Abstract
Background
Thiazide diuretics are among the most commonly prescribed antihypertensives. However, <50% of thiazide treated patients achieve blood pressure (BP) control. Herein, we used different omics (genomics and transcriptomics) to identify novel biomarkers of thiazide diuretics BP response.
Methods and Results
Genome-wide analysis included 228 white hypertensives with BP determined at baseline and after 9 weeks of hydrochlorothiazide (HCTZ). Single nucleotide polymorphisms (SNPs) with p<5×10−5 were prioritized according to their biological function, using RegulomeDB, haploreg and GWAVA. The results from the prioritization approach revealed rs10995 as the most likely functional SNP, among SNPs tested, that has been associated with HCTZ BP response. The rs10995 G-allele was associated with better BP response to HCTZ vs. non-carriers (ΔSBP/ΔDBP:−12.3/−8.2 vs. −6.8/−3.5 mmHg, respectively, ΔSBP p=3×10−4, ΔDBP p=5×10−5). This association was replicated in independent participants treated with chlorthalidone (CLT). In addition, rs10995 G-allele was associated with increased mRNA expression of VASP, encoding vasodilator-stimulated phosphoprotein. Moreover, baseline expression of the VASP mRNA was significantly higher in 25 good-responders to HCTZ compared to 25 poor-responders (p=0.01). This finding was replicated in independent participants treated with CLT (p=0.04). Lastly, allelic-specific expression analysis revealed a significant but modest imbalance with rs10995 and rs10156, a SNP in high linkage disequilibrium (r2=0.7) with rs10995, which both could contribute to the observed genetic effects by affecting VASP mRNA expression.
Conclusions
This study highlights the strength of using different omics to identify novel biomarkers of drug response and suggests VASP as a potential determinant of thiazide diuretics BP response.
Clinical Trial Information
ClinicalTrials.gov; Unique Identifiers: NCT00246519, NCT01203852.
Keywords: pharmacogenetics, hypertension, blood pressure, transcriptome, genomics, pharmacogenomics, antihypertensive agent, personalized medicine, RNA-Seq, thiazide diuretics
Introduction
Hypertension (HTN) is the most common chronic disease and the primary cause of cardiovascular morbidity and mortality globally.1 Studies have shown that the risk for coronary diseases and stroke doubles with every 20 mmHg increase in systolic BP (SBP) or 10 mmHg increase in diastolic BP (DBP).2 Accordingly, using anti-hypertensive medications for controlling BP substantially reduces cardiovascular risk and the overall mortality associated with HTN.3 Thiazide diuretics, including hydrochlorothiazide (HCTZ), are ranked among the most commonly prescribed drugs for the treatment of HTN in the U.S.,4 and are highly recommended as first-line agents for most patients with uncomplicated essential HTN.5 Although they have been used as anti-hypertensives for more than half a century, the mechanism by which thiazide diuretics chronically lower BP has not been fully elucidated yet.6 Additionally, studies have shown that < 50% of thiazide treated patients achieve BP control.7, 8 Even with the use of other anti-HTN medications, acting on a variety of BP regulatory systems, only 44% of HTN treated patients achieve BP control.9 Given these facts, it is clear that the current approach for selecting anti-HTN medications is suboptimal and more work is still needed to optimize the use of these drugs.
In the past decade, application of genome-wide association studies (GWAS) has advanced our understanding of the potential role of genetics in variable response to drugs.10 Recently, we conducted a GWAS analysis in which we identified novel genetic regions associated with the BP response to HCTZ, such as protein kinase C, alpha (PRKCA) and G-protein alpha subunit-endothelian-3 (GNAS-EDN3).11 In this study, we propose that there are additional true genetic contributors that have not met the stringent genome-wide statistical threshold, but they are difficult to ascertain statistically using genomics data alone, particularly with the small sample sizes of the globally available HTN pharmacogenetics datasets. Thus, it is critical to leverage other information to effectively prioritize GWAS signals, increase replication rates and better understand the mechanism underlying HCTZ BP response.
Recent studies have shown that GWAS single nucleotide polymorphisms (SNPs) associated with complex traits are more likely to be expression quantitative trait loci (eQTLs).12, 13 Additionally, studies have demonstrated that the majority (~93%) of previously conducted GWAS findings lay in non-coding regions,14 and that these findings are significantly enriched in the regions that harbor functional elements, such as transcriptional factor binding sites (TFBSs), histone modification marked regions, DNase I hypersensitive sites (DHSs) and eQTLs.15, 16 Accordingly, we hypothesized that prioritizing the GWAS output based on regulatory functional signals that perturb gene expression might elucidate novel genetic signals affecting HCTZ BP response. Investigating genes where these signals are involved might open new avenues for a better understanding of the molecular determinants of the BP response. Thus, we aimed in the current study to use data from the Encyclopedia of DNA Elements (ENCODE) project, like transcriptional factor CHIP-seq, histone CHIP-seq, and DNase I hypersensitivity site data, along with publically available eQTL data, to prioritize and highlight novel genetic variants affecting the BP response to HCTZ. Additionally, we sought to use transcriptomics data to further confirm our genetic finding.
Methods
Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) study
The primary analysis included samples and data from participants recruited as part of the PEAR trial (clinicaltrials.gov # NCT00246519).17 In brief, PEAR was a prospective, randomized, multi-center study with one of its primary aims was to evaluate the role of genetics on the BP response of HCTZ treated participants. PEAR recruited mild to moderate hypertensive participants, defined as participants with untreated DBP ≤ 110 and SBP ≤ 180. After enrollment, all participants had an average of 4 weeks washout period followed by a randomization to either a monotherapy of 12.5 mg/daily HCTZ or 50 mg/daily atenolol (β-1-selective blocker) for a duration of three weeks, with the dose titrated upwards for additional 6 weeks (i.e. HCTZ 25 mg/daily or atenolol 100 mg/daily) in participants with BP > 120/70 mmHg.
