC. These molecular aberrations occur more often in nonsquamous cell histology and light or never smokers, but can also be found to a lesser extent in (former) smokers and sporadically in squamous cell histology. The mutations offer promising new therapeutic approaches and the use of targeted drugs has changed the way lung cancer is being treated [Cancer Genome Atlas Research Network, 2014].
Clinical trials evaluating drugs that target several genetic alterations are ongoing in patients whose tumor exhibits a HER2 exon 20 insertion, ROS1 translocation, RET fusion, MET amplification and exon 14 splicing alteration, or BRAF exon 15 V600 mutation. In addition to these sensitizing genomic alterations, lung adenocarcinoma is characterized by high rates of somatic mutations and genomic rearrangements that are not currently clinically actionable. Examples are tumor suppressor gene abnormalities such as TP53 and STK11, but also activating KRAS mutations and non-V600E BRAF mutations and BRAF fusion products.
These genetic alterations are not cancer type specific and tend to occur throughout the cancer landscape with variable frequency. BRAF mutations, for example, can be seen in 50% of melanoma cases, but only in 1–4% of NSCLC cases [Ali et al. 2015; Brustugun et al. 2014; Cardarella et al. 2013; Ilie et al. 2013; Kinno et al. 2014; Litvak et al. 2014; Luk et al. 2015; Marchetti et al. 2011; Paik et al. 2011; Tissot et al. 2016; Villaruz et al. 2015]. In this review we will focus on BRAF alterations and the therapeutic approach for patients with NSCLC whose tumor harbors a genetic aberration in the BRAF gene.
Distribution of BRAF aberrations in lung cancer
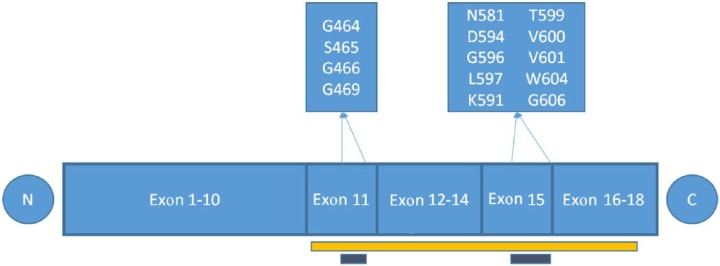
The BRAF gene is mutated in ∼7% of human cancers, including melanoma, colorectal, papillary thyroid, and NSCLC [Davies et al. 2002; Dhomen and Marais, 2007] with variable frequency and allelic distribution. All somatic mutations in the BRAF gene are found in the glycines of the G-loop in exon 11 or in the activation segment in exon 15 (Figure 1).
Figure 1.
BRAF mutations are found in exons 11 and 15, which are both within the kinase domain (yellow bar). The conserved glycine motif (G-loop) in exon 11 and the activation segment in exon 15 are indicated with a grey bar. All BRAF mutations are within these two areas.
Somatic mutations in BRAF are encountered in 40–60% of cutaneous melanomas with the valine substitution at residue 600 (V600E) accounting for >90% of the BRAF mutations [Long et al. 2011; Wellbrock and Hurlstone, 2010].
Somatic mutations in BRAF are observed in 1–4% of NSCLC cases [Ali et al. 2015; Brustugun et al. 2014; Cardarella et al. 2013; Ilie et al. 2013; Kinno et al. 2014; Litvak et al. 2014; Luk et al. 2015; Marchetti et al. 2011; Paik et al. 2011; Tissot et al. 2016; Villaruz et al. 2015]. Lung adenocarcinoma is known for its high rate of somatic mutations and genomic rearrangements, challenging identification of driver gene alterations because of a large burden of passenger events per tumor genome [Ding et al. 2008; Govindan et al. 2012; Imielinski et al. 2012]. Therefore, it is important to characterize the downstream effects of these alterations. The kinase activity of many of the BRAF mutants has been shown to be enhanced compared with wild type (WT) and result in increased downstream ERK phosphorylation, but some BRAF mutations are kinase-dead or have lower kinase activity compared with WT BRAF [Wan et al. 2004]. BRAF-mutant NSCLC frequently harbors the V600E allele (∼55%). Other highly frequent activating BRAF variants include G469A (∼35%) and D594G (∼10%) [Brose et al. 2002; Imielinski et al. 2012; Marchetti et al. 2011; Paik et al. 2011].
RAF fusions and truncations in which part of the amino-terminal domain of RAF is deleted occur sporadically in a number of malignancies including NSCLC [Jang et al. 2015] and these fusion proteins may also increase ERK signaling [Yao et al. 2015].
The phenotype of the BRAF-positive patients differs from that of sensitizing EGFR mutations and ALK translocations. In different patient cohorts, the percentage of smoking patients varies between 57–100%, with smoking being more prevalent in the population with non-V600E BRAF mutations [Brustugun et al. 2014; Cardarella et al. 2013; Ilie et al. 2013; Litvak et al. 2014; Luk et al. 2015; Marchetti et al. 2011; Paik et al. 2011; Tissot et al. 2016; Villaruz et al. 2015].
Signaling of the BRAF receptor
The MAPK/ERK pathway (also known as the RAS-RAF-MEK-ERK pathway) is a chain of proteins that communicates receptor signaling to the DNA in the nucleus of the cell.
The pathway’s general structure includes RAS (a small G protein) and three protein kinases (RAF, MEK, ERK). The protein kinases communicate by adding phosphate groups to a neighboring protein, which acts as an ‘on’ or ‘off’ switch. The signal starts when a growth factor binds to the extracellular domain of a transmembrane receptor tyrosine kinase (RTK). The resulting signaling cascade with RAS, RAF, MEK and ERK results in translocation of ERK to the nucleus, where it activates transcription factors that results in the expression of genes that control cell cycle progression, differentiation, protein synthesis, metabolism, cell survival, cell migration, and invasion and senescence [Yoon and Seger, 2006].
Mutations in RAS and RAF family members are important driver mutations in >30% of NSCLC [Imielinski et al. 2012; Kortum and Morrison, 2015] because they result in sustained ERK stimulation.
RAS proteins are GTPases (small G proteins) involved in the MAPK signaling cascade as well as the phosphatidylinositol-3-kinase (PI-3K) pathway. Among the three RAS proteins (HRAS, KRAS, and NRAS), KRAS is the most frequently encountered mutation in NSCLC (~30% of lung adenocarcinomas) [Imielinski et al. 2012]. Under physiological circumstances, upon receptor activation by ligand binding, RAS is activated by binding to guanosine triphosphate (GTP) [Luo et al. 1996; Rajakulendran et al. 2009]. Via RAF and MEK, ERK activation leads to a number of negative regulatory events that serve to inhibit the pathway upstream (negative feedback loop) [Lito et al. 2012].
Mutations in codons 12 and 13 of KRAS exon 2 lead to impaired cycling between GTP and GDP and therefore accumulation of GTP, and these mutations are therefore activating mutations that result in constitutive activation of downstream RAF kinases, independent of upstream ligand binding to the RTK and avoiding the negative feedback loop [Parada et al. 1982; Santos et al. 1982; Taparowsky et al. 1982].
