Abstract
Perineuronal nets (PNNs) are lattice-like supramolecular assemblies of extracellular glycoproteins that surround subsets of neuronal cell bodies in the mammalian telencephalon. PNNs emerge at the end of the critical period of brain development, limit neuronal plasticity in the adult brain, and are lost in a variety of complex brain disorders diseases, including schizophrenia. The link between PNNs and schizophrenia led us to question whether neuronally expressed extracellular matrix (ECM) molecules associated with schizophrenia contribute to the assembly of these specialized supramolecular ECM assemblies. We focused on collagen XIX—a minor, nonfibrillar collagen expressed by subsets of telencephalic interneurons. Genetic alterations in the region encoding collagen XIX have been associated with familial schizophrenia, and loss of this collagen in mice results in altered inhibitory synapses, seizures, and the acquisition of schizophrenia-related behaviors. Here, we demonstrate that loss of collagen XIX also results in a reduction of telencephalic PNNs. Loss of PNNs was accompanied with reduced levels of aggrecan (Acan), a major component of PNNs. Despite reduced levels of PNN constituents in collagen XIX-deficient mice (col19a1−/−), we failed to detect reduced expression of genes encoding these ECM molecules. Instead, we discovered a widespread upregulation of extracellular proteases capable of cleaving Acan and other PNN constituents in col19a1−/− brains. Taken together, these results suggest a mechanism by which the loss of collagen XIX speeds PNN degradation and they identify a novel mechanism by which the loss of collagen XIX may contribute to complex brain disorders.
Keywords: aggrecan, cerebral cortex, collagen, extracellular matrix, hippocampus, perineuronal net
Introduction
Extracellular matrix (ECM) is a noncellular component of all tissues, composed of a dense mixture of proteins and polysaccharides. Outside of the nervous system, major constituents of ECM include laminins, fibrillar and nonfribrillar (i.e., unconventional) collagens, fibronectins, nidogens, tenascins, and proteoglycans (i.e., proteins with one or more covalently linked glycosaminoglycan side chains) (LeBleu et al., 2007; Frantz et al., 2010). Interactions between these ECM constituents lead to their assembly into supramolecular structures, which include basal laminas, basement membranes, filaments, and fibrils. These supramolecular ECM structures provide structural support for adjacent cells, act as reservoirs for growth factors, morphogens, and extracellular proteases, and impact nearly all biological functions outside of the brain.
Despite our vast knowledge of ECM assemblies in most tissues, our understanding of their role in brain development and function remains limited. This is surprising given that a significant portion of brain parenchyma comprises extracellular space. In the juvenile brain, when neural circuits are still being assembled, extracellular space accounts for ∼40% of brain parenchyma (Nicholson and Sykova, 1998). Although there is a dramatic reduction in the volume of extracellular space as the mammalian brain matures, a significant fraction of adult brain parenchyma remains occupied by extracellular space (∼20%; Nicholson and Sykova, 1998; Sykova and Nicholson, 2008; Korogod et al., 2015; Pallotto et al., 2015). Despite this, there has been little attention paid to the presence and roles of supramolecular ECM assemblies in the mammalian brain. This likely stems from a general lack of fibrillar assemblies of ECM constituents in the healthy brain and a limited presence of basal laminas or basement membranes. Basal lamina-like structures in the brain are limited to vascular structures and to “fractones,” specialized ECM assemblies associated with the subventricular zone (Mercier et al., 2002; Mercier, 2016). However, it is becoming increasing clear that many ECM constituents are in fact generated by neurons and glia in the developing, adult, and injured brain and these constituents play essential roles in neuronal migration, neural circuit formation, and synaptic plasticity (Dityatev et al., 2010; Dityatev and Rusakov, 2011; Frischknecht and Gundelfinger, 2012; Risher and Eroglu, 2012; Heikkinen et al., 2014; Smith et al., 2015). But, at present, we lack a clear understanding of how these factors are assembled in the extracellular space surrounding neurons and glia in the brain. The one exception to this is the perineuronal net (PNN)—a specialized, lattice-like supramolecular assembly of ECM constituents that surrounds the cell bodies and proximal neurites of select classes of interneurons (Celio and Blumcke, 1994; Celio et al., 1998; Yamaguchi, 2000). PNNs emerge late in postnatal brain development, coinciding with closing of the critical period of brain development. For this reason it has been hypothesized that these specialized matrices limit the neuroplastic potential of adult brains (Rhodes and Fawcett, 2004; Faissner et al., 2010; Wang and Fawcett, 2012). PNNs have garnered attention not only for their potential role in developmental plasticity but also because they appear disrupted in a number of psychiatric and complex brain disorders.
Interestingly, the molecular composition of PNNs differs from supramolecular assemblies of ECM constituents outside the brain. PNNs are composed primarily of lecticans (a family of chondroitin sulfate proteoglycans [CSPGs]), tenascins, hyaluranonin and proteoglycan binding link proteins (HAPLNs), and hyaluran (Zimmermann and Dours-Zimmermann, 2008; Frischknecht and Seidenbecher, 2012; Morawski et al., 2012). More traditional ECM constituents, such as laminins, nidogens, fibronectin, and fibrillar and nonfibrillar collagens, are either absent from PNNs or their roles remain unclear. However, we have recently become interested in the link between collagen XIX and PNNs. Collagen XIX is a nonfibrillar, unconventional collagen generated by telencephalic interneurons in the postnatal rodent brain (Sumiyoshi et al., 1997; Su et al., 2010; Su et al., 2016). Collagen XIX-expressing neurons are present in regions densely packed with PNNs. And, just as PNNs have been linked to schizophrenia and complex brain disorders (Berretta, 2012; Berretta et al., 2015; Bitanihirwe et al., 2016), genetic alterations in the region encoding collagen XIX have been associated with schizophrenia (Liao et al., 2012). Furthermore, targeted deletion of collagen XIX in mice leads to the acquisition of schizophrenia-related behaviors and seizures (Su et al., 2016). Based upon these similarities, we tested whether collagen XIX contributes to the assembly or maintenance of PNNs by assessing PNN morphology and distribution in targeted mouse mutants lacking this collagen (col19a1−/−). Our data revealed a significant reduction in the number of PNNs in regions of mouse brain that normally generate collagen XIX. Loss of PNNs was accompanied by a widespread reduction in protein levels of lecticans in these regions of col19a1−/− mutant brains. However, we failed to detect reduced levels of lectican or PNN constituent mRNA in mutants, suggesting that the reduced number of PNNs in collagen XIX-deficient brains did not result from diminished lectican transcription. Finally, we tested whether the loss of this unconventional neuronally expressed collagen altered the expression of extracellular proteases that degrade lecticans and PNNs. To our surprise, we discovered a widespread upregulation in the transcription of matrix metalloproteinases (MMPs) and a distintegrin and metalloproteinase with thrombospondin motif proteases (ADAMTSs) in these regions of col19a1−/− mutant brains.
Materials and Methods
Animals
CD1 and C57BL/6 mice were obtained from Charles River Laboratories (Wilmington, MA, USA). The generation of collagen XIX null mice (col19a1−/−; previously referred to as N19) was previously described (Sumiyoshi et al., 2004). Col19a1−/− mice were backcrossed for more than 10 generations on C57BL/6 mice. Parv-cre and thy1-stop-yfp (line 15) mice were obtained from Jackson Laboratories (stock numbers 008069 and 005630, respectively). Genomic DNA was isolated from tail using the HotSHOT method (Truett et al., 2000), and genotyping was performed with the following primers: lacZ 5′-TTC ACT GGC CGT CGT TTT ACA ACGTCG TGA-3′ and 5′-ATG TGA GCG AGT AAC AAC CCG TCG GAT TCT-3′; col19a1 (exon4) 5′-CTTCGC AAA ACG CAT GCC TCA GA-3′ and 5′-TTG TTC GTT TGT TTG TTT TTA ATC AAT CAA-3′; yfp 5′-AAG TTC ATC TGC ACC ACC G-3′ and 5′-TCC TTG AAG AAG ATG GTG CG; cre 5′-TGC ATG ATC TCC GGT ATT GA-3′ and 5’-CGT ACT GAC GGT GGG AGA AT-3′. The following cycling conditions were used on an Eppendorf Mastercycler EP: 95℃ for 5 min, followed by 35 cycles of amplification (95℃ for 30 s, 52℃ for 30 s, and 72℃ for 45 s), and 10 min at 72℃. All analyses conformed to National Institutes of Health (NIH) guidelines and protocols, approved by the Virginia Polytechnic Institute and State University Institutional Animal Care and Use Committees.
Reagents and Antibodies
The following reagents and antibodies were purchased: Biotinylated Wisteria Floribunda Lectin (diluted 1:1000 for IHC; Vector Laboratories, Burlingame, CA, USA), mouse anti-CSPG protein core epitope (Cat315, aggrecan [Acan]; Matthews et al., 2002; Brooks et al., 2013) (diluted 1:5000 for IHC; 1:10000 for WB; Millipore, Billerica, MA), mouse anti-actin (diluted 1:10000 for WB; Millipore, Billerica, MA), rabbit anti-GFP (diluted 1:500; Life Technologies, Carlsbad, CA), rabbit anti-ADAMTS4 (diluted 1:1000 for WB; Abcam, Cambridge, MA), mouse anti-NeuN (diluted 1:100 for IHC; Millipore, Billerica, MA), and Alexa Fluor-488 Streptavidin conjugate (diluted 1:1000 for IHC; Life Technologies, Carlsbad, CA). All peroxidase-conjugated anti-mouse, -rabbit, or -sheep antibodies were from Jackson ImmunoResearch Inc. (diluted 1: 5000 for WB), and all fluorescent secondary antibodies for IHC were from Life Technologies (diluted 1:1000). All other reagents are from Fisher unless otherwise noted.
