Abstract
We developed a computational method to infer the complementarity determining region 3 (CDR3) sequences of tumor infiltrating T-cells in 9,142 RNA-seq samples across 29 cancer types. We identified over 600 thousand CDR3 sequences, including 15% with full-length. CDR3 sequence length distribution and amino acid conservation, as well as variable gene usage of infiltrating T-cells in many tumors, except brain and kidney cancers, resembled those in the peripheral blood of healthy donors. We observed a strong association between T-cell diversity and tumor mutation load, and predicted SPAG5 and TSSK6 as putative immunogenic cancer/testis antigens in multiple cancers. Finally, we identified 3 potential immunogenic somatic mutations based on their co-occurrence with CDR3 sequences. One of them, PRAMEF4 F300V, was predicted to bind strongly to both MHC-I and MHC-II, with matched HLA types in its carriers. Our analyses have the potential to simultaneously identify immunogenic neoantigens and the tumor-reactive T-cell clonotypes.
Introduction
T cell receptor (TCR) consists of a heterodimer of two chains (α, β or γ, δ), both of which are products of V(D)J recombination1. This somatic rearrangement only occurs in the T cell genome and produces an extremely diverse repertoire of TCRs. The most variable region in TCR is the complementarity determining region 3 (CDR3), which plays a critical role in antigen recognition2. The lower limit of distinct TCRs in the peripheral blood of a healthy individual is around 1.1 million3, and the theoretical diversity for αβ T cells, the most abundant T cell type in humans, is up to1016 types4. This large repertoire of T cells with structurally divergent TCRs is required to recognize cells expressing foreign or mutated proteins, including neoantigens in cancer cells. Therefore, characterizing the repertoire of tumor-infiltrating T cells can help identify the tumor-reactive T cell clones and facilitate the clinical practice of cancer immunotherapies.
The current common strategy for characterizing CDR3 is TCR profiling which amplifies the cDNA or gDNA β-CDR3 locus using predesigned PCR primers followed by deep sequencing. Recent developments of cancer immunotherapies5–7 have seen TCR sequencing applied to monitor T cell repertoire changes before and after the therapy in humans or animal models8–11. While these studies revealed exciting mechanisms of tumor immunity and the pharmacology of checkpoint blockade drugs, they were limited by small sample size thus had low power to detect important features shared among individuals. Efforts have been made to study the repertoire of T/B cells using unselected RNA-seq data in liquid or solid tumors12,13, which can potentially scale up to large cohorts. However, these studies adopt computational methods not specifically designed for unselected RNA-seq data14–16, resulting in poor CDR3 calls and limited power in the downstream characterization of the tumor-infiltrating T cell repertoire on the cohorts.
In this study, we developed a novel computational method for de novo assembly of CDR3 regions using paired-end RNA-seq data, and applied it on 9,142 samples from the Cancer Genome Atlas (TCGA). Compared to a previous RNA-seq based analysis13, we assembled an order of magnitude more distinct CDR3 sequences, which gave us enough power to perform deeper analyses on the TCR repertoire of the tumor microenvironment. We observed interesting interactions between tumor and the host immune system and identified potential therapeutic targets that might be useful for multiple immunotherapies.
Results
De novo assembly of CDR3 sequences and method validation
We developed a de novo method to assemble the CDR3 sequences produced by TCR transcripts using paired-end RNA-seq data (Supplementary Fig. 1 and Methods). In brief, this method first maps the reads to the human genome and searches for read pairs with one mate properly mapped to a TCR gene and the other mate unmappable to the genome, potentially due to V(D)J recombination. It then initiates pairwise comparison of the unmapped reads and constructs a read-overlap matrix, represented by an undirected graph, with each node representing a read and an edge for partial sequence overlap between two connected reads. This graph is further divided into disjoint cliques to represent potentially different CDR3 sequences. Finally, the method assembles all the reads in each clique to obtain contigs of DNA sequences and annotates them with information such as amino acid sequence, associated variable (V) and joining (J) genes. Contigs not annotated as CDR3 regions were discarded to reduce false positive calls (Methods). Counts of reads and contigs kept at each step of the method for an example was summarized in Supplementary Fig. 2.
To validate the above approach, we first took 3 kidney renal clear cell carcinoma (KIRC) samples from TCGA with available RNA-seq data, extract the genomic DNA from formalin-fixed and parrffin-embedded (FFPE) tumors, and sent for TCRβ sequencing (immunoSEQ). Although the number of CDR3 sequences assembled from RNA-seq is much smaller than that from immunoSEQ, over 60% of CDR3 from RNA-seq was also observed in immunoSEQ, which partially validates the accuracy of our method. It is worth noting that due to DNA fragmentation in the FFPE samples, only a subpopulation (~25–50%) of the infiltrating T cells can be recovered, so it is not surprising that a fraction of our assemblies were not contained in the immunoSEQ results. Also as expected, CDR3 assemblies from RNA-seq data were enriched for the abundant T cell clones, recovering over 50% of the most abundant (99.9% quantile) clones (Supplementary Fig. 3 and Methods).