Downstream of RAS, RAF protein kinases include ARAF, BRAF and CRAF. RAS stimulates and recruits the RAF proteins to the cell membrane where they are activated by RAS [Garnett and Marais, 2004].
RAF proteins are subject to complex regulation, which is reflected by the presence of numerous phosphorylation sites that are distributed throughout the protein. Some of the sites are conserved in all three isoforms, which indicates common mechanisms of regulation, but others are not universal, which indicates that these proteins can be independently regulated [Wellbrock et al. 2004]. RAF protein structure consists of an amino terminus that contains the regulatory domain, an activation loop, and a carboxyl terminus that contains the kinase domain. The regulatory domain is located within exons 1–10 in the amino (N) terminus, whereas the kinase domain is located within exons 11–18 at the carboxyl (C) terminus (Figure 1). RAF is activated by phosphorylation of two amino acids within the activation segment of the kinase domain [Zhang and Guan, 2000].
The three RAF proteins are not equal in their ability to activate MEK. ARAF is a poor MEK activator, while BRAF has been identified as the major MEK activator, displaying higher affinity for MEK1 and MEK2 than CRAF [Papin et al. 1998].
The differential regulation of the RAF isoforms lies in phosphorylation of the negative-charge regulatory region (N-region). CRAF and ARAF must be phosphorylated for maximal activation, but BRAF carries a constitutive negative charge and therefore, N region phosphorylation is not required, making BRAF primed for activation [Wellbrock et al. 2004].
Under normal signaling conditions, RAF dimerization is an obligatory step in RAF activation and this is mediated by RAS [Freeman et al. 2013]. Homodimers as well as heterodimers can be formed. BRAF/CRAF and ARAF/BRAF heterodimers, as well as BRAF/BRAF and CRAF/CRAF homodimers have been observed.
Activating BRAF mutations and fusion products constitutively activate downstream signaling. The mechanism of action is well described for the most common BRAF mutation in lung cancer, a thymidine to adenosine transversion at nucleotide 1799 at exon 15 (T1799A), which results in a valine to glutamate substitution at codon 600 (V600E). This mutation appears to mimic regulatory phosphorylation and increases BRAF activity approximately 500-fold compared with WT [Wan et al. 2004]. Also, the mutation enables BRAF to function as a monomer, independent of RAS, leading to continuous hyperactivation of ERK, despite negative feedback inhibition of RAS [Lito et al. 2012; Poulikakos et al. 2011; Yao et al. 2015]. Due to the negative feedback loop and subsequent RAS inhibition, activating BRAF alterations bypass and suppress RAS mediated dimerization.
The kinase activity of most non-V600E BRAF mutations is increased compared with WT BRAF receptor activity, but some BRAF mutations are kinase-dead or have lower activity than WT BRAF [Wan et al. 2004; Yao et al. 2015]. The nonactivating BRAF mutations marginally activate ERK signaling and do not inhibit RAS [Yao et al. 2015].
In contrast with the V600 mutants (V600E/K/D/R), all other activating BRAF mutations function as RAS-independent homodimers and do not exhibit monomer activity [Yao et al. 2015]. BRAF fusions are tumor-derived fusion proteins in which the N-terminal domain of BRAF has been replaced by a fusion partner. The fusions also activate ERK signaling in a RAS-independent manner [Yao et al. 2015]. Thus, with the exception of V600 BRAF mutants that function as an active monomer, all other activating BRAF mutations, translocations and fusions bypass ERK-dependent negative feedback by dimerization that is RAS independent.
Targeting BRAF V600E mutation positive NSCLC
To date, two selective BRAF inhibitors have been approved by the United States Food and Drug Administration (FDA) and European Medicines Agency (EMA) for the treatment of BRAF V600-positive melanoma, and a third is currently being tested in clinical trials. Two phase III studies with the approved drugs vemurafenib and dabrafenib showed the efficacy of BRAF TKI treatment in BRAF V600-positive melanoma with an objective response rate (ORR) of 48% and 50% and a median PFS of 5.3 and 5.1 months for vemurafenib and dabrafenib, respectively [Chapman et al. 2011; Hauschild et al. 2012]. Single-agent MEK TKI activity was shown in a phase III study with trametinib with an ORR of 22% and a median PFS of 4.8 months [Flaherty et al. 2012]. A multicenter phase III study evaluated the combination of dabrafenib and trametinib versus dabrafenib monotherapy in melanoma patients [Long et al. 2015]. An ORR of 69% was observed for the combination versus 53% for dabrafenib monotherapy. The PFS was also better for the combination with a median of 11.0 months versus 8.8 months for dabrafenib monotherapy.
In the lung, preclinical studies showed that single-agent BRAF inhibition is effective in BRAF-V600-mutated NSCLC [Joshi et al. 2015; Lin et al. 2014]. The MEK TKI trametinib was also effective as a single agent in V600E BRAF-mutated cells [Joshi et al. 2015]. The combination of vemurafenib and trametinib caused a small but significant increase in apoptosis when compared with either single agent in this in vitro study.
Several clinical case reports have been published with partial and complete responses to vemurafenib [Gautschi et al. 2012; Peters et al. 2013; Robinson et al. 2014; Schmid et al. 2015] and dabrafenib [Falchook et al. 2012; Rudin et al. 2013]. However, as expected from the melanoma setting, the responses to single-agent BRAF TKI are short lived due to the development of resistance (typically 5 months for single-agent BRAF TKI treatment) [Gautschi et al. 2015]. Sorafenib, a weak RAF TKI, may also be active against BRAF-mutated NSCLC. In the European cohort study, one patient received sorafenib treatment and experienced a partial response (PR) according to RECIST [Gautschi et al. 2015; Sereno et al. 2015]. In this retrospective multicenter cohort study in Europe, patients with advanced BRAF-mutant lung cancer received treatment with either vemurafenib (n = 29), dabrafenib (n = 9), or sorafenib (n = 1). Of the BRAF mutations, 83% was V600E. Rapid and marked tumor responses were observed in patients with heavy pretreatment and advanced age. Overall, 2 patients (6%) had a complete response, 16 (47%) had a partial response, 11 (32%) had stable disease, and 4 (11%) had progressive disease. The ORR was 53% and the PFS and overall survival (OS) were 5 and 10.8 months, respectively. Duration of BRAF therapy was evaluable in 34 patients, with a median of 17 weeks (4.3 months; range 2–164 weeks), showing durable responses in some patients.
In a vemurafenib basket trial (VE-BASKET), patients with various BRAF V600 mutation-positive nonmelanoma tumors were enrolled in 6 prespecified cancer cohorts, including a NSCLC cohort with 20 patients [Hyman et al. 2015]. The primary endpoint was response rate and secondary endpoints included PFS and OS. A total of 19 NSCLC patients were evaluable for response. Overall, one patient was treatment naive and 45% of patients received two or more lines of therapy prior to study inclusion. The ORR was 42% [95% confidence interval (CI) 20–67]. Tumor regression to any extent was observed in the majority of the patients (74%). Median PFS was 7.3 months (95% CI 3.5–10.8) and median OS not yet reached. The 1-year PFS and OS rates were 23% (95% CI 6–46) and 66% (95% CI 36–85), respectively, with 15 of 20 patients censored for OS estimate. At the time of cutoff date, five patients were still receiving treatment.
