Abstract
Modifications of histones play important roles in balancing transcriptional output. The discovery of acyl marks, besides histone acetylation, has added to the functional diversity of histone modifications. Since all modifications use metabolic intermediates as substrates for chromatin modifying enzymes, the prevalent landscape of histone modifications in any cell type is a snapshot of its metabolic status. Here we review some of the current findings of how differential use of histone acylations regulates gene expression as response to metabolic changes and differentiation programs.
Keywords: Histone acylation, YEATS domain, bromodomain, metabolism
In all eukaryotes, nucleosomes are formed by DNA wrapped around histone octamers that are composed of two copies each of the four core histones, H2A, H2B, H3, and H4. Organization into this chromatin structure helps in DNA compaction and plays important roles in dynamic regulation of transcription, DNA repair and recombination. Changes in chromatin organization are dependent primarily on modification of amino acid residues in histones, particularly within the N-terminal histone tails. The expanding list of histone modifications and the complexity of their actions has been an area of intense study, with recent efforts directed toward understanding their roles in nuclear processes and their effects on cellular homeostasis.
Histone lysine acetylation was one of the first histone modifications to be discovered, where it was shown to correlate with RNA synthesis (Allfrey et al., 1964). The link between histone acetylation, changes in chromatin structure, and altered transcription was later established in parallel groundbreaking reports. These studies revealed a role for histone lysine acetylation in transcription silencing at yeast telomeres (Aparicio et al., 1991), regulation of the yeast mating type loci (Johnson et al., 1990; Megee et al., 1990), and the presence of distinct domains of histone acetylation on polytene chromosomes in flies (Turner et al., 1992). The pivotal link between transcription and histone acetylation came with the discovery of an orthologue of yeast Gcn5 in Tetrahymena that acetylated histones (Brownell and Allis, 1995; Brownell et al., 1996) and a bovine orthologue of yeast Rpd3 that aided in removal of histone acetyl marks (Taunton et al., 1996). Following the initial discovery of the Gcn5 histone acetyltransferase (HAT) (Brownell et al., 1996) and the Rpd3 histone deacetylase (HDAC) (Rundlett et al., 1996), studies have identified several members of both enzyme families. The HATs include CREB-binding protein (CBP), E1a-binding protein p300, and the MYST family of proteins (MOZ, YBF2/SAS3, SAS2 and TIP60) (Lee and Workman, 2007; Marmorstein and Zhou, 2014). HDACs include Rpd3-related proteins, HDAC1-HDAC3 and HDAC8 (Class I HDACs), Hda1-related proteins, HDAC4–HDAC7, and HDAC9 (Class II HDACs) and Sir family proteins (Class III HDACs), HDAC10 and HDAC11 (Delcuve et al., 2012). Biochemical purification of several of these enzymes showed that they function within large multi-subunit complexes that are conserved from yeast to humans. For example, the Gcn5 HAT was shown to be part of SAGA, SLIK and ADA complexes in yeast, and part of SAGA and ATAC in flies and humans (Brand et al., 1999; Eberharter et al., 1999; Grant et al., 1997; Guelman et al., 2006; Kusch et al., 2003; Martinez et al., 1998; Pray-Grant et al., 2002). Similarly, histone deacetylases HDAC1/HDAC2 are part of chromatin modifying complexes Sin3, NURD and CoREST (Kelly and Cowley, 2013).
Since the 1990’s, an extensive number of studies have uncovered proteins with enzymatic activities that work in opposition to each other through modifying histones. These studies demonstrate that acetylation of lysine residues is a highly dynamic process regulated by HATs and HDACs. HATs use acetyl-CoA to catalyze addition of the acetyl group onto the ε-amino group of lysine side chains, and HDACs remove this histone mark (Anne-Lise Steunou, 2013). Neutralization of the positive charge on lysine residues by acetylation weakens interactions between histones and DNA and may allow access to the transcription machinery. The stimulatory effect of histone acetylation on transcription through nucleosomes has been demonstrated by elegant in vitro studies in which addition of HATS, including yeast SAGA, Nua3 and Nua4 complexes or human CBP/P300, facilitated transcription of RNA polymerase thorough nucleosomal templates (Kundu et al., 2000; Steger et al., 1998). While opening of chromatin by acetylation has a stimulatory effect on transcription through nucleosomes, HDACs like Rpd3, play opposing roles in suppressing spurious transcription by deacetylation of histones both at promoters and in gene bodies (Burgess et al., 1999; Carrozza et al., 2005; Deckert and Struhl, 2002; Li et al., 2007).