Since immunoSEQ from FFPE samples cannot retrieve the complete infiltrating repertoire, we conducted in silico simulations to systematically evaluate the performance of our method. To this end, we generated pseudo tumor RNA-seq samples by in silico mixing of TCR transcript reads from a deeply sequenced immunoSEQ sample3 and RNA-seq reads from a TCR negative cancer cell line (K562) (Supplementary Fig. 4 and Methods). This procedure demonstrated that our method achieved high accuracy in CDR3 sequence assembly over a large range of T cell infiltration levels (Supplementary Fig. 5a). Our results also confirmed the above observation that the assembled CDR3 sequences were enriched for clonotypes with high frequency (Supplementary Fig. 5b–c).
In both the immunoSeq validation and in silico mixing simulation, our method assembled a small fraction (0.5%–5%) of the total CDR3 repertoire (Supplementary Fig. 3 and 5). This is because the actual coverage for the TCR region in an RNA-seq sample is estimated to be as low as 0.04 (Methods). We introduced additional simulations to investigate how CDR3 assembly rate changes with sequence depth (Supplementary Fig. 4 and Methods). Surprisingly, we found that at coverage 1 (library size 5 billion reads), our method achieved 33% recall of the simulated true CDR3 transcripts with high precision (97.2%), while for a competing method iSSAKE16 the recall was only 0.7% with precision 7.1% (Supplementary Fig. 6). The above results indicate that our method is a highly sensitive and accurate CDR3 assembler for tumor RNA-seq data. It outperforms competing method by at least an order of magnitude, making it statistically powerful to analyze the immune repertoire of large-scale RNA-seq sample cohorts.
Distribution of TCR gene usage and T cell type abundance
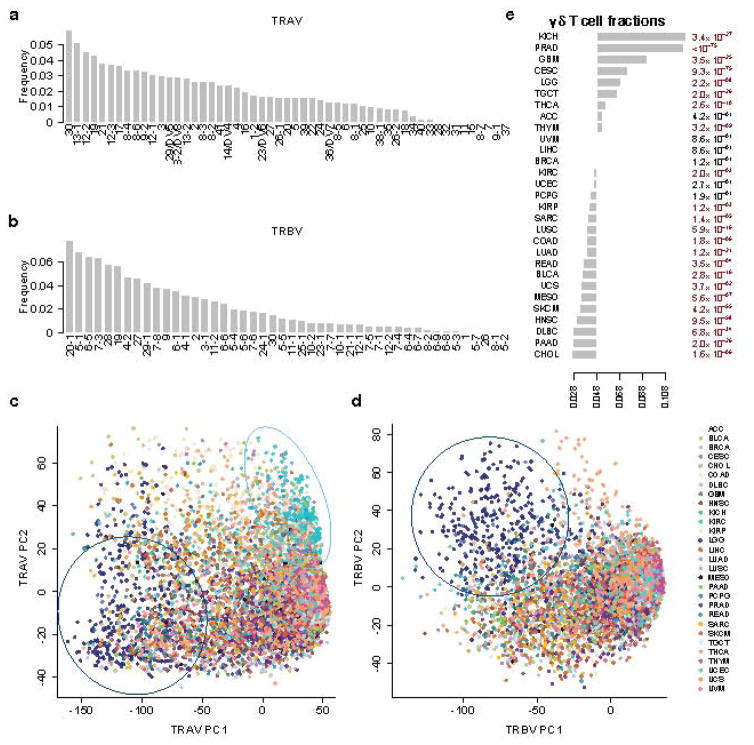
We applied the CDR3 assembly method to study 9,142 samples from 29 TCGA cancers, and the resulting CDR3 sequences are available in Supplementary Data Set 1. We first used mapped reads to estimate the usage of different TCRα variable (TRAV) and β variable (TRBV) genes across all tumor samples. The top three genes are 30, 13-1 and 12-2 for TRAV and 20-1, 5-1 and 6-5 for TRBV (Fig. 1a–b). While there are few studies on TRAV usage to validate our estimates, our observation on TRBV is consistent with previous reports17,18 using the peripheral blood from healthy donors16,17. This result suggests that the TRBV usage in tumor-infiltrating T cells do not deviate significantly from that in the peripheral blood. In most cancer types, TRAV or TRBV usage distributions among the tumor samples are similar, except those in the brain and kidney cancers (Fig. 1c–d). Brain cancer displays very different patterns of both TRAV and TRBV usage, while kidney cancer is only different in TRAV usage, compared to the majority of TCGA tumors (Fig. 1a–b and Supplementary Fig. 7). These differences might be due to potentially different immune regulations in brain cancer and expression of endogenous retrovirus in kidney cancer19.
Figure 1.