Pharmacogenomic Evaluation of Antihypertensive Responses 2 (PEAR-2) Study
We also used clinical data and biological samples from PEAR-2 participants to validate and replicate our findings from the PEAR primary analysis. In brief, PEAR-2 was a prospective, multi-center, sequential monotherapy study (clinicaltrials.gov # NCT01203852)18 with one of its primary aims being to evaluate the role of genetics on the BP response to chlorthalidone (CLT; a thiazide-like diuretic). PEAR-2 recruited mild to moderate hypertensive participants, aged 18-65 years, from the University of Florida (Gainesville, FL), Emory University (Atlanta, GA), and the Mayo Clinic (Rochester, MN). All participants had an average 4 week washout period then they started on a monotherapy of 15 mg/daily CLT for a duration of two weeks, with the dose titrated upwards for additional 6 weeks (i.e. CLT 25 mg/daily) in participants with BP still > 120/70 mmHg. The Institutional Review Board approved both PEAR and PEAR-2 studies at each study site, and written informed consent was obtained from all participants.
Thiazide Blood Pressure Response Measurement
The primary analysis of the current study included 228 European American (white) participants from the PEAR study with their BP measured pre-HCTZ (baseline) and 9 weeks post-HCTZ therapy. In PEAR, BP at home, office and ambulatory daytime and night-time were measured, as previously described17 and as explained in the Supplement. For the analysis of the PEAR HCTZ treated participants included in this study, we used a composite weighted average of the home, office and ambulatory daytime and nighttime, which we showed that it represents a more accurate measurement of BP response with a better signal-to-noise ratio and more power to identify genetic predictors of BP response.19
The SNP replication analysis within this study included 185 white participants from the PEAR-2 study with their BP measured pre-CLT (baseline) and 8 weeks post-CLT therapy. Both home and office BP were measured, as previously described.18 Please check the Supplemental Material for more details.
Genotyping
Genome-wide genotyping for the PEAR participants was performed as previously described11 and as explained in the Supplement. For PEAR-2, genotyping was conducted using Human Omni2.5 S BeadChip (Illumina, San Diego CA). Participants from PEAR or PEAR-2 were excluded if sample genotype call rates were below 95%. Additionally, SNPs with a genotype call rates below 95% were also excluded.
Gene Expression
Gene expression was measured using RNA-Seq analysis on both PEAR (discovery) and PEAR-2 (replication) white participants treated with HCTZ and CLT, respectively. For PEAR RNA-Seq analysis, we collected whole blood samples at baseline from 50 white participants treated with HCTZ. Participants were selected from the upper and lower quartiles of HCTZ home DBP response (25 good BP responders and 25 poor BP responders). Similarly, for PEAR-2 gene expression, whole blood samples were collected at baseline from 50 white participants that were selected from the upper and lower quartiles of home DBP response to CLT (25 good BP responders and 25 poor BP responders) to conduct RNA-Seq analysis on these samples. Home DBP response was used for selection of participants for RNA-Seq analyses since it exhibits less variability and superior reproducibility compared to office BP response.20 Of note, out of the 50 samples used for the RNA-Seq analysis in PEAR, one sample was removed because it did not pass quality control.
Details for the RNA-Seq analysis for both PEAR and PEAR-2 samples are described in the Supplement. In brief, a strand-specific protocol was used to prepare libraries, as previously described.21 For data quality control purposes, read duplicates removal was implemented using Picard (http://picard.sourceforge.net) Mark Duplicates option. The 100 bp reads generated in the paired-end RNA sequencing were uniquely mapped to the human reference genome (hg19) using TopHat v2.0.10 allowing for four reads mismatches, read edit distance of six, one mismatch in the anchor region of a spliced read, and a maximum of five multi-hits. Transcript assembly was performed using Cufflinks (v2.0.2) and gene expression values, reported in fragments per kilobase per million reads (FPKM), were calculated using cuffdiff (v2.2.1).22 Alternative tools for investigating differential gene expression were used to adjust for covariates such as age, gender and baseline BP. First, we removed duplicate BAM files from TopHat alignments, as an input to the function htseq-count,23 to count the number of reads for each gene (Gencode gene annotation release 18). Counts were modeled to a negative binomial distribution using a generalized linear model implemented in edgeR24 (Bioconductor R Package).
Statistical Analyses
Genomics Analysis
GWAS analysis was previously conducted11 to test the association between dosage imputed genotype data with composite SBP and composite DBP responses to HCTZ in 228 white participants from the PEAR study. PLINK software was used to run the analysis with adjustment for age, gender, pre-HCTZ BP and population substructure by considering the first and second principal components (PC1 and PC2) in all our analysis. In this study, we investigated SNPs with a p-value <5×10−5 from the GWAS of HCTZ BP response, selected on the basis of quantile-quantile (Q-Q) plots showing that SNPs with p-values <5×10−5 deviated from expectation under the null hypothesis of no relationship between SNPs and HCTZ BP response in PEAR white participants treated with HCTZ (Supplementary Figure S1). Deviation from Hardy-Weinberg Equilibrium (HWE) for the selected SNPs was tested using exact test, and SNPs with HWE p-value <1×10−5 were removed. The statistical analyses conducted in this study were carried out with SAS (version 9.3; SAS Institute) and SPSS software (version 17.0; SPSS Inc.).