Immunohistochemistry
Fluorescent immunohistochemistry (IHC) was performed on 16 µm cryosectioned paraformaldehyde (PFA)-fixed brain tissue as described previously (Fox et al., 2007; Su et al., 2012). Briefly, tissue slides were allowed to air dry for 15 min before being incubated with blocking buffer (2.5% normal goat serum, 2.5% bovine serum albumin, and 0.1% Triton X-100 in PBS) for 30 min. Primary antibodies were diluted in blocking buffer and incubated on tissue sections for overnight at 4℃. On the following day, tissue slides were washed in PBS and secondary antibodies diluted 1:1000 in blocking buffer were applied to slides for 1 hr at room temperature. After thoroughly washing in PBS, tissue slides were coverslipped with VectaShield (Vector Laboratories, Burlingame, CA, USA). Images were acquired on a Zeiss Examiner Z1 LSM 710 confocal microscope or a Zeiss LSM 700 confocal microscope (Oberkochen, Germany). When comparing between genotypes, images were acquired with identical parameters and analyzed blind. A minimum of three animals (per genotype and per age) were compared in all IHC experiments. IHC images were analyzed and quantified as previously described (Su et al., 2010; Levy et al., 2015; Su et al., 2016). For PNN counts in hippocampus, all images were obtained with identical parameters, and to ensure similar regions were analyzed, we only analyzed coronal sections of hippocampus that also contained central regions of visual thalamus—which includes both dorsal and ventral lateral geniculate nuclei (dLGN and vLGN, respectively). For quantification purposes, all WFA and Acan-labeled PNNs in the entire hippocampus of these sections were counted manually and at least four sections were analyzed per animal (from both hemispheres; n = 4 animals per genotype). Limiting analysis to identical regions of hippocampus (and identically sized hippocampi) reduced potential errors from counting different sized regions of hippocampi. Likewise, all images and analysis of PNN distribution in cerebral cortex were obtained from region- and size-matched sagittal sections of wild-type (WT) and littermate mutant mice. PNNs is all layers of motor cortex (mCtx), prefrontal cortex (pfCtx), and primary visual cortex (vCTX) were counted in sagittal sections from mutants and controls (n = 4 animals per genotype). Cortical regions in mutant and control sections were identified blind to the genotype using the Allen Brain Atlas references atlas (http://mouse.brain-map.org/static/atlas). Quantified data were analyzed with a Student’s t-test to determine statistical significance.
Western Blot
Mouse brains were perfused with PBS, tissue was dissected in ice-cold PBS, and tissue was lysed in modified loading buffer containing 50 mmol/L Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate (SDS), 10% glycerol, and protease inhibitors (1 mmol/L PMSF). Samples were homogenized, boiled for 10 min, centrifuged at 12,000 g for 10 min, and insoluble material was removed. Equal amounts of protein were loaded and separated by SDS-PAGE and transferred to PVDF membranes (Bio-Rad). SDS-PAGE was performed with 7% polyacrylamide gels (7% polyacrylamide [Fisher], 0.1% SDS [Fisher], 0.05% ammonium persulfate [APS; Fisher], 0.001% N,N,N′,N′-tetramethylethylene-diamine [TEMED; Bio-Rad] in 375 mM Tris-HCl pH 8.8) prepared with 4% stacking gels (4% polyacrylamide, 0.1% SDS, 0.05% APS, 0.001% TEMED in 125 mM Tris-HCl pH 6.8). After blocking in 5% nonfat milk in PBS (containing 0.05% Tween20), PVDF membranes (Bio-Rad, Hercules, CA) were incubated with appropriate primary antibodies, followed by HRP-conjugated secondary antibodies. Immunoblotted proteins were detected with the enhanced chemiluminescent detection system (ECL Plus, Amersham Pharmacia Biotech, Piscataway, NJ) and a Gel-Doc imaging system (Bio-Rad). Densitometry of images was performed in ImageJ, and all quantified data were normalized to actin levels.
Quantitative PCR
RNA was isolated using the BioRad Total RNA Extraction from Fibrous and Fatty Tissue kit (BioRad, Hercules, CA). cDNAs were generated from 500 ng of RNA with the Superscript II Reverse Transcriptase First Strand cDNA Synthesis kit (Invitrogen, La Jolla, CA). The following primers were used: acan-qF: 5′ CCC TCA GAG TCA CAA AGA CCA 3′, acan-qR: 5′ TTC GCA GGG ATA AAG GAC TG 3′; bcan-qF: 5′ AAC ATA GGC AGC GGA AAC C 3′, bcan-qR: 5′ GTG GAG TGG CTG TGG CTC 3′; tnr-qF: 5′ AAT TTC TTG GGT CAG GGA GG 3′, tnr-qR: 5′ GAA CCG ACT GTG CTA AGG CT 3′; hapln1-qF: 5′ ATC AGC ACC AGG AGA AGC AG 3′, hapln1-qR: 5′ CTC CAG CTC CCA GCT AAG TG 3′; hapln2-qF: 5′ CTG CTT GGC ATG ATG GTG T 3′, hapln2-qR: 5′ CTC GAT TTG CCT CTG AGG AG 3′; hapln3-qF: 5′ ACA AGG AAC AGC AGG CTC AT 3′, hapln3-qR: 5′ CGA AGA GCT TTC TGG GGA C 3′; hapln4-qF: 5′ GGG TGA TAG GGG AAG ACC AC 3′; hapln4-qR: 5′ GTC ACC CTT CAG GAC TAC GG 3′; has1-qF: 5′ AAG ATG ATC GTG AGT GCT CG 3′, has1-qR: 5′ GAG AGA ATC CAG GAG GAC CC 3′; has2-qF: 5′ ACA GAT GAG GCA GGG TCA AG 3′, has2-qR: 5′ TGGG GTG GAA AGA GAG AAG T 3′; has3-qF: 5′ AGC ACT ACC AGG GCA AAC AG 3′; has3-qR: 5′ CCC TTG GCG TTC AGA AGA T 3′; mmp3-qF: 5′ AGC CTT GGC TGA GTG GTA GA 3′, mmp3-qR: 5′ CGA TGA TGA ACG ATG GAC AG-3′; mmp7-qF: 5′ GCA TTT CCT TGA GGT TGT CC 3′, mmp7-qR: 5′ CAC ATC AGT GGG AAC AGG C 3′; mmp8-qF: 5′ AGA CCG GAA TTG ATT GCT TG 3′, mmp8-qR: 5′ CCC AGT ATC TGA ACA CCT GGA 3′; mmp9-qF: 5′ CTG TCG GCT GTG GTT CAG T-3′, mmp9-qR: 5′ AGA CGA CAT AGA CGG CAT CC 3′; mmp12-qF: 5′ TTT GGA TTA TTG GAA TGC TGC 3′, mmp12-qR: 5′ ATG AGG CAG AAA CGT GGA CT 3′; mmp13-qF: 5′ GGT CCT TGG AGT GAT CCA GA 3′; mmp13-qR: 5′ TGA TGA AAC CTG GAC AAG CA 3′; adamts1-qF: 5′ GGA CAC AAA TCG CTT CTT CC 3′, adamts1-qR: 5′ CAG AAC ACC CGG AAC CAG T 3′; adamts4-qF: 5′ GTC ATG GCT CCT GTC ATG G 3′, adamts4-qR: 5′ CCG GTT TGT CTA AGA GGC AG 3′; adamts5-qF: 5′ GTC ACA TGA ATG ATG CCC AC 3′; adamts5-qR: 5′ CAA ATG GCA GCA CCA ACA TA 3′; adamts8-qF: 5′ ATC ACC GTG AGG ATG TGG TT 3′, adamts8-qR: 5′ CAA GAG GTT TGT GTC CGA GG 3′; adamts9-qF: 5′ TGT GGT GTT GGA GTG ATG CAG AGA 3′, adamts9-qR: 5′ TCT GGC TTC AGA TCA GTG TGG CAT 3′; adamts15-qF: 5′ ACA CTG CCA TCC TCT TCA CC 3′, adamts15-qR: 5′ TCT TGG GGT CAC ACA TGG TA 3′. Quantitative PCR (qPCR) primers were designed over introns. iTaq Universal SYBR Green Supermix is from Bio-Rad. The following cycling conditions were used with 10 ng of RNA: 95℃ for 30 s, followed by 40 cycles of amplification (95℃ for 5 s, 60℃ for 30 s, 55℃ for 60 s, read plate), and a melting curve analysis. Relative quantities of RNA were determined using the ΔΔ-CT method. A minimum of n = 3 experiments (each in triplicate) was run for each gene. All data were normalized to gapdh levels.
In Situ Hybridization
In situ hybridization (ISH) was performed on 16 µm coronal cryosectioned tissues as previously described. The antisense riboprobe generation of full length of adamts4 and syt1 were described previously (Su et al., 2010; Levy et al., 2015). Briefly, riboprobes were synthesized using digoxigenin (DIG)-labeled UTP (Roche, Mannheim, Germany) and the MAXIscript In Vitro Transcription Kit (Ambion, Austin, TX, USA). Probes were hydrolyzed to 500 nt. Sagittal brain sections were prepared and hybridized at 65℃ as previously described (Su et al., 2010), and bound riboprobes were detected by horseradish peroxidase (POD)-conjugated anti-DIG antibodies and fluorescent staining with Tyramide Signal Amplification (TSA) systems (PerkinElmer, Shelton, CT, USA). Images were obtained on a Zeiss Axio Imager A2 fluorescent microscope or a Zeiss Examiner Z1 LSM 700 confocal microscope. A minimum of three animals per genotype and age was compared in ISH experiments.