Distribution of αβ T cell variable gene usage and γδ T cell abundance in multiple cancer types. a–b: Proportions of TRAV and TRBV genes in decreasing order. IMGT functional genes were selected in the display. c–d. PCA analysis on TRAV and TRBV usage across different cancer types. For TRAV, PC1 was driven by the difference between brain cancer (LGG) and other tumors, while PC2 was driven by kidney cancer (KIRC). Dark blue circle: LGG samples; cyan circle: KIRC samples. e. γδ T cell fractions (labeled in the x-axis) in multiple cancer types in decreasing order. The mean γδ T cell fraction across all samples was 4.8%. For each cancer, we used Binomial test with expected probability 0.048 to calculate the statistical significance. We applied Benjamini-Hochberg adjusted P values for FDR. The numbers listed on the right margin of the plot are q values. Disease abbreviations: ACC: adenocortical carcinoma, BLCA: bladder carcinoma, BRCA: breast carcinoma, CESC: cervical squamous carcinoma, CHOL: cholangiocarcinoma, COAD: colon adenocarcinoma, DLBC: diffusive large B-cell lymphoma, GBM: glioblastoma multiforme, HNSC: head and neck carcinoma, KICH: kidney chromophobe, KIRC: kidney renal clear cell carcinoma, KIRP: kidney renal papillary cell carcinoma, LGG: lower grade glioma, LIHC: liver hepatocellular carcinoma, LUAD: lung adenocarcinoma, LUSC: lung squamous carcinoma, MESO: mesothelioma, PAAD: pancreatic adenocarcinoma, PCPG: pheochromocytoma and paraganglioma, PRAD: prostate adenocarcinoma, READ: rectum adenocarcinoma, SARC: sarcoma, SKCM: skin cutaneous melanoma, TGCT: testicular germ cell tumors, THCA: thyroid carcinoma, THYM: thymoma, UCEC: uterine corpus endometrial carcinoma, UCS: uterine carsinosarcoma, UVM: uveal melanoma.
We next investigated the CDR3 assemblies. Here CDR3 regions are defined as all amino acids between the last cysteine of variable gene and the phenylalanine in joining gene motif FGXG as was previously described17. In total, we identified 683,418 sequences, including 650,496 from αβ T cells and 32,922 from γδ T cells. Of all the CDR3 assemblies, the vast majority (95.8%) has read counts smaller than 10, with a median of 1.7 (Supplementary Fig. 8). Of these, 77,060 β-CDR3 and 1,060 δ-CDR3 are complete sequences harboring the conserved N terminal 4 amino acids and C terminal phenylalanine. Based on sequence count, γδ T cells account for ~4.8% of the total T cell population, consistent with previous observations20. However, this γδ T cell fraction can vary among cancer types (Fig 1e), potentially due to different neoantigens presented on different tumor cells.
Features of β and δ chain CDR3 sequences
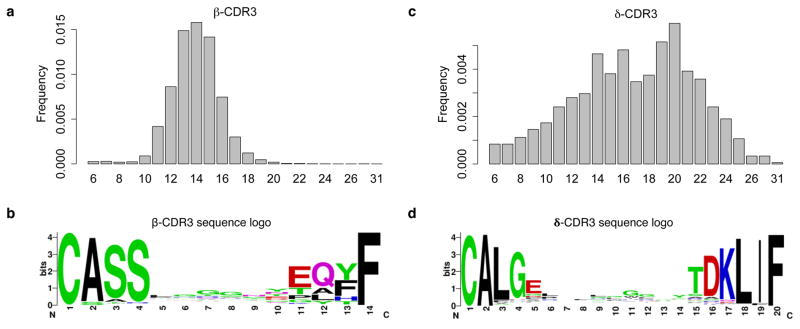
The TCR β and δ chains have undergone V(D)J recombination and are responsible for most of the antigen recognition. β-CDR3 has sequence length ranging from 6 to 31, with a median of 14 amino acids (Fig. 2a). The sequence pattern21 from the most frequent 14-amino acid CDR3 sequences (Fig. 2b) is very similar to that from the peripheral blood of healthy donors by TCR sequencing17 with minor differences in the first 4 residues. This difference is potentially due to reduced TRBV20-1 abundance in our data, an observation consistent with previous studies18,22.
Figure 2.
Length and amino acid conservation of β and δ chain CDR3 sequences in tumor-infiltrating T cells. Length distribution of complete CDR3 calls was estimated using histogram for β and δ chains (a and c). Length 14 β-CDR3 and length 20 δ-CDR3 sequences were selected for weblogo analysis (b and d). The y-axis in the sequence logo plot was the conservation score. For a given locus, the height of a letter reflects the relative frequency of that amino acid.
δ-CDR3 sequences also have length ranging from 6 to 31 amino acids, although a longer median of 17 (Fig. 2c). Besides the larger median, δ-CDR3 also has larger length variation (SD=4.6) than β-CDR3 (SD=1.9). The more constrained distribution of β-CDR3 lengths potentially reflects the functional requirement for TCR β chain to contact the peptide major histocompatibility complex (pMHC), which is not required for γδ T cells. These observations agree with previous reports20,23, supporting the validity of our CDR3 calls. In addition, β-CDR3 and δ-CDR3 show no strong overrepresentation of specific amino acids except the small glycine prevalence in the middle of the sequence logo (Fig. 2d).