Single Nucleotide Polymorphisms Prioritization Approach
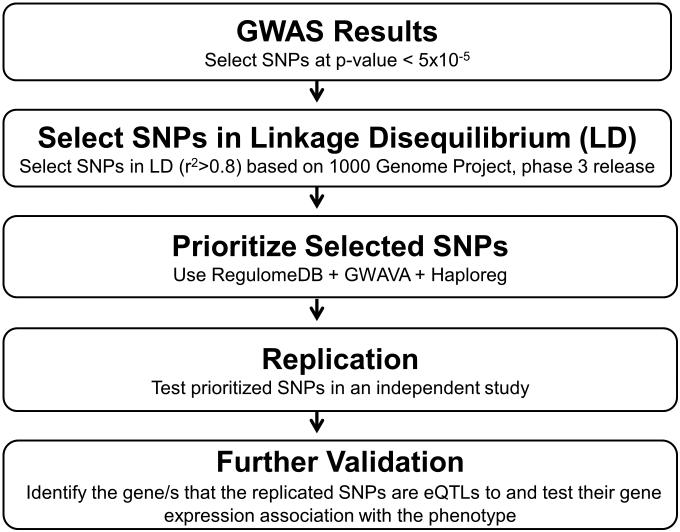
To address the aims of the current study, a SNP prioritization approach was conducted as shown in Figure 1. For the prioritization approach, we selected 103 SNPs (Supplementary Table S1), representing 41 independent genetic signals, with p-values < 5×10−5 from the GWAS analyses of both HCTZ SBP and DBP responses conducted on PEAR whites. It is likely that among the 103 selected SNPs, true positive SNPs might be in linkage disequilibrium (LD) with functional variants that are responsible for the significant association with HCTZ BP response. Thus, we used rAggr webserver (http://raggr.usc.edu/) to select all SNPs in high LD (r2>0.8, according to Northern Europeans from Utah (CEU) population) with the 103 SNPs, within a distance of 500kb, based on 1000 Genomes Project, phase 3 release. This step yielded 1,082 SNPs which were then annotated and prioritized using three analytical tools: (1) RegulomeDB25 (http://www.regulomedb.org/), (2) Genome-Wide Annotation of Variants (GWAVA)26 (https://www.sanger.ac.uk/sanger/StatGen Gwava) and (3) Haploreg v4.127 webserver (http://www.broadinstitute.org/mammals/ haploreg/haplo reg. php). These three tools annotate SNPs based on high-throughput data sets from the ENCODE Project, along with other publically available Expression Quantitative Trait Loci (eQTL) data sets, computational predictions and manual annotations. More details about each tool are described in the Supplement.
Figure 1.
The overall framework of the experimental approaches used in this study. GWAS, genome-wide association study; SNPs, single nucleotide polymorphisms; eQTLs, expression quantitative trait loci.
In brief, the RegulomeDB has a scoring system that categorizes SNPs, ranging from 1 to 6, based on the degree of experimental or computational evidence and regulatory functional consequences of tested SNPs. For prioritization using RegulomeDB, we focused on SNPs with a score of 1 and 2, since the lower score indicates stronger evidence for a SNP to be located in a potentially functional region. Using GWAVA, SNPs were prioritized based on their TSS score (calculated based on nearest transcription start site). We used GWAVA TSS score to prioritize our signals because it takes into account different regulatory annotations and not influenced by any single annotation. Additionally, we selected SNPs with a TSS score ≥ 0.5, which represents the threshold used by GWAVA’s original authors to classify SNPs as functional.26 Using Haploreg v4.1, we selected all SNPs that had been previously shown to affect gene expression (eQTLs). SNPs that overlap between the three annotation tools were then moved forward to be tested for replication in PEAR-2 whites treated with CLT, as explained below.
Replication
A general linear model was used to test the association between prioritized SNPs and CLT BP response in PEAR-2 whites, with adjustment for age, gender, pre-CLT BP and PC1 and 2.
Further Validation
For SNPs replicated in PEAR-2, we used Haploreg v4.1 to check whether these SNPs had been shown to be significantly associated with the mRNA expression levels of any genes. We then selected these genes and tested differences in their mRNA expression levels between PEAR HCTZ good responders and poor responders. We further replicated our findings by testing the differences in mRNA expression levels, of the genes of interest, between PEAR-2 CLT good responders and poor responders.
We also tested replicated SNPs for allelic mRNA expression imbalance (AEI). SNP calls and allele-specific read counts were generated with SaMtools mpileup from the duplicate removed alignment file.28 Analysis was conducted in individuals heterozygous for the relevant transcript SNPs in both PEAR and PEAR-2 thiazide diuretics treated participants. Chi-square Goodness-of-Fit was applied to test if the allele ratio (minor allele/major allele) deviates from the expected ratio 0.5 (i.e. when the two alleles are expressed equally).29 Read counts for SNPs in high LD with each other were combined to yield a more accurate estimate of allelic ratios.
Lastly, for SNPs located in the 3’UTR of the gene, we tested them to check if they interfere with miRNA target site using in silico tools like MirSNP (http://bioinfo.bjmu.edu.cn/mirsnp/search/)30 and PolymiRTS database 3.0 (http://compbio.uthsc.edu/miRSNP/).31 More details about those tools and the analysis approach are present in the Supplement.
Results
Characteristics of Study Participants and Thiazide Diuretics Blood Pressure Response
Characteristics and thiazide diuretic BP responses of participants, included in the discovery and replication genetic analyses, are presented in Table 1. We found that baseline characteristics such as age, gender, body mass index (BMI) and baseline BP were similar among PEAR and PEAR-2 participants. However, CLT treated participants in PEAR-2 had greater BP lowering effects compared to HCTZ treated participants in PEAR, likely due to the fact that CLT is approximately 1.5 to 2.0 times as potent as HCTZ therapy.32
Table 1.
Characteristics of PEAR and PEAR-2 white participants
| Characteristics | PEAR HCTZ monotherapy (N=228) |
PEAR-2 CLT monotherapy (N=185) |
|---|---|---|
| Age, mean (SD) years | 50 ± 9.5 | 51.1 ± 8.9 |
| Women, N (%) | 91 (40) | 79 (42.5) |
| BMI, mean (SD) kg*m−2 | 30.30 ± 4.90 | 30.66 ± 4.95 |
| Pre-treatment home SBP, mean (SD) mmHg | 146 ± 9.96 | 147.43 ± 10.31 |
| Pre-treatment home DBP, mean (SD) mmHg | 93.61 ± 5.59 | 90.28 ± 5.04 |
| Home SBP response, mean (SD) mmHg | −7.68 ± 8.1 | −12.25 ± 9.1 |
| Home DBP response, mean (SD) mmHg | −4.23 ± 5.32 | −6.83 ± 5.43 |
| Composite SBP response, mean (SD) mmHg | −8.50 ± 7.02 | NA |
| Composite DBP response, mean (SD) mmHg | −4.68 ± 4.79 | NA |
PEAR, pharmacogenomic evaluation of antihypertensive responses; BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure; NA, not applicable. For PEAR, composite BP response was generated using the weighted average of the home, office, ambulatory daytime and nighttime BP responses.