In addition to altering chromatin structure, acetylated lysines function as docking sites for protein domains, which are therefore referred to as “readers” of the histone marks. Bromodomains recognize acetylated histones and are present in many nuclear proteins, including ATP dependent chromatin remodelers, HATs, transcriptional co-activators, methyltransferases and the BET (bromodomain and extraterminal) family of proteins (Marmorstein and Zhou, 2014; Yun et al., 2011). For example, the Gcn5 HAT bromodomain recognizes histones that it has actually acetylated itself (Li and Shogren-Knaak, 2009). This property could, in principle, act as in a positive feedback loop to recruit and stabilize Gcn5 containing complexes. Other functions of histone acetylation at gene promoters include recruitment of chromatin remodelers like Swi/Snf and RSC via bromodomains in subunits of these complexes (Cairns et al., 1999; Dutta et al., 2014; Hassan et al., 2001; Kasten et al., 2004) which, in turn stimulate nucleosome remodeling. In addition, acetylated histones can act as a docking platform for transcription initiation factor TFIID via interaction with Taf1 double bromodomains (Jacobson et al., 2000), which promotes transcription initiation.
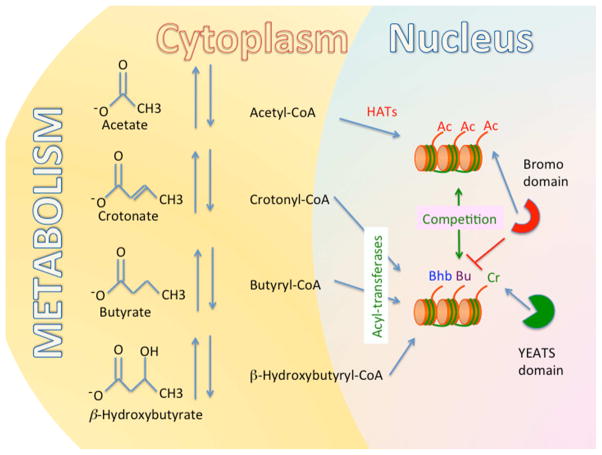
Though histone acetylation has been an area of extensive investigation for the last quarter century, the variety of different histone acylations has continued to expand. As early as 1992, studies showing that histone succinylation (KSu) facilitates transcription through nucleosomal cores hinted at roles for other histone lysine acylations (Pineiro et al., 1992). Proteomic approaches have broadened the landscape of histone acylation to include lysine propionylation (KPr) and butyrylation (KBu) (Chen et al., 2007), and recent studies have shown that histone lysines are extensively decorated with crotonylation (KCr), β-hydroxybutyrylation (KBhu), succinylation (KSu), malonylation (KMa) and glutarylation (KGlu) (Figure 1) (Dai et al., 2014; Li et al., 2016; Tan et al., 2011; Tan et al., 2014; Xie et al., 2012; Xie et al., 2016; Zhang et al., 2009). These recent studies have shown that the newly discovered histone acylations can have a stimulatory effect on transcription at several genomic loci, akin to histone acetylation. In the remainder of this perspective we highlight the roles of differential histone lysine acylations working in consort or in opposition to help respond to varying cellular conditions.
Figure 1.