Identification of public and private β-CDR3 sequences
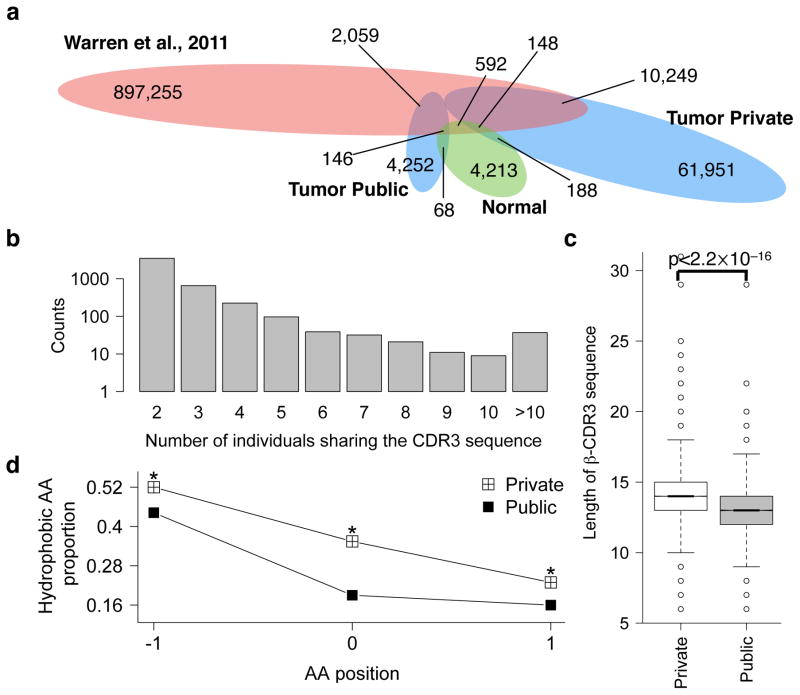
Despite the extreme amino acid sequence diversity, 4,252 of the complete β-CDR3 sequences appeared in more than one tumors (Fig. 3a) and the number of individuals sharing these sequences can be as high as 65 (Fig. 3b). A large fraction of the shared, or public, CDR3 sequences are potentially products of convergent recombination in the thymus24. We compared our CDR3 calls to the peripheral blood repertoire of healthy individuals by deep TCR sequencing3, and 2,059 of TCGA shared CDR3 sequences are also present in this dataset (Fig. 3a). This result suggests that a large fraction of the shared sequences might not be related to the tumor antigens but derived from public T cells with potential role in responses to common antigens such as persistent viral infections24. Interestingly, 10,249 CDR3 sequences private to individual TCGA tumor also overlap with the peripheral blood CDR3 repertoire. These sequences are likely also shared but missed in other TCGA tumor(s) due to the limited power of identifying T cell clonotypes with low abundance from the RNA-seq data. Therefore, we merged these 10,249 sequences with the previous 4,252 ones as the final set of shared β-CDR3 sequences. It is worth noting that a significant fraction of our final set of private sequences in the TCGA data may still contain a significant number of truly public β-CDR3 sequences.
Figure 3.
Public and private β-CDR3 amino acid sequences have different lengths and hydrophobicity. a. β-CDR3 sharing between TCGA tumor, TCGA normal samples and peripheral blood repertoire displayed in Venn diagram. Colors indicated difference tissues: red: peripheral blood; green: TCGA normal samples; blue: TCGA tumor samples. The numbers of sharing labeled inside the eclipses were the overall calls of that category. Numbers labeled outside and connected to a colored region is the counts of the overlapped calls between categories. b. Distribution of public β-CDR3 frequency. Sequence sharing was determined using only TCGA data, not including the 10,249 sequences shared with blood repertoire. c. Comparison of β-CDR3 lengths of private and public sequences. P value was calculated using Wilcoxon test. Box includes data between the 25th and 75th percentiles, with horizontal line indicates the median. There are 14,443 and 51,583 sequences in the public and private group respectively. d. Hydrophobicity analysis of the middle 3 amino acids (see main text for details) in the private and public CDR3 sequences. In each position, Binomial test was applied to estimate the significance of the difference in hydrophobic amino acid fraction between groups, using the fraction in the public group as expected probability. All three positions were significant at FDR=0.05.
Previous study reported that β-CDR3 sequences of private T cells are significantly longer than those of public T cells in the peripheral blood3 and we observed the same in tumor-infiltrating T cells (Fig. 3c). As CDR3 sequences contain highly conserved four amino acid sequence in the N terminus and phenylalanine in the C terminus (Fig. 2b), we defined “CDR3 motif” as the amino acid sequence in between these conserved regions of the complete CDR3 sequence. Interestingly, the middle 3 amino acids of the private CDR3 motifs contained significantly higher fraction of hydrophobic residues than those of the public motifs. A recent study reported that hydrophobicity is a hallmark of immunogenic neoepitopes25, and hence our results suggest that private CDR3 sequences might have higher potential for tumor-antigen recognition.