Characteristics of participants selected from either PEAR or PEAR-2 for the RNA-Seq analyses are presented in Table 2 and 3, respectively. Additionally, histograms describing the distribution of BP response among HCTZ participants in PEAR and CLT participants in PEAR-2, who were included in the RNA-Seq analyses, are presented in the data Supplement (Supplementary Figure S2). For PEAR RNA-Seq selected participants, baseline characteristics such as age, gender, BMI and baseline BP were not significantly different between good responders and poor responders to HCTZ. However, in PEAR-2 RNA-Seq selected participants, age was marginally significant, and gender and baseline BP were both significantly different between CLT good responders and poor responders. Accordingly, we adjusted for age, gender and baseline DBP in the analyses conducted using PEAR-2 RNA-Seq data. Of note, the mapping statistics of the RNA-Seq runs are presented in the Supplement (Supplementary Table S2).
Table 2.
Characteristics of PEAR white participants included in the RNA-Seq analysis
| Characteristics | PEAR Good Responders (N=24)* |
PEAR Poor Responders (N=25) |
P-value |
|---|---|---|---|
| Age, mean (SD) years | 48 ± 11.94 | 47.5 ± 8.70 | 0.85 |
| Women, N (%) | 9 (36.0) | 12 (50.0) | 0.46 |
| BMI, mean (SD) kg*m−2 | 29.3 ± 5.13 | 31.8 ± 5.90 | 0.12 |
| Pre-treatment home SBP, mean (SD) mmHg | 145.6 ± 10.2 | 144.1 ± 9.82 | 0.60 |
| Pre-treatment home DBP, mean (SD) mmHg | 93.8 ± 5.0 | 94.1 ± 4.37 | 0.81 |
| Home SBP response, mean (SD) mmHg | −11.80 ± 6.85 | −0.9 ± 5.85 | 3E-07 |
| Home DBP response, mean (SD) mmHg | −8.36 ± 5.57 | 0.08 ± 3.65 | 1E-07 |
| Composite SBP response, mean (SD) mmHg | −12 ± 6.44 | −4 ± 5.60 | 2E-05 |
| Composite DBP response, mean (SD) mmHg | −7.56 ± 4.84 | −1.6 ± 3.69 | 1E-05 |
PEAR, pharmacogenomic evaluation of antihypertensive responses; BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure. For PEAR, composite BP response was generated using the weighted average of the home, office, ambulatory daytime and night time BP responses.
In PEAR good responders, one sample failed the quality control procedures and was not included in the study.
Table 3.
Characteristics of PEAR-2 white participants included in the RNA-Seq analysis
| Characteristics | PEAR-2 Good Responders (N=25) |
PEAR-2 Poor responders (N=25) |
P-value |
|---|---|---|---|
| Age, mean (SD) years | 53.36 ± 7.80 | 48.48 ± 10.23 | 0.064 |
| Women, N (%) | 15 (60) | 5 (20) | 4E-03 |
| BMI, mean (SD) kg*m−2 | 32.60 ± 5.14 | 30.59 ± 5.31 | 0.18 |
| Pre-treatment home SBP, mean (SD) mmHg | 152.23 ± 10.95 | 143.90 ± 8.99 | 5E-03 |
| Pre-treatment home DBP, mean (SD) mmHg | 96.78 ± 6.59 | 92.87 ± 5.02 | 0.023 |
| Home SBP response, mean (SD) mmHg | −21.66 ± 7.67 | −1.56 ± 5.06 | 1.3E-14 |
| Home DBP response, mean (SD) mmHg | −14.25 ± 4.22 | −0.26 ± 2.24 | 2.4E-19 |
PEAR-2, pharmacogenomic evaluation of antihypertensive responses-2; BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure.
Single Nucleotide Polymorphisms Prioritization Approach
Out of the 103 SNPs, none of them deviated from HWE (Supplementary Table S1). Selecting SNPs in LD (r2>0.8) with the 103 SNPs provided a list of 1,082 SNPs. Of the 1,082 SNPs, nineteen displayed a RegulomeDB score less than 3 (Supplementary Table S3). Out of those nineteen, rs10995 located in the 3’UTR of the vasodilator-stimulated phosphoprotein gene (VASP) was the top prioritized SNP with a RegulomeDB score of 1d. Testing the 1,082 SNPs in GWAVA identified 29 SNPs with a TSS score ≥ 0.5 (Supplementary Table S4). Again, GWAVA identified rs10995 as the top prioritized SNP with a score of 0.87. Using Haploreg v4.1, we were able to annotate 290 SNPs out of the 1,082 SNPs as eQTLs. Visualizing the combined results of these different tools (Supplementary Figure S3, Supplementary Table S5) highlights rs10995 as the only overlapping and top prioritized signal associated with HCTZ BP response. Thus, we tested the rs10995 association for replication in PEAR-2 whites treated with CLT.