The levels and types of short chain histone lysine acylations are influenced by metabolism. The levels of short chain acyl metabolic intermediates increase or decrease (arrows) dependent on metabolic state. These include acetate, crotonate, butyrate, β-hydroxybutyrate, and more. When conjugated to coenzyme A (CoA) these short acyl-CoAs function in fatty acid metabolism and can be used by acyl-transferases to modify lysines on histones within nucleosomes (disks). The levels of each type of histone acylation (e.g. acetylation Ac, crotonylation Cr, butyrylation Bu and β-hydroxybutyrylation Bhb) are influenced by the levels of these metabolites in the cell. Thus, metabolism affects the type and levels of histone acylation in chromatin and chromatin can in principle serve as a reservoir of these metabolites. Recent work suggests this will have epigenetic effects on chromatin function. Bromodomain-containing chromatin modifying complexes interact (arrow) with nucleosomes containing acetylated histones (Ac). By contrast, histone butyrylation (Bu), and perhaps other acylations, reduce or resist (blocked line) bromodomain containing protein complex interaction with nucleosomes. Histone acetylation and histone butyrylation are in competition (double arrow) with each other leading to dynamic interactions of bromodomain-containing complexes with nucleosomes. Acylations other than acetylation may have reader domain proteins that prefer those modifications. For example YEATS domain containing chromatin modifying complexes prefer to interact (arrow) with nucleosomes containing crotonylated (Cr) histones.
In order to mark histones post-translationally, chromatin modifiers use metabolic intermediates such as acetyl-CoA, the key metabolite used as a cofactor by HATs, and S-adenosylmethionine, the cofactor for histone methyltransferases. Thus, histone modifications may reflect the metabolic status of cells and be influenced by the concentrations of these metabolites. The delicate balance between metabolic processes and the levels of their intermediates is key to cellular homeostasis, and misregulation of this balance can trigger a variety of diseases. Moreover, metabolites and metabolic enzymes can directly regulate transcription, as shown by several studies involving the glycolytic enzyme PKM2, which is responsible for pyruvate synthesis. PKM2 phosphorylates histone H3T11 (Yang et al., 2012), and the epidermal growth factor receptor (EGFR) mediated activation of PKM2-dependent H3T11 phosphorylation is required for H3K9 acetylation (Yang et al., 2011). Further, a complex of PKM2 and β-catenin functions to dissociate HDAC3 from gene promoters (Yang et al., 2011). Pyk1, the yeast PKM2 orthologue, forms a novel complex termed SESAME that includes other metabolic enzymes involved in serine and acetyl-CoA synthesis. In addition to Pyk1, SESAME contains SAM (S-adenosylmethionine) synthetases and an acetyl-CoA synthetase, and it regulates crosstalk between H3K4 methylation and H3T11 phosphorylation at gene promoters (Li et al., 2015). However, unlike the human PKM2, yeast Pyk1 does not affect histone acetylation, specifically at histone H3 lysine 9 (Li et al., 2015). The enzyme 5′ adenosine monophosphate (AMP)-activated protein kinase (AMPK) also influences histone modification and gene expression. AMPK acts as a sensor of cellular ATP/ADP levels in cells and promotes β-oxidation of fatty acids (Suter et al., 2006). It is phosphorylated in response to several stress conditions, including low concentrations of glucose. Phosphorylated AMPK, in turn, activates P53 responsive genes by phosphorylating histone H2BS36 (Bungard et al., 2010).
Acetyl-CoA is produced from pyruvate through the tricarboxylic acid (TCA) cycle or by β-oxidation of fatty acids. Growth of yeast in nutrient-limiting conditions causes cells to oscillate between oxidative and reductive growth (Tu et al., 2007). This rhythmic pattern results in changes in acetyl-CoA levels and concomitant fluctuations in the level of acetylation on both histones H3 and H4 (Cai et al., 2011). The change in histone acetylation status then results in differential gene expression during these two phases of growth (Tu et al., 2007). In mammalian systems, undifferentiated embryonic stem cells have higher levels of acetyl-CoA than differentiating cells (Wang et al., 2011). This difference results, in part, from changes in expression of threonine dehydrogenase (TDH), which catalyzes the conversion of threonine into glycine and acetyl-CoA (Alexander et al., 2011). The drop in levels of TDH during differentiation can directly affect gene expression by regulating acetyl-CoA levels.