Results
Loss of WFA-Labeled PNNs in the Cerebral Cortex and Hippocampus of Collagen XIX-Deficient Mice
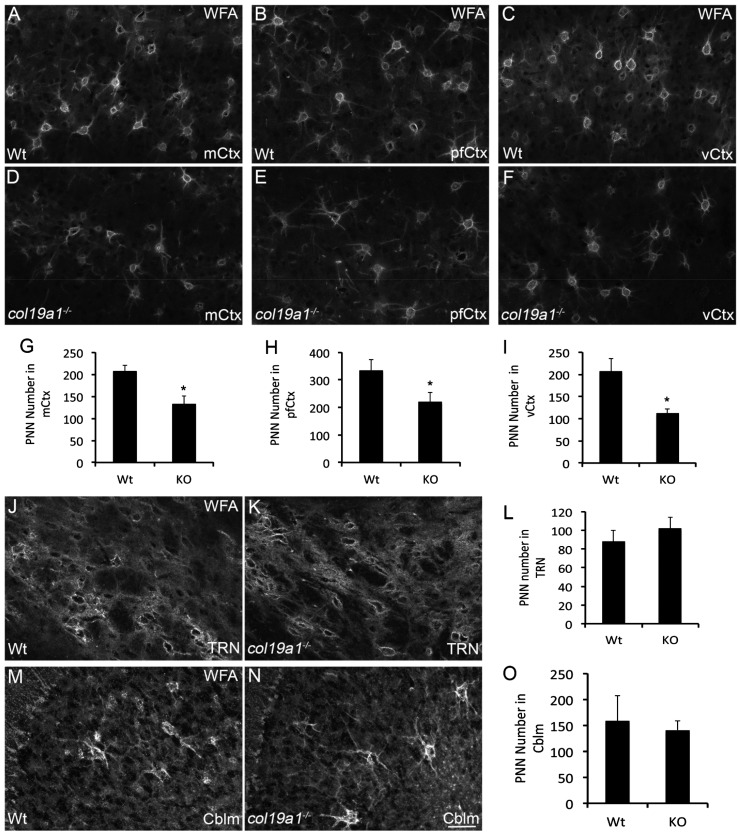
We previously reported that collagen XIX is generated by subsets of interneurons in the mammalian cerebral cortex (Su et al., 2016). To test whether collagen XIX is necessary for the formation or maintenance of PNNs, we assessed the presence of PNNs in several regions of the cerebral cortex of col19a1−/− mutant brains. PNNs were labeled with biotin-conjugated Wisteria floribunda agglutinin (WFA), a lectin that specifically binds N-acetylgalactosamines (Hartig et al., 1992; Figure 1). In adult wild-type mice, WFA labeled a large set of PNNs that surround the somas and proximal neurites of subsets of interneurons in all regions of cortex (Figure 2(a) to (c)). Similarly, in littermate col19a1−/− mutants WFA-labeling revealed PNNs in all regions of cortex and these specialized ECM assemblies appeared similar in morphology and size to those in controls (Figure 2(d) to (f) and data not shown), although the intensity of WFA-labeling appeared reduced compared to controls. However, we observed far fewer PNNs in the absence of collagen XIX (Figure 2(a) to (f)). We quantified PNN numbers in three regions of the cerebral cortex (pfCtx, mCtx, and vCtx) and discovered statistically significant reductions in WFA-labeled PNNs in all three regions of col19a1−/− brains (Figure 2(g) to (i)).
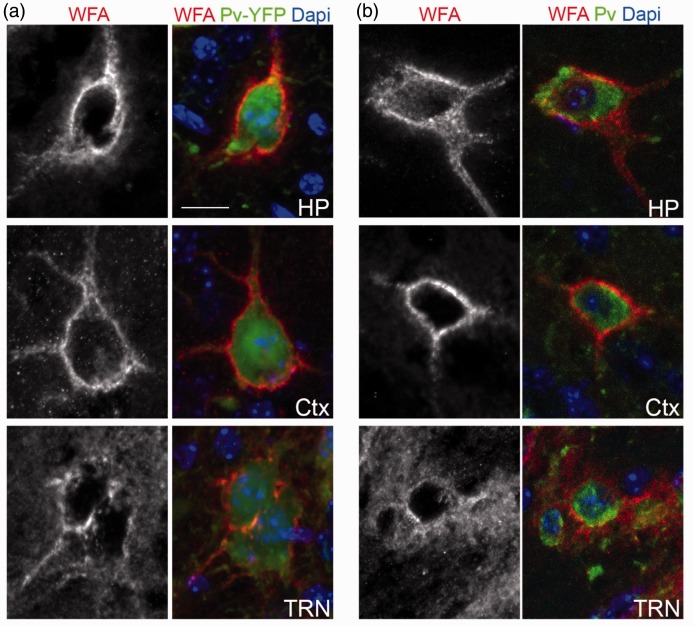
Figure 1.
PNNs ensheath the cell bodies of Parv-expressing neurons in adult mouse brain. (a) WFA-labeled PNN were imaged in hippocampus (HP), cortex (Ctx), and the thalamic reticular nucleus (TRN) of P56 parv-cre::thy1-stop-yfp15 transgenic reporter mice. Parv-expressing interneurons are genetically labeled with YFP in these mice. (b) WFA-labeled PNN were imaged in hippocampus, cortex, and the thalamic reticular nucleus (TRN) of P56 wild-type mice. Parv-expressing interneurons were labeled by IHC. Scale bar = 20 µm.
Figure 2.
Loss of WFA-labeled PNNs in collagen XIX-deficient cerebral cortex. WFA-labeled PNN in P56 wild-type (WT; (a)–(c)) and col19a1−/− mutant (KO; (d)–(f))—motor cortex (mCtx; (a) and (d)), prefrontal cortex (pfCtx; (b) and (e)), and primary visual cortex (vCtx; (c) and (f)). Quantification of WFA-labeled PNN number in P56 WT and KO mCtx (g), pfCtx (h), and vCtx (i). Data represent means ± standard deviation (SD). * indicates differs from littermate WT by p < 0.05 by Student’s t-test. WFA-labeled PNN in the thalamic reticular nucleus of P56 WT (j) and col19a1−/− mutant (k). (l) Quantification of WFA-labeled PNN number in P56 WT and KO TRN. Data represent means ± SD. Data were not statistically significant. WFA-labeled PNN in the cerebellum (Cblm) of P56 WT (m) and col19a1−/− mutant (n). (o) Quantification of WFA-labeled PNN number in P56 WT and KO Cblm. Data represent means ± SD. Data were not statistically significant. Scale bar in N = 50 µm for (a)–(f), (j), (k), (m), and (n).
The distribution of WFA-labeled PNNs in mouse brains is far more extensive than the distribution of col19a1-expressing interneurons (Bruckner et al., 1994; Hartig et al., 1994; Bruckner et al., 2003; Su et al., 2010; Su et al., 2016). To test whether the loss of PNNs in col19a1−/− mutants was the result of a local role for collagen XIX or the result of general changes in mice that globally lack collagen XIX, we assessed PNNs in the thalamic reticular nucleus (TRN). TRN is a region of ventral thalamus containing PNN-ensheathed inhibitory projection neurons (Figure 2(j)), but that lacks collagen XIX expression (Su et al., 2016). The number and density of WFA-labeled PNNs in control and col19a1−/− mutant TRN were indistinguishable (Figure 2(j) to (l)). Likewise, the number of PNNs in cerebellum (Cblm), another region of mouse brain that lacks col19a1-expressing cells (Su et al., 2010), appeared similar control and col19a1−/− mutant mice (Figure 2(m) to (o)). Taken together, these results suggest a local role for collagen XIX in the assembly or maintenance of WFA-labeled PNNs.
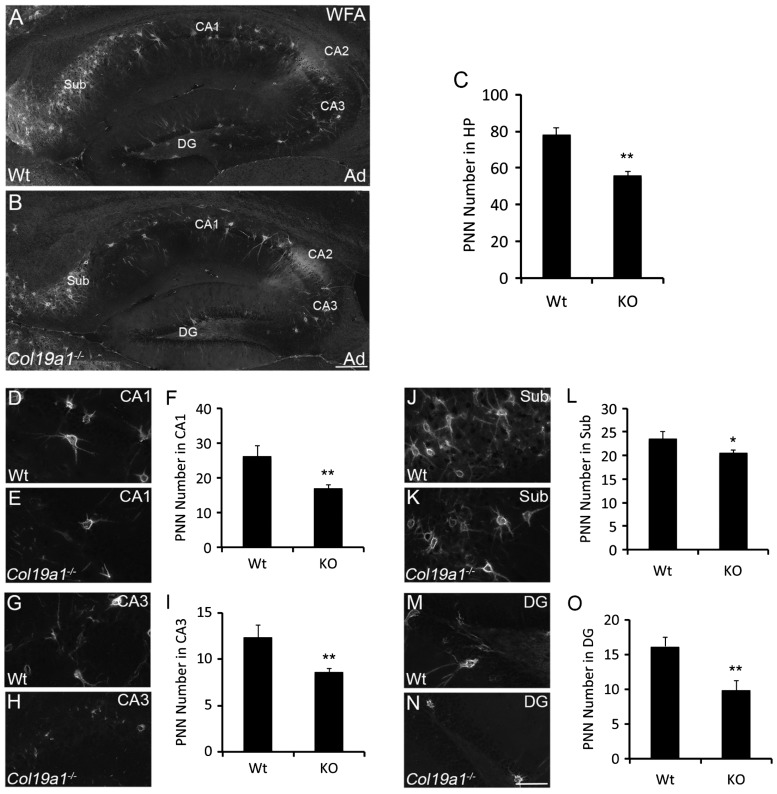
To confirm this hypothesis, we addressed whether the number of WFA-labeled PNNs was altered in the hippocampus of col19a1−/− mutant mice. The hippocampus, a region within the telencephalon and a component of the limbic system, contains an abundance of PNNs (Figure 3(a); Bruckner et al., 2003) and interneurons that generate collagen XIX (Su et al., 2010). Indeed, there was a statistically significant reduction in PNN number in col19a1−/− hippocampi (Figure 3(b) and (c)). The hippocampus can be subdivided into at least five different areas—cornu ammonis (CA) areas 1–3, the dentate gyrus (DG), and the subiculum (Sub). In control mice, we observed prominent WFA-labeled PNNs in all five of these regions, with the highest density in Sub and CA2 (Figure 3(a)). We observed significant reductions in PNN numbers in four of these areas of collagen XIX-deficient hippocampi: CA1, CA3, DG, and Sub (Figure 3(d) to (o)). It is noteworthy that we failed to detect a significant reduction in WFA-labeling in CA2 in the absence of collagen XIX (Figure 3(a) and (b)). Sparing of PNNs in CA2 of collagen XIX-deficient hippocampi may reflect the distinct molecular architecture and ECM composition of this region (Bruckner et al., 2003; Carstens et al., 2016; Dudek et al., 2016) and the relative lack of collagen XIX expression by cells in this region (Su et al., 2010).
Figure 3.
Loss of WFA-labeled PNNs in collagen XIX-deficient hippocampus. WFA-labeled PNN in hippocampus of P56 wild-type (WT; (a)) and col19a1−/− mutant (KO; (b)). (c) Quantification of WFA-labeled PNN number in P56 WT and KO hippocampus. Data represent means ± standard deviation (SD). ** indicates differs from littermate WT by p < 0.0001 by Student’s t-test. Distribution of WFA-labeled PNNs was assessed in subregions of P56 WT ((d), (g), (j), and (m)) and col19a1−/− mutant ((e), (h), (k), and (n)) hippocampi: Regions of analysis included conru ammonis area (CA) 1 ((d)–(f)), CA3 ((g)–(i)), subiculum (Sub; (j)–(l)), and dentate gyrus (DG; (m)–(o)). Quantification of WFA-labeled PNN number in these subregions are shown in (f), (i), (l), and (o). Data represent means ± SD. * indicates differs from littermate WT by p < 0.05 by Student’s t-test. ** indicates differs from littermate WT by p < 0.0001 by Student’s t-test. Scale bar in (b) = 300 µm for (a) and (b) and in (n) = 50 µm for (d), (e), (g), (h), (j), (k), (m), and (n).