Association of T cell diversity with neoantigen load
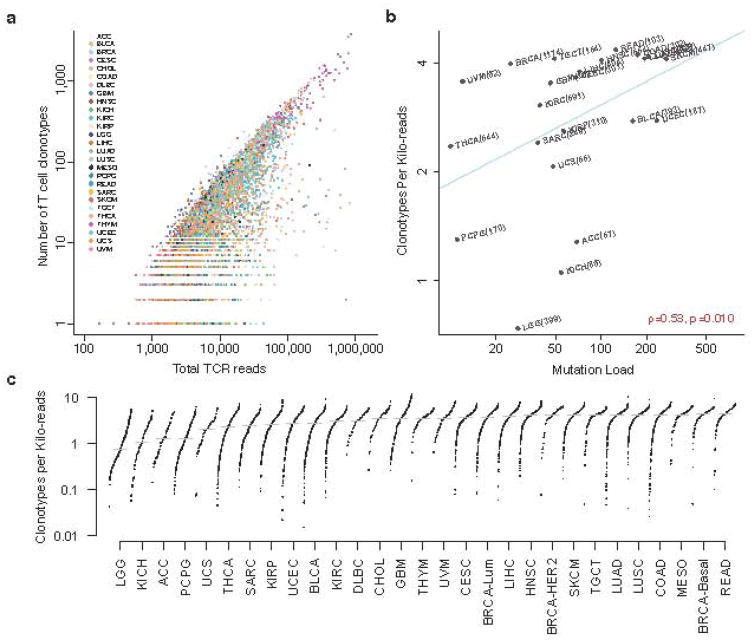
Clonotype diversity of T cell repertoire is an important property of the immune system and is closely related to the capacity for T cells to recognize antigens. As each T cell clone possesses a unique TCR, CDR3 sequences are often used as proxies to represent clonotype diversity. In our data, the number of unique CDR3 calls in each tumor is linearly correlated with total TCR reads (Fig. 4a), an expected observation since tumors with higher T-cell infiltrates have more TCR reads to assemble more CDR3 sequences. We therefore used the number of unique CDR3 calls in each sample normalized by the total read count in the TCR region, which we called clonotypes per kilo-reads (CPK), as a measure of clonotype diversity (Methods). In kidney, lung, and pancreatic cancers, female patients have significantly higher CPK than male patients, consistent with the long-standing knowledge of elevated immune responses in females26 (Supplementary Table 1). In addition, expression level of granzyme A, an indicator of immune mediated cytolysis18,25, is also positively correlated with CPK even after correcting for tumor purity19,27 (Supplementary Fig. 9). These observations support the validity of using CPK as a measure of immune responses.
Figure 4.
The diversity of T cell clonotypes positively associates with cancer somatic mutation load. a. Scatter plot of the number of CDR3 calls in each sample against the total reads extracted from the 3 TCR regions. Prostate and pancreatic cancers were excluded due to high expression of non-TCR genes in the region (Methods). b. Clonotypes per kilo-reads (CPK) was positively associated with tumor mutation load. Median CPK and median somatic mutation load for each cancer type were displayed on the scatter plot. Cancers with <50 samples were excluded. Significance was estimated using Spearman’s correlation test. c. Distributions of CPK across all cancer types. PAM50 subtypes of breast cancer46 were displayed to show the inter-tumor heterogeneity in this disease.
We next calculated CPK for each tumor sample and observed a strong positive association between the CPK value and the load of nonsynonymous somatic mutations (Fig. 4b and Supplementary Fig. 10). When ranking the cancer types by their median CPK, we found that breast cancer showed surprisingly high inter-tumor heterogeneity, where the CPK of basal breast cancer is 1.2-fold that of the luminal subtypes. Besides basal breast cancer, testicular cancer (TGCT) also has unusually high CPK, which might be related to the high level of alternative splicing during spermatogenesis28. The remaining cancers with highest T cell clonotype diversity include colorectal cancers, non-small cell lung carcinomas, mesothelioma, and melanoma (Fig. 4c). These cancers are known to be associated with external stimuli, such as microbiota, smoking, carcinogen, and UV exposure, respectively. There are at least two explanations to our observation: 1) tumor genomes with higher mutation load present more neoantigens to the immune system, which recruit antigen-specific infiltrating-T cells; 2) external stimuli such as UV or carcinogen directly interact with the immune system to increase the T cell repertoire diversity. If the second explanation were valid, we would expect a higher fraction of public β-CDR3 sequences in these cancer types, due to the presence of public T cells in response to the common stimuli. Among the above cancers, this is true only in melanoma (Supplementary Fig. 11), suggesting that the diversity of infiltrating T cells in most cancers might be regulated through tumor-specific somatic mutations.
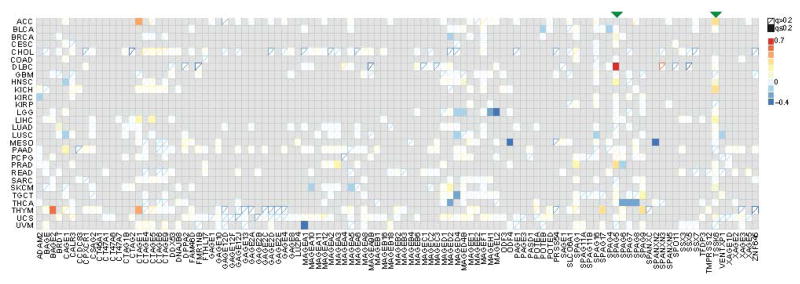
Cancer/testis (CT) antigens are a family of genes with normal expression restricted to germ cells, but can also be expressed in tumors due to epigenetic instability. CT antigens do not have thymus tolerance as genes expressed in other tissues and can be recognized as foreign antigens by the immune system. Efforts have been made to explore the possibility of using CT antigens as cancer vaccine targets29,30 and clinical trials have been conducted using a number of CT antigens31. We examined whether the expression of CT antigens are associated with infiltrating T cell diversity. Among the 109 known CT antigens19,32, SPAG5 and TSSK6 expression levels positively correlate with CPK in multiple cancers (Fig. 5). We further analyzed all the 9 amino acid peptides in the SPAG5 and TSSK6 protein sequences for their MHC-I binding affinity using NetMHC4.033 and identified 25 strong (rank < 0.5%) binding sites for SPAG5 and 7 for TSSK6 for common HLA alleles (Supplementary Fig. 12). Together, these evidences support SPAG5 and TSSK6 as potential vaccine targets in multiple cancer types.