Interim results of an ongoing prospective multicenter single arm phase II study of dabrafenib monotherapy, or combination therapy with trametinib in patients with BRAF V600E-mutated metastatic NSCLC (BRF113928) were presented at the 2015 American Society of Clinical Oncology annual meeting [Planchard et al. 2015]. A two-stage design was applied. The first stage evaluated the efficacy of dabrafenib monotherapy in a cohort of predominantly pretreated subjects. Among the first 20 subjects, there were 8 patients (40%) with a partial response, 4 patients (20%) with stable disease, 6 patients (30%) with progressive disease, and 2 patients (10%) were not evaluable [Planchard et al. 2013]. With a response rate of 40% (8/20) the primary endpoint (⩾3 of first 20 patients) was met and the trial proceeded with stage 2. In the second stage of the trial, three cohorts were evaluated; dabrafenib as single agent (cohort A), or in combination with trametinib in pretreated (cohort B) and treatment-naïve (cohort C) patients. The primary objective of the second stage was ORR as well, with PFS and OS being secondary objectives. Enrollment of the three cohorts has been completed and the final results of cohorts A (dabrafenib monotherapy) and B (dabrafenib and trametinib in pretreated patients) were reported recently [Planchard et al. 2016a, 2016b]. With dabrafenib monotherapy, (n = 78), the ORR was 33% (n = 26, all partial responses; 95% CI 23–45) and disease control rate (DCR) for >12 weeks was 58% (95% CI 46–67). Median duration of response was 9.6 months (95% CI 5.4–15.2) [Planchard et al. 2016b]. Male/female distribution was equal and most patients had adenocarcinoma (96%) and were (former) smokers (63% in total and 31% ⩾30 pack years). After a protocol amendment, six patients who had received no previous systemic anticancer therapy for metastatic disease were enrolled in cohort A. However, after discussions with regulatory agencies, enrolment of first-line patients was delayed until results were obtained in cohorts B and C because it was expected that dabrafenib and trametinib combination therapy would be superior to dabrafenib monotherapy. In these six patients, four achieved a partial response. PFS in these four patients was 4.5, 8.6, 11.0 and 16.6 months, with corresponding durations of response of 3.2, 7.2, 9.6 and 12.5 months, respectively. The two patients without a response had PFS of 4.0 and 8.1 months [Planchard et al. 2016b].
With combination dabrafenib and trametinib therapy in pretreated patients (cohort B, n = 57), the ORR was 63% (n = 36; 95% CI 49–76) and DCR for >12 weeks was 79% (95% CI 66–89) [Planchard et al. 2016a]. Male and female patients in the combination arm were equally distributed, most patients were White (86%), had adenocarcinoma (98%) and were former smokers (61%), confirming the observation that BRAF mutations are more associated with (cigarette) smoking than other activating molecular alterations. Responses were generally observed after 6 weeks, revealing that tumor response is rapid. Results of cohort C have not yet been reported. As study enrolment has been completed, the full study report of cohort is awaited.
BRAF TKIs also showed to be active in patients with brain metastases. Robinson and colleagues reported on a BRAF V600E-positive NSCLC patient with multiple supra- and infratentorial brain metastases that decreased in size 1 month after vemurafenib initiation [Robinson et al. 2014]. In the prospective phase II BRF113928 study, patients with untreated brain metastases were allowed as long as they were asymptomatic and <1 cm in the longest diameter.
Toxicity of BRAF and MEK inhibition
In NSCLC, toxicity seems to be similar to that observed in the melanoma setting with BRAF and MEK TKI treatment [Chapman et al. 2011; Hauschild et al. 2012; Long et al. 2015]. In the BRF113928 study, the most common adverse events (AEs) in the dabrafenib monotherapy arm were pyrexia (35%), asthenia (30%), hyperkeratosis (30%), loss of appetite (28%), nausea (27%), cough (26%), fatigue (26%) and alopecia (21%). Of 84 patients, 45 (54%) experienced adverse events of grade 2 or worse. The most frequent grade 3 or worse AEs were the development of cutaneous squamous cell carcinoma (n = 10, 12%) and basal cell carcinoma (n = 4, 5%) and asthenia (n = 4, 5%). Overall, 5 (6%) patients had AEs that led to dabrafenib discontinuation (blistering, deterioration of general health, intracranial hemorrhage, malaise, and palmar–plantar erythrodysesthesia syndrome (all n = 1) and 36 (43%) patients had a treatment-related dose interruption. The most common AEs leading to dose interruption were pyrexia in nine (11%) patients, chills in five (6%), and vomiting in four (5%) [Planchard et al. 2016b]. In the combination arm of the BRF113928 study, most common (>20%) AEs were pyrexia, nausea, vomiting, diarrhea, decreased appetite, asthenia, dry skin, peripheral edema, chills and cough. Most were grade 1 or 2. Grade 3 AEs occurred in 49% of patients; most frequent were neutropenia (9%), hyponatremia (7%), and anemia (5%).
AEs led to permanent discontinuation of study drugs in seven patients (12%). AEs leading to a dose interruption or delay were reported in 35 (61%) patients and a dose reduction was needed in 20 (35%) patients. Secondary cutaneous squamous cell carcinoma occurred in two patients (4%) [Planchard et al. 2016a].
The toxicity profile of vemurafenib and dabrafenib shows similarities, as well as differences. Skin toxicity, arthralgia and pyrexia are seen with both inhibitors, while photosensitivity is a typical side effect of vemurafenib and rare with dabrafenib. Fever and chills are common with dabrafenib but rare with vemurafenib [Welsh and Corrie, 2015].
Caution should be employed in combining concurrent BRAF inhibitor therapy and radiotherapy (especially to large treatment fields) due to apparent radiosensitization of normal tissues and an increased risk of AEs [Boussemart et al. 2013; Merten et al. 2014; Satzger et al. 2013]. Forschner and colleagues reported on two patients that developed radiation recall pneumonitis [Forschner et al. 2014]. Early application of systemic corticosteroids enabled these investigators to continue BRAF TKI treatment without dose reduction.
In melanoma, MEK inhibition is associated with a 2–3% incidence of pneumonitis [Flaherty et al. 2012]. In our own experience with combination dabrafenib and trametinib treatment in NSCLC patients (n = 14) we observed one case with pneumonitis that was not radiotherapy-associated and one case with radiation recall pneumonitis, demonstrating that close monitoring of pulmonary symptoms is warranted.