Loss of Acan-Labeled PNNs in col19a1−/− Cerebral Cortex and Hippocampus
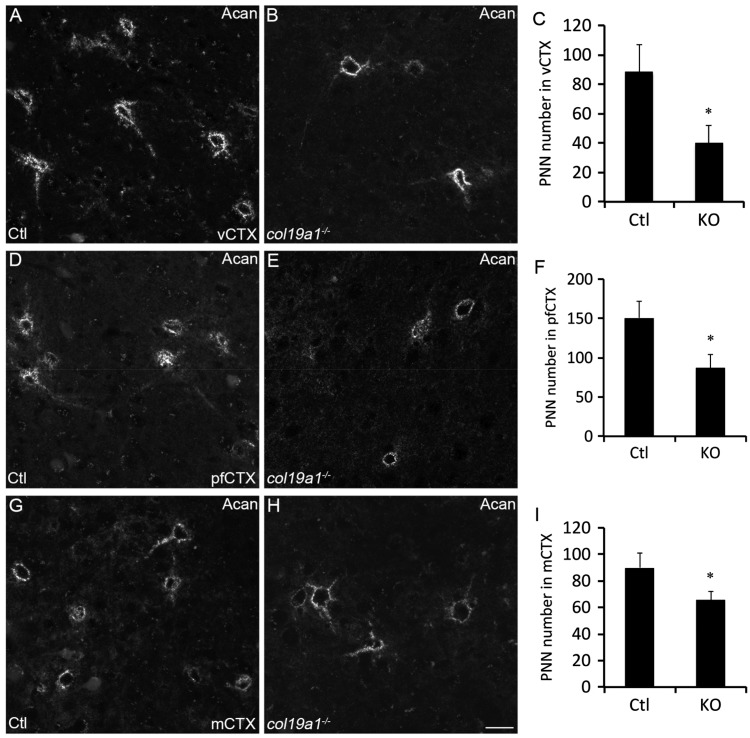
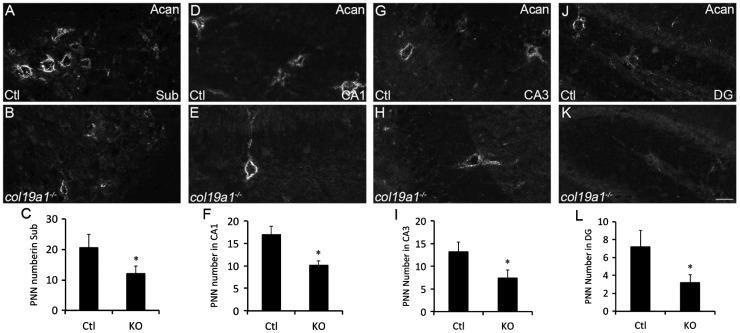
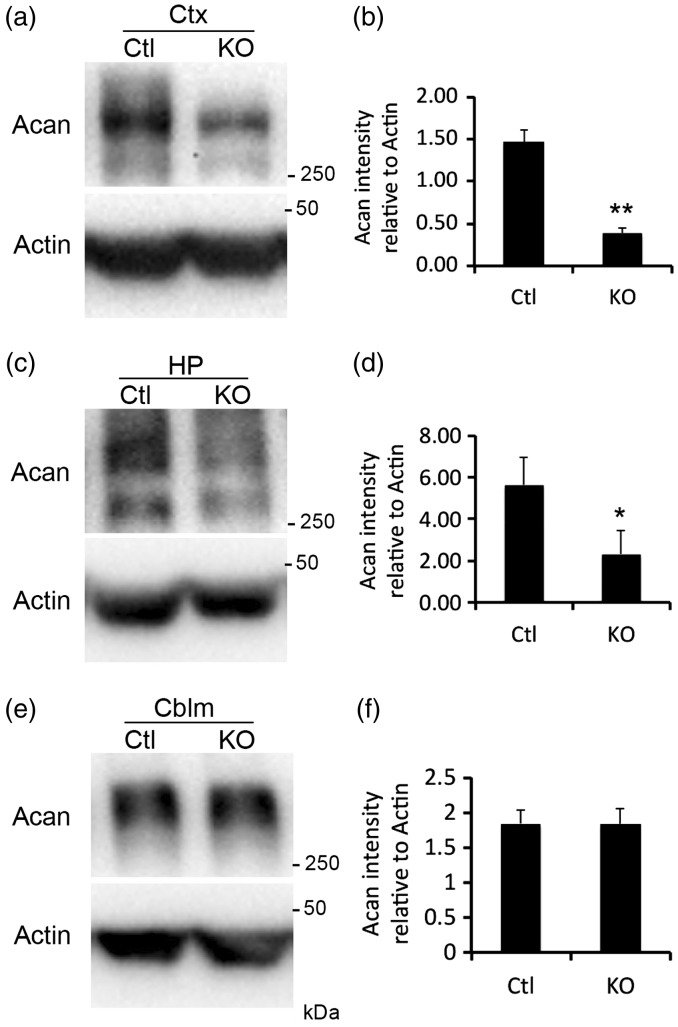
To confirm the loss of PNNs in collagen XIX-deficient cerebral cortex and hippocampus (rather than the loss of WFA-binding sites with these PNNs), we took an alternative approach to label and quantify PNN number; we immunostained sections of control and col19a1−/− mutant brains with antibodies directed against Acan (Cat315; Matthews et al., 2002; Brooks et al., 2013), a lectican present in most PNNs (Zimmermann and Dours-Zimmermann, 2008). Acan-immunolabeled a similar, but not identical, population of PNNs as WFA in mouse cortex and hippocampus (Figures 4 and 5; Bruckner et al., 2003; Morawski et al., 2012). Similar to the results described above for WFA labeling, we detected a statistically significant reduction in the number of Acan-containing PNNs in pfCtx, mCtx, and vCTX (Figure 4) and in CA1, CA3, DG, and Sub (Figure 5) of col19a1−/− mutants. Based upon the reduced Acan-immunolabeling, we hypothesized that total telencephalic Acan levels were reduced in the absence of collagen XIX. To test this, protein extracts were isolated from the cerebral cortex or hippocampus of postnatal Day 67 (P67) control or col19a1−/− mutant mice. Western blots revealed a significant reduction in Acan levels in mutant cortex and hippocampus (Figure 6(a) to (d)). Reduced levels of Acan in collagen XIX-deficient mutants were highly specific to regions of mouse brain that normally generate collagen XIX, since levels appeared normal in cerebellum (Figure 6(e) and (f)). Taken together, these results demonstrate that the local loss of collagen XIX leads to significant reductions in both PNNs and PNN constituents in regions of brain that normally generated this unconventional collagen.
Figure 4.
Loss of aggrecan-containing PNNs in collagen XIX-deficient cerebral cortex. Aggrecan (Acan)-immunolabeled PNNs in P56 wild-type (Ctl; (a), (d), and (g)) and col19a1−/− mutant (KO; (b), (e), and (h)). Images depict Acan-rich PNNs in primary visual cortex (vCtx; (a) and (b)), prefrontal cortex (pfCtx; (d) and (e)), and motor cortex (mCtx; (g) and (h)). Quantification of Acan-immunolabeled PNNs in P56 WT and KO vCtx (c), pfCtx (f), and mCtx (i). Data represent means ± standard deviation (SD). * indicates differs from littermate WT by p < 0.05 by Student’s t-test. Scale bar in (h) = 25 µm for (a), (b), (d), (e), (g), and (h).
Figure 5.
Loss of aggrecan-containing PNNs in collagen XIX-deficient hippocampus. Aggrecan (Acan)-immunolabeled PNNs in subregions of P56 wild-type (Ctl; (a), (d), (g), and (j)) and col19a1−/− mutant (KO; (b), (e), (h), and (k)) hippocampus. Images depict Acan-rich PNNs in Sub ((a)–(c)), CA1 ((d)–(f)), CA3 ((g)–(i)), and DG ((j)–(l)). Quantification of Acan-immunolabeled PNNs in P56 WT and KO in these subregions are shown in (c), (f), (i), and (l). Data represent means ± standard deviation (SD). * indicates differs from littermate WT by p < 0.05 by Student’s t-test. Scale bar in (k) = 25 µm for (a), (b), (d), (e), (g), (h), (i), and (k).
Figure 6.
Reduced levels of aggrecan (Acan) in collagen XIX-deficient cerebral cortex and hippocampus. Western blots demonstrate reduced levels of Acan in protein extracts derived from P67 control (Ctl) or col19a1−/− mutant (KO) cortex (a), hippocampus (HP; (c)), and cerebellum (Cblm; (e)). Levels of Actin protein were used as loading controls. Quantification of levels of Acan detected by Western blot and normalized to Actin levels. Data shown are mean relative intensities normalized to Actin ± SD (n = 3). * indicates differs from littermate WT by p < 0.05 by Student’s t-test. ** indicates differs from littermate WT by p < 0.0001 by Student’s t-test.
