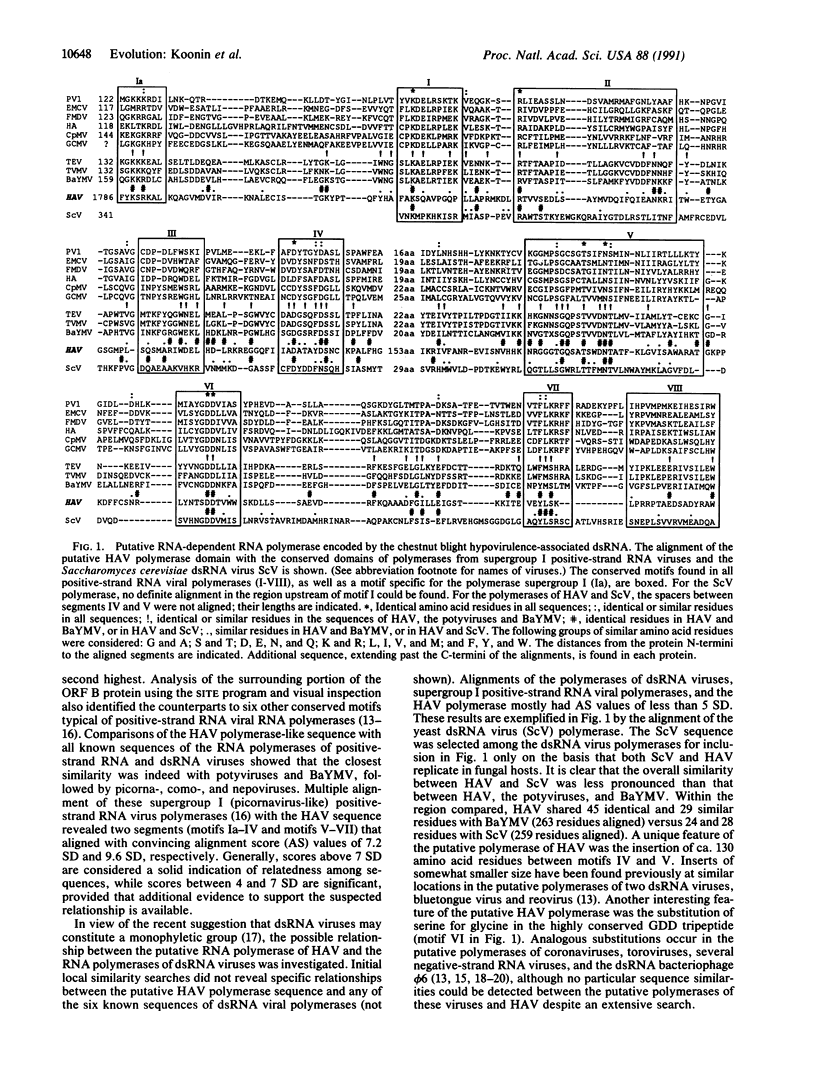
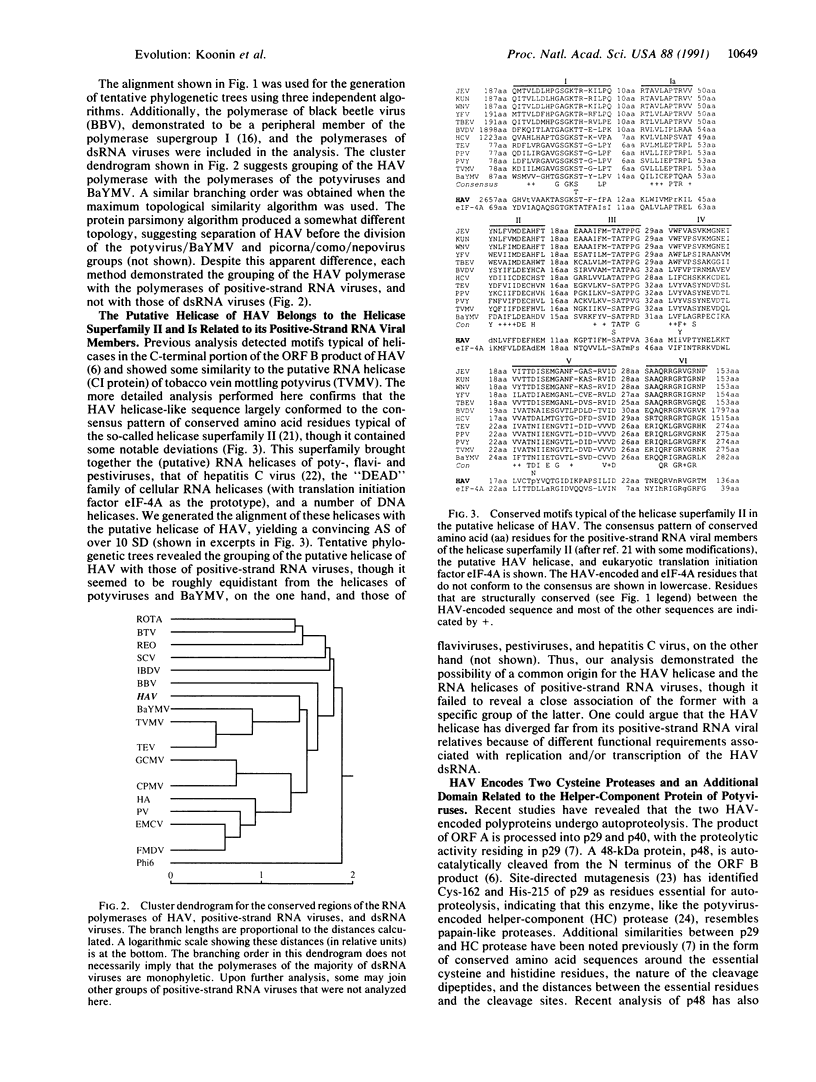
Abstract
Computer-assisted analysis of the putative polypeptide products encoded by the two open reading frames present in a large virus-like double-stranded RNA, L-dsRNA, associated with hypovirulence of the chestnut blight fungus, Cryphonectria parasitica, revealed five distinct domains with significant sequence similarity to previously described conserved domains within plant potyvirus-encoded polyproteins. These included the putative RNA-dependent RNA polymerase, RNA helicase, two papain-like cysteine proteases related to the potyvirus helper-component protease, and a cysteine-rich domain of unknown function similar to the N-terminal portion of the potyvirus helper-component protein. Phylogenetic trees derived from the alignment of the polymerase domains of L-dsRNA, a subset of positive-stranded RNA viruses, and double-stranded RNA viruses, using three independent algorithms, suggested that the hypovirulence-associated dsRNA and potyvirus genomes share a common ancestry. However, comparison of the organization of the conserved domains within the encoded polyproteins of the respective viruses indicated that the proposed subsequent evolution involved extensive genome rearrangement.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostakis S. L. Biological control of chestnut blight. Science. 1982 Jan 29;215(4532):466–471. doi: 10.1126/science.215.4532.466. [DOI] [PubMed] [Google Scholar]
- Bredenbeek P. J., Pachuk C. J., Noten A. F., Charité J., Luytjes W., Weiss S. R., Spaan W. J. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 1990 Apr 11;18(7):1825–1832. doi: 10.1093/nar/18.7.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruenn J. A. Relationships among the positive strand and double-strand RNA viruses as viewed through their RNA-dependent RNA polymerases. Nucleic Acids Res. 1991 Jan 25;19(2):217–226. doi: 10.1093/nar/19.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G. H., Pawlyk D. M., Nuss D. L. The autocatalytic protease p29 encoded by a hypovirulence-associated virus of the chestnut blight fungus resembles the potyvirus-encoded protease HC-Pro. Virology. 1991 Aug;183(2):747–752. doi: 10.1016/0042-6822(91)91004-z. [DOI] [PubMed] [Google Scholar]