Since many of the same histone lysine residues can be modified by several types of acylation, changes in the concentrations of intermediary metabolites seem likely to affect histone modifications. For example, upon depletion of carbohydrates during prolonged starvation, cells use fatty acid oxidation as source of energy. Much of the acetyl CoA produced in this process is converted into ketone bodies consisting of β-hydroxybutyrate, acetoacetate and acetone (Laffel, 1999). A recent study by Zhao and colleagues has identified that β-hydroxybutyrylation (KBhb), a new histone lysine acylation, is induced by starvation and is present on all four core histones (Figure 1) (Xie et al., 2016). β-hydroxybutyrylation is present with the activating marks H3K4me3 and H3K9ac at many gene promoters. Also, a different group of genes specifically upregulated by starvation contained only H3K9Bhb, completely replacing H3K9ac at these genes. Interestingly, β-hydroxybutyrate, the substrate for this new histone mark, is elevated in untreated diabetes (Laffel, 1999). β-hydroxybutyrate is used as a source of energy by the brain and heart during starvation (Cahill, 2006). It is also used as a protective agent to treat neurological disorders, including epilepsy (McNally and Hartman, 2012), Parkinson’s and Alzheimer’s (Kashiwaya et al., 2000). Thus, positive regulation of transcription by lysine β-hydroxybutyrylation provides an entry into the mechanism by which histone modifications regulate specific subsets of genes, and how a failure to produce optimal concentrations of key cellular metabolites may result in diseased states. Future studies should allow us to determine metabolite concentrations in normal vs diseased tissues, and explore whether histone modifications influenced by these differing concentrations impact the disease phenotypes. It then remains only to evaluate whether exogenously provided cellular metabolites correct histone modification defects and, in turn, impact the diseased state.
The finding that histone acylation changes during metabolic stress leads to the question of whether dynamic regulation of differential histone acylation occurs in response to other cellular changes. Such mechanisms could play important roles during development in eukaryotes, where temporal and spatial control of gene expression is a key determinant. Spermatogenesis, for example, involves a specialized differentiation program and requires a program of gene expression aimed at producing haploid cells that carry the male genome. Goudarzi and colleagues have now explored histone acylation during spermatogenesis, and uncovered competing roles for histone lysine butyrylation (KBu) and histone lysine acetylation (KAc) (Goudarzi et al., 2016). These studies have revealed that levels of histone H4K5Bu and H4K8Bu increase as spermatocytes differentiate into elongating spermatids and are associated with a concomitant loss of acetylation on the same lysine residues. Moreover, increased histone butyrylation corresponds well with genes that increase in expression as spermatocytes differentiate. In addition to histone lysine butyrylation (KBu), histone lysine crotonylation (KCr) marks testis-specific genes on sex chromosomes and is enriched in male germ cells following meiosis (Tan et al., 2011). The increased use of butyrate and crotonate in the male germ line hints at a metabolic flux during spermatogenesis that changes levels of these metabolites. It seems likely that the metabolic profiles of other differentiating tissues will be similarly reflected in their pattern of histone-acyl marks, and that these changing marks will guide specific cellular differentiation programs. As noted above, histone H4K5 can be either acetylated or butyrylated during spermatogenesis (Goudarzi et al., 2016). Change in these histone H4K5 marks regulates the affinity of the testis specific bromodomain protein Brdt1 for nucleosomes. While the Brdt1 bromodomain readily binds acetylated H4K5, butyrylation of H4K5 blocks this binding. Structurally, a steric clash within the Brdt1 bromodomain binding pocket prevents binding to the bulkier butyryl group. The authors hypothesize that alternating H4K5Ac and H4K5Bu could regulate Brdt1 recruitment and dissociation and, in turn, stimulate waves of transcription initiation by RNA polymerase II (RNAPII) recruitment. In fact, many bromodomains can accommodate only smaller acyl groups such as acetyl and propionyl in their binding pockets, and fail to bind bulky succinyl marks (Flynn et al., 2015).