Loss of Collagen XIX Leads to Altered Expression of Lectican-Degrading Proteases
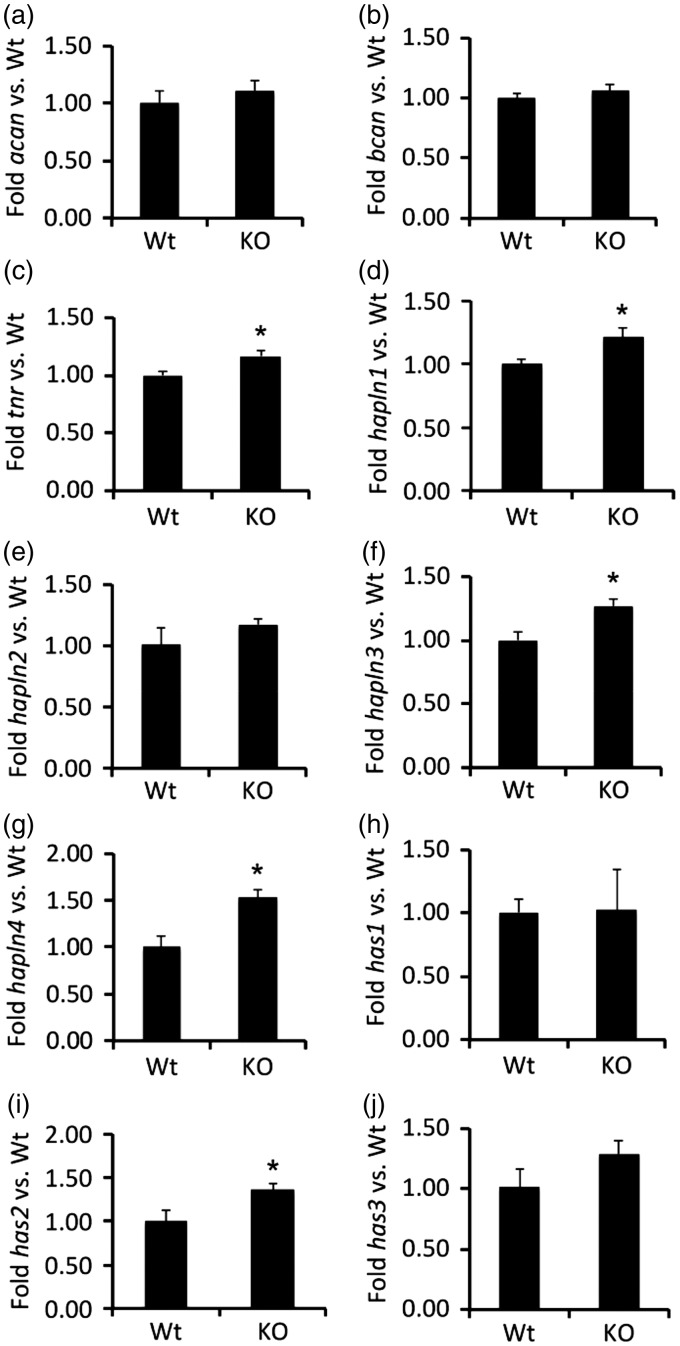
The significant reduction in Acan protein led us to explore whether the loss of collagen XIX altered Acan transcription. To test this, RNA was isolated from the cerebral cortex of P56 control and col19a1−/− mutant mice and levels of acan mRNA were assessed by qPCR. We failed to detect significant alterations in acan mRNA levels in col19a1−/− mutant cortex (Figure 7(a)) suggesting the reduced Acan levels was not the result of altered transcription.
Figure 7.
Alteration of PNN gene transcription in the absence of collagen XIX. Quantitative PCR (qPCR) analysis was performed on RNA extracted from P56 control (Ctl) or col19a1−/− mutant (KO) cortex. Levels of target gene expression were normalized to levels of gapdh. Data represent mean fold change (±SD) in mRNA expression in KO compared to levels in control samples. * indicates differs from littermate WT by p < 0.05 by Student’s t-test. Note that despite reduced levels of Acan protein levels in KO, qPCR revealed Acan transcription was unaltered in KO cortex. PNN genes analyzed: acan = aggrecan; bcan = brevican; tnr = tenascin R; hapln1 = hyaluranonin and proteoglycan binding link protein 1; hapln2 = hyaluranonin and proteoglycan binding link protein 2; hapln3 = hyaluranonin and proteoglycan binding link protein 3; hapln4 = hyaluranonin and proteoglycan binding link protein 4; has1 = hyaluronan synthase 1; has2 = hyaluronan synthase 2; has3 = hyaluronan synthase 3.
In fact, the same was true for other components of PNNs. We tested whether the mRNA expression of other integral components of PNNs, including other lecticans (like brevican), tenascin-R, HAPLN family members (HAPLN1-4), or Hyaluronan Synthases (Has1-3) were altered in col19a1−/− mutant cortex. Previous studies have revealed that partial or complete loss of any of these PNN constituents leads to reduced number or altered morphology of cortical PNNs (Weber et al., 1999; Bruckner et al., 2000; Carulli et al., 2010; Kwok et al., 2010). We found that the loss of Collagen XIX failed to reduce the transcription of mRNAs of any of these PNN constituents (Figure 7), and in several cases increased expression levels were detected (e.g., tenascin-R, HAPLN1, HAPLN3, HAPLN4, and Has2). This increased expression of PNN-associated genes may represent a homeostatic mechanism activated in collagen XIX-deficient mutants to counteract the loss of this collagen and PNNs.
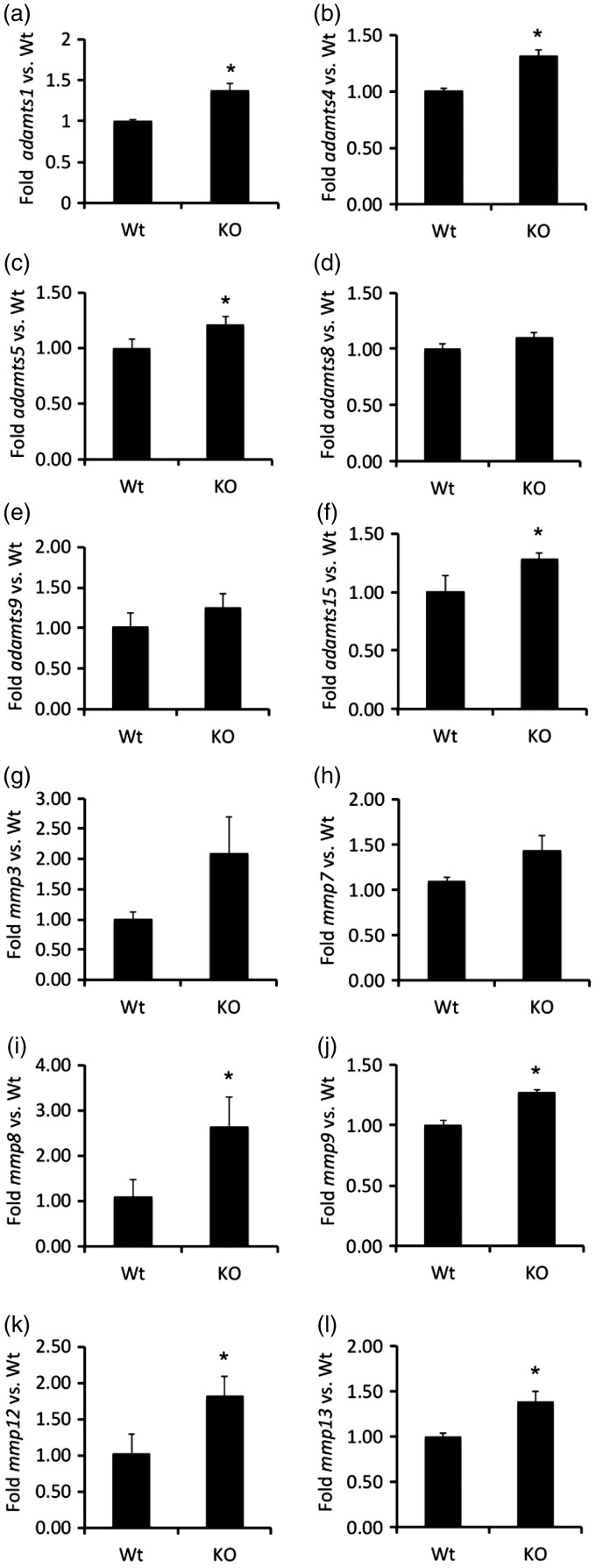
The increase in expression of tenascin-R, HAPLN1, HAPLN3, HAPLN4, and Has2 in col19a1−/− mutant cortex failed to provide insight into the reduced levels of PNNs and Acan protein in these mutants. However, the increase production of these PNN constituents led us to speculate that collagen loss may also lead to an upregulation of extracellular proteases capable of degrading PNN components. PNN components are degraded by several families of metalloproteinases, including ADAMTSs and MMPs. Many ADAMTSs (including ADAMTS1, 4, 5, 8, 9, 11, and 15) are classified as lecticanases and not only have a high affinity and activity for cleaving Acan (Abbaszade et al., 1999; Tortorella et al., 1999; Tang, 2001; Porter et al., 2005), but are expressed in mouse cortex (Gottschall and Howell, 2015; Levy et al., 2015). Likewise, many MMPs are expressed in mouse brain and have been shown to cleave PNN constituents (Nagel et al., 2005; Rankin-Gee et al., 2015). We explored whether expression of 12 members of the ADAMTS and MMP families were altered in col19a1−/− mutant cortex. Eight of these proteases were significantly increased in collagen XIX-deficient cortex (Figure 8), including ADAMTS1, ADAMTS4, ADAMTS5, ADAMTS15, MMP8, MMP9, MMP12, and MMP13.
Figure 8.
Alteration of protease gene transcription in the absence of collagen XIX. Quantitative PCR (qPCR) analysis was performed on RNA extracted from P56 control (Ctl) or col19a1−/− mutant (KO) cortex. Levels of protease gene expression were normalized to levels of gapdh. Data represent mean fold change (±SD) in mRNA expression in KO compared to levels in control samples. * indicates differs from littermate WT by p < 0.05 by Student’s t-test. Note that eight genes encoding PNN-degrading proteases from the ADAMTS and MMP families of proteases were upregulated in col19a1−/− mutant (KO) cortex.