Figure 5.
Association of T cell diversity and expression of cancer/testis antigens reveals SPAG5 and TSSK6 as vaccine targets. Gray entries indicated the gene was not overexpressed in the tumor cells, as suggested by correlative analysis with tumor purity. Association between CPK and the CT antigen expression was evaluated using partial Spearman correlation corrected for tumor purity. Solid boxes indicated significant associations at FDR=0.2.
Joint prediction of neoantigen and tumor-reactive T cell
According to the mechanism of immunoediting34, tumors sharing the same immunogenic somatic mutation might harbor tumor-reactive T cells with similar antigen-recognizing TCR domains. To explore the recurrent patterns in the CDR3 sequences, we studied the CDR3 motifs as above defined. From the complete β-CDR3 sequences, we identified a total of 64,824 unique motifs. For each motif, we searched all the 683,418 CDR3 calls for sequences containing the motif and documented the corresponding individuals. Extremely short motifs are not informative, as their recurrences can be random. Also, highly recurrent motifs may come from public T cell, which is not the interest of this analysis. Therefore, we focused on CDR3 motifs with recurrence in 5 to 20 tumors and longer than 5 amino acids, resulting in 5,347 high quality motifs. Next, we obtained the somatic mutation profiles from the exome-sequencing data of each tumor. We filtered 5’UTR, 3’UTR, nonsense and nonstop mutations because they don’t result in altered peptides or potential neoantigens in the tumors. The remaining 2,353 nonsynonymous (NS) mutations occurring in more than 3 tumors were kept for downstream analyses.
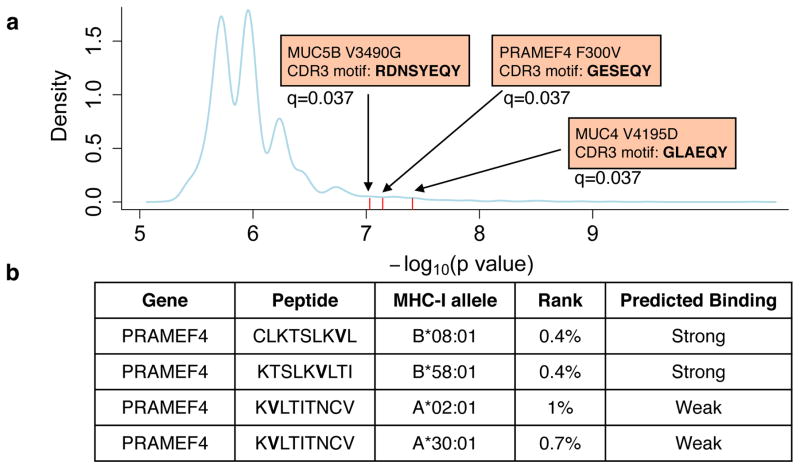
We then examined the co-occurrence of CDR3 motifs and NS mutations in cancer patients, using Fisher’s exact test to estimate statistical significance. We applied a heuristic method to find the most promising CDR3-mutation pairs and used permutation test to correct for false discovery rate (FDR, Methods). Based on over 1.5 million null permutation tests and a stringent selection criterion, we identified top 3 pairs to be statistically significant (FDR=0.05) involving mutations on MUC4, PRAMEF4, and MUC5B genes (Fig. 6a). Examining the 9 amino acid peptides containing these mutations for MHC-I binding with NetMHC4.035, we found that all three mutations have at least one predicted binding peptide (Supplementary Table 2). We compared the mutated peptides with their wild type (WT) sequence and excluded mutated peptides with lower binding affinity than the WT ones. The remaining peptides were all produced by PRAMEF4 F300V (Fig. 6b). Interestingly, PRAMEF4 F300V is also predicted by NetMHC-II2.233,36 to produce high affinity MHC-II binding peptide (Supplementary Fig. 13). PRAMEF4 F300V co-occurs with CDR3 motif GESEQY in three patients: TCGA-4K-AA1I, TCGA-2G-AAGE and TCGA-2G-AAKO, all of whom with testicular germ cell tumors. We did not find the CDR3 motif in two other tumors carrying the F300V mutation, and one possible reason is that CDR3 motif was present but failed to be identified from RNA-seq data. PRAMEF4 is expressed in cholangiocarcinoma, liver, ovarian, endometrial and testicular cancers, but is almost silent in other cancers and normal tissues (Supplementary Fig. 14). The three individuals with PRAMEF4 F300V mutation all have tumor PRAMEF4 expression, so mutant PRAMEF4 peptides are produced in these tumors. We next annotated the HLA type information of the above three individuals using POLYSOLVER27 (Supplementary Table 3). One has allele A*30:01, the exact type predicted to bind peptide KVLTITNCV. Two of them, TCGA-2G-AAGE and TCGA-2G-AAKO also have allele B*08:01, which is predicted to bind CLKTSLKVL. Therefore, all three harbored at least one HLA allele binding the mutated peptides. These results supported PRAMEF4 F300V as a potential immunogenic mutation in testicular cancer. In addition, our analysis suggested that if F300V is truly immunogenic, the corresponding tumor-reactive T cell clonotypes are likely to carry the GESEQY motif in the CDR3 sequences.
Figure 6.