Molecular basis of increased activity and decreased skin toxicity of BRAF and MEK TKI combination treatment when compared with BRAF TKI monotherapy
The unique observation of dual therapy being more active and less toxic to the skin than monotherapy has a biological explanation. BRAF inhibitors mediate a curious paradox. The enhanced rate of secondary skin malignancies seen during BRAF TKI treatment alone is due to BRAF TKI-induced activation of the MAPK pathway in cells expressing WT BRAF and especially in cells with co-expression of activating RAS mutations [Su et al. 2012]. BRAF TKIs do not initiate tumorigenesis but rather accelerate the progression of pre-existing subclinical cancerous lesions due to paradoxical MAPK pathway activation, which can be inhibited by adding a MEK inhibitor. In BRAF WT cells, BRAF inhibitors do not result in marked ERK inhibition because WT BRAF is not active as a monomer and the current BRAF inhibitors are not able to block RAF dimers. Therefore, RAS is not suppressed by ERK and is still active. Upon physiological RAS activation, BRAF and CRAF kinases form homo- and heterodimers and this process is exaggerated by activating RAS mutations [Poulikakos et al. 2010; Rushworth et al. 2006; Wan et al. 2004; Weber et al. 2001]. In vitro models show that BRAF inhibitors have an intrinsic ability to activate RAF kinases that function as dimers. Drug binding to one member of RAF dimers inhibits one protomer, but results in transactivation of the drug-free protomer through scaffolding or conformational functions, consequently stimulating MEK and ERK hyperactivation [Hatzivassiliou et al. 2010; Heidorn et al. 2010; Poulikakos et al. 2010]. This effect is further attenuated by increased upstream signaling caused by activating RAS mutations and rare cases of induced mutant-RAS leukemia [Callahan et al. 2012] and colon cancer [Andrews et al. 2013] have been reported in patients with advanced melanoma treated with BRAF inhibitors alone. Likely, these tumors were occult present at the time of BRAF inhibition and that the BRAF TKI treatment accelerated tumor growth.
Targeting non-V600E BRAF mutation-positive NSCLC
Preclinical cell line studies show that single-agent BRAF TKI is effective in BRAF-V600-mutated NSCLC, but not in non-V600 BRAF mutated (lung) cancer [Joshi et al. 2015; Yao et al. 2015]. BRAF V600-mutated cells are activated monomers when RAS activity is low, while all other activating BRAF-mutated cells function as constitutive RAS-independent dimers. RAF inhibitors effectively inhibit mutant monomers, but not dimers. With BRAF dimers, BRAF TKIs bind to one site of the dimer, thereby significantly reducing their affinity for the second and failing downstream ERK inhibition [Poulikakos et al. 2011; Yao et al. 2015]. Tumors with a non-V600 BRAF mutation are therefore insensitive to BRAF inhibitors.
In the European cohort study, six patients with non-V600E BRAF mutated NSCLC were treated with BRAF inhibitors [Gautschi et al. 2015]. All tumors with non-V600E mutations located outside of the activation segment of the BRAF kinase domain (codon 596 through 600) were refractory to BRAF therapy. However, one patient with a G596V mutation achieved a partial response to vemurafenib. The authors concluded that mutations located within the activation loop (codon 596 through 600) are potentially sensitive to BRAF inhibition. On the contrary, in vitro experiments in NSCLC cell lines harboring another non-V600E mutation within the activation loop (L597) showed insensitivity to BRAF inhibition [Yao et al. 2015].
In another in vitro study, trametinib was effective as a single agent in BRAF-mutated NSCLC cells, either V600E or non-V600E. Adding vemurafenib was not more effective at inhibiting cell growth, but did cause a small but significant increase in apoptosis [Joshi et al. 2015]. Interestingly, treatment with single-agent trametinib caused upregulation of AKT signaling in BRAF non-V600E cells only, suggesting a potential mechanism of resistance to treatment with single-agent MEK inhibition. The AKT pathway did not appear to be upregulated when BRAF non-V600E cells were treated with the combination of trametinib and vemurafenib, suggesting that the combination therapy may be superior to trametinib single agent for non-V600E BRAF-mutated NSCLC [Joshi et al. 2015].
In melanoma, patients with non-V600E BRAF mutations did not respond to BRAF inhibitors (as expected) [Falchook et al. 2012; Kim et al. 2014; Yang et al. 2010], but responses were observed on MEK inhibition [Dahlman et al. 2012; Kim et al. 2013]. To our knowledge, no results have been published of MEK-targeted treatment for BRAF non-V600-mutated NSCLC.
Non-V600E BRAF mutations often co-occur with oncogenic RAS mutations [Tissot et al. 2016; Zheng et al. 2015]. In case of kinase-dead BRAF mutations, RAS mutations are actually required for the BRAF mutation to be oncogenic [Heidorn et al. 2010]. Kinase-dead BRAF mutations mimic the effects of BRAF inhibitors. Both drive hyperactivation of ERK in the presence of activating KRAS mutations by forming BRAF and CRAF homo- and heterodimers.
Resistance to BRAF TKI treatment
Resistance patterns have been published for melanoma, but not for NSCLC. Primary and secondary resistance have been described and patterns are far more diverse than described for sensitizing EGFR mutations where, upon progression, ~50% of tumors are characterized by a targetable secondary EGFR exon 20 T790M mutation. Resistance can occur due to a secondary MAPK alteration or due to activation of bypass tracks. Secondary MAPK alterations are responsible for the majority of resistance in melanoma and result in reactivation of ERK signaling [Johnson et al. 2015; Shi et al. 2014]. These include NRAS or KRAS mutations, BRAF splice variants, BRAF V600E amplification and MEK1/2 mutations [Johnson et al. 2015]. As explained above, upstream RAS mutations promote BRAF and CRAF homo- and heterodimer formation, insensitive to current BRAF inhibitors. In addition, BRAF inhibitors have an intrinsic ability to activate RAF kinases. BRAF V600E amplification and BRAF splice variants cause acquired resistance by increasing levels of BRAF V600E homodimers [Poulikakos et al. 2011; Yao et al. 2015]. MEK1/2 mutations confer resistance to both BRAF and MEK inhibitors [Emery et al. 2009; Villanueva et al. 2013; Wagle et al. 2011].
Despite the hope that dual BRAF and MEK inhibition would limit or overcome BRAF TKI resistance, treatment failure still occurs and resistance mechanisms observed to combined BRAF and MEK inhibition mimic that of BRAF inhibitor monotherapy. A preliminary genetic analysis of five patients failing dabrafenib and trametinib treatment revealed similar mechanisms of resistance to those seen in patients treated with BRAF-inhibitor monotherapy [Wagle et al. 2014]. It is therefore perhaps not surprising that failure on BRAF-inhibitor therapy also confers resistance to MEK inhibition, with minimal clinical activity (ORR 13%) being seen on combined dabrafenib and trametinib in patients failing dabrafenib monotherapy [Johnson et al. 2014].
Bypass tracks that have been described to cause resistance are aberrations in the PI3K-PTEN-AKT pathway [Krepler et al. 2015; Shi et al. 2014; Van Allen et al. 2014], MET amplification [Van Allen et al. 2014] and upregulation of other RTKs [Marusiak et al. 2014; Nazarian et al. 2010].
Overcoming resistance to BRAF inhibitors
Vemurafenib and dabrafenib are classified as type I inhibitors that bind to the active ‘DGF-in’ kinase conformation [Kortum and Morrison, 2015] and binding of these drugs to one protomer in the dimer significantly allosterically transactivates the other protomer [Hatzivassiliou et al. 2010; Heidorn et al. 2010; Poulikakos et al. 2010] and reduces the affinity for binding of type I inhibitors to the second protomer. Therefore, these drugs are ineffective against resistance mechanisms that result in the formation of BRAF dimers, such as BRAF V600E amplification and splice variants.