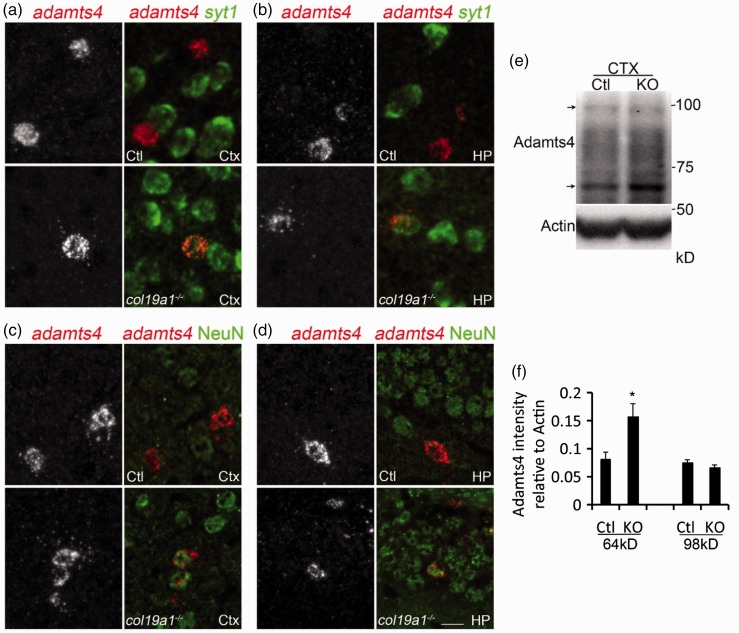
One of the mRNAs increased in collagen XIX-deficient cortex encodes the most well-studied endogenous Acan-cleaving enzyme in the brain—ADAMTS4 (also called aggrecanase 1; Tang, 2001; Porter et al., 2005). We previously showed that adamts4 mRNA expression was restricted to oligodendrocytes in the mouse cortex and hippocampus (Levy et al., 2015). Here, we discovered that adamts4 mRNA becomes expressed in a subset of neurons in col19a1−/− mutant cortex and hippocampus (Figure 9(a) to (d)), suggesting that the significant increase in adamts4 mRNA was the result of an upregulation of this gene in a cell-type that normally does not express it. Moreover, we assessed whether protein levels of ADAMTS4 were also altered in col19a1−/− mutant cortex. ADAMTS4 is produced as a pro-form (∼98 kDa) that undergoes cleavage to become a mature, active peptide (∼64 kDa)(Tortorella et al., 1999; Vistnes et al., 2014). In mutant and control cortex, we detected similar levels of the pro-form of ADAMTS4; however, increased levels of active ADAMTS4 were present in col19a1−/− mutant cortex (Figure 9(e)). Densitometry revealed that this increase in active ADAMTS4 was statistically significant (Figure 9(f)). Thus, these results reveal ectopic expression of adamts4 mRNA in mutant brains and an increase in active ADAMTS4 peptides and therefore suggest that increased production and activity of Acan-degrading proteases contribute to the loss of PNNs in collagen XIX-deficient brains.
Figure 9.
Ectopic neuronal expression of adamts4 mRNA and increased active ADMATS4 peptides in the absence of collagen XIX. ((a) and (b)) In situ hybridization for adamts4 and syt1 mRNA in adult control or col19a1−/− mutant cortex (Ctx) and hippocampus (HP). Note the co-expression of adamts4 and syt1 mRNA in neurons in mutant Ctx and HP. ((c) and (d)) In situ hybridization for adamts4 and IHC for NeuN in adult control or col19a1−/− mutant cortex (Ctx) and hippocampus (HP). Note the ectopic co-expression of adamts4 and NeuN in neurons in mutant Ctx and HP. (e) Western blots of ADAMTS4 in protein extracts from control or col19a1−/− mutant Ctx. The pro- and active forms of ADAMTS4 are indicated with arrows (pro-form – 98 kDa; active form – 64 kDa). (f) Quantification of amounts of pro- and active forms of ADAMTS4 in control and col19a1−/− mutant Ctx. Data shown are mean relative intensities (normalized to actin) ± SD (n = 3). * indicates differ from controls by p < 0.05 by Student’s t-test. Scale bar = 20 µm.
Discussion
A recent study identified a novel micro-deletion at chromosome 6q12-13 in a cohort of patients with familial schizophrenia, a complex brain disorder characterized by alterations in cognitive function, acquisition of behaviors not observed in healthy individuals (such as hallucinations), and neglect and loss of apathy (Liao et al., 2012). This genomic region contains the coding sequence for collagen XIX. In support of the association of collagen XIX mutation or deletion with schizophrenia, we recently demonstrated that loss of collagen XIX leads to the acquisition of schizophrenia-related behaviors and seizures in mice (Su et al., 2016). These collagen-deficient mutant mice display a striking reduction in the formation of inhibitory axo-somatic synapses in the cerebral cortex and hippocampus (Su et al., 2010; Su et al., 2016), a class of synapse implicated in the mechanisms underlying schizophrenia, seizures, and other neurodevelopmental disorders (Gonzalez-Burgos et al., 2011; Lewis et al., 2011; Nakazawa et al., 2012; Wohr et al., 2015). Taken together, the loss of axo-somatic inhibitory synapses in col19a1−/− mutant mice and schizophrenic patients suggests a possible mechanism of how mutation, reduction or complete loss of collagen XIX may contribute to schizophrenia. However, there are other cellular and molecular alterations in the brains of patients with schizophrenia in addition to the loss of axo-somatic inhibitory synapses. One such alteration is the loss of interneurons (Zhang and Reynolds, 2002; Penschuck et al., 2006; Enwright et al., 2016). Previously we assessed interneuron number and distribution in cerebral cortex and hippocampus of collagen XIX-deficient brains and found no appreciable affect on interneuron number or distribution in the absence of this collagen in mice (Su et al., 2010; Su et al., 2016). Another alteration in patients with schizophrenia is PNN loss (Berretta et al., 2015; Bitanihirwe et al., 2016). Here, we set out to assess whether the loss of collagen XIX altered the number or distribution of PNNs. Our results demonstrate for the first time that the loss of a neuronally expressed collagen results in a significant reduction in the specialized lattice-like ECM that surround cortical and hippocampal interneurons. Therefore, these results identify a second mechanism by which the loss of collagen XIX may contribute to the development of neuropsychiatric disorders, such as schizophrenia.
Does Collagen XIX Play a Direct role in PNN Assembly or Maintenance?
It is possible that interneuron-derived collagen XIX acts as a nucleating factor for PNNs. Outside of the nervous system, nonfibrillar collagens bind and anchor numerous ECM constituents into supramolecular assemblies (Ricard-Blum, 2011). Collagen XIX is a relatively recently identified, minor collagen (Myers et al., 1997; Sumiyoshi et al., 1997) and therefore the extent to which it binds other ECM components remains to be determined. In addition to its numerous collagenous domains, collagen XIX contains an N-terminal thrombospondin domain which may interact with other components of the brain ECM (Ricard-Blum, 2011). It also contains a C-terminal noncollagenous domain that interacts with a variety of RGD-dependent integrins (Oudart et al., 2015, 2016; Su et al., 2016) which are known to be expressed by interneurons in the cerebral cortex and hippocampus (Bi et al., 2001; Su et al., 2016). At present it remains unclear whether collagen XIX itself is present in PNNs. While collagen XIX is generated by subsets of cortical and hippocampal interneurons and PNNs preferentially ensheath interneurons in these brain regions, it is important to note that as many as 20 different classes of interneurons exist in these brain regions (Klausberger and Somogyi, 2008). Collagen XIX is generated by classes of interneurons that are defined neurochemically by their expression of calbindin, somatostatin, or neuropeptide Y and only a small fraction of these cells are ensheathed by PNNs (Su et al., 2010; Su et al., 2016). In contrast, PNNs preferentially ensheath classes of telencephalic interneurons that generate parvalbumin (Hartig et al., 1994) but only a small fraction of these cells appear to generate Collagen XIX (<10% of col19a1-expressing cells co-express Parv in cerebral cortex; Su et al., 2016). Therefore, while results presented here indicate a local role for collagen XIX in the assembly of PNNs, it is likely that many interneurons ensheathed by PNNs do not generate this collagen. An alternative possibility is that collagen XIX is secreted by one class of interneurons and aggregates around different classes of neighboring cells. This is the case for PNN constituents that are produced by glial cells (John et al., 2006; Giamanco and Matthews, 2012). And this type of paracrine mechanism appears to be how collagen XIX influences the development of inhibitory axo-somatic synapses in the cerebral cortex and hippocampus (Su et al., 2016). Of course, to test this hypothesis it will be essential to localize collagen XIX protein in PNNs in vivo, something that has not been possible with the current set of molecular tools to interrogate this collagen.
Although collagens may act as nucleating factors in basal laminas, it is entirely possible that the same is not true in PNNs and loss of PNNs may not be the direct result of the absence of this unconventional collagen. This possibility is supported by transcriptional changes of extracellular metalloproteinases known to degrade PNN constituents in col19a1−/− mutants. Many proteases upregulated in the cerebral cortex of col19a1−/− mutant mice are known to be generated by cortical interneurons that are coated by PNNs (Levy et al., 2015). So how might the loss of this collagen alter MMP or ADAMTS expression? A likely possibility is that this collagen plays a direct role in regulating transcription factors that act upstream of extracellular protease genes. Certainly the influence of ECM in regulating transcription in this fashion has been well documented outside of the brain (Bissell and Barcellos-Hoff, 1987; Spencer et al., 2007). Alternatively, the loss of collagen XIX is known to lead to molecular or cellular changes in the telencephalon (Su et al., 2010; Su et al., 2016) that may lead to altered protease production. While the number and distribution of PNN coated interneurons does not appear to change in col19a1−/− mutant brains, synapses onto these cells are altered in these mutants (Su et al., 2016). Perturbing connectivity in the brain is known to lead to alterations in protease production (Zhang et al., 1998; Jourquin et al., 2003), just as perturbing protease expression can alter connectivity (Huntley, 2012). In addition to structural changes in circuitry, the acquisition of behavior phenotypes and seizures in col19a1−/− mutant mice may lead to altered extracellular protease production. Prolonged seizure induction upregulates MMP and ADAMTS expression and the subsequent loss of PNNs in rodent models (Rankin-Gee et al., 2015). Likewise, an upregulation of ADAMTS15, which degrades Acan and other chondroitin sulfate proteoglycans and which we show is upregulated in cortex of col19a1−/− mutant mice, is upregulated in the hippocampi of patients suffering from medial lobe epilepsy (Venugopal et al., 2012). Thus, it is possible that altered connectivity and activity in col19a1−/− mutant mice leads to changes in protease production and a loss of PNNs.