- Choi G. H., Shapira R., Nuss D. L. Cotranslational autoproteolysis involved in gene expression from a double-stranded RNA genetic element associated with hypovirulence of the chestnut blight fungus. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1167–1171. doi: 10.1073/pnas.88.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A. D., Pröls M., Schell J., Steinbiss H. H. The nucleotide sequence of RNA 2 of barley yellow mosaic virus. J Gen Virol. 1991 Apr;72(Pt 4):989–993. doi: 10.1099/0022-1317-72-4-989. [DOI] [PubMed] [Google Scholar]
- Dolja V. V., Boyko V. P., Agranovsky A. A., Koonin E. V. Phylogeny of capsid proteins of rod-shaped and filamentous RNA plant viruses: two families with distinct patterns of sequence and probably structure conservation. Virology. 1991 Sep;184(1):79–86. doi: 10.1016/0042-6822(91)90823-t. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Blinov V. M., Donchenko A. P., Koonin E. V. An NTP-binding motif is the most conserved sequence in a highly diverged monophyletic group of proteins involved in positive strand RNA viral replication. J Mol Evol. 1989 Mar;28(3):256–268. doi: 10.1007/BF02102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Donchenko A. P., Blinov V. M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989 Jun 26;17(12):4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy W. R., Strauss J. H. Processing the nonstructural polyproteins of sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J Virol. 1989 Nov;63(11):4653–4664. doi: 10.1128/jvi.63.11.4653-4664.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwazaki S., Minobe Y., Hibino H. Nucleotide sequence of barley yellow mosaic virus RNA 2. J Gen Virol. 1991 Apr;72(Pt 4):995–999. doi: 10.1099/0022-1317-72-4-995. [DOI] [PubMed] [Google Scholar]
- Kashiwazaki S., Minobe Y., Omura T., Hibino H. Nucleotide sequence of barley yellow mosaic virus RNA 1: a close evolutionary relationship with potyviruses. J Gen Virol. 1990 Dec;71(Pt 12):2781–2790. doi: 10.1099/0022-1317-71-12-2781. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Gorbalenya A. E., Chumakov K. M. Tentative identification of RNA-dependent RNA polymerases of dsRNA viruses and their relationship to positive strand RNA viral polymerases. FEBS Lett. 1989 Jul 31;252(1-2):42–46. doi: 10.1016/0014-5793(89)80886-5. [DOI] [PubMed] [Google Scholar]
- Koonin E. V. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol. 1991 Sep;72(Pt 9):2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- Kunin E. V., Gorbalenia A. E., Chumakov K. M., Donchenko A. P., Blinov V. M. Evoliutsiia RNK-zavisimykh RNK-polimeraz pozitivnykh ribovirusov. Mol Gen Mikrobiol Virusol. 1987 Jul;(7):27–39. [PubMed] [Google Scholar]