Recently, next generation RAF inhibitors were identified and characterized in vitro and in vivo in xenografts [Girotti et al. 2015; Peng et al. 2015; Yao et al. 2015; Zhang et al. 2015]. These drugs are called paradox-breaking RAF inhibitors because they suppress mutant BRAF cells without activating the MAPK pathway in cells bearing WT BRAF or upstream activation by mutated RAS.
Zhang and colleagues screened vemurafenib analogues against a panel of RAS and BRAF V600E mutated cell lines and discovered two compounds (PLX7904 and PLX8394) that were only subtly different from vemurafenib based on their chemical structure and potently inhibited phosphorylated ERK (pERK) in BRAF V600E cells, but showed essentially no pERK activation in RAS mutant cell lines [Zhang et al. 2015]. The compounds showed preferential inhibition of mutated-BRAF V600E over WT BRAF and CRAF, comparable with that of vemurafenib. PLX7904 and PLX8394 were also active against BRAF V600E amplification and splice variants and PLX8394 is currently in phase I/II clinical evaluation.
Yao and colleagues discovered BGB659, a second generation RAF inhibitor with equivalent efficacy against both the monomeric and homodimeric BRAF mutants. Like PLX7904 and PLX8394, BGB659 shows preferential inhibition of mutated BRAF monomers and dimers over other WT RAF dimers with or without a RAS mutation, therefore in theory limiting toxicity [Yao et al. 2015]. Inhibition of mutated RAF was shown to be independent of co-existing RAS mutations. However, in contrast with PLX7904 and PLX8394, BGB659 causes marked, dose-dependent, induction of BRAF/CRAF heterodimers in RAF WT cells. However, BGB659 does not result in paradoxal ERK activation because the drug inhibits dimer activity. In BRAF V600 cells, induction of RAF dimerization does not appear because the process is RAS-dependent and RAS-GTP levels are too low.
BGB283, a weaker type II RAF inhibitor than BGB659 [Yao et al. 2015], entered clinical evaluation and results of a phase I study in patients with RAF or RAS-mutated solid tumors were presented at the 2016 AACR meeting [Desai et al. 2016]. Among 29 evaluable patients, 3 had a partial response, including 1 patient with KRAS-mutated NSCLC. Furthermore, 14 patients achieved prolonged stable disease with 3 patients for over 300 days. 18F-FDG uptake was measured at baseline and at the end of cycle 1 and showed a partial metabolic response in 43% of patients [Desai et al. 2016]. Phase Ib expansion cohorts in molecularly-defined tumors are ongoing.
Peng and colleagues and Girotti and colleagues characterized new pan-RAF inhibitors with minimal paradoxical pathway activation in BRAF WT or RAS mutant cells [Girotti et al. 2015; Peng et al. 2015]. These pan-RAF compounds inhibit ARAF, BRAF and CRAF isoforms with high affinity and are active in BRAF mutant cells, first generation BRAF TKI resistant cells, as well as RAS mutant cells with or without a co-occurring BRAF mutation. Although in mice, toxicity of the pan-RAF inhibitors CCT196969 and CCT241161 was minimal [Girotti et al. 2015], the concern for pan-RAF inhibitors is that blocking MAPK signaling in normal tissues will cause toxicity.
In melanoma cell lines, broad upregulation of ErbB family members was seen in response to PLX4720 treatment, a vemurafenib analogue. Adding lapatinib, a pan-EGFR TKI, to PLX4720, resulted in cytotoxicity in 6 of 12 PLX4720-resistant cell lines [Capaldo et al. 2015].
Krepler and colleagues established 12 PDX models from melanoma patients that progressed on BRAF inhibitors and identified alterations in NRAS and MEK1, BRAF amplification and aberrant PTEN [Krepler et al. 2015]. At the protein level, reactivation of the MAPK pathway predominated, with parallel activation of PI3K in some others. In these cases, second-line efficacy was confirmed with the addition of the pan-PI3K inhibitor BKM120 to either BRAF or MEK inhibitor. Amplification of MET was observed in three PDX models. Importantly, MET amplification alone did not predict sensitivity to the MET-inhibitor, capmatinib, only when MET amplification was present together with increased levels of phosphorylated MET. Second line efficacy was confirmed with the addition of capmatinib to BRAF/MEK combination therapy.
Other systemic therapies
In small series no differences were seen in the prognosis of BRAF-mutated and BRAF WT patients treated with platinum-doublet chemotherapy [Cardarella et al. 2013; Tissot et al. 2016; Villaruz et al. 2015]. In a small retrospective study, the value of BRAF inhibitor rechallenge was evaluated in patients with BRAF V600-positive melanoma. A total of 5 out of 10 patients showed a partial response on BRAF rechallenge after a median drug holiday of 7.25 months (range 2–18 months) [Roux et al. 2015].
Conclusion
BRAF V600 mutation is a driver mutation in patients with NSCLC that is actionable with currently available BRAF inhibitors such as dabrafenib and vemurafenib with or without the addition of a MEK inhibitor. Randomized controlled trials have not been performed in NSCLC. A single-arm phase II study with dabrafenib ± trametinib in patients with BRAF V600E-positive NSCLC finished accrual and final results of two cohorts were published. The analyses of this study and the results of other retrospective and prospective studies show that BRAF-inhibitor monotherapy is associated with an ORR of 32–53%, a PFS of 5–7.3 months and a median duration of response of 11.8 months [Gautschi et al. 2015; Hyman et al. 2015; Planchard et al. 2014, 2016b]. The combination of dabrafenib and trametinib results in an ORR of 63% and a DCR of 79% [Planchard et al. 2016a] in pretreated patients. The results of the first-line cohort are awaited.
Mimicking the melanoma setting, brain activity of BRAF inhibitors has been shown, although the number of patients with reported results are too small to draw general conclusions on this point.
As is the case for other driver mutations, resistance develops in all patients. Patterns of resistance have been described for melanoma patients and are yet unknown for NSCLC. It is likely that the situation in NSCLC will mimic that of melanoma in the way that multiple resistance patterns occur. The majority are secondary MAPK alterations for which second generation RAF inhibitors already entered early clinical trials. Other resistance mechanisms involve bypass tracks such as MET amplification or alterations in the PI3K pathway.
The second generation RAF inhibitors are active against BRAF monomers as well as dimers and are therefore not only active in the BRAF V600-resistance setting, but also in patients with activating BRAF non-V600E mutant NSCLC.
Footnotes
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Conflict of interest statement: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Contributor Information
Adrianus J. de Langen, Department of Pulmonary Diseases, VU University Medical Center, De Boelelaan 1117, 1007 MB Amsterdam, The Netherlands.