Recent studies have revealed that several non-acetyl histone acylations occupy promoters of highly expressing genes, in a pattern similar to that observed for histone acetyl marks (Goudarzi et al., 2016; Li et al., 2016; Xie et al., 2016). Acetylation of lysine residues plays a dual roles in decompacting chromatin and acting as a docking site for proteins containing bromodomains. While the direct effects of various other histone acylations on chromatin structure have not been studied, their presence at active genes raises the speculation that they regulate chromatin decompaction in a similar manner as acetylation. In addition, while recognition of acetyl lysine by bromodomains is well documented, readers of bulkier acyl groups are only beginning to be described. Histone lysine crotonylation, which is also associated with active transcription, binds selectively to the bromodomains in Brd9, Taf1 and Cecr2, albeit at a lower affinity than their binding to acetyl-lysine (Flynn et al., 2015). However, a recent study has shown that YEATS domains in AF9, Yaf9, Taf14, ENL and YEATS2 preferentially bind histone lysine crotonyl marks with higher affinities than acetyl marks (Andrews et al., 2016; Li et al., 2016; Zhao et al., 2016). This differential binding is due, in part, to the architecture of acyl group binding pockets in bromodomains versus YEATS domains. While a smaller acetyl group can be accommodated in the center of the four helix bundle within the binding pocket of a bromodomain, bulkier acyl groups would have to be bent to be accommodated, which would require significant energy cost for binding. On the other hand, a more open and flat binding pocket in the YEATS domain can easily fit a bulky rigid acyl group like crotonyl. As predicted by increased binding of AF9 to crotonyl marks, treatment of cells with crotonate increases AF9 occupancy at crotonate-responsive genes. AF9 is a part of both the Super Elongation Complex (SEC) and DOT1C (Biswas et al., 2011; Bitoun et al., 2007; Smith et al., 2011), and recruitment of the SEC to promoters allows for release of polymerases from paused state to one of productive elongation. In higher organisms, many genes that are responsive to changes in environmental cues and developmental signals contain paused polymerases at their promoters. Histone crotonylation mediated recruitment of AF9 to promoters of these genes can thus allow robust transcription by directly stimulating release of paused polymerases into productive transcription elongation. The identification of the YEATS domain as a histone crotonylation reader is the first in what is likely to be an expanding number of novel domains that binding an ever increasing number of histone modifications, especially bulky acyl modifications.
The identification of a varied repertoire of acyl modifications, many of which can occur on the same lysine residues, promotes the hypothesis that cells use alternate strategies to stimulate transcription when the balance between metabolite levels changes (Figure 1). These changes could be either a response to or a consequence of altered metabolism. Readers with domains that recognize only one acyl mark or prefer one mark over another allows for specific targeting of transcription complexes to regulate defined spatio-temporal output. For example the SAGA complex contains a bromodomain on the acetyl transferase subunit Gcn5, while Taf14 in TFIID and the Nua3 acetyltransferase complex contains a YEATS domain. In yeast, genes prefer to use either the SAGA or TFIID complexes to regulate transcription initiation (Huisinga and Pugh, 2004). The presence of YEATS domain containing Taf14 in TFIID and TFIIF, a general transcription factor, could alter transcription initiation programs in response to cellular accumulation of acyl group precursors. On the other hand, chromatin modifying complexes such as Swi/Snf, that are critical for gene regulation under metabolic and other stress conditions, have both the bromodomain containing Snf2 subunit and the YEATS domain containing Taf14 subunit. Thus, Swi/Snf could be recruited by different histone lysine acyl marks to carry out similar or distinct functions at gene promoters. In addition to influencing the recruitment and binding of chromatin remodelers, different histone lysine acylations could trigger different activities of these complexes. For example, histone acetylation stimulates both the binding of Swi/Snf to nucleosomes (Hassan et al., 2001) and enhances its ability to displace histones (Chandy et al., 2006).