- Lee H. J., Shieh C. K., Gorbalenya A. E., Koonin E. V., La Monica N., Tuler J., Bagdzhadzhyan A., Lai M. M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991 Feb;180(2):567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. H., Purcell R. H. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov SYu, Dolja V. V., Atabekov J. G. Probable reassortment of genomic elements among elongated RNA-containing plant viruses. J Mol Evol. 1989 Jul;29(1):52–62. doi: 10.1007/BF02106181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh C. S., Carrington J. C. Identification of essential residues in potyvirus proteinase HC-Pro by site-directed mutagenesis. Virology. 1989 Dec;173(2):692–699. doi: 10.1016/0042-6822(89)90582-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poch O., Sauvaget I., Delarue M., Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989 Dec 1;8(12):3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira R., Choi G. H., Hillman B. I., Nuss D. L. The contribution of defective RNAs to the complexity of viral-encoded double-stranded RNA populations present in hypovirulent strains of the chestnut blight fungus Cryphonectria parasitica. EMBO J. 1991 Apr;10(4):741–746. doi: 10.1002/j.1460-2075.1991.tb08005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira R., Choi G. H., Nuss D. L. Virus-like genetic organization and expression strategy for a double-stranded RNA genetic element associated with biological control of chestnut blight. EMBO J. 1991 Apr;10(4):731–739. doi: 10.1002/j.1460-2075.1991.tb08004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira R., Nuss D. L. Gene expression by a hypovirulence-associated virus of the chestnut blight fungus involves two papain-like protease activities. Essential residues and cleavage site requirements for p48 autoproteolysis. J Biol Chem. 1991 Oct 15;266(29):19419–19425. [PubMed] [Google Scholar]
- Snijder E. J., den Boon J. A., Bredenbeek P. J., Horzinek M. C., Rijnbrand R., Spaan W. J. The carboxyl-terminal part of the putative Berne virus polymerase is expressed by ribosomal frameshifting and contains sequence motifs which indicate that toro- and coronaviruses are evolutionarily related. Nucleic Acids Res. 1990 Aug 11;18(15):4535–4542. doi: 10.1093/nar/18.15.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia J., Paul C. P., Fulbright D. W., Nuss D. L. Structural properties of double-stranded RNAs associated with biological control of chestnut blight fungus. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9109–9113. doi: 10.1073/pnas.83.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]