A Role for Collagen XIX in Closing the Critical Period of Neural Circuit Development
We now report two morphological phenotypes in the telencephalon of mice lacking collagen XIX—the loss of axo-somatic, inhibitory synapses onto Parvalbumin-expressing interneurons (Su et al., 2016) and the loss of PNNs from this same class of interneurons. Our data indicate that alterations in the formation of inhibitory axo-somatic synapses appear well before the first PNNs begin to assemble in the developing cerebral cortex or hippocampus (Su et al., 2010; Su et al., 2016). However, it is very likely that these phenotypes are interconnected, since they involve the same cell type and both morphological phenotypes are associated with a variety of complex brain disorders. Relatively late in development and after a period of activity-dependent refinement of neural circuits, PNNs emerge and are thought to form a physical barrier to prevent either the loss of established axo-somatic connections or the formation of new connections (Wang and Fawcett, 2012). The emergence of PNNs coincides with the closing of the “critical period” in circuit development, a period in which neural activity and experience are critical for shaping neural circuit refinement. To test the link between PNNs, axo-somatic synapse and the critical period of circuit development, previous laboratories have enzymatically degraded PNNs in vCTX by delivering chondroitinaseABC, an enzyme that degrades CSPGs and PNNs. Experimental degradation of PNNs in vCTX led to both the re-emergence of ocular dominance plasticity (Pizzorusso et al., 2002) and enhanced motility of dendritic spines and increased structural and functional neuroplasticity in visual cortical circuits (de Vivo et al., 2013). Taken together with results here, and the temporal expression of collagen XIX in the postnatal brain (Sumiyoshi et al., 1997; Su et al., 2010; Su et al., 2016), we posit that loss of collagen XIX may stall the brain in an immature, plastic state and that its expression may contribute to the closing of the critical period in the developing cortex. While total loss of collagen XIX is detrimental due to the emergence of schizophrenia-related behaviors and seizures, our results raise the intriguing possibility that local manipulation of this collagen may be a useful therapeutic approach to modulate critical periods of cortical plasticity.
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported in part by a NARSAD Independent Investigator Award from the Brain and Behavior Research Foundations (to M. A. F.), the National Institutes of Health (NIH) Grant EY021222 (to M. A. F.), and the National Institutes of Health (NIH) Grant AI124677 (to M. A. F.).
References
- Abbaszade I., et al. (1999) Cloning and characterization of ADAMTS11, an aggrecanase from the ADAMTS family. Journal of Biological Chemistry 274: 23443–23450. [DOI] [PubMed] [Google Scholar]
- Berretta S. (2012) Extracellular matrix abnormalities in schizophrenia. Neuropharmacology 62: 1584–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretta S., Pantazopoulos H., Markota M., Brown C., Batzianouli E. T. (2015) Losing the sugar coating: Potential impact of perineuronal net abnormalities on interneurons in schizophrenia. Schizophrenia Research 167: 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Lynch G., Zhou J., Gall C. M. (2001) Polarized distribution of alpha5 integrin in dendrites of hippocampal and cortical neurons. Journal of Comparative Neurology 435: 184–193. [DOI] [PubMed] [Google Scholar]
- Bissell M. J., Barcellos-Hoff M. H. (1987) The influence of extracellular matrix on gene expression: Is structure the message? Journal of Cell Science – Supplement 8: 327–343. [DOI] [PubMed] [Google Scholar]
- Bitanihirwe B. K., Mauney S. A., Woo T. U. (2016) Weaving a net of neurobiological mechanisms in schizophrenia and unraveling the underlying pathophysiology. Biological Psychiatry 80: 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J. M., Su J., Levy C., Wang J. S., Seabrook T. A., Guido W., Fox M. A. (2013) A molecular mechanism regulating the timing of corticogeniculate innervation. Cell Reports 5: 573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckner G., Grosche J., Hartlage-Rubsamen M., Schmidt S., Schachner M. (2003) Region and lamina-specific distribution of extracellular matrix proteoglycans, hyaluronan and tenascin-R in the mouse hippocampal formation. Journal of Chemical Neuroanatomy 26: 37–50. [DOI] [PubMed] [Google Scholar]
- Brückner G., Grosche J., Schmidt S., Härtig W., Margolis R. U., Delpech B., Seidenbecher C. I., Czaniera R., Schachner M. (2000) Postnatal development of perineuronal nets in wild-type mice and in a mutant deficient in tenascin-R. Journal of Comparative Neurology 428: 616–629. [DOI] [PubMed] [Google Scholar]
- Bruckner G., Seeger G., Brauer K., Hartig W., Kacza J., Bigl V. (1994) Cortical areas are revealed by distribution patterns of proteoglycan components and parvalbumin in the Mongolian gerbil and rat. Brain Research 658: 67–86. [DOI] [PubMed] [Google Scholar]
- Carstens K. E., Phillips M. L., Pozzo-Miller L., Weinberg R. J., Dudek S. M. (2016) Perineuronal nets suppress plasticity of excitatory synapses on CA2 pyramidal neurons. Journal of Neuroscience 36: 6312–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carulli D., Pizzorusso T., Kwok J. C., Putignano E., Poli A., Forostyak S., Andrews M. R., Deepa S. S., Glant T. T., Fawcett J. W. (2010) Animals lacking link protein have attenuated perineuronal nets and persistent plasticity. Brain 133: 2331–2347. [DOI] [PubMed] [Google Scholar]
- Celio M. R., Blumcke I. (1994) Perineuronal nets—A specialized form of extracellular matrix in the adult nervous system. Brain Research Reviews 19: 128–145. [DOI] [PubMed] [Google Scholar]
- Celio M. R., Spreafico R., De Biasi S., Vitellaro-Zuccarello L. (1998) Perineuronal nets: Past and present. Trends in Neurosciences 21: 510–515. [DOI] [PubMed] [Google Scholar]
- de Vivo L., Landi S., Panniello M., Baroncelli L., Chierzi S., Mariotti L., Spolidoro M., Pizzorusso T., Maffei L., Ratto G. M. (2013) Extracellular matrix inhibits structural and functional plasticity of dendritic spines in the adult visual cortex. Nature Communications 4: 1484. [DOI] [PubMed] [Google Scholar]
- Dityatev A., Rusakov D. A. (2011) Molecular signals of plasticity at the tetrapartite synapse. Current Opinion in Neurobiology 21: 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dityatev A., Schachner M., Sonderegger P. (2010) The dual role of the extracellular matrix in synaptic plasticity and homeostasis. Nature Reviews Neuroscience 11: 735–746. [DOI] [PubMed] [Google Scholar]
- Dudek S. M., Alexander G. M., Farris S. (2016) Rediscovering area CA2: Unique properties and functions. Nature Reviews Neuroscience 17: 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enwright J. F., Sanapala S., Foglio A., Berry R., Fish K. N., Lewis D. A. (2016) Reduced labeling of parvalbumin neurons and perineuronal nets in the dorsolateral prefrontal cortex of subjects with schizophrenia. Neuropsychopharmacology 41: 2206–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faissner A., Pyka M., Geissler M., Sobik T., Frischknecht R., Gundelfinger E. D., Seidenbecher C. (2010) Contributions of astrocytes to synapse formation and maturation—Potential functions of the perisynaptic extracellular matrix. Brain Research Reviews 63: 26–38. [DOI] [PubMed] [Google Scholar]
- Fox M. A., Sanes J. R., Borza D. B., Eswarakumar V. P., Fassler R., Hudson B. G., John S. W., Ninomiya Y., Pedchenko V., Pfaff S. L., Rheault M. N., Sado Y., Segal Y., Werle M. J., Umemori H. (2007) Distinct target-derived signals organize formation, maturation, and maintenance of motor nerve terminals. Cell 129: 179–193. [DOI] [PubMed] [Google Scholar]
- Frantz C., Stewart K. M., Weaver V. M. (2010) The extracellular matrix at a glance. Journal of Cell Science 123: 4195–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischknecht R., Gundelfinger E. D. (2012) The brain’s extracellular matrix and its role in synaptic plasticity. Advances in Experimental Medicine and Biology 970: 153–171. [DOI] [PubMed] [Google Scholar]
- Frischknecht R., Seidenbecher C. I. (2012) Brevican: A key proteoglycan in the perisynaptic extracellular matrix of the brain. The International Journal of Biochemistry & Cell Biology 44: 1051–1054. [DOI] [PubMed] [Google Scholar]
- Giamanco K. A., Matthews R. T. (2012) Deconstructing the perineuronal net: Cellular contributions and molecular composition of the neuronal extracellular matrix. Neuroscience 218: 367–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Burgos G., Fish K. N., Lewis D. A. (2011) GABA neuron alterations, cortical circuit dysfunction and cognitive deficits in schizophrenia. Neural Plasticity 2011: 723184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschall P. E., Howell M. D. (2015) ADAMTS expression and function in central nervous system injury and disorders. Matrix Biology 44–46: 70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig W., Brauer K., Bigl V., Bruckner G. (1994) Chondroitin sulfate proteoglycan-immunoreactivity of lectin-labeled perineuronal nets around parvalbumin-containing neurons. Brain Research 635: 307–311. [DOI] [PubMed] [Google Scholar]
- Hartig W., Brauer K., Bruckner G. (1992) Wisteria floribunda agglutinin-labelled nets surround parvalbumin-containing neurons. NeuroReport 3: 869–872. [DOI] [PubMed] [Google Scholar]
- Heikkinen A., Pihlajaniemi T., Faissner A., Yuzaki M. (2014) Neural ECM and synaptogenesis. Progress in Brain Research 214: 29–51. [DOI] [PubMed] [Google Scholar]
- Huntley G. W. (2012) Synaptic circuit remodelling by matrix metalloproteinases in health and disease. Nature Reviews Neuroscience 13: 743–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John N., Krugel H., Frischknecht R., Smalla K. H., Schultz C., Kreutz M. R., Gundelfinger E. D., Seidenbecher C. I. (2006) Brevican-containing perineuronal nets of extracellular matrix in dissociated hippocampal primary cultures. Molecular and Cellular Neuroscience 31: 774–784. [DOI] [PubMed] [Google Scholar]
- Jourquin J., Tremblay E., Decanis N., Charton G., Hanessian S., Chollet A. M., Le Diguardher T., Khrestchatisky M., Rivera S. (2003) Neuronal activity-dependent increase of net matrix metalloproteinase activity is associated with MMP-9 neurotoxicity after kainate. European Journal of Neuroscience 18: 1507–1517. [DOI] [PubMed] [Google Scholar]