In summary, HATs and HDACs that acetylate and deacetylate various histone residues have been extensively studied. However, a variety of approaches over the last few years have now identified additional histone acyl modifications. We lack a clear understanding of the writers and erasers of these non-acetyl acyl groups. CBP/P300 is the only HAT in higher eukaryotes that has been shown to acylate histones with bulkier acyl groups (Chen et al., 2007; Goudarzi et al., 2016; Tan et al., 2014). Also. only HDAC3 (Madsen and Olsen, 2012), Sirt1, Sirt2 and Sirt3 (Bao et al., 2014; Feldman et al., 2013) have been shown to decrotonylate histones. Moreover, though the CBP/P300 HAT is not present in organisms like yeast, the yeast Rtt109 is structurally orthologous to CBP/P300 (Tang et al., 2008) and a recent study has shown that yeast HATs Rtt109 and Gcn5 may play roles in non-acetyl histone acylations, as loss of these HATs in yeast reduces H3K9 crotonylation. The time is now ripe for the discovery of both writers and erasers of these differential acyl marks, and their identification and description of functional roles will influence this expanding field for years to come.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander PB, Wang J, McKnight SL. Targeted killing of a mammalian cell based upon its specialized metabolic state. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15828–15833. doi: 10.1073/pnas.1111312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allfrey VG, Faulkner R, Mirsky AE. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews FH, Shinsky SA, Shanle EK, Bridgers JB, Gest A, Tsun IK, Krajewski K, Shi X, Strahl BD, Kutateladze TG. The Taf14 YEATS domain is a reader of histone crotonylation. Nature chemical biology. 2016;12:396–398. doi: 10.1038/nchembio.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne-Lise Steunou DR, Jacques Côté. Regulating Chromatin by Histone Acetylation. Fundamentals of Chromatin. 2013:147–212. [Google Scholar]
- Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Bao X, Wang Y, Li X, Li XM, Liu Z, Yang T, Wong CF, Zhang J, Hao Q, Li XD. Identification of ‘erasers’ for lysine crotonylated histone marks using a chemical proteomics approach. eLife. 2014;3 doi: 10.7554/eLife.02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D, Milne TA, Basrur V, Kim J, Elenitoba-Johnson KS, Allis CD, Roeder RG. Function of leukemogenic mixed lineage leukemia 1 (MLL) fusion proteins through distinct partner protein complexes. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15751–15756. doi: 10.1073/pnas.1111498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Human molecular genetics. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- Brand M, Yamamoto K, Staub A, Tora L. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. The Journal of biological chemistry. 1999;274:18285–18289. doi: 10.1074/jbc.274.26.18285. [DOI] [PubMed] [Google Scholar]
- Brownell JE, Allis CD. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Bungard D, Fuerth BJ, Zeng PY, Faubert B, Maas NL, Viollet B, Carling D, Thompson CB, Jones RG, Berger SL. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SM, Ajimura M, Kleckner N. GCN5-dependent histone H3 acetylation and RPD3-dependent histone H4 deacetylation have distinct, opposing effects on IME2 transcription, during meiosis and during vegetative growth, in budding yeast. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:6835–6840. doi: 10.1073/pnas.96.12.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill GF., Jr Fuel metabolism in starvation. Annual review of nutrition. 2006;26:1–22. doi: 10.1146/annurev.nutr.26.061505.111258. [DOI] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Molecular cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Molecular cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Chandy M, Gutierrez JL, Prochasson P, Workman JL. SWI/SNF displaces SAGAacetylated nucleosomes. Eukaryotic cell. 2006;5:1738–1747. doi: 10.1128/EC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Molecular & cellular proteomics : MCP. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Peng C, Montellier E, Lu Z, Chen Y, Ishii H, Debernardi A, Buchou T, Rousseaux S, Jin F, et al. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nature chemical biology. 2014;10:365–370. doi: 10.1038/nchembio.1497. [DOI] [PubMed] [Google Scholar]