Non-synonymous mutations co-occur with CDR3 motif. a. Three pairs of NS mutation and CDR3 motifs co-occurred more often than random, with statistical significance (FDR=0.05) based on permutation test (Methods). MHC-I binding predictions were performed on all the possible 9 amino acid peptides derived from the above three mutations using NetMHC4.0, and those with mutated peptides binding stronger than wild type peptides were displayed (b). Bold letters indicated mutated amino acids.
Discussion
Understanding the crosstalk between cancer antigens and the host adaptive immunity is critical to finding therapeutic targets and developing effective immunotherapies. Improved characterization of tumor-infiltrating T cell repertoire is highly desirable yet has been limited to small scale of samples due to technical and cost barriers. In this study, we developed a computational method to extract the TCR sequence from unselected tumor RNA-seq data, and applied it to over nine thousands TCGA samples across 29 cancer types. To our best knowledge, this work is among the first to analyze infiltrating T cell repertoire on a large cancer cohort. Compared to a similar work relying on reads that covering the complete CDR3 region13, our method assembled an order of magnitude more CDR3 sequences, leading to stronger associations and improved statistical power.
Our observations on variable gene usage, CDR3 sequence length, amino acid conservation, γδ T cells and features of public/private T cells were similar as previously reported on peripheral blood repertoire3,17. These results suggest that the population of infiltrating T cells maintained a large fraction of public clonotypes, which are also present in the peripheral repertoire of healthy donors. Comparing to private T cells, the CDR3 regions of the public clonotypes were shorter and less likely to bind neoepitopes according to the hydrophobicity analyses25. Future efforts are needed to elucidate the potential functional impact of public T cells in the tumor microenvironment.
According to our results, the presence of cancer antigens, including somatic mutations and cancer/testis genes might increase the diversity of infiltrating T cell repertoire. Specifically, we identified SPAG5 and TSSK6 as candidates of cancer vaccine targets based on their associations with CPK, a metric for T cell clonotype diversity. It must be emphasized that our CDR3 calls only represent prevalent clonotypes in infiltrating T cells due to limited detection power from RNA-seq data. Therefore, CPK is a simplified diversity measure of abundant T cell clonotypes, which is potentially the reason that CPK was not associated with patient survival (Supplementary Table 1).
Our analysis identified 3 strong co-occurrences between recurrent tumor mutations and CDR3 sequence motifs, and provided HLA typing evidence to support at least one of them, PRAMEF4 F300V, as a putative immunogenic mutation. Our analysis also identified the corresponding antigen-recognizing CDR3 motifs. Unfortunately we could not conduct TCR sequencing on the remaining mutation positive but CDR3 motif negative samples or other experimental validation at this point to further substantiate the association. Our computational approach is potentially useful for cancer vaccines, adoptive T cell5 as well as chimeric antigen receptor T cell therapies37,38. One possible reason that we were not able to find more significant pairs is our limited CDR3 detection power. Another reason is that TCRs with different CDR3s might be able to bind the same neoepitope and the same somatic mutation may be recognized by multiple CDR3 motifs. Therefore, future efforts might be made to group different CDR3 sequences with similar biochemical properties and match the groups to somatic mutations to identify more immunogenic somatic mutations.
In this study, we demonstrated the feasibility of using unselected RNA-seq data to characterize the tumor-infiltrating T cell repertoire. Although the scale and power of our analysis are sometimes still limited by low coverage and insufficient sample size, we were able to observe interesting associations between T cell repertoire and tumor clinical and molecular features in the TCGA cohort. With the rapid decrease of sequencing cost and increase of tumor profiling efforts, we anticipate further analyses of tumor-immune interactions on more high-quality RNA-seq data to yield better biological insights in the near future.
Supplementary Material
Acknowledgments
We thank Dr. Gordon Freeman for helpful discussion during manuscript preparation. We also acknowledge the following funding sources for supporting our work: NCI 1U01 CA180980, National Natural Science Foundation of China 31329003 and Chinese Scholarship Council Fellowship.
Footnotes
URLs
Cancer Genomics Hub: https://cghub.ucsc.edu
TCGA data portal: https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp
GDAC Firehose: https://gdac.broadinstitute.org/
simNGS: http://www.ebi.ac.uk/goldman-srv/simNGS/
ImmunoSeq data accession: https://adaptivebiotech.com/pub/Liu-2016-NatGenetics
Data Accession
ImmunoSeq data generated in this study is accessible through Adaptive Biotechnologies. Link to the data is provided in the URLs section.
Author Contribution
B.L. conceived this project, developed the CDR3 calling method, processed the datasets and performed statistical analysis. T.L. performed statistical analysis, generated a subset of figures and helped to write the manuscript. B.W., J.W. and R.D. helped with the analysis on CDR3 sequences. S.S. performed analyses using POLYSOLVER. Q.C helped analyze the data. JC.P, S.S and T.C conducted experimental validation. F.S.H, C.W and N.H. conceived some of the analyses and contributed to the manuscript. X.S.L and J.S.L supervised the whole study and wrote the manuscript with B.L..
Competing financial interests: the authors declared no competing financial interest.