Replication and Further Validation
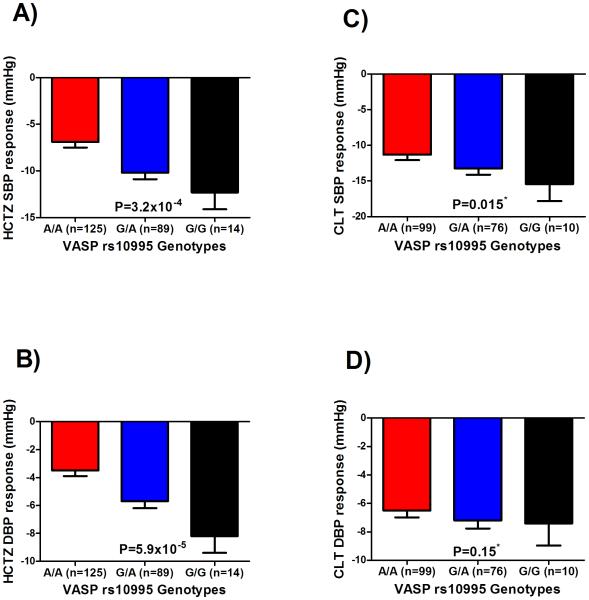
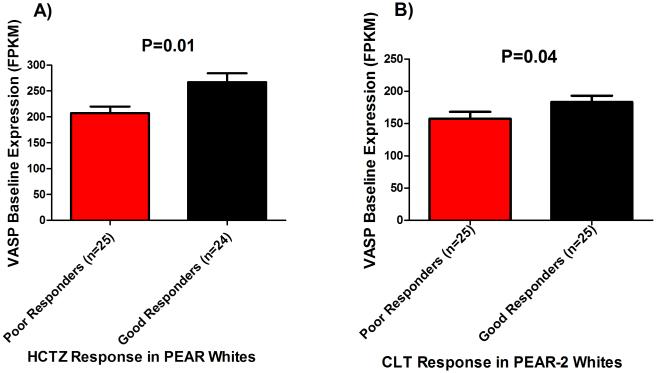
The minor allele frequency of rs10995 G>A SNP in PEAR and PEAR-2 whites was 25.6% and 25.9%, respectively. Testing the rs10995 SNP in PEAR-2 whites revealed a significant association with SBP response to CLT, and a trend toward significance with DBP response (Figure 2), with adjustment for baseline BP, age, gender and PC1 and 2. Although we adjusted for baseline BP in our analysis, we also tested the association between rs10995 and baseline BP in PEAR and PEAR-2 participants included in this study. Our results showed no association between the rs10995 and baseline BP measured in PEAR (DBP=0.42, SBP=0.22) and PEAR-2 (DBP=0.17, SBP=0.88). Additionally, the in silico analysis using Haploreg v4.1 shows that rs10995 is associated with the mRNA expression levels of four genes (VASP, FOSB, GPR4 and KLC3; Supplementary Table S6). Therefore, we tested baseline expression levels of those genes between HCTZ good responders and poor responders in PEAR participants. Out of the four genes, only VASP displayed a significantly different mRNA levels between HCTZ good responders and poor responders (p=0.01; Figure 3A, Supplementary Table S7). We found that HCTZ good responders had higher VASP mRNA levels at baseline compared to poor responders. This association was replicated in the same direction in PEAR-2 participants treated with CLT (p=0.04; Figure 3B, Supplementary Table S7). However, after adjustment for age, gender and baseline BP using EdgeR, the association weakened (adjusted p=0.12) (Supplementary Table S8).
Figure 2.
Effect of the rs10995 polymorphism on the blood pressure response of whites treated with thiazide diuretics in the PEAR and PEAR-2 studies. Blood pressure responses were adjusted for baseline blood pressure, age, gender, and PC1 and 2. P-values represented are for contrast of adjusted means between different genotype groups. Error bars represent standard error of the mean. *One sided p-value based on a one-sided hypothesis tested in the replication study. A)systolic blood pressure response to hydrochlorothiazide in the PEAR study. B)diastolic blood pressure response in the PEAR study. C)systolic blood pressure response to chlorthalidone in the PEAR-2 study. D)diastolic blood pressure response to chlorthalidone in the PEAR-2 study.
Figure 3.
Plots showing the difference in the VASP baseline mRNA expression levels between thiazide diuretics good responders and poor responders. A)PEAR study. B)PEAR-2 study.
We also found a significant association between rs10995 and VASP mRNA expression at baseline in PEAR-2 (p=5×10−3), and a trend toward significance in PEAR participants (p=0.06). Our analysis showed that the rs10995 G/G genotype carriers (with better response to HCTZ) had higher expression levels of VASP mRNA at baseline compared to G/A and A/A genotype carriers (Supplementary Figure S4). This finding was supported by publically available eQTL data showing the same effect of the rs10995 G-allele on the VASP mRNA expression in several tissues (Supplementary Table S6). We also sought to test whether allelic VASP mRNA expression is higher in the presence of the rs10995 G-allele compared to the A-allele. For rs10995, of 39 heterozygous samples tested from PEAR and PEAR-2, two samples had G/A-allele ratios greater than 2 fold, evidence of AEI, and eighteen samples displayed a ≥ 1.2 fold change (Supplementary Figure S5), while the remaining sample did not show allelic ratios suggestive of AEI. Because of inherent noise in allelic RNA ratio analysis, it is not possible to assign the presence of AEI unambiguously.33 Thus, we tested the SNPs in high LD (r2>0.8) with rs10995 (Supplementary Table S9) and all SNPs in the exon region of the VASP gene with coverage >50 reads (Supplementary Table S10). From this analysis, we only focused on SNPs that have been shown in at least two heterozygous samples to have an AEI fold change >1.3. According to this criterion, we found two SNPs, rs10156 and rs546716916 with samples that had a change between their reference and variant alleles counts >1.3. The latter SNP is a rare SNP located 78 base pairs away from the rs10995 SNP, but it is not in LD with the rs10995 SNP (Supplementary Figure S6). While for the rs10156 SNP, we found that it is in high LD with the rs10995 SNP (r2=0.7, D’=1) and laying 173 base pairs away from the rs10995 signal. rs10156 T>C SNP had higher read counts and displayed allelic ratios in the same direction as rs10995 in the vast majority of samples (Supplementary Figure S7), suggesting the copy of the gene containing the reference GT-alleles is expressed more than the AC-alleles with the two SNPs combined with a mean log2 allelic expression ratio of 0.48 (p=0.004). These results support a finding of AEI, while we cannot ascertain whether rs10995 or rs10156 or both, alter mRNA expression.
Lastly, since the rs10995 SNP is located in the 3’UTR of the VASP gene, we tested whether rs10995 SNP interferes with a miRNA binding site. Both MirSNP and PolymiRTS database 3.0 databases indicate that the rs10995 G-allele creates a new binding site for miRNA 3127-3p, which enhance its binding to VASP mRNA compared to the A-allele (Supplementary Table S11). This result suggests an additional mechanism of action that could account at least in part for the observed clinical associations.