Egbert F. Smit, Department of Pulmonary Diseases, VU University Medical Center, and Department of Thoracic Oncology, Netherlands Cancer Institute-Antoni van Leeuwenhoek Hospital, Amsterdam, The Netherlands
References
- Ali S., Wang K., Johnson A., Suh J., Heilmann A., Lipson D., et al. (2015) Comprehensive genomic profiling characterizes the spectrum of non-V600E activating BRAF alterations including BRAF fusions in lung cancer. Eur J Cancer 51(Suppl. 3): abstract 3007. [Google Scholar]
- Andrews M., Behren A., Chionh F., Mariadason J., Vella L., Do H., et al. (2013) BRAF inhibitor-driven tumor proliferation in a KRAS-mutated colon carcinoma is not overcome by MEK1/2 inhibition. J Clin Oncol 31: e448–e451. [DOI] [PubMed] [Google Scholar]
- Borghaei H., Paz-Ares L., Horn L., Spigel D., Steins M., Ready N., et al. (2015) Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med 373: 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussemart L., Boivin C., Claveau J., Tao Y., Tomasic G., Routier E., et al. (2013) Vemurafenib and radiosensitization. JAMA Dermatol 149: 855–857. [DOI] [PubMed] [Google Scholar]
- Brahmer J., Reckamp K., Baas P., Crino L., Eberhardt W., Poddubskaya E., et al. (2015) Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med 373: 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose M., Volpe P., Feldman M., Kumar M., Rishi I., Gerrero R., et al. (2002) BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res 62: 6997–7000. [PubMed] [Google Scholar]
- Brustugun O., Khattak A., Tromborg A., Beigi M., Beiske K., Lund-Iversen M., et al. (2014) BRAF-mutations in non-small cell lung cancer. Lung Cancer 84: 36–38. [DOI] [PubMed] [Google Scholar]
- Callahan M., Rampal R., Harding J., Klimek V., Chung Y., Merghoub T., et al. (2012) Progression of RAS-mutant leukemia during RAF inhibitor treatment. N Engl J Med 367: 2316–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. (2014) Comprehensive molecular profiling of lung adenocarcinoma. Nature 511: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldo B., Roller D., Axelrod M., Koeppel A., Petricoin E., Slingluff C., et al. (2015) Systems analysis of adaptive responses to MAP kinase pathway blockade in BRAF mutant melanoma. PLoS One 10: e0138210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardarella S., Ogino A., Nishino M., Butaney M., Shen J., Lydon C., et al. (2013) Clinical, pathologic, and biologic features associated with BRAF mutations in non-small cell lung cancer. Clin Cancer Res 19: 4532–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman P., Hauschild A., Robert C., Haanen J., Ascierto P., Larkin J., et al. (2011) Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 364: 2507–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman K., Xia J., Hutchinson K., Ng C., Hucks D., Jia P., et al. (2012) BRAF(L597) mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discov 2: 791–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H., Bignell G., Cox C., Stephens P., Edkins S., Clegg S., et al. (2002) Mutations of the BRAF gene in human cancer. Nature 417: 949–954. [DOI] [PubMed] [Google Scholar]
- Desai J., Gan H., Barrow C., Jameson M., McArthur G., Tran B., et al. (2016) Phase I study of RAF dimer inhibitor BGB-283 in patients with B-RAF or K-RAS/N-RAS mutated solid tumors. Clin Cancer Res 76(Suppl.): abstract CT005. [Google Scholar]
- Dhomen N., Marais R. (2007) New insight into BRAF mutations in cancer. Curr Opin Genet Dev 17: 31–39. [DOI] [PubMed] [Google Scholar]
- Ding L., Getz G., Wheeler D., Mardis E., McLellan M., Cibulskis K., et al. (2008) Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery C., Vijayendran K., Zipser M., Sawyer A., Niu L., Kim J., et al. (2009) MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci USA 106: 20411–20416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falchook G., Long G., Kurzrock R., Kim K., Arkenau T., Brown M., et al. (2012) Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: a phase 1 dose-escalation trial. Lancet 379: 1893–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty K., Robert C., Hersey P., Nathan P., Garbe C., Milhem M., et al. (2012) Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med 367: 107–114. [DOI] [PubMed] [Google Scholar]
- Forschner A., Zips D., Schraml C., Rocken M., Iordanou E., Leiter U., et al. (2014) Radiation recall dermatitis and radiation pneumonitis during treatment with vemurafenib. Melanoma Res 24: 512–516. [DOI] [PubMed] [Google Scholar]
- Freeman A., Ritt D., Morrison D. (2013) Effects of RAF dimerization and its inhibition on normal and disease-associated RAF signaling. Mol Cell 49: 751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett M., Marais R. (2004) Guilty as charged: B-RAF is a human oncogene. Cancer Cell 6: 313–319. [DOI] [PubMed] [Google Scholar]
- Gautschi O., Milia J., Cabarrou B., Bluthgen M., Besse B., Smit E., et al. (2015) Targeted therapy for patients with BRAF-mutant lung cancer: results from the European EURAF Cohort. J Thorac Oncol 10: 1451–1457. [DOI] [PubMed] [Google Scholar]
- Gautschi O., Pauli C., Strobel K., Hirschmann A., Printzen G., Aebi S., et al. (2012) A patient with BRAF V600E lung adenocarcinoma responding to vemurafenib. J Thorac Oncol 7: e23–e24. [DOI] [PubMed] [Google Scholar]
- Girotti M., Lopes F., Preece N., Niculescu-Duvaz D., Zambon A., Davies L., et al. (2015) Paradox-breaking RAF inhibitors that also target SRC are effective in drug-resistant BRAF mutant melanoma. Cancer Cell 27: 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan R., Ding L., Griffith M., Subramanian J., Dees N., Kanchi K., et al. (2012) Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 150: 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzivassiliou G., Song K., Yen I., Brandhuber B., Anderson D., Alvarado R., et al. (2010) RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 464: 431–435. [DOI] [PubMed] [Google Scholar]
- Hauschild A., Grob J., Demidov L., Jouary T., Gutzmer R., Millward M., et al. (2012) Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet 380: 358–365. [DOI] [PubMed] [Google Scholar]
- Heidorn S., Milagre C., Whittaker S., Nourry A., Niculescu-Duvas I., Dhomen N., et al. (2010) Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell 140: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman D., Puzanov I., Subbiah V., Faris J., Chau I., Blay J., et al. (2015) Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med 373: 726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilie M., Long E., Hofman V., Dadone B., Marquette C., Mouroux J., et al. (2013) Diagnostic value of immunohistochemistry for the detection of the BRAFV600E mutation in primary lung adenocarcinoma Caucasian patients. Ann Oncol 24: 742–748. [DOI] [PubMed] [Google Scholar]