- Deckert J, Struhl K. Targeted recruitment of Rpd3 histone deacetylase represses transcription by inhibiting recruitment of Swi/Snf, SAGA, and TATA binding protein. Molecular and cellular biology. 2002;22:6458–6470. doi: 10.1128/MCB.22.18.6458-6470.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcuve GP, Khan DH, Davie JR. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clinical epigenetics. 2012;4:5. doi: 10.1186/1868-7083-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Gogol M, Kim JH, Smolle M, Venkatesh S, Gilmore J, Florens L, Washburn MP, Workman JL. Swi/Snf dynamics on stress-responsive genes is governed by competitive bromodomain interactions. Genes & development. 2014;28:2314–2330. doi: 10.1101/gad.243584.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Sterner DE, Schieltz D, Hassan A, Yates JR, 3rd, Berger SL, Workman JL. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Molecular and cellular biology. 1999;19:6621–6631. doi: 10.1128/mcb.19.10.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. The Journal of biological chemistry. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn EM, Huang OW, Poy F, Oppikofer M, Bellon SF, Tang Y, Cochran AG. A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications. Structure. 2015;23:1801–1814. doi: 10.1016/j.str.2015.08.004. [DOI] [PubMed] [Google Scholar]
- Goudarzi A, Zhang D, Huang H, Barral S, Kwon OK, Qi S, Tang Z, Buchou T, Vitte AL, He T, et al. Dynamic Competing Histone H4 K5K8 Acetylation and Butyrylation Are Hallmarks of Highly Active Gene Promoters. Molecular cell. 2016;62:169–180. doi: 10.1016/j.molcel.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes & development. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Guelman S, Suganuma T, Florens L, Swanson SK, Kiesecker CL, Kusch T, Anderson S, Yates JR, 3rd, Washburn MP, Abmayr SM, et al. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Molecular and cellular biology. 2006;26:871–882. doi: 10.1128/MCB.26.3.871-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Molecular cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- Johnson LM, Kayne PS, Kahn ES, Grunstein M. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwaya Y, Takeshima T, Mori N, Nakashima K, Clarke K, Veech RL. D-betahydroxybutyrate protects neurons in models of Alzheimer’s and Parkinson’s disease. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5440–5444. doi: 10.1073/pnas.97.10.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. The EMBO journal. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RD, Cowley SM. The physiological roles of histone deacetylase (HDAC) 1 and 2: complex co-stars with multiple leading parts. Biochemical Society transactions. 2013;41:741–749. doi: 10.1042/BST20130010. [DOI] [PubMed] [Google Scholar]
- Kundu TK, Palhan VB, Wang Z, An W, Cole PA, Roeder RG. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Molecular cell. 2000;6:551–561. doi: 10.1016/s1097-2765(00)00054-x. [DOI] [PubMed] [Google Scholar]
- Kusch T, Guelman S, Abmayr SM, Workman JL. Two Drosophila Ada2 homologues function in different multiprotein complexes. Molecular and cellular biology. 2003;23:3305–3319. doi: 10.1128/MCB.23.9.3305-3319.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffel L. Ketone bodies: a review of physiology, pathophysiology and application of monitoring to diabetes. Diabetes/metabolism research and reviews. 1999;15:412–426. doi: 10.1002/(sici)1520-7560(199911/12)15:6<412::aid-dmrr72>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nature reviews Molecular cell biology. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes & development. 2007;21:1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Shogren-Knaak MA. The Gcn5 bromodomain of the SAGA complex facilitates cooperative and cross-tail acetylation of nucleosomes. The Journal of biological chemistry. 2009;284:9411–9417. doi: 10.1074/jbc.M809617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Swanson SK, Gogol M, Florens L, Washburn MP, Workman JL, Suganuma T. Serine and SAM Responsive Complex SESAME Regulates Histone Modification Crosstalk by Sensing Cellular Metabolism. Molecular cell. 2015;60:408–421. doi: 10.1016/j.molcel.2015.09.024. [DOI] [PubMed] [Google Scholar]