Discussion
Thiazide diuretics have been the mainstay anti-HTN therapy for years and are currently ranked among the most commonly prescribed first-line anti-HTN in the US. Despite their widespread use, a wide inter-individual variability in response to thiazide diuretics has been reported, which has stimulated interest in identifying predictors that can be used for optimizing the BP response to this therapy. Over the past years, results from GWAS have advanced our understanding of the potential role of genetics on the inter-individual variability in response to different drugs, including thiazide diuretics.6, 11 However, GWAS stringent statistical thresholds hinder the discovery of additional true genetic variants, particularly with the small sample sizes of the globally available HTN pharmacogenetic studies. Thus, in this study, we sought to identify additional novel genetic variants associated with thiazide diuretics BP response by leveraging functional data generated from the ENCODE project and other publically available eQTL datasets. We hypothesized that this approach would help prioritize the genetic signals from the GWAS and increase the chances of refining and identifying additional true signals that might be missing.
In this study, out of the 1,082 SNPs tested in our prioritization analysis, we identified and replicated the rs10995 in the VASP gene as a novel genetic signal with clinically relevant effects on the BP response of thiazide diuretics. Additionally, using RNA-Seq data from thiazide diuretics treated individuals showed that the rs10995 SNP represents a VASP eQTL. Moreover, we found that HCTZ good BP responders had higher baseline expression levels of the VASP gene compared to poor responders, which suggests that VASP might be a potential predictor of thiazide diuretics BP response. This finding was replicated in PEAR-2 participants treated with CLT, where CLT good BP responders had significantly higher VASP mRNA levels at baseline compared to poor responders. While this association weakened after adjustment for age, gender and baseline BP, this might be attributed to the small sample size used for the RNA-Seq analysis.
For further validation, we also tested whether rs10995 SNP and an adjacent SNP in high LD, rs10156, display allele-specific VASP mRNA expression. Taken together, the allelic ratio results demonstrate the presence of AEI as a marker of a genetic variant, most likely rs10995 or rs10156, or both. Of note, we did not have the rs10156 among the genotyped or imputed SNPs in our GWAS data, thus we were not able to test its association with thiazide diuretics BP response. Future studies should consider testing this SNP to check its association with HCTZ BP response. Lastly, in silico testing reveals that rs10995 minor allele enhances the binding of miRNA 3127-3p in the 3’UTR region. Although these results are intriguing, more experimental work is needed to test the interaction between rs10995 SNP and miRNA 3127-3p, and its clinical relevance.
VASP is highly expressed in vascular endothelial cells, smooth muscle cells and platelets.34, 35 As a member of the ENA/VASP protein family, VASP is involved in actin polymerization, actin cytoskeleton regulation and intracellular pathways of integrin-extracellular matrix (ECM) interaction36 - pathways involved in the mechanism underlying smooth muscle contraction and BP regulation.37, 38 VASP has also been well characterized as a substrate for cAMP- and cGMP-dependent protein kinases,39 known for their role in regulating contraction of vascular smooth muscle.40 Additionally, VASP phosphorylation has been implicated in various cellular responses ranging from endothelial cell permeability and angiogenesis41, 42 to platelet aggregation and secretion.43, 44 Moreover, it serves as a biochemical marker for monitoring nitric oxide stimulated soluble guanylyl cyclase/cGMP-dependent protein kinase type I pathway,45 which is involved in BP regulation, vascular remodeling and platelet, cardiac and kidney function.46-48 Collectively, the results of this study along with literature evidence support the hypothesis that VASP might be involved in the BP lowering mechanism of thiazides, and might serve as an important determinant of thiazide diuretics BP response. Future replication of the findings presented in this study in large well-designed studies is needed to confirm their utility in guiding the selection of thiazide diuretics for a better control on BP response. Additionally, conducting mechanistic studies on the VASP gene might provide insight into the mechanism underlying thiazide diuretics BP response.
Our study has several limitations. First, the small sample sizes used for the genetic and transcriptomics analyses may have limited our power to identify additional novel markers and replicate some of our genetic and transcriptomics signals. Secondly, we used whole blood-based RNA for the RNA-Seq analyses, which might not be the most relevant tissue for thiazide BP response. However, response to anti-HTN drugs might arise from a variety of target tissues. Thus, it is very difficult to select a specific tissue for testing the expression of BP genes as BP genes might be expressed in blood, heart, brain, kidney or any other specific tissue. Additionally, such tissues are essentially impossible to obtain from otherwise healthy hypertensive individuals. Lastly, whole blood samples can have different ratios of cell types, which may result in artifacts of differences in expression levels of genes that are differentially expressed between cell types. However, we did not measure differences in cell type distributions in blood for our RNA-seq runs. Thus, we were not able to account for possible changes in cell type composition. Although, it is possible that differences in cell type distribution could affect gene expression levels, however, this happens particularly with a disease state or drug treatment suspected of changing cell type content, which is not the case with the drug treatment regimen used in this study.
Our study also has several strengths. The prioritization approach used in this study was successful in identifying a novel genetic signal that we were not able to identify using traditional approaches for analyzing the GWAS output. Our findings from this approach support the hypothesis that not all SNPs are equal,49 and mandate the importance of utilizing functional annotations to better prioritize the GWAS output and increase the chances of identifying and replicating true signals associated with variability in drug response. We believe that using such an approach could help us to take forward the large investment in GWAS and convert the output of this approach to identify additional genetic variants, and biologically relevant pathways associated with drug response.
In summary, to our knowledge, this is the first study to highlight the association between the VASP gene and thiazide diuretics BP response. This study illustrates the power of utilizing the publically available ENCODE data and eQTL datasets to prioritize the GWAS output and increase the probability of identifying novel genetic variants underlying drug response. Future use of these publically available data might give us more insight into the mechanism underlying BP response, which might facilitate the development of new drugs and therapeutic approaches that can be utilized for optimizing anti-hypertensive BP response.
Supplementary Material
Clinical Perspective.