- Klausberger T., Somogyi P. (2008) Neuronal diversity and temporal dynamics: The unity of hippocampal circuit operations. Science 321: 53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korogod N., Petersen C. C., Knott G. W. (2015) Ultrastructural analysis of adult mouse neocortex comparing aldehyde perfusion with cryo fixation. eLife 4: e05793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok J. C., Carulli D., Fawcett J. W. (2010) In vitro modeling of perineuronal nets: Hyaluronan synthase and link protein are necessary for their formation and integrity. Journal of Neurochemistry 114: 1447–1459. [DOI] [PubMed] [Google Scholar]
- LeBleu V. S., Macdonald B., Kalluri R. (2007) Structure and function of basement membranes. Experimental Biology and Medicine 232: 1121–1129. [DOI] [PubMed] [Google Scholar]
- Levy C., Brooks J. M., Chen J., Su J., Fox M. A. (2015) Cell-specific and developmental expression of lectican-cleaving proteases in mouse hippocampus and neocortex. Journal of Comparative Neurology 523: 629–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis D. A., Fish K. N., Arion D., Gonzalez-Burgos G. (2011) Perisomatic inhibition and cortical circuit dysfunction in schizophrenia. Current Opinion in Neurobiology 21: 866–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H. M., Chao Y. L., Huang A. L., Cheng M. C., Chen Y. J., Lee K. F., Fang J. S., Hsu C. H., Chen C. H. (2012) Identification and characterization of three inherited genomic copy number variations associated with familial schizophrenia. Schizophrenia Research 139: 229–236. [DOI] [PubMed] [Google Scholar]
- Matthews R. T., Kelly G. M., Zerillo C. A., Gray G., Tiemeyer M., Hockfield S. (2002) Aggrecan glycoforms contribute to the molecular heterogeneity of perineuronal nets. Journal of Neuroscience 22: 7536–7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier F. (2016) Fractones: Extracellular matrix niche controlling stem cell fate and growth factor activity in the brain in health and disease. Cellular and Molecular Life Sciences 73: 4661–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier F., Kitasako J. T., Hatton G. I. (2002) Anatomy of the brain neurogenic zones revisited: Fractones and the fibroblast/macrophage network. Journal of Comparative Neurology 451: 170–188. [DOI] [PubMed] [Google Scholar]
- Morawski M., Bruckner G., Arendt T., Matthews R. T. (2012) Aggrecan: Beyond cartilage and into the brain. The International Journal of Biochemistry & Cell Biology 44: 690–693. [DOI] [PubMed] [Google Scholar]
- Myers J. C., Li D., Bageris A., Abraham V., Dion A. S., Amenta P. S. (1997) Biochemical and immunohistochemical characterization of human type XIX defines a novel class of basement membrane zone collagens. American Journal of Pathology 151: 1729–1740. [PMC free article] [PubMed] [Google Scholar]
- Nagel S., Sandy J. D., Meyding-Lamade U., Schwark C., Bartsch J. W., Wagner S. (2005) Focal cerebral ischemia induces changes in both MMP-13 and aggrecan around individual neurons. Brain Research 1056: 43–50. [DOI] [PubMed] [Google Scholar]
- Nakazawa K., Zsiros V., Jiang Z., Nakao K., Kolata S., Zhang S., Belforte J. E. (2012) GABAergic interneuron origin of schizophrenia pathophysiology. Neuropharmacology 62: 1574–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson C., Sykova E. (1998) Extracellular space structure revealed by diffusion analysis. Trends in Neurosciences 21: 207–215. [DOI] [PubMed] [Google Scholar]
- Oudart J. B., Brassart-Pasco S., Vautrin A., Sellier C., Machado C., Dupont-Deshorgue A., Brassart B., Baud S., Dauchez M., Monboisse J. C., Harakat D., Maquart F. X., Ramont L. (2015) Plasmin releases the anti-tumor peptide from the NC1 domain of collagen XIX. Oncotarget 6: 3656–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudart J. B., Monboisse J. C., Maquart F. X., Brassart B., Brassart-Pasco S., Ramont L. (2016) Type XIX collagen: A new partner in the interactions between tumor cells and their microenvironment. Matrix Biology. S0945-053X(16)30111-1. [DOI] [PubMed] [Google Scholar]
- Pallotto M., Watkins P. V., Fubara B., Singer J. H., Briggman K. L. (2015) Extracellular space preservation aids the connectomic analysis of neural circuits. eLife 4: e08206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penschuck S., Flagstad P., Didriksen M., Leist M., Michael-Titus A. T. (2006) Decrease in parvalbumin-expressing neurons in the hippocampus and increased phencyclidine-induced locomotor activity in the rat methylazoxymethanol (MAM) model of schizophrenia. European Journal of Neuroscience 23: 279–284. [DOI] [PubMed] [Google Scholar]
- Pizzorusso T., Medini P., Berardi N., Chierzi S., Fawcett J. W., Maffei L. (2002) Reactivation of ocular dominance plasticity in the adult visual cortex. Science 298: 1248–1251. [DOI] [PubMed] [Google Scholar]
- Porter S., Clark I. M., Kevorkian L., Edwards D. R. (2005) The ADAMTS metalloproteinases. Biochemical Journal 386: 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin-Gee E. K., McRae P. A., Baranov E., Rogers S., Wandrey L., Porter B. E. (2015) Perineuronal net degradation in epilepsy. Epilepsia 56: 1124–1133. [DOI] [PubMed] [Google Scholar]
- Rhodes K. E., Fawcett J. W. (2004) Chondroitin sulphate proteoglycans: preventing plasticity or protecting the CNS? Journal of Anatomy 204: 33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard-Blum S. (2011) The collagen family. Cold Spring Harbor Perspectives in Biology 3: a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risher W. C., Eroglu C. (2012) Thrombospondins as key regulators of synaptogenesis in the central nervous system. Matrix Biology 31: 170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. D., Coulson-Thomas V. J., Foscarin S., Kwok J. C., Fawcett J. W. (2015) “GAG-ing with the neuron”: The role of glycosaminoglycan patterning in the central nervous system. Experimental Neurology 274: 100–114. [DOI] [PubMed] [Google Scholar]
- Spencer V. A., Xu R., Bissell M. J. (2007) Extracellular matrix, nuclear and chromatin structure, and gene expression in normal tissues and malignant tumors: A work in progress. Advances in Cancer Research 97: 275–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Chen J., Lippold K., Monavarfeshani A., Carrillo G. L., Jenkins R., Fox M. A. (2016) Collagen-derived matricryptins promote inhibitory nerve terminal formation in the developing neocortex. Journal of Cell Biology 212: 721–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Gorse K., Ramirez F., Fox M. A. (2010) Collagen XIX is expressed by interneurons and contributes to the formation of hippocampal synapses. Journal of Comparative Neurology 518: 229–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Stenbjorn R. S., Gorse K., Su K., Hauser K. F., Ricard-Blum S., Pihlajaniemi T., Fox M. A. (2012) Target-derived matricryptins organize cerebellar synapse formation through alpha3beta1 integrins. Cell Reports 2: 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyoshi H., Inoguchi K., Khaleduzzaman M., Ninomiya Y., Yoshioka H. (1997) Ubiquitous expression of the alpha1(XIX) collagen gene (Col19a1) during mouse embryogenesis becomes restricted to a few tissues in the adult organism. Journal of Biological Chemistry 272: 17104–17111. [DOI] [PubMed] [Google Scholar]
- Sumiyoshi H., Mor N., Lee S. Y., Doty S., Henderson S., Tanaka S., Yoshioka H., Rattan S., Ramirez F. (2004) Esophageal muscle physiology and morphogenesis require assembly of a collagen XIX-rich basement membrane zone. Journal of Cell Biology 166: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykova E., Nicholson C. (2008) Diffusion in brain extracellular space. Physiological Reviews 88: 1277–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B. L. (2001) ADAMTS: A novel family of extracellular matrix proteases. The International Journal of Biochemistry & Cell Biology 33: 33–44. [DOI] [PubMed] [Google Scholar]
- Tortorella M. D., et al. (1999) Purification and cloning of aggrecanase-1: A member of the ADAMTS family of proteins. Science 284: 1664–1666. [DOI] [PubMed] [Google Scholar]
- Truett G. E., Heeger P., Mynatt R. L., Truett A. A., Walker J. A., Warman M. L. (2000) Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29: 52, 54. [DOI] [PubMed] [Google Scholar]
- Venugopal A. K., Sameer Kumar G. S., Mahadevan A., Selvan L. D., Marimuthu A., Dikshit J. B., Tata P., Ramachandra Y., Chaerkady R., Sinha S., Chandramouli B., Arivazhagan A., Satishchandra P., Shankar S., Pandey A. (2012) Transcriptomic profiling of medial temporal lobe epilepsy. Journal of Proteomics & Bioinformatics 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vistnes M., Aronsen J. M., Lunde I. G., Sjaastad I., Carlson C. R., Christensen G. (2014) Pentosan polysulfate decreases myocardial expression of the extracellular matrix enzyme ADAMTS4 and improves cardiac function in vivo in rats subjected to pressure overload by aortic banding. PLoS One 9: e89621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fawcett J. (2012) The perineuronal net and the control of CNS plasticity. Cell and Tissue Research 349: 147–160. [DOI] [PubMed] [Google Scholar]
- Weber P., Bartsch U., Rasband M. N., Czaniera R., Lang Y., Bluethmann H., Margolis R. U., Levinson S. R., Shrager P., Montag D., Schachner M. (1999) Mice deficient for tenascin-R display alterations of the extracellular matrix and decreased axonal conduction velocities in the CNS. Journal of Neuroscience 19: 4245–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohr M., Orduz D., Gregory P., Moreno H., Khan U., Vörckel K. J., Wolfer D. P., Welzl H., Gall D., Schiffmann S. N., Schwaller B. (2015) Lack of parvalbumin in mice leads to behavioral deficits relevant to all human autism core symptoms and related neural morphofunctional abnormalities. Translational Psychiatry 5: e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y. (2000) Lecticans: Organizers of the brain extracellular matrix. Cellular and Molecular Life Sciences 57: 276–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. W., Deb S., Gottschall P. E. (1998) Regional and differential expression of gelatinases in rat brain after systemic kainic acid or bicuculline administration. European Journal of Neuroscience 10: 3358–3368. [DOI] [PubMed] [Google Scholar]
- Zhang Z. J., Reynolds G. P. (2002) A selective decrease in the relative density of parvalbumin-immunoreactive neurons in the hippocampus in schizophrenia. Schizophrenia Research 55: 1–10. [DOI] [PubMed] [Google Scholar]
- Zimmermann D. R., Dours-Zimmermann M. T. (2008) Extracellular matrix of the central nervous system: From neglect to challenge. Histochemistry and Cell Biology 130: 635–653. [DOI] [PubMed] [Google Scholar]