- Li Y, Sabari BR, Panchenko T, Wen H, Zhao D, Guan H, Wan L, Huang H, Tang Z, Zhao Y, et al. Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain. Molecular cell. 2016;62:181–193. doi: 10.1016/j.molcel.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen AS, Olsen CA. Profiling of substrates for zinc-dependent lysine deacylase enzymes: HDAC3 exhibits decrotonylase activity in vitro. Angewandte Chemie. 2012;51:9083–9087. doi: 10.1002/anie.201203754. [DOI] [PubMed] [Google Scholar]
- Marmorstein R, Zhou MM. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harbor perspectives in biology. 2014;6:a018762. doi: 10.1101/cshperspect.a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Kundu TK, Fu J, Roeder RG. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. The Journal of biological chemistry. 1998;273:23781–23785. doi: 10.1074/jbc.273.37.23781. [DOI] [PubMed] [Google Scholar]
- McNally MA, Hartman AL. Ketone bodies in epilepsy. Journal of neurochemistry. 2012;121:28–35. doi: 10.1111/j.1471-4159.2012.07670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee PC, Morgan BA, Mittman BA, Smith MM. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science. 1990;247:841–845. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- Pineiro M, Hernandez F, Palacian E. Succinylation of histone amino groups facilitates transcription of nucleosomal cores. Biochimica et biophysica acta. 1992;1129:183–187. doi: 10.1016/0167-4781(92)90485-i. [DOI] [PubMed] [Google Scholar]
- Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, Cook RG, Workman JL, Yates JR, 3rd, Grant PA. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Molecular and cellular biology. 2002;22:8774–8786. doi: 10.1128/MCB.22.24.8774-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes & development. 2011;25:661–672. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Eberharter A, John S, Grant PA, Workman JL. Purified histone acetyltransferase complexes stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12924–12929. doi: 10.1073/pnas.95.22.12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter M, Riek U, Tuerk R, Schlattner U, Wallimann T, Neumann D. Dissecting the role of 5′-AMP for allosteric stimulation, activation, and deactivation of AMP-activated protein kinase. The Journal of biological chemistry. 2006;281:32207–32216. doi: 10.1074/jbc.M606357200. [DOI] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell metabolism. 2014;19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Holbert MA, Wurtele H, Meeth K, Rocha W, Gharib M, Jiang E, Thibault P, Verreault A, Cole PA, et al. Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP. Nature structural & molecular biology. 2008;15:738–745. doi: 10.1038/nsmb.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM, Birley AJ, Lavender J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- Wang J, Alexander P, McKnight SL. Metabolic specialization of mouse embryonic stem cells. Cold Spring Harbor symposia on quantitative biology. 2011;76:183–193. doi: 10.1101/sqb.2011.76.010835. [DOI] [PubMed] [Google Scholar]
- Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y, Boeke JD, Zhao Y. Lysine succinylation and lysine malonylation in histones. Molecular & cellular proteomics : MCP. 2012;11:100–107. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Zhang D, Chung D, Tang Z, Huang H, Dai L, Qi S, Li J, Colak G, Chen Y, et al. Metabolic Regulation of Gene Expression by Histone Lysine beta-Hydroxybutyrylation. Molecular cell. 2016;62:194–206. doi: 10.1016/j.molcel.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell research. 2011;21:564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Chen Y, Zhang Z, Zhao Y. Identification and verification of lysine propionylation and butyrylation in yeast core histones using PTMap software. Journal of proteome research. 2009;8:900–906. doi: 10.1021/pr8005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Guan H, Zhao S, Mi W, Wen H, Li Y, Zhao Y, Allis CD, Shi X, Li H. YEATS2 is a selective histone crotonylation reader. Cell research. 2016;26:629–632. doi: 10.1038/cr.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]