Thiazide diuretics have been the cornerstone in managing hypertension for more than half a century, yet when used as monotherapy, only about half of thiazide treated patients achieve their target blood pressure (BP) goal. Thus, identifying biomarkers to predict variability in BP response to thiazides may have future clinical utility. Herein, we used publically available ENCODE data along with expression quantitative trait loci (eQTL) data to prioritize for further investigation of genetic signals from a GWAS of BP response to hydrochlorothiazide. Our prioritization approach identified rs10995 variant in the vasodilator-stimulated phosphoprotein (VASP) as the most functional genetic signal associated with BP response. This genetic variant was then replicated in an independent group of hypertensives treated with chlorthalidone. We found rs10995 G-allele carriers had better BP response to thiazides vs. non-carriers, and the G-allele had higher mRNA expression of VASP. We also identified that baseline expression of VASP mRNA was significantly higher in people with a better BP response to thiazide diuretics. This finding was also replicated in an independent group of patients treated with chlorthalidone. The results from this study may help advance a personalized approach to antihypertensive therapy. The rs10995 variant should be tested in other independent cohorts to confirm its utility in guiding the selection of thiazide therapy. Additionally, future work on the VASP association with the BP response to thiazides may provide more insights into the chronic BP lowering mechanism underlying thiazide diuretics and may open new avenues for identifying novel antihypertensive drug targets.
Acknowledgments
We appreciate the valuable contributions of the Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) and PEAR-2 study participants, support staff and study physicians.
Sources of Funding: PEAR was supported by the National Institute of Health Pharmacogenetics Research Network grant U01 GM074492 and the National Center for Advancing Translational Sciences under the award number UL1 TR000064 (University of Florida); UL1 TR000454 (Emory University) and UL1 TR000135 (Mayo Clinic). PEAR was also supported by funds from the Mayo Foundation. Additional support for this work includes: U01 GM092655 (W.S.) and AHA pre-doctoral fellowship award #14PRE20460115 (M.H.S.).
Footnotes
Journal Subject Terms: Genetic, Association Studies; Hypertension; High Blood Pressure; Genetics; Gene Expression and Regulation
Disclosures: None
References
- 1.Oparil S, Schmieder RE. New approaches in the treatment of hypertension. Circ Res. 2015;116:1074–1095. doi: 10.1161/CIRCRESAHA.116.303603. [DOI] [PubMed] [Google Scholar]
- 2.Lewington S, Clarke R, Qizilbash N, Peto R, Collins R, Prospective Studies C Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet. 2002;360:1903–1913. doi: 10.1016/s0140-6736(02)11911-8. [DOI] [PubMed] [Google Scholar]
- 3.Mancia G. Blood pressure reduction and cardiovascular outcomes: past, present, and future. Am J Cardiol. 2007;100:3J–9J. doi: 10.1016/j.amjcard.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 4.IMS Institue for Healthcare Informatics Medicines Use and Spending Shifts: A Review of the Use of Medicines in the U.S. 2014 Accessed October 4th, 2016. [Google Scholar]
- 5.Weber MA, Schiffrin EL, White WB, Mann S, Lindholm LH, Kenerson JG, et al. Clinical practice guidelines for the management of hypertension in the community a statement by the American Society of Hypertension and the International Society of Hypertension. J Hypertens. 2014;32:3–15. doi: 10.1097/HJH.0000000000000065. [DOI] [PubMed] [Google Scholar]
- 6.Shahin MH, Johnson JA. Mechanisms and pharmacogenetic signals underlying thiazide diuretics blood pressure response. Curr Opin Pharmacol. 2016;27:31–37. doi: 10.1016/j.coph.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Materson BJ. Variability in response to antihypertensive drugs. Am J Med. 2007;120:S10–20. doi: 10.1016/j.amjmed.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Materson BJ, Reda DJ, Cushman WC, Massie BM, Freis ED, Kochar MS, et al. Single-drug therapy for hypertension in men. A comparison of six antihypertensive agents with placebo. The Department of Veterans Affairs Cooperative Study Group on Antihypertensive Agents. N Engl J Med. 1993;328:914–921. doi: 10.1056/NEJM199304013281303. [DOI] [PubMed] [Google Scholar]
- 9.Thoenes M, Neuberger HR, Volpe M, Khan BV, Kirch W, Bohm M. Antihypertensive drug therapy and blood pressure control in men and women: an international perspective. J Hum Hypertens. 2010;24:336–344. doi: 10.1038/jhh.2009.76. [DOI] [PubMed] [Google Scholar]
- 10.Perera MA, Cavallari LH, Limdi NA, Gamazon ER, Konkashbaev A, Daneshjou R, et al. Genetic variants associated with warfarin dose in African-American individuals: a genome-wide association study. Lancet. 2013;382:790–796. doi: 10.1016/S0140-6736(13)60681-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner ST, Boerwinkle E, O'Connell JR, Bailey KR, Gong Y, Chapman AB, et al. Genomic association analysis of common variants influencing antihypertensive response to hydrochlorothiazide. Hypertension. 2013;62:391–397. doi: 10.1161/HYPERTENSIONAHA.111.00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stranger BE, Stahl EA, Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187:367–383. doi: 10.1534/genetics.110.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson JA, Boerwinkle E, Zineh I, Chapman AB, Bailey K, Cooper-DeHoff RM, et al. Pharmacogenomics of antihypertensive drugs: rationale and design of the Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) study. Am Heart J. 2009;157:442–449. doi: 10.1016/j.ahj.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamadeh IS, Langaee TY, Dwivedi R, Garcia S, Burkley BM, Skaar TC, et al. Impact of CYP2D6 polymorphisms on clinical efficacy and tolerability of metoprolol tartrate. Clin Pharmacol Ther. 2014;96:175–181. doi: 10.1038/clpt.2014.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner ST, Schwartz GL, Chapman AB, Beitelshees AL, Gums JG, Cooper-Dehoff RM, et al. Power to identify a genetic predictor of antihypertensive drug response using different methods to measure blood pressure response. J Transl Med. 2012;10:47. doi: 10.1186/1479-5876-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stergiou GS, Baibas NM, Gantzarou AP, Skeva II, Kalkana CB, Roussias LG, et al. Reproducibility of home, ambulatory, and clinic blood pressure: implications for the design of trials for the assessment of antihypertensive drug efficacy. Am J Hypertens. 2002;15:101–104. doi: 10.1016/s0895-7061(01)02324-x. [DOI] [PubMed] [Google Scholar]