References
- 1.Alt FW, et al. VDJ recombination. Immunol Today. 1992;13:306–14. doi: 10.1016/0167-5699(92)90043-7. [DOI] [PubMed] [Google Scholar]
- 2.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 3.Warren RL, et al. Exhaustive T-cell repertoire sequencing of human peripheral blood samples reveals signatures of antigen selection and a directly measured repertoire size of at least 1 million clonotypes. Genome Res. 2011;21:790–7. doi: 10.1101/gr.115428.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robins HS, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg SA, Restifo NP, Yang JC, Morgan RA, Dudley ME. Adoptive cell transfer: a clinical path to effective cancer immunotherapy. Nat Rev Cancer. 2008;8:299–308. doi: 10.1038/nrc2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma P, Wagner K, Wolchok JD, Allison JP. Novel cancer immunotherapy agents with survival benefit: recent successes and next steps. Nat Rev Cancer. 2011;11:805–12. doi: 10.1038/nrc3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savage PA, et al. Recognition of a ubiquitous self antigen by prostate cancer-infiltrating CD8+ T lymphocytes. Science. 2008;319:215–20. doi: 10.1126/science.1148886. [DOI] [PubMed] [Google Scholar]
- 9.Obenaus M, et al. Identification of human T-cell receptors with optimal affinity to cancer antigens using antigen-negative humanized mice. Nat Biotechnol. 2015;33:402–7. doi: 10.1038/nbt.3147. [DOI] [PubMed] [Google Scholar]
- 10.Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–71. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Twyman-Saint Victor C, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015;520:373–7. doi: 10.1038/nature14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blachly JS, et al. Immunoglobulin transcript sequence and somatic hypermutation computation from unselected RNA-seq reads in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2015;112:4322–7. doi: 10.1073/pnas.1503587112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown SD, Raeburn LA, Holt RA. Profiling tissue-resident T cell repertoires by RNA sequencing. Genome Med. 2015;7:125. doi: 10.1186/s13073-015-0248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolotin DA, et al. MiTCR: software for T-cell receptor sequencing data analysis. Nat Methods. 2013;10:813–4. doi: 10.1038/nmeth.2555. [DOI] [PubMed] [Google Scholar]
- 15.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–52. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warren RL, Nelson BH, Holt RA. Profiling model T-cell metagenomes with short reads. Bioinformatics. 2009;25:458–64. doi: 10.1093/bioinformatics/btp010. [DOI] [PubMed] [Google Scholar]
- 17.Freeman JD, Warren RL, Webb JR, Nelson BH, Holt RA. Profiling the T-cell receptor beta-chain repertoire by massively parallel sequencing. Genome Res. 2009;19:1817–24. doi: 10.1101/gr.092924.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Heijst JW, et al. Quantitative assessment of T cell repertoire recovery after hematopoietic stem cell transplantation. Nat Med. 2013;19:372–7. doi: 10.1038/nm.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chien YH, Hampl J. Antigen-recognition properties of murine gamma delta T cells. Springer Semin Immunopathol. 2000;22:239–50. doi: 10.1007/pl00006752. [DOI] [PubMed] [Google Scholar]
- 21.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dean J, et al. Annotation of pseudogenic gene segments by massively parallel sequencing of rearranged lymphocyte receptor loci. Genome Med. 2015;7:123. doi: 10.1186/s13073-015-0238-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rock EP, Sibbald PR, Davis MM, Chien YH. CDR3 length in antigen-specific immune receptors. J Exp Med. 1994;179:323–8. doi: 10.1084/jem.179.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venturi V, Price DA, Douek DC, Davenport MP. The molecular basis for public T-cell responses? Nat Rev Immunol. 2008;8:231–8. doi: 10.1038/nri2260. [DOI] [PubMed] [Google Scholar]
- 25.Chowell D, et al. TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes. Proc Natl Acad Sci U S A. 2015;112:E1754–62. doi: 10.1073/pnas.1500973112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuurs AH, Verheul HA. Effects of gender and sex steroids on the immune response. J Steroid Biochem. 1990;35:157–72. doi: 10.1016/0022-4731(90)90270-3. [DOI] [PubMed] [Google Scholar]
- 27.Shukla SA, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol. 2015;33:1152–1158. doi: 10.1038/nbt.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He C, et al. Genome-wide detection of testis- and testicular cancer-specific alternative splicing. Carcinogenesis. 2007;28:2484–90. doi: 10.1093/carcin/bgm194. [DOI] [PubMed] [Google Scholar]
- 29.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–25. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 30.Caballero OL, Chen YT. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci. 2009;100:2014–21. doi: 10.1111/j.1349-7006.2009.01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drake CG, Lipson EJ, Brahmer JR. Breathing new life into immunotherapy: review of melanoma, lung and kidney cancer. Nat Rev Clin Oncol. 2014;11:24–37. doi: 10.1038/nrclinonc.2013.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silina K, et al. Sperm-associated antigens as targets for cancer immunotherapy: expression pattern and humoral immune response in cancer patients. J Immunother. 2011;34:28–44. doi: 10.1097/CJI.0b013e3181fb64fa. [DOI] [PubMed] [Google Scholar]
- 33.Andreatta M, Schafer-Nielsen C, Lund O, Buus S, Nielsen M. NNAlign: a web-based prediction method allowing non-expert end-user discovery of sequence motifs in quantitative peptide data. PLoS One. 2011;6:e26781. doi: 10.1371/journal.pone.0026781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–8. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 35.Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen M, Lundegaard C, Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinformatics. 2007;8:238. doi: 10.1186/1471-2105-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grupp SA, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368:1509–18. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365:725–33. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.