- Imielinski M., Berger A., Hammerman P., Hernandez B., Pugh T., Hodis E., et al. (2012) Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150: 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J., Lee A., Li J., Liyanage H., Yang Y., Guo L., et al. (2015) Common oncogene mutations and novel SND1-BRAF transcript fusion in lung adenocarcinoma from never smokers. Sci Rep 5: 9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D., Flaherty K., Weber J., Infante J., Kim K., Kefford R., et al. (2014) Combined BRAF (Dabrafenib) and MEK inhibition (Trametinib) in patients with BRAFV600-mutant melanoma experiencing progression with single-agent BRAF inhibitor. J Clin Oncol 32: 3697–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D., Menzies A., Zimmer L., Eroglu Z., Ye F., Zhao S., et al. (2015) Acquired BRAF inhibitor resistance: a multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur J Cancer 51: 2792–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi M., Rice S., Liu X., Miller B., Belani C. (2015) Trametinib with or without vemurafenib in BRAF mutated non-small cell lung cancer. PLoS One 10: e0118210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Kefford R., Pavlick A., Infante J., Ribas A., Sosman J., et al. (2013) Phase II study of the MEK1/MEK2 inhibitor trametinib in patients with metastatic BRAF-mutant cutaneous melanoma previously treated with or without a BRAF inhibitor. J Clin Oncol 31: 482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Nowroozi S., Kim K., Davies M., Routbort M., Lazar A., et al. (2014) Clinical characteristics of patients with non-V600 BRAF mutant melanomas. J Clin Oncol 32(Suppl.): abstract 9100. [Google Scholar]
- Kinno T., Tsuta K., Shiraishi K., Mizukami T., Suzuki M., Yoshida A., et al. (2014) Clinicopathological features of nonsmall cell lung carcinomas with BRAF mutations. Ann Oncol 25: 138–142. [DOI] [PubMed] [Google Scholar]
- Kortum R., Morrison D. (2015) Path forward for RAF therapies: inhibition of monomers and dimers. Cancer Cell 28: 279–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krepler C., Xiao M., Spoesser K., Brafford P., Shannan B., Beqiri M., et al. (2015) Personalized pre-clinical trials in BRAF inhibitor resistant patient derived xenograft models identify second line combination therapies. Clin Cancer Res 22: 1592–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Asthana S., Chan E., Bandyopadhyay S., Martins M., Olivas V., et al. (2014) Mapping the molecular determinants of BRAF oncogene dependence in human lung cancer. Proc Natl Acad Sci USA 111: E748–E757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lito P., Pratilas C., Joseph E., Tadi M., Halilovic E., Zubrowski M., et al. (2012) Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell 22: 668–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak A., Paik P., Woo K., Sima C., Hellmann M., Arcila M., et al. (2014) Clinical characteristics and course of 63 patients with BRAF mutant lung cancers. J Thorac Oncol 9: 1669–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long G., Menzies A., Nagrial A., Haydu L., Hamilton A., Mann G., et al. (2011) Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol 29: 1239–1246. [DOI] [PubMed] [Google Scholar]
- Long G., Stroyakovskiy D., Gogas H., Levchenko E., de B., Larkin J., et al. (2015) Dabrafenib and trametinib versus dabrafenib and placebo for Val600 BRAF-mutant melanoma: a multicentre, double-blind, phase 3 randomised controlled trial. Lancet 386: 444–451. [DOI] [PubMed] [Google Scholar]
- Luk P., Yu B., Ng C., Mercorella B., Selinger C., Lum T., et al. (2015) BRAF mutations in non-small cell lung cancer. Transl Lung Cancer Res 4: 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Tzivion G., Belshaw P., Vavvas D., Marshall M., Avruch J. (1996) Oligomerization activates c-Raf-1 through a Ras-dependent mechanism. Nature 383: 181–185. [DOI] [PubMed] [Google Scholar]
- Lynch T., Bell D., Sordella R., Gurubhagavatula S., Okimoto R., Brannigan B., et al. (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139. [DOI] [PubMed] [Google Scholar]
- Marchetti A., Felicioni L., Malatesta S., Grazia S., Guetti L., Chella A., et al. (2011) Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol 29: 3574–3579. [DOI] [PubMed] [Google Scholar]
- Marusiak A., Edwards Z., Hugo W., Trotter E., Girotti M., Stephenson N., et al. (2014) Mixed lineage kinases activate MEK independently of RAF to mediate resistance to RAF inhibitors. Nat Commun 5: 3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merten R., Hecht M., Haderlein M., Distel L., Fietkau R., Heinzerling L., et al. (2014) Increased skin and mucosal toxicity in the combination of vemurafenib with radiation therapy. Strahlenther Onkol 190: 1169–1172. [DOI] [PubMed] [Google Scholar]
- Nazarian R., Shi H., Wang Q., Kong X., Koya R., Lee H., et al. (2010) Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468: 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez J., Janne P., Lee J., Tracy S., Greulich H., Gabriel S., et al. (2004) EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304: 1497–1500. [DOI] [PubMed] [Google Scholar]
- Paik P., Arcila M., Fara M., Sima C., Miller V., Kris M., et al. (2011) Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol 29: 2046–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W., Miller V., Zakowski M., Doherty J., Politi K., Sarkaria I., et al. (2004) EGF receptor gene mutations are common in lung cancers from ‘never smokers’ and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA 101: 13306–13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papin C., Denouel-Galy A., Laugier D., Calothy G., Eychene A. (1998) Modulation of kinase activity and oncogenic properties by alternative splicing reveals a novel regulatory mechanism for B-RAF. J Biol Chem 273: 24939–24947. [DOI] [PubMed] [Google Scholar]
- Parada L., Tabin C., Shih C., Weinberg R. (1982) Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus RAS gene. Nature 297: 474–478. [DOI] [PubMed] [Google Scholar]
- Peng S., Henry J., Kaufman M., Lu W., Smith B., Vogeti S., et al. (2015) Inhibition of RAF isoforms and active dimers by LY3009120 leads to anti-tumor activities in RAS or BRAF mutant cancers. Cancer Cell 28: 384–398. [DOI] [PubMed] [Google Scholar]
- Peters S., Michielin O., Zimmermann S. (2013) Dramatic response induced by vemurafenib in a BRAF V600E-mutated lung adenocarcinoma. J Clin Oncol 31: e341–e344. [DOI] [PubMed] [Google Scholar]
- Planchard D., Besse B., Groen H., Souquet P., Quoix E., Baik C., et al. (2016a) Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol 17: 984–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchard D., Kim T., Mazieres J., Quoix E., Riely G., Barlesi F., et al. (2016b) Dabrafenib in patients with BRAF-positive advanced non-small-cell lung cancer: a single-arm, multicentre, open-label, phase 2 trial. Lancet Oncol 17: 642–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchard D., Groen H., Kim T., Rigas J., Souquet P., Baik C., et al. (2015) Interim results of a phase II study of the BRAF inhibitor (BRAFi) dabrafenib (D) in combination with the MEK inhibitor trametinib (T) in patients (pts) with BRAF V600E mutated (mut) metastatic non-small cell lung cancer (NSCLC). J Clin Oncol 33(Suppl.): abstract 8006. [Google Scholar]
- Planchard D., Kim T., Mazieres J., Quoix E., Riely G., Barlesi F., et al. (2014) Dabrafenib in patients with BRAF V600E-mutant advanced non-small cell lung cancer (NSCLC): a multicenter, open-label, phase II trial (BRF113928). Ann Oncol 25(Suppl. 5): abstract LBA38_PR. [Google Scholar]
- Planchard D., Mazieres J., Riely G., Rudin C., Barlesi F., Quoix E., et al. (2013) Interim results of phase II study BRF113928 of dabrafenib in BRAF V600E mutation-positive non-small cell lung cancer (NSCLC) patients. J Clin Oncol 31(Suppl.): abstract 8009. [Google Scholar]
- Poulikakos P., Persaud Y., Janakiraman M., Kong X., Ng C., Moriceau G., et al. (2011) RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 480: 387–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos P., Zhang C., Bollag G., Shokat K., Rosen N. (2010) RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464: 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakulendran T., Sahmi M., Lefrancois M., Sicheri F., Therrien M. (2009) A dimerization-dependent mechanism drives RAF catalytic activation. Nature 461: 542–545. [DOI] [PubMed] [Google Scholar]
- Robinson S., O’Shaughnessy J., Cowey C., Konduri K. (2014) BRAF V600E-mutated lung adenocarcinoma with metastases to the brain responding to treatment with vemurafenib. Lung Cancer 85: 326–330. [DOI] [PubMed] [Google Scholar]
- Roux J., Pages C., Malouf D., Basset S., Madjlessi N., Baccard M., et al. (2015) BRAF inhibitor rechallenge in patients with advanced BRAF V600-mutant melanoma. Melanoma Res 25: 559–563. [DOI] [PubMed] [Google Scholar]
- Rudin C., Hong K., Streit M. (2013) Molecular characterization of acquired resistance to the BRAF inhibitor dabrafenib in a patient with BRAF-mutant non-small-cell lung cancer. J Thorac Oncol 8: e41–e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushworth L., Hindley A., O’Neill E., Kolch W. (2006) Regulation and role of Raf-1/B-Raf heterodimerization. Mol Cell Biol 26: 2262–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos E., Tronick S., Aaronson S., Pulciani S., Barbacid M. (1982) T24 human bladder carcinoma oncogene is an activated form of the normal human homologue of BALB- and Harvey-MSV transforming genes. Nature 298: 343–347. [DOI] [PubMed] [Google Scholar]
- Satzger I., Degen A., Asper H., Kapp A., Hauschild A., Gutzmer R. (2013) Serious skin toxicity with the combination of BRAF inhibitors and radiotherapy. J Clin Oncol 31: e220–e222. [DOI] [PubMed] [Google Scholar]
- Scagliotti G., Parikh P., von P., Biesma B., Vansteenkiste J., Manegold C., et al. (2008) Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol 26: 3543–3551. [DOI] [PubMed] [Google Scholar]
- Schmid S., Siano M., Joerger M., Rodriguez R., Muller J., Fruh M. (2015) Response to dabrafenib after progression on vemurafenib in a patient with advanced BRAF V600E-mutant bronchial adenocarcinoma. Lung Cancer 87: 85–87. [DOI] [PubMed] [Google Scholar]
- Sereno M., Moreno V., Moreno R., Gomez-Raposo C., Garcia S., Hernandez J., et al. (2015) A significant response to sorafenib in a woman with advanced lung adenocarcinoma and a BRAF non-V600 mutation. Anticancer Drugs 26: 1004–1007. [DOI] [PubMed] [Google Scholar]
- Shi H., Hugo W., Kong X., Hong A., Koya R., Moriceau G., et al. (2014) Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov 4: 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda M., Choi Y., Enomoto M., Takada S., Yamashita Y., Ishikawa S., et al. (2007) Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448: 561–566. [DOI] [PubMed] [Google Scholar]
- Su F., Viros A., Milagre C., Trunzer K., Bollag G., Spleiss O., et al. (2012) RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med 366: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taparowsky E., Suard Y., Fasano O., Shimizu K., Goldfarb M., Wigler M. (1982) Activation of the T24 bladder carcinoma transforming gene is linked to a single amino acid change. Nature 300: 762–765. [DOI] [PubMed] [Google Scholar]
- Tissot C., Couraud S., Tanguy R., Bringuier P., Girard N., Souquet P. (2016) Clinical characteristics and outcome of patients with lung cancer harboring BRAF mutations. Lung Cancer 91: 23–28. [DOI] [PubMed] [Google Scholar]
- Van Allen E., Wagle N., Sucker A., Treacy D., Johannessen C., Goetz E., et al. (2014) The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov 4: 94–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J., Infante J., Krepler C., Reyes-Uribe P., Samanta M., Chen H., et al. (2013) Concurrent MEK2 mutation and BRAF amplification confer resistance to BRAF and MEK inhibitors in melanoma. Cell Rep 4: 1090–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villaruz L., Socinski M., Abberbock S., Berry L., Johnson B., Kwiatkowski D., et al. (2015) Clinicopathologic features and outcomes of patients with lung adenocarcinomas harboring BRAF mutations in the Lung Cancer Mutation Consortium. Cancer 121: 448–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N., Emery C., Berger M., Davis M., Sawyer A., Pochanard P., et al. (2011) Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol 29: 3085–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N., Van Allen E., Treacy D., Frederick D., Cooper Z., Taylor-Weiner A., et al. (2014) MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov 4: 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan P., Garnett M., Roe S., Lee S., Niculescu-Duvaz D., Good V., et al. (2004) Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 116:855–867. [DOI] [PubMed] [Google Scholar]
- Weber C., Slupsky J., Kalmes H., Rapp U. (2001) Active RAS induces heterodimerization of cRaf and BRaf. Cancer Res 61: 3595–3598. [PubMed] [Google Scholar]
- Wellbrock C., Hurlstone A. (2010) BRAF as therapeutic target in melanoma. Biochem Pharmacol 80: 561–567. [DOI] [PubMed] [Google Scholar]
- Wellbrock C., Karasarides M., Marais R. (2004) The RAF proteins take centre stage. Nat Rev Mol Cell Biol 5: 875–885. [DOI] [PubMed] [Google Scholar]
- Welsh S., Corrie P. (2015) Management of BRAF and MEK inhibitor toxicities in patients with metastatic melanoma. Ther Adv Med Oncol 7: 122–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Higgins B., Kolinsky K., Packman K., Go Z., Iyer R., et al. (2010) RG7204 (PLX4032), a selective BRAFV600E inhibitor, displays potent antitumor activity in preclinical melanoma models. Cancer Res 70: 5518–5527. [DOI] [PubMed] [Google Scholar]
- Yao Z., Torres N., Tao A., Gao Y., Luo L., Li Q., et al. (2015) BRAF mutants evade ERK-dependent feedback by different mechanisms that determine their sensitivity to pharmacologic inhibition. Cancer Cell 28: 370–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S., Seger R. (2006) The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors 24: 21–44. [DOI] [PubMed] [Google Scholar]
- Zhang B., Guan K. (2000) Activation of B-RAF kinase requires phosphorylation of the conserved residues Thr598 and Ser601. EMBO J 19: 5429–5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Spevak W., Zhang Y., Burton E., Ma Y., Habets G., et al. (2015) RAF inhibitors that evade paradoxical MAPK pathway activation. Nature 526: 583–586. [DOI] [PubMed] [Google Scholar]
- Zheng G., Tseng L., Chen G., Haley L., Illei P., Gocke C., et al. (2015) Clinical detection and categorization of uncommon and concomitant mutations involving BRAF. BMC Cancer 15: 779. [DOI] [PMC free article] [PubMed] [Google Scholar]