- 21.Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritchie GR, Dunham I, Zeggini E, Flicek P. Functional annotation of noncoding sequence variants. Nat Methods. 2014;11:294–296. doi: 10.1038/nmeth.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li G, Bahn JH, Lee JH, Peng G, Chen Z, Nelson SF, et al. Identification of allele-specific alternative mRNA processing via transcriptome sequencing. Nucleic Acids Res. 2012;40:e104. doi: 10.1093/nar/gks280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C, Zhang F, Li T, Lu M, Wang L, Yue W, et al. MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA-related SNPs in GWAS SNPs and eQTLs. BMC Genomics. 2012;13:661. doi: 10.1186/1471-2164-13-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharya A, Ziebarth JD, Cui Y. PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014;42:D86–91. doi: 10.1093/nar/gkt1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ernst ME, Carter BL, Goerdt CJ, Steffensmeier JJ, Phillips BB, Zimmerman MB, et al. Comparative antihypertensive effects of hydrochlorothiazide and chlorthalidone on ambulatory and office blood pressure. Hypertension. 2006;47:352–358. doi: 10.1161/01.HYP.0000203309.07140.d3. [DOI] [PubMed] [Google Scholar]
- 33.Smith RM, Webb A, Papp AC, Newman LC, Handelman SK, Suhy A, et al. Whole transcriptome RNA-Seq allelic expression in human brain. BMC Genomics. 2013;14:571. doi: 10.1186/1471-2164-14-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markert T, Krenn V, Leebmann J, Walter U. High expression of the focal adhesion- and microfilament-associated protein VASP in vascular smooth muscle and endothelial cells of the intact human vessel wall. Basic Res Cardiol. 1996;91:337–343. doi: 10.1007/BF00788712. [DOI] [PubMed] [Google Scholar]
- 35.Haffner C, Jarchau T, Reinhard M, Hoppe J, Lohmann SM, Walter U. Molecular cloning, structural analysis and functional expression of the proline-rich focal adhesion and microfilament-associated protein VASP. EMBO J. 1995;14:19–27. doi: 10.1002/j.1460-2075.1995.tb06971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- 37.Yamin R, Morgan KG. Deciphering actin cytoskeletal function in the contractile vascular smooth muscle cell. J Physiol. 2012;590:4145–4154. doi: 10.1113/jphysiol.2012.232306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gunst SJ, Zhang W. Actin cytoskeletal dynamics in smooth muscle: a new paradigm for the regulation of smooth muscle contraction. Am J Physiol Cell Physiol. 2008;295:C576–587. doi: 10.1152/ajpcell.00253.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walter U, Eigenthaler M, Geiger J, Reinhard M. Role of cyclic nucleotide-dependent protein kinases and their common substrate VASP in the regulation of human platelets. Adv Exp Med Biol. 1993;344:237–249. doi: 10.1007/978-1-4615-2994-1_19. [DOI] [PubMed] [Google Scholar]
- 40.Hofmann F, Feil R, Kleppisch T, Schlossmann J. Function of cGMP-dependent protein kinases as revealed by gene deletion. Physiol Rev. 2006;86:1–23. doi: 10.1152/physrev.00015.2005. [DOI] [PubMed] [Google Scholar]
- 41.Comerford KM, Lawrence DW, Synnestvedt K, Levi BP, Colgan SP. Role of vasodilator-stimulated phosphoprotein in PKA-induced changes in endothelial junctional permeability. FASEB J. 2002;16:583–585. doi: 10.1096/fj.01-0739fje. [DOI] [PubMed] [Google Scholar]
- 42.Chen H, Levine YC, Golan DE, Michel T, Lin AJ. Atrial natriuretic peptide-initiated cGMP pathways regulate vasodilator-stimulated phosphoprotein phosphorylation and angiogenesis in vascular endothelium. J Biol Chem. 2008;283:4439–4447. doi: 10.1074/jbc.M709439200. [DOI] [PubMed] [Google Scholar]
- 43.An D, Rodrigues B. Role of changes in cardiac metabolism in development of diabetic cardiomyopathy. Am J Physiol Heart Circ Physiol. 2006;291:H1489–1506. doi: 10.1152/ajpheart.00278.2006. [DOI] [PubMed] [Google Scholar]
- 44.Aszodi A, Pfeifer A, Ahmad M, Glauner M, Zhou XH, Ny L, et al. The vasodilator-stimulated phosphoprotein (VASP) is involved in cGMP- and cAMP-mediated inhibition of agonist-induced platelet aggregation, but is dispensable for smooth muscle function. EMBO J. 1999;18:37–48. doi: 10.1093/emboj/18.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oelze M, Mollnau H, Hoffmann N, Warnholtz A, Bodenschatz M, Smolenski A, et al. Vasodilator-stimulated phosphoprotein serine 239 phosphorylation as a sensitive monitor of defective nitric oxide/cGMP signaling and endothelial dysfunction. Circ Res. 2000;87:999–1005. doi: 10.1161/01.res.87.11.999. [DOI] [PubMed] [Google Scholar]
- 46.Feil R, Lohmann SM, de Jonge H, Walter U, Hofmann F. Cyclic GMP-dependent protein kinases and the cardiovascular system: insights from genetically modified mice. Circ Res. 2003;93:907–916. doi: 10.1161/01.RES.0000100390.68771.CC. [DOI] [PubMed] [Google Scholar]
- 47.Warner TD, Mitchell JA, Sheng H, Murad F. Effects of cyclic GMP on smooth muscle relaxation. Adv Pharmacol. 1994;26:171–194. doi: 10.1016/s1054-3589(08)60054-x. [DOI] [PubMed] [Google Scholar]
- 48.Buechler WA, Ivanova K, Wolfram G, Drummer C, Heim JM, Gerzer R. Soluble guanylyl cyclase and platelet function. Ann N Y Acad Sci. 1994;714:151–157. doi: 10.1111/j.1749-6632.1994.tb12039.x. [DOI] [PubMed] [Google Scholar]
- 49.Schork AJ, Thompson WK, Pham P, Torkamani A, Roddey JC, Sullivan PF, et al. All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet. 2013;9:e1003449. doi: 10.1371/journal.pgen